In Caenorhabditis elegans, the lysosomal cathepsin protease CPL-1 is indispensable for clearance of apoptotic cells by playing a leading role in destruction of cell corpses in phagolysosomes.
Abstract
During programmed cell death, the clearance of apoptotic cells is achieved by their phagocytosis and delivery to lysosomes for destruction in engulfing cells. However, the role of lysosomal proteases in cell corpse destruction is not understood. Here we report the identification of the lysosomal cathepsin CPL-1 as an indispensable protease for apoptotic cell removal in Caenorhabditis elegans. We find that loss of cpl-1 function leads to strong accumulation of germ cell corpses, which results from a failure in degradation rather than engulfment. CPL-1 is expressed in a variety of cell types, including engulfment cells, and its mutation does not affect the maturation of cell corpse–containing phagosomes, including phagosomal recruitment of maturation effectors and phagosome acidification. Of importance, we find that phagosomal recruitment and incorporation of CPL-1 occurs before digestion of cell corpses, which depends on factors required for phagolysosome formation. Using RNA interference, we further examine the role of other candidate lysosomal proteases in cell corpse clearance but find that they do not obviously affect this process. Collectively, these findings establish CPL-1 as the leading lysosomal protease required for elimination of apoptotic cells in C. elegans.
INTRODUCTION
Appropriate clearance of cell corpses generated by apoptosis is critical to animal development and tissue homeostasis, defects in which contribute to many human diseases, including autoimmune diseases and neuronal disorders (Elliott and Ravichandran, 2010). The elimination of cell corpses requires that they be recognized and internalized by engulfing cells followed by fusion with lysosomes for degradation. The regulatory mechanisms underlying these cellular processes are essentially conserved among diverse species ranging from the nematode Caenorhabditis elegans to mammals. In the lifetime of a C. elegans hermaphrodite, apoptotic cells generated during embryogenesis are engulfed and digested by neighboring cells, whereas cell corpses derived from oogenesis in the germ line are removed by germline sheath cells. The exposure of the eat-me signal phosphatidylserine (PtdSer) on the surface of cell corpses is coordinately regulated by the Xk-related protein CED-8, the scramblase SCRM-1, and the P-type adenosine triphosphatase TAT-1 (Wang et al., 2007; Darland-Ransom et al., 2008; Chen et al., 2013b; Suzuki et al., 2013). For corpse engulfment, two major signaling pathways have been identified that mediate engulfment signals redundantly to induce cytoskeletal rearrangement of engulfing cells (Reddien and Horvitz, 2004). In one pathway, the evolutionarily conserved phagocytic receptor CED-1 recognizes PtdSer with the help of CED-7, an ABC transporter, and two PtdSer-binding proteins, TTR-52 and NRF-5 (Wu and Horvitz, 1998a; Zhou et al., 2001b; Wang et al., 2010; Mapes et al., 2012; Zhang et al., 2012). By further interacting with the CED-6 adaptor protein clathrin and its adaptors, CED-1 mediates engulfment signals to induce the rearrangement of the actin cytoskeleton of engulfing cells for cell corpse internalization (Chen et al., 2013a; Shen et al., 2013). After formation of phagosomes, CED-1 is released and recycled back to the plasma membrane by the retromer complex (Chen et al., 2010b). In the other pathway, the interaction among several intracellular proteins, including CED-2/CrKII, CED-5/DOCK180, and CED-12/ELMO, activates the CED-10/Rac1 GTPase, which in turn triggers the cytoskeleton rearrangement required for engulfment (Wu and Horvitz, 1998b; Wu et al., 2001; Reddien and Horvitz, 2000; Gumienny et al., 2001; Zhou et al., 2001a). Other molecules, including the PtdSer receptor PSR-1, INA-1/integrin, UNC-73/Trio, and MOM-5, are also reported to act through this pathway to promote cell corpse engulfment (Wang et al., 2003; deBakker et al., 2004; Cabello et al., 2010; Hsu and Wu, 2012). In addition, the ABL-1 protein kinase, the phosphatidylinositol 3-phosphatase MTM-1, and the Rho GTPase activating protein SRGP-1 negatively regulate engulfment of cell corpses in embryos (Hurwitz et al., 2009; Zou et al., 2009; Neukomm et al., 2011a, b).
After entry into the engulfing cell, cell corpses contained in phagosomes are destined to fuse with lysosomes for destruction. This process, termed phagosome maturation, involves multiple factors that are shared by endocytic trafficking. Recent findings suggest that clathrin and its adaptor AP2 complex are required for initiation of phagosome maturation in addition to a role in cell corpse engulfment. This is achieved by interacting with the large GTPase DYN-1/dynamin and the sorting nexin LST-4/Snx9/18/33 (Chen et al., 2013a). DYN-1 also forms a complex with the phosphatidylinositol-3 kinase VPS-34 and the RAB-5 small GTPase to promote the generation of phosphatidylinositol-3 phosphate (PI3P) required for phagosome maturation (Kinchen et al., 2008). Other Rab and ARL small GTPases and their regulators or effectors, including TBC-2, RAB-14, RAB-2, RAB-7, ARL-8, and the HOPS complex, act sequentially and cooperatively to regulate phagosome maturation (Lu et al., 2008; Mangahas et al., 2008; Yu et al., 2008; Li et al., 2009; Xiao et al., 2009; Guo et al., 2010; Sasaki et al., 2013). In turn, phagosome maturation is accompanied by progressive acidification of phagosomal lumen, which is critical for activation of lysosomal enzymes needed for digesting phagosomal contents (Kinchen and Ravichandran, 2008).
In contrast to the in-depth mechanisms governing cell corpse engulfment and phagosome maturation that are being uncovered, little is known regarding how lysosomal factors such as lysosomal enzymes contribute to degradation of cell corpses in phagolysosomes. As the major degradation sites in the cell, lysosomes contain many acidic hydrolases, including proteases, nucleases, glycosidases, sulfatases, and lipases. However, it is not known whether individual lysosome enzymes play a major role in clearance of cell corpses. In this study, we perform a genetic screen to look for mutants defective in cell corpse clearance and find that CPL-1, a C. elegans homologue of mammalian cathepsin L, is an essential lysosomal protease for cell corpse degradation. Using live-cell imaging analysis, we find that phagosomal incorporation of CPL-1 occurs at the terminal step of phagolysosome formation, failure in which causes defective cell corpse clearance. We also find that CPL-1 is important for degradation of autophagic and endocytic cargoes. Furthermore, our findings show that inactivation of other lysosomal proteases individually does not obviously impair cell corpse clearance. These findings establish CPL-1 as the major lysosomal protease required for at least initiating phagosomal degradation of apoptotic cells.
RESULTS
Loss of cpl-1 function leads to accumulation of apoptotic cells in the germ line
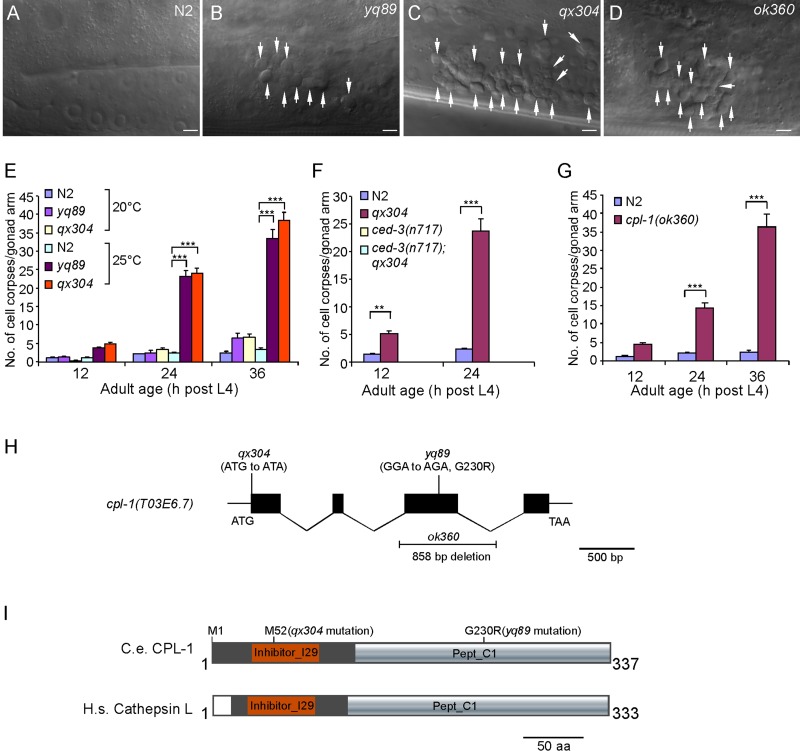
In C. elegans hermaphrodites, germ cell corpses generated by apoptosis are swiftly removed by sheath cells. Thus only a few cell corpses can be observed in germ lines of adult animals. To identify factors affecting the cell death program, we performed ethyl methanesulfonate (EMS) mutagenesis to search for mutants that displayed a significant increase in apoptotic cells in germ lines. Two mutants, yq89 and qx304, were found to accumulate germ cell corpses in a temperature-sensitive manner. When cultured at 20°C, yq89 and qx304 mutants developed normally and did not show an obvious cell corpse phenotype in germ lines compared with wild type. However, these animals exhibited an age-dependent accumulation of germ cell corpses when grown at 25°C (Figure 1, A–C and E). yq89 and qx304 mutants failed to complement one another, indicating that they affected the same gene (unpublished data). Cell corpses were rarely observed in the double mutants of qx304 with n717, a strong loss-of-function mutant of the CED-3 caspase required for the majority of apoptosis in C. elegans (Figure 1F), indicating that cell corpses seen in qx304 and yq89 animals indeed resulted from programmed cell death.
FIGURE 1:
Loss-of-function mutation of cpl-1 caused accumulation of apoptotic cells in C. elegans germ lines. (A–D) Representative DIC images of germ cell corpses in N2 (wild type) and yq89, qx304, and ok360 mutants. Cell corpses are indicated by arrows. Bars, 5 μm. (E) Quantification of germ cell corpses in N2, yq89, and qx304 animals. Cell corpses in one gonad arm of each animal were scored in ≥15 animals at every time point as indicated. Error bars represent SEM. (F) Quantification of germ cell corpses in N2 and qx304, ced-3(n717), and ced-3(n717);qx304 double-mutant animals. Cell corpse analyses were performed as in E. (G) Quantification of germ cell corpses in ok360 deletion mutants as in E. For cell corpse analysis, N2 and cpl-1(ok360) animals were cultured at 20°C, and yq89, qx304, and ced-3(n717) single and double mutants were grown at 25°C. In E–G, comparisons were performed between N2 and cpl-1 mutants using an unpaired t test. *p < 0.05, **p < 0.01, ***p < 0.001. (H) Schematic representation of the cpl-1 gene. Solid boxes represent exons, and wavy lines indicate introns. yq89 and qx304 point mutations and the ok360 deletion are indicated. (I) Schematic representation of C. elegans CPL-1 (C.e. CPL-1) and human cathepsin L (H.S. cathepsin L). Protein domains were predicted by using the SMART program (http://smart.embl-heidelberg.de/). Amino acid changes caused by qx304 and yq89 mutations are indicated.
We mapped yq89 and qx304 to the linkage group V and found that they affected the cpl-1 gene that encodes the lysosomal protease cathepsin L (Figure 1H). C. elegans CPL-1 is highly homologous to human and Drosophila cathepsin L (Hashmi et al., 2002; Britton and Murray, 2004). Using the SMART program, we found that CPL-1 contains a cathepsin propeptide inhibitor domain (Inhibitor_I29, amino acids 32–91), which functions to prevent access of substrate to the active site, and a pept_C1 domain (amino acids 120–336), which is shared by papain-family cysteine proteases (http:://smart.embl-heidelberg.de/; Figure 1I). In qx304 mutants, the start codon ATG of the cpl-1 gene was mutated to ATA, leading to a predicated translation initiation at methionine 52 relative to the wild-type protein; this results in a CPL-1 protein lacking the first 51 amino acids (Figure 1, H and I). In yq89 mutants, a change of GGA to AGA was identified in the cpl-1 gene, causing a G230R mutation in the pept_C1 domain of the encoded protein (Figure 1, H and I). The cpl-1 mutations in yq89 and qx304 mutants were further confirmed by sequencing cpl-1 cDNAs obtained from these mutants.
The cpl-1(ok360) deletion mutant contains a deletion of 858 base pairs in the cpl-1 genomic DNA, representing a null mutation of the cpl-1 gene (Figure 1H; Britton and Murray, 2004). cpl-1(ok360) mutants are embryonically lethal, which is rescued by maternally provided CPL-1. Homozygous cpl-1(ok360) animals generated by heterozygous mothers developed normally to adults due to maternal contribution of wild-type CPL-1, but their embryos were lethal; these cpl-1(ok360) animals also exhibited an obvious accumulation of cell corpses in the germ line in an age-dependent manner at 20°C (Figure 1, D and G), confirming that loss of cpl-1 function caused defects in the cell death program.
Cell corpse degradation is defective in cpl-1 mutants
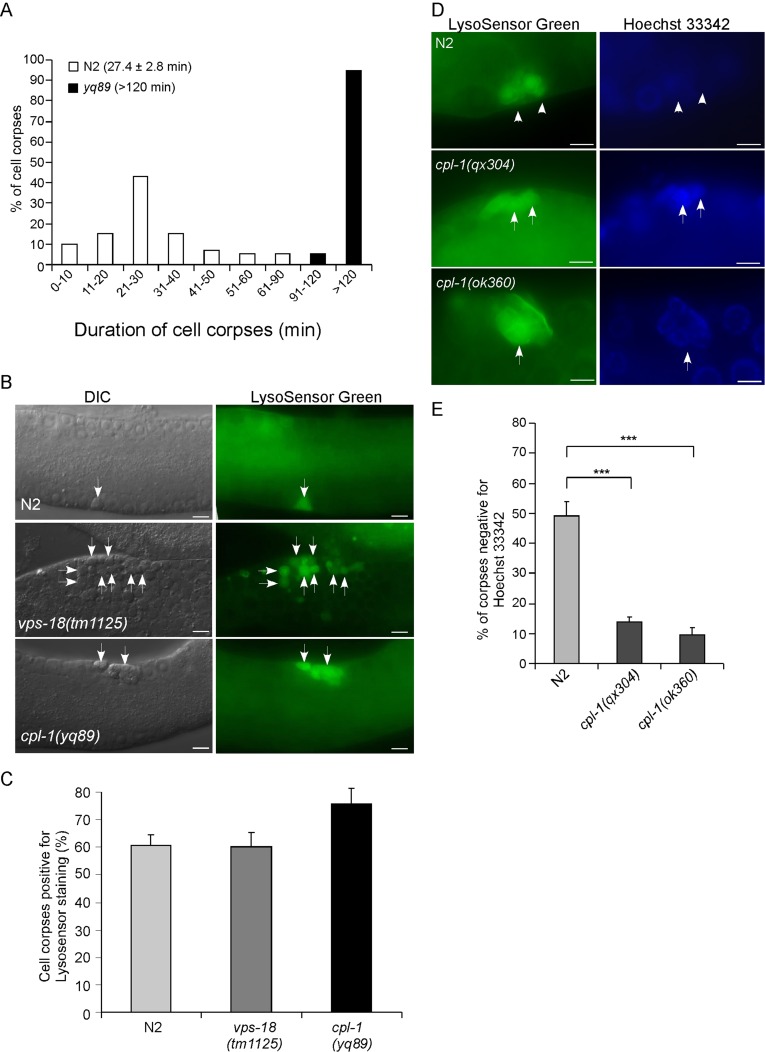
To understand how the cell death program is affected by loss of cpl-1 function, we measured the duration of germ cell corpse persistence in cpl-1 mutants using four-dimensional microscopy. In wild-type animals, the majority of germ cell corpses normally disappear within 30 min based on their appearance as disk-like structures, with an average of 27.4 ± 2.8 (mean ± SEM) min. In cpl-1(yq89) mutants, the duration of germ cell corpse persistence was much longer than in wild type, with ≥90% of cell corpses persisting >120 min (Figure 2A). These findings indicate that loss of cpl-1 function affected the clearance of cell corpses.
FIGURE 2:
cpl-1 mutations caused defective cell corpse clearance. (A) Four-dimensional analysis of germ cell corpse persistence in N2 and cpl-1(yq89) animals. Thirty germ cell corpses from ≥5 animals were analyzed for each genotype. The numbers in parentheses indicate the average duration of cell corpses (mean SEM). (B) LysoSensor Green staining of compartments containing germ cell corpses in N2, vps-18(tm1125), and cpl-1(yq89) animals. Arrows indicate some of the cell corpses stained by LysoSensor Green. Bars, 5 μm. (C) Quantification of germ cell corpses positive for LysoSensor Green staining as shown in B. We scored ≥100 germ cell corpses from ≥15 animals for each genotype; data were derived from three repeats. (D, E) Representative images of germ cell corpses stained by LysoSensor Green and Hoechst 33342 in cpl-1 mutants (D) and quantification of cell corpses stained by Hoechst 33342 (E). Arrows and arrowheads indicate cell corpses positive and negative for Hoechst 33342 staining, respectively. Bars, 5 μm. We analyzed ≥100 germ cell corpses from ≥20 animals for each genotype; data were derived from three repeats. ***p < 0.001. For cell corpse analysis, N2, vps-18(tm1125), and cpl-1(ok360) animals were cultured at 20°C, and cpl-1(yq89) and cpl-1(qx304) mutants were grown at 25°C.
We next asked whether the persistence of cell corpses in cpl-1 mutants resulted from a failure in acidification of phagosomes containing cell corpses. In wild-type germ lines, cell corpses are quickly engulfed by sheath cells, and the resulting phagosomes are progressively acidified during phagosome maturation. Thus most germ cell corpses were stained by the acidophilic dye LysoSensor Green DND-189. In vps-18(tm1125) mutants deficient in the HOPS subunit VPS-18 that is required for phagosome-lysosome fusion (Xiao et al., 2009), germ cell corpse–containing phagosomes were also acidified as in wild type. We found that cpl-1(yq89) mutants exhibited normal LysoSensor Green staining of germ cell corpses compared with wild-type and vps-18(tm1125) animals (Figure 2, B and C). Thus loss of cpl-1 did not impair the acidification of cell corpse–containing phagosomes. We further used Hoechst 33342 dye to examine the existence of apoptotic DNA in the persisting cell corpses. In wild type, ∼50% of germ cell corpses did not show obvious DNA staining by Hoechst 33342, indicative of efficient degradation of apoptotic DNA. In cpl-1(qx304) and cpl-1(ok360) mutants, however, only ∼10% of cell corpses showed no staining by Hoechst 33342 (Figure 2, D and E). Taken together, these findings suggest that loss of cpl-1 function caused defective degradation of cell corpses.
CPL-1 is not required for phagosomal recruitment of phagosome maturation effectors
To further understand the defects of cell corpse clearance caused by cpl-1 mutation, we examined phagosomal recruitment of factors required for phagosome maturation.
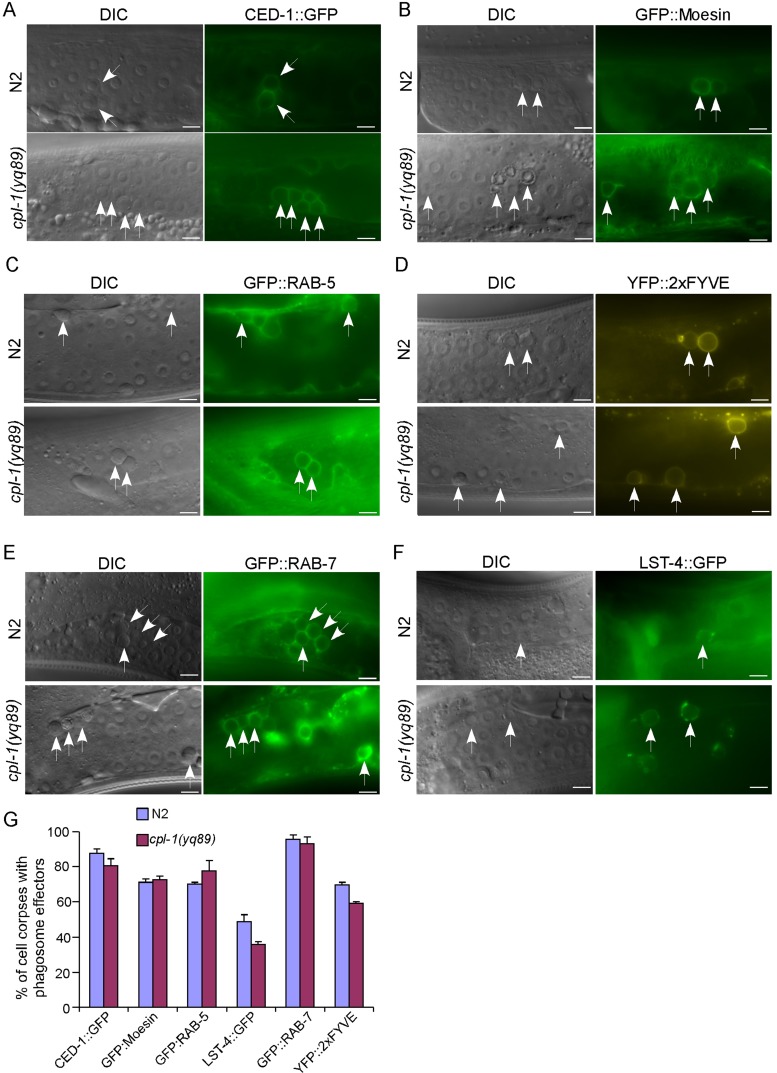
First we confirmed that cell corpses persisting in cpl-1(yq89) mutants were phagocytosed by engulfing cells because they were well surrounded by green fluorescent protein (GFP)–fused CED-1 (CED-1::GFP), the engulfment receptor, and GFP-tagged moesin (GFP::moesin), a Drosophila actin-binding protein (Figure 3, A, B, and G; Chen et al., 2013a). Next we introduced integrated arrays expressing GFP- or yellow fluorescent protein (YFP)–tagged phagosome markers into cpl-1(yq89) mutants, including RAB-5 (GFP::RAB-5), a small GTPase specific for early endosomes and phagosomes; 2xFYVE (YFP::2xFYVE), an indicator of PI3P; LST-4 (LST-4::GFP), a factor required for initiation of phagosome maturation; and RAB-7 (GFP::RAB-7), a small GTPase required for late endosome–phagosome fusion (Kinchen et al., 2008; Guo et al., 2010; Chen et al., 2013a). We found that phagosomes in cpl-1(yq89) germ lines are positive for these phagosome maturation factors at a similar level to that in wild type (Figure 3, C–G), indicating that loss of cpl-1 did not affect the phagosome maturation process. Thus CPL-1 should function at the stage of cell corpse digestion in phagolysosomes.
FIGURE 3:
Characterization of phagosome maturation in cpl-1 mutants. Representative images of cell corpse labeling by CED-1::GFP (A), GFP::moesin (B), GFP::RAB-5 (C), YFP::2xFYVE (D), GFP::RAB-7 (E), and LST-4::GFP (F). Cell corpses are indicated by arrows. Bars, 5 μm. (G) Quantification of the labeling of cell corpses by different phagosomal markers shown in A–F. We analyzed ≥100 cell corpses from ≥20 animals for each marker, and data were derived from three repeats. Comparisons were performed between N2 and cpl-1(yq89) mutants for each marker using an unpaired t test. All points have p > 0.05.
Expression and localization of CPL-1
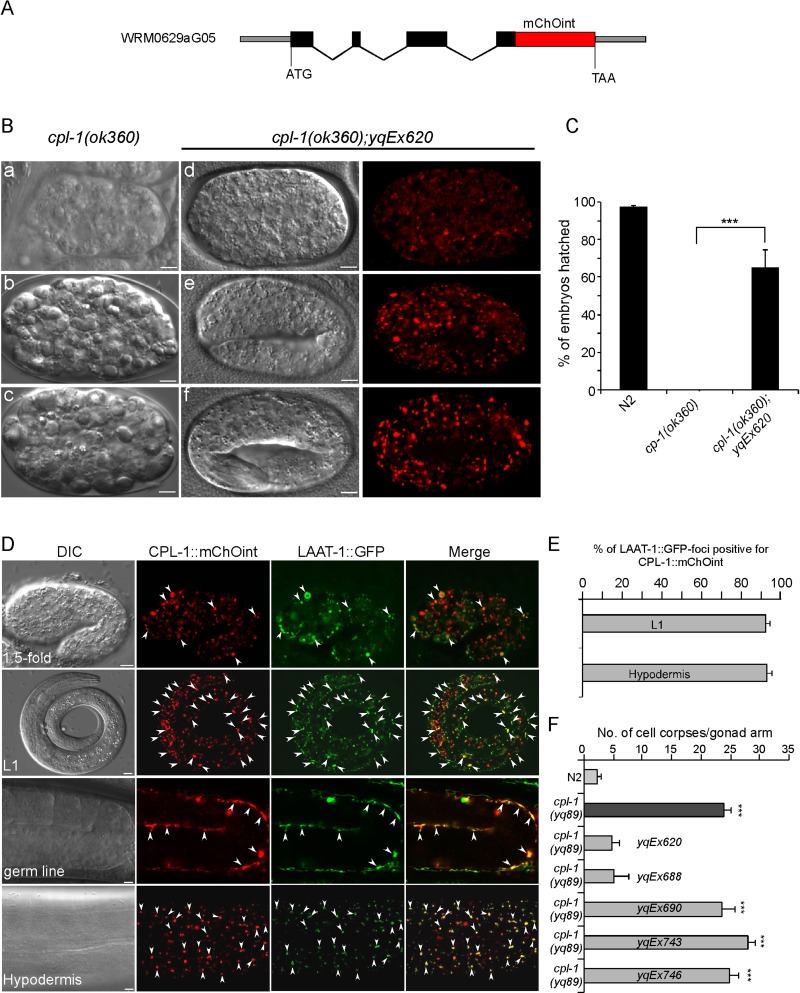
The foregoing findings show that CPL-1 plays an essential role in degradation of cell corpses generated by programmed cell death. To understand how CPL-1 acts in this process, we sought to determine the expression and localization of CPL-1 in a developmental context. First we generated a cpl-1 fosmid reporter by tagging a fluorescence protein, mChOint, at the C-terminus of CPL-1 (Figure 4A; Tursun et al., 2009). We then introduced the extrachromosomal arrays (e.g., yqEx620) expressing the cpl-1 fosmid reporter into cpl-1(ok360) deletion mutants. yqEx620 rescued the embryonic lethality of cpl-1(ok360) mutants. Whereas cpl-1(ok360) embryos lain by homozygous animals arrested at early stages and could rarely hatch, ∼70% of embryos generated by cpl-1(ok360);yqEx620 animals hatched and developed normally (Figure 4, B and C). CPL-1::mChOint is expressed ubiquitously from embryos to adults, including in germ line sheath cells and hypodermal cells, which function as engulfing cells for removing cell corpses (Figure 4D). The punctate localization of CPL-1::mChOint overlapped very well with the GFP-tagged lysosomal arginine and lysine transporter LAAT-1 (LAAT-1::GFP) driven by the promoter of the engulfing cell-specific ced-1 gene (Liu et al., 2012), indicating that CPL-1 is delivered to lysosomes (Figure 4, D and E). Of importance, transgenic arrays expressing CPL-1 also rescued the cell corpse phenotype in cpl-1 mutants. For example, cpl-1(yq89) mutants containing yqEx620 showed a similar level of germ cell corpses to that of wild-type animals. Similarly, another transgenic array, yqEx688 expressing CPL-1 under the control of its own promoter, also rescued the cell corpse phenotype of cpl-1(yq89) mutants (Figure 4F). In contrast, transgenic animals (yqEx690) expressing CPL-1 containing a point mutation similar to that in the qx304 mutants, in which the N-terminal 51 amino acid residues were deleted, failed to rescue the cell corpse phenotype in yq89 mutants (Figure 4F). Similarly, no rescuing effect was observed in cpl-1(yq89) mutants either expressing CPL-1 with a G230R mutation (yqEx746) as caused by the yq89 mutation or expressing CPL-1 with a C144A mutation (Hashmi et al., 2002) affecting the amino acid residue essential for CPL-1 activity (yqEx743) (Figure 4F), although the expression and localization patterns of these mutant proteins were similar to those of wild-type CPL-1 (Supplemental Figure S1).These findings indicate that the CPL-1 mutations in qx304 and yq89 mutants indeed damaged the normal function of CPL-1.
FIGURE 4:
Expression and subcellular localization of CPL-1::mChOint and its rescuing activity. (A) Schematic representation of the fosmid reporter of the cpl-1 gene fused in frame with mChOint. Solid boxes indicate exons, and wavy lines indicate introns. The red box indicates the mChOint ORF. (B) Representative images showing the rescue of embryonic lethality of cpl-1(ok360) mutants by the transgenic array yqEx620 expressing the cpl-1 fosmid reporter. (a–c) DIC images showing cpl-1(ok360) embryos that arrested at early embryonic stages. (d–f) DIC and fluorescence images of cpl-1(ok360) mutants carrying yqEx620, which developed normally. Bars, 5 μm. (C) Quantification of the rescuing effect of yqEx620 on the embryonic lethality of cpl-1(ok360) mutants grown at 20°C. We analyzed ≥100 embryos for each genotype, and data were derived for three repeats. ***p < 0.001, unpaired t test. (D) Expression and subcellular localization of CPL-1::mChOint driven by the cpl-1 promoter. Images of DIC, CPL-1::mChOint, and LAAT-1::GFP (driven by the ced-1 promoter) and merged images of CPL-1::mChOint with LAAT-1::GFP are shown for a 1.5-fold embryo, an L1 larvae, the germ line, and the hypodermis. Bars, 5 μm. Arrowheads indicate some of the foci with colocalization of CPL-1::mChOint and LAAT-1::GFP. (E) Quantification of the colocalization of LAAT-1::GFP driven by the ced-1 promoter and CPL-1::mChOint driven by the cpl-1 promoter. (F) Rescuing effect of CPL-1::mChOint transgenic arrays yqEx620 (cpl-1 fosmid reporter), yqEx688 (Pcpl-1cpl-1::mChOint), yqEx690 (Pcpl-1cpl-1(del51)::mChOint), yqEx743 (Pcpl-1cpl-1(C144A)::mChOint), and yqEx746 (Pcpl-1cpl-1(G230R)::mChOint) on cell corpse phenotype in cpl-1(yq89) mutants. cpl-1 mutants carrying transgenic arrays were grown at 25°C, and cell corpses were scored in animals at 24 h post L4 (n ≥ 15 for each genotype). Comparisons were performed between N2 and cpl-1(yq89) mutants with or without transgenic arrays as indicated using an unpaired t test. ***p < 0.001.
CPL-1 acts in phagolysosomes for cell corpse digestion
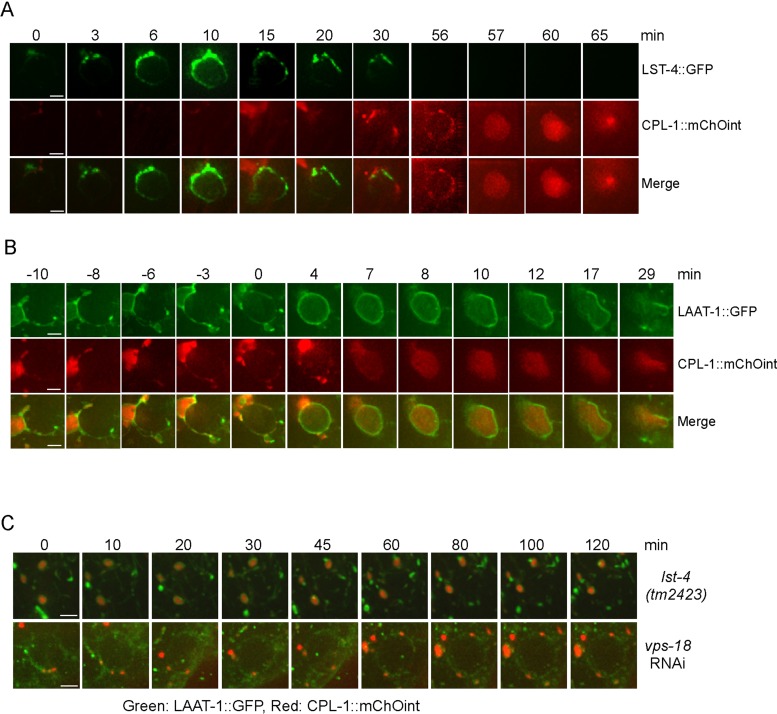
The capacity of CPL-1::mChOint to rescue the defective cell corpse clearance in cpl-1 mutants enabled us to monitor the dynamic recruitment of CPL-l to phagosomes during cell corpse removal. First, we compared the dynamic association of CPL-1 with LST-4, the C. elegans homologue of mammalian Snx9/18/33, which acts in a very early stage to initiate the phagosome maturation process (Almendinger et al., 2011; Chen et al., 2013a). No obvious overlapping of GFP-tagged LST-4 (LST-4::GFP) with CPL-1::mChOint was observed on phagosomes. Instead, CPL-1::mChOint was found to be recruited to phagosomes after LST-4 dissociation, indicating that CPL-1 acts at a late stage in the process of cell corpse clearance (Figure 5A). Next we examined whether CPL-1 and the lysosomal membrane protein LAAT-1 were corecruited to cell corpse–containing phagosomes. We found that LAAT-1 and CPL-1 were simultaneously docking on phagosomes (Figure 5B, −10 min). Surprisingly, CPL-1 remained mostly enriched at the same docking site, whereas LAAT-1 progressively encircled the phagosome (Figure 5B, −10 to 4 min). A few minutes after LAAT-1 fully encircled the phagosome, CPL-1 was incorporated into the phagosome (Figure 5B, 4–8 min). These findings suggest that the incorporation of CPL-1 into phagosomes appears to lag behind that of lysosomal membrane proteins during phagosome–lysosome fusion.
FIGURE 5:
Analysis of dynamic recruitment of CPL-1::mChOint to cell corpse–containing phagosomes. (A) Time-lapse analysis of the dynamic association of LST-4::GFP and CPL-1::mChOint with phagosomes. The time point that LST-4::GFP started to appear on the phagosome was set as 0 min. Bars, 2 μm. (B) Time-lapse monitoring of phagosome recruitment of LAAT-1::GFP and CPL-1::mChOint. The time point that LAAT-1::GFP mostly encircled the phagosome was set as 0 min. Bars, 2 μm. (C) Time-lapse monitoring of phagosomal association of both LAAT-1::GFP (green) and CPL-1::mChOint (red) in germ lines of lst-4(tm2423) and vps-18(RNAi) animals. Bars, 2 μm. Animals were cultured at 25°C for imaging analysis.
We also examined the requirement of phagosome maturation effectors for incorporation of CPL-1 to phagosomes. In lst-4(tm2423) deletion mutants, in which phagosome maturation arrests at early stage, lysosomes labeled by both CPL-1::mChOint and LAAT-1::GFP were found to dock on the periphery of phagosomes but did not fuse with phagosomes (Figure 5C). RNA interference (RNAi) depletion of vps-18 also led to a failure of CPL-1::mChOint– and LAAT-1::GFP–positive lysosomes to fuse with phagosomes, although these lysosomes were also found to dock on phagosomes (Figure 5C). These findings indicate that phagosome maturation factors are required for phagolysosome formation, defects of which lead to the failure of phagosomal incorporation of CPL-1 and likely other lysosomal proteases.
Role of other cathepsin proteases in cell corpse clearance
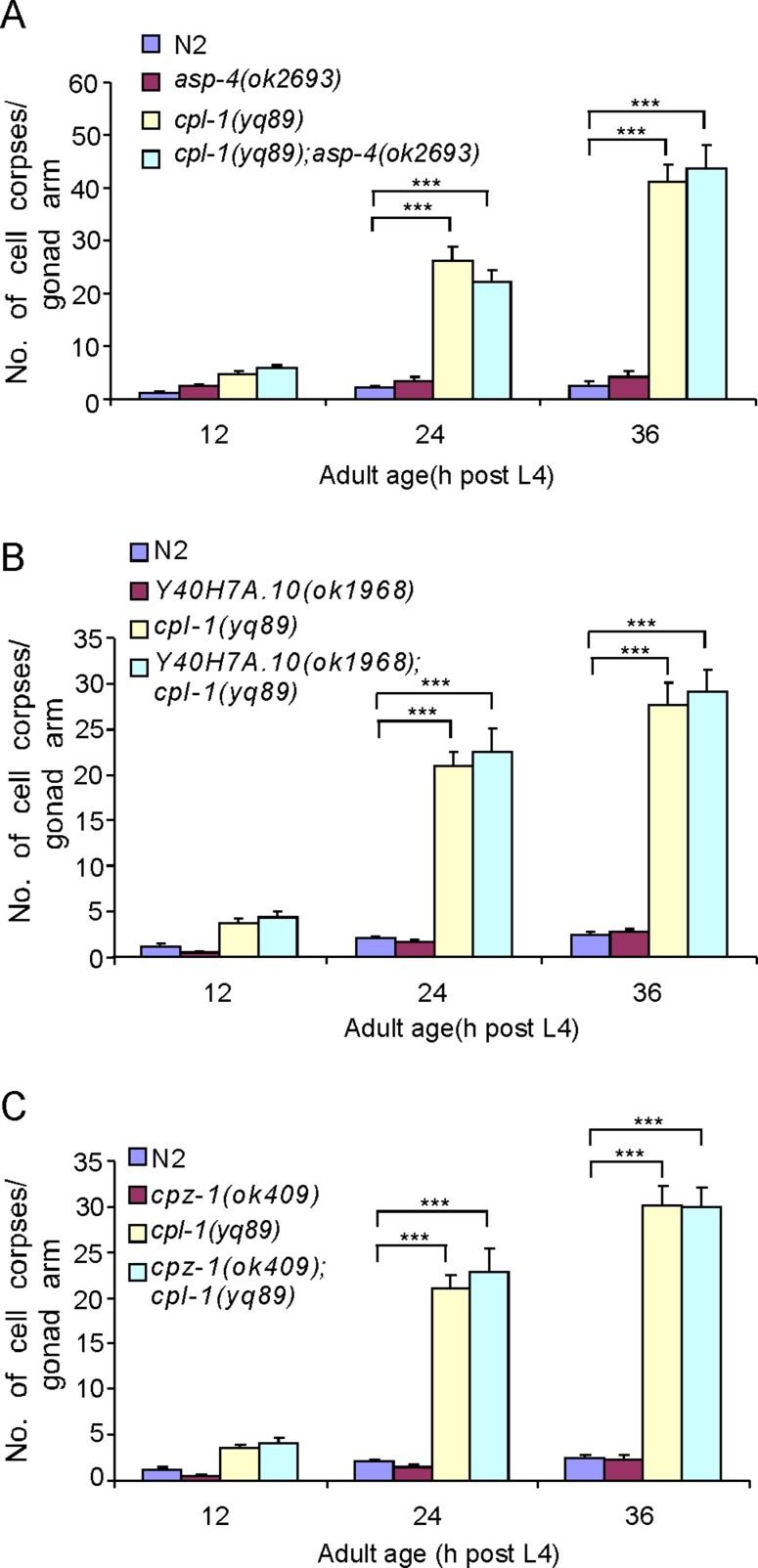
To determine whether CPL-1 is the major cathepsin required for cell corpse clearance in C. elegans, we evaluated the role of other lysosomal cathepsins in cell corpse removal by performing RNAi to deplete homologues of mammalian cathepsins. The effect of RNAi was confirmed by measuring the reduction of mRNA levels of several cathepsins selected randomly using real-time reverse-transcription PCR (Supplemental Figure S2). RNAi of individual genes in the families of cathepsin A, B, D, E, H, F, S, and Z did not result in an obvious accumulation of cell corpses in the germ line (Table 1). Similarly, RNAi of the other three members in addition to CPL-1(T03E6.7) of the cathepsin L family did not induce any discernible increase in germ cell corpses, except for RNAi of cpl-1, which gave similar cell corpse accumulation as the germ line null allele (Table 1). These results indicate that inactivation of single member of these cathepsin proteases was not sufficient to perturb cell corpse degradation. To determine whether these enzymes act synergistically with CPL-1 to promote cell corpse degradation, we inactivated each of these genes in cpl-1(yq89) mutants. Compared with control RNAi treatment, RNAi of individual cathepsins did not further enhance the cell corpse phenotype of cpl-1(yq89) mutants (Table 1). In addition, we also examined deletion mutants of asp-4(ok2693), Y40H7A.10(ok1968), and cpz-1(ok409) but observed no obvious cell corpse phenotype. Furthermore, in double mutants of cpl-1(yq89) with these deletion mutants, the numbers of cell corpses remained the same as for cpl-1(yq89) single mutants (Figure 6). Together these findings suggest that CPL-1 plays a leading role among lysosomal cathepsins in degradation of apoptotic cells.
TABLE 1:
Analysis of germ cell corpses in animals treated with RNAi of candidate lysosomal cathepsins in C. elegans.
| Human cathepsin | C. elegans cathepsin genes (RNAi) | Number of cell corpses in rrf-3(pk1426) mutants (mean ± SEM) | Number of germ cell corpses in cpl-1(yq89) mutants (mean ± SEM) |
|---|---|---|---|
| Control RNAi | 1.4 ± 0.3 | ||
| Control RNAi | 28.9 ± 1.5 | ||
| Cathepsin A | F13D12.6 | 1.7 ± 0.5 | 26.4 ± 1.7 |
| F32A5.3 | 1.9 ± 0.5 | 25.4 ± 2.2 | |
| F41C3.5 | 2.2 ± 0.3 | 27.4 ± 1.5 | |
| K10B2.2 | 1.8 ± 0.4 | 30.1 ± 1.7 | |
| Y16B4A.2 | 1.9 ± 0.5 | 29.3 ± 1.6 | |
| Y32F6A.5 | 1.7 ± 0.2 | 30.1 ± 1.9 | |
| K10C2.1 | 1.7 ± 0.4 | 33.9 ± 1.7 | |
| Y40D12A.2 | 2.6 ± 0.3 | 29.6 ± 2.2 | |
| Cathepsin D | asp-1(Y39B6A.20) | 2.9 ± 0.4 | 32.5 ± 1.8 |
| asp-3(H22K11.1) | 1.8 ± 0.4 | 28.7 ± 2.4 | |
| asp-4(R12H7.2) | 1.5 ± 0.5 | 32.9 ± 2.1 | |
| Cathepsin B | cpr-1(C52E4.1) | 1.1 ± 0.3 | 29.1 ± 1.7 |
| cpr-2(F36D3.9) | 1.4 ± 0.2 | 27.8 ± 2.3 | |
| cpr-3(T10H4.12) | 1.8 ± 0.4 | 32.3 ± 2.2 | |
| cpr-4(F44C4.3) | 1.7 ± 0.4 | 32.4 ± 1.6 | |
| cpr-5(W07B8.5) | 2.3 ± 0.4 | 30.1 ± 1.5 | |
| cpr-6(C25B8.3) | 2.2 ± 0.3 | 30.3 ± 2.2 | |
| F32H5.1 | 2.3 ± 0.4 | 32.9 ± 2.0 | |
| F57F5.1 | 2.5 ± 0.3 | 30.7 ± 2.0 | |
| W07B8.1 | 2.0 ± 0.2 | 27.6 ± 2.4 | |
| W07B8.4 | 2.0 ± 0.3 | 29.0 ± 2.5 | |
| Y65B4A.2 | 1.9 ± 0.2 | 27.9 ± 1.9 | |
| Cathepsin E | asp-5(F21F8.3) | 2.6 ± 0.5 | 29.1 ± 2.3 |
| asp-6(F21F8.7) | 2.1 ± 0.3 | 30.1 ± 1.7 | |
| asp-9(C11D2.2) | 1.7 ± 0.5 | 31.0 ± 1.9 | |
| asp-10(C15C8.3) | 2.1 ± 0.4 | 27.2 ± 1.9 | |
| asp-12(F21F8.4) | 2.2 ± 0.4 | 32.4 ± 2.6 | |
| asp-19(ZK384.6) | 1.6 ± 0.3 | 33.4 ± 1.7 | |
| Cathepsin F | R07E3.1 | 1.7 ± 0.3 | 31.1 ± 2.3 |
| R09F10.1 | 3.0 ± 0.6 | 24.7 ± 1.6 | |
| F41E6.6 | 1.7 ± 0.3 | 29.2 ± 1.6 | |
| Cathepsin H | K02E7.10 | 2.3 ± 0.6 | 29.8 ± 2.1 |
| tag-329(C50F4.3) | 2.5 ± 0.3 | 31.2 ± 1.9 | |
| Cathepsin L | Y51A2D.1 | 2.3 ± 0.6 | 30.4 ± 1.5 |
| Y51A2D.8 | 2.3 ± 0.4 | 26.9 ± 1.4 | |
| Y71H2AR.2 | 2.4 ± 0.4 | 30.4 ± 2.0 | |
| cpl-1(T03E6.7) | 25.0 ± 1.6 | 29.1 ± 2.1 | |
| Cathepsin S | C32B5.7 | 1.4 ± 0.2 | 29.4 ± 2.2 |
| F15D4.4 | 2.8 ± 0.6 | 26.8 ± 2.5 | |
| Y40H7A.10 | 1.9 ± 0.2 | 33.1 ± 1.6 | |
| Y71H2AM.25 | 1.4 ± 0.3 | 29.8 ± 1.3 | |
| Cathepsin Z | cpz-1(F32B5.8) | 1.3 ± 0.3 | 32.0 ± 1.9 |
| cpz-2(M04G12.2) | 1.7 ± 0.3 | 28.8 ± 1.5 |
Germ cell corpses were analyzed in rrf-3(pk1462) adult animals (24 h post L4) grown on RNAi plates at 20°C and in cpl-1(yq89) animals that were grown to L4 molt on RNAi plates at 20°C and then transferred to 25°C for further culture of 24 h. The identity of each RNAi clone was confirmed by sequencing.
FIGURE 6:
(A) Quantification of germ cell corpses in asp-4(ok2693) and cpl-1(yq89) single and double mutants. (B) Quantification of germ cell corpses in Y40H7A.10(ok1968) and cpl-1(yq89) single and double mutants. (C) Quantification of germ cell corpses in cpz-1(ok409) and cpl-1(yq89) single and double mutants. Animals were grown at 25°C, and germ cell corpses were scored and analyzed as described in Figure 1. Comparisons were performed between N2 and mutants and between cpl-1(yq89) and its double mutants with asp-4(ok2693), Y407A.10(ok1968), and cpz-1(ok409) using an unpaired t test. ***p < 0.001. All other points have p > 0.05.
CPL-1 is required for degradation of endocytic and autophagic cargoes
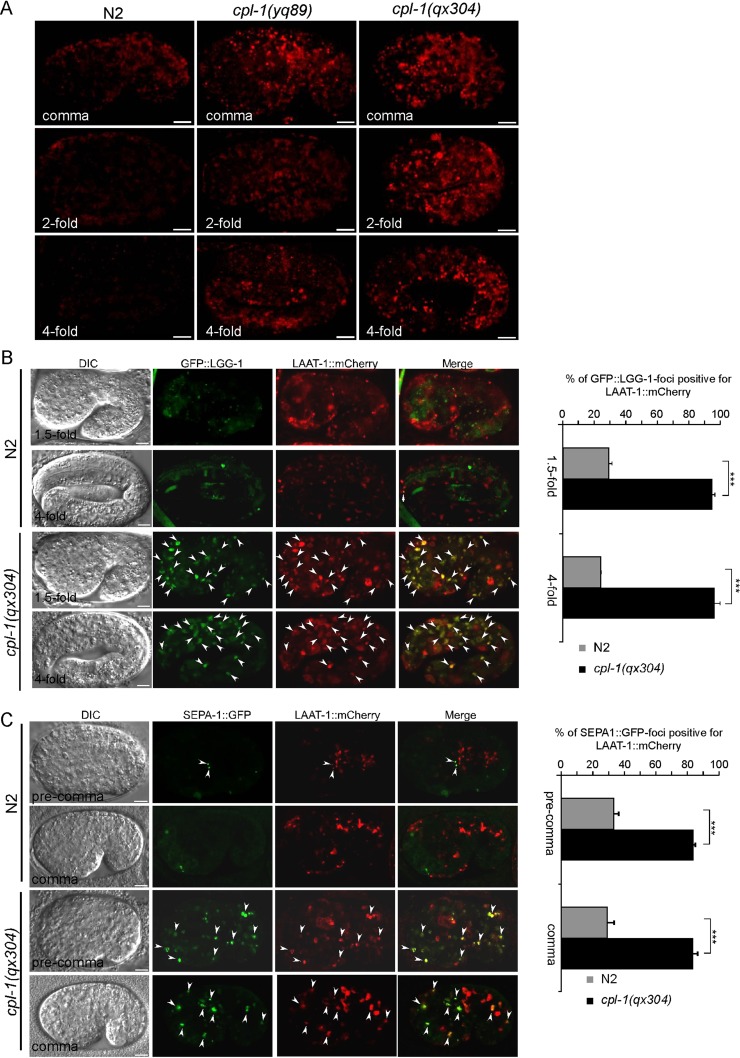
Embryos of cpl-1(ok360) deletion mutants are lethal, indicating that CPL-1 is required for embryonic development. To understand the role of CPL-1 in this process, we examined whether yolk proteins are properly degraded in cpl-1 mutant embryos, because yolk is the major source of nutrition required for embryogenesis. In wild type, yolk is present at a high level in early-stage embryos but is progressively reduced in later-stage embryos, as indicated by immunostaining (Figure 7A). In cpl-1(yq89) and cpl-1(qx304) mutants, however, yolk was found to be abundant at all embryonic stages examined (Figure 7A), consistent with previous observations in cpl-1(ok360) mutants (Britton and Murray, 2004). Together these findings indicate that CPL-1 is required for yolk degradation during embryogenesis.
FIGURE 7:
Loss of cpl-1 function impaired lysosomal degradation of endocytic and autophagic cargoes. (A) Immunostaining of yolk in embryos of N2, cpl-1(yq89) and cpl-1(qx304) embryos. Embryos of comma, twofold, and fourfold stages are shown for each genotype. Bars, 5 μm. (B) Representative images of DIC, GFP::LGG-1, and LAAT-1::mCherry and merged images of GFP::LGG-1 and LAAT-1::mCherry in N2 and cpl-1(qx304) embryos. A 1.5- and 4-fold embryo are shown for both N2 and cpl-1(qx304) mutants. Arrowheads indicate some of the foci with GFP::LGG-1 and LAAT-1::mCherry colocalization. Bars, 5 μm. Right, quantification of colocalization of GFP::LGG-1 with LAAT-1::mCherry. (C) Representative images of DIC, SEPA-1::GFP, and LAAT-1::mCherry and merged images of SEPA-1::GFP with LAAT-1::mCherry of N2 and cpl-1(qx304) animals. A precomma and a comma embryo are shown for both N2 and cpl-1(qx304) mutants. Arrowheads indicate some of the foci with SEPA-1::GFP and LAAT-1::mCherry colocalization. Bars, 5 μm. Right, quantification of colocalization of SEPA-1::GFP with LAAT-1::mCherry. All animals were raised at 25°C for analysis. In B, right, and C, right, three embryos were analyzed at each developmental stage for each genotype. Comparisons were performed using an unpaired t test. ***p < 0.001.
To determine whether CPL-1 is also essential for degrading cargoes resulting from autophagy, we examined the accumulation of autophagosomes marked by GFP-fused LGG-1, a mammalian LC3 homologue, and GFP-fused SEPA-1, a receptor of selective autophagy (Zhang et al., 2009), in cpl-1 mutant embryos. As shown in Figure 7, B and C, GFP::LGG-1 and SEPA-1::GFP puncta accumulated in cpl-1(qx304) embryos, which overlapped with lysosomes positive for LAAT-1::mCherry. This is in contrast to the finding that few GFP::LGG-1 and SEPA-1::GFP foci existed and that they did not overlap with LAAT-1::mCherry-positive lysosomes in wild type. These observations indicate that CPL-1 is indispensable for lysosomal degradation of autophagic cargoes during embryogenesis.
DISCUSSION
The fusion of phagosomes with lysosomes is required for degradation of cell corpses in engulfing cells. However, it has not been documented whether a single lysosomal protease plays a major role in cell corpse degradation after phagolysosome formation. Here we identify the cysteine protease CPL-1 as the major protease required for cell corpse degradation in the phagolysosome. Our genetic screen identified two hypomorphic alleles of cpl-1, yq89 and qx304, both of which exhibited temperature-sensitive accumulation of cell corpses in germ lines. yq89 caused a G230R mutation in the pept_C1 domain, and qx304 resulted in deletion of the N-terminal 51 amino acids in the CPL-1 protein, respectively. Our data suggest that these mutations reduced but did not abolish CPL-1 activity. It is likely that the rate of germ cell corpse clearance is lower at 20 than at 25°C. Thus the activity of these mutant CPL-1 proteins is sufficient for cell corpse clearance at lower temperature. At 25°C, however, the activity of these CPL-1 mutant proteins is not strong enough for a higher rate of cell corpse digestion, leading to accumulation of germ cell corpses in mutant animals. Consistently, cpl-1(ok360) deletion mutants exhibited a strong accumulation of germ cell corpses at all temperatures. By generating transgenes expressing mChOint-fused CPL-1 under the control of the cpl-1 gene promoter, we found that CPL-1 is expressed in multiple tissues and cell types, including the engulfing cells. The cell corpse phenotype in cpl-1 mutants was rescued by transgenes expressing wild-type but not mutant CPL-1, further confirming that cpl-1 is required for clearance of cell corpses.
In cpl-1 mutants, germ cell corpses are labeled by the phagocytic receptor CED-1, similar to wild type. The rearrangement of the actin cytoskeleton around cell corpses also appears normal, indicating that loss of cpl-1 does not impair the engulfment of cell corpses. Furthermore, phagosomes containing cell corpses normally recruit factors required for phagosome maturation in cpl-1 mutants. For example, LST-4 and RAB-5, two factors acting on early phagosomes, and RAB-7, a small GTPase specific for late phagosomes, are similarly observed on phagosomes in germ lines in both wild-type and cpl-1 mutant animals. These facts indicate that CPL-1 plays a role in phagolysosomes rather than phagosome maturation. In support of this, our time-course chase of the dynamic recruitment of CPL-1 revealed that CPL-1 was recruited and incorporated into phagosomes after the dissociation of LST-4, an essential factor for initiation of phagosome maturation. It is very interesting to observe that the incorporation of CPL-1 into phagosomes occurs slightly later than that of the lysosomal membrane protein LAAT-1, which suggests that lysosomal components likely fuse with the phagosome in a sequential manner during phagolysosome formation. Moreover, we found that CPL-1 failed to be incorporated in phagosomes in loss-of-function mutants of factors essential for phagosome maturation, including LST-4 and the HOPS complex. Together our findings demonstrate that CPL-1 acts in phagolysosomes for cell corpse degradation. Mutations in CPL-1 or failure to incorporate CPL-1 in phagosomes lead to defective cell corpse degradation.
In mammalian cells, the lysosome contains three major groups of proteases based on the catalytic residues in the active sites, including aspartic (cathepsin D and E), serine (cathepsin A and G), and cysteine (cathepsin B, C, F, H, K, L, O, S, V, W, and Z) cathepsins (Reiser et al., 2010). Our data indicated that RNAi depletion of individual homologues of these cathepsins, except CPL-1, did not result in a discernible increase in germ cell corpses in C. elegans, suggesting that these cathepsins on their own do not affect cell corpse degradation; instead, they may need to act coordinately during cell corpse degradation. In addition, RNAi depletion of other cathepsins did not enhance cell corpse phenotype in cpl-1 weak loss-of-function mutants, suggesting that these cathepsins do not play a similar role to CPL-1 in cell corpse digestion. These findings further suggest that CPL-1 is the major lysosomal protease required for degrading cell corpses after formation of phagolysosome. It is possible that CPL-1 is required for breaking down cell corpses, whereas other cathepsins are needed for further digestion of cell corpse debris, depending on their enzymatic specificities.
Previous studies revealed that CPL-1 is required for yolk processing during embryogenesis in C. elegans (Britton and Murray, 2004). In our study, we also observed defective yolk processing in the cpl-1 weak loss-of-function mutants yq89 and qx304. In addition, we found that degradation of autophagosomes/autophagic cargoes was also impaired in cpl-1 mutants. For example, foci of GFP::LGG-1, an autophagosome marker, and SEPA-1::GFP, an autophagic substrate, accumulated in cpl-1(qx304) embryos. The GFP::LGG-1 and SEPA-1::GFP foci colocalized with lysosomes, indicating that they resulted from defective lysosomal degradation. The requirement of CPL-1 in clearance of autophagic cargoes in C. elegans may resemble the role of cathepsin L in autophagy in mammalian cells. For example, in HeLa and Huh-7 cells, specific inhibition of cathepsin L by a chemical inhibitor prevented degradation of LC3-II and GABARAP, mammalian homologues of C. elegans LGG-1 (Ueno and Takahashi, 2009). In comparison, inhibition of cathepsin B did not have an obvious effect on these two proteins. Thus mammalian cathepsin L appears to have certain substrate specificity during autophagosome degradation (Ueno and Takahashi, 2009). It remains to be determined whether CPL-1 has similar substrate specificity to human cathepsin L for the degradation of both phagocytic and autophagic cargoes in C. elegans.
MATERIALS AND METHODS
C. elegans strains and genetics
The Bristol strain N2 was used as wild type. cpl-1(ok360) deletion mutants were obtained from the C. elegans Genetics Center (University of Minnesota, Minneapolis, MN). Other mutant alleles used in this study are listed by linkage groups: LG I: ced-1(e1735) and cpz-1(ok409). LG II: vps-18(tm1125) and rrf-3(pk1426). LG IV: Y40H7A.10 (ok1968) and lst-4(tm2423). LG V: unc-76(e911). LG X: asp-4 (ok2693). All mutants were outcrossed at least four times with N2. Integrated arrays used in this study are as follows: adIs2122 (Plgg-1gfp::lgg-1) and bpIs131 (Psepa-1sepa-1::gfp; provided by Hong Zhong, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China), opIs334 (Pced-1yfp::2×fyve; provided by K. S. Ravichandran, University of Virginia, Charlottesville, VA), qxIs66 (Pced-1gfp::rab-7), qxIs352 (Pced-1laat-1::mCherry), qxIs354 (Pced-1laat-1::gfp), qxIs408 (Pced-1gfp::rab-5), smIs34 (Pced-1ced-1::gfp), yqIs114 (Plst-4lst-4(cDNA)::gfp), and yqIs121 (Pced-1gfp::Moesin). Animals carrying stably integrated array were outcrossed with the N2 strain four times. C. elegans cultures and genetic crosses were performed essentially according to standard procedures (Brenner, 1974).
Mutant screen and gene mapping
To screen for mutants with germ cell corpse accumulation, synchronized L4-stage animals were treated with EMS (50 mM) for 4 h. The F2 progeny of EMS-treated animals were grown to the age of 24-48 h after the L4 molt at 25°C and observed under differential interference contrast (DIC) microscopes. This allowed us to obtain hypomorphic mutants showing temperature-sensitive accumulation of germ cell corpses. Mapping of mutations was performed by using single-nucleotide polymorphism mapping as described by Davis et al. (2005).
Expression constructs
To make the cpl-1 fosmid reporter construct, mChOint was inserted into the fosmid WRM0629aG05 containing the cpl-1 gene using the assay as described by Tursun et al. (2009). To make the Pcpl-1cpl-1::mChOint construct, a DNA fragment of 5762 base pairs containing the promoter region (1761 base pairs), open reading frame (ORF), and 3′-untranslated region (UTR; 505 base pairs) of the cpl-1 gene and the ORF of mChOint was amplified from the cpl-1 fosmid reporter construct by PCR and cloned into the pEASY-T1 Simple vector (TransGen, Beijing, China). The Pcpl-1cpl-1(del51)::mChOint construct was generated by changing the first ATG of the cpl-1 ORF to ATA with a PCR-based mutagenesis assay, using the Pcpl-1cpl-1::mChOint vector as the template and a pair of oligos as follows (mutation site is underlined): sense, 5′-ATAAAATTCCAGAATAAACCGATTCATTCTTCTGGCACTG-3′; antisense, 5′-CAGTGCCAGAAGAATGAATCGGTTTATTCTGGAATTTTAT-3′. Pcpl-1cpl-1(G230R)::mChOint and Pcpl-1cpl-1(C144A)::mChOint constructs were generated by PCR-based mutagenesis using Pcpl-1cpl-1::mChOint as template and confirmed by sequencing.
cpl-1 transgenic arrays
The extrachromosomal array yqEx620 is one transgenic line from three lines generated by injecting the cpl-1 fosmid reporter into unc-76(e911) animals. yqEx688 is a representative extrachromosomal array of several similar transgenic lines expressing Pcpl-1cpl-1::mChOint construct in which CPL-1 expression was controlled by its own promoter and 3′-UTR. yqEx690 is a representative transgenic array expressing the Pcpl-1cpl-1(del51)::mChOint construct with the first 51 amino acids of CPL-1 deleted. yqEx743 and yqEx746 are independent transgenic arrays expressing Pcpl-1cpl-1(C144A)::mChOint and Pcpl-1cpl-1(G230R)::mChOint, respectively.
RNAi experiments
RNAi experiments were performed by using bacterial feeding assays as described previously (Chen et al., 2013a). In most cases, L4-stage animals of rrf-3(pk1426) or cpl-1(yq89) were transferred to plates seeded with bacteria expressing either control double-stranded RNA (dsRNA; L4440 empty vector; Control RNAi) or dsRNA corresponding to the ORFs of genes of interest. Germ cell corpses were observed in adults of the progeny at different time points.
Germ cell corpse analysis
Animals synchronized to different adult stages were scored for germ cell corpses under Nomarski optics. Animals were grown at 20°C unless otherwise indicated. For cell corpse analysis in yq89 and qx304 mutants or their double mutants with other genes, animals were shifted to 25°C after growing to the L4 molt at 20°C and scored for germ cell corpses 12, 24, and 36 h later. At each time point, germ cell corpses in the meiotic region of one gonad arm were counted for every animal, and ≥15 animals were analyzed. Germ cell corpses in cpl-1(ok360) animals were scored and analyzed at 20°C.
LysoSensor Green and Hoechst 33342 staining
Adult animals at 36 h after the L4 molt were dissected in gonad dissection buffer (60 mM NaCl, 32 mM KCl, 3 mM Na2HPO4, 2 mM MgCl2, 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 50 μg/ml penicillin, 50 μg/ml streptomycin, 100 μg/ml neomycin, 10 mM glucose, 33% fetal calf serum, and 2 mM CaCl2) containing 1 μM LysoSensor Green DND-189 (Invitrogen, Carlsbad, CA) or 1 μM Hoechst33342 (Invitrogen). The staining of cell corpses was examined with fluorescence microscopy.
Time-lapse imaging
Time-lapse imaging was performed under 100× objectives at 20°C by using the DeltaVision imaging system (GE Healthcare, Little Chalfont, United Kindom). Animals were anesthetized with 1 mM levamisole in M9 buffer (1 l contains 3 g of KH2PO4, 6 g of Na2HPO4, 5 g of NaCl, and 1 mM MgSO4) and mounted on 2% agar pads. Time-lapse images of CPL-1::mChOint, LAAT-1::GFP, and LST-4::GFP were captured every 30 s for 120 min with a Z-series of 1 μm/section for a total of 20 sections for each time point. The excitation filters used for GFP and mChOint are 488 and 559 nm, respectively.
Immunostaining
Embryos with mixed stages were fixed with methanol for 5 min and blocked in phosphate-buffered saline (PBS) containing 0.3% Triton X-100 and 5% normal goat serum for 1 h at room temperature. The samples were then incubated with VIT-2 antibody (Chen et al., 2010a) in blocking buffer at 4°C overnight. After extensive washing with PBS containing 0.3% Triton X-100, the samples were incubated with Cy3-conjugated secondary antibody for 2 h at room temperature. The samples were washed again as described and mounted on slides for visualizing using an inverted confocal microscope (100× objectives; Olympus, Tokyo, Japan).
Supplementary Material
Acknowledgments
We thank the C. elegans Genetic Center and Hong Zhang, K. S. Ravichandran, and M. O. Hengartner for strains and reagents. This research was supported by grants from the National Science Foundation of China (31230043 and 31025015), the National Basic Research Program of China (2013CB910102 and 2011CB910102), and the Chinese Academy of Sciences (KJZD-EW-L08).
Abbreviations used:
- DIC
differential interference contrast
- EMS
ethyl methanesulfonate
- GFP
green fluorescent protein
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-01-0015) on May 14, 2014.
REFERENCES
- Almendinger J, Doukoumetzidis K, Kinchen JM, Kaech A, Ravichandran KS, Hengartner MO. A conserved role for SNX9-family members in the regulation of phagosome maturation during engulfment of apoptotic cells. PLoS One. 2011;6:e18325. doi: 10.1371/journal.pone.0018325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton C, Murray L. Cathepsin L protease (CPL-1) is essential for yolk processing during embryogenesis in Caenorhabditis elegans. J Cell Sci. 2004;117:5133–5143. doi: 10.1242/jcs.01387. [DOI] [PubMed] [Google Scholar]
- Cabello J, Neukomm LJ, Gunesdogan U, Burkart K, Charette SJ, Lochnit G, Hengartner MO, Schnabel R. The Wnt pathway controls cell death engulfment, spindle orientation, and migration through CED-10/Rac. PLoS Biol. 2010;8:e1000297. doi: 10.1371/journal.pbio.1000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Jiang Y, Zeng S, Yan J, Li X, Zhang Y, Zou W, Wang X. Endocytic sorting and recycling require membrane phosphatidylserine asymmetry maintained by TAT-1/CHAT-1. PLoS Genet. 2010a;6:e1001235. doi: 10.1371/journal.pgen.1001235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, et al. Clathrin and AP2 are required for phagocytic receptor-mediated apoptotic cell clearance in Caenorhabditis elegans. PLoS Genet. 2013a;9:e1003517. doi: 10.1371/journal.pgen.1003517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Xiao H, Zhang K, Wang B, Gao Z, Jian Y, Qi X, Sun J, Miao L, Yang C. Retromer is required for apoptotic cell clearance by phagocytic receptor recycling. Science. 2010b;327:1261–1264. doi: 10.1126/science.1184840. [DOI] [PubMed] [Google Scholar]
- Chen YZ, Mapes J, Lee ES, Skeen-Gaar RR, Xue D. Caspase-mediated activation of Caenorhabditis elegans CED-8 promotes apoptosis and phosphatidylserine externalization. Nat Commun. 2013b;4:2726. doi: 10.1038/ncomms3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darland-Ransom M, Wang X, Sun CL, Mapes J, Gengyo-Ando K, Mitani S, Xue D. Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science. 2008;320:528–531. doi: 10.1126/science.1155847. [DOI] [PubMed] [Google Scholar]
- Davis MW, Hammarlund M, Harrach T, Hullett P, Olsen S, Jorgensen EM. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics. 2005;6:118. doi: 10.1186/1471-2164-6-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deBakker CD, et al. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol. 2004;14:2208–2216. doi: 10.1016/j.cub.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Elliott MR, Ravichandran KS. Clearance of apoptotic cells: implications in health and disease. J Cell Biol. 2010;189:1059–1070. doi: 10.1083/jcb.201004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny TL, et al. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Guo P, Hu T, Zhang J, Jiang S, Wang X. Sequential action of Caenorhabditis elegans Rab GTPases regulates phagolysosome formation during apoptotic cell degradation. Proc Natl Acad Sci USA. 2010;107:18016–18021. doi: 10.1073/pnas.1008946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashmi S, Britton C, Liu J, Guiliano DB, Oksov Y, Lustigman S. Cathepsin L is essential for embryogenesis and development of Caenorhabditis elegans. J Biol Chem. 2002;277:3477–3486. doi: 10.1074/jbc.M106117200. [DOI] [PubMed] [Google Scholar]
- Hsu TY, Wu YC. Engulfment of apoptotic cells in C. elegans is mediated by integrin alpha/SRC signaling. Curr Biol. 2012;20:477–486. doi: 10.1016/j.cub.2010.01.062. [DOI] [PubMed] [Google Scholar]
- Hurwitz ME, Vanderzalm PJ, Bloom L, Goldman J, Garriga G, Horvitz HR. Abl kinase inhibits the engulfment of apoptotic [corrected] cells in Caenorhabditis elegans. PLoS Biol. 2009;7:e99. doi: 10.1371/journal.pbio.1000099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen JM, Doukoumetzidis K, Almendinger J, Stergiou L, Tosello-Trampont A, Sifri CD, Hengartner MO, Ravichandran KS. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zou W, Zhao D, Yan J, Zhu Z, Lu J, Wang X. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 2009;136:2445–2455. doi: 10.1242/dev.035949. [DOI] [PubMed] [Google Scholar]
- Liu B, Du H, Rutkowski R, Gartner A, Wang X. LAAT-1 is the lysosomal lysine/arginine transporter that maintains amino acid homeostasis. Science. 2012;337:351–354. doi: 10.1126/science.1220281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Zhang Y, Hu T, Guo P, Li W, Wang X. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development. 2008;135:1069–1080. doi: 10.1242/dev.016063. [DOI] [PubMed] [Google Scholar]
- Mangahas PM, Yu X, Miller KG, Zhou Z. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J Cell Biol. 2008;180:357–373. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapes J, Chen YZ, Kim A, Mitani S, Kang BH, Xue D. CED-1, CED-7, and TTR-52 regulate surface phosphatidylserine expression on apoptotic and phagocytic cells. Curr Biol. 2012;22:1267–1275. doi: 10.1016/j.cub.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm LJ, et al. Loss of the RhoGAP SRGP-1 promotes the clearance of dead and injured cells in Caenorhabditis elegans. Nat Cell Biol. 2011a;13:79–86. doi: 10.1038/ncb2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neukomm LJ, et al. The phosphoinositide phosphatase MTM-1 regulates apoptotic cell corpse clearance through CED-5-CED-12 in C. elegans. Development. 2011b;138:2003–2014. doi: 10.1242/dev.060012. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat Cell Biol. 2000;2:131–136. doi: 10.1038/35004000. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Horvitz HR. The engulfment process of programmed cell death in Caenorhabditis elegans. Annu Rev Cell Dev Biol 20, 193–221. 2004 doi: 10.1146/annurev.cellbio.20.022003.114619. [DOI] [PubMed] [Google Scholar]
- Reiser J, Adair B, Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest. 2010;120:3421–3431. doi: 10.1172/JCI42918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, et al. Arl8/ARL-8 functions in apoptotic cell removal by mediating phagolysosome formation in Caenorhabditis elegans. Mol Biol Cell. 2013;24:1584–1592. doi: 10.1091/mbc.E12-08-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, He B, Lu N, Conradt B, Grant BD, Zhou Z. Phagocytic receptor signaling regulates clathrin and epsin-mediated cytoskeletal remodeling during apoptotic cell engulfment in C. elegans. Development. 2013;140:3230–3243. doi: 10.1242/dev.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki J, Denning DP, Imanishi E, Horvitz HR, Nagata S. Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science. 2013;341:403–406. doi: 10.1126/science.1236758. [DOI] [PubMed] [Google Scholar]
- Tursun B, Cochella L, Carrera I, Hobert O. A toolkit and robust pipeline for the generation of fosmid-based reporter genes in C. elegans. PLoS One. 2009;4:e4625. doi: 10.1371/journal.pone.0004625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno T, Takahashi K. A cathepsin L-specific inhibitor preferentially inhibits degradation of autophagosomal LC3 and GABARAP in HeLa and Huh-7 cells. Autophagy. 2009;5:878–879. doi: 10.4161/auto.9097. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat Cell Biol. 2010;12:655–664. doi: 10.1038/ncb2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, et al. C. elegans mitochondrial factor WAH-1 promotes phosphatidylserine externalization in apoptotic cells through phospholipid scramblase SCRM-1. Nat Cell Biol. 2007;9:541–549. doi: 10.1038/ncb1574. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12. Science. 2003;302:1563–1566. doi: 10.1126/science.1087641. [DOI] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. The C. elegans cell corpse engulfment gene ced-7 encodes a protein similar to ABC transporters. Cell. 1998a;93:951–960. doi: 10.1016/s0092-8674(00)81201-5. [DOI] [PubMed] [Google Scholar]
- Wu YC, Horvitz HR. C. elegans phagocytosis and cell-migration protein CED-5 is similar to human DOCK180. Nature. 1998b;392:501–504. doi: 10.1038/33163. [DOI] [PubMed] [Google Scholar]
- Wu YC, Tsai MC, Cheng LC, Chou CJ, Weng NY. C. elegans CED-12 acts in the conserved crkII/DOCK180/Rac pathway to control cell migration and cell corpse engulfment. Dev Cell. 2001;1:491–502. doi: 10.1016/s1534-5807(01)00056-9. [DOI] [PubMed] [Google Scholar]
- Xiao H, Chen D, Fang Z, Xu J, Sun X, Song S, Liu J, Yang C. Lysosome biogenesis mediated by vps-18 affects apoptotic cell degradation in Caenorhabditis elegans. Mol Biol Cell. 2009;20:21–32. doi: 10.1091/mbc.E08-04-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang H, Kage-Nakadai E, Mitani S, Wang X. C. elegans secreted lipid-binding protein NRF-5 mediates PS appearance on phagocytes for cell corpse engulfment. Curr Biol. 2012;22:1276–1284. doi: 10.1016/j.cub.2012.06.004. [DOI] [PubMed] [Google Scholar]
- Zhang Y, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Caron E, Hartwieg E, Hall A, Horvitz HR. The C. elegans PH domain protein CED-12 regulates cytoskeletal reorganization via a Rho/Rac GTPase signaling pathway. Dev Cell. 2001a;1:477–489. doi: 10.1016/s1534-5807(01)00058-2. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001b;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- Zou W, Lu Q, Zhao D, Li W, Mapes J, Xie Y, Wang X. Caenorhabditis elegans myotubularin MTM-1 negatively regulates the engulfment of apoptotic cells. PLoS Genet. 2009;5:e1000679. doi: 10.1371/journal.pgen.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.