Abstract
Although a big deal of dental research is being focused to the understanding of early stages of tooth development, a huge gap exist on our knowledge on how the dental hard tissues are formed and how this process is controlled daily in order to produce very complex and diverse tooth shapes adapted for specific functions. Emerging evidence suggests that clock genes, a family of genes that controls circadian functions within our bodies, regulate also dental mineralized tissues formation. Enamel formation, for example, is subjected to rhythmical molecular signals that occur on short (24 hour) periods and control the secretion and maturation of the enamel matrix. Accordingly, gene expression and ameloblast functions are also tightly modulated in regular daily intervals. This review summarizes the current knowledge on the circadian controls of dental mineralized tissues development with a special emphasis on amelogenesis.
Keywords: Ameloblasts, Odontoblasts, Circadian Rhythms, Clock Genes, Cementum, Bone
Introduction to the circadian clock
Circadian rhythms are generated within an organism by endogenous biological clocks driven by cyclic events [1]. The key requirement for the description of a rhythm as circadian is to show that the rhythm persists under constant conditions (i.e. the rhythm must continue within an approximate 24 hour period when all external time cues are removed from the environment), thus demonstrating the endogenous existence of a time-keeping mechanism. Circadian rhythms are controlled by the body’s “central” clock located in the brain at the suprachiasmatic nucleus (SCN). Body functions are also regulated directly by “peripheral” clocks located in several tissues. The central clock is light responsive and can be entrained by light/dark cycles. The peripheral clocks can be entrained by the central clock or independently by other physiological stimuli such as feeding [2].
At the molecular level, the circadian clock is regulated by differential expression of ~20 transcription factors called clock genes. The central clock located in the brain is composed of about 20,000 neurons, all of which express clock genes that oscillate in synchrony [3, 4]. Clock genes are defined by a set of criteria that include rhythm in activity or amount as well as molecular evidence of a feedback mechanism [5, 6]. The key mammalian clock genes are named Aryl hydrocarbon receptor nuclear translocator-like (Arntl or Bmal1), circadian locomoter output cycles kaput (Clock), Period1 (Per1), Period2 (Per2), Period 3 (Per3), and Cryptochrome 1 and 2 (Cry1, Cry2). Clock genes functions are regulated by feedback mechanisms consisting of an auto-regulatory ability in which specific clock proteins negatively regulate their own synthesis. Clock genes transmit output signals that drive rhythms of gene expression in central and peripheral tissues.
Evidence for a role of clock genes in circadian physiology and clock controlled gene expression comes from the studies of clock gene knockout mice. Wild-type mice will increase their locomotor activity at night as measured by wheel-running activity. This characteristic is used to assess circadian phenotypes of knockout mice. For example, mice deficient in Per1 lose wheel-running rhythmicity when placed in constant darkness [7]. This observation is much more pronounced in Per1/Per2 double knockout mice, which display an immediate loss of wheel-running rhythmicity when placed in constant darkness. Moreover, Per1/Per2 double knockout mice have no circadian rhythms in central and peripheral (i.e. hepatic) clocks, or clock-controlled gene expression [8, 9]. On the other hand, deletion of Clock and Bmal1 genes results not only in circadian disturbances, but also in metabolic abnormalities of lipid and glucose homeostasis [10–12]. Thus, clock gene knockout mice have profound changes in circadian rhythmicity and offer a unique experimental genetic model to analyze the link between circadian gene networks and organ physiology.
The most direct mechanism by which clock genes drive circadian gene expression is through regulation of promoter activity of clock-controlled genes (CCG) [13]. In a given tissue, approximately 10–20% of the organ specific genes are under circadian control [14]. Recent studies in humans confirm and further specify the role of clock genes in human diseases [1]. Exciting links between peripheral organs (such as the gut) and the brain are being discovered [2]. Regulation of stem cells behavior is also being linked to clock genes opening a whole new area of fascinating research [15]. Most importantly, understanding how the circadian systems work may facilitate innovative treatment options for patients with untreatable diseases such as autoimmune diseases and cancer as well as for psychiatric conditions [16].
In oral health, the possibilities of clock genes involvement into patho-physiology of oral and craniofacial tissues remain largely unexplored [17]. This review summarizes the potential roles of clock genes in dental tissues formation with a special focus on enamel development. We also present hypotheses regarding the potential connections between dysregulated clock gene expression and mineralized tissues formation in general. The recently hypothesized role of clock genes in regulating stem cells properties is also briefly mentioned.
Expression and potential roles of the circadian clock in mineralized tissues
Bone
The diurnal variation in the synthesis of type I collagen and osteocalcin are well known [18, 19] supporting the hypothesis that synthesis and secretion of the matrix proteins are under circadian control. More recent studies have confirmed that osteoblast contain a peripheral clock mechanism that regulates bone volume [20, 21]. Per1, Per2, Cry1, Bmal1, and Clock all showed robust, rhythmic expression over a 24hr period in osteoblasts, and their expression was decreased and became arrhythmic in Per1−/−; Per2m/m bones, indicating that bone has a peripheral clock. It has also been documented that the molecular clock, specifically the Per genes, inhibits bone formation by preventing osteoblast proliferation [21]. It has been also found that Per2 mutant mice and mice lacking Cry2(−/−) displayed significantly increased bone volume at 12 weeks of age, when bone turnover is high [22]. Of interest, Cry2 influences mostly the osteoclastic cellular component of bone while Per2 acts on osteoblast parameters [22]. Furthermore, it is recently shown that mineralization in developing bone tissue is regulated by a local circadian oscillator mechanism [23]. Thus, we can postulate that bone deposition and mineralization are under direct circadian controls. It remains to clarify how the potential interplay between the “central” and “peripherals” clocks regulates bone homeostasis.
The clinical relevance of these cyclic events controlled by clock genes is currently being studied for several bone related diseases and this new knowledge may influence novel treatment options [24]. For example, the circadian clock plays a key role in obesity and diabetes which may be controlled by osteblasts released hormones [25]. In dentistry, studies elucidating the role of clock genes in alveolar bone homeostasis warrantee novel exciting discoveries in the field of orthodontics tooth movement and periodontal tissue regeneration.
Enamel
Dental enamel is formed during two distinct developmental stages: the secretory stage (when the full thickness of enamel is completed) and the maturation stage (when the enamel organic material is replaced by hydroxyapatite crystals) [26–28]. Ameloblasts, the cells responsible for making enamel, are specialized epithelial cells with distinct morphological characteristics that change at each developmental stage. Secretory and maturation ameloblasts are characterized by expression of enamel stage-specific genes that perform stage-specific functions. Besides the stage-dependent developmental control of enamel formation several observations of the enamel matrix suggest that enamel is formed incrementally. In human dental enamel for instance, there are two regularly occurring incremental markers: daily cross-striations and long-period striae of Retzius (SR). These lines correspond to what was, at precise points in time during the secretory stage of amelogenesis, the enamel surface. Between the SR and running parallel with them are distinct growth lines known as cross-striations (Fig. 1). Cross-striations delineate the amount of enamel deposited in a single day [29, 30]. Enamel cross-striations occur because of a circadian rhythm of ameloblast activity [27, 31]. Similar growth lines have been showed recently in mouse enamel as well [32]. This rate, and the distance between daily incremental lines, averages 12 μm in rodents [27].
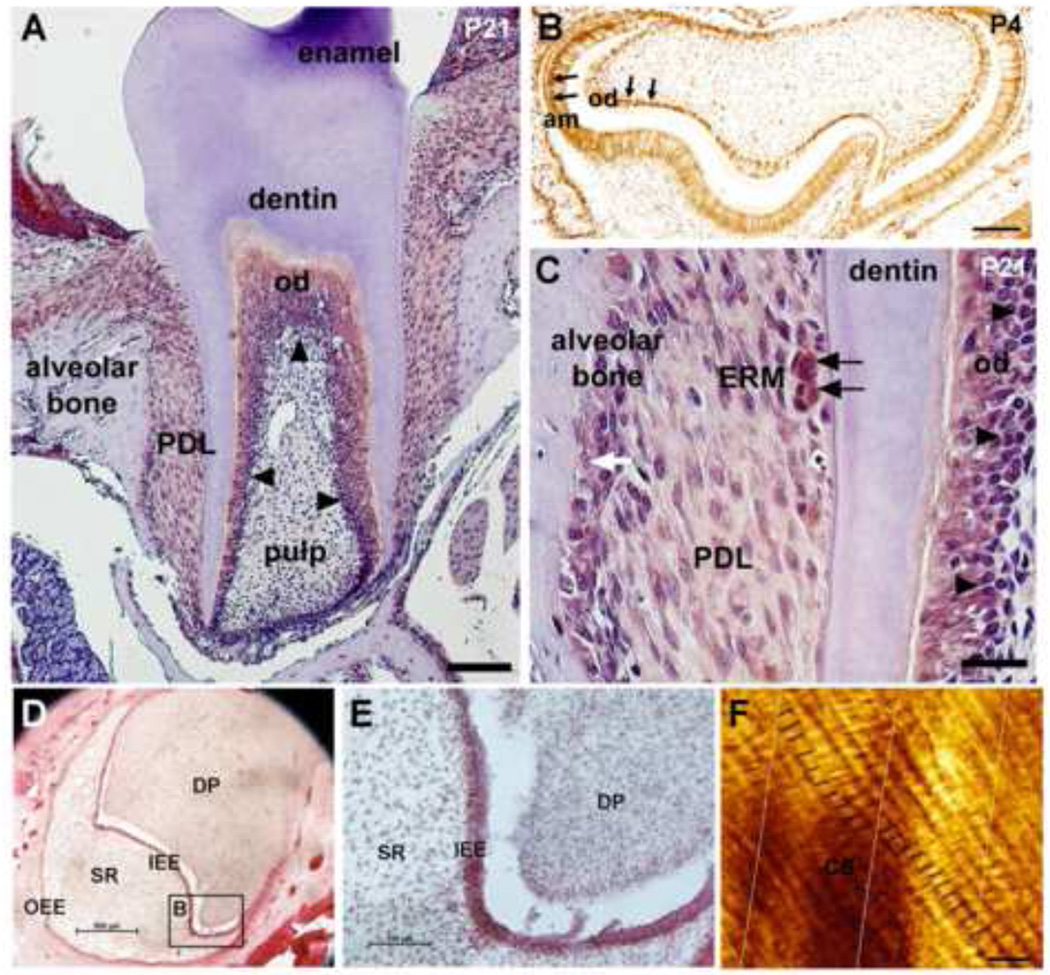
Figure 1.
PER2 expression in mouse molar at PN21 stage (A). Dental pulp is devoid of staining in both incisors and molars (A). Odontoblasts in first molars are strongly stained for PER2 (A, black arrowheads). PER2 shows relatively weak expression in periodontal dental ligament (PDL) cells when compared with odontoblast expression (A and C). Epithelial rests of Malassez (ERM) show strong expression of PER2 within the PDL space (C, black arrows). PER2 proteins are also detected in the nucleus of osteoblasts and osteoclasts in the alveolar bone (C, white arrows). On P4, both ameloblast and odontoblast show strong expression of PER2 in the nucleus (B). In human embryonic teeth, PER2 is detected in inner enamel epithelium (IEE) and outer enamel epithelium (OEE) but not in stellate reticulum (SR) and dental pulp (D and E). Black lines in figure F indicate the enamel cross striations in human tooth enamel. Ten cross striations are visualized between two successive Striae of Retzius (perpendicular lines). Am, ameloblast; DP, dental pulp; ERM, epithelial rests of Malassez; od, odontoblasts; Scale bars = 200 µm in A, 100 µm in B-E, 10 µm in F.
At the cellular level, we have shown that clock proteins are strongly expressed in pre-ameloblasts and differentiated ameloblasts throughout amelogenesis both in human embryonic teeth (Fig. 1) and mouse embryonic and post-natal teeth [8] (Fig. 1). Clock genes products are detected specifically in ameloblasts and their expression levels and localization alternates following circadian rhythms. In addition, melatonin receptors, key regulators of the circadian functions, are also differentially expressed in ameloblasts [33].
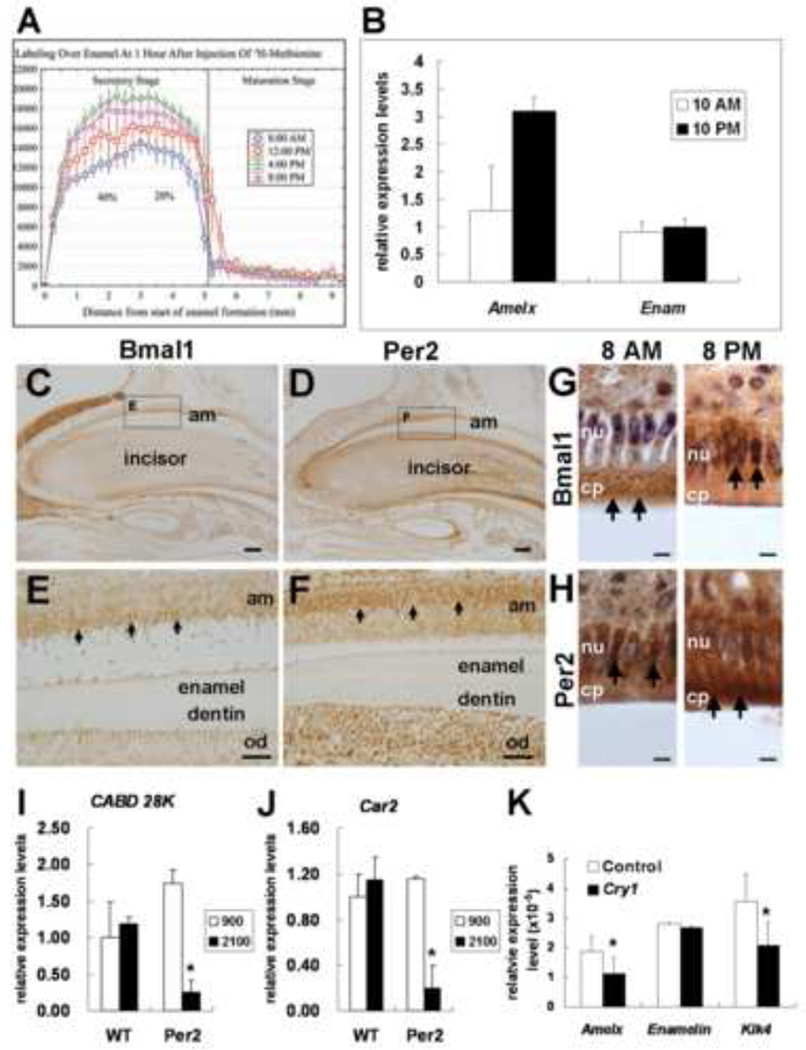
We have also recently published that there are daily variations in the rate of production and secretion of enamel proteins between early morning and late afternoon suggesting that enamel protein secretion is under circadian control [27] (Fig. 2). Enamel proteins secretion peaks at around 8pm and follows a circadian pattern. We have also evaluated RNA oscillations of clock genes and amelogenin and enamelin in teeth of mice kept under normal LD cycles at two time points: 10am and 10pm (Fig. 2). This preliminary data shows that Per2 and amelogenin expression peaks at 10pm with minimum expression at 10am. In contrast, we have not detected any significant difference for enamelin expression. It is possible that enamelin is not under circadian control. Of significance, amelogenin expression still oscillates under DD conditions, however in less extend and with a slight shift in peak times [31]. In vitro studies have also confirmed that amelogenin expression is modulated by clock genes [31, 34, 35] (Fig. 2). Consistent with the above findings, our preliminary analysis of Per2 knock-out teeth show abnormal enamel matrix formation (unpublished). Our data also suggest that Kallikrein 4 (Klk4), a key gene that regulates enamel maturation, might be under circadian control [31]. Taken together the above-discussed data, we postulate that enamel matrix production and maturation is closely controlled by selectively regulating some of enamel matrix proteins encoding genes.
Figure 2.
Different amounts of proteins secreted in rat enamel are measured at different time points (A) Amelogenin RNA expression levels in developing mouse molars are higher at night time compare to day time. In contrast, Enamelin RNA levels do not show significant differences between night and day (B). Immunohistochemistry results show that BMAL1 (C) and PER2 (D) are differentially localized in ameloblasts at a given time point (black arrows). On serial sections, BMAL1 is mostly expressed in the cytoplasm (E) while PER2 is localized mainly in the nucleus (F). Most of BMAL1 proteins are localized in the cytoplasm at 8 am and then in the nucleus of ameloblasts at 8 pm (G). In contrast, PER2 proteins are found in the nucleus at 8 am and in the cytoplasm of ameloblasts at 8 pm (H). Relative mRNA levels are evaluated by qRT-PCR between wild type and knock-out mice (I-J) and in transfected HAT-7 cells (H). Calbidin-D28K RNA expression is altered in Per2 knockout mice (I). Similar trend is be found in Car2 RNA expression level (J). Over-expression of Cry1 results in the down-regulation of Amelx and Klk4 expression levels in HAT-7 cells (H). am, ameloblast; od, odontoblast; nu, nucleus; cp, cytoplasm. Scale bars = 20 µm in A, 200 µm in C and D, 40 µm in E and F, 10 µm in G and H.
Clock genes expression is also found synchronized in two other epithelial tissues with high similarities with ameloblasts, i.e. duodenum, and kidney. We also found that genes involved in the calcium and phosphate metabolism such as calbidin-D28K (a calcium binding protein), carbonic anhydrase 2 (Car2), and alkaline phospatase oscillate in regular daily intervals in kidney cells suggesting that these genes might be functional targets of clock genes. Furthermore, expression of calbidin-D28K and alkaline phospatase is altered in Per2 knock-out mice kidney (Fig. 2). It is then logical to hypothesize that these genes may also be targets of the circadian clock (CCG) in ameloblasts. Consistently with our hypothesis clock genes knockout mice (Per2 KO) show also defects of enamel mineralization (unpublished data). However, the precise mechanisms of calcium regulation in ameloblasts remain to be elucidated.
CCG control of enamel related genes is suggested to be direct or indirect through the circadian regulation of key transcription factors as we showed for Runx2 [34]. Recently, another group showed that Runx2 is under circadian control in the suprachiasmatic nucleus and functions in the control of rhythmic behavior [36]. More studies are needed to fully comprehend the complex circadian controls that regulate enamel formation. Nevertheless, we can postulate that the ameloblast circadian clock plays significant roles in enamel secretion, maturation and mineralization.
Dentin
Similar to enamel, dentin is formed incrementally suggesting the involvement of a circadian clock mechanism during dentinogenesis. Daily incremental lines in dentin are termed von Ebner’s lines. von Ebner’s lines delineate the amount of mineral deposited in a single day [37]. The numbers of short-period incremental lines observed in enamel or dentin between sequentially administered dyes correspond to the number of days between successive administrations [38]. Circadian rhythms have been demonstrated using 3H-proline tracers that label collagen in dentin formation [39]. Twice as much collagen is secreted during the daylight 12 h as during the night time 12 h.
At the cellular level, we recently showed evidence that odontoblasts express clock proteins [8]. Consistently, Ohtsuka-Isoya et al. [40] presented convincing evidence that SCN is related to the production of daily lines in dentin, as rats in which the SCN is fully or partially lesioned show respective permanent or temporary loss of circadian dentin increments. These studies suggest that dentin, similar to bone and enamel, is controlled by a circadian clock mechanism. However, very little is known on the molecular circadian mechanisms that regulate dentin formation. It will be interesting to elucidate the differential controls of dentin formation by the master clock in the brain and/or a peripheral clock located in odontoblasts.
Cementum, and Periodontal Ligament
It has been described that human cementum is formed incrementally [41] and we found that periodontal ligament (PDL) cells differentially express clock proteins [8] (Fig. 1). Mouse cementoblasts lack of staining (Fig. 1) but we do not know if this is a generalized characteristic of cementoblasts. It is possible, that once cementum formation is completed clock expression is turned off in cementoblasts. Alternatively, species or teeth differences may exist. Of interest, Epithelial Rest of Mallasez (ERMs) strongly express clock proteins (Fig. 1). ERM are hypothesized to play a role in periodontal ligament regeneration. Further studies are needed to elucidate the potential links between clock genes and periodontal maintenance and turn over.
Clock Genes and Stem Cells
A surpassingly finding of our analysis of clock gene expression in tooth is a strong expression of clock genes in areas rich in stem cells. For example, ERM cells, which are considered as dormant progenitor cells [42], show the strongest staining among all other cells in PDL (Fig. 1). Clock proteins are also strongly expressed by dental epithelium stem cells at the incisor cervical root area [8]. Of interest, clock genes mutants show variations in incisor size, being always smaller in Per2 knockout mice (unpublished).
Similar to dental epithelia, hair cells express clock genes [43]. Indeed, clock genes modulate the human hair cycle clock independently of the brain suggesting that a peripheral hair clock also exists [44, 45]. Clock genes and their products are also co-expressed with stem cells markers in hair and perturbations of circadian clocks result in changes in hair follicle renewal [46]. Of great interest, this study also reveals that disruption of clock equilibrium, through deletion of Bmal1 or Per1/2, resulted in a progressive accumulation or depletion of dormant stem cells, respectively [46]. Hair and dental epithelium share common pathways [47]. We postulate that at least part of the developmental alterations observed in the enamel and hair of clock genes mutant mice are due to changes of stem cells behavior in these tissues. However, the potential role of the circadian clock in regulating stem cells behavior in tooth and hair remains elusive.
Conclusions
Both, the secretory and maturation stages of enamel formation are characterized by cyclic events that control cell morphology, gene expression and protein secretion and removal. In order to understand how enamel development is overall controlled, studying the circadian control of ameloblast differentiation is necessary. The concrete benefits of this research will provide the basis for a better understanding of enamel abnormalities and differences among the population. Once such knowledge becomes available, there is a promise that diseases associated with enamel formation can be better understood and potentially diagnosed and treated. The time spent each day by ameloblasts to perform unique functions as well as the time spent into the secretory versus the maturation stages are critical for the final product, the enamel. Enamel may vary considerably between individuals, due to differences in circadian profiles. Therefore, alteration of the circadian network would result in significant differences of enamel morphology, thickness and mineral content, predisposing teeth to disease or protecting them against it.
Equally important emerging evidence confirms that, bone, dentin and cementum are also under circadian controls. In the Per2 ko mice both bone and teeth are affected. These data suggest that a synchronized formation of teeth and bone may be controlled by the central and/or peripheral circadian body clocks. This perspective opens new avenues of research that would study the potential communications between bone and teeth and/or their coordinated regulation by the circadian central and peripheral clocks. It is anticipated that circadian clock research will contribute to broader understanding of how mineralized tissues are formed. A system’s biology approach having as “skeleton” the circadian system may help us understand how mineralized tissues are developed in synchrony [17]. New inputs such as mastication or bone load could be established in the circadian system equation increasing our understanding of this complex communication between the brain and the peripheral organs.
Although recent studies have shown that the behavior of some types of stem cells is circadian, we are still far from understanding how circadian rhythmicity contributes to their function. Indeed, additional emerging evidence [15] and our own unpublished data suggests an important role of the circadian clock in regulating aging, cancer, and potentially tissue repair but we are far way of understanding the complexity of this system. Future studies on the relationship between clock genes and stem cells are expected to elucidate the contribution of the timing as a “fourth dimension” on the study of stem cells [48] in the years to come.
Acknowledgements
PP was supported by NIH grant DE018878 and the University of Michigan (OVPR) funds. SP was supported by funds from the University of Michigan Department of Otolaryngology – Head and Neck Surgery.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Richards J, Gumz ML. Mechanism of the circadian clock in physiology. Am J Physiol Regul Integr Comp Physiol. 2013;304:R1053–R1064. doi: 10.1152/ajpregu.00066.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Konturek PC, Brzozowski T, Konturek SJ. Gut clock: implication of circadian rhythms in the gastrointestinal tract. J Physiol Pharmacol. 2011;62:139–150. [PubMed] [Google Scholar]
- 3.Ikeda M. Calcium dynamics and circadian rhythms in suprachiasmatic nucleus neurons. Neuroscientist. 2004;10:315–324. doi: 10.1177/10738584031262149. [DOI] [PubMed] [Google Scholar]
- 4.Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 5.Myers EM, Yu J, Sehgal A. Circadian control of eclosion: interaction between a central and peripheral clock in Drosophila melanogaster. Curr Biol. 2003;13:526–533. doi: 10.1016/s0960-9822(03)00167-2. [DOI] [PubMed] [Google Scholar]
- 6.Williams JA, Sehgal A. Molecular components of the circadian system in Drosophila. Annu Rev Physiol. 2001;63:729–755. doi: 10.1146/annurev.physiol.63.1.729. [DOI] [PubMed] [Google Scholar]
- 7.Masubuchi S, Kataoka N, Sassone-Corsi P, Okamura H. Mouse Period1 (mPER1) acts as a circadian adaptor to entrain the oscillator to environmental light/dark cycles by regulating mPER2-protein. J Neurosci. 2005;25:4719–4724. doi: 10.1523/JNEUROSCI.4761-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 9.Jang YS, Kang YJ, Kim TJ, Bae K. Temporal expression profiles of ceramide and ceramide-related genes in wild type and mPer1/mPer2 double knockout mice. Mol Biol Rep. 2012;39:4215–4221. doi: 10.1007/s11033-011-1207-2. [DOI] [PubMed] [Google Scholar]
- 10.Husse J, Hintze SC, Eichele G, Lehnert H, Oster H. Circadian clock genes Per1 and Per2 regulate the response of metabolism associated transcripts to sleep disruption. PLoS One. 2012;7:e52983. doi: 10.1371/journal.pone.0052983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee YJ, Han DH, Pak YK, Cho SH. Circadian regulation of low density lipoprotein receptor promoter activity by CLOCK/BMAL1, Hes1-and Hes6. Exp Mol Med. 2012;44:642–652. doi: 10.3858/emm.2012.44.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, Bradfield CA, Vaughan CH, Eiden M, Masoodi M, Griffin JL, Wang F, Lawson JA, Fitzgerald GA. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–1777. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1-circadian complex in transcriptional regulation. Faseb J. 2006;20:530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
- 14.Kumaki Y, Ukai-Tadenuma M, Uno KD, Nishio J, Masumoto KH, Nagano M, Komori T, Shigeyoshi Y, Hogenesch JB, Ueda HR. Analysis and synthesis of high-amplitude Cis elements in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2008;105:14946–14951. doi: 10.1073/pnas.0802636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janich P, Toufighi K, Solanas G, Luis NM, Minkwitz S, Serrano L, Lehner B, Benitah SA. Human Epidermal Stem Cell Function Is Regulated by Circadian Oscillations. Cell Stem Cell. 2013 doi: 10.1016/j.stem.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Schroeder AM, Colwell CS. How to fix a broken clock. Trends Pharmacol Sci. 2013 doi: 10.1016/j.tips.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papagerakis S, Zheng L, Schnell S, Sartor MA, Somers E, Marder W, McAlpin B, Kim D, McHugh J, Papagerakis P. The Circadian Clock in Oral Health and Diseases. J Dent Res. 2013 doi: 10.1177/0022034513505768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gundberg CM, Markowitz ME, Mizruchi M, Rosen JF. Osteocalcin in human serum: a circadian rhythm. J Clin Endocrinol Metab. 1985;60:736–739. doi: 10.1210/jcem-60-4-736. [DOI] [PubMed] [Google Scholar]
- 19.Simmons DJ, Nichols G. Jr., Diurnal periodicity in the metabolic activity of bone tissue. Am J Physiol. 1966;210:411–418. doi: 10.1152/ajplegacy.1966.210.2.411. [DOI] [PubMed] [Google Scholar]
- 20.Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 21.Fu L, Patel MS, Karsenty G. The circadian modulation of leptin controlled bone formation. Prog Brain Res. 2006;153:177–188. doi: 10.1016/S0079-6123(06)53010-9. [DOI] [PubMed] [Google Scholar]
- 22.Maronde E, Schilling AF, Seitz S, Schinke T, Schmutz I, van der Horst G, Amling M, Albrecht U. The clock genes Period 2-and Cryptochrome 2-differentially balance bone formation. PLoS One. 2010;5:e11527. doi: 10.1371/journal.pone.0011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McElderry JD, Zhao G, Khmaladze A, Wilson CG, Franceschi RT, Morris MD. Tracking circadian rhythms of bone mineral deposition in murine calvarial organ cultures. J Bone Miner Res. 2013;28:1846–1854. doi: 10.1002/jbmr.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iimura T, Nakane A, Sugiyama M, Sato H, Makino Y, Watanabe T, Takagi Y, Numano R, Yamaguchi A. A fluorescence spotlight on the clockwork development and metabolism of bone. J Bone Miner Metab. 2012;30:254–269. doi: 10.1007/s00774-011-0295-3. [DOI] [PubMed] [Google Scholar]
- 25.DiGirolamo DJ, Clemens TL, Kousteni S. The skeleton as an endocrine organ. Nat Rev Rheumatol. 2012;8:674–683. doi: 10.1038/nrrheum.2012.157. [DOI] [PubMed] [Google Scholar]
- 26.Hu JC, Chun YH, Al Hazzazzi T, Simmer JP. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. 2007;186:78–85. doi: 10.1159/000102683. [DOI] [PubMed] [Google Scholar]
- 27.Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, Hu JC. Regulation of dental enamel shape and hardness. J Dent Res. 2010;89:1024–1038. doi: 10.1177/0022034510375829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CE. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 29.Dean JA, Avery DR, Swartz ML. Attachment of anterior tooth fragments. Pediatr Dent. 1986;8:139–143. [PubMed] [Google Scholar]
- 30.Risnes S, Moinichen CB, Septier D, Goldberg M. Effects of accelerated eruption on the enamel of the rat lower incisor. Adv Dent Res. 1996;10:261–269. doi: 10.1177/08959374960100022401. [DOI] [PubMed] [Google Scholar]
- 31.Zheng L, Seon YJ, Mourao MA, Schnell S, Kim D, Harada H, Papagerakis S, Papagerakis P. Circadian rhythms regulate amelogenesis. Bone. 2013;55:158–165. doi: 10.1016/j.bone.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sehic A, Nirvani M, Risnes S. Incremental lines in mouse molar enamel. Arch Oral Biol. 2013;58:1443–1449. doi: 10.1016/j.archoralbio.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Kumasaka S, Shimozuma M, Kawamoto T, Mishima K, Tokuyama R, Kamiya Y, Davaadorj P, Saito I, Satomura K. Possible involvement of melatonin in tooth development: expression of melatonin 1a receptor in human and mouse tooth germs. Histochem Cell Biol. 2010;133:577–584. doi: 10.1007/s00418-010-0698-6. [DOI] [PubMed] [Google Scholar]
- 34.Athanassiou-Papaefthymiou M, Kim D, Harbron L, Papagerakis S, Schnell S, Harada H, Papagerakis P. Molecular and circadian controls of ameloblasts. Eur J Oral Sci. 2011;119(Suppl 1):35–40. doi: 10.1111/j.1600-0722.2011.00918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacruz RS, Hacia JG, Bromage TG, Boyde A, Lei Y, Xu Y, Miller JD, Paine ML, Snead ML. The circadian clock modulates enamel development. J Biol Rhythms. 2012;27:237–245. doi: 10.1177/0748730412442830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reale ME, Webb IC, Wang X, Baltazar RM, Coolen LM, Lehman MN. The transcription factor Runx2 is under circadian control in the suprachiasmatic nucleus and functions in the control of rhythmic behavior. PLoS One. 2013;8:e54317. doi: 10.1371/journal.pone.0054317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dean MC. Comparative observations on the spacing of short period (von Ebner's) lines in dentine. Arch Oral Biol. 1998;43:1009–1021. doi: 10.1016/s0003-9969(98)00069-7. [DOI] [PubMed] [Google Scholar]
- 38.Dean MC, Scandrett AE. The relation between long-period incremental markings in dentine and daily cross striations in enamel in human teeth. Arch Oral Biol. 1996;41:233–241. doi: 10.1016/0003-9969(95)00137-9. [DOI] [PubMed] [Google Scholar]
- 39.Ohtsuka M, Saeki S, Igarashi K, Shinoda H. Circadian rhythms in the incorporation and secretion of 3H-proline by odontoblasts in relation to incremental lines in rat dentin. J Dent Res. 1998;77:1889–1895. doi: 10.1177/00220345980770110501. [DOI] [PubMed] [Google Scholar]
- 40.Ohtsuka-Isoya M, Hayashi H, Shinoda H. Effect of suprachiasmatic nucleus lesion on circadian dentin increment in rats. Am J Physiol Regul Integr Comp Physiol. 2001;280:R1364–R1370. doi: 10.1152/ajpregu.2001.280.5.R1364. [DOI] [PubMed] [Google Scholar]
- 41.Kvaal SI, Solheim T, Bjerketvedt D. Evaluation of preparation, staining and microscopic techniques for counting incremental lines in cementum of human teeth. Biotech Histochem. 1996;71:165–172. doi: 10.3109/10520299609117155. [DOI] [PubMed] [Google Scholar]
- 42.Oka K, Morokuma M, Imanaka-Yoshida K, Sawa Y, Isokawa K, Honda MJ. Cellular turnover in epithelial rests of Malassez in the periodontal ligament of the mouse molar. Eur J Oral Sci. 2012;120:484–494. doi: 10.1111/eos.12003. [DOI] [PubMed] [Google Scholar]
- 43.Tanioka M, Yamada H, Doi M, Bando H, Yamaguchi Y, Nishigori C, Okamura H. Molecular clocks in mouse skin. J Invest Dermatol. 2009;129:1225–1231. doi: 10.1038/jid.2008.345. [DOI] [PubMed] [Google Scholar]
- 44.Al-Nuaimi Y, Hardman JA, Biro T, Haslam IS, Philpott MP, Toth BI, Farjo N, Farjo B, Baier G, Watson RE, Grimaldi B, Kloepper JE, Paus R. A Meeting of Two Chronobiological Systems: Circadian Proteins Period1-and BMAL1-Modulate the Human Hair Cycle Clock. J Invest Dermatol. 2013 doi: 10.1038/jid.2013.366. [DOI] [PubMed] [Google Scholar]
- 45.Watabe Y, Tomioka M, Watabe A, Aihara M, Shimba S, Inoue H. The clock gene brain and muscle Arnt-like protein-1 (BMAL1) is involved in hair growth. Arch Dermatol Res. 2013;305:755–761. doi: 10.1007/s00403-013-1403-0. [DOI] [PubMed] [Google Scholar]
- 46.Janich P, Pascual G, Merlos Suarez A, Batlle E, Ripperger J, Albrecht U, Cheng HY, Obrietan K, Di Croce L, Benitah SA. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- 47.Pispa J, Thesleff I. Mechanisms of ectodermal organogenesis. Dev Biol. 2003;262:195–205. doi: 10.1016/s0012-1606(03)00325-7. [DOI] [PubMed] [Google Scholar]
- 48.Gimble JM, Floyd ZE, Bunnell BA. The 4th dimension and adult stem cells: Can timing be everything? J Cell Biochem. 2009;107:569–578. doi: 10.1002/jcb.22153. [DOI] [PMC free article] [PubMed] [Google Scholar]