Abstract
As increasing amounts of anthropogenic chemicals are released into the environment, it is vital to human health and the preservation of ecosystems to evaluate the fate of these chemicals in the environment. It is useful to predict whether a particular compound is biodegradable and if alternate routes can be engineered for compounds already known to be biodegradable. In this work, we describe a computational framework (called BNICE) that can be used for the prediction of novel biodegradation pathways of xenobiotics. The framework was applied to 4-chlorobiphenyl, phenanthrene, γ-hexachlorocyclohexane, and 1,2,4-trichlorobenzene, compounds representing various classes of xenobiotics with known biodegradation routes. BNICE reproduced the proposed biodegradation routes found experimentally, and in addition, it expanded the biodegradation reaction networks through the generation of novel compounds and reactions. The novel reactions involved in the biodegradation of 1,2,4-trichlorobenzene were studied in depth, where pathway and thermodynamic analyses were performed. This work demonstrates that BNICE can be applied to generate novel pathways to degrade xenobiotic compounds that are thermodynamically feasible alternatives to known biodegradation routes and attractive targets for metabolic engineering.
Keywords: bioremediation, complex networks, metabolic engineering, network analysis, reaction pathway analysis
Introduction
The amount and number of anthropogenic chemicals, xenobiotics, released into the environment has led to a growing interest in bioremediation, the use of microorganisms to degrade these recalcitrant contaminants (Dua et al., 2002; Jain et al., 2005; Watanabe, 2001). Bioremediation is an environmentally friendly and economic alternative to commonly used pollution treatment methods such as incineration, landfilling, and air-stripping (Dua et al., 2002; Paul et al., 2005; Pieper and Reineke, 2000). While there are millions of organic compounds believed to be biodegradable, information on the reactions through which microbial biodegradation occurs is largely incomplete (Wackett and Ellis, 1999), and in most cases, it is not feasible to perform all of the experiments needed to investigate the microbial metabolism of each substrate thought to be biodegradable. Additionally, a growing number of pollutants contain structural features that are not common in nature, and natural microorganisms are slow to evolve the necessary metabolic pathways needed to degrade such compounds. Genetic engineering is a viable tool used to design organisms capable of degrading xenobiotics (Chen et al., 2005), and the use of computational tools to predict the biodegradation of xenobiotics can aid in identifying the reactions needed to degrade these compounds, providing insight into the fate of xenobiotic compounds in the environment.
Current biodegradation prediction methods are rule-based and rely on extensive databases. One such method is the Pathway Prediction System (PPS), which utilizes the University of Minnesota Biocatalysis/Biodegradation Database (UM-BBD) (Ellis and Wackett, 2006), a database of biodegradation reactions and pathways compiled from scientific literature. The PPS is a user-guided system that uses metabolic rules describing the transformation of chemical functional groups. In this system, a biotransformation rule is created if there is an example of the metabolism in the UM-BBD or if it is known to occur in the environment (Ellis et al., 2008; Hou et al., 2003). Another prediction method is META, initially developed to predict mammalian metabolism and toxicity of drug compounds using dictionaries containing biotransformation rules, or transforms, created based on published data for mammalian enzymes (such as cytochrome P450s). META is an expert system used to predict all of the possible metabolites from a given precursor and prioritize the metabolites based on biological reactivity, biodisposition, and tissue distribution (Klopman et al., 1994). The META program has been used to predict metabolism of xenobiotics in mammals, biodegradation pathways of polycyclic aromatic hydrocarbons, aerobic and anaerobic biodegradation, and photodegradation. Other tools have been developed to predict the biodegradability of organic compounds and reactions involved in biodegradation, including BDPServer, which determines the fate of a metabolite based on the correlation between its structure and the known capacity of microorganisms to degrade pollutants (Gomez et al., 2007); and CATABOL, MetabolExpert, and METEOR (Darvas, 1987; Greene et al., 1999; Jaworska et al., 2002), which rely on expert rules to predict biotransformations. More recently, Oh et al. (2007) used the KEGG RPAIR database containing structure transformation patterns, called RDM patterns, to predict bacterial biodegradation pathways. This prediction method utilizes a large database of RDM patterns and limits the possible compounds produced from the query compound based on substrate and reaction specificity.
We propose an alternative method for the prediction of biodegradation that employs the Biochemical Network Integrated Computational Explorer (BNICE), a framework developed to generate every possible biochemical reaction based on a set of enzyme reaction rules and starting compounds (Hatzimanikatis et al., 2004, 2005; Li et al., 2004). BNICE utilizes NetGen, an automated network generation method developed by Broadbelt et al. (1994), and a set of enzyme reaction rules based on the enzyme commission (EC) classification system. BNICE has been used to generate novel metabolic pathways (Hatzimanikatis et al., 2005; Li et al., 2004) and to study the combinatorial nature of polyketide synthesis (Gonzalez-Lergier et al., 2005), and it can provide a systematic framework for bridging enzyme chemistry and reactive sites of metabolic compounds (Hatzimanikatis et al., 2004). Recently, BNICE was employed to determine the percentage of the reactions in Escherichia coli metabolism in the KEGG database that can be reproduced using a set of carefully defined reaction rules (Henry, 2007).
We have extended the BNICE framework for the prediction of biodegradation pathways of xenobiotics and applied BNICE on compounds that represent various classes of xenobiotics, including chlorinated aliphatics, polychlorinated biphenyls, and polycyclic aromatic hydrocarbons. Using BNICE, we reproduced the proposed biodegradation routes found experimentally and generated large reaction networks consisting of novel compounds and reactions. We exemplified the type of analyses that can be performed given a reaction network generated by BNICE using the metabolism of 1,2,4-trichlorobenzene, and illustrated how the BNICE methodology can be applied to the analysis and design of biodegradation pathways of recalcitrant, toxic compounds.
Methods
BNICE Formalism and Definition of Reaction Rules
The BNICE methodology utilizes a systematic formulation of enzyme reaction rules based on the EC classification system, where each enzyme is assigned a four-digit number: EC i. j. k. l. The first level of classification, i, designates the type of chemistry; the second level, j, designates the functional group being acted upon; the third level, k, designates the cofactors the enzyme uses; and the fourth level, l, designates the specific substrates and reactants participating in the reaction. Thus, the first three levels of classification describe the action of an enzyme and can be used to define the generalized enzyme reaction or reaction rule. The reaction rules are implemented using a matrix called the reaction operator that indicates which bonds are formed and broken according to the generalized enzyme reaction it encodes, and they are iteratively applied to a set of starting compounds. For the application of these rules, each starting compound is evaluated to determine if it contains the functional group acted upon by the reaction rules, generating all possible products. The rules are then applied to the products from the previous generation, and this process is repeated in successive generations until no new compounds are created or the maximum number of iterations has been reached.
Hatzimanikatis et al. (2004, 2005) and Li et al. (2004) developed a set of reaction operators based on the curation of biochemical reactions catalogued in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2006) and the iJR904 genome-scale model of E. coli metabolism (Reed et al., 2003). In this curation, the reaction rules associated with a particular third-level enzyme class were defined by examining all of the known reactions assigned to that enzyme class. Priority was given to EC classes with reactions important in the production of organic chemicals, those involved in E. coli metabolism, and small molecule metabolism. Thirty-three EC classes were examined in the curation, resulting in the definition of 86 reaction operators (Henry, 2007).
Reaction Mapping
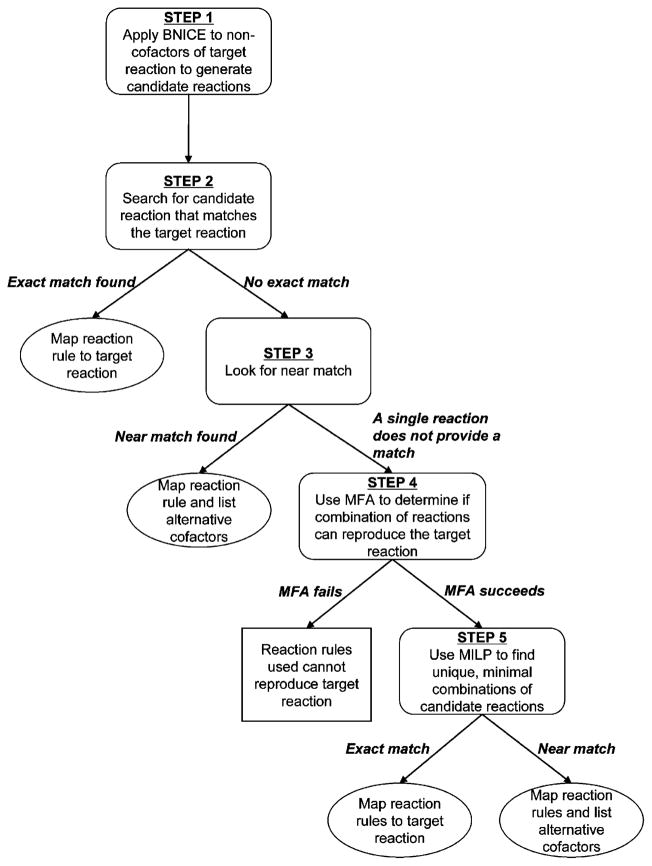
A mapping algorithm has been developed to determine whether a set of reaction operators can reproduce a given reaction, the target reaction (Henry, 2007). Reaction mapping has been used to assign EC classes to reactions for which the third-level EC number is unknown, identify and correct errors in reaction cofactor stoichiometry, and map the reactant atoms to the product atoms in biochemical reactions (Henry, 2007). In the mapping algorithm (described in Fig. 1), the automated network generation method from BNICE is applied in an iterative manner to the non-cofactors of the target reaction. The result is a set of candidate reactions that includes reactions generated from the reactants and products of the target reaction. Metabolic Flux Analysis (MFA) is used to first determine if a subset of the candidate reactions can be combined to reproduce the target reaction. Next, mixed integer linear programming (MILP) is applied to find the minimal set of candidate reactions that combine to reproduce the target reaction. The reaction rules associated with the candidate reactions in the solution are assigned (“mapped”) to the target reaction.
Figure 1.
Flow chart of the reaction mapping algorithm. BNICE is used to generate a set of candidate reactions. If one of the candidate reactions is an exact match, the algorithm stops and maps the reaction rule used to generate the match to the target reaction. If no exact match is found, the algorithm searches for a near match. If a single reaction does not match the target reaction, metabolic flux analysis (MFA) is used to determine if a set of candidate reactions can combine to reproduce the target reaction. If the MFA procedure succeeds, mixed integer linear programming (MILP) is used to identify all unique, minimal combinations of candidate reactions that combine to match the target reaction. Reaction rule(s) used to generate the candidate reactions are mapped to the target reaction.
Results and Discussion
Definition of Enzyme Reaction Rules Relevant to Biodegradation
In the present work, the reactions catalogued in the UM-BBD were examined in order to develop reaction rules required to predict biodegradation. We used the following two criteria in defining the reaction rules for BNICE: (1) the rule is consistent with its assigned third-level EC classification number, that is, there exists a set of specific, fourth-level EC enzymes that follow the same rule, and (2) the reaction rule describes at least two biochemical reactions in the UM-BBD. The formulation of the reaction rules was an iterative process, where a rule was first defined based on the reaction pattern observed in enzymes from the same third-level enzyme class, and it was then used in the mapping algorithm. Based on the mapping results, the reaction rule was refined. The mapping may have indicated that a reaction rule could be expanded to capture a broader set of enzyme actions, for example, the rule should act on both benzene rings and heteroaromatic rings. In some instances, the reaction operator was modified to include spectator atoms, neighboring atoms whose bonds are not changed during the reaction, but must be present for the reaction to occur.
In total, twenty third-level enzyme classes were studied, resulting in the definition of 68 reaction operators (Table I). These enzyme classes contained 600 UM-BBD reactions, which is 58% of the reactions in the database that have been assigned a third-level enzyme classification number. The number of reaction rules defined was more than three times the number of EC classes curated, indicating that in most instances several reaction rules were required to describe the chemistry of a single enzyme class. A single reaction operator was defined for five of the EC classes examined. In contrast, 12 and 8 operators were defined for EC 1.14.13 and 1.13.11, respectively. These two enzyme classes, which also contained the largest number of UM-BBD reactions, involved the most diverse set of enzymatic reactions.
Table I.
Curated third-level enzyme classes that appear in the UM-BBD.
| Third level EC number | Total reactionsa | Reaction operatorsb | Mapped reactions
|
|
|---|---|---|---|---|
| Using biodegradation reaction rulesc | Using all BNICE reaction rulesd (total percent) | |||
| 1.3.1 | 54 | 1 | 5 | 46 (85%) |
| 1.7.1 | 19 | 3 | 16 | 16 (84%) |
| 1.13.11 | 78 | 8 | 62 | 68 (87%) |
| 1.13.12 | 17 | 1 | 16 | 17 (100%) |
| 1.14.12 | 58 | 5 | 50 | 53 (91%) |
| 1.14.13 | 96 | 12 | 83 | 86 (90%) |
| 1.14.15 | 19 | 3 | 9 | 15 (79%) |
| 1.97.1 | 24 | 3 | 22 | 22 (92%) |
| 2.1.1 | 12 | 2 | 9 | 9 (75%) |
| 2.3.1 | 15 | 4 | 15 | 15 (100%) |
| 2.8.3 | 10 | 1 | 8 | 10 (100%) |
| 3.1.1 | 18 | 2 | 2 | 17 (94%) |
| 3.7.1 | 28 | 1 | 6 | 28 (100%) |
| 3.8.1 | 19 | 3 | 17 | 18 (95%) |
| 4.1.1 | 28 | 3 | 15 | 26 (93%) |
| 4.2.1 | 46 | 7 | 23 | 42 (91%) |
| 4.5.1 | 13 | 2 | 12 | 12 (92%) |
| 5.3.99 | 7 | 2 | 6 | 7 (100%) |
| 5.5.1 | 16 | 4 | 12 | 13 (81%) |
| 6.2.1 | 23 | 1 | 19 | 23 (100%) |
Number of UM-BBD reactions assigned to the given third level EC classification number.
Number of distinct reaction rules defined based on curation of given third level EC classification number.
Number of reactions that could be reproduced using the BNICE enzyme reaction rules defined in this work.
Number of reactions that could be reproduced using the reaction rules from this work plus those defined by Henry, 2007).
Validation of Enzyme Reaction Rules
Reaction mapping was used to quantify how much of the chemistry in the UM-BBD the reaction operators encode. We first determined how well the 68 operators covered the 600 reactions in the third-level enzyme classes curated. The operators reproduced 68% (407) of the reactions. We repeated the same analysis using the complete set of reaction operators (the 68 operators defined in this work and the 86 defined by Henry, 2007) to reproduce reactions in the curated enzyme classes, and the operators were able to reproduce 91% (543) of the reactions. Lastly, we attempted to map all of the 1,205 reactions in the UM-BBD using the complete set of reaction operators. In this case, the operators were able to reproduce 72% (878) of the reactions. Sixty five percent of the database could be reproduced using a single operator, and 7% was mapped using the sequential application of two operators. Less than 1% of the reactions required the sequential application of three operators. It is important to compare this mapping with the coverage obtained using only the 86 reaction operators defined by Henry (2007), which reproduced 24% of the reactions in the UM-BBD. Thus, by defining reaction rules relevant to biodegradation, we were able to reproduce nearly three times as many reactions (878 reactions compared to 294 reactions).
We have compared the BNICE rules with those utilized in the University of Minnesota Pathway Prediction System (PPS). The PPS biotransformation rules are defined if there is an example of the metabolism in the UM-BBD, or if it is known to occur in the environment. As of June 2008, there are 235 biotransformation rules compiled in the PPS, which describe 903 reactions (75% of the reactions in the database). Based on the mapping results, we identified 756 reactions that could be described using both the PPS and the BNICE framework. The PPS method requires 150 biotransformation rules to describe the chemistry of these reactions, while a total of 95 BNICE reaction rules were required to reproduce the reactions, indicating that the generalized enzyme operator may be a more concise way in which to organize known biodegradation reactions.
We have defined reaction rules relevant to biodegradation using the two guiding principles described above. We have enriched these rules with a set of reaction rules defined based on a larger training set consisting of the generalized enzyme chemistry of small molecule metabolism and the metabolic reactions involved in E. coli. By utilizing the complete set of reaction rules, we can capture existing reactions, add new connections between known compounds, and generate novel reactions that metabolize dead-end compounds. Therefore, in addition to being a compact way to describe biochemical reactions, the generalized enzyme reactions allow us to better navigate the chemistry of biodegradation, ultimately achieving broad coverage of known biodegradation reactions, as well as proposing novel reactions.
Application of BNICE Methodology
We have applied the BNICE framework to generate a reaction network for four xenobiotic compounds: 4-chlorobiphenyl, phenanthrene, γ-hexachlorocyclohexane, and 1,2,4-trichlorobenzene. 4-Chlorobiphenyl (CBP) serves as a model for polychlorinated biphenyls (PCBs). PCBs, once widely used because of their thermal stability, have been banned in the United States (Unterman, 1996) and are among the top 10% of the EPA’s most toxic chemicals (Kline et al., 2008). Phenanthrene is a member of a class of chemicals called polycyclic aromatic hydrocarbons (PAHs). These compounds, which consist of more than one aromatic ring, usually persist in natural environments and may be carcinogenic. While phenanthrene is non-carcinogenic, it serves as a model for PAHs (Mueller et al., 1996). γ-Hexachlorocyclohexane (GHCH) is a halogenated organic insecticide that is prohibited in the United States. Although GHCH is rapidly metabolized anaerobically, it is persistent in upland soil (Ellis and Wackett, 2006; Imai et al., 1991). 1,2,4-Trichlorobenzene (1,2,4-TCB) is one of the most widely used chlorobenzenes (Wang et al., 2006) with many industrial uses, including acting as a degreaser, lubricant, and solvent. Chlorobenzenes have toxic effects in humans and animals (den Besten et al., 1991; Zhang et al., 2005). All four compounds studied here are included on the list of Priority Chemicals, as designated by the Environmental Protection Agency (EPA) (http://www.epa.gov/epawaste/hazard/wastemin/priority.htm). Additionally, the UM-BBD contains a proposed pathway for the biodegradation of these compounds based on reactions found experimentally or observed in the environment (Supplementary Fig. 1), which can be compared to the novel pathways obtained from BNICE.
For each of the four compounds, we utilized the mapping algorithm to determine the subset of the complete set of reaction operators required to reproduce the proposed biodegradation pathway (Table II), and only those reaction rules were used in the BNICE framework to generate novel routes to “termination compounds,” compounds listed in the UM-BBD with known intermediary metabolism. For 4-chlorobiphenyl and phenanthrene, the number of applications of the reaction rules was equal to the length of the known pathway. In the cases of γ-hexachlorocyclohexane and 1,2,4-trichlorobenzene, there were instances where one of the known reactions was decomposed into more than one generalized enzyme reaction, and therefore, the number of applications of the rules was greater than the length of the known pathway. A summary of the BNICE results, including the number of compounds and reactions generated for each compound studied, is given in Table III.
Table II.
Set of reaction operators utilized in each biodegradation pathway.
| Reaction operatora | CBPb | PHEc | GHCHd | 1,2,4-TCBe
|
|
|---|---|---|---|---|---|
| Aerobic | Anaerobic | ||||
| 1.2.1.a | X | X | |||
| 1.3.1.a | X | X | X | ||
| 1.3.-1.a | X | X | X | ||
| 1.3.1.c | X | ||||
| 1.3.1.e | X | ||||
| 1.13.11.a | X | X | X | X | X |
| 1.13.11.b | X | ||||
| 1.14.12.a | X | X | X | X | |
| 1.14.13.a | X | ||||
| 1.14.13.b | X | ||||
| 1.97.1.a | X | X | X | ||
| 2.3.1.b | X | ||||
| 2.8.3.a | X | ||||
| 3.1.1.a | X | X | |||
| 3.1.2.a | X | ||||
| 3.7.1.d | X | ||||
| 3.7.1.e | X | ||||
| 3.8.1.a | X | X | X | ||
| 4.2.1.j | X | ||||
| 4.5.1.a | X | ||||
| 5.3.2.a | X | X | X | ||
| 5.3.99.a | X | ||||
| 5.5.1.a | X | X | |||
| 5.5.1.b | X | ||||
| 6.2.1.a | X | ||||
| Total | 8 | 8 | 10 | 10 | 9 |
Description of operators is given in Supplementary Table I.
4-Chlorobiphenyl.
Phenanthrene.
γ-Hexachlorocyclohexane.
1,2,4-Trichlorobenzene.
Table III.
Compounds and reactions in the reaction networks generated by BNICE.
| CBPa | PHEc | GHCHb | 1,2,4-TCBd
|
||
|---|---|---|---|---|---|
| Aerobic | Anaerobic | ||||
| Total reactions | 297,332 | 35,903 | 73,490 | 3,760 | 734 |
| Total compounds | 60,285 | 13,512 | 1,089 | 1,031 | 273 |
| CAS compounds | 228 | 78 | 372 | 344 | 105 |
| KEGG compounds | 47 | 22 | 64 | 75 | 35 |
| UM-BBD compounds | 30 | 24 | 53 | 49 | 26 |
| Termination compounds | 8 | 3 | 8 | 9 | 2 |
| Novel compounds (total percent) | 60,051 (99.6%) | 13,433 (99.4%) | 710 (65.1%) | 687 (66.6%) | 166 (60.8%) |
4-Chlorobiphenyl.
Phenanthrene.
γ-Hexachlorocyclohexane.
1,2,4-Trichlorobenzene.
Case 1: 4-Chlorobiphenyl (CBP)
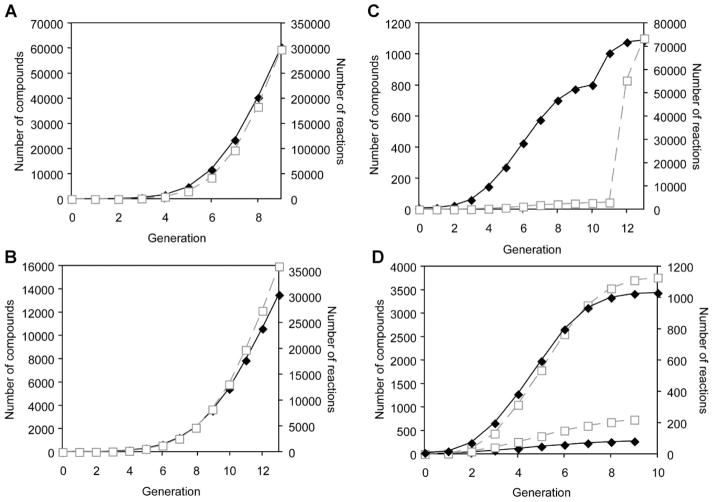
The known CBP biodegradation pathway is comprised of nine reaction steps and yields oxopent-4-enoate and either 3-carboxy-cis,cis-muconate or 2-hydroxy-4-carboxymuco-nate semialdehyde. Eight reaction rules were utilized to reproduce the reported CBP degradation pathway: four reaction rules encoded for oxidoreductases, three rules encoded for hydrolases, and one rule encoded a ligase reaction. The iterative application of these eight reaction rules produced a network of 60,285 compounds and 297,332 reactions. The number of compounds and reactions increased exponentially with each application of the reaction rules (Fig. 2A). Very few reaction rules were responsible for the exponential expansion of the generated networks. The reaction rule for EC 1.14.13 corresponded to a mono-xygenase that can act upon any carbon atom bound to at least one hydrogen atom. This rule applied to a wide range of compounds, and it generated more than half of the reactions in the CBP reaction network.
Figure 2.
Number of compounds (solid diamonds) and reactions (open squares) at each application of the reaction rules. A: 4-chlorobiphenyl; (B) phenanthrene; (C) γ-hexachlorocyclohexane; (D) 1,2,4-trichlorobenzene.
In addition to examining the size of the reaction network, we have studied the profile of compounds generated and used the number of termination compounds obtained to characterize the reaction network. Here, BNICE generated the three endpoints of the known pathway and five additional termination compounds (formate, gentisate, 2-hydroxymuconate, cis,cis-muconate, and oxalate), indicating that the framework can be used to generate routes to additional compounds not produced in the known pathway.
Case 2: Phenanthrene (PHE)
The known biodegradation pathway of phenanthrene to cis,cis-muconate and pyruvate involves 13 reaction steps. Eight reaction rules were required to reproduce the known pathway, and included six rules encoding oxidoreductases and one rule each for EC 4.2.1 and EC 5.3.99. The reaction rules involved in the phenanthrene pathway were applied for 13 successive generations, resulting in a reaction network consisting of 13,512 compounds and 35,903 reactions (Fig. 2B). The number of compounds and reactions increased exponentially with each application of the reaction rules, and the reaction rule EC 1.14.12, which corresponded to the first reaction in the original pathway, generated the largest number of reactions. This enzyme added two hydroxyl groups to two adjacent carbon atoms in an aromatic ring. Due to the structure of phenanthrene, EC 1.14.12 generated a wide array of hydroxylated PAHs, as evidenced by the size of the reaction network.
Nearly all of the compounds generated were novel and were not catalogued in the CAS Registry, KEGG database or UM-BBD (99.4% of the compounds); however, the profile of products did include three termination compounds. Cis,cis-muconate and pyruvate, the termination compounds observed in the known pathway involving 13 reaction steps, appeared after 12 and 5 generations, respectively. This indicates that BNICE generated novel pathways to both endpoints that are shorter than the known route. Additionally, acetate, another compound with known intermediary metabolism, was produced in the sixth generation.
Case 3: γ-Hexachlorocyclohexane (GHCH)
The UM-BBD shows that GHCH is degraded to form acetyl-CoA and succinate (Ellis and Wackett, 2006). This biodegradation pathway involves 12 reaction steps and one of them, the 4-formylbenzenesulfonate dehydrogenase reaction, was decomposed into two generalized enzyme reactions. Thus, 13 applications of the reaction rules were required to reproduce the known biodegradation pathway. After 13 applications of the rules, 73,490 reactions and 1,089 compounds were generated, including 64 KEGG compounds and 8 termination compounds. Only 11 generations were required to produce acetyl-CoA and succinate, compared to 13 reaction steps in the known pathway. Six additional termination compounds were generated: acetaldehyde, acetate, 2-hydroxymuconate, cis,cis-muconate, 3-oxoadipate, and succinyl-CoA.
We further examined how the number of compounds and reactions grew with each application of the reaction rules (Fig. 2C). The number of compounds initially increased exponentially and then began to level off after eight generations. In the 11th generation, the number of compounds exhibited a slight increase before leveling off again. Similarly, the number of reactions increased exponentially through eight generations, leveled off and then exhibited a sharp increase in the 12th generation. The change in the growth rate of the compounds and reactions was due to EC 2.8.3, which encoded the CoA-transferase enzyme. EC 2.8.3 was able to act in the 12th generation, after the ring had been hydroxylated and decyclized. In total, the EC 2.8.3 operator generated more than 70,000 reactions, but it did not significantly affect the number of compounds; there were many compounds that contained the necessary functional groups to undergo the reaction, but the products of these reactions had been created in a previous generation. Thus, in the 12th generation, the number of reactions increased dramatically, while the number of compounds only showed a slight increase.
Case 4: 1,2,4-Trichlorobenzene (1,2,4-TCB)
The metabolism of 1,2,4-TCB has been shown to occur under aerobic and anaerobic conditions, utilizing different enzymes to initialize the degradation under different conditions (Ellis and Wackett, 2006). Glycolate and succinate are the terminal compounds in the aerobic metabolism of 1,2,4-TCB, whereas the anaerobic biodegradation pathway terminates with the production of 3-oxoadipate. A dioxygenase enzyme (EC 1.14.12) catalyzes the first reaction in the aerobic pathway, while a reductive dehalogenase (EC 1.97.1) performs the first reaction in the anaerobic pathway. A total of ten and nine reaction rules were required for the aerobic and anaerobic pathways, respectively. In both pathways, the carboxymethylenebutenolidase reaction (EC 3.1.1.45) was decomposed into two generalized enzyme reactions, EC 3.1.1 and EC 5.3.2, and two generalized enzyme reactions, EC 1.3.-1 and 1.97.1, were required to reproduce the maleylacetate reductase reaction (EC 1.3.1.32) present in the aerobic pathway.
We encoded the individual sets of reaction rules involved in aerobic and anaerobic metabolism and applied the BNICE framework to 1,2,4-TCB (Fig. 2D). We generated 1,031 compounds and 3,760 reactions using the reaction rules from the aerobic metabolism of 1,2,4-TCB, and 273 compounds and 734 reactions using the reaction rules from the anaerobic metabolism of 1,2,4-TCB. The aerobic metabolism of 1,2,4-TCB yielded a larger reaction network compared to anaerobic metabolism due to different types of reaction rules utilized in each pathway. EC 3.8.1, a hydrolase involved only in the aerobic pathway, acted on carbon-halide compounds, generating 996 reactions, which accounted for much of the difference between the aerobic and anaerobic metabolism of 1,2,4-TCB.
In the aerobic pathway, nine termination compounds were generated from 1,2,4-TCB, including glycolate and succinate, the endpoints of the known pathway, and cis,cis-muconate, formate, fumarate, 2-hydroxymuconate, lactate, malate, and propanoate. Contrastingly, the anaerobic metabolism only generated one termination compound, 3-oxoadipate, which is the endpoint of the known biodegradation route.
Novel 1,2,4-Trichlorobenzene Biodegradation Routes
Pathway Analysis
We studied the aerobic degradation of 1,2,4-TCB in greater detail in order to identify novel biodegradation pathways and evaluate their energetic feasibility. BNICE generates a network of reactions, where the compounds are the nodes and reactions are the directed edges. Using this network, we searched for pathways from a given source to a target compound (Ma and Zeng, 2003). The pathway search algorithm provided two types of output: (1) pathway—the set of distinct reactions required to obtain desired products and (2) overall reaction of a pathway—the input and output of the pathway; different pathways can share the same overall reaction. Using the pathway search algorithm, we postulated novel biodegradation routes based on the individual reactions that comprise the pathway, as well as the pathway stoichiometry, which provided a way to classify the pathways.
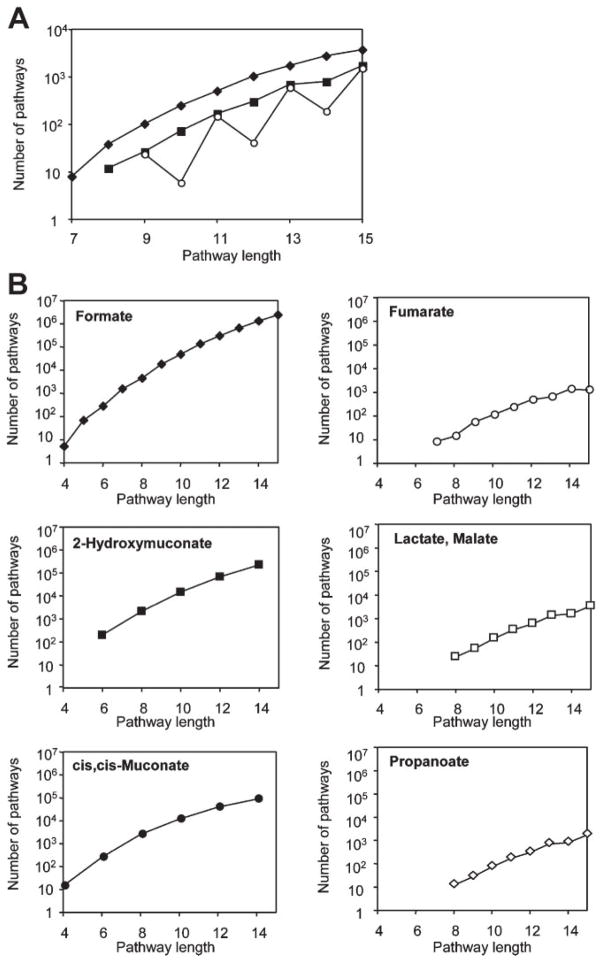
We searched for pathways leading to glycolate and succinate, the endpoints of the known aerobic biodegradation route. In addition to reproducing the known pathway, we discovered thousands of pathways of varying length from 1,2,4-TCB to glycolate, succinate, or both of these compounds, and the number of these pathways increased exponentially with the pathway length (Fig. 3A). Nearly all of the succinate pathways of odd length also generated glycolate, because the last reaction step for those pathways required EC 3.7.1, which produced both glycolate and succinate. We also identified novel pathways that consumed 1,2,4-TCB and produced compounds whose metabolism was known (Fig. 3B). These pathways segregated into two classes and could be distinguished based on the number of carbon atoms in the termination compound. BNICE generated more pathways to larger organic compounds with more carbon atoms. Consider pathways of length 10; there were O(104) pathways to 2-hydroxymuconate or cis,cis-muconate, compared to O(102) pathways to smaller compounds such as glycolate or fumarate. Therefore, we can estimate the number of novel pathways BNICE will generate, given the size of the final product we would like to produce. Pathways that produced formate are the exception to this trend. All of the reactions that generated formate, a simple organic acid, also produced another organic compound, which must be further metabolized in order to obtain a pathway where the final products have known intermediary metabolism.
Figure 3.
Distribution of pathways from 1,2,4-trichlorobenzene to termination compounds. A: Pathways to endpoints of the known pathways: glycolate (solid diamonds), succinate (solid squares), and both glycolate and succinate (open circles); (B) pathways to additional termination compounds.
We classified these pathways according to the overall reaction, and the pathways whose non-cofactor products were termination compounds corresponded to the following reactions:
1,2,4-TCB + (2) O2 + NADH + (3) H2O + (2) e−→ glycolate + succinate + NAD + + (3) H + + (3) Cl−
1,2,4-TCB + (2) O2 + (3) H2O + (2) e−→glycolate + fumarate + (4) H + + (3) Cl−
1,2,4-TCB + (2) O2 + NADH + (4) H2O → glycolate + malate + NAD + + (5) H + + (3) Cl−
1,2,4-TCB + (2)O2 + (4)H + + (6)e−→cis,cis-muconate + (3) Cl−
1,2,4-TCB + O2 + (2)H2O + (2)e−→cis,cis-muconate + (3) H + + (3) Cl−
1,2,4-TCB + (2) O2 + H2O + (4) e−→2-hydroxymuconate + H + + (3) Cl−
The cofactors involved were the distinguishing factors among these overall reactions. Although molecular oxygen was required in all of the pathways, NADH only served as an electron donor in the pathways that corresponded to overall reactions [1] and [3]. With the exception of the pathways with overall reaction [4], all of the pathways required water. Additionally, all of the overall reactions, with the exception of reaction [3], included at least one reaction catalyzed by a reductive dehalogenase enzyme (EC 1.97.1) to remove the chlorine atoms. Each dechlorination step required two electrons, and an electron donor must be available in order for the reaction to proceed. Alternatively, in the pathways corresponding to overall reaction [3], only halidohydrolases (EC 3.8.1), which utilize water, were used to remove chlorine atoms.
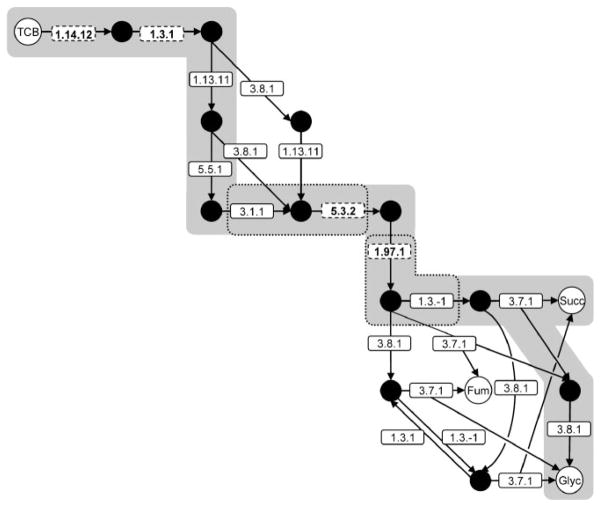
We examined in-depth the pathways which were shorter than or equal to the length of the known pathway and whose endpoints were compounds with known intermediary metabolism. This criterion assumed that the organism performing the biodegradation could utilize the products of the pathway to generate cellular components required for cell maintenance and growth, given that their intermediary metabolism was known. For the purposes of this study, we have focused on the pathways that generated at least one of the compounds of the known pathway; however, the same analysis can be extended to any of the novel pathways generated by BNICE. Lastly, we identified pathways that involved the fewest number of novel intermediates (compounds not found in the CAS, KEGG, or UM-BBD databases). These pathways would require the least amount of genetic engineering to enhance an organism such that it has the enzymes needed to degrade the compound. Based on these criteria, we selected novel pathways involving no more than 10 reaction steps and no novel intermediates. Fifteen novel pathways emerged from this selection process (Fig. 4). All of the pathways began with the same two reaction steps and 11 (69%) of the pathways were shorter than the known biodegradation pathway. The shortest novel pathways, which corresponded to overall reaction [2], consisted of eight reaction steps. Most of the divergence of the alternative pathways from the known pathway occurred at the end of the pathways and resulted from the action of EC 3.7.1 and EC 3.8.1.
Figure 4.
Fifteen novel biodegradation routes of 1,2,4-trichlorobenzene (TCB). Pathways shown were shorter than or equal in length compared to the known pathway and did not involve any novel intermediates. Fum, fumarate; Glyc, glycolate; Succ, succinate. Shading indicates the known pathway and portions of the pathway outlined in dotted boxes show reactions that were decomposed into two enzyme reaction rules. Enzyme classes in dashed boxes denote the reactions common to all of the pathways.
Thermodynamic Analysis
We performed a thermodynamic analysis to evaluate potential biodegradation routes. We used a group contribution method (Jankowski et al., 2008) to estimate the Gibbs free energy of the individual reactions and calculated the cumulative energy of the pathway. This method provides a means of estimating the thermodynamic properties of biochemical reactions and has been shown to be a viable tool to provide a priori estimates of the thermodynamic feasibility of biodegradation pathways (Finley et al., 2009). We have previously shown how to treat biodegradation reactions involving oxygenase and reductive dechlorination reactions (Finley et al., 2009). Since much of the free energy released in oxygenase reactions is associated with the reduction of oxygen to water and is not coupled to the generation of electron carriers, we only report the free energy change that is available for cell mass maintenance and growth. Additionally, we have used acetate as the electron donor for pathways that involve reductive dechlorination reactions. We can use the thermodynamic properties to classify the pathways as thermodynamically favorable, based on the overall free energy change of the pathway and the individual reactions that comprise the pathway, as described below.
Pathways are said to be thermodynamically favorable if the free energy of the overall reaction is less than zero. Overall reaction [1], which corresponded to the known pathway, was found to be highly thermodynamically favorable (Table IV). The pathways corresponding to overall reaction [4] were estimated to have a more negative cumulative free energy change than that of the known pathway. The cumulative free energies of the remaining overall reactions had negative values that were 7–35% higher than that of the known pathway. Therefore, when considering the transformation from 1,2,4-trichlorobenzene to products with known intermediary metabolism, all of the pathways generated by BNICE were estimated to be thermodynamically favorable.
Table IV.
Pathway and thermodynamic analysis of novel 1,2,4-TCB biodegradation pathways.
| Overall reactiona | Number of pathways
|
Cumulative free energy change (kcal/mol)
|
||
|---|---|---|---|---|
| Total | Meet selection criteriab (total percent) | As written | With electron donorc | |
| [1] 1,2,4-Trichlorobenzene + (2) O2 + NADH + (3) H2O + (2) e− → glycolate + succinate + NAD + + (3) H + + (3) Cl− | 1,368 | 8 (0.6%) | −117.7 | −144.6 |
| [2] 1,2,4-Trichlorobenzene + (2) O2 + (3) H2O + (2) e− → glycolate + fumarate + (4) H + + (3) Cl− | 1,374 | 8 (0.6%) | −102.7 | −129.6 |
| [3] 1,2,4-Trichlorobenzene + (2) O2 + NADH + (4) H2O → glycolate + malate + NAD + + (5) H + + (3) Cl− | 2,736 | 0 | −118.4 | n/a |
| [4] 1,2,4-Trichlorobenzene + (2) O2 + (4) H + + (6) e− →cis,cis-muconate + (3) Cl− | 5,331 | 855 (16%) | −81.4 | −161.8 |
| [5] 1,2,4-Trichlorobenzene + O2 + (2) H2O + (2) e− →cis,cis-muconate + (3) H + + (3) Cl− | 9,253 | 504 (5%) | −76.9 | −103.7 |
| [6] 1,2,4-Trichlorobenzene + (2) O2 + H2O + (4) e− →2-hydroxymuconate + H + + (3) Cl− | 17,434 | 2,047 (12%) | −82.4 | −136.0 |
The overall reactions corresponding to pathways whose non-cofactor products were compounds with known intermediary metabolism.
Pathways that: (i) did not involve any novel intermediates and (ii) were shorter than or equal to the length of the known pathway.
Acetate is used as the electron donor for pathways that involved reductive dechlorination reactions.
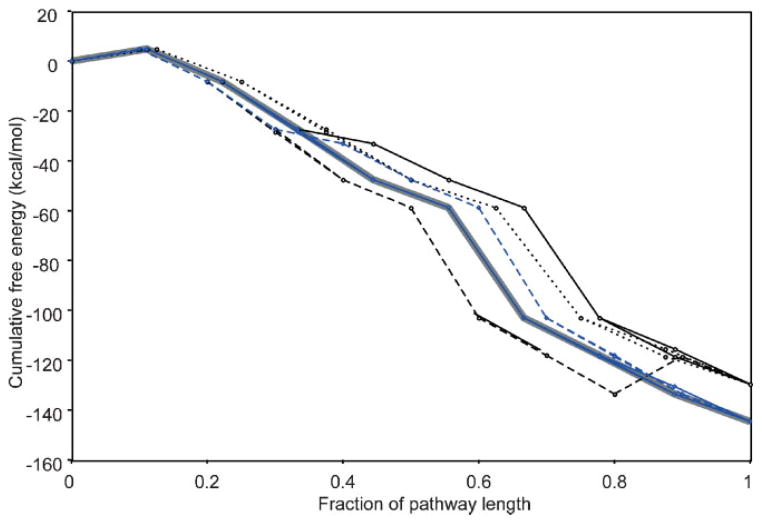
Pathways with the same overall reaction had the same estimated cumulative free energy; however, the free energy landscape of the biodegradation pathways can vary depending on the individual reactions that comprise the pathway. Therefore, in addition to estimating the cumulative free energy of the pathway, we also examined the free energy landscape of the pathway. This type of analysis can identify the specific reactions that account for the range of cumulative free energy values of the overall reactions. Additionally, the free energy landscape provides another criterion for pathways to be classified as thermodynamically favorable: the pathway does not involve reaction steps with a significantly positive free energy change. We calculated the free energy landscape of the 15 alternative pathways discussed above (Fig. 5), and found that all pathways corresponding to overall reaction [1] involved a thermodynamically favorable step (ΔrG′° =−15.0 kcal/mol), leading to the more negative cumulative free energy value compared to overall reaction [2]. This same reaction, EC 1.3.1, was involved in one of the pathways corresponding to overall reaction [2]; however, that particular pathway also included the reverse reaction (ΔrG′° =15.0 kcal/mol), resulting in a net free energy change of zero. With the exception of this pathway, all of the pathways studied were classified as thermodynamically favorable. All 15 pathways shared the same first reaction step, which was slightly uphill (4.8 kcal/mol), and all other reactions had a negative free energy change ranging from −44.2 kcal/mol to −5.8 kcal/mol.
Figure 5.
Thermodynamic landscape of alternative novel biodegradation pathways for 1,2,4-TCB. The fifteen pathways examined in-depth are shown: dotted line, 8 reaction steps; solid line, 9 reaction steps; dashed line, 10 reaction steps. Blue lines represent pathways that correspond to overall reaction [1]; Black lines represent pathways with overall reaction [2]. Shaded gray line indicates the free energy landscape of the known pathway.
Conclusions
We have introduced a biodegradation prediction method that utilizes the BNICE computational framework (Hatzimanikatis et al., 2004, 2005; Li et al., 2004), which is able to generate every possible compound and reaction from a set of starting compounds and reaction rules, creating a large reaction network that includes novel compounds and reactions. The reaction rules implemented in BNICE were derived directly from the EC classification system and corresponded to biochemical reactions known to occur in cellular processes, allowing for broad application of the prediction method to many different biodegradation systems.
We have developed a set of reaction rules to encode the chemistry of biodegradation and applied the BNICE framework to generate novel biodegradation routes for various classes of xenobiotic compounds. The complete set of operators reproduced at least 90% of the biodegradation reactions for 14 of the 20 reaction classes examined. This included six enzyme classes that were fully reproduced by the reaction rules. Although the enzyme reaction rules were not able to fully describe all of the reactions in the reaction classes examined, we have defined reaction rules that follow the given third-level EC classification system and have attempted to capture the observed reaction patterns. We have demonstrated that the application of these reaction rules results in a wealth of novel biodegradation reactions, and further curation of the enzyme classes will expand the diversity of reactions generated by the BNICE framework.
Pathway and thermodynamic analyses were used to identify novel biodegradation pathways for 1,2,4-trichlorobenzene that are thermodynamically feasible alternatives to the known biodegradation route. Here we have considered both the overall transformation to products with known intermediary metabolism and the thermodynamic topology of the individual reactions in the pathway. The predicted biodegradation routes were screened to identify those that only involved known intermediates, assuming that pathways involving fewer novel intermediates will require a lesser amount of genetic engineering in order to implement them into a host organism. This screening method can be applied to all of the biodegradation reaction networks generated by BNICE. In the case of the reaction network for CBP, although more than 99% of the compounds were novel, BNICE did generate 234 known compounds. Using the pathway search algorithm, it is still possible to identify novel biodegradation routes involving these known compounds, ultimately predicting new linkages between known compounds. The predicted reaction pathways can be further screened by estimating how implementation of the pathway into a host organism will influence the existing metabolism of the cell, where metabolic flux analysis (Varma and Palsson, 1993) and thermodynamics-based metabolic flux analysis (Henry et al., 2007) can be used to provide an estimate of the cellular feasibility. Additionally, we can prune the reaction network using toxicity estimation tools, where newly created compounds with a high toxicity value are prevented from undergoing subsequent transformations.
The reaction networks obtained by BNICE differed in size due to inherent differences in the structure of the compounds and the types of reaction rules implemented in each network. Reaction rules encoding oxygenases, enzymes required for metabolism of aromatic compounds, generated the greatest number of reactions when applied to 4-chlorobiphenyl and phenanthrene, compounds containing multiple aromatic rings. Additionally, CoA-transferases, used in small molecule metabolism, generated a wealth of biochemical reactions when applied to γ-hexachlorocyclohexane. These operators generated sizable reaction networks and offered more flexibility in the types of compounds they acted upon. Reaction operators such as these are useful in predicting the biodegradation route of a compound for which there is no prior information about the reactions required to degrade the compound. In this way, BNICE can be used to address the information gap between the number of compounds thought to be biodegradable and the reactions needed to carry out this biodegradation. When combined with thermodynamic analysis, BNICE offers a pathway prediction method that generates energetically feasible biodegradation routes, and it can serve as a tool to determine the fate of compounds in the environment.
Supplementary Material
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Broadbelt LJ, Stark SM, Klein MT. Computer generated pyrolysis modeling: On-the-fly generation of species, reactions, and rates. Ind Eng Chem Res. 1994;33:790–799. [Google Scholar]
- Chen W, Mulchandani A, Deshusses MA. Environmental biotechnology: Challenges and opportunities for chemical engineering. AICHE J. 2005;51(3):690–695. [Google Scholar]
- Darvas F. MetabolExpert, an expert system for predicting metabolism of substances. In: Kaiser K, editor. QSAR in environmental toxicology. Riedel; Dordrecht: 1987. pp. 71–81. [Google Scholar]
- den Besten C, Vet JJ, Besselink HT, Kiel GS, van Berkel BJ, Beems R, van Bladeren PJ. The liver, kidney, and thyroid toxicity of chlorinated benzenes. Toxicol Appl Pharmacol. 1991;111:69–81. doi: 10.1016/0041-008x(91)90135-2. [DOI] [PubMed] [Google Scholar]
- Dua M, Singh A, Sethunathan N, Johri AK. Biotechnology and bioremediation: Successes and limitations. Appl Microbiol Biotechnol. 2002;59:143–152. doi: 10.1007/s00253-002-1024-6. [DOI] [PubMed] [Google Scholar]
- Ellis LB, Wackett LP. The University of Minnesota Biocatalysis/Biodegradation Database: The first decade. Nucleic Acids Res. 2006;34:D517–D521. doi: 10.1093/nar/gkj076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis LB, Gao J, Fenner K, Wackett L. The University of Minnesota pathway prediction system: Predicting metabolic logic. Nucleic Acids Res. 2008;36:W427–W432. doi: 10.1093/nar/gkn315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley SD, Broadbelt LJ, Hatzimanikatis V. Thermodynamic analysis of biodegradation pathways. Biotechnol Bioeng. 2009;103(3):532–541. doi: 10.1002/bit.22285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez MJ, Pazos F, Guijarro FJ, de Lorenzo V, Valencia A. The environmental fate of organic pollutants through the global microbial metabolism. Mol Syst Biol. 2007;3:1–11. doi: 10.1038/msb4100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lergier J, Broadbelt LJ, Hatzimanikatis V. Theoretical considerations and computational analysis of the complexity in poly-ketide synthesis pathways. J Am Chem Soc. 2005;127:9930–9938. doi: 10.1021/ja051586y. [DOI] [PubMed] [Google Scholar]
- Greene NPN, Judson PN, Langowski JJ, Marchant CA. Knowledge-based expert systems for toxicity and metabolism prediction: DEREK, StAR and METEOR. SAR QSAR Environ Res. 1999;10:299–314. doi: 10.1080/10629369908039182. [DOI] [PubMed] [Google Scholar]
- Hatzimanikatis V, Li C, Ionita JA, Broadbelt LJ. Metabolic networks: Enzyme function and metabolite structure. Curr Opin Struct Biol. 2004;14:300–306. doi: 10.1016/j.sbi.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Hatzimanikatis V, Li C, Ionita JA, Henry CS, Jankowski MD, Broadbelt LJ. Exploring the diversity of complex metabolic networks. Bioinformatics. 2005;21(8):1603–1609. doi: 10.1093/bioinformatics/bti213. [DOI] [PubMed] [Google Scholar]
- Henry CS. Computational thermodynamic and biosynthetic analysis of genome-scale metabolic models. Evanston, IL: Northwestern University; 2007. p. 227. [Google Scholar]
- Henry CS, Broadbelt LJ, Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys J. 2007;92:1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou BK, Wackett LP, Ellis LB. Microbial pathway prediction: A functional group approach. J Chem Inf Comput Sci. 2003;43(3):1051–1057. doi: 10.1021/ci034018f. [DOI] [PubMed] [Google Scholar]
- Imai R, Nagata Y, Fukuda M, Takagi M, Yano K. Molecular cloning of a Pseudomonas paucimobilis gene encoding a 17-kiloDalton poly-peptide that eliminates HCl molecules from gamma-hexachlorocyclohexane. J Bacteriol. 1991;173(21):6811–6819. doi: 10.1128/jb.173.21.6811-6819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain RK, Kapur M, Labana S, Lal BPMS, Bhattacharya D, Thakur IS. Microbial diversity: Application of microorganisms for the biodegradation of xenobiotics. Curr Sci. 2005;89:101–112. [Google Scholar]
- Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys J. 2008;95:1487–1499. doi: 10.1529/biophysj.107.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworska J, Dimitrov S, Nikolova N, Mekenyan O. Probabilistic assessment of biodegradability based on metabolic pathways: Catabol system. SAR QSAR Environ Res. 2002;13(2):307–323. doi: 10.1080/10629360290002794. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Sawashima S, Katayama T, Araki M, Harakawa M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline B, Owen T, Lesser B. National Priority Chemicals Trends Report (2001–2005) U.S. Environmental Protection Agency; 2008. [Google Scholar]
- Klopman G, Dimayuga M, Talafous J. META. 1. A program for the evaluation of metabolic transformation of chemicals. J Chem Inf Comput Sci. 1994;34(6):1320–1325. doi: 10.1021/ci00022a014. [DOI] [PubMed] [Google Scholar]
- Li C, Henry CS, Jankowski MD, Ionita JA, Hatzimanikatis V, Broadbelt LJ. Computational discovery of biochemical routes to specialty chemicals. Chem Eng Sci. 2004;59:5051–5060. [Google Scholar]
- Ma H, Zeng A-P. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics. 2003;19(2):270–277. doi: 10.1093/bioinformatics/19.2.270. [DOI] [PubMed] [Google Scholar]
- Mueller JG, Cerniglia CE, Prichard PH. Bioremediation of environments contaminated by polycyclic aromatic hydrocarbons. In: Crawford RL, Crawford DL, editors. Bioremediation: Principles and applications. New York, NY: Cambridge University Press; 1996. pp. 125–194. [Google Scholar]
- Oh M, Yamada T, Hattori M, Goto S, Kanehisa M. Systematic analysis of enzyme-catalyzed reaction patterns and prediction of microbial biodegradation pathways. J Chem Inf Model. 2007;47:1702–1712. doi: 10.1021/ci700006f. [DOI] [PubMed] [Google Scholar]
- Paul D, Pandey G, Pandey J, Jain RK. Accessing microbial diversity for bioremediation and environmental restoration. Trends Biotechnol. 2005;23(3):135–142. doi: 10.1016/j.tibtech.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Pieper DH, Reineke W. Engineering bacteria for bioremediation. Curr Opin Biotechnol. 2000;11:262–270. doi: 10.1016/s0958-1669(00)00094-x. [DOI] [PubMed] [Google Scholar]
- Reed JL, Vo TD, Schilling CH, Palsson BO. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR) Genome Biol. 2003;4:54.1–54.12. doi: 10.1186/gb-2003-4-9-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterman R. A history of PCB biodegradation. In: Crawford DL, Crawford RL, editors. Bioremediation: Principles and applications. New York, NY: Cambridge University Press; 1996. pp. 209–253. [Google Scholar]
- Varma A, Palsson BO. Metabolic capabilities of Escherichia coli: I. Synthesis of biosynthetic precursors and cofactors. J Theor Biol. 1993;165:477–502. doi: 10.1006/jtbi.1993.1202. [DOI] [PubMed] [Google Scholar]
- Wackett LP, Ellis LB. Predicting biodegradation. Environ Microbiol. 1999;1(2):119–124. [PubMed] [Google Scholar]
- Wang FW, Grundmann S, Schmid M, Dorfler U, Roherer S, Munch JC, Hartmann A, Jiang X, Schroll R. Isolation and characterization of 1,2,4-trichlorobenzene mineralizing Bordetella sp. and its bioremediation potential in soil. Chemosphere. 2006;67(5):896–902. doi: 10.1016/j.chemosphere.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Watanabe K. Microorganisms relevant to bioremediation. Curr Opin Biotechnol. 2001;12:237–241. doi: 10.1016/s0958-1669(00)00205-6. [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhao W, Pan J, Qiu L, Zhu Y. Tissue-dependent distribution and accumulation of chlorobenzenes by vegetables in urban area. Environ Int. 2005;31:855–860. doi: 10.1016/j.envint.2005.05.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.