SUMMARY
Antigen specificity is critical in immune response and requires integration of antigen-specific signals with antigen-nonspecific signals such as those provided by cytokines. The mechanism integrating these pathways is incompletely understood. We report here that antigen-specific proliferative responses of CD4+ T cells required downmodulation of tumor suppressor p53. In the absence of T cell receptor (TCR) signal, IL-2 induced sustained increase in p53 protein, which prevented proliferative responses despite strong signaling through the IL-2 receptor. In contrast, TCR signaling resulted in early termination of p53 protein expression by decreasing p53 mRNA as well as strong transcriptional induction of the p53-regulating protein Mdm2. Downmodulation of p53 in response to antigen stimulation was in fact critical for antigen-specific T cell proliferation, and preventing p53 degradation by inhibiting Mdm2 resulted in sustained p53 protein and prevented antigen-specific T cell proliferation. It is thus termination of p53 by TCR signaling that allows proliferative responses, enforcing antigen specificity.
INTRODUCTION
The physiologic stimuli that signal activation of T cells include antigen-specific stimuli delivered through the T cell receptor (TCR) (Smith-Garvin et al., 2009) and antigen-nonspecific signals such as those provided by cytokines (Schluns and Lefrançois, 2003). These classes of T cell signals can be interactive, for example through the ability of TCR engagement to upregulate cytokine receptors (Kim and Leonard, 2002), resulting in cooperativity between antigenic and cytokine stimuli in the induction of proliferative and differentiative responses (Boyman and Sprent, 2012; Constant and Bottomly, 1997; Yamane and Paul, 2013). However, the mechanisms that regulate cooperative interactions and determine the responsiveness of T cells to these diverse stimuli are incompletely understood.
In the adaptive immune system, T and B lymphocytes proliferate extensively after recognition of antigen via TCR or BCR, respectively, increasing the number of antigen-specific T or B lymphocytes, a process of clonal expansion that allows the immune system to rapidly respond to antigenic challenges (Jenkins et al., 2001; McHeyzer-Williams and McHeyzer-Williams, 2005). Antigen-nonspecific cytokines cooperate with antigen receptor signals in these responses to support proliferation and differentiation of antigen-specific cells (Boyman and Sprent, 2012; Schluns and Lefrançois, 2003). After the encounter of a naive or antigen-inexperienced T cell with specific antigen, initial clonal expansion is followed by the appearance of differentiated memory T cells (Harty and Badovinac, 2008; van Leeuwen et al., 2009), which retain antigen specificity and have acquired the capacity for rapid reactivation, proliferation, and expression of effector activity. Memory T cells proliferate in the periphery, and this self-renewal of memory T cells is a mechanism for maintaining their pool size for long periods of time, supporting persistence of immunological memory (Surh and Sprent, 2008). The specific contributions of cytokine and TCR-driven signals in naive and memory cell responses and homeostasis remain uncertain.
In the present study, we have identified a critical role of p53 in antigen-specific responses of CD4+ T cells. p53 is well known as a tumor suppressor that functions to prevent tumor development and growth through induction of cell cycle arrest, senescence, and/or apoptosis in response to abnormal oncogene activation or DNA damage (Kruse and Gu, 2009; Vousden and Prives, 2009). Less is known about the physiological role of p53 in regulating proliferation of normal cells in response to diverse signals. We found that p53 had a profound impact on CD4+ T cell proliferation and that this impact was highly selective. Both primary and memory antigen-specific proliferative responses of CD4+ T cells required downmodulation of p53. Stimulation with interleukin-2 (IL-2) in the absence of concomitant antigen-specific TCR stimulation induced sustained increases in p53 protein expression, and proliferation did not occur under this condition. In contrast, TCR stimulation suppressed p53 mRNA and induced expression of the p53-specific ubiquitin ligase Mdm2, thus limiting the duration of p53 protein expression and allowing only antigen-specific T cell proliferation. This downregulation of p53 was necessary for antigen-specific responses of naive and antigen-primed peripheral T cells and T cell clones. These findings indicate that p53 plays a critical and previously unappreciated role in integrating growth signals to selectively support antigen-specific T cell proliferation.
RESULTS
p53 Inhibits IL-2-Driven Proliferation in the Absence of Antigen-Specific Stimulus
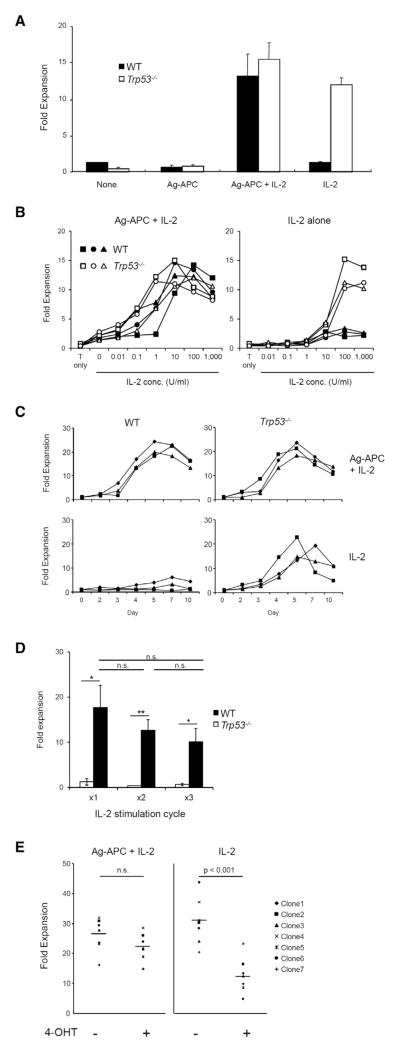
An effective immune system requires a high degree of antigen specificity in responses of T cells to specific antigens. However, T cells can also be driven to proliferate by antigen-nonspecific signals such as those provided by cytokines. Integration of these signals is therefore essential to maintain antigen specificity of response. We have tested the possibility that tumor suppressor p53, a regulator of cell cycle and cell survival, might play a role in the responses of antigen-specific T cells. To characterize the role of p53 in regulating antigen-specific and antigen-nonspecific cytokine-driven T cell responses, we first analyzed antigen-specific CD4+ T cell clones generated from Trp53−/− and littermate Trp53 WT mice. Proliferation of Trp53−/− and WT clones was equivalent after stimulation with specific antigen (KLH) plus antigen-presenting cells (APCs) in the presence of recombinant IL-2 (Figure 1A). Exogenous IL-2 was required for antigen-specific proliferation because of the limiting amount of endogenous IL-2 produced by these clones. Although WT and Trp53−/− clones were equivalently responsive to IL-2 in the presence of antigen-specific signals (Figures 1A and 1B, left), a striking difference was seen in their responses to IL-2 in the absence of antigen (Figures 1A and 1B, right). Trp53−/− clones proliferated strongly to higher concentrations of IL-2 alone at a magnitude of response comparable to their response to Ag-APC + IL-2, whereas WT clones exhibited very limited expansion (Figures 1A and 1B). Similar patterns were observed for multiple KLH-specific WT and Trp53−/− clones (Figures 1A and 1B) as well as for uncloned OVA-specific WT and Trp53−/− CD4+ T cell lines (Figure S1A available online). This differential effect of p53 on T cell responses was sustained over a time course of proliferative response (Figure 1C). These results indicate that WT CD4+ cells proliferate in response to IL-2 only in the presence of TCR signals, whereas Trp53−/− clones respond to IL-2 in either presence or absence of antigen. This difference in responsiveness of WT and Trp53−/− T cells was maintained for multiple IL-2 stimulation cycles (Figure 1D), indicating that recent Ag stimulation was not required for IL-2 responsiveness of Trp53−/− T cells.
Figure 1. Antigen-Specific Trp53−/−, but Not WT, CD4+ T Cell Clones Proliferate in Response to IL-2 in the Absence of Antigenic Stimulation.
(A) WT and Trp53−/− KLH-specific CD4+ T cell clones were cultured for 10 days under indicated conditions. Fold expansion was calculated as (recovered cell number/initial cell number). Mean ± SEM is shown (three clones per genotype). The data are representative of at least three independent experiments.
(B) IL-2 dose response for cell expansion of WT and Trp53−/− CD4+ T clones. Three clones each of WT and Trp53−/− were stimulated under indicated conditions for 10 days and fold expansion was calculated. The graph is representative of two independent experiments.
(C) Kinetic analysis of fold expansion of CD4+ T clones. Three clones each of WT and Trp53−/− were stimulated with Ag-APC + IL-2 or IL-2 alone, and fold expansion at indicated time points was calculated. The graph is representative of three independent experiments.
(D) Three clones each of WT and Trp53−/− were stimulated with IL-2 alone for 10 days in 3 consecutive cycles without Ag-APC stimulation. Fold expansion was calculated at the end of each cycle. The graph is mean ± SEM and representative of at least three independent experiments.
(E) KLH-specific CD4+ T cell clones established from p53ER-TAM mice were stimulated with Ag-APC + IL-2 or IL-2 alone in the presence or absence of 4-hydoroxy-tamoxifen (4-OHT) for 10 days and fold expansion was calculated. The data are representative of three independent experiments.
*p < 0.05; **p < 0.01. See also Figure S1.
The observed differences between WT and constitutively Trp53−/− clones could reflect the direct effect of p53 absence or the secondary consequences of long-term p53 deficiency, for example through accumulation of mutations or epigenetic changes secondary to absence of p53. To address this possibility, we generated T cell clones from genetically targeted p53ER-TAM mice (Christophorou et al., 2005) that express a p53-estrogen receptor fusion protein, p53ER, which can translocate to the nucleus and exert its p53 functions only in the presence of 4-hydroxy-tamoxifen (4-OHT, TAM). The proliferative responses of p53ER-TAM clones to antigen stimulation were similar in the presence of 4-OHT (p53 active) or in its absence (p53 inactive) (Figure 1E, left). In contrast, addition of 4-OHT (p53 active) resulted in suppression of T cell proliferation in response to IL-2 alone (Figure 1E, right). 4-OHT had no effect on the responses of constitutive Trp53−/− clones to IL-2 alone, confirming the specificity of its effect (Figure S1B). It is thus the presence of functional p53 that acts to prevent cytokine-driven antigen-nonspecific T cell proliferation but does not inhibit responses to IL-2 in the presence of specific antigen stimulus.
p53 Does Not Affect Proximal IL-2 Receptor Signaling
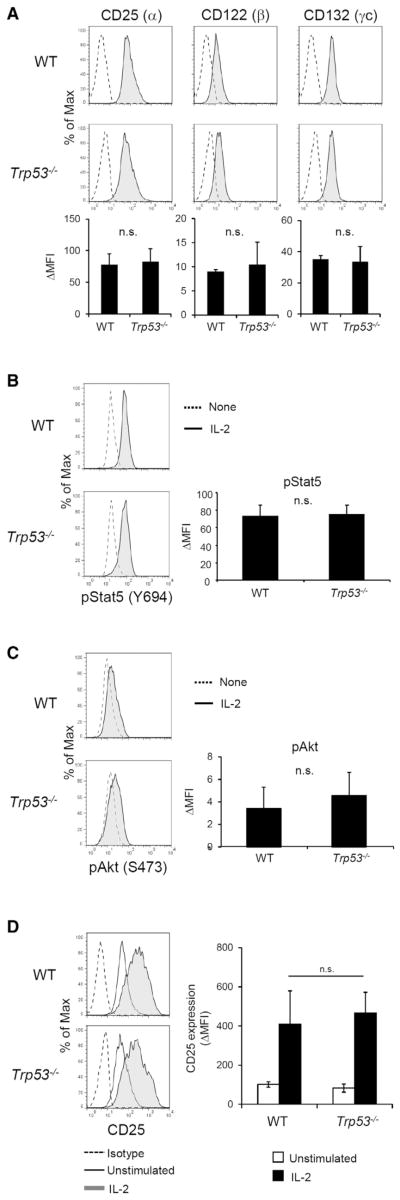
We next pursued the mechanism by which p53 prevents antigen-independent IL-2-induced T cell proliferation. We first asked whether the difference in response to IL-2 by WT and Trp53−/− cells could be explained by differences in IL-2 receptor (IL-2R) complex expression or proximal signaling. The IL-2R complex is composed of CD25 (α), CD122 (β), and CD132 (γc) molecules. Cell surface expression of CD25, CD122, and CD132 was not significantly different between WT and Trp53−/− T cells prior to IL-2 stimulation (Figure 2A). IL-2R signaling proceeds through two known proximal pathways mediated by Stat5 and Akt, respectively (Liao et al., 2013). Phosphorylation of Stat5 and Akt after stimulation with IL-2 was measured by flow cytometric quantitation and was found to be comparable between WT and Trp53−/− (Figures 2B and 2C). CD25 induction after IL-2 stimulation was also analyzed because CD25 expression is known to be a target of IL-2R signaling (Kim et al., 2001). CD25 upregulation was not significantly different between WT and Trp53−/− clones (Figure 2D). These results indicate that proximal IL-2R signaling pathways are not affected by the presence of p53 and that differences in IL-2R signaling therefore do not account for the differential proliferative response of Trp53−/− but not WT T cells to IL-2 stimulation.
Figure 2. p53 Does Not Affect Proximal IL-2 Receptor Signaling.
(A) IL-2 receptor expression of CD4+ T cell clones at resting state was analyzed by flow cytometry. In histograms, solid lines indicate specific staining and dashed lines indicate isotype control staining. The graphs are mean ± SEM of three clones each of WT and Trp53−/− and are representative of three independent experiments. n.s., no significant difference.
(B) Induction of Stat5 phosphorylation upon IL-2 stimulation was analyzed by flow cytometry. Resting WT and Trp53−/−clones were stimulated with IL-2 (100 U/ml) for 15 min and then analyzed. In histograms, solid lines indicate pStat5 (Y694) staining of IL-2-stimulated cells and dashed lines indicate staining of unstimulated cells. The graph presents mean ± SEM of three clones each of WT and Trp53−/− and is representative of three independent experiments.
(C) Induction of Akt phosphorylation upon IL-2 stimulation was analyzed by flow cytometry. Resting WT and Trp53−/− clones were stimulated with IL-2 for 15 min and then analyzed. In histogram, solid lines indicate pAkt (S473) staining of IL-2-stimulated cells and dashed lines indicate staining of unstimulated cells. The graph presents mean ± SEM of three clones each of WT and Trp53−/− and is representative of three independent experiments.
(D) Induction of CD25 expression upon IL-2 stimulation. Resting WT and Trp53−/− clones were stimulated with IL-2 for 24 hr and then CD25 expression was analyzed by flow cytometry. Gray filled histograms indicate CD25 staining of IL-2-stimulated cells, open histograms indicate staining of unstimulated cells, and dashed lines indicate isotype control staining. The graph presents mean ± SEM of three clones each of WT and Trp53−/− and is representative of three independent experiments.
p53 Blocks Cell Cycle Progression in Response to IL-2
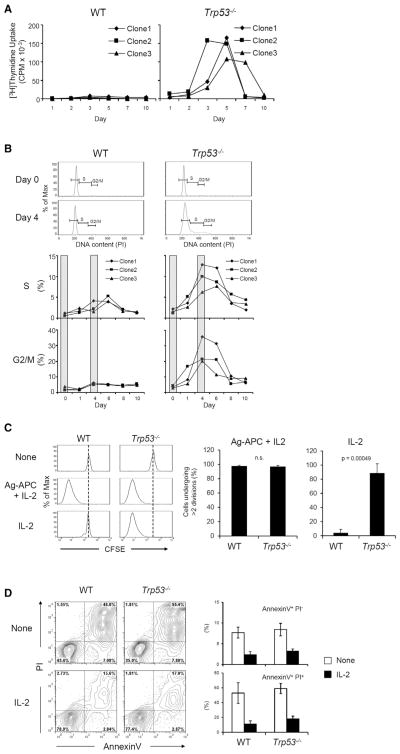
Because p53 is known to prevent cell cycle progression and to induce apoptosis in response to DNA damage and oncogenic signals, we tested whether these mechanisms are also involved in prevention of IL-2-induced T cell expansion. First, DNA synthesis was evaluated by [3H]thymidine uptake. Whereas WT clones showed no significant DNA synthesis at any time point, Trp53−/− clones showed substantial DNA synthesis during days 3–7 after IL-2 stimulation (Figure 3A). Similar results were obtained for cell cycle analysis measured by DNA content, with Trp53−/− clones showing increasing proportions of cells in S and G2-M phase after IL-2 stimulation, with peaks at day 4–8, whereas WT clones showed only small increases of cells in S phase (Figure 3B). Analysis of cell division by CFSE dye dilution similarly showed extensive and equivalent cell division in WT and Trp53−/− clones in response to Ag-APC + IL-2, whereas Trp53−/− but not WT clones underwent extensive division to IL-2 alone (Figure 3C). We also used AnnexinV-PI staining to measure T cell death after IL-2 stimulation (Figure 3D). IL-2 stimulation protected WT as well as Trp53−/− cell from apoptosis that occurred in the absence of added cytokine, and there was no significant difference in proportion of apoptotic cells between WT and Trp53−/− clones in response to IL-2 (Figure 3D). These results indicate that p53 inhibits IL-2-driven proliferation by cell cycle blockade.
Figure 3. p53 Suppresses Cell Cycle Progression in Response to Stimulation with IL-2 Alone.
(A) DNA synthesis of CD4+ clones upon IL-2 stimulation. WT and Trp53−/− clones were stimulated with IL-2 (100 U/ml), and DNA synthesis at indicated time points was analyzed by measuring [3H]thymi-dine incorporation. The result is representative of three independent experiments.
(B) Cell cycle progression of CD4+ clones upon IL-2 stimulation. WT and Trp53−/− clones were stimulated with IL-2, and DNA content at indicated time points was analyzed by propidium iodide (PI) staining. The result is representative of two independent experiments.
(C) Cell division of CD4+ clones upon IL-2 stimulation. WT and Trp53−/− clones were stained with CFSE and cultured under indicated conditions for 10 days. CFSE dilution was analyzed to evaluate cell division by flow cytometry. The graph indicates percent of cells dividing more than two times as mean ± SEM of three clones each of WT and Trp53−/−. The result is representative of three independent experiments.
(D) Cell death of CD4+ clones upon IL-2 stimulation. WT and Trp53−/− clones were stimulated with IL-2 for 48 hr. Percent dead cells were evaluated with AnnexinV and PI staining by flow cytometry. The graphs shown are mean ± SEM of three clones each of WT and Trp53−/− and representative of two independent experiments.
p53 Protein Expression Is Differentially Regulated by Antigen and IL-2 Stimulation
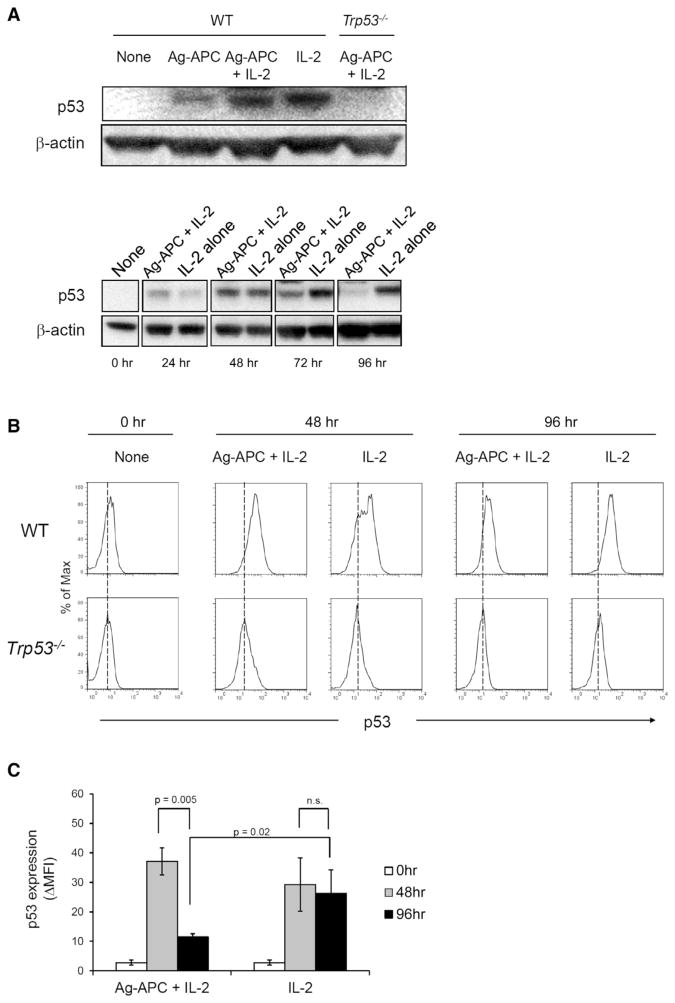
The observations to this point indicated that intact p53 prevents the proliferative response of CD4+ T cells to IL-2 in the absence of antigenic stimulation but does not inhibit responses to IL-2 in the presence of antigenic stimulation. To address the mechanism underlying this differential effect, we asked whether p53 protein expression was differentially regulated in CD4+ T cell clones after stimulation with IL-2 alone, Ag-APC, or Ag-APC + IL-2. After 48 hr, p53 protein was increased in each of these stimulation conditions (Figure 4A, top). Notably, however, although early induction of p53 protein was similar in response to all stimuli, p53 protein decreased substantially by 72 hr and was nearly returned to prestimulation amounts by 96 hr in Ag-APC + IL-2-stimulated clones, whereas induced p53 protein expression was sustained after stimulation with IL-2 alone through 96 hr (Figure 4A, bottom). Similar results were obtained when p53 protein expression was examined at the single cell level by flow cytometry (Figures 4B and 4C). These results indicate that p53 protein is differentially regulated by antigen-specific and IL-2-mediated stimulation, with TCR-mediated antigen stimulation terminating p53 expression.
Figure 4. Antigen Stimulation Downmodulates p53 Expression Induced by IL-2.
(A) WT clones were stimulated for 48 hr (top) and for the indicated times up to 96 hr (bottom), and p53 protein was analyzed by immunoblotting. β-actin was used as internal control. The data are representative of three independent experiments.
(B) WT and Trp53−/− clones were stimulated as indicated for 96 hr and p53 protein expression was analyzed by flow cytometry. Trp53−/− clones were used as a negative control for p53 staining. Results are representative of three independent experiments.
(C) Statistical analysis of the results shown in (B). The graph presents means ± SEM of three clones each of WT and Trp53−/− and representative of three independent experiments.
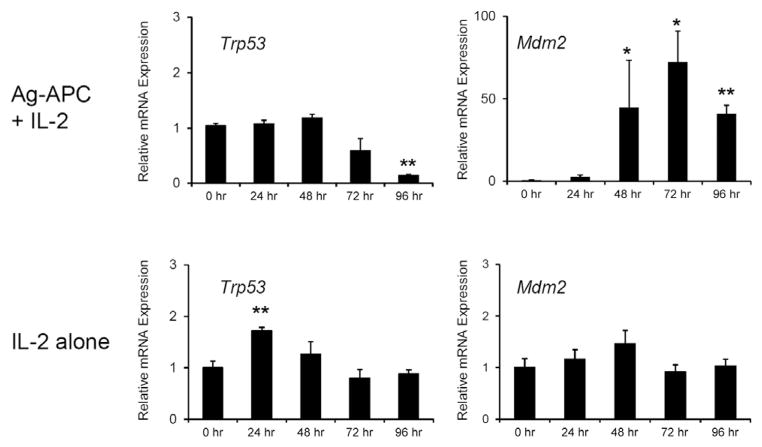
To assess the basis for p53 regulation in response to T cell stimulation, we first measured p53 mRNA by quantitative PCR. p53 mRNA was transiently elevated 24 hr after stimulation with IL-2 alone and then returned to prestimulation baseline. In contrast, after Ag-APC + IL-2 stimulation, p53 mRNA expression was not elevated and by 96 hr was significantly lower than that of prestimulation cells (Figure 5). It is known that p53 protein expression is also regulated posttranscriptionally after DNA damage or oncogene stress by modulation of Mdm2-mediated ubiquitination and proteasomal degradation (Kruse and Gu, 2009). We therefore examined the effect of IL-2 stimulation, in the presence or absence of antigenic stimulus, on expression of Mdm2. Ag-APC + IL-2 stimulation but not IL-2 alone strongly induced Mdm2 mRNA expression to approximately 50 times that of prestimulation cells (Figure 5). This effect was seen by 48 hr and persisted through at least 96 hr. These results suggest that downmodulation of p53 protein upon Ag-APC + IL-2 stimulation was the consequence of both decreased p53 mRNA and increased Mdm2-mediated proteolysis.
Figure 5. Antigen Stimulation Induces Mdm2 and Termination of p53 Expression.
Trp53 and Mdm2 mRNA expression upon stimulation with Ag-APC + IL-2 or IL-2 alone. KLH-specific clones were stimulated as indicated. mRNA expression was measured by real-time qPCR and normalized to Gapdh mRNA expression. The graphs are mean ± SEM of three WT clones and representative of three independent experiments.
Downmodulation of p53 Is Required for Antigen-Specific T Cell Proliferation
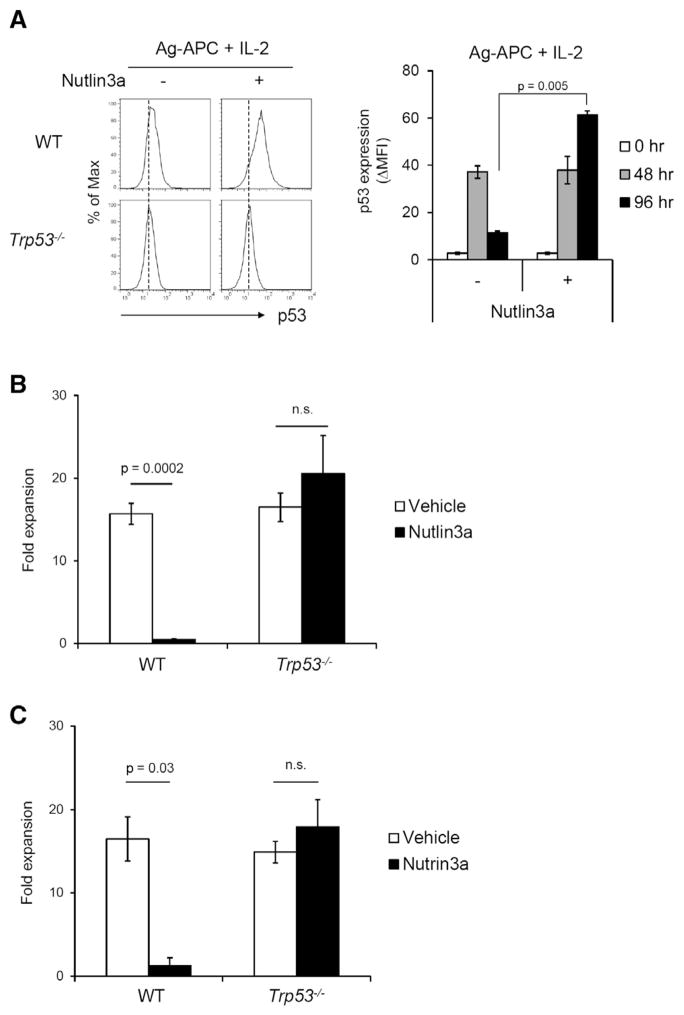
The finding that p53 protein was downmodulated by stimulation with Ag-APC + IL-2 but persisted after stimulation with IL-2 alone suggested that this downmodulation of p53 protein might be critical to induction of antigen-specific proliferation in WT T cells. We therefore tested whether inhibition of Mdm2 would prevent downregulation of p53 protein after Ag-APC + IL-2 stimulation, allowing assessment of the importance of p53 downregulation for T cell proliferative responses. Although p53 downregulation occurred in Ag-APC + IL-2-stimulated T cells by 96 hr (Figures 4B and 4C), in the presence of Nutlin3a, which inhibits Mdm2-p53 interaction and prevents Mdm2-mediated p53 protein degradation (Vassilev et al., 2004), downmodulation of p53 protein in Ag-APC + IL-2-stimulated clones was not observed at 96 hr (Figure 6A). We therefore tested whether preventing p53 downmodulation after Ag-APC + IL-2 stimulation would indeed prevent T cell proliferation.
Figure 6. Nutlin3a Prevents Downmodulation of p53 and Inhibits Antigen-Specific Proliferation of WT but Not Trp53−/− T Cell Clones.
(A) Nutlin3a inhibits downmodulation of p53 protein expression after Ag-APC + IL-2 stimulation. KLH-specific WT CD4+ clones were stimulated with Ag-APC + IL-2 with or without Nutlin3a (5 μM) for 96 hr, and p53 protein expression was measured by flow cytometry.
(B) The effect of Nutlin3a on cell expansion of KLH-specific CD4+ T clones. Three WT and three Trp53−/− clones were stimulated with Ag-APC + IL-2 for 8 days in the presence of Nutlin3a (5 μM) and fold expansion was calculated. The graph is mean ± SEM of three clones each of WT and Trp53−/− and is representative of three independent experiments.
(C) The effect of Nutlin3a on cell expansion of OVA-specific CD4+ T lines. WT and Trp53−/− lines were stimulated with OVA323-339-APC + IL-2 with or without Nutlin3a (5 μM) for 8 days and fold expansion was calculated. The result is mean ± SEM of pool data of two independent experiments.
*p < 0.05; **p < 0.01.
Strikingly, Nutlin3a strongly inhibited the proliferation of KLH-specific WT clones stimulated with KLH-APC + IL-2 but had no effect on the responses of Trp53−/− clones to the same stimulation (Figure 6B). A similar effect was observed when WT or Trp53−/− OVA-specific T cell lines were stimulated with OVA323-339-APC + IL-2 in the presence or absence of Nutlin3a (Figure 6C). These results indicate that downmodulation of p53 is critical for T cell proliferation and that the ability of antigen-specific stimulation to induce IL-2 responsiveness and antigen-specific proliferation requires termination of p53 expression.
Downmodulation of p53 Is Required for Antigen-Specific Proliferation of Naive or In Vivo Primed T Cells
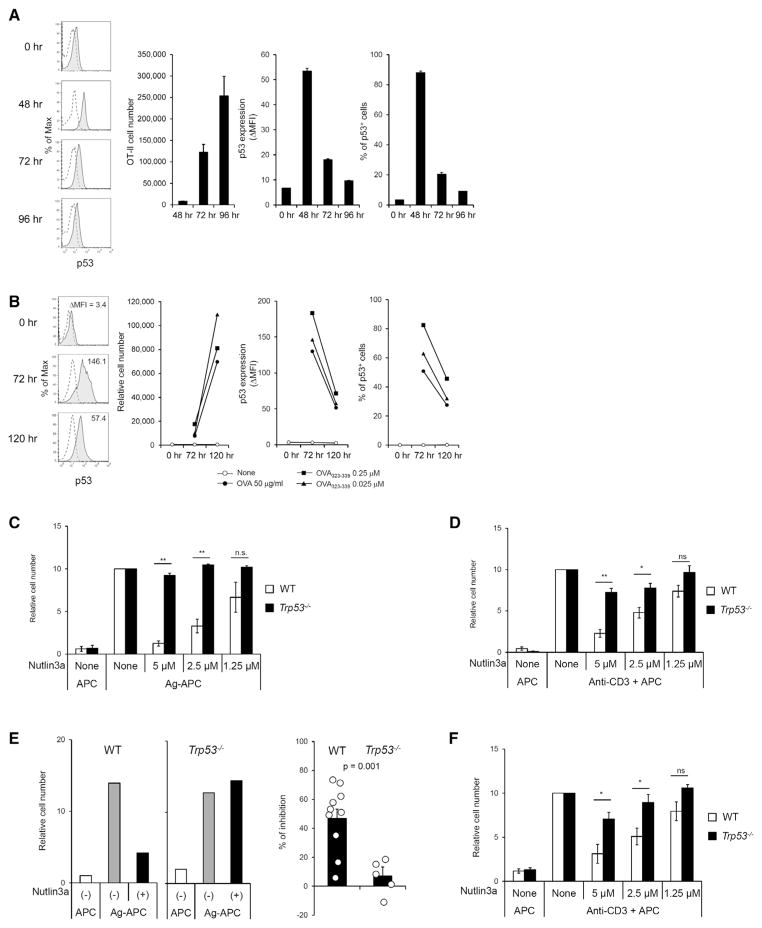
These results suggested that regulation of p53 might play a critical role in the physiologic responses of T cells to TCR-mediated antigen-specific challenge. It was therefore important to determine whether p53 plays a role in regulating antigen-specific proliferative responses of naive or in vivo antigen-primed memory CD4+ T cells, as was shown for cell lines. Kinetic patterns of p53 expression and proliferation of CD4+ T cells were first analyzed in response to in vivo Ag stimulation. OVA-specific OT-II cells were adoptively transferred to congenic CD45.1 mice followed by NP-OVA-Alum immunization. Upregulation of p53 protein in OT-II cells was observed on day 2 after immunization, but p53 was drastically downmodulated at day 3–4, during which time extensive OT-II clonal expansion occurred (Figure 7A), consistent with the pattern observed in KLH-specific CD4+ T cell clones (Figures 4B–4D). Similarly, upregulation of p53 expression in in vitro Ag-stimulated naive OT-II cells was observed at day 3, followed by downmodulation of p53 by day 5, during which time extensive clonal expansion of OT-II cells occurred (Figure 7B). Therefore we next determined whether the primary proliferative responses of unprimed T cells are dependent upon downmodulation of p53. Responses of unprimed p53 WT OT-II cells to specific antigen were strongly inhibited by Nutlin3a, whereas responses of Trp53−/− cells were unaffected (Figure 7C). Nutlin3a also inhibited proliferation of p53 WT CD4+ T cells in response to anti-CD3 TCR-specific stimulation (Figure 7D).
Figure 7. Downmodulation of p53 Is Necessary for Antigen-Specific Proliferation by Naive or Memory CD4+ T Cells.
(A) p53 expression is initially upregulated, followed by downmodulation after in vivo antigen stimulation of CD4+ T cells. OT-II CD4+ T cells were adoptively transferred to CD45.1 mice followed by NP-OVA-Alum immunization. Cell number and p53 expression of OT-II cells were analyzed at the indicated time points. The graph is mean ± SEM (n = 3) and is representative of two independent experiments (total n = 5 mice analyzed individually).
(B) p53 expression is initially upregulated, followed by downmodulation after in vitro antigen stimulation of naive OT-II CD4+ T cells. Naive OT-II CD4+ T cells were stimulated with OVA323-339-APC in vitro, and then cell number and p53 expression of OT-II cells were analyzed at the indicated time points. The data are representative of three independent experiments.
(C) Naive WT or Trp53−/− OT-II CD4+ T cells were stimulated with OVA323-339-APC for 96 hr. Nutlin3a was added to the culture as indicated concentration at 24 hr after stimulation. Recovered live cell numbers are shown. The result is mean ± SEM of pool data of four independent experiments.
(D) Naive B6 CD4+ T cells were stimulated with anti-CD3-APC for 96 hr. Nutlin3a was added to culture as indicated concentration at 24 hr after stimulation. Recovered live cell numbers are shown. The result is mean ± SEM of pool data of three independent experiments.
(E) OT-II memory T cells were generated in vivo as described in Experimental Procedures and restimulated with OVA323-339-APC for 96 hr. Nutlin3a (2.5 μM) was added to the culture at 24 hr after stimulation. Representative result of recovered cell numbers is shown after stimulation with antigen in the presence or absence of Nutlin3a (left). Inhibitory effect of Nutlin3a for antigen-specific memory T cell proliferation is shown as percent inhibition (right). The results shown are mean ± SEM of pool data of three independent experiments (total WT; n = 10, Trp53−/−; n = 5).
(F) Peripheral naturally occurring memory phenotype C57BL/6 CD4+ T cells (CD44hiCD62LloCD25−) were stimulated with anti-CD3-APC for 96 hr. Nutlin3a was added to the culture as indicated at 24 hr after stimulation. Recovered cell numbers are shown. The result is mean ± SEM of pool data of three independent experiments.
*p < 0.05; **p < 0.01. See also Figure S2.
We next determined whether proliferative responses of in vivo primed CD4+ T cells are similarly dependent upon downmodulation of p53. Antigen-primed T cells were generated by adoptive transfer of Trp53−/− or p53-intact OVA-specific OT-II TCR transgenic T cells into Rag1−/− mice and priming with OVA-Alum 1 day after transfer. Three to four weeks later, splenic CD4+ T cells were recovered and challenged in vitro with OVA in the presence or absence of Nutlin3a. OT-II T cells were at this point uniformly CD44hiCD62LloCD25− memory phenotype (Figure S2). In the absence of Nutlin3a, Trp53−/− and p53-intact memory OT-II T cells proliferated vigorously and equivalently in response to OVA (Figure 7E). Addition of Nutlin3a had no effect on the response of Trp53−/− T cells but strongly inhibited responses of p53-intact OT-II cells. Similar results were obtained for proliferation of naturally occurring nontransgenic memory phenotype (CD44hiCD62LloCD25−) peripheral CD4+ T cells in response to anti-CD3 TCR-specific stimulation (Figure 7F). Thus, the antigen-specific responses as well as anti-CD3-induced TCR-specific responses of unprimed and in vivo primed CD4+ T cells are dependent upon downmodulation of p53.
DISCUSSION
The studies reported here identify a central role of p53 in regulating the antigen-specific responses of T lymphocytes. The ability of wild-type naive or memory CD4+ T cells or T cell clones to proliferate in response to TCR stimulation was strictly dependent upon downregulation of p53 protein. This was accomplished by the ability of TCR signaling to induce termination of an initial increase of p53 protein expression by both decreasing p53 mRNA and markedly increasing transcription of Mdm2, which mediates posttranscriptional inactivation of p53 (Kruse and Gu, 2009). Nutlin3a prevents p53 downmodulation by inhibiting p53-Mdm2 interaction and resulted in inhibition of antigen-specific and TCR-mediated proliferation by p53 wild-type but not p53-deficient T cells. The unique ability of TCR signaling to downregulate p53 acts to prevent proliferative responses to antigen-nonspecific signals in the absence of specific antigen and to thereby enforce the antigen specificity of T cell proliferation and clonal expansion that is a hallmark of the adaptive immune system.
The work reported here indicates a central role of p53 in enforcing the antigen specificity of T cell proliferative responses, even when these responses are cytokine dependent. Wild-type CD4+ T cell clones, with intact p53, failed to proliferate in response to IL-2 in the absence of antigen-specific TCR stimulus, despite the presence of strong proximal IL-2 receptor signaling. However, in the presence of a TCR stimulus, these T cells gave strong IL-2-dependent proliferative responses. We found that this TCR dependence of CD4+ T cell clone responses to IL-2 is mediated by p53. Trp53−/− and WT T cells proliferated comparably in response to IL-2 in concert with antigen-specific TCR signals. In marked contrast, p53-deficient but not WT T cell lines were capable of strong responses to IL-2 alone, without TCR stimulus. These findings suggested that p53 inhibits T cell proliferative responses to cytokine, but that this inhibition is overcome by antigen-specific TCR signaling. We indeed found that stimulation with IL-2 alone induced a strong and sustained increase in p53 protein expression. In contrast, stimulation with Ag-APC + IL-2 induced only transient increases in p53 protein, terminated by marked downregulation to prestimulation baseline. These findings suggested that sustained expression of p53 prevents responses of WT T cells to IL-2 and that antigen-specific TCR stimulation overcomes this block by causing termination of the p53 response. Consistent with this model, the proliferative responses of naive CD4+ T cells to TCR stimulation, as well as the responses of in vivo primed T cells or T cell lines to specific antigen, were strongly inhibited when downregulation of p53 was prevented and upregulated p53 was maintained by inhibition of Mdm2. It is thus the termination of p53 elevation by TCR-dependent signaling that allows proliferative response to occur, favoring antigen specificity over nonspecific bystander responses.
The role of p53 in immune responses, and specifically in T cell responses, has been unclear. It was reported that antigen-specific T cell clonal expansion in responses to foreign antigen, such as lymphocytic choriogmeningitis virus (LCMV) infection or sheep red blood cell (SRBC) immunization, was not substantially affected by the absence of p53 in Trp53−/− mice (Grayson et al., 2001; Ohkusu-Tsukada et al., 1999). Other reports showed that Trp53−/− mice exhibited exaggerated phenotypes in T-cell-dependent autoimmune models (Okuda et al., 2003; Simelyte et al., 2005; Zheng et al., 2005) and enhanced antitumor response (Singh et al., 2010), although the T-cell-intrinsic role of p53 was not extensively addressed in these studies. Recently it has been reported that aged T-cell-specific Trp53−/− mice develop an autoimmune phenotype (Kawashima et al., 2013), indicating a role of p53 for regulation of T cell response. Our results reported here demonstrate that p53 expression and function are tightly regulated during antigen-specific T cell proliferative responses, because of the ability of TCR signaling to terminate p53 expression and consequently permit antigen-specific clonal expansion. It has been reported that p53 expression is downregulated in germinal center (GC) B cells, a B cell subset that undergoes massive clonal expansion and class switch- and somatic hypermutation-related DNA break responses in the process of antigen-driven selection (Phan and Dalla-Favera, 2004). In the case of GC B cells, the transcription factor Bcl6, which is highly expressed in GC B cells, represses p53 transcription and downregulates p53 expression (Phan and Dalla-Favera, 2004). We found that TCR-dependent stimulation of CD4+ T cells with specific antigen resulted in decreased p53 mRNA, as well as a very large increase in Mdm2 mRNA. These results suggest that Mdm2-dependent posttranslational mechanisms as well as suppression of p53 transcription contribute to the termination of elevated p53 protein expression by TCR stimulation.
In addition to Mdm2, it is known that the Mdm2-related gene product Mdm4 (also known as Mdmx) interacts with p53 protein to inhibit p53 function (Wade et al., 2010, 2013). However, Nutlin3a is known to more effectively inhibit p53-Mdm2 interaction than p53-Mdm4 interaction because Nutlin3a binds with higher affinity to Mdm2 than to Mdm4 (Wade and Wahl, 2009). Thus, the nearly complete inhibition by Nutlin3a of proliferation of T cell clones and naive CD4+ T cells suggests that the role of Mdm4 is limited in these cells. However, a contribution of Mdm4 cannot be ruled out, especially in memory CD4 response, where inhibition of proliferation by Nutlin3a was less complete. Similarly, p73 and p63 proteins are known p53-related gene products that show transcriptional activity similar to that of p53 (Dötsch et al., 2010; Yang and McKeon, 2000). p73 interacts with Mdm2 and Mdm4 with an affinity similar to that of p53, whereas p63 binding affinity to Mdm2 and Mdm4 is much weaker than that of p53 and p73 (Zdzalik et al., 2010). It is thus possible that Nutlin3a could inhibit p73-Mdm2 interaction and induce increases in p73 in addition to p53 activity. However, the contribution of p73 to inhibition of T cell proliferation, if any, seems very limited in the conditions that we have studied, because Nutlin3a showed only limited effect on Trp53−/− T cells under conditions in which WT T cell proliferation was effectively inhibited (Figures 7C–7F). Therefore, although we cannot completely rule out possible involvement of Mdm4 for p53 regulation and/or p73 for suppression of T cell proliferation in this study, Mdm2-mediated p53 regulation appears to be of predominant importance for controlling antigen-specific T cell proliferation.
Factors in addition to Mdm2 might play a role in regulation of p53 protein expression or function. It is known, for example, that ATM kinase, activated upon DNA damage response, mediates phosphorylation of Ser15 and Ser20 residues of p53, affecting Mdm2 interaction with p53 (Kruse and Gu, 2009). Arf (p19) protein, an alternative reading frame product of the Cdkn2a locus, is another upstream molecule known to activate p53 in the presence of aberrant activation of oncogenes such as c-myc, Ras, or E2F (Chen et al., 2010; Sherr, 2006). Arf binds to Mdm2 and inhibits Mdm2 E3 ubiquitin ligase function resulting in p53 stabilization (Sherr, 2006). Thus it was possible that IL-2 signals might induce Arf as a mechanism for upregulating p53 protein expression. However, KLH-specific Atm−/− or Cdkn2a−/− (Arf-specific ablation) CD4+ T cell clones failed to recapitulate the Trp53−/− phenotype of long-term proliferative response to IL-2 alone (unpublished data). Therefore, in this experimental setting, ATM or Arf is not essential for functional activity of p53. It will similarly be of interest to determine the mechanisms downstream of p53 that are involved in prevention of IL-2-induced T cell proliferation. We found that KLH-specific CD4+ T cell clones deficient in p21 (Cdkn1a−/−), p27 (Cdkn1b−/−), or Ink4a (Cdkn2a−/−; Ink4a-specific ablation), known to be p53 target genes and/or regulators of cell cycle, failed to recapitulate the Trp53−/− T cell phenotype of responsiveness to IL-2 in absence of antigenic stimulus (unpublished data). These results may reflect the presence of redundant pathways that compensate for loss of single genes that act downstream of p53 to mediate p53 function. Indeed, none of the gene-ablated strains that we have tested here, when characterized by others, recapitulate the phenotype of Trp53−/− mice for susceptibility to spontaneous tumor development (Deng et al., 1995; Fero et al., 1996; Krimpenfort et al., 2001; Sharpless et al., 2001). It has been recently proposed that the p53 target genes responsible for tumor suppression in vivo are actually different from the classical p53 target genes that are induced and have been characterized upon acute DNA damage responses (Brady et al., 2011).
Our data indicate that p53 has a role in preventing CD4+ T cell proliferation in response to IL-2 in the absence of antigen-specific TCR signals, potentially limiting bystander proliferation of nonrelevant T cells. Cytokine-driven bystander proliferation of memory T cells requires regulation because unnecessary proliferation could result in accelerated exhaustion of memory T cells as well as competition with naive T cells or with antigen-specific T cells during an immune response, for example through competition for limited availability of cytokines essential for survival. Our results showed that p53 induced by IL-2 suppressed cell cycle progression but did not exhibit a proapoptotic effect. Therefore, in response to IL-2 and potentially other cytokine growth factors, p53 may function to minimize bystander proliferation of nonrelevant memory CD4+ T cells, without affecting cell survival, in order to preserve functional memory T cells without their exhaustion. The ability of TCR signaling to induce Mdm2 and to downmodulate p53 protein expression may also be relevant to the role of p53 as a tumor suppressor. It has been reported that a number of T cell lymphomas show evidence of ongoing TCR signaling pathway activity (Warner et al., 2013), and it is possible that this represents a mechanism for suppressing p53 as one component of growth dysregulation and malignant transformation, a possibility that requires further experimental testing.
The studies presented here provide insight into mechanisms that integrate TCR and cytokine signals to determine the outcome of T cell response, identifying a central role of p53 in this process. IL-2 and antigen-specific signals interact through regulation of p53 protein expression. The mechanism mediating this interaction involves the ability of TCR-mediated antigenic signals, but not IL-2, to induce decreased p53 transcription as well as vigorous upregulation of Mdm2. These effects mediate the termination of a transient elevation in p53 and are required for antigen-specific proliferative responses, as evidenced by the demonstration that inhibition of Mdm2 results in a sustained increase in p53 protein and a p53-dependent inhibition of TCR-dependent primary T cell responses as well as antigen-specific response of in vivo primed T cells or T cell lines. Regulation of p53 thus plays a central role in preferentially allowing proliferative responses by those T cells that encounter specific antigenic challenge in association with growth factor stimulus, resulting in antigen-specific T cell clonal expansion.
EXPERIMENTAL PROCEDURES
Mice
C57BL/6 p53 genetically ablated heterozygous (Trp53+/−) mice were purchased from Jackson laboratory and maintained in our animal facility to generate WT (Trp53+/+) and p53-deficient (Trp53−/−) mice used in the studies reported here. p53ER-TAM genetically targeted mice were obtained from G. Evan (Christophorou et al., 2005). Mice were maintained in accordance with US NIH guidelines. All animal experiments were approved by the NCI Animal Care and Use Committee.
Generation of KLH-Specific CD4+ T Cell Clones
Mice were immunized with KLH (Calbiochem) mixed with Alum (Imject Alum, Pierce) i.p. and 3 weeks later splenocytes were restimulated with KLH (50 μg/ml) plus irradiated splenocytes. Cells were restimulated with KLH plus irradiated splenocytes every 10 days to establish KLH-specific CD4+ T cell lines. KLH-specific CD4+ T cell clones were then isolated by limiting dilution with KLH-APC (irradiated splenocytes) in the presence of recombinant human IL-2 (10 units/ml). TCR Vβ usage of each clone was analyzed by flow cytometry to identify and select independent clones expressing different TCR Vβ for use in experiments. To maintain KLH-specific CD4+ T cell clones, cells were restimulated with KLH (50 μg/ml) plus irradiated splenocytes in the presence of recombinant human IL-2 (10 units/ml) every 10 days. KLH-specific p53ER-TAM CD4+ T cell clones were similarly established and maintained, with the exception that 4-hydroxy tamoxifen (4-OHT) (SIGMA) (200 nM) was added to culture medium every other day. OVA-specific T cell lines were established by repeated stimulation of OT-II Tg CD4+ T cell with OVA323-339 plus irradiated splenocytes every 10 days.
Preparation of T Cell Populations
To analyze mouse primary CD4+ T cells, naive (CD62LhiCD44lo) or memory (CD62LloCD44hiCD25−) phenotype CD4+ T cells were prepared from spleen via MACS (CD4+ T cell isolation kit II and CD62L microbeads [Miltenyi]) and/or flow cytometric cell sorting. To generate OVA-specific memory CD4+ T cells, CD4+ T cells from OVA-specific OT-II TCR Tg mice were utilized. Naive WT or Trp53−/− OT-II CD4+ T cells (CD45.2) were purified and 5 × 105 cells were adoptively transferred (i.v.) to CD45.1 or Rag1−/− mice. Recipient mice were immunized with OVA-alum (i.p.) 1 day after cell transfer. Three to four weeks later, CD44hiCD62LloCD25− memory phenotype CD4+ T cells (CD45.2) were isolated by MACS from splenocytes of the immunized mice.
Flow Cytometry
Cells were washed with FACS buffer (HBSS containing 0.2% BSA and 0.05% Azide) and treated with anti-FcR (24G2), then stained with specific Abs. Anti-CD3, CD4, CD8, CD25, CD122, CD132, pStat5, pAkt, isotype control Abs, and AnnexinV-FITC kits were purchased from BD Biosciences. Anti-p53 (1C12)-Alexa647 Ab was purchased from Cell Signaling. Propidium iodide (PI) was purchased from SIGMA. For pStat5 and pAkt staining, IL-2-stimulated cells were fixed and permeabilized with 4% paraformaldehyde and ice-cold methanol and acetone solution and incubated with Abs for 1 hr at room temperature. For p53 staining, cells were fixed and permeabilized with Foxp3 staining buffer (eBioscience) according to manufacturer’s instructions and incubated with Ab for 30 min at 4°C. Data were collected with FACS Calibur II, LSR III, or Fortessa (BD Biosciences) and analyzed with FlowJo software (Tree Star). Cell sorting was performed with FACS Aria III (BD Biosciences).
Measurement of T Cell Proliferation
To evaluate fold cell expansion of T cell clones, T cells (1 × 105) were stimulated under the indicated stimulation conditions for 10 days and recovered live cell number was microscopically counted. Fold expansion was calculated as (recovered live cell number)/(initial cell number). For cell division analysis, cells were stained with calboxyfluorescein succinimide ester (CFSE) (Molecular Probes) according to manufacturer’s instructions and analyzed by flow cytometry. To evaluate the effect of Nutlin3a (Cayman Chemical) on T cell proliferation, Nutlin3a (1.25–5 μM) or ethanol (vehicle control) was added to cultures.
Immunoblot
Cells were lysed with RIPA buffer (Boston Bioproducts) containing proteinase inhibitor (Clystalgen). Cell lysate were resolved with NuPAGE Bis-Tris SDS-Gel (Invitrogen) and transferred to 0.2 μm PVDF membrane (Invitrogen). Membranes were blotted with specific first Ab followed by HRP-conjugated second Ab. Enhanced chemiluminescence (ECL) (Millipore) was used for developing. Anti-p53 (1C12) Ab was purchased from Cell Signaling. Anti-β-actin Ab was purchased from SIGMA and HRP-labeled anti-mouse-IgG Ab was purchased from Promega.
Quantitative RT-PCR
mRNA was extracted with RNeasy mini kit (QIAGEN). Reverse transcriptase reaction was performed with SuperScript III First-Strand Synthesis System (Invitrogen) with Oligo(dT) primer. The synthesized cDNA was subjected to quantitative PCR with QantiTect SYBR Green PCR kit (QIAGEN). The data were collected and analyzed with 7900HT Real-Time PCR system (Applied Biosystems). Primers used for PCR were the following. Trp53-F: 5′-ACG CTTCTCCGAAGACTGG-3′, Trp53-R: 5′-AGGGAGCTCGAGGCTGATA-3′, Mdm2-F: 5′-CCAGGCCAATGTGCAATACCAACA-3′, Mdm2-R: 5′-TGCGCTC CAACGGACTTTAACAAC-3′, Gadph-F: 5′-CAACTACATGGTCTACATGT TC-3′, Gapdh-R: 5′-CACCAGTAGACTCCACGAC-3′.
Statistical Analysis
Student’s t test with two-tailed distributions was performed for statistical analyses. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank A. Singer, C. Harris, and M. Perry for critical reading of the manuscript and insightful comments during the course of these studies. This work was supported by the Intramural Research Program of the NIH. M.W. was supported by the Uehara Memorial Foundation.
Footnotes
Supplemental Information includes two figures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2014.04.006.
AUTHOR CONTRIBUTIONS
All authors contributed to research planning and experimental design and to analysis of results. M.W., K.M., M.S.V., and K.S.H. carried out experiments. M.W. and R.J.H. wrote the manuscript with review and comments by all authors.
References
- Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Kenzelmann Broz D, Basak S, Park EJ, McLaughlin ME, et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell. 2011;145:571–583. doi: 10.1016/j.cell.2011.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature. 2010;464:624–627. doi: 10.1038/nature08820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christophorou MA, Martin-Zanca D, Soucek L, Lawlor ER, Brown-Swigart L, Verschuren EW, Evan GI. Temporal dissection of p53 function in vitro and in vivo. Nat Genet. 2005;37:718–726. doi: 10.1038/ng1572. [DOI] [PubMed] [Google Scholar]
- Constant SL, Bottomly K. Induction of Th1 and Th2 CD4+ T cell responses: the alternative approaches. Annu Rev Immunol. 1997;15:297–322. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- Dötsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2:a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- Grayson JM, Lanier JG, Altman JD, Ahmed R. The role of p53 in regulating antiviral T cell responses. J Immunol. 2001;167:1333–1337. doi: 10.4049/jimmunol.167.3.1333. [DOI] [PubMed] [Google Scholar]
- Harty JT, Badovinac VP. Shaping and reshaping CD8+ T-cell memory. Nat Rev Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- Jenkins MK, Khoruts A, Ingulli E, Mueller DL, McSorley SJ, Reinhardt RL, Itano A, Pape KA. In vivo activation of antigen-specific CD4 T cells. Annu Rev Immunol. 2001;19:23–45. doi: 10.1146/annurev.immunol.19.1.23. [DOI] [PubMed] [Google Scholar]
- Kawashima H, Takatori H, Suzuki K, Iwata A, Yokota M, Suto A, Minamino T, Hirose K, Nakajima H. Tumor suppressor p53 inhibits systemic autoimmune diseases by inducing regulatory T cells. J Immunol. 2013;191:3614–3623. doi: 10.4049/jimmunol.1300509. [DOI] [PubMed] [Google Scholar]
- Kim HP, Leonard WJ. The basis for TCR-mediated regulation of the IL-2 receptor alpha chain gene: role of widely separated regulatory elements. EMBO J. 2002;21:3051–3059. doi: 10.1093/emboj/cdf321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HP, Kelly J, Leonard WJ. The basis for IL-2-induced IL-2 receptor alpha chain gene regulation: importance of two widely separated IL-2 response elements. Immunity. 2001;15:159–172. doi: 10.1016/s1074-7613(01)00167-4. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- Ohkusu-Tsukada K, Tsukada T, Isobe K. Accelerated development and aging of the immune system in p53-deficient mice. J Immunol. 1999;163:1966–1972. [PubMed] [Google Scholar]
- Okuda Y, Okuda M, Bernard CC. Regulatory role of p53 in experimental autoimmune encephalomyelitis. J Neuroimmunol. 2003;135:29–37. doi: 10.1016/s0165-5728(02)00428-9. [DOI] [PubMed] [Google Scholar]
- Phan RT, Dalla-Favera R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 2004;432:635–639. doi: 10.1038/nature03147. [DOI] [PubMed] [Google Scholar]
- Schluns KS, Lefrançois L. Cytokine control of memory T-cell development and survival. Nat Rev Immunol. 2003;3:269–279. doi: 10.1038/nri1052. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, Bardeesy N, Lee KH, Carrasco D, Castrillon DH, Aguirre AJ, Wu EA, Horner JW, DePinho RA. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- Simelyte E, Rosengren S, Boyle DL, Corr M, Green DR, Firestein GS. Regulation of arthritis by p53: critical role of adaptive immunity. Arthritis Rheum. 2005;52:1876–1884. doi: 10.1002/art.21099. [DOI] [PubMed] [Google Scholar]
- Singh N, Huang L, Qin H. Defective T-cell receptor-induced apoptosis of T cells and rejection of transplanted immunogenic tumors in p53(−/−) mice. Eur J Immunol. 2010;40:559–568. doi: 10.1002/eji.200939736. [DOI] [PubMed] [Google Scholar]
- Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- van Leeuwen EM, Sprent J, Surh CD. Generation and maintenance of memory CD4(+) T cells. Curr Opin Immunol. 2009;21:167–172. doi: 10.1016/j.coi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Wade M, Wahl GM. Targeting Mdm2 and Mdmx in cancer therapy: better living through medicinal chemistry? Mol Cancer Res. 2009;7:1–11. doi: 10.1158/1541-7786.MCR-08-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner K, Weit N, Crispatzu G, Admirand J, Jones D, Herling M. T-cell receptor signaling in peripheral T-cell lymphoma - a review of patterns of alterations in a central growth regulatory pathway. Curr Hematol Malig Rep. 2013;8:163–172. doi: 10.1007/s11899-013-0165-2. [DOI] [PubMed] [Google Scholar]
- Yamane H, Paul WE. Early signaling events that underlie fate decisions of naive CD4(+) T cells toward distinct T-helper cell subsets. Immunol Rev. 2013;252:12–23. doi: 10.1111/imr.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol. 2000;1:199–207. doi: 10.1038/35043127. [DOI] [PubMed] [Google Scholar]
- Zdzalik M, Pustelny K, Kedracka-Krok S, Huben K, Pecak A, Wladyka B, Jankowski S, Dubin A, Potempa J, Dubin G. Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73. Cell Cycle. 2010;9:4584–4591. doi: 10.4161/cc.9.22.13871. [DOI] [PubMed] [Google Scholar]
- Zheng SJ, Lamhamedi-Cherradi SE, Wang P, Xu L, Chen YH. Tumor suppressor p53 inhibits autoimmune inflammation and macrophage function. Diabetes. 2005;54:1423–1428. doi: 10.2337/diabetes.54.5.1423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.