Abstract
Crystal structures of methyl, ethyl, propyl and butyl isocyanide bound to sperm whale myoglobin (Mb) reveal two major conformations. In the in conformer, His(E7) is in a “closed” position, forcing the ligand alkyl chain to point inward. In the out conformer, His(E7) is in an “open” position, allowing the ligand side chain to point outward. A progressive increase in the population of the out conformer is observed with increasing ligand length in P21 crystals of native Mb at pH 7.0. This switch from in to out with increasing ligand size also occurs in solution as measured by the decrease in the relative intensity of the low (~2075 cm 1) versus high frequency (~2125 cm 1) isocyano bands. In contrast, all four isocyanides in P6 crystals of wild type recombinant Mb occupy the in conformation. However, mutating either His64 to Ala, creating a “hole” to solvent, or Phe46 to Val, freeing rotation of His64, causes bound butyl isocyanide to point completely outward in P6 crystals. Thus, the unfavorable hindrance caused with crowding a large alkyl side chain into the distal pocket appears to be roughly equal to that for pushing open the His(E7) gate and is easily affected by crystal packing. This structural conclusion supports the “side path” kinetic mechanism for O2 release, in which the dissociated ligand first moves toward the protein interior and then encounters steric resistance, which is roughly equal to that for escaping to solvent through the His(E7) channel.
Keywords: myoglobin, alkyl isocyanide, isonitrile, X-ray crystallography, FTIR, infrared, vibrational spectroscopy, iron coordination
Mammalian myoglobin (Mb1) is used as a simple model for understanding the ligand binding properties of more complex and less well-characterized globins, from the α and β subunits of adult human hemoglobin to the newly discovered animal neuroglobins and plant non-symbiotic hemoglobins (1-7). However, there is still an active debate regarding the route of ligand entry into the myoglobin binding pocket, which is enclosed and protected from solvent in crystal structures.
Perutz (8) first suggested that ligands enter the distal pocket through outward rotation of the distal histidine, creating a short “open” channel between solvent and the iron atom, and in subsequent work this route has been called the His(E7) gate pathway (9, 10). The discovery of cavities within sperm whale myoglobin, which can bind Xe gas, and of channels between them have led many workers to suggest alternate, multiple routes through the protein interior (11, 12). These apolar pathways appear to be dominant in molecular dynamics simulations of ligand trajectories (13-16) and movements of photodissociated CO between the distal pocket and the Xe1 and Xe4 cavities are well documented by time resolved X-ray crystallography (17-23). However, extensive mutagenesis mapping of all these possible pathways has suggested strongly that ligands primarily enter and leave Mb through the distal histidine, His(E7), gate and not the Xe cavities (9). This same experimental approach has shown that small ligands traverse a long apolar channel to bind to the heme iron in Cerebratulus lacteus hemoglobin (24), and therefore mutagenesis mapping can identify alternative pathways, if they exist. Thus, there still is a significant discrepancy between the kinetic results from libraries of Mb mutants and molecular dynamics simulations of ligand trajectories.
In the 1980s, we began using long chain alkyl isocyanides to probe ligand binding channels within hemoglobins and myoglobins (25-29), using a strategy that Hans Frauenfelder suggested was analogous to Ariadne's thread3. We hoped to find the route into and out of Mb by determining the position of the long alkyl side chain attached to the bound isocyanide. Between 1989 and 1999, we determined the crystal structures of 15 different isocyanide-myoglobin complexes (PDB IDs in Table 1; (30-33)). The distal histidine “gate” is propped open in the published structures of Mb with either imidazole (34) or a phenyl group (35) bound to the heme iron. However, alkyl isocyanides (CNRs) are conformationally more flexible than the rigid aromatic ring ligands and may offer a more relevant probe of the resistance to outward rotation of the His(E7) side chain for dissociating diatomic ligands.
Table 1.
X-ray data collection and refinement statistics.a
| CNR Complex | pH | dmin (Å) | Comp. (%) | Rsym (%) | Rfinal (%) | Rfree (%) | PDB ID |
|---|---|---|---|---|---|---|---|
| native MbCNC1 | 7.0 | 1.9 | 80.5 | 5.9 | 13.6 | NC | 2myb |
| native MbCNC2 | 5.6 | 1.7 | 95.9 | 6.5 | 16.0 | NC | 2mye |
| native MbCNC2 | 7.0 | 2.0 | 88.9 | 18.5 | 15.9 | NC | 2mya |
| native MbCNC3 | 7.0 | 1.8 | 84.5 | 7.1 | 13.3 | NC | 2myd |
| native MbCNC4 | 7.0 | 1.7 | 92.0 | 4.0 | 15.4 | 22.5 | 104m |
| native MbCNC4 | 9.0 | 2.0 | 90.7 | 7.1 | 17.4 | 26.8 | 105m |
| wt MbCNC1 | 9.0 | 1.8 | 88.2 | 5.2 | 15.6 | 21.5 | 110m |
| wt MbCNC2 | 9.0 | 1.8 | 80.7 | 4.4 | 14.7 | 21.3 | 109m |
| wt MbCNC3 | 9.0 | 2.3 | 87.6 | 12.9 | 15.0 | 23.9 | 112m |
| wt MbCNC4 | 9.0 | 1.9 | 98.7 | 5.3 | 16.2 | 22.5 | 111m |
| F46V MbCNC4 | 9.0 | 2.1 | 99.9 | 7.0 | 15.7 | 20.2 | 101m |
| H64A MbCNC4 | 9.0 | 2.1 | 98.4 | 7.2 | 15.8 | 21.6 | 103m |
| V68F MbCNC4 | 7.0 | 2.7 | 99.0 | 9.8 | 12.4 | 22.6 | 108m |
| V68F MbCNC4 | 9.0 | 2.1 | 91.7 | 6.9 | 16.7 | 23.0 | 107mb |
The pH listed is that of the crystal liquor during data collection. dmin is the data resolution. Comp.= completeness. NC (not calculated) is listed for Rfree for the structures refined at a time before this statistic was routinely determined.
A V68F Mb-ethyl isocyanide complex has been deposited into the PDB as 106m, but it is not discussed in this manuscript.
Mb arranges into P21 or P6 crystallographic space groups depending upon its amino acid sequence and the pH of the mother liquor. Native sperm whale Mb was isolated from the skeletal muscle of the animal before the 1986 international whaling ban treaty. Wild-type (wt) sperm whale Mb was first expressed recombinantly in E. coli with a D122N substitution due to an error in the original sequence (36) and an additional N terminal methionine (37). Both residues are distant from the heme and do not significantly affect the rate and equilibrium parameters for ligand binding to Mb in solution. However, as a result of these two surface residue differences, native Mb crystallizes in a P21 arrangement at pH 7.0, whereas wt Mb, and most of its mutant derivatives only crystallize in the P6 crystal form at pH 9.0 (38-40).
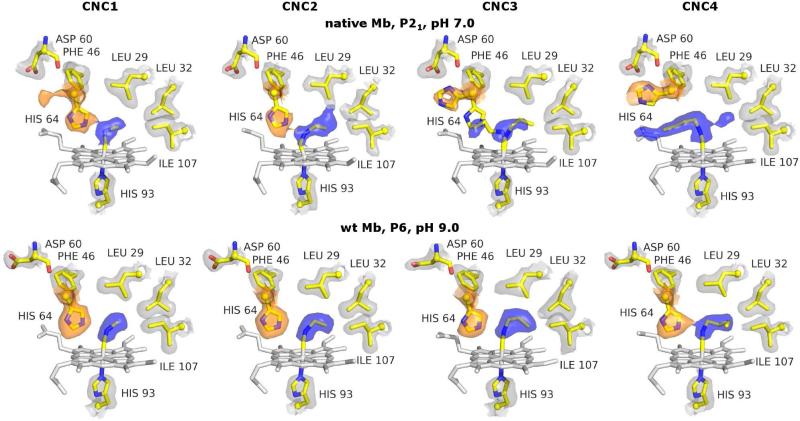
When the X-ray crystal structures of the native MbCNR and the wt MbCNR complexes were initially determined, the ligand conformations were not consistent between the P21 and P6 crystals (Fig. 1, top four vs. bottom four structures; (30-33)). In the P21 crystal form, the ligand conformation is strongly dependent on its size (Fig. 1, top four structures; (30)). The alkyl side chain of CNC2 bound to native Mb points into the interior portion of the distal pocket and resides in the same location as the initial nanosecond state of photodissociated CO. In contrast, the alkyl side chain in native MbCNC4 occupies an out conformation and lies in the channel created by outward movement of the His64(E7) side chain. However, when structures were determined for wt MbCNC1 through MbCNC4 in the P6 crystal form, all of the ligand side chains were found to point inward (Fig. 1, bottom four structures; (33)). There was no obvious explanation for this discrepancy, and at the time, we could not determine the preferred CNR conformation in solution. In effect, Ariadne's thread was pointing in two opposing directions.
Figure 1.
X-ray crystal structures of native Mb CNC1 through CNC4 (top row) and wt Mb CNC1 through -CNC4 (bottom row). For native Mb, in P21 crystals at pH 7.0, an increase in the isocyanide length corresponds to a greater occupancy of the open conformation of His64 (orange 2Fo-Fc electron density surface) and the out conformation of the ligand (blue 2Fo-Fc electron density surface). However, for wt Mb in P6 crystals at pH 9.0, little conformational variation with CNR chain length is observed. Stick representations show the heme (white) and key amino acid side chains (CPK with yellow carbons) of the refined structures. 2Fo-Fc electron density surfaces are shown in orange, blue, and gray for the His64, ligand, and other side chains, respectively.
Resolution of this crystallographic problem has come from FTIR measurements of the stretching frequencies of bound isocyanides in solution, which are described in the preceding paper (41). When the ligand points inward, a hydrogen bond is donated by His64(E7) to the isocyano group, which acts to strengthen Fe-C back bonding, weaken the C≡N triple bond, and decrease the isocyano stretching frequency (νCN). When the ligand is pointing outward, its isocyano group is in an apolar environment, and the C-N bond order (~2.9) and νCN are increased. Thus, the relative heights of the low and high frequency νCN bands allow a direct measure of the relative amounts of the in and out ligand conformers, respectively (41).
In the case of bound CNC4, the low and high frequency νCN bands are present in roughly equal amounts, suggesting that both crystallographic conformers are present in solution (Fig. 2B and 3B). These solution FTIR results have provided the key for interpreting the deposited MbCNR structures in terms of their relevance to proposed models for diatomic ligand movement into and out of myoglobin (42). Our conclusion is that the Ariadne's thread analogy does apply, but in a more complex way than Frauenfelder and we originally envisioned. Dissociated diatomic ligands do appear to move in opposite directions depending on the time scale examined. The ligands first move rapidly to the protein interior but then come back again to the active site on longer nanosecond time scales to escape through the His(E7) gate, following paths defined by the two positions of the longer side chains of bound alkyl isocyanides. The position of the ligand side chain in the out conformation of native MbCNC4 provides the first experimental structural model of the transition state for diatomic ligand movement through the E7 channel of Mb.
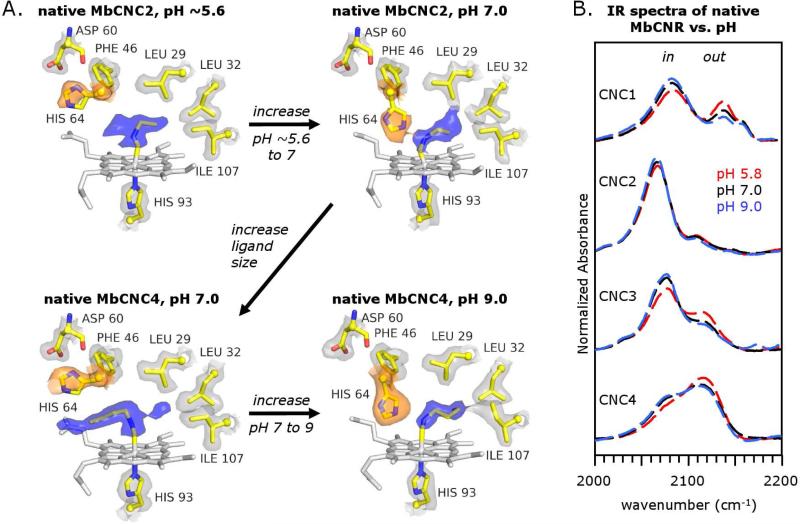
Figure 2.
A. Dependence of the His64 conformation on the CNR length and pH for native Mb in the P21 crystal structure. The 2Fo-Fc and stick color schemes are given in Fig. 1. B. The conformations of MbCNRs in solution are relatively invariant with pH between ~6 and 9.
Figure 3.
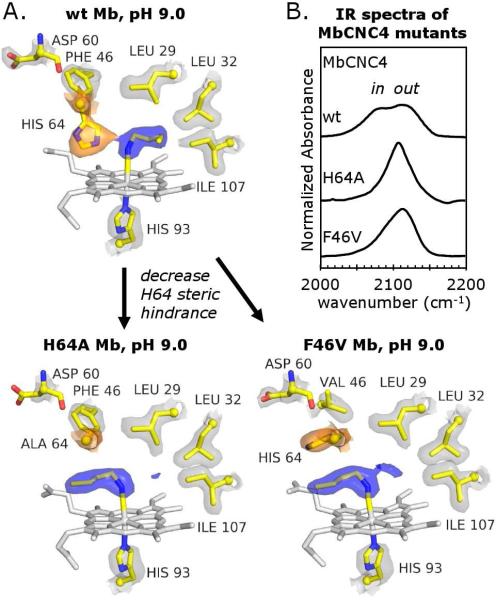
A. The effects of mutations that reduce steric hindrance by His64 on bound CNC4. In P6 crystals at pH 9.0, the bound ligand changed from the in conformation for wt MbCNC4 to the out conformation for both the H64A (bottom left) and F46V (bottom right) MbCNC4 complexes. B. The effects of the same mutations on the νCN bands in FTIR spectra. The IR spectra were collected at pH 7.0 in 0.1 M phosphate buffer.
MATERIALS AND METHODS
Crystallization of Native and Recombinant Sperm Whale Myoglobin
All of the MbCNR structures described in this paper were determined between 1989 and 1999, and the coordinates and structure factors were deposited previously in the Protein Data Bank with the accession codes listed in Table 1. Summaries of the conditions, data collection, and refinement are given below and in Johnson (30) and Smith (33).
Native sperm whale Mb (ferric) was purchased from Sigma before the ban on whale products and crystallized without further purification according to Takano (43). The crystals grew within two weeks and were > 0.5 mm in the longest dimension. Recombinant sperm whale Mb was prepared as described in Rohlfs et al. (37, 44) and crystallized according to Quillin et al. (39). The crystals grew within four to six weeks and were ~1 mm in the longest dimension.
Preparation of the isocyanide derivatives
The isocyanides were prepared as described in Reisberg and Olson (27, 45, 46). The reduced Mb complexes were prepared by: (a) placing two or three metmyoglobin crystals in 5 ml of 3.2 M (NH4)2SO4 and either 100 mM sodium phosphate buffer, 1 mM EDTA, pH 7.0 for native Mb, or 100 mM Tris base, 1 mM EDTA pH 9.0 for the recombinant myoglobins; (b) gently bubbling this solution with nitrogen in a sealed container; (c) injecting 10 μl of neat isocyanide; and (d) after a few minutes, slowly injecting 25 mg sodium dithionite dissolved in 2 ml of the appropriate N2-bubbled ammonium sulfate/buffer solution over a period of several minutes. All of these manipulations were carried out in an anaerobic glove box to prevent oxygen contamination.
The first crystal generated was native MbCNC2 buffered with 0.010 M phosphate solution. The metMb crystal was reduced with a 6-fold greater sodium dithionite concentration to remove residual O2 and reduce the heme iron atoms. After removal of the crystals, the pH of the mother liquor was measured as quickly as possible with brief exposure to air and found to be 5.6. Replication of these low phosphate conditions without crystals also resulted in a solution pH ≤ 6.0. To avoid this drop in pH, the buffer concentration was raised to 0.1 M phosphate and much less dithionite was used. Under these conditions, the pH remained at 7.0 after crystallization.
To verify that an isocyanide-Mb complex was formed, crystals were smashed between glass slides, sealed with wax in an anaerobic chamber, and then transferred to a spectrophotometer for analysis. To change the pH from 7.0 to 9.0 for native Mb crystals and from 9.0 to 7.0 for V68F recombinant Mb P6 crystals, the 0.1 M phosphate and Tris buffers were exchanged in steps before the addition of butyl isocyanide and sodium dithionite.
Crystal mounting, data collection, and refinement
All manipulations were carried out in an anaerobic chamber. A crystal in mother liquor was transferred to a 1 mm internal diameter quartz capillary tube, followed by addition of anaerobic mineral oil containing the appropriate alkyl isocyanide. The capillary was then sealed with wax.
Except as noted, diffraction data were collected at room temperature utilizing a Siemens rotating Cu anode source and the following detectors: for native MbCNC1 and native MbCNC3, a Siemens multi-wire area detector; for native MbCNC2 at pH 5.6, a Nicolet P21 serial diffractometer on a sealed-tub X-ray source; for native MbCNC2 at pH 7, a Siemens P4 serial diffractometer; and for native MbCNC4 (pH 7 and 9), and for the recombinant Mb complexes, a Rigaku R-AXIS IIC imaging-plate detector. The native myoglobin crystals grown at pH 7.0 and the recombinant myoglobin crystals grown at pH 9.0 had P21 and P6 symmetry, respectively.
All data sets were reduced and merged with XDS and XSCALE (47), except those collected on the Nicolet and Siemens serial diffractometers where PROCESS (48) and Siemens software were used, respectively. All data sets were phased by molecular replacement. Refinement of the low pH MbCNC2 data was initially done with the program PROFFT (49, 50) and then completed with XPLOR (A. T. Brunger) version 2.3. The latter program was used exclusively in refinement of the native Mb-CNC1, -CNC2, and -CNC3 structures, whereas XPLOR ver. 3.851 was used for the remaining structures. The range of variations of bond lengths and bond angles from ideal values was 0.01 to 0.03 Å and 1.2 to 2.2 degrees, respectively for all structures. The number of Ramachandran plot outliers was either zero or one for all structures. Completeness in the last shell of data was typically over 90% with R-syms below 0.35. The first structure refined with two conformations for the distal histidine and ligand side chains was that for MbCNC3 (Fig. 1, upper row, third panel). These side chains were retained using grouped occupancy for all refinement iterations. Subsequent models were derived with these groups omitted during the first iterations, then added back for two final rounds.
FTIR Measurements
The samples were prepared as described in the previous paper of this series (51). Each concentrated MbCNR solution was transferred by pipet to the center of a 40 μm well of a calcium fluoride cell. Spectra were recorded in a N2-purged Nicolet Nexus 470 FTIR spectrometer (Nicolet Instrument Corp., Madison, WI) and analyzed as described in Blouin and Olson (51).
Molecular modeling
The molecular graphics program PyMOL from DeLano Scientific, copyright 2006, which incorporates Open-Source PyMOL versions 0.99rc6 and 0.99rev8, was used to generate figures, density maps, crystal symmetry mates, virtual residue replacements, vacuum electrostatic potential surfaces, distance and angle values, and structure alignments. The “align” function minimizes the RMSD distance between the selected atoms followed by two cycles of refinement where outliers are dropped from the minimization.
RESULTS
The CNR Orientation is Length Dependent in P21 Crystals of Native Mb, but not in P6 Crystals of wt Mb
As described in the introduction, the conformation of the distal histidine and the bound CNR shows a marked dependence on the length of the ligand side chain in native Mb P21 crystals at pH 7.0 (Fig. 1, top row). In the native MbCNC1 structure the ligand bends inward toward the protein interior. The His64 side chain in this structure is modeled in a closed position with Cε and Nε near the heme plane and bound ligand, respectively. However, the presence of a minor secondary out conformation is indicated by the unmodeled “bleb” of electron density located near the Asp60 side chain (Fig. 1, top left). Maurus et al. (52) found that replacement of the distal histidine with a smaller threonine in horse heart metMb allowed bound azide to adopt alternate conformations that are similar to those observed for the bound ligand in native sperm whale MbCNC1. In solution, the occupancy of the out conformation for CNC1 bound to Mb is ~35% as determined from its FTIR spectrum (Fig. 2B; Table 2; (41)).
Table 2.
Ligand and His64 conformations and occupancies for the MbCNR structures.
| CNR complex | space group | crystal pH | resolution (Å) | His64 rotomer | His64 occupancya | His64-Nε to CNR-N (Å) | His64-Nδ to Asp60-O′ (Å) | CNR rotomer | CNR occupancyb | FTIR Finc |
|---|---|---|---|---|---|---|---|---|---|---|
| native MbCNC1 | P21 | 7.0 | 1.9 | closed | 0.84 | 2.4 | >4.0 | in | 0.86 | 0.65 |
| native MbCNC2 | P21 | 5.6 | 1.7 | open | 0.53 | >4.0 | 2.6 | in | 0.89 | 0.83 |
| native MbCNC2 | P21 | 7.0 | 2.0 | closed | 0.79 | 2.6 | >4.0 | in | 1.00 | 0.85 |
| native MbCNC3 | P21 | 7.0 | 1.8 | closed / open | 0.26 / 0.68 | 3.4 (closed) | 2.7 (open) | in / out | 0.33 / 0.69 | 0.71 |
| native MbCNC4 | P21 | 7.0 | 1.7 | open | 0.53 | >4.0 | 2.7 | out | 0.70 | 0.44 |
| native MbCNC4 | P21 | 9.0 | 2.0 | closed | 1.00 | 3.0 | >4.0 | in | 0.54 | 0.45 |
| wt MbCNC1 | P6 | 9.0 | 1.8 | closed | 1.00 | 2.9 | >4.0 | in | 0.80 | 0.60d |
| wt MbCNC2 | P6 | 9.0 | 1.8 | closed | 1.00 | 3.2 | >4.0 | in | 0.84 | 0.82d |
| wt MbCNC3 | P6 | 9.0 | 2.3 | closed | 1.00 | 3.0 | >4.0 | in | 0.68 | 0.69d |
| wt MbCNC4 | P6 | 9.0 | 1.9 | closed | 1.00 | 2.8 | >4.0 | in | 0.51 | 0.47d |
| F46V MbCNC4 | P6 | 9.0 | 2.1 | open | 1.00 | >4.0 | 3.7f | out | 0.68 | 0.26d |
| H64A MbCNC4 | P6 | 9.0 | 2.1 | NA | NA | NA | NA | out | 0.62 | NA |
| V68F MbCNC4 | P6 | 7.0 | 2.7 | closed / open | 0.45 / 0.53 | 2.7 (closed) | 3.1 (open) | in / out | 0.45 / 0.53 | 0.61 |
| V68F MbCNC4 | P6 | 9.0 | 2.1 | closed | 1.00 | 2.9 | >4.0 | in | 0.61 | 0.59 |
Occupancies are for the His64 imidazole.
Occupancies are for the CNR alkyl chain.
Solution based measurement. The fraction of complexes in the in conformation, Fin = (in peak Abs)/(in peak Abs + out peak Abs) (56).
Measured at pH 7.0.
e Although His64 occupies the open conformation, this distance is larger than expected due to rotation about its χ2 dihedral angle away from Asp60 to fill the vacant space left by the F46V mutation.
The solution FTIR spectrum of native MbCNC2 indicates the presence of only 15% out conformers (Table 2) in spite of the potential of this larger ligand to sterically hinder His64 in its closed conformation. Predominance of the in conformer is also seen in both the native P21 and wt P6 MbCNC2 crystal structures (Fig. 1), which show that the ethyl group of the ligand points into the back of the distal pocket with a well-ordered and closed histidine gate.
The increase in the fraction of in conformers for bound CNC2 compared to CNC1 is explained by the inductive effect, which alters the isocyano bond order and C-N-C geometry (53). Alkyl groups act as electron donors that promote the neutral sp2 form of the nitrogen atom in isocyanides. A single methyl group is a poorer electron donor than ethyl and other substituted alkyl side chains. Thus, there is less electron density on the isocyano N atom of CNC1, a greater C-NR bond order, and a more upright geometry for the first three atoms of the FeCNCH3 complex than for the FeCNCH2CH3 complex. The more upright conformer tends to force the His64 side chain outward and the lesser electron density on the ligand N atom weakens hydrogen bond donation from His64. The occurrence of a greater bond order for bound CNC1 compared to the larger CNRs is verified by the higher νCN values in the FTIR spectra of CNC1 bound to model heme and Mb mutants with apolar binding pockets (51).
The 2Fo-Fc electron density map for native MbCNC3 indicates the presence of both in and out conformations (Fig. 1), whereas the map for native MbCNC4 shows only the out ligand conformation (Fig. 1, top right). small amounts of electron density are apparent in the back of the distal pocket of native MbCNC4, suggesting the presence of a small fraction of in conformation. The electron density trailing off the end of the CNC4 butyl chain and into the solvent phase cannot be assigned to alternate rotomers of the ligand and may be due to crystallographic waters.
In contrast to the native MbCNR structures, the conformations of CNRs bound to wt recombinant Mb in P6 crystals at pH 9.0 depend little on ligand length (Fig. 1, bottom row). The distal histidine is in the closed position and the ligand side chain is in the back of the distal pocket in all four structures with well-defined electron density for most of the ligand atoms. The C≡N-C fragment of each isocyanide ligand leads from the iron atom toward Leu32 due to steric pressure from Val68, which is not shown. This ligand geometry is also observed for MbO2 and MbN3 complexes. The fourth alkyl carbon of CNC4 and the Cδ of Ile107 show less well defined electron densities, suggesting steric clashes and conformational disorder (Fig. 1, bottom right).
In solution, there is a progressive decrease in the proportion of ligands in the in conformation from 82% for MbCNC2 to 47% for MbCNC4, which is observed as a smaller intensity for the low frequency or in νCN band and greater intensity for the high frequency or out νCN band (Fig. 2B). In solution there are roughly equal populations of the in and out conformers for MbCNC4 at either pH 7 or 9 (Fig. 2B, bottom right). Thus, there are significant discrepancies between the conformer distributions both between the two crystal forms and between each crystal form and those observed in solution.
Ligand Length and pH Act Synergistically to Change the in/out Ratio in Mb P21 crystals
Recombinant wt myoglobin has an N-terminal initiator methionine (Met0) and an N122D mutation (37, 38), which do not significantly alter the protein structure or function, as assessed by unfolding studies (54) and ligand binding measurements (44), but do require adjustment of the crystallization conditions to pH 9.0 in Tris buffer (38). As a result, wt Mb crystals pack with P6 symmetry instead of the P21 symmetry observed for native Mb at pH 7.0 (38). In spite of these crystallization differences, the positions of the backbone atoms, distal residues, heme, and the bound diatomic ligands are very similar (38, 39). The same identity is observed for refined models of wt MbCNC2 at pH 9.0 and native MbCNC2 at pH 7.0 (Fig. 1).
However, the crystal structures of wt versus native MbCNC3 or MbCNC4 are markedly different (Fig. 1), and changes in protonation of His64 could provide an explanation. Yang and Phillips found that in native MbCO crystals at pH 4.0, His64 is rotated outward (55). This effect was attributed to protonation of His64 and its movement outward into solvent for better solvation of the positively charged imidazolium cation (55). In this rotomer, a proton on the Nδ of His64 donates a strong hydrogen bond to the backbone carbonyl oxygen of Asp60. Raising the pH of these native MbCO crystals from 4.0 to 5.0 causes the His64 side chain to rotate into the closed conformation (55). The apparent pKa of ~4.5 for this transition in the P21 crystals is most likely due to protonation of the distal histidine.
In the native MbCNC2 structure determined at pH 5.6, the His64(E7) conformation is similar to that found for the native MbCO structure at pH 4.0 (Fig. 2A, top left; Fig. 2B). The distal histidine side chain is clearly rotated outward and away from the bound ligand, creating an opening between the solvent and the binding pocket. The distance between His64 Nδ and Asp60 O' in the low pH native MbCNC2 structure is 2.6 Å (Table 1), indicating either hydrogen bonding or favorable Coulombic interactions, as was observed in the pH 4.0 MbCO structure (55). The CNC2 ligand was modeled with an inward conformation, but the observed electron density near the iron atom indicates conformational heterogeneity (Fig. 2A, top left), especially when compared to the ligand electron density of the native MbCNC2 complex determined at pH 7.0 (Fig. 2A, top right). For the pH 7 complex, His64 is rotated into the distal pocket (Fig. 2A, top right) and donates a hydrogen bond to the CNC2 isocyano group.
The His64 side chain occupancies for the three MbCNC2 complexes listed in Table 2 reveal a trend from low to high pH of more to less disorder in its position. In the low-pH native MbCNC2 structure, His64 is modeled in the open position with an occupancy for the imidazole ring of ~0.5. A lobe of density contiguous with that assigned to the ligand is observed in the direction of the heme propionates (Fig. 2A, top left). This density coincides with the positions of the Nε and Cδ atoms in the closed His64 conformation, and could represent partial occupancy of this rotomer. The closed His64 positions in the native structure at pH 7.0 and the wt structure at pH 9.0 have occupancies of 0.84-1.00 (Table 2). The occupancies of CNC2 are similar in all three MbCNC2 structures and have values of 0.89 (low pH/native Mb), 1.00 (pH 7/native Mb), and 0.84 (pH 9/wt Mb).
Increasing the ligand length has a similar effect on the His64 conformation as decreasing the pH. In the structures of CNC3 or CNC4 bound to native Mb in P21 crystals at pH 7.0, steric hindrance from the back of the distal pocket is sufficient to “push” the ligand alkyl group into the His64 side chain and force it outward into the open conformation. The electron density for CNC4 bound to native Mb in P21 crystals at pH 7.0 is well ordered and points primarily outward, leading from the coordinating iron atom and out into solvent (Fig. 1, top right; Fig. 2A, bottom left). This increase in the out population caused by the larger CNC4 ligand can be reversed by raising the pH of the P21 crystals of native MbCNC4 from 7.0 to 9.0 (Fig. 2A, bottom right). Remarkably, the bound CNC4 ligand in P21 crystals at pH 9.0 is rotated by 180° compared to its position at pH 7.0 and points inward with His64 rotated into its closed conformation. No other significant structural changes for the amino acids within the distal pocket were resolved that could account for this pH effect. However, structural changes outside the binding site were found including a switch from a type I turn at pH 7.0 to a type II turn at pH 9.0 in the GH-corner and globally larger B-factors for the pH 9.0 structure. It is possible that delocalized structural changes result in a small overall expansion of the distal pocket that is not detectable at the observed data resolution.
The effect of pH on P6 crystals of wt MbCNC4
As shown in Figure 1, bound CNC4 displaces the His(E7) side chain from the distal pocket of native Mb in P21 crystals at pH 7.0, but not when it is bound to wt Mb in P6 crystals at pH 9.0. Increasing the pH of the native MbCNC4 complexes from 7.0 to 9.0, resulted in the loss of the out conformation and the appearance of the same in conformation observed in the pH 9.0 wt MbCNC4 complex (Fig. 2A). We attempted to bring wt MbCNC4 P6 crystals from pH 9.0 to pH 7.0, but in each experiment the crystals became disordered and no diffraction data could be collected. However, we were able to obtain P6 V68F MbCNC4 crystals at pH 9.0, which show a dominant in conformation (PDB ID 107m), and then drop the pH of these mutant crystals to pH 7.0. The resolution of the resulting structure (2.7 Å; Table 1; PDB ID 108m) was too poor for a detailed analysis, and the presence of the large phenyl side chain at the E11 position complicates comparisons with the wt and native forms of Mb. In spite of these limitations, the electron density pattern clearly indicated the presence of both the in and out conformers in the P6 structure of V68F MbCNC4 at pH 7.0 (Table 2). It is unclear if the reduction in pH altered intramolecular or intermolecular structures within the crystal. No such pH-dependent conformational change of the bound CNC4 was observed by FTIR for V68F Mb in solution (Table 2, Fin values).
n-Butyl isocyanide points outward when His64 is removed or made more flexible
At acidic pH, protonation of His64 favors opening of the E7 gate to facilitate hydration of its imidazole cation. However at neutral and basic pHs, which are well above the pKa of the His(E7) side chain, the cause and effect relationship between pH and the in/out equilibrium in Mb crystals is less clear. In solution, the fraction of in versus out conformers does not vary between pH 7 and 9 (Fig. 2B). It is possible that an increase in alkalinity of the crystal environment may either force the His64 side chain into the distal pocket or make the distal pocket less sterically restrictive, and therefore significantly lower the free energy of the in CNR conformation. To help differentiate between the two above scenarios, the His(E7) gate was removed by the H64A mutation, and, separately, made less restrictive by a F46V replacement. In P6 pH 9 MbCNC4 crystals of both mutants, the ligand alkyl group points out into solvent with ~60 to 80% occupancy in the structures, and an out fraction of greater than 70% is observed in the solution FTIR spectra (Fig. 3A, bottom row; Table 2). Thus, the out conformer can occur in P6 crystals at high pH if the His(E7) gate is made significantly smaller or more flexible.
In H64A MbCNC4, there is clearly resolved electron density in the 2Fo-Fc map for the ligand in the outward-pointing orientation (Figure 3A, bottom left). A very small bleb of residual density is observed in the back of the distal pocket, which may represent a minor fraction of the ligand in that space. There are no large deviations of side chain rotomers from those observed for the wt MbCNC4 complex, although, the loss of the H64 side chain does causes the E-helix to move ~0.5 Å toward the CD corner. The H64A MbCNC4 crystal structure clearly indicates that the butyl group of CNC4 is sterically restricted in the back of the distal pocket and will point outward toward solvent, if this movement is not inhibited by a closed distal histidine conformation. The FTIR spectrum does not report on the solution conformation of H64A MbCNC4 since there is no distal histidine and therefore no conformationally-dependent hydrogen bond donation to the bound isocyano group. One peak is observed for this complex at 2108 cm −1 and is indicative of an apolar environment, which would occur in either in or out conformations in the absence of the distal histidine (Fig. 3B).
The crystal structure of F46V MbCNC4 shows results similar to those observed for H64A MbCNC4. In this Mb mutant, substitution of the smaller valine residue for the highly conserved Phe at the CD4 position creates an open space adjacent to the distal histidine. Without steric hindrance from Phe46(CD4), the adjacent E-helix bends by ~1 Å toward the CD-corner, and the His64(E7) side chain can easily rotate about its Cα-Cβ bond (χ1) into the open conformation and about its Cβ-Cγ bond (χ2) by ~46° so that the edge of the imidazole ring partially fills the space left by the missing Phe46 benzyl side chain. The latter Cβ-Cγ rotation creates a larger opening into the distal pocket in the crystal structure (Fig. 3A, bottom right), and probably in solution (56), than is observed in the other Mb structures with an open His64 conformation. Unlike the wt and native MbCNC4 complexes, the conformer distribution in the F46V MbCNC4 crystal structure appears to approximate that found in solution. In the FTIR spectrum of F46V MbCNC4, there is a major out peak at 2112 cm −1 with only a slight shoulder at the lower in peak in the 2070-2080 cm −1 region (Fig. 3B, lower spectrum).
DISCUSSION
In the past, large alkyl isocyanides were used to amplify and highlight steric constraints encountered by the smaller diatomic ligands in their reactions with Mbs, Hbs, and P450s (27, 57-66). Our motivation for solving the crystal structures of straight-chain CNRs bound to Mb was to use the alkyl side chain to visualize ligand pathways from the heme iron atom out into solvent. Two ligand pathways were identified, one through the distal histidine gate and one into the back of the distal pocket (Fig 1, in and out conformers of MbCNC4). The free energies of the individual bound CNR conformations have a complex dependence on the solution and crystallographic environments (Figs 1-4). There is no clear mechanism to explain the conformational differences observed for the P21 versus P6 MbCNC3 and MbCNC4 crystal structures. Our FTIR solution measurements indicate that all of the bound CNC3 and CNC4 conformations identified separately by X-ray crystallography are present in solution, and in the case of MbCNC4, the in and out conformers are found in roughly equal populations in the pH range from 7 to 9. (Figs. 2B and 3B; Table 2, Fin values; (51)).
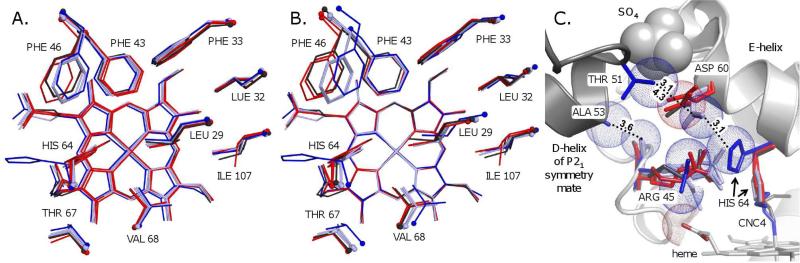
Figure 4.
The effect of the crystal form (P21 and P6) on the distal pocket structures of MbCO and MbCNC4. A. The binding pocket residues and heme groups of Mb structures aligned by all protein Cα atoms, including native Mb (P21 crystals, pH ~7.0) complexed to CO (light blue; PDB IDs 1bzr, 1vxf and 1mbc) and CNC4 (dark blue; this work), and wt Mb (P6 crystals, pH 9.0) complexed to CO (red; PDB IDs 2mgk and 1jw8) and CNC4 (black; this work). The ligands are not shown. B. The same structures were aligned by superimposing the atoms of the heme plane. This alignment emphasizes crystal lattice-dependent variations in the positions of the distal pocket amino acid side chains relative to the ligand heme complex. C. Intermonomer contacts and sulfate binding at the CD-corner of Mb that are present in P21 but not P6 crystals. The structures, colors and alignment are as in panel A. Spheres are shown with van der Waals radii for the native MbCNC4 Thr51-Cγ; Asp60-Cγ, Ala53-Cβ, His64-Cε, Arg45-O', -Nη1 and -Nη2, and sulfate atoms, and for the wt MbCNC4 Asp60-Cγ and heme propionate O1D atoms.
Phillips' group showed that the solution dynamics of Mb are most accurately represented by a composite of Mb X-ray structures from four different crystal forms due to packing constraints (67, 68). Our results show that composite crystal structures are also required to describe the solution properties of MbCNC4, which has roughly equal populations of alternate in versus out ligand conformers. These conformers are “frozen out” separately in the P21 and P6 crystal forms. The pH of the mother liquor also has a large effect on the free energy penalty for opening the histidine gate versus that for sequestering CNC4 within the binding pocket. As shown in Fig. 2, raising the pH of native MbCNC4 crystals from 7.0 to 9.0 results in an inward movement of the ligand side chain. However, in solution, there are no changes in the distribution of in versus out conformers between pH 7 and 9, and an increase in the amount of out conformer does not occur until the pH is lowered to < 6.0 (Fig. 3; (51)).
Comparisons of the Binding Pocket Structures for Mb in P21 versus P6 crystals
Several MbCO structures have been deposited in the PDB library and were used to try to isolate any shifts in side chain placements due to differences in crystal space group packing and pH conditions from those for large ligand-induced movements (Fig. 4). There is no dependence on crystallographic space group for the distal pocket volumes, calculated with the web based CASTp server (69) after deletion of the ligand atoms, nor for the total protein volumes, calculated with Blue Star Sting Millennium (70, 71), for either the MbCO or the MbCNR structures shown in Fig. 4. (data not shown).
MbCO and MbCNC4 structures were aligned in Pymol by their Cα atoms to compare the positions of the distal pocket side chains (Fig. 4A). There are consistent differences between the group of three native MbCO structures and the pair of wt MbCO structures in side chain and heme locations, but they are subtle and imprecise given the resolutions and the potential model bias for the crystal structures. A second comparison was made by aligning the atoms in the heme pyrroles and the carbon atoms directly attached to them (Fig 4B). This frame of reference better represents the steric environment encountered by a ligand, but hides crystallographically induced shifts of the heme groups. For example, the Cα alignment in Fig. 4A shows that the heme group of the native MbCNC4 complex at pH 7 lies more toward solvent (at left) relative to the wt Mb structures at pH 9. In the heme aligned structures in Fig. 4B, the displacements of the distal pocket side chains are slightly larger between the native (light blue) and wt (red) MbCO structures.
The heme alignment also emphasizes the relative change in position of the distal pocket residues in response to steric clashes with bound CNC4, as compared to CO. The Val68 Cγ2 directly contacts and sterically hinders the first two ligand atoms bound at the heme iron (40, 72, 73). The Cγ2 atom is positioned laterally just to the front of (left) and just to the back of (right) the iron atom in P6 and P21 MbCNC4 crystal structures, respectively (Fig. 4B). Thus, in the P21 crystals (pH 7.0) of MbCNC4, this atom may be restricting the in conformation and favoring outward movement of the ligand butyl group, whereas in the P6 crystal structure of wt MbCNC4, the Val68 Cγ2 is closer to the solvent side of the iron, perhaps favoring the in conformation (black structure in Fig. 4B).
The largest side chain shift occurs for Phe46(CD4). Mutation of this residue to the smaller amino acid valine causes the His(E7) gate to open in P6 crystals of both MbCO (74) and F46V MbCNC4, confirming that Phe46 sterically controls movement of the His64 side chain (Fig. 3). The phenyl ring is further from the iron atom in all of the P6 structures and should facilitate the in conformation for His64 and bound CNC4. In the P21 structures the Phe46 side chain is closer to the iron, reduces the space available to the His64 imidazole ring in its closed conformation, and promotes its outward rotation, particularly under the steric pressure exerted by large bound isocyanides.
The effects of pH, crystal packing, and electrostatic interactions on the crystal structures of MbCNC4
The in versus out ratio for MbCNC4 conformers in solution does not change between pH 7.0 and 9.0 (Fig. 2B; Table 2). If a significant proportion of His64 in the out conformer were protonated at pH 7.0, then increasing the pH to 9.0 would result in an decrease in the absorbance of the out peak at 2083 cm −1 and an increase in the intensity of the in peak at 2113 cm −1 (Fig. 2B). However, there is no change in the in to out ratio, which is ~0.5 at both neutral and high pH. Only when the solution pH is lowered to 5.8 does the relative intensity of the out peak increase significantly suggesting that the pKa of His64 in solution is below 6.0 for MbCNR complexes. This conclusion is supported previous work with MbCO, both in crystals and in solution, where the pKa for the imidazole group has been estimated to be 3.8 for the closed conformation and 6.0 for the open conformation (75), with a net value of 4.5 for the overall transition (76).
In addition to the positions of Val68 and Phe46, another cause for the difference between the solution, P6, and P21 MbCNC4 structures is shown in Fig. 4C. In the P21 crystal form, adjacent Mb molecules pack closely together at the D helix and CD corner regions, and part of this interface contains a well-defined sulfate anion. As a result, the Asp60 side chain is forced toward His64 (χ1 ≈ −160° ± 10°) by the Thr51 methyl group of an adjacent Mb molecule and by repulsion from the bound sulfate anion (Fig. 4C, blue sticks). In P6 crystals of Mb, this crystal contact and the sulfate anion are absent (Fig. 4C, red sticks), and the Asp60 side chain is allowed to rotate outward (χ1 ≈ −80° +/− 10°) and adopt the preferred χ1 rotomer for an aspartate side chain on an α-helix (77). In solution, neither a protein protein interface nor a bound sulfate are present to influence the Asp60 side chain conformation.
In the out conformation for P21 crystals of MbCNC4 at pH 7, the Asp60 carboxylate and the His64 imidazole are in van der Waals contact (Fig. 4C, blue sticks), This proximity should raise the His64 pKa markedly and stabilize the out conformation by favoring an electrostatic interaction between the His64 imidizolium cation and the Asp60 carboxylate anion. Raising the pH of these crystals would eventually deprotonate His64, favoring the in conformation, which is observed in the structure of native MbCNC4 in P21 crystals at pH 9.0. The P21 crystallographic interface shown in Fig. 4C also includes an Ala53-Cβ/Arg45-O' contact that may alter the conformations of Arg45 and Phe46, and secondarily His64 (74, 78-80). However, the crystal structures do not offer direct evidence that these structural elements are interdependent.
Correlations with the His(E7) Gate, Side Pathway Mechanism for Diatomic Ligand Binding
Regardless of the exact interpretations, it is clear that the free energies of the in versus out conformations are very similar for bound CNC3 and CNC4, and small crystal packing perturbations can easily change the relative populations. Significant amounts of both conformers are observed in the solution FTIR spectra, and poor occupancies of CNC3, CNC4, and the His64 side chain are observed in the crystal structures, implying significant conformational disorder. Presumably the flexible butyl group of bound CNC4 finds low energy pathways or channels near the Mb active site. One pathway points into the back of the distal pocket and the other leads out of the protein through the E7 channel (Figs. 1-3). These ligand pathways are very similar to those predicted for movement of photodissociated O2 in the E7-gate/side-path model supported by the mutagenesis mapping of Scott et al. (9, 81) and for the movement of CO in time resolved crystallographic experiments (17-23). In both measurements, dissociated ligands appear to first move into the back of the distal pocket but only escape the protein by moving back over the heme and out through the His(E7) gate.
The latter movement through the E7 channel has never been “seen” or detected directly by time resolved spectroscopy or crystallography because there are no stable transient states along this short pathway. Even in molecular dynamics simulations the ligand is not “found” to any great extent in the E7 channel. The crystal structures for the long-chain isocyanide complexes provide the first direct experimental structures of plausible transition states for diatomic ligands entering Mb through the E7 gate and then being captured in the back of the distal pocket.
Our conclusion that the in and out ligand side chain conformers of bound CNRs represent diatomic ligand pathways is supported by the structures of the F46V and H64A MbCNC4 complexes. Compared to wt Mb, both of these mutants have markedly increased fractions of CNC4 out conformers (Fig. 4), which correlate with increased bimolecular rate constants and decreased fractions of geminate recombination for diatomic gases. Thus, Frauenfelder's idea that long chain isocyanides are analogous to Ariadne's thread has proved to be correct, even if the route of escape involves initial movement into the protein interior and then reversal of direction back over the heme and out through the E7 gate.
BRIEFS.
Alkyl isocyanides bind to myoglobin in two major conformations, one pointing through the histidine “gate” and one occupying state “B” in the back of the binding pocket.
Footnotes
Supported by U.S. Public Health Service Grants GM 35649 (J.S.O.), HL 47020 (J.S.O.) and AR 40252 (G.N.P.), and Grants C 612 (J.S.O.) and C 1142 (G.N.P.) from the Robert A. Welch Foundation. G. C. B. was the recipient of a traineeship from The Houston Area Molecular Biophysics Predoctroral Training Grant GM08280. The coordinates and structure factors for all of the myoglobin alkyl isocyanide structures determined in this paper have been deposited in the Protein Data Bank with the accession codes listed in Table 1.
Abbreviations: Mb, myoglobin; wt, wild type; CNR, alkyl isocyanide, where R = C1, C2, C3, C4 for methyl, ethyl, n-propyl and n-butyl groups; FTIR, Fourier transform infrared spectroscopy; νCN, isocyano group stretching frequency; Fin, Fraction of CNRs that point into the Mb binding pocket as measured by FTIR; k', association rate; k, dissociation rate; Ka, equilibrium association constant calculated as k'/k; Fgem, fraction of geminate recombination; PDB, Protein Data Bank.
Frauenfelder made this suggestion in 1981. In Greek mythology, Ariadne, King Minos' daughter, gave her lover Theseus a skein of thread to unravel as he entered the labyrinth of the Minotaur under the royal palace on the island of Crete. After slaying the beast, Theseus was able to find the path for escaping out of the maze by following the thread (Bulfinch, T. (1865) The Age of Fable, Tilden, Boston). The Ariadne's thread metaphor is also used in the field of logic to describe methods for solving problems with multiple solution pathways.
REFERENCES
- 1.Dou Y, Maillett DH, Eich RF, Olson JS. Myoglobin as a model system for designing heme protein based blood substitutes. Biophys Chem. 2002;98:127–148. doi: 10.1016/s0301-4622(02)00090-x. [DOI] [PubMed] [Google Scholar]
- 2.Bellelli A, Brunori M, Miele AE, Panetta G, Vallone B. The allosteric properties of hemoglobin: insights from natural and site directed mutants. Curr Protein Pept Sci. 2006;7:17–45. doi: 10.2174/138920306775474121. [DOI] [PubMed] [Google Scholar]
- 3.Hankeln T, Ebner B, Fuchs C, Gerlach F, Haberkamp M, Laufs TL, Roesner A, Schmidt M, Weich B, Wystub S, Saaler-Reinhardt S, Reuss S, Bolognesi M, De Sanctis D, Marden MC, Kiger L, Moens L, Dewilde S, Nevo E, Avivi A, Weber RE, Fago A, Burmester T. Neuroglobin and cytoglobin in search of their role in the vertebrate globin family. J Inorg Biochem. 2005;99:110–119. doi: 10.1016/j.jinorgbio.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Hoy JA, Hargrove MS. The structure and function of plant hemoglobins. Plant Physiol Biochem. 2008;46:371–379. doi: 10.1016/j.plaphy.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Hoy JA, Robinson H, Trent JT, 3rd, Kakar S, Smagghe BJ, Hargrove MS. Plant hemoglobins: a molecular fossil record for the evolution of oxygen transport. J Mol Biol. 2007;371:168–179. doi: 10.1016/j.jmb.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 6.Kundu S, Trent JT, 3rd, Hargrove MS. Plants, humans and hemoglobins. Trends Plant Sci. 2003;8:387–393. doi: 10.1016/S1360-1385(03)00163-8. [DOI] [PubMed] [Google Scholar]
- 7.Trent JT, 3rd, Watts RA, Hargrove MS. Human neuroglobin, a hexacoordinate hemoglobin that reversibly binds oxygen. J Biol Chem. 2001;276:30106–30110. doi: 10.1074/jbc.C100300200. [DOI] [PubMed] [Google Scholar]
- 8.Perutz MF, Matthews FS. An X-ray study of azide methaemoglobin. Journal of Molecular Biology. 1966;21:199–202. doi: 10.1016/0022-2836(66)90088-x. [DOI] [PubMed] [Google Scholar]
- 9.Scott EE, Gibson QH, Olson JS. Mapping the pathways for O2 entry into and exit from myoglobin. J Biol Chem. 2001;276:5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 10.Olson JS, Phillips GN., Jr. Kinetic Pathways and Barriers for Ligand Binding to Myoglobin. J Biol Chem. 1996;271:17596. [PubMed] [Google Scholar]
- 11.Tilton RF, Jr., Kuntz ID, Jr., Petsko GA. Cavities in proteins: Structure of a metmyoglobin-xenon complex solved to 1.9 Å. Biochemistry. 1984;23:2849–2857. doi: 10.1021/bi00308a002. [DOI] [PubMed] [Google Scholar]
- 12.Cohen J, Arkhipov A, Braun R, Schulten K. Imaging the migration pathways for O2, CO, NO, and Xe inside myoglobin. Biophys J. 2006;91:1844–1857. doi: 10.1529/biophysj.106.085746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elber R, Gibson QH. Toward quantitative simulations of carbon monoxide escape pathways in myoglobin. J Phys Chem B. 2008;112:6147–6154. doi: 10.1021/jp0769779. [DOI] [PubMed] [Google Scholar]
- 14.Elber R, Karplus M. Enhanced sampling in molecular dynamics: use of the time dependent Hartree approximation for simulation of carbon monoxide diffusion through myoglobin. J. Am. Chem. Soc. 1990;112:9161–9175. [Google Scholar]
- 15.Ruscio JZ, Kumar D, Shukla M, Prisant MG, Murali TM, Onufriev AV. Atomic level computational identification of ligand migration pathways between solvent and binding site in myoglobin. Proc Natl Acad Sci U S A. 2008;105:9204–9209. doi: 10.1073/pnas.0710825105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golden SD, Olsen KW. Use of the conjugate peak refinement algorithm for identification of ligand-binding pathways in globins. Methods Enzymol. 2008;437:417–437. doi: 10.1016/S0076-6879(07)37021-3. [DOI] [PubMed] [Google Scholar]
- 17.Brunori M, Vallone B, Cutruzzola F, Travaglini-Allocatelli C, Berendzen J, Chu K, Sweet RM, Schlichting I. The role of cavities in protein dynamics: crystal structure of a photolytic intermediate of a mutant myoglobin. Proc Natl Acad Sci U S A. 2000;97:2058–2063. doi: 10.1073/pnas.040459697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nienhaus K, Ostermann A, Nienhaus GU, Parak FG, Schmidt M. Ligand migration and protein fluctuations in myoglobin mutant L29W. Biochemistry. 2005;44:5095–5105. doi: 10.1021/bi047513t. [DOI] [PubMed] [Google Scholar]
- 19.Ostermann A, Waschipky R, Parak FG, Nienhaus GU. Ligand binding and conformational motions in myoglobin. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 20.Schotte F, Soman J, Olson JS, Wulff M, Anfinrud PA. Picosecond time resolved X-ray crystallography: probing protein function in real time. J Struct Biol. 2004;147:235–246. doi: 10.1016/j.jsb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 21.Srajer V, Ren Z, Teng TY, Schmidt M, Ursby T, Bourgeois D, Pradervand C, Schildkamp W, Wulff M, Moffat K. Protein conformational relaxation and ligand migration in myoglobin: a nanosecond to millisecond molecular movie from time-resolved Laue X-ray diffraction. Biochemistry. 2001;40:13802–13815. doi: 10.1021/bi010715u. [DOI] [PubMed] [Google Scholar]
- 22.Aranda R, IV, Levin EJ, Schotte F, Anfinrud PA, Phillips GN., Jr. Time dependent atomic coordinates for the dissociation of carbon monoxide from myoglobin. Acta Crystallogr D Biol Crystallogr. 2006;62:776–783. doi: 10.1107/S0907444906017318. [DOI] [PubMed] [Google Scholar]
- 23.Bourgeois D, Schotte F, Brunori M, Vallone B. Time-resolved methods in biophysics. 6. Time-resolved Laue crystallography as a tool to investigate photo-activated protein dynamics. Photochem Photobiol Sci. 2007;6:1047–1056. doi: 10.1039/b704249c. [DOI] [PubMed] [Google Scholar]
- 24.Salter MD, Nienhaus K, Nienhaus GU, Dewilde S, Moens L, Pesce A, Nardini M, Bolognesi M, Olson JS. The apolar channel in Cerebratulus lacteus hemoglobin is the route for O2 entry and exit. J Biol Chem. 2008;283:35689–35702. doi: 10.1074/jbc.M805727200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reisberg PI, Olson JS. Rates of isonitrile binding to the isolated alpha and beta subunits of human hemoglobin. J Biol Chem. 1980;255:4151–4158. [PubMed] [Google Scholar]
- 26.Reisberg PI, Olson JS. Kinetic and cooperative mechanisms of ligand binding to hemoglobin. J Biol Chem. 1980;255:4159–4169. [PubMed] [Google Scholar]
- 27.Reisberg PI, Olson JS. Equilibrium binding of alkyl isocyanides to human hemoglobin. J Biol Chem. 1980;255:4144–4130. [PubMed] [Google Scholar]
- 28.Mims MP, Olson JS, Russu IM, Miura S, Cedel TE, Ho C. Proton nuclear magnetic resonance studies of isonitrile-heme protein complexes. J Biol Chem. 1983;258:6125–6134. [PubMed] [Google Scholar]
- 29.Mims MP, Porras AG, Olson JS, Noble RW, Peterson JA. Ligand binding to heme proteins. An evaluation of distal effects. J Biol Chem. 1983;258:14219–14232. [PubMed] [Google Scholar]
- 30.Johnson K. Ph. D. Dissertation. Rice University; Houston,TX: 1993. High resolution X-ray structures of myoglobin and hemoglobin alkyl isocyanide complexes. [Google Scholar]
- 31.Johnson KA, Olson JS, Phillips GN., Jr. Structure of myoglobin-ethyl isocyanide: histidine as a swinging door for ligand entry. J. Mol. Biol. 1989;207:459–463. doi: 10.1016/0022-2836(89)90269-6. [DOI] [PubMed] [Google Scholar]
- 32.Eich RF, Li T, Lemon DD, Doherty DH, Curry SR, Aitken JF, Mathews AJ, Johnson KA, Smith RD, Phillips GN, Jr., Olson JS. Mechanism of NO-induced oxidation of myoglobin and hemoglobin. Biochemistry. 1996;35:6976–6983. doi: 10.1021/bi960442g. [DOI] [PubMed] [Google Scholar]
- 33.Smith RD. Biochemistry & Cell Biology. Rice University; Houston, TX: 1999. p. 203. [Google Scholar]
- 34.Lionetti C, Guanziroli MG, Frigerio F, Ascenzi P, Bolognesi M. X-ray crystal structure of the ferric sperm whale myoglobin: imidazole complex at 2.0 A resolution. J Mol Biol. 1991;217:409–412. doi: 10.1016/0022-2836(91)90744-q. [DOI] [PubMed] [Google Scholar]
- 35.Ringe D, Petsko GA, Kerr DE, Ortiz de Montellano PR. Reaction of myoglobin with phenylhydrazine: a molecular doorstop. Biochemistry. 1984;23:2–4. doi: 10.1021/bi00296a001. [DOI] [PubMed] [Google Scholar]
- 36.Edmundson AB. The Amino-Acid Sequence of Sperm Whale Myoglobin. Nature. 1965;205:883–887. doi: 10.1038/190663a0. [DOI] [PubMed] [Google Scholar]
- 37.Springer BA, Sligar SG. High-level expression of sperm whale myoglobin in Escherichia coli. Proc Natl Acad Sci U S A. 1987;84:8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips GN, Jr., Arduini RM, Springer BA, Sligar SG. Crystal structure of myoglobin from a synthetic gene. Proteins. 1990;7:358–365. doi: 10.1002/prot.340070407. [DOI] [PubMed] [Google Scholar]
- 39.Quillin ML, Arduini RM, Olson JS, Phillips GN., Jr. High-resolution crystal structures of distal histidine mutants of sperm whale myoglobin. J Mol Biol. 1993;234:140–155. doi: 10.1006/jmbi.1993.1569. [DOI] [PubMed] [Google Scholar]
- 40.Quillin ML, Li T, Olson JS, Phillips GN, Jr., Dou Y, Ikeda-Saito M, Regan R, Carlson M, Gibson QH, Li H, et al. Structural and functional effects of apolar mutations of the distal valine in myoglobin. J Mol Biol. 1995;245:416–436. doi: 10.1006/jmbi.1994.0034. [DOI] [PubMed] [Google Scholar]
- 41.Blouin GC, Olson JS. The Stretching Frequencies of Bound Alkyl Isocyanides Indicate Two Distinct Ligand Orientations within the Distal Pocket of Myoglobin. Biochemistry. 2009 doi: 10.1021/bi100172c. [to be co-published with this article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blouin GC, Schweers RL, Olson JS. Alkyl Isocyanides Serve as Transition State Analogs for Ligand Entry and Exit in Myoglobin. Biochemistry. 2009 doi: 10.1021/bi1001745. [to be co-published with this article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takano T. Structure of myoglobin refined at 2-0 A resolution. I. Crystallographic refinement of metmyoglobin from sperm whale. J Mol Biol. 1977;110:537–568. doi: 10.1016/s0022-2836(77)80111-3. [DOI] [PubMed] [Google Scholar]
- 44.Rohlfs RJ, Mathews AJ, Carver TE, Olson JS, Springer BA, Egeberg KD, Sligar SG. The effects of amino acid substitution at position E7 (residue 64) on the kinetics of ligand binding to sperm whale myoglobin. J. Biol. Chem. 1990;265:3168–3176. [PubMed] [Google Scholar]
- 45.Casanova J, Jr., Schuster RE, Werner ND. Synthesis of aliphatic isocyanides. Journal of the Chemical Society. 1963:4280–4281. [Google Scholar]
- 46.Reisberg PI. Ph. D. Dissertation. Rice University; Houston, TX: 1980. [Google Scholar]
- 47.Kabsch W. Evaluation of single-crystal x-ray diffraction data from a position-sensitive detector. J. Appl. Crystallogr. 1988;21:916–924. [Google Scholar]
- 48.Saper MA, Quiocho FA. Leucine, isoleucine, valine-binding protein from Escherichia coli. Structure at 3.0-A resolution and location of the binding site. J Biol Chem. 1983;258:11057–11062. [PubMed] [Google Scholar]
- 49.Hendrickson WA, Konnert JH. Indian Academy of Sciences for the International Union of Crystallography. Vol. 13. Bangalore: 1980. pp. 13.01–13.23. [Google Scholar]
- 50.Finzel BC. Incorporation of fast Fourier transforms to speed restrained least-squares refinement of protein structures. J. Appl. Crystallogr. FIELD Full Journal Title:Journal of Applied Crystallography. 1987;20:53–55. [Google Scholar]
- 51.Blouin GC, Olson JS. The Stretching Frequencies of Bound Alkyl Isocyanides Indicate Two Distinct Ligand Orientations within the Distal Pocket of Myoglobin. Biochemistry. 2010 doi: 10.1021/bi100172c. [to be co-published with this article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maurus R, Bogumil R, Nguyen NT, Mauk AG, Brayer G. Structural and spectroscopic studies of azide complexes of horse heart myoglobin and the His-64-->Thr variant. Biochem J. 1998;332(Pt 1):67–74. doi: 10.1042/bj3320067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stephany RW, De Bie MJA, Drenth W. Carbon-13 NMR and infrared study of isocyanides and their complexes. Organic Magnetic Resonance. 1974;6:45–47. [Google Scholar]
- 54.Hargrove MS, Krzywda S, Wilkinson AJ, Dou Y, Ikeda-Saito M, Olson JS. Stability of myoglobin: a model for the folding of heme proteins. Biochemistry. 1994;33:11767–11775. doi: 10.1021/bi00205a012. [DOI] [PubMed] [Google Scholar]
- 55.Yang F, Phillips GN., Jr. Crystal structures of CO-, deoxy- and met-myoglobins at various pH values. J. Mol. Biol. 1996;256:762–774. doi: 10.1006/jmbi.1996.0123. [DOI] [PubMed] [Google Scholar]
- 56.Blouin GC, Schweers RL, Olson JS. Alkyl Isocyanides Serve as Transition State Analogs for Ligand Entry and Exit in Myoglobin. Biochemistry. 2010 doi: 10.1021/bi1001745. [to be co-published with this article] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.St George RC, Pauling L. The combining power of hemoglobin for alkyl isocyanides, and the nature of the heme-heme interactions in hemoglobin. Science. 1951;114:629–634. doi: 10.1126/science.114.2972.629. [DOI] [PubMed] [Google Scholar]
- 58.Lein A, Pauling L. The Combining Power of Myoglobin for Alkyl Isocyanides and the Structure of the Myoglobin Molecule. Proc Natl Acad Sci U S A. 1956;42:51–54. doi: 10.1073/pnas.42.2.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Olson JS, Gibson QH. The reaction of n-butyl isocyanide with human hemoglobin. I. Determination of the kinetic parameters involved in the last step in ligand binding. J Biol Chem. 1971;246:5241–5253. [PubMed] [Google Scholar]
- 60.Talbot B, Brunori M, Antonini E, Wyman J. Studies on the reaction of isocyanides with haemproteins. I. Equilibria and kinetics of the binding to the isolated chains of human haemoglobin. J Mol Biol. 1971;58:261–276. doi: 10.1016/0022-2836(71)90245-2. [DOI] [PubMed] [Google Scholar]
- 61.Brunori M, Talbot B, Colosimo A, Antonini E, Wyman J. Studies on the reaction of isocyanides with haemproteins. II. Binding to normal and modified human haemoglobins. J Mol Biol. 1972;65:423–434. doi: 10.1016/0022-2836(72)90199-4. [DOI] [PubMed] [Google Scholar]
- 62.Olson JS, Gibson QH. The reaction of n-butyl isocyanide with human hemoglobin. II. The ligand-binding properties of the and chains within deoxyhemoglobin. J Biol Chem. 1972;247:1713–1726. [PubMed] [Google Scholar]
- 63.Stetzkowski F, Cassoly R, Banerjee R. Binding of alkylisocyanides with soybean leghemoglobin. Comparisons with sperm whale myoglobin. J Biol Chem. 1979;254:11351–11356. [PubMed] [Google Scholar]
- 64.Sommer JH, Henry ER, Hofrichter J. Geminate recombination of n-butyl isocyanide to myoglobin. Biochemistry. 1985;24:7380–7388. doi: 10.1021/bi00346a053. [DOI] [PubMed] [Google Scholar]
- 65.Jongeward KA, Magde D, Taube DJ, Marsters JC, Traylor TG, Sharma VS. Picosecond and nanosecond geminate recombination of myoglobin with CO, O2, NO, and isocyanides. J. Amer. Chem. Soc. 1988;110:380–387. [Google Scholar]
- 66.Lee DS, Park SY, Yamane K, Obayashi E, Hori H, Shiro Y. Structural characterization of n-butyl-isocyanide complexes of cytochromes P450nor and P450cam. Biochemistry. 2001;40:2669–2677. doi: 10.1021/bi002225s. [DOI] [PubMed] [Google Scholar]
- 67.Phillips GN., Jr. Comparison of the dynamics of myoglobin in different crystal forms. Biophysical Journal. 1990;57:381–383. doi: 10.1016/S0006-3495(90)82540-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kondrashov DA, Zhang W, Aranda R. t., Stec B, Phillips GN., Jr Sampling of the native conformational ensemble of myoglobin via structures in different crystalline environments. Proteins. 2008;70:353–362. doi: 10.1002/prot.21499. [DOI] [PubMed] [Google Scholar]
- 69.Binkowski TA, Naghibzadeh S, Liang J. CASTp: Computed Atlas of Surface Topography of proteins. Nucleic Acids Res. 2003;31:3352–3355. doi: 10.1093/nar/gkg512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Higa RH, Togawa RC, Montagner AJ, Palandrani JC, Okimoto IK, Kuser PR, Yamagishi ME, Mancini AL, Neshich G. STING Millennium Suite: integrated software for extensive analyses of 3d structures of proteins and their complexes. BMC Bioinformatics. 2004;5:107. doi: 10.1186/1471-2105-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neshich G, Togawa RC, Mancini AL, Kuser PR, Yamagishi ME, Pappas G, Jr., Torres WV, Fonseca e Campos T, Ferreira LL, Luna FM, Oliveira AG, Miura RT, Inoue MK, Horita LG, de Souza DF, Dominiquini F, Alvaro A, Lima CS, Ogawa FO, Gomes GB, Palandrani JF, dos Santos GF, de Freitas EM, Mattiuz AR, Costa IC, de Almeida CL, Souza S, Baudet C, Higa RH. STING Millennium: A web-based suite of programs for comprehensive and simultaneous analysis of protein structure and sequence. Nucleic Acids Res. 2003;31:3386–3392. doi: 10.1093/nar/gkg578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Egeberg KD, Springer BA, Sligar SG, Carver TE, Rohlfs RJ, Olson JS. The role of Val68(E11) in ligand binding to sperm whale myoglobin. Site-directed mutagenesis of a synthetic gene. J Biol Chem. 1990;265:11788–11795. [PubMed] [Google Scholar]
- 73.Carver TE, Rohlfs RJ, Olson JS, Gibson QH, Blackmore RS, Springer BA, Sligar SG. Analysis of the kinetic barriers for ligand binding to sperm whale myoglobin using site-directed mutagenesis and laser photolysis techniques. J. Biol. Chem. 1990;265:20007–20020. [PubMed] [Google Scholar]
- 74.Lai HH, Li T, Lyons DS, Phillips GN, Jr., Olson JS, Gibson QH. Phe-46(CD4) orients the distal histidine for hydrogen bonding to bound ligands in sperm whale myoglobin. Proteins. 1995;22:322–339. doi: 10.1002/prot.340220404. [DOI] [PubMed] [Google Scholar]
- 75.Morikis D, Champion PM, Springer BA, Sligar SG. Resonance Raman investigations of site-directed mutants of myoglobin: Effects of distal histidine replacement. Biochemistry. 1989;28:4791–4800. doi: 10.1021/bi00437a041. [DOI] [PubMed] [Google Scholar]
- 76.Muller JD, McMahon BH, Chien EY, Sligar SG, Nienhaus GU. Connection between the taxonomic substates and protonation of histidines 64 and 97 in carbonmonoxy myoglobin. Biophys J. 1999;77:1036–1051. doi: 10.1016/s0006-3495(99)76954-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 78.Carver TE, Olson JS, Smerdon SJ, Krzywda S, Wilkinson AJ, Gibson QH, Blackmore RS, Ropp JD, Sligar SG. Contributions of residue 45(CD3) and heme-6-propionate to the biomolecular and geminate recombination reactions of myoglobin. Biochemistry. 1991;30:4697–4705. doi: 10.1021/bi00233a009. [DOI] [PubMed] [Google Scholar]
- 79.Li T, Quillin ML, Phillips GN, Jr., Olson JS. Structural determinants of the stretching frequency of CO bound to myoglobin. Biochemistry. 1994;33:1433–1446. doi: 10.1021/bi00172a021. [DOI] [PubMed] [Google Scholar]
- 80.Oldfield TJ, Smerdon SJ, Dauter Z, Petratos K, Wilson KS, Wilkinson AJ. High-resolution X-ray structures of pig metmyoglobin and two CD3 mutants: Mb(Lys45----Arg) and Mb(Lys45----Ser). Biochemistry. 1992;31:8732–8739. doi: 10.1021/bi00152a008. [DOI] [PubMed] [Google Scholar]
- 81.Scott EE, Gibson QH. Ligand migration in sperm whale myoglobin. Biochemistry. 1997;36:11909–11917. doi: 10.1021/bi970719s. [DOI] [PubMed] [Google Scholar]