Abstract
Introduction
Interleukin (IL)-33 is a proinflammatory cytokine contributing to the pathogenesis of rheumatoid arthritis (RA). The gene encoding IL-33 may serve as a genetic factor and be associated with the risk of RA. To investigate the potential association between IL33 and RA, we performed a case–control study based on Chinese Han population.
Methods
A three-stage case–control study was performed. Two tag single-nucleotide polymorphisms (SNPs) (rs7044343 and rs10975514), mapping to the IL33 gene, were first genotyped in the discovery population. We further genotyped rs7044343 and rs10975514 in the validation and replication population. The associations between the two tag SNPs and phenotypic subgroups of RA and levels of serum IL-33 were assessed with a logistic regression model.
Results
In the discovery population, the CC genotype of rs7044343 was associated with RA patients (odds ratio (OR) = 0.777, 95% confidence interval (CI), 0.611 to 0.988; P = 0.040). After anti-citrullinated peptide antibody (ACPA) stratification, the CC genotype of rs7044343 was also shown to be a protective genotype in RA without ACPA (OR = 0.610; 95% CI, 0.379 to 0.982; P = 0.042). In the validation population and replication population, the association between rs7044343 and RA, especially ACPA-negative RA, was still significant. A meta-analysis of discovery, validation, and replication panels confirmed the association between CC genotype of rs7044343 and RA (Pcombined = 0.0004; ORcombined = 0.77; 95% CI, 0.67 to 0.89). No evidence was found for heterogeneity between three sample sets (Phet = 0.99; I2 = 0%). Similar results were also obtained in ACPA-negative RA (Pcombined = 0.0002; ORcombined = 0.57; 95% CI, 0.43 to 0.77). No association was detected between rs10975514 polymorphism and RA susceptibility in the discovery and validation population. The serum levels of IL-33 were significantly lower in the patients with the rs7044343 CC genotype.
Conclusion
The CC genotype of rs7044343 in IL33 is associated with RA patients and downregulates IL-33 expression in RA.
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease characterized by chronic inflammation of synovial joints without precisely known pathogenesis. Genetic factors contribute to the development of RA; it was estimated that the total heritability of RA is approximately 66% [1]. Current genome-wide association studies (GWASs) have identified 46 genetic loci, such as HLA, PTPN22, and CTLA4 associated with RA [2]; however, the identified risk loci of RA have modest effect sizes (odds ratios in the range of 0.78 to 2.78) and can explain only about 16% of the RA heritability [3]. Therefore, more population-based studies are needed to find the genetic basis of RA.
Interleukin (IL)-33 was recently identified as a member of the IL-1 family and a ligand for the IL-1 family receptor ST2 [4]. In patients with RA, immunohistochemistry and in situ hybridization have identified IL-33 residing in the synovial cells of inflamed joints [5]. The level of IL-33 was elevated in both serum and synovial fluid and associated with autoantibody production, bone erosion, and interstitial lung disease [6,7]. In a murine model, IL-33 could exacerbate collagen-induced arthritis (CIA) and elevate the production of proinflammatory cytokines and anticollagen antibodies [8]. The ST2 antibody that blocks IL-33 signaling could attenuate the severity of CIA [9]. These studies suggest that IL-33 plays an important role in the pathogenesis of RA and indicate that the IL33 genetic variants associated with RA merit further investigation.
Methods
Patients and controls
In total, 1,952 patients with established RA and 1,755 unrelated healthy controls of Northern Han Chinese origin were included in this study. The discovery population of 700 cases and 598 controls was assembled from Northern Han Chinese; the validation population of 586 cases and 456 controls was assembled from southern China; and the replication population of 666 cases and 701 controls was assembled from both northern and southern China. All patients fulfilled the revised criteria of the American College of Rheumatology for RA [10]. The healthy control subjects were defined as healthy individuals without inflammatory arthritis by medical history, general examinations, and laboratory examinations and were individually matched to RA cases on the basis of sex, ethnicity, and local residential region. The study was approved by the ethics committee of Peking University People’s Hospital (FWA00001384), and informed consents were obtained from all participants.
After oral and written informed consents had been obtained, genomic DNA and serum samples were extracted from peripheral blood of patients and healthy controls. The following clinical data were recorded for ascertainment of the clinical phenotype of RA patients: age at RA onset, sex, disease duration, and rheumatoid factor (RF) and ACPA status.
Genotyping methods
Haplotypes from the HapMap database (Han Chinese in Beijing, CHB) [11] were used to select tag SNPs (r2 = 0.8) residing in IL33 (dbSNP data (build129)). In total, 16 SNPs were identified in CHB. All SNPs were in one block and in high linkage disequilibrium (LD) (D’ > 0.92; Figure 1). Tag SNPs were selected according to the following principles: (a) linkage disequilibrium (LD) between SNPs according to Haploview (v. 4.2) based on HapMap CHB data sets with the thresholds of r2 > 0.8 and D’ > 0.7 to reduce the redundancy; (b) potential functional sites predicted by bioinformatics (Promoter and Genevar); and (c) a minor allele frequency (MAF) threshold of >0.05. Data were excluded if the allele call rate was <95%.
Figure 1.
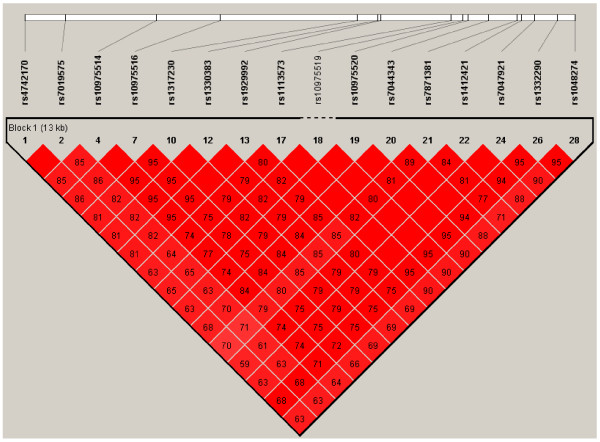
Information of IL33 SNPs from HapMap CHB database. All IL33 SNPs are in one block and in high linkage disequilibrium (LD) (D’ > 0.92).The numbers present the r2 between the SNPs. Two SNPs (rs7044343 and rs10975514) were selected in this study. SNP, single-nucleotide polymorphism; CHB, Han Chinese in Beijing.
Genotyping of rs7044343 and 10975514 was performed by using predesigned Taqman SNP Genotyping Assays (C_31083545_10; Applied Biosystems, Foster City, CA, USA). Allelic discrimination was performed in an ABI 7300 Real-Time PCR system (Applied Biosystems). The successful genotyping rate was 99.0%.
The genotypes obtained were subsequently validated and confirmed by sequencing the PCR products with an ABI 3700 automated sequencer (Applied Biosystems). In brief, 130 individuals of cases and controls were sequenced for confirmation of the rs7044343 and rs10975514. The results of DNA sequence analysis are shown in Additional file 1: Figure S1. The confirmation rate was 100%.
Meta-analysis was performed by using Review Manager 5 software and carried out with the Mantel-Haenszel method. A significant I2 statistic (I2 > 30%; P < 0.05) indicated heterogeneity for OR across studies. The fixed-effects model was applied in current meta-analyses.
Two genetic markers selected for the study, rs10975514 and rs7044343, were genotyped in the discovery population. Then we further genotyped rs7044343 and 10975514 in the validation population and replication population.
Assay for serum levels of IL-33
Serum IL-33 was measured with enzyme-linked immunosorbent assay (ELISA), according to the manufacturer’s instructions (R&D Systems, Minneapolis, MN, USA).
Statistical analyses
Allele frequencies were calculated and tested for agreement with Hardy-Weinberg equilibrium (HWE) by using a χ2 goodness-of-fit test. Differences in allele frequencies and genotype distribution of the polymorphism between RA cases and control subjects were compared by using a 2 × 2 contingency χ2 test with 1 degree of freedom, and calculated odds ratios (ORs) with 95% confidence intervals (CIs), adjusting for age and sex. Associations between genetic variations and susceptibility or phenotype features of RA were analyzed by using the Mann–Whitney U test. All statistical analyses were conducted by using SPSS 16.0 (SPSS Inc., Chicago, IL, USA). A P value of <0.05 was considered statistically significant in the expression of IL-33.
Results
Population characteristics
The demographic distribution of the population under study is shown in Table 1. The mean age in the case group was 53.6 ± 13.3 years, and 81.3% were women. The mean age at disease onset was 46.4 ± 15.6 years. The positive rates of rheumatoid factor (RF) and ACPA were 81.2% and 72.5%, respectively. The mean age of healthy controls was 40.4 ± 10.3 years, and 80.6% were women (Table 1).
Table 1.
Clinical characteristics of the study cohorts
| Discovery population | Validation population | Replication population | |
|---|---|---|---|
| Number of cases |
700 |
586 |
666 |
| Ethnicity |
Northern Han Chinese |
Southern Han Chinese |
Northern and Southern |
| Female, % |
79.9 |
81.2 |
82.8 |
| Age, mean ± SD years |
54.0 ± 13.1 |
52.4 ± 13.9 |
54.3 ± 12.8 |
| Age at RA onset, mean ± SD years |
46.4 ± 16.0 |
46.3 ± 15.0 |
46.6 ± 15.4 |
| Disease duration, mean ± SD years |
8.1 ± 8.1 |
7.8 ± 7.3 |
9.0 ± 10.3 |
| ACPA status, % positive |
72.6 |
75.3 |
68.9 |
| RF status, % positive | 81.6 | 78.3 | 84.0 |
ACPA, anti-citrullinated proteins antibodies; RF, rheumatoid factor.
Association analysis of rs7044343 with RA
In HapMap CHB (data resulted from sequencing of 42 individuals), the allele distributions for rs7044343 T and C were 0.526 and 0.474 [11]. In our study, we found the allele frequencies for rs7044343 T and C were 0.452 and 0.548 in the control subjects (n = 1,755). Based on the assumption of a population prevalence of disease of 0.2%, α = 0.05, minor allele frequencies of 0.452, and linkage disequilibrium of r2 = 0.8, the study had 99.7% power to detect an additive association with an additive OR of 1.26.
Allelic and genotype frequencies of rs7044343 SNP in RA patients and controls are shown in Tables 2, 3, and 4. In the discovery population, the CC genotype of rs7044343 was associated with (OR = 0.777; 95% CI, 0.611 to 0.988; P = 0.040) RA patients. When we divided the RA patients into different subgroups by ACPA and RF status, the results showed that the CC genotype of rs7044343 was associated with RA patients without ACPA (OR = 0.610; 95% CI, 0.379 to 0.982; P = 0.042). No association was found between rs7044343 and RA after gender stratification.
Table 2.
Association analysis of rs7044343 with RA in the discovery population
| Characteristic ( n ) |
Allele frequency (%) |
Genotype frequency (%) |
T versus C |
CC versus TT + TC |
CC + TC versus TT |
|||
|---|---|---|---|---|---|---|---|---|
| T | C | TT | TC | CC |
OR (95% CI) |
OR (95% CI) |
OR (95% CI) |
|
| P value | P value | P value | ||||||
| Control (598) |
545 (45.6) |
651 (54.4) |
137 (22.9) |
271 (45.3) |
190 (31.8) |
|
|
|
| RA (700) |
641 (45.8) |
759 (54.2) |
127 (18.1) |
387 (55.3) |
186 (26.6) |
1.009 |
0.777 |
1.354 |
| (0.864-1.178) |
(0.611-0.988) |
(1.033-1.774) |
||||||
| 0.912 |
0.040* |
0.028* |
||||||
| RF + (418) |
377 (45.1) |
459 (54.9) |
78 (18.7) |
221 (52.9) |
119 (28.5) |
0.981 |
0.855 |
1.308 |
| (0.822-1.172) |
(0.650-1.123) |
(0.958-1.785) |
||||||
| 0.833 |
0.260 |
0.091 |
||||||
| RF- (94) |
86 (45.7) |
102 (54.3) |
17 (18.1) |
52 (55.3) |
25 (26.6) |
1.007 |
0.778 |
1.359 |
| (0.740-1.371) |
(0.477-1.269) |
(0.777-2.376) |
||||||
| 0.964 |
0.314 |
0.282 |
||||||
| ACP (300) |
281 (46.8) |
319 (53.2) |
62 (20.7) |
157 (52.3) |
81 (27.0) |
1.052 |
0.794 |
1.152 |
| (0.864-1.281) |
(0.584-1.080) |
(0.821-1.615) |
||||||
| 0.612 |
0.142 |
0.413 |
||||||
| ACPA - (113) |
108 (47.8) |
118 (52.2) |
20 (17.7) |
68 (60.2) |
25 (22.1) |
1.093 |
0.610 |
1.395 |
| (0.822-1.453) |
(0.379-0.982) |
(0.830-2.345) |
||||||
| 0.539 |
0.042* |
0.209 |
||||||
| Female control (465) |
432 (46.5) |
498 (53.5) |
106 (22.8) |
220 (47.3) |
139 (29.9) |
|
|
|
| Female RA (482) |
426 (44.2) |
538 (55.8) |
85 (17.6) |
256 (53.1) |
141 (29.3) |
0.913 |
0.970 |
1.396 |
| (0.762-1.094) |
(0.734-1.282) |
(1.015-1.920) |
||||||
| 0.323 |
0.829 |
0.040 |
||||||
| Female RF+ (333) |
300 (45.0) |
366 (55.0) |
62 (18.6) |
176 (52.9) |
95 (28.5) |
0.945 |
0.936 |
1.306 |
| (0.774-1.154) |
(0.687-1.276) |
(0.920-1.855) |
||||||
| 0.578 |
0.676 |
0.135 |
||||||
| Female RF- (75) |
64 (42.7) |
86 (57.3) |
12 (16.0) |
40 (53.3) |
23 (30.7) |
0.858 |
1.037 |
1.569 |
| (0.606-1.215) |
(0.611-1.761) |
(0.816-3.018) |
||||||
| 0.388 |
0.892 |
0.177 |
||||||
| Female ACPA+ (243) |
225 (46.3) |
261 (53.7) |
50 (20.6) |
125 (51.4) |
68 (28.0) |
0.994 |
0.911 |
1.154 |
| (0.798-1.238) |
(0.647-1.285) |
(0.790-1.685) |
||||||
| |
|
|
0.956 |
0.596 |
0.459 |
|||
| Female ACPA- (84) |
77 (45.8) |
91 (54.2) |
15 (17.9) |
47 (56.0) |
22 (26.2) |
0.975 |
0.832 |
1.375 |
| (0.701-1.356) |
(0.492-1.407) |
(0.756-2.502) |
||||||
| 0.882 |
0.493 |
0.297 |
||||||
| Male CON (105) |
99 (47.1) |
111 (52.9) |
26 (24.8) |
47 (44.8) |
32 (30.5) |
|
|
|
| Male RA (121) |
116 (47.9) |
126 (52.1) |
24 (19.8) |
68 (56.2) |
29 (24.0) |
1.032 |
0.719 |
1.330 |
| (0.713-1.495) |
(0.399-1.296) |
(0.709-2.496) |
||||||
| 0.867 |
0.272 |
0.374 |
||||||
| Male RF+ (84) |
75 (44.6) |
93 (55.4) |
15 (17.9) |
45 (53.6) |
24 (28.6) |
0.904 |
0.913 |
1.514 |
| (0.602-1.359) |
(0.486-1.713) |
(0.742-3.088) |
||||||
| 0.628 |
0.776 |
0.254 |
||||||
| Male RF- (19) |
22 (57.9) |
16 (42.1) |
5 (26.3) |
12 (63.2) |
2 (10.5) |
1.542 |
0.268 |
0.922 |
| (0.767-3.100) |
(0.059-1.231) |
(0.303-2.805) |
||||||
| 0.222 |
0.090 |
0.886 |
||||||
| Male ACPA+ (57) |
56 (49.1) |
58 (50.9) |
12 (21.1) |
32 (56.1) |
13 (22.8) |
1.083 |
0.674 |
1.234 |
| (0.686-1.709) |
(0.320-1.420) |
(0.568-2.681) |
||||||
| 0.733 |
0.300 |
0.595 |
||||||
| Male ACPA- (27) | 29 (53.7) | 25 (46.3) | 5 (18.5) | 19 (70.4) | 3 (11.1) | 1.301 |
0.285 |
1.448 |
| (0.714-2.369) |
(0.080-1.016) |
(0.498-4.211) |
||||||
| 0.390 | 0.053 | 0.497 | ||||||
*P < 0.05 versus controls; ACPAs, anti-citrullinated proteins antibodies; CI, confidence interval.
CON, healthy control; OR, odds ratio; RA, rheumatoid arthritis; RF, rheumatoid factor.
Table 3.
Association analysis of rs7044343 with RA in the validation population
| Characteristic ( n ) |
Allele frequency (%) |
Genotype frequency (%) |
T vs. C |
CC vs. TT + TC |
CC + TC vs. TT |
|||
|---|---|---|---|---|---|---|---|---|
| T | C | TT | TC | CC |
OR (95% CI) |
OR (95% CI) |
OR (95% CI) |
|
| P value | P value | P value | ||||||
| Control (456) |
409 (44.8) |
503 (55.2) |
100 (21.9) |
209 (45.8) |
147 (32.2) |
|
|
|
| RA (586) |
548 (46.8) |
624 (53.2) |
118 (20.1) |
312 (53.2) |
156 (26.6) |
1.080 |
0.763 |
1.114 |
| (0.908-1.285) |
(0.583-0.998) |
(0.825-1.504) |
||||||
| 0.385 |
0.048* |
0.480 |
||||||
| RF + (354) |
322 (45.5) |
386 (54.5) |
67 (18.9) |
188 (53.1) |
99 (28.0) |
1.026 |
0.816 |
1.203 |
| (0.842-1.250) |
(0.602-1.106) |
(0.851-1.701) |
||||||
| 0.799 |
0.190 |
0.295 |
||||||
| RF- (98) |
100 (51.0) |
96 (49.0) |
25 (25.5) |
50 (51.0) |
23 (23.5) |
1.281 |
0.645 |
0.820 |
| (0.941-1.745) |
(0.388-1.070) |
(0.495-1.360) |
||||||
| 0.116 |
0.089 |
0.442 |
||||||
| ACPA + (281) |
257 (45.7) |
305 (54.3) |
51 (18.1) |
155 (55.2) |
75 (26.7) |
1.036 |
0.765 |
1.267 |
| (0.839-1.280) |
(0.551-1.064) |
(0.870-1.845) |
||||||
| 0.741 |
0.111 |
0.217 |
||||||
| ACPA- (92) |
94 (51.1) |
90 (48.9) |
21 (22.8) |
52 (56.5) |
19 (20.7) |
1.284 |
0.547 |
0.950 |
| (0.935-1.764) |
(0.318-0.940) |
(0.556-1.622) |
||||||
| 0.121 |
0.029* |
0.850 |
||||||
| Female control (363) |
319 (43.9) |
407 (56.1) |
75 (20.7) |
169 (46.6) |
119 (32.8) |
|
|
|
| Female RA (441) |
413 (46.8) |
469 (53.2) |
87 (19.7) |
239 (54.2) |
115 (26.1) |
1.124 |
0.723 |
1.060 |
| (0.922-1.369) |
(0.533-0.982) |
(0.750-1.497) |
||||||
| 0.247 |
0.038 |
0.743 |
||||||
| Female RF+ (291) |
264 (45.4) |
318 (54.6) |
53 (18.2) |
158 (54.3) |
80 (27.5) |
1.059 |
0.777 |
1.169 |
| (0.851-1.319) |
(0.554-1.090) |
(0.791-2.730) |
||||||
| 0.607 |
0.144 |
0.433 |
||||||
| Female RF- (75) |
77 (51.3) |
73 (48.7) |
20 (26.7) |
37 (49.3) |
18 (24.0) |
1.346 |
0.648 |
0.716 |
| (0.946-1.914) |
(0.365-1.149) |
(0.404-1.268) |
||||||
| 0.098 |
0.137 |
0.252 |
||||||
| Female ACPA+ (234) |
217 (46.4) |
251 (53.6) |
43 (18.4) |
131 (56.0) |
60 (25.6) |
1.103 |
0.707 |
1.157 |
| (0.873-1.393) |
(0.490-1.020) |
(0.762-1.755) |
||||||
| 0.410 |
0.064 |
0.494 |
||||||
| Female ACPA- (73) |
74 (50.7) |
72 (49.3) |
18 (24.7) |
38 (52.1) |
17 (23.3) |
1.311 |
0.622 |
0.796 |
| (0.919-1.872) |
(0.347-1.118) |
(0.441-1.435) |
||||||
| 0.135 |
0.112 |
0.448 |
||||||
| Male CON (89) |
86 (48.3) |
92 (51.7) |
24 (27.0) |
38 (42.7) |
27 (30.3) |
|
|
|
| Male RA (102) |
98 (48.0) |
106 (52.0) |
23 (22.5) |
52 (51.0) |
27 (26.5) |
0.956 |
0.827 |
1.268 |
| (0.615-1.486) |
(0.440-1.553) |
(0.656-2. 452) |
||||||
| 0.840 |
0.554 |
0.480 |
||||||
| Male RF+ (63) |
58 (46.0) |
68 (54.0) |
14 (22.2) |
30 (47.6) |
19 (30.2) |
0.827 |
0.992 |
1.292 |
| (0.527-1.299) |
(0.491-2.002) |
(0.607-2.753) |
||||||
| 0.410 |
0.981 |
0.506 |
||||||
| Male RF- (22) |
23 (52.3) |
21 (47.7) |
5 (22.7) |
13 (59.1) |
4 (18.2) |
1.172 |
0.510 |
1.255 |
| (0.605-2.268) |
(0.158-1.651) |
(0.417-3.777) |
||||||
| 0.638 |
0.261 |
0.686 |
||||||
| Male ACPA+ (47) |
40 (42.6) |
54 (57.4) |
8 (17.0) |
24 (51.1) |
15 (31.9) |
0.792 |
1.076 |
1.800 |
| (0.479-1.311) |
(0.502-2.306) |
(0.737-4.397) |
||||||
| 0.365 |
0.850 |
0.197 |
||||||
| Male ACPA- (18) | 19 (52.8) | 17 (47.2) | 3 (16.7) | 13 (72.2) | 2 (11.1) | 0.376 |
0.287 |
1.846 |
| (0.207-0.686) |
(0.062-1.336) |
(0.491-6.946) |
||||||
| 0.001 | 0.112 | 0.364 | ||||||
*P < 0.05 versus controls; ACPA anti-citrullinated proteins antibodies; CI confidence interval CON healthy controls; OR Odds ratio RA rheumatoid arthritis; RF rheumatoid factor.
Table 4.
Association analysis of rs7044343 with RA in the replication population
| Characteristic ( n ) |
Allele frequency (%) |
Genotype frequency (%) |
T vs. C |
CC vs. TT + TC |
CC + TC vs. TT |
|||
|---|---|---|---|---|---|---|---|---|
| T | C | TT | TC | CC |
OR (95% CI) |
OR (95% CI) |
OR (95% CI) |
|
| P -value | P .-value | P -value | ||||||
| Control (701) |
631 (45.0) |
771 (55.0) |
154 (22.0) |
323 (46.1) |
224 (32.0) |
|
|
|
| RA (666) |
632 (47.4) |
700 (52.6) |
144 (21.6) |
344 (51.7) |
178 (26.7) |
1.103 |
0.777 |
1.021 |
| (0.949-1.282) |
(0.615-0.981) |
(0.789-1.320) |
||||||
| 0.201 |
0.034* |
0.877 |
||||||
| RF + (325) |
300 (46.2) |
350 (53.8) |
73 (22.5) |
154 (47.4) |
98 (30.2) |
1.047 |
0.919 |
0.972 |
| (0.869-1.262) |
(0.691-1.223) |
(0.709-1.333) |
||||||
| 0.627 |
0.563 |
0.860 |
||||||
| RF-(62) |
54 (43.5) |
70 (56.5) |
10 (16.1) |
34 (54.8) |
18 (29.0) |
0.943 |
0.871 |
1.464 |
| (0.651-1.365) |
(0.492-1.542) |
(0.727-2.948) |
||||||
| 0.754 |
0.636 |
0.286 |
||||||
| ACPA + (224) |
192 (42.9) |
256 (57.1) |
39 (17.4) |
114 (50.9) |
71 (31.7) |
0.916 |
0.988 |
1.335 |
| (0.739-1.136) |
(0.715-1.365) |
(0.905-1.970) |
||||||
| 0.425 |
0.943 |
0.2145 |
||||||
| ACPA-(101) |
103 (51.0) |
99 (49.0) |
23 (22.8) |
57 (56.4) |
21 (20.8) |
1.271 |
0.559 |
0.955 |
| (0.946-1.708) |
(0.337-0.927) |
(0.580-1.571) |
||||||
| 0.111 |
0.024* |
0.856 |
||||||
| Female Control (519) |
474 (45.7) |
564 (54.3) |
114 (22.0) |
246 (47.4) |
159 (30.6) |
|
|
|
| Female RA (458) |
410 (44.8) |
506 (55.2) |
92 (20.1) |
226 (49.3) |
140 (30.6) |
1.040 |
0.997 |
1.120 |
| (0.869-1.246) |
(0.759-1.309) |
(0.822-1.525) |
||||||
| 0.668 |
0.982 |
0.473 |
||||||
| Female RF+ (256) |
237 (46.3) |
275 (53.7) |
56 (21.9) |
125 (48.8) |
75 (29.3) |
1.025 |
0.938 |
1.005 |
| (0.829-1.268) |
(0.676-1.302) |
(0.700-1.444) |
||||||
| 0.817 |
0.703 |
0.977 |
||||||
| Female RF- (53) |
46 (43.4) |
60 (56.6) |
8 (15.1) |
30 (56.6) |
15 (28.3) |
0.912 |
0.894 |
1.583 |
| (0.610-1.365) |
(0.478-1.672) |
(0.726-3.455) |
||||||
| 0.655 |
0.725 |
0.248 |
||||||
| Female ACPA+ (158) |
133 (42.1) |
183 (57.9) |
27 (17.1) |
79 (50.0) |
52 (32.9) |
0.865 |
1.111 |
1.366 |
| (0.670-1.115) |
(0.759-1.625) |
(0.859-2.171) |
||||||
| 0.263 |
0.589 |
0.187 |
||||||
| Female ACPA- (76) |
72 (47.4) |
80 (52.6) |
14 (18.4) |
44 (57.9) |
18 (23.7) |
1.071 |
0.703 |
1.247 |
| (0.761-1.506) |
(0.401-1.231) |
(0.673-2.308) |
||||||
| 0.694 |
0.217 |
0.483 |
||||||
| Male CON (130) |
125 (48.1) |
135 (51.9) |
30 (23.1) |
65 (50.0) |
35 (26.9) |
|
|
|
| Male RA (95) |
75 (39.5) |
115 (60.5) |
16 (16.8) |
43 (45.3) |
36 (37.9) |
0.704 |
1.656 |
1.481 |
| (0.482-1.029) |
(0.939-2.921) |
(0.754-2. 908) |
||||||
| 0.070 |
0.081 |
0.254 |
||||||
| Male RF+ (54) |
42 (38.9) |
66 (61.1) |
10 (18.5) |
22 (40.7) |
22 (40.7) |
0.687 |
1.866 |
1.320 |
| (0.435-1.085) |
(0.958-3.636) |
(0.594-2.934) |
||||||
| 0.107 |
0.067 |
0.496 |
||||||
| Male RF- (9) |
8 (44.4) |
10 (55.6) |
2 (22.2) |
4 (44.4) |
3 (33.3) |
0.864 |
1.357 |
1.050 |
| (0.330-2.259) |
(0.322-5.723) |
(0.207-5.325) |
||||||
| 0.765 |
0.677 |
0.953 |
||||||
| Male ACPA+ (44) |
29 (33.0) |
59 (67.0) |
4 (9.1) |
21 (47.7) |
19 (43.2) |
0.531 |
2.063 |
3.000 |
| (0.320-0.881) |
(1.013-4.202) |
(0. 993–9.065) |
||||||
| 0.014 |
0.046 |
0.052 |
||||||
| Male ACPA- (11) | 13 (59.1) | 9 (40.9) | 5 (45.5) | 3 (27.3) | 3 (27.3) | 1.560 |
1.018 |
0.360 |
| (0.644-3.776) |
(0.255-4.055) |
(0.103-1.263) |
||||||
| 0.321 | 0.980 | 0.111 | ||||||
*P < 0.05 vs controls; RA rheumatoid arthritis; CON healthy controls; RF rheumatoid factor; ACPA anti-citrullinated proteins antibodies; OR Odds ratio; CI confidence interval.
In the validation population and replication population, the associations between rs7044343 and RA, especially ACPA-negative RA, were still significant (OR = 0.763; 95% CI, 0.583 to 0.998; P = 0.048; OR = 0.547, 95% CI 0.318-0.940, P = 0.029; OR = 0.777, 95% CI 0.615 to 0.981, P = 0.034; OR = 0.559, 95% CI, 0.337 to 0.927, P = 0.024). A meta-analysis of discovery, validation, and replication panels confirmed the association between CC genotype of rs7044343 and RA (Pcombined = 0.0004; ORcombined = 0.77; 95% CI, 0.67 to -0.89). No evidence was noted for heterogeneity between the three sample sets (Phet = 0.99; I2 = 0). Similar results were also obtained in ACPA-negative RA (Pcombined = 0.0002; ORcombined = 0.57; 95% CI, 0.43 to 0.77) (Figure 2). No association was detected between rs10975514 polymorphism and RA susceptibility in the discovery and validation population (see Additional file 2: Table S1, Table S2, and Table S3).
Figure 2.

The allele model meta-analysis for rs7044343. A meta-analysis of discovery, validation, and replication panels confirmed the association between the CC genotype of rs7044343 and RA (Pcombined = 0.0004; ORcombined = 0.77; 95% CI, 0.67 to 0.89). No evidence was found for heterogeneity between three sample sets (Phet = 0.99; I2 = 0) (a). Similar results were also obtained in ACPA-negative RA (Pcombined = 0.0002; ORcombined = 0.57; 95% CI, 0.43 to 0.77) (b).
No association was found between rs7044343 and erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), or 28-joint-count disease activity score (DAS28) in RA (data not shown).
Patients with CC genotype of rs7044343 display lower IL-33 serum levels
To evaluate whether IL33 polymorphism is associated with dysregulation of IL-33 at the protein level, we conducted ELISA assays of 141 sera of DMARDs-naïve RA patients. The results showed that serum levels of IL-33 were significantly lower in RA patients with the rs7044343 CC genotype (n = 28) compared with the patients with rs7044343 TT and TC genotypes (Figure 3b), indicating that the CC genotype of rs7044343 downregulates IL-33 expression in RA. When we divided the patients into ACPA-positive and -negative groups, the same result was obtained in the ACPA-positive group (Figure 3c). In the ACPA-negative group, patients with the CC genotype also showed lower serum IL-33 level than did patients with TC and TT genotypes (21.8 ± 40.5 versus 91.5 ± 209.5 versus 125.1 ± 331.0 ng/ml); however, no statistical difference was found. Moreover, IL-33 expression was significantly higher in the dominant model (TT + TC) (P = 0.008; Figure 3d).
Figure 3.
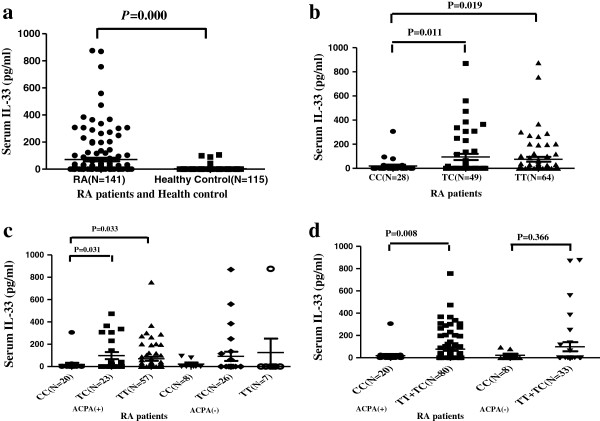
Comparison of serum IL-33 levels among IL33 genotypes. The IL-33 levels were tested in 141 DMARDs-naïve RA patients and 115 healthy controls (HCs). Serum IL-33 concentrations were significantly higher in patients with RA than in healthy controls (P = 0.000 (a). Patients with the CC genotype showed significantly lower serum IL-33 levels than did patients with TC and TT genotypes (b). When we divided the patients into ACPA-positive and ACPA-negative groups, the same result was obtained in the ACPA-positive group (c). In the ACPA-negative group (n = 41), patients with the CC genotype also showed lower serum IL-33 levels than did patients with TC and TT genotypes (21.8 ± 40.5 versus 91.5 ± 209.5 versus 125.1 ± 331.0 ng/ml); however, no statistical difference was found. IL-33 expression was significantly higher in the dominant model (TT + TC) (P = 0.008 (d). TT, TC, and CC represent genotypes of IL33 rs7044343.
Discussion
In the present study, we demonstrated that the CC genotype of rs7044343 in IL33 is associated with RA patients, especially in the ACPA-negative subset. Results also showed that the CC genotype of rs7044343 was associated with lower serum IL-33 levels. Genetic factors contribute to the development of ACPA-negative RA as much as ACPA-positive RA [12]. Only a few susceptibility alleles of HLA and non-HLA genes have been associated with ACPA-negative RA [13]. From the present data, gene polymorphism of IL33 is a new marker related to susceptibility to ACPA-negative RA.
The relation between the genotype of IL33 and the production of autoantibodies is worth studying. Based on the present study, the protective role provided by the CC genotype of rs7044343 in RA patients may be due to the impaired expression of IL-33 protein. The mechanisms by which IL-33 is associated with RA and its specific antibodies production are still obscure. It was deduced that IL-33 drove production of Th2-associated cytokines, including IL-5 and IL-13, which could promote B-cell function, such as autoantibody production [4]. IL-33 may also contribute to the antibody production by inducing mast cell activation [6].
Genetic variants in IL33 were reported to be associated with decreased risk of Alzheimer disease (AD) [14,15]. Chapuis et al.[14] showed that three intronic rs1157505, rs11792633, and rs7044343 SNPs within IL33 decrease the risk of AD. In this study, we found that a tag SNP-rs7044343 in IL33 was also associated with RA. This indicated that rs7044343 in IL33 decreased the risks of both RA and AD. The inverse relation between AD and RA was challenged. Simmons et al.[16] found that 17 alleles associated with increased RA risk were not associated with reduced AD risk, and three RA-associated SNPs increased the risk of AD, indicating that RA genetics did not underlie the inverse relation between RA and AD, but rather may promote AD [17].
GWAS studies have shown that rs1342326 [16], rs3939286 [18], and rs2381416 [19] SNPs within IL33 seem to be an asthma-susceptibility gene. IL33 encodes a cytokine belonging to the IL1 superfamily, and is the natural ligand for the IL1RL1 receptor [4], which has been previously implicated in asthma, inflammation, and a number of immune disorders [20].
The present study suggests a lack of association between IL33 polymorphism and RA disease activity index (including DAS 28 score, acute-phase reactant, and so on), although the IL33 7044343 CC genotype was associated with lower serum IL-33 levels. Whether the serum levels of IL-33 were related to RA disease activity shows conflicting evidence. Matsuyama et al. [21] reported that serum IL-33 levels were significantly higher in RA patients and correlated with disease activity. However, we found that serum IL-33 levels were not correlated to inflammation markers, such as ESR and CRP, but to autoantibodies in RA patients, which was consistent with the present finding.
Some limitations exist in our study. First, a larger sample size in different populations, especially in other ethnic populations, is required to validate the results. Second, although the results showed that the rs7044343 CC genotype of IL33 is associated with lower serum IL-33 levels, more large-scale population study and functional experiments are needed to elucidate the association. Third, the rs7044343-polymorphism genotype distribution was not in accordance with the HWE in RA patients and discovery controls. Theoretically, cases do not need to be in HWE [22]. Rs7044343 was not in HWE in discovery controls, but was in HWE in validation and replication. This might be due to the smaller sample size or to other uncertain factors. We sequenced 130 cases and controls, and double checked the primer and probe sequences of SNPs and genotype plots of TaqMan to confirm that no genotyping bias existed.
Conclusion
To our knowledge, we first found that the rs7044343 in IL33 is associated with RA patients, and the CC genotype of rs7044343 is associated with lower serum IL-33 levels. These findings, together with our previous studies, suggest that IL-33, both in gene and in protein levels, plays an important role in the pathogenesis of RA.
Abbreviations
ACPA: Anti-citrullinated peptide antibody; AD: Alzheimer disease; CHB: Han Chinese in Beijing; CI: confidence interval; CIA: collagen-induced arthritis; CRP: C-reactive protein; CTLA: cytotoxic T-lymphocyte antigen; DAS28: 28-joint-count disease activity score; DMARD: disease-modifying antirheumatic drug; DNA: deoxyribonucleic acid; ELISA: enzyme-linked immunosorbent assay; ESR: erythrocyte sedimentation rate; GWAS: genome-wide association study; HLA: human leukocyte antigen; HWE: Hardy–Weinberg equilibrium; IL: interleukin; LD: linkage disequilibrium; MAF: minor allele frequency; OR: odds ratio; PCR: polymerase chain reaction; PTPN22: protein tyrosine phosphatase N22; RA: rheumatoid arthritis; RF: rheumatoid factor; SNP: single-nucleotide polymorphism.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CL, data collection and analysis, manuscript writing, and final approval of the manuscript. RM, conception and design, critical revision, final approval, and responsibility for the manuscript. JPG, analysis and interpretation, critical revision, and final approval of the manuscript. XYW, conception and design and data collection, analysis, and interpretation, manuscript writing, and final approval of the manuscript. XT, analysis, interpretation, critical revision, and final approval of the manuscript. XL, data collection and interpretation, manuscript writing, and final approval of the manuscript. FLH, data collection and critical revision, manuscript writing, and final approval of the manuscript. SWG, data collection, critical revision, and final approval of the manuscript. JXZ, data collection, manuscript writing, and final approval of manuscript. HJX, interpretation, critical revision, and final approval of the manuscript. ZGL, conception and design, data collection and analysis, financial support, manuscript writing, final approval, and responsibility for the manuscript. All authors read and approved the final manuscript.
Supplementary Material
DNA sequence analysis of the three different genotypes of the rs7044343 and rs10975514. (a) The TT genotype of rs7044343. (b) The CT genotype of rs7044343. (c) The CC genotype of rs7044343. (d) The GG genotype of rs10975514. (e) The AG genotype of rs10975514. (f) The AA genotype of rs10975514.
Association analysis of rs10975514 with RA in the discovery population. Table S2. Association analysis of rs10975514 with RA in the validation population. Table S3. Association analysis of rs10975514 with RA in the replication population.
Contributor Information
Chun Li, Email: fiona_leechun@163.com.
Rong Mu, Email: murongster@gmail.com.
Jianping Guo, Email: jianpingguo@pkuph.edu.cn.
Xinyu Wu, Email: wuxinyufidence@yahoo.com.cn.
Xin Tu, Email: xtu@mail.hust.edu.cn.
Xu Liu, Email: liuxu0201@yahoo.com.cn.
Fanlei Hu, Email: fanleihu@gmail.com.
Shiwei Guo, Email: shiweiashiwei@126.com.
Jiaxin Zhu, Email: jxzhu252@gmail.com.
Huji Xu, Email: xuhuji@smmu.edu.cn.
Zhanguo Li, Email: li99@bjmu.edu.cn.
Acknowledgements
We thank all patients and healthy controls who took part in this study. We also thank Dr. Xiaoying Zhang and Dr. Tianjiao Duan for their work in DNA extraction.
Funding
Supported by Major State Basic Research Development Program of China (973 Program) (No.2010CB529100 to ZG. L) , the National Natural Science Foundation of China (No.81072401 to R.M), and the Program for New Century Excellent Talents in University of China (NCET-11-0181 to X.T).
References
- MacGregor AJ, Snieder H, Rigby AS, Koskenvuo M, Kaprio J, Aho K, Silman AJ. Characterizing the quantitative genetic contribution to rheumatoid arthritis using data from twins. Arthritis Rheum. 2000;43:30–37. doi: 10.1002/1529-0131(200001)43:1<30::AID-ANR5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Kawaguchi T, Stahl EA, Kurreeman FA, Nishida N, Ohmiya H, Myouzen K, Takahashi M, Sawada T, Nishioka Y, Yukioka M, Matsubara T, Wakitani S, Teshima R, Tohma S, Takasugi K, Shimada K, Murasawa A, Honjo S, Matsuo K, Tanaka H, Tajima K, Suzuki T, Iwamoto T, Kawamura Y. et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet. 2012;44:511–516. doi: 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- Stahl EA, Raychaudhuri S, Remmers EF, Xie G, Eyre S, Thomson BP, Li Y, Kurreeman FA, Zhernakova A, Hinks A, Guiducci C, Chen R, Alfredsson L, Amos CI, Ardlie KG, Barton A, Bowes J, Brouwer E, Burtt NP, Catanese JJ, Coblyn J, Coenen MJ, Costenbader KH, Criswell LA, Crusius JB, Cui J, de Bakker PI, De Jager PL, Ding B. Consortium BIRAC et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Carriere V, Roussel L, Ortega N, Lacorre DA, Americh L, Aguilar L, Bouche G, Girard JP. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci U S A. 2007;104:282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu R, Huang HQ, Li YH, Li C, Ye H, Li ZG. Elevated serum interleukin 33 is associated with autoantibody production in patients with rheumatoid arthritis. J Rheumatol. 2010;37:2006–2013. doi: 10.3899/jrheum.100184. [DOI] [PubMed] [Google Scholar]
- Xiangyang Z, Lutian Y, Lin Z, Liping X, Hui S, Jing L. Increased levels of interleukin-33 associated with bone erosion and interstitial lung diseases in patients with rheumatoid arthritis. Cytokine. 2012;58:6–9. doi: 10.1016/j.cyto.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Xu D, Jiang HR, Kewin P, Li Y, Mu R, Fraser AR, Pitman N, Kurowska-Stolarska M, McKenzie AN, McInnes IB, Liew FY. IL-33 exacerbates antigen-induced arthritis by activating mast cells. Proc Natl Acad Sci U S A. 2008;105:10913–10918. doi: 10.1073/pnas.0801898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer G, Talabot-Ayer D, Lamacchia C, Toy D, Seemayer CA, Viatte S, Finckh A, Smith DE, Gabay C. Inhibition of interleukin-33 signaling attenuates the severity of experimental arthritis. Arthritis Rheum. 2009;60:738–749. doi: 10.1002/art.24305. [DOI] [PubMed] [Google Scholar]
- Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, Medsger TA Jr, Mitchell DM, Neustadt DH, Pinals RS, Schaller JG, Sharp JT, Wilder RL, Hunder GG. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmura K, Terao C, Maruya E, Katayama M, Matoba K, Shimada K, Murasawa A, Honjo S, Takasugi K, Tohma S, Matsuo K, Tajima K, Yukawa N, Kawabata D, Nojima T, Fujii T, Yamada R, Saji H, Matsuda F, Mimori T. Anti-citrullinated peptide antibody-negative RA is a genetically distinct subset: a definitive study using only bone-erosive ACPA-negative rheumatoid arthritis. Rheumatology (Oxford) 2010;49:2298–2304. doi: 10.1093/rheumatology/keq273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terao C, Ohmura K, Kochi Y, Ikari K, Maruya E, Katayama M, Shimada K, Murasawa A, Honjo S, Takasugi K, Matsuo K, Tajima K, Suzuki A, Yamamoto K, Momohara S, Yamanaka H, Yamada R, Saji H, Matsuda F, Mimori T. A large-scale association study identified multiple HLA-DRB1 alleles associated with ACPA-negative rheumatoid arthritis in Japanese subjects. Ann Rheum Dis. 2011;70:2134–2139. doi: 10.1136/annrheumdis-2011-200353. [DOI] [PubMed] [Google Scholar]
- Chapuis J, Hot D, Hansmannel F, Kerdraon O, Ferreira S, Hubans C, Maurage CA, Huot L, Bensemain F, Laumet G, Ayral AM, Fievet N, Hauw JJ, DeKosky ST, Lemoine Y, Iwatsubo T, Wavrant-Devrièze F, Dartigues JF, Tzourio C, Buée L, Pasquier F, Berr C, Mann D, Lendon C, Alpérovitch A, Kamboh MI, Amouyel P, Lambert JC. Transcriptomic and genetic studies identify IL-33 as a candidate gene forAlzheimer's disease. Mol Psychiatry. 2009;14:1004–1016. doi: 10.1038/mp.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JT, Song JH, Wang ND, Wu ZC, Zhang Q, Zhang N, Zhang W, Xuan SY, Tan L. Implication of IL-33 gene polymorphism in Chinese patients with Alzheimer's disease. Neurobiol Aging. 2012;33:1014. doi: 10.1016/j.neurobiolaging.2010.07.003. e11-4. [DOI] [PubMed] [Google Scholar]
- Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von ME, Farrall M, Lathrop M, Cookson WO. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CR, Zou F, Younkin SG, Estus S. Rheumatoid arthritis-associated polymorphisms are not protective against Alzheimer's disease. Mol Neurodegener. 2011;6:33. doi: 10.1186/1750-1326-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudbjartsson DF, Bjornsdottir US, Halapi E, Helgadottir A, Sulem P, Jonsdottir GM, Thorleifsson G, Helgadottir H, Steinthorsdottir V, Stefansson H, Williams C, Hui J, Beilby J, Warrington NM, James A, Palmer LJ, Koppelman GH, Heinzmann A, Krueger M, Boezen HM, Wheatley A, Altmuller J, Shin HD, Uh ST, Cheong HS, Jonsdottir B, Gislason D, Park CS, Rasmussen LM, Porsbjerg C. et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- Torgerson DG, Ampleford EJ, Chiu GY, Gauderman WJ, Gignoux CR, Graves PE, Himes BE, Levin AM, Mathias RA, Hancock DB, Hansel NN, Hernandez RD, Israel E, Salam MT, Galanter J, Avila PC, Avila L, Rodriquez-Santana JR, Chapela R, Rodriguez-Cintron W, Diette GB, Adkinson NF, Abel RA, Ross KD, Shi M, Faruque MU, Dunston GM, Watson HR, Mantese VJ, Ezurum SC. et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American populations. Nat Genet. 2011;43:887–892. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew FY, Pitman NI, McInnes IB. Disease-associated functions of IL-33: the new kid in the IL-1 family. Nat Rev Immunol. 2010;10:103–110. doi: 10.1038/nri2692. [DOI] [PubMed] [Google Scholar]
- Matsuyama Y, Okazaki H, Tamemoto H, Kimura H, Kamata Y, Nagatani K, Nagashima T, Hayakawa M, Iwamoto M, Yoshio T, Tominaga S, Minota S. Increased levels of interleukin 33 in sera and synovial fluid from patients with active rheumatoid arthritis. J Rheumatol. 2010;37:18–25. doi: 10.3899/jrheum.090492. [DOI] [PubMed] [Google Scholar]
- Lee WC. Searching for disease-susceptibility loci by testing for Hardy-Weinberg disequilibrium in a gene bank of affected individuals. Am J Epidemiol. 2003;158:397–400. doi: 10.1093/aje/kwg150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequence analysis of the three different genotypes of the rs7044343 and rs10975514. (a) The TT genotype of rs7044343. (b) The CT genotype of rs7044343. (c) The CC genotype of rs7044343. (d) The GG genotype of rs10975514. (e) The AG genotype of rs10975514. (f) The AA genotype of rs10975514.
Association analysis of rs10975514 with RA in the discovery population. Table S2. Association analysis of rs10975514 with RA in the validation population. Table S3. Association analysis of rs10975514 with RA in the replication population.


