Abstract
Gene-targeting vectors derived from mobile group II introns capable of forming a ribonucleoprotein (RNP) complex containing excised intron lariat RNA and an intron-encoded protein (IEP) with reverse transcriptase (RT), maturase, and endonuclease (En) activities have been described. RmInt1 is an efficient mobile group II intron with an IEP lacking the En domain. We performed a comprehensive study of the rules governing RmInt1 target site recognition based on selection experiments with donor and recipient plasmid libraries, with randomization of the elements of the intron RNA involved in target recognition and the wild-type target site. The data obtained were used to develop a computer algorithm for identifying potential RmInt1 targets in any DNA sequence. Using this algorithm, we modified RmInt1 for the efficient recognition of DNA target sites at different locations in the Sinorhizobium meliloti chromosome. The retargeted RmInt1 integrated efficiently into the chromosome, regardless of the location of the target gene. Our results suggest that RmInt1 could be efficiently adapted for gene targeting.
Keywords: group II introns, gene targeting, ribozyme, RmInt1, Sinorhizobium meliloti
Introduction
Group II introns are self-splicing catalytic RNAs that act as mobile retroelements. It is thought that both nuclear spliceosomal introns and non-long-terminal repeat retrotransposons evolved from mobile group II introns.1-4 Group II introns were initially identified in the mitochondrial and chloroplast genomes of lower eukaryotes and plants, and have subsequently been described in bacteria and archaea.5-9 Group II introns consist of a structured RNA that fold into a conserved three-dimensional structure organized into six double-helical domains, DI to DVI.10 Most bacterial group II introns have an open reading frame (ORF) encoding an intron-encoded protein (IEP) in DIV. Group II IEPs have an N-terminal reverse transcriptase (RT) domain, followed by a putative RNA-binding domain with RNA splicing or maturase activity (X domain), and a C-terminal DNA-binding (D) region followed by a DNA endonuclease (En) domain.11,12 Three main classes of group II introns (IIA, IIB, and IIC), based on the structure of the ribozyme, have been described, together with subclasses of IIA and IIB introns displaying specific structural variations (IIA1, IIA2, IIB1, and IIB2).8 Phylogenetic analyses of group II intron IEPs have resulted in their classification into several groups (A, B, C, D, E, F, CL1 [chloroplast-like 1], CL2 [chloroplast-like 2], and ML [mitochondrion-like]). All these classes of group II intron IEPs are present in bacteria, whereas only the CL and ML classes are present in organelles. The A, C, D, E, and F introns encode proteins with no En domain.13-16 Class F has recently been revised, with the identification of new varieties of intron ORF.16
The movement of group II intron is mediated by a ribonucleoprotein (RNP) complex consisting of the IEP encoded by the ORF and the spliced intron lariat RNA, which remains associated with the IEP. These RNP complexes recognize intron targets through both the IEP and the intron lariat RNA. The central part of the target, containing the intron-insertion site, is recognized by base pairing of the lariat RNA via the exon-binding site (EBS) and the complementary region in the DNA target, known as the intron-binding site (IBS).5 The target flanking regions are recognized by the IEP.17-19 The EBS/IBS interaction differs between intron classes. Group IIA and IIB introns recognize the 5′exon through EBS2/IBS2 and EBS1/IBS1 base pairing. Group IIC introns use EBS1/IBS1 pairing and there is no recognizable EBS2/IBS2 interaction. Recognition of the 3′ exon by group IIA introns involves a δ/δ’ interaction encompassing three nucleotides, whereas IIB and IIC introns use EBS3/IBS3 interaction, encompassing only one nucleotide.19-21 Recognition of the distal part of the 5′ and 3′ exons by the IEPs also involves specific bases in particular positions. For example, T-23, G-21, A-20, and T+5 are Ll.LtrB target site positions recognized by LltrA.17 In the target site of the EcI5 intron, the IEP recognizes C-18, C-17, A-15, A-14, and T+5.22 For RmInt1, the two most critical positions have been identified as T-15 and G+4,18 which are probably recognized by the IEP. The mechanism by which group II introns recognize their target sites, mostly by base pairing between the EBS and IBS sequences, the possibility of modifying this interaction, and the small number and flexibility of interactions between IEPs and target sites open up possibilities for using these introns as biotechnological tools. The Ll.LtrB intron from Lactococcus lactis has been used to develop gene targeting in both Gram-positive23-25 and Gram-negative26,27 bacteria. In addition, EcI5, an Escherichia coli intron, has been retargeted for insertion in different positions within the lacZ gene. The IEPs of Ll.LtrB and EcI5 both have D and En domains. We have previously shown that the RmInt1 intron, the IEP of which has no D or En domain, can be retargeted for the invasion of chromosomal genes in E. coli. In this study, we analyzed the sequence requirements for each position in the RmInt1 target site and used the nucleotide frequencies within active target sites to develop a computer algorithm predicting optimal target sites in any DNA.
Results
Distal 5′-exon and 3′-exon nucleotide sequence requirements for RmInt1 retrohoming
For identification of the positions and essential nucleotides required for RmInt1 retrohoming in the distal exons of its target site sequence, we used the two-plasmid retrohoming assay already described for RmInt1.28 The intron donor plasmid was pKGEMA4T7, which contains an RmInt1-ΔORF intron and short flanking exons (-20/+5), with a phage T7 promoter inserted into domain IV and expression of the IEP from a position upstream from the 5′ exon. As the intron recipient, we used a plasmid library in pACE,28 in which RmInt1 target sites with randomized nucleotide residues from positions -20 to -14 and +2 to +5 were inserted upstream from a promoter-less tetR gene.
E. coli HMS174 (DE3), harboring both the intron donor plasmid and the recipient plasmid library, was plated on LB agar containing ampicillin and tetracycline. The colonies growing on this medium necessarily displayed integration of the intron into a random target, driving expression of the tetR gene by the cargo T7 promoter. Colony PCR was performed to amplify target sites with inserted introns, and the PCR products were sequenced to determine the sequences of the target sites invaded. We determined the sequences of 105 active target sites and 105 recipient plasmids from the initial pool, to correct for possibly nucleotide frequency biases upon randomization (Fig. S1). Figure 1 shows the nucleotide frequencies (percentages) at each randomized position in the selected target sites, already corrected for biases in the initial pool, and results are summarized in WebLogo format.
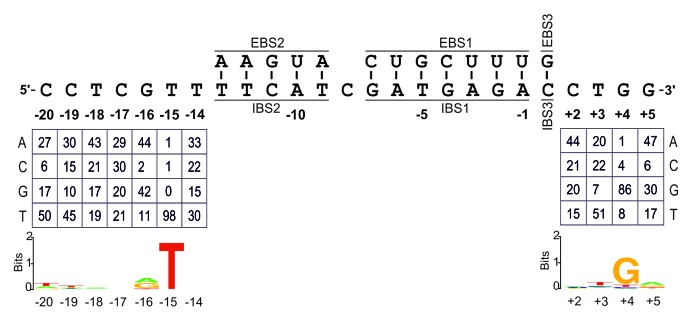
Figure 1. Identification of critical nucleotide residues in the distal 5′- and 3′-exon regions of the RmInt1 DNA target sequence. EBS/IBS interactions are shown. Nucleotide frequencies in the randomized positions of the intron-inserted plasmids corrected for biases in the initial pool, are summarized below. WebLogo representation illustrating the nucleotide frequencies shown above.
The distal 5′-exon displayed strong selection for the T-15 residue, which was found in 98% of all active target sites. Other positions (-20, -19, and -16) displayed some selection for specific nucleotides. In the distal 3′-exon, only one nucleotide residue was identified as highly conserved in active target sites, G+4, with positions +3 and +5 displaying mild selection for certain nucleotides. Our data also suggested that there was selection against C residues in positions -20, -16, and +5 and against G residues in position +3. Surprisingly, the wild-type intron target site has a C residue in position -20. We have also previously reported selection for T-15 and G+4, and mutations at these positions block and reduce homing efficiency, respectively.18
Rules for EBS/IBS interactions
EBS2/IBS2 pairing
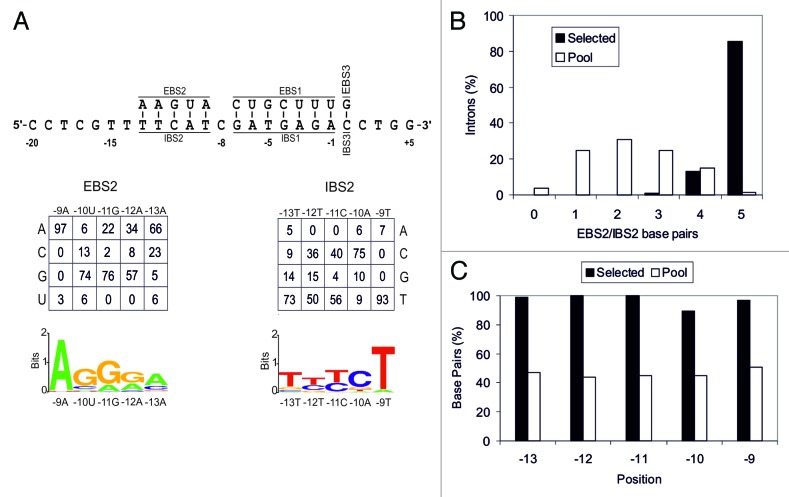
We investigated the rules governing the EBS2/IBS2 interaction (Fig. 2A), by performing selection experiments in which the EBS2 of RmInt1 and the corresponding positions in the target site IBS2 (-9 to -13) were randomized. In addition, the donor intron plasmid had randomized IBS2 nucleotide positions in the 5′ exon, to provide complementary nucleotide combinations to facilitate RNA splicing. We obtained sequences for 95 intron insertion events, along with 97 unselected donor plasmids and 107 unselected recipient plasmids, to correct for nucleotide frequency biases in the pools (Fig. S2). In this region, 86% of the insertion events involved the pairing of all five bases, whereas most of the randomly chosen intron-target site combinations had between one and three paired bases (Fig. 2B). Data shown in Figure 2C indicate strong selection for RNA/DNA base paring between EBS2/IBS2 with all positions, but -10, base paired in ≥ 95% of the insertion events (Fig. 2C). For this interaction, we detected strong selection for specific nucleotides, particularly in position -9, where there is a strong selection for the T residue present in 98% of invasion events (Fig. S2). In mobility assays with a wild-type RmInt1 target and RmInt1 with a single nucleotide modification in the EBS2 region (A-9G), mobility rates were two orders of magnitude lower than for the wild-type RmInt1 (data not shown). This mutation does not prevent RmInt1 from recognizing its target, because the wild-type base pair at position -9 A-T is changed to G-T. This selection for specific nucleotide residues in the EBS2 region probably reflects restrictions on intron RNA structure resulting in preferences for specific RNA-DNA base pair interactions. We also observed selection at position -10 in this region, for a G-C base pair rather than the wild-type U-A, suggesting that RmInt1 does not have the optimal nucleotide at this position in its natural target within ISRm2011-2.
Figure 2. Analysis of EBS2/IBS2 interaction. Panel A shows EBS/IBS interaction, with the nucleotide frequencies at each randomized position of the selected plasmids, already corrected for biases in the initial pool, shown below. A WebLogo representation of the corrected nucleotide frequencies at each randomized position is also shown. (B) Percentage of introns with different numbers of EBS2/IBS2 base pairs formed with the target site in the selected invasion events (black bars) and randomly paired donor and recipient plasmids from the initial pools (white bars). (C) Percentage of base pairs at each EBS2/IBS2 position in the selected invasion events (black bars) and randomly paired donor and recipient plasmids from the initial pools (white bars)
EBS1/IBS1 pairing
We investigated the rules governing the pairing of EBS1 and IBS1 by carrying out experiments similar to those described above. We first studied the whole EBS1/IBS1 interaction. However, the frequency of insertion of the RmInt1 intron with randomized EBS1 sites into its targets with randomized IBS1 sites was very low. This may be due to the small number of introns able to splice from the donor plasmid, because 14 positions were randomized and there were only a very small number of donor plasmids with complementary EBS1 and IBS1. We solved this problem by focusing on EBS1/IBS1 pairing in two regions. We first analyzed the pairing of the 5′-end region of IBS1 (positions -7 to -5, 5′-GAT-3′) with the 3′-end of EBS1 (5′-GUC-3′). We then studied pairing of the 3′-end region of IBS1 (positions -4 to -1; 5′-GAGA-3′) and the 5′-end of EBS1 (5′-UUUC-3′).
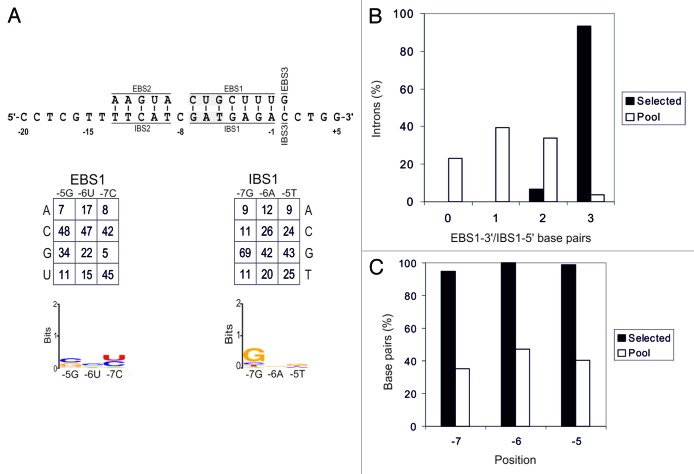
We determined the frequencies of EBS1-3′/IBS1-5′ pairing, correcting for biases in the initial pools (Fig. 3A). Data were obtained from sequences for 95 intron-integration events, together with 107 unselected recipient plasmids, and 97 unselected donor plasmids (Fig. S3). In this region, 93% of the inserted introns had three EBS1-3′ nucleotides paired with the IBS1-5′, indicating that selection for base pairing in this region was stronger than that for EBS2/IBS2 (Fig. 3B). Individually, positions -5, -6, and -7 displayed base pairing in 99%, 100%, and 95% of retrohoming events, respectively (Fig. 3C). Position -7 of IBS1 displayed moderate selection for G nucleotide, as for the wild-type RmInt1. No selection for any particular nucleotide was noted at the other two positions (Fig. 3A).
Figure 3. Analysis of EBS1-3′/IBS1-5′ interaction. Panel A shows EBS/IBS interaction (the randomized nucleotides are highlighted in gray), with the nucleotide frequencies obtained at each randomized position of intron-invaded plasmids, corrected for biases in the original pool, shown below. A WebLogo representation of corrected nucleotide frequencies at each randomized position of the selected target sites is also shown. (B) Percentage of introns with different numbers of EBS2/IBS2 base pairs formed with the target site in the selected invasion events (black bars) and randomly paired donor and recipient plasmids from the initial pools (white bars). (C) Percentage of base pairs at each EBS2/IBS2 position in the selected invasion events (black bars) and randomly paired donor and recipient plasmids from the initial pools (white bars).
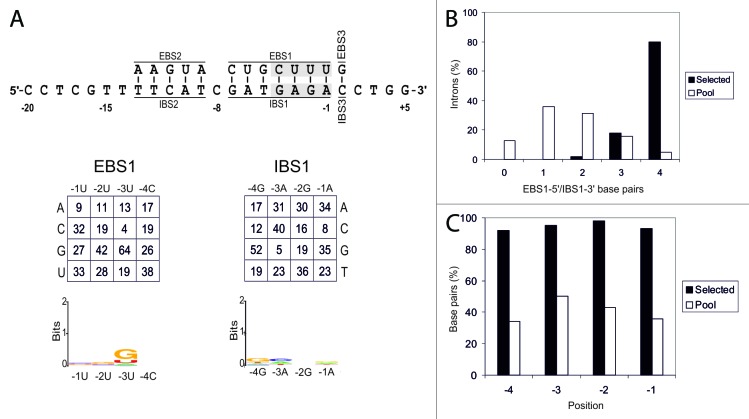
The results of selection experiments for EBS1-5′/IBS1-3′ are shown in Figure 4. This region displayed weaker selection for base pairing (Fig. 4B) than EBS1-3′/IBS1-5′ (80% and 93%, respectively), although all positions were base paired in ≥ 92% of intron insertion events (Fig. 4C). In this region, we observed mild selection for a G residue in position -3 of EBS1. This is not the nucleotide present in the wild-type RmInt1 (Fig. 4A), suggesting that RmInt1 does not have the optimal nucleotide at this position.
Figure 4. Analysis of EBS1-5′/IBS1-3′ interaction. Panel A shows EBS/IBS interaction (the randomized nucleotides are highlighted in gray) and nucleotide frequencies for randomized positions of intron-inserted plasmids corrected for biases in the initial pool. A WebLogo representation of nucleotide frequencies at each randomized position of the selected target sites, corrected for biases in the initial pool, is also shown. (B) Percentage of introns with different numbers of EBS2/IBS2 base pairs formed with the target site in the selected invasion events (black bars) and randomly paired donor and recipient plasmids from the initial pools (white bars). (C) Percentage of base pairs at each EBS2/IBS2 position in the selected invasion events (black bars) and randomly paired donor and recipient plasmids from the initial pools (white bars).
EBS3/IBS3 pairing
Nucleotide preferences for the EBS3/IBS3 interaction were also determined by selection experiments in which the nucleotide in position +1, in both the IBS3 of the recipient plasmids and the EBS3 of the donor plasmids, was randomized (Fig. 5). Previous work on RmInt1 showed that the mispairing of EBS3/IBS3 in the precursor RNA had only a slight effect on RNA splicing,29 but we nevertheless also randomized this position in EBS3 to decrease the impact of possible mispairing on splicing. We observed selection for RNA/DNA base-pairing in 100% of the selected target sites, vs 38% in the original pool. Only Watson–Crick combinations were found in the 88 selected target sites and clear selection was observed for only one of the four Watson–Crick combinations, the wild-type G–C base pair. This bias for selection of the wild-type base pair may reflect constraints on RNA structure.
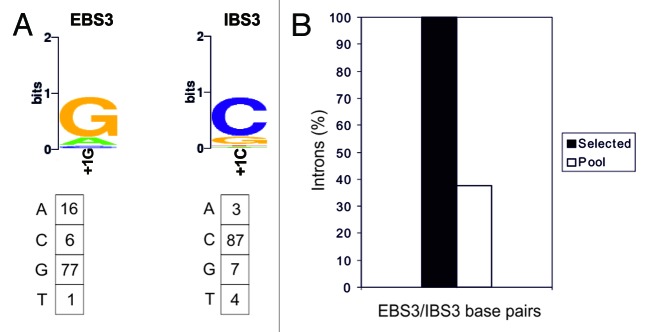
Figure 5. Analysis of EBS3/IBS3 interaction. (A) WebLogo representation of nucleotide frequencies in the integration events, corrected for biases in the initial pool. Nucleotide frequencies at the randomized position in the integration events corrected for biases in the initial pool are summarized below. (B) Percentage of intron base pairing at EBS3/IBS3 in the selected events and for randomly paired donor and recipient plasmids from the initial pools.
Design and testing of an algorithm for predicting RmInt1 targets
Using the RmInt1 target site recognition rules and data obtained in the selection experiments described above, we developed a computerized algorithm for identifying the best potential RmInt1 targets present in a given DNA sequence. As for Ll.LtrB and EcI5,22,30 we used a model equivalent to a zero-order Markov model, assuming that a nucleotide residue in the DNA target sequence occurs at random and that the likelihood of its occurrence is independent of all other residues in the DNA sequence.31 This algorithm scores a DNA sequence across a 25 bp sliding window, moving by 1 bp at a time, through the use of a log-odds score assessing whether the probability that the sequence generated by the model M, P(seq|M), is greater than the probability P(seq) that the sequence was generated by chance (null model). P(seq|M) and P(seq) are calculated as the products of individual probabilities P(np|M) and P(np) that a nucleotide n at position p in the target sequence was generated by the model and by chance, respectively, with the latter calculated from the frequencies and for a nucleotide in selected target sites and the initial pool, respectively, according to the following equation:
A positive score means that the model predicts the potential RmInt1 intron target sequence better than the null model.30
We tested the performance of the algorithm by scanning the lacZ gene for RmInt1 target sites. RmInt1 was retargeted for insertion into the predicted target sites. The RmInt1 intron-less strain RMO17 contains no lacZ gene, so we integrated a copy of this gene into its chromosome. Two orientations of lacZ were used in these analyses. In strain RMO17Zs, the lacZ gene was integrated such that the sense strand acted as the lagging template strand during chromosome replication (LAG). In RMO17Zas, lacZ was in the opposite orientation (LEAD). The gene was integrated into a region of the RMO17 chromosome 100% identical (unpublished data) to the chromosome of the sequenced reference strain S. meliloti 1021.32 In 1021, this region is located 10 kb away from the oriC between genes SMc2781 and gabD1. Figure 6 shows the predicted targets in lacZ, the base-pairing interactions with the retargeted RmInt1 and the score calculated for each target. The modifications of RmInt1 span EBS1, EBS2, and EBS3, to ensure base-pairing with the selected targets. In the donor plasmids, we modified not only the EBS, but also IBS1, IBS2, and IBS3 in the 5′ and 3′ exons, making them complementary to the modified EBS1, EBS2, and EBS3, to facilitate efficient RmInt1 splicing. All retargeted introns were assayed for retrohoming intro both RMO17Zs and RMO17Zas. The donor plasmid was introduced into RMO17Zs or RMO17Zas by triparental mating and transconjugants were plated on TY agar supplemented with 5-bromo-4-chloro-3indolyl-D-galactopyranoside (X-gal). The blue colonies on these plates had no lacZ gene invasion, whereas the white colonies had RmInt1 insertions into the lacZ gene. For each retargeted intron, we performed blue/white scoring for 500–5000 colonies. Intron insertion at the desired target site was confirmed by colony PCR on at least 30 white colonies and further sequencing of the PCR products.
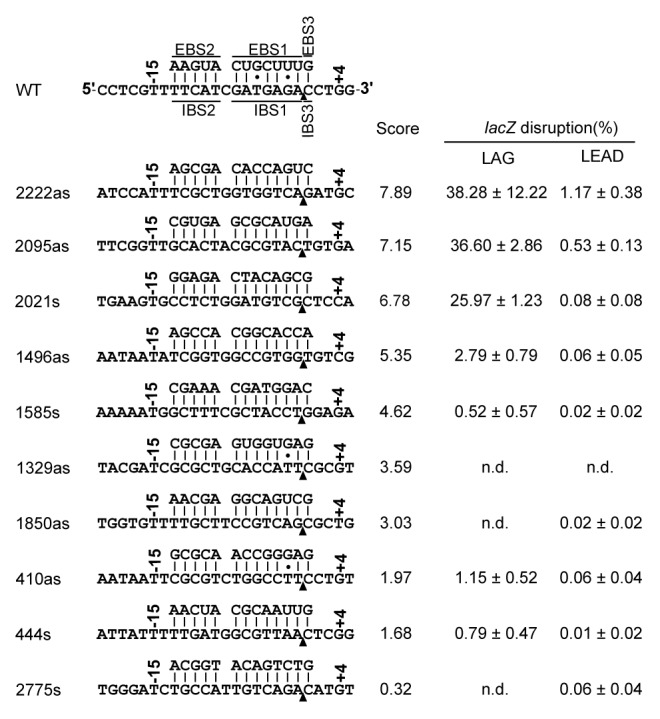
Figure 6. Target site sequences and base-pairing interactions for the insertion of retargeted RmInt1 at different positions in the lacZ gene. Retargeted introns are denoted by a number corresponding to the nucleotide position of the intron insertion site in the target gene coding sequence. “s” and “as” indicate the sense and antisense strands, respectively. The arrowhead indicates the intron insertion site. The score obtained with the computer algorithm is indicated, together with the targeting frequency, which was determined by counting blue and white colonies in both the lagging and leading orientations with respect to the replication origin. The values shown are means + standard deviations for three independent determinations. (n.d.) not detectable.
The targeting frequencies obtained for each target indicated that all target sites had a low invasion frequency when in the LEAD orientation, regardless of the score attributed by the computer algorithm. This reflects the reported bias of RmInt1 for invading the nascent lagging strand during replication.33 For targets in the LAG orientation, efficiently invaded targets (disruption frequencies 25.97–38.28%) had algorithm scores of more than 5.35. The model used for the algorithm included target positions -20 to -14, -10, -9, -8, +2, +3, and +5. Note that other models, including IBS1, IBS2 positions -11, -12, and -13, as well as position +4 of the 3′ distal exon do not discriminate efficient targets from inefficient ones, even though the latter was present in 86% of the selected targets, which suggest that other still unknown rules might be involved in target selection.
We used the same algorithm to identify potential RmInt1 targets in a modified sacB gene34 conferring sucrose sensitivity. The sacB gene was integrated into the RMO17 chromosome together with the lacZ gene. Four potential target sites with a score of more than 5.35 within the sacB gene were predicted by the algorithm. RmInt1 was modified to recognize the target sacB661s, located in the sacB sense strand and with a score of 6.91. Invasion experiments were performed in RMO17RZas, in which sacB661s is in the LAG orientation. An invasion frequency of 81.31% was obtained, confirming the ability of the algorithm to predict potential RmInt1 target sites in different genes.
We assessed the insertion specificity of the retargeted introns, by carrying out Southern hybridization on total DNA isolated from cells harboring lacZ disrupted by the more efficient retargeted RmInt1 introns (2222as, 2021s, and 2095as), with an intron probe (Fig. 7). All three of the disruptants obtained had a band corresponding to the intron-invaded lacZ and a band corresponding to the intron donor plasmid, which was still present in the cells. However, disruptants 2021s and 2222as had at least one additional band, indicating off-target integration. Sequence analyses revealed that some of the extra bands in 2222as corresponded to tandem insertions of the intron into both the target site and the intron donor plasmid, due to the pairing of the first nucleotide of the intron (G) to EBS3 (C). The single extra band appearing in disruptant 2021s corresponded to an insertion into a target site displaying perfect base-pairing with the retargeted intron EBS sequences. These results indicate that the retargeted RmInt1 is highly specific but that care is required to prevent unwanted off-target insertion, by shortening the intron expression time and choosing target sites with unique IBSs in the corresponding genomes, for example.
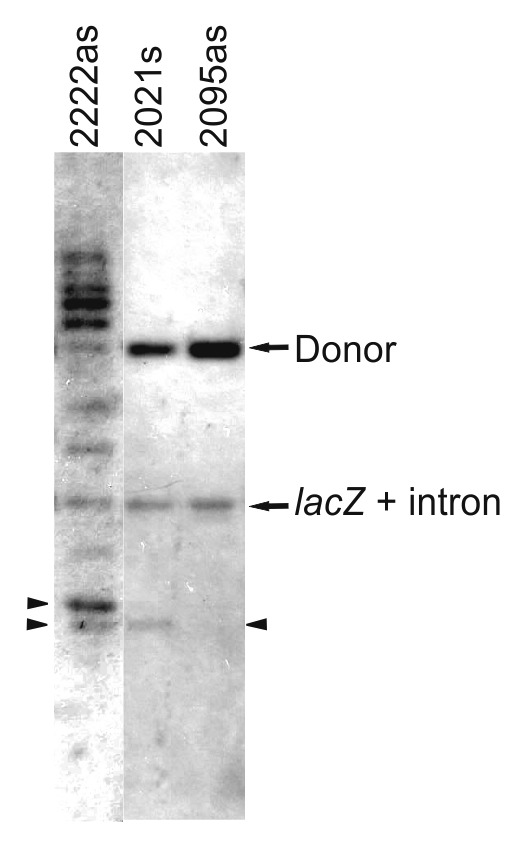
Figure 7. Southern hybridization. Total DNA was isolated from RMO17 and the indicated lacZ disruptants. DNA was digested with BamHI to release the lacZ gene with the inserted intron. Donor plasmids still present are also shown. The arrowhead indicates additional intron insertion. The lanes presented here come from different gels.
Retrohoming of RmInt1 into the S. meliloti chromosome
An Ll.LtrB library with randomized target site recognition sequences has been reported to display higher rates of invasion in a short region close to the chromosomal origin of replication in the E. coli genome.35 We investigated whether retargeted RmInt1 displayed a bias for efficient insertion at the replication origin in S. meliloti, by integrating the lacZ gene into four additional sites in the chromosome of strain RMO17 (Fig. 8). For lacZ integrations, the target site was placed on the lagging template strand (LAG), except for location 4, which is on the leading strand template (LEAD), and location 3 which is on both strand templates (LAG and LEAD). The reprogrammed intron used was 2095as, a retargeted RmInt1 that inserts at nucleotide 2095 in the lacZ antisense strand. Targeting efficiency (Table 1) was scored by blue-white screening. We found that the retargeted RmInt1 invaded genes located on different regions of the S. meliloti chromosome. In the LAG orientation, the frequency of intron insertion into the same target site was higher close to the replication origin, nonetheless it still inserts efficiently out of this region.
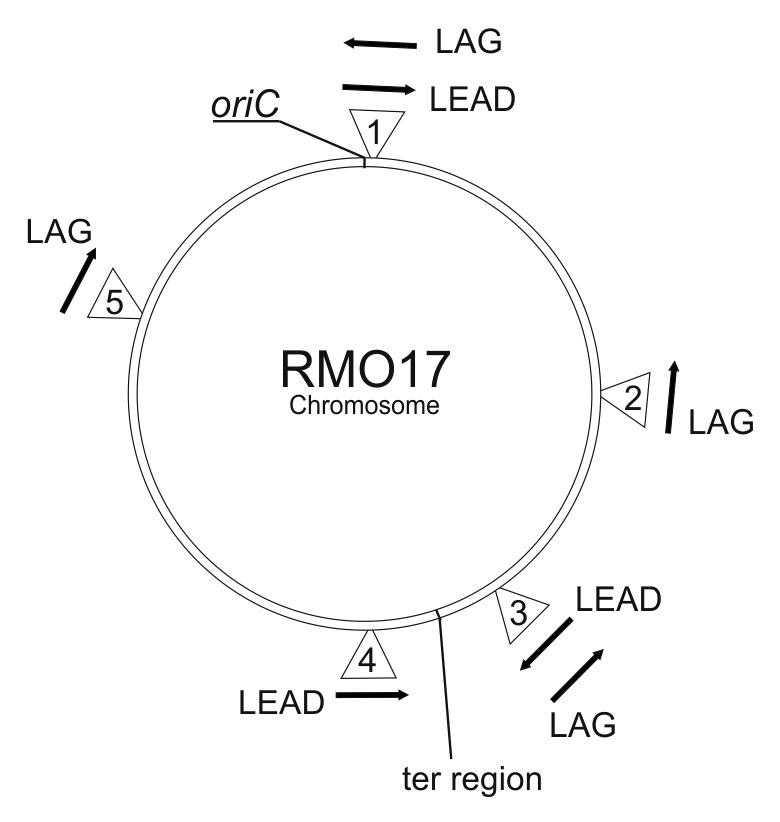
Figure 8. Insertion of lacZ genes into different locations on the RMO17 chromosome. Numbers within triangles represent the insertions and their location in RMO17. Insertion 1 in the LAG and LEAD orientations is the same as for 2095as in Figure 6. Arrows indicate the direction of lacZ relative to the replication origin. LAG and LEAD indicate that the target site is on the lagging or leading template strands, respectively. The location of oriC is shown and the ter region is shown to indicate the point at which GCskew changes from + to - on the S. meliloti 1021 chromosome.
Table 1. Frequencies of lacZ gene invasion at various sites on the RMO17 chromosome.
| Insertion | Invasion frequency (%) | |
|---|---|---|
| LAG | LEAD | |
| 1 | 36.60 ± 2.86 | 0.53 ± 0.13 |
| 2 | 22.86 ± 3.26 | - |
| 3 | 26.83 ± 2.06 | 2.80 ± 1.14 |
| 4 | - | 7.53 ± 0.81 |
| 5 | 24.56 ± 1.06 | - |
Data are means ± standard deviations for at least three independent experiments. -, not determined.
In the LEAD orientation, targeting efficiency was greater closer to the terminal region (insertion 3LEAD and 4LEAD) than in the region close to the replication origin (insertion 1LEAD). One possible explanation for these results is the replication of a percentage of the target sites, located close to the terminal region of the S. meliloti chromosome, as lagging strand templates depending on whether the clockwise or anticlockwise replication fork is the first to arrive at the terminal region.36
Discussion
We describe here the rules governing DNA target site recognition for the mobile group II intron RmInt1, which belongs to the structural subclass IIB. This intron has an IEP with no recognizable D or En domain. We used the derived rules to reprogram this intron to disrupt genes integrated at different locations on the chromosome of S. meliloti. Like mesophilic targetrons based on Ll.ltrB (IIA) and EcI5 (IIB), the IEPs of which contain D and En domains, the reprogrammed RmInt1 was able to disrupt chromosomal genes highly efficiently. However, it differed from these other targetrons in the recognition of a smaller number of nucleotides by the IEP. This increases the number of potential insertion sites and, hence, the number of retargeted RmInt1 introns that could be tested for each target gene. This work thus extends the range of mesophilic introns currently available for use in group II intron gene targeting technology.
Like other group II introns, RmInt1 recognizes its target sequence through both the IEP and base pairing between the intron RNA and the DNA target. For class IIB introns, interactions with the 5′ exon involve EBS2 and EBS1 of the intron and the 3′ exon is recognized by EBS3. We investigated the interaction EBS1/IBS1 of RmInt1 and its targets, by splitting this region in two and studying the two parts separately. However, a combined analysis of the data obtained showed that positions -5, -6, and -7 were paired in 93% of active introns and positions -1, -2, -3, and -4 were paired in 80% of inserted introns, suggesting that the pairing of the 5′ part of this region was less demanding. Nevertheless, in the presence of seven base pairs in the EBS1/IBS1 interaction in 80% of active introns indicates that the whole EBS1/IBS1 region should base pair for retrohoming. Other IIB introns, such as EcI5, the EBS1/IBS1 interactions of which involve only five nucleotides, has less stringent requirements for this interaction, as only about 50% of active introns have all five bases paired.22 Despite our finding that RmInt1 required base pairing in this region, we observed no strong selection for any specific sequence. There was some selection for G at EBS1 position -3, although this is not the nucleotide present in the wild-type intron.
For the EBS2/IBS2 region, selection experiments revealed strong selection for pairing of the nucleotides involved in the EBS2/IBS2 interaction, with 86% of the active introns displaying pairing of the five bases, a percentage similar to that reported for the EcI5 intron.22 This region displays stronger selection for a specific sequence than was observed for the EBS1/IBS1 interaction, particularly at position -9 of EBS2/IBS2, for which an A–T interaction was observed in 98% of invasion events. Replacement of the A residue in position -9 of EBS2 with a G decreased the frequency with which RmInt1 invaded its wild-type targets by two orders of magnitude, although the G–T interaction did not entirely prevent target recognition. In particular, a G–C pair was found to be selected at position -10, rather than the A–T present in the interaction of wild-type RmInt1 and its target. This preference for specific nucleotides at EBS2 may reflect constraints on the structure of the intron RNA, leading to the selection of complementary nucleotide residues in IBS2.
Selection experiments investigating the EBS3/IBS3 interaction showed that 100% of invaded targets displayed base pairing of the Watson–Crick type, with no wobble pairing observed in any invasion event. The G–C pair, corresponding to the wild-type nucleotide pair observed in this position, was selected. This selection for specific bases may indicate certain constraints on RNA structure, but the detection of only G–C pairs, with no G–T observed, indicates that other as-yet-unidentified factors probably play a role in this process.
Group II IEPs recognize sequences in both the distal 5′ and 3′ exons, but different IEPs interact with different nucleotides in different positions. Thus, for EcI5, the most strongly conserved distal 5′ exon nucleotide residues are C-18, C-17, A-15, and A-14, with A-14 the most critical of these residues. For Ll.LtrB, the most critical nucleotide residues in this region are T-23, G-21, A-20, T-19, and G-15. In the 3′ exon, for both introns, the most critical residue is T+5, which is required for endonuclease-dependent retrohoming.37 For RmInt1, the most critical positions are T-15 and, to a lesser extent, G-16, in the distal 5′exon region, and G+4 in the distal 3′exon region. The RmInt1 IEP plays a smaller role in target site recognition than the IEPs of other mesophilic targetrons, probably because it is not required to promote DNA melting or to cut the opposite strand to generate a primer for reverse transcription after intron RNA insertion.
The need for EBS pairing with the target over a total of 13 base pairs ensures sufficient specificity during target recognition by RmInt1. However, the sequence requirements of the EBSs are flexible enough to allow changes in EBS sequences, facilitating the reprogramming of RmInt1 for insertion into any target site desired. Nevertheless, off-target integration was observed for some of the highly efficient retargeted RmInt1 introns. Such off-target integrations have also been reported for the thermotargetron derived from a mobile group II intron of structural subclass IIB from the thermophilic cyanobacterium Thermosynechococcus elongatus,38 the specificity of which is also dependent principally on intron RNA base pairing. The IEP of this intron has D and En domains, but it recognizes smaller numbers of target site positions than Ll.ltrB or EcI5.39 The number of secondary RmInt1 intron insertions could be minimized by using a less stable intron donor plasmid vector or controlling intron donor expression and carefully selecting the target site.
One possible drawback is that En- introns, such as RmInt1, have a bias favoring insertion into lagging strand templates. However, the intron can also be reprogrammed to invade target sites on the leading strand template. We have also shown that RmInt1 can be retargeted for insertion into genes located anywhere in the genome, with no substantial bias. By contrast to what has been reported for LtrA,40 the bacterial RmInt1 IEP has been shown to be homogeneously distributed in S. meliloti and E. coli cells.41 It therefore seems likely that RmInt1 would behave similarly in other bacterial genomes. Finally, retargeted RmInt1 may also have the potential advantage of ensuring the control of specific processes associated with DNA replication, such as cell proliferation and differentiation. All of these characteristics of RmInt1 in target recognition make this intron suitable for gene targeting applications.
Materials and Methods
Bacterial strains, media, and growth conditions
S. meliloti strain RMO17 (RmInt1 intron-less strain) was grown at 28 °C on TY or defined minimal medium (MM).42 The E. coli DH5α and DH10B strains were used for the cloning and maintenance of plasmid constructs. The E. coli HMS174(DE3) strain carrying the T7 RNA polymerase gene was used in selection experiments with the pACE system. E. coli strains were routinely grown at 37 °C or, when specified, at 28 °C, on Luria-Bertani (LB) medium.43 Antibiotics were added to the medium as required, at the following concentrations: kanamycin at 200 μg/ml for S. meliloti and at 50 μg/ml for E. coli, ampicillin at 200 μg/ml, tetracycline at 10 μg/ml and gentamicin at 50 μg/ml for S. meliloti, and 10 μg/ml for E. coli.
Selection experiments
For selection experiments, HMS174(DE3) cells were electroporated with donor and recipient plasmids or libraries and transformants were allowed to recover in 1 ml of SOC medium at 28 °C for 1 h. Electroporated cells were diluted 1/200 in LB medium containing kanamycin plus ampicillin and grown at 28 °C overnight. The culture were then serially diluted and plated on LB agar containing tetracycline plus ampicillin or ampicillin only, for culture at 28 °C. The colonies obtained were analyzed by colony PCR and sequencing. From colonies, grown on tetracycline plus ampicillin containing medium we obtained data corresponding to “Selected.” Data denoted as “Pool” were obtained from colonies grown on medium containing ampicillin alone. Colony PCR was performed as previously described.44
The nucleotide frequency at each of the randomized positions was corrected for biases in the initial pool, as described by Zhuang et al.,22 by calculating the ratio of the frequency of each nucleotide n at that position p in the selected introns to its frequency in the initial pool as follows:
The ratio at each position was then normalized to 1, with the following equation:
where is the normalized ratio. The normalized frequencies were used to generate a sample set of 100 active target sites, displayed in “WebLogo” format.45
Plasmid library construction
The recipient plasmid libraries used in the experiments consisted of plasmids containing the minimal target site for RmInt1 -20/+518 with random bases at different positions. Targets were constructed by annealing primers and filling in the gaps with the Klenow fragment of DNA polymerase I. The annealed primers added PstI and XbaI sites, facilitating insertion into pACELAG.28 The primers used to generate the recipient plasmid library used in Figure 1 were RmInt1-5′N7 and RmInt1-3′N4; those for the IBS2 library were RmInt1 IBS2-A and RmInt1IBS-B; those for the IBS1-3′ library, IBS1R-A1 and RmInt1IBS-B; those for the IBS1-5′ library, IBS1R-A2 and RmInt1IBS-B; and those for the IBS3 library were RmInt1-IBS3-A and RmInt1-IBS3-B (Table S1).
Following their construction, the randomized donor libraries were used to electroporate E. coli DH10B cells, which were then plated on LB plus ampicillin and incubated overnight at 37 °C. The number of clones integrating randomized libraries was estimated by plating the transformed cells on LB ampicillin plates. It was found to be 1.712 × 105 for positions -20 to -14/+2 to +5; 4 × 105 for the IBS2 library; 5.3 × 104 for the IBS1-5′ library; 7.5 × 105 for the IBS1-3′ library and 1.02 × 104 for the IBS3 library.
Donor plasmid libraries were constructed in pKGEMA4T7Cm, a derivative of pKGEMA4T7.28 We digested pKGEMA4T7 with PmlI and XhoI, and the resulting fragment containing part of the RmInt1 ribozyme and the 5′ distal exon was replaced with the camR gene, yielding pKGEMA4T7Cm. This vector was used to prevent the contamination of libraries with the wild-type intron donor plasmid. The donor plasmid libraries contained randomized nucleotides at EBS2 positions -9 to -3 (EBS2 library), positions -5 to -7 of EBS1 in the EBS1-5′ library, positions -1 to -4 of EBS1 in the EBS1-3′ library and EBS3 position +1 in the EBS3 library. All the donor libraries also contained randomized nucleotides in the corresponding IBS positions within the 5′ exon or 3′ exon, to provide complementary sequences able to base pair with the randomized EBS, to ensure that the RNA could be spliced. Libraries were constructed by two-step PCR, with pKGEMA4 as the template and primers introducing the random nucleotides at specified positions. In the first step, two PCRs were performed in parallel to generate two partially overlapping PCR products. The primers used were as follow: for the EBS2 library: RmInt1-IBS2-Nsac + RmInt1-EBS2-N and RmInt1-EBS1-wt + OLAL-A; for the EBS1-3′ library, RmInt1-IBS1D-5′N + RmInt1-EBS2-wt and RmInt1-EBS1D-3′N and OLAL-A; for the EBS1-5′ library, RmInt1-IBS1D-3′N + RmInt1-EBS2-wt and RmInt1-EBS1D-5N’ + OLAL-A; for the EBS3 library Sac-Bbr + RmInt1-EBS3-wt and RmInt1-EBS3-N + RmInt1-IBS3-N (Table S1). In the second step, the two PCR products, purified by gel filtration with the MicroSpin S-300HR system (GE Healthcare), were mixed and amplified with the outer primers (underlined). The amplicons for the EBS2 and EBS1 libraries were digested with SacI and XhoI and used to replace the corresponding fragment of pKGEMA4T7Cm. The amplicon for the EBS3 library was treated in the same way, except that SacI and AvrII were the restriction enzymes used.
E. coli DH10B was electroporated with libraries and plated on LB medium supplemented with kanamycin. The number of clones in the libraries was 1.4 × 104 for the EBS2 library; 1.2 × 104 for the EBS3 library; 1.38 × 104 for the EBS1-3′ library, and 5.82 × 104 for the EBS1-5′ library.
Southern hybridization
Southern blotting was performed on total genomic DNA isolated with the Real-Pure genomic DNA extraction kit (Real-Durviz, SLU) according to the manufacturer’s protocols. We digested 2 μg of DNA with BamHI, subjected it to electrophoresis in a 1% TAE agarose gel, and vacuum blotted it onto nylon filters (Pall Corp.) according to the manufacturer’s instructions. A DNA probe for RmInt1 was obtained by PCR amplification in the presence of digoxigenin-11-dUTP (DIG; Roche), with pKGEMA4 as the template and the Epsilon and 18R0 primers. Hybridization was performed at high stringency; the membranes were washed, and hybridization signals were detected according the manufacturer’s instructions (Roche).
Retargeting RmInt1 for insertion into the lacZ and sacB genes
The RmInt1-ΔORF intron was retargeted by a two-step PCR, with the plasmids pKGEMA4-A, -T, -C, and -G as the template. These plasmids are variants of pKGEMA446 in which EBS3 is an A, T, C, or G residue and IBS3 is the complementary nucleotide. The plasmid chosen as the template depended on the EBS3 nucleotide, which was chosen so as to be complementary to the IBS3 of the target sequence. In the first step, two independent PCRs were performed, to amplify two partially overlapping segments. In one of these PCRs, primers P1 and P2 were used. These primers introduced the necessary modifications into IBS2 and IBS1 (primer P1) and EBS2 (primer P2). The second PCR used primer P3 to modify EBS1 and a fixed primer (OLAL-A) for all retargeted introns. The products of these two PCRs were gel-purified and mixed for use as the template for the second-step PCR with primers P1 and OLAL-A. This final fragment was gel-purified, digested with SacI and XhoI, and inserted into the pGmS4S-A, -T, -C, and -G vectors digested with the same enzymes. pGmS4S-A, -T, -C, and -G are derivatives of pBBR1MCS-547 equivalent to pKGEMA4-A, -T, -C, and -G but with a gentamicin resistance marker instead of a kanamycin resistance marker and the RmInt1-ΔORF expressed under the control of the Psyn promoter48 rather than the kanamycin promoter.
Integration of lacZ into the S. meliloti RMO17 chromosome
We inserted the lacZ gene under the control of the Psyn promoter, as a XbaI fragment, into the XbaI site of pK18mobsacB.49 We introduced a DNA fragment homologous to the chromosomal region into which we wished to insert lacZ into the PstI site of this plasmid. These fragments were amplified by PCR, with total DNA from RMO17 as the template and the following primers: for integration into the region close to the replication origin, primers 1ins-A and 1ins-B; for the second integration, 2ins-A and 2ins-B; for the third integration, 3ins-A and 3ins-B; for the fourth integration, 4ins-A and 4ins-B, and for the fifth integration, 5ins-A and 5ins-B (Table S1). All insertions took place in intergenic regions according to S. meliloti 1021 chromosome sequence. Primers were designed for region with synteny between 1021, AK83, Bl225C, SM11, and GR4 chromosomes. Plasmids were transferred into S. meliloti RMO17 by triparental mating. Selected clones were Kmr, LacZ+, and sensitive to sucrose.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by research grants CSD 2009-0006 from the Consolider-Ingenio program, and BIO2011-24401 from the Ministerio de Economía y Competitividad, including ERDF (European Regional Development Funds).
Glossary
Abbreviations:
- RNP
ribonucleoprotein
- IEP
intron-encoded protein
- RT
reverse transcriptase
- ORF
open reading frame
- EBS
exon-binding site
- IBS intron-binding site
References
- 1.Sharp PA. On the origin of RNA splicing and introns. Cell. 1985;42:397–400. doi: 10.1016/0092-8674(85)90092-3. [DOI] [PubMed] [Google Scholar]
- 2.Cech TR. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell. 1986;44:207–10. doi: 10.1016/0092-8674(86)90751-8. [DOI] [PubMed] [Google Scholar]
- 3.Cavalier-Smith T. Intron phylogeny: a new hypothesis. Trends Genet. 1991;7:145–8. doi: 10.1016/0168-9525(91)90102-V. [DOI] [PubMed] [Google Scholar]
- 4.Eickbush TH. Origins and evolutionary relationships of retroelements. In: Morse SS, editor. The evolutionary biology of viruses. New York, NY: Raven Press, Inc.; 1994. p.121-157. [Google Scholar]
- 5.Michel F, Ferat JL. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–61. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 6.Dai L, Zimmerly S. ORF-less and reverse-transcriptase-encoding group II introns in archaebacteria, with a pattern of homing into related group II intron ORFs. RNA. 2003;9:14–9. doi: 10.1261/rna.2126203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toro N. Bacteria and Archaea Group II introns: additional mobile genetic elements in the environment. Environ Microbiol. 2003;5:143–51. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- 8.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 9.Toro N, Jiménez-Zurdo JI, García-Rodríguez FM. Bacterial group II introns: not just splicing. FEMS Microbiol Rev. 2007;31:342–58. doi: 10.1111/j.1574-6976.2007.00068.x. [DOI] [PubMed] [Google Scholar]
- 10.Michel F, Costa M, Westhof E. The ribozyme core of group II introns: a structure in want of partners. Trends Biochem Sci. 2009;34:189–99. doi: 10.1016/j.tibs.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Mohr G, Perlman PS, Lambowitz AM. Evolutionary relationships among group II intron-encoded proteins and identification of a conserved domain that may be related to maturase function. Nucleic Acids Res. 1993;21:4991–7. doi: 10.1093/nar/21.22.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.San Filippo J, Lambowitz AM. Characterization of the C-terminal DNA-binding/DNA endonuclease region of a group II intron-encoded protein. J Mol Biol. 2002;324:933–51. doi: 10.1016/S0022-2836(02)01147-6. [DOI] [PubMed] [Google Scholar]
- 13.Zimmerly S, Hausner G, Wu Xc Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–50. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toro N, Molina-Sánchez MD, Fernández-López M. Identification and characterization of bacterial class E group II introns. Gene. 2002;299:245–50. doi: 10.1016/S0378-1119(02)01079-X. [DOI] [PubMed] [Google Scholar]
- 15.Simon DM, Clarke NA, McNeil BA, Johnson I, Pantuso D, Dai L, Chai D, Zimmerly S. Group II introns in eubacteria and archaea: ORF-less introns and new varieties. RNA. 2008;14:1704–13. doi: 10.1261/rna.1056108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toro N, Martínez-Abarca F. Comprehensive phylogenetic analysis of bacterial group II intron-encoded ORFs lacking the DNA endonuclease domain reveals new varieties. PLoS One. 2013;8:e55102. doi: 10.1371/journal.pone.0055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh NN, Lambowitz AM. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J Mol Biol. 2001;309:361–86. doi: 10.1006/jmbi.2001.4658. [DOI] [PubMed] [Google Scholar]
- 18.Jiménez-Zurdo JI, García-Rodríguez FM, Barrientos-Durán A, Toro N. DNA target site requirements for homing in vivo of a bacterial group II intron encoding a protein lacking the DNA endonuclease domain. J Mol Biol. 2003;326:413–23. doi: 10.1016/S0022-2836(02)01380-3. [DOI] [PubMed] [Google Scholar]
- 19.Robart AR, Seo W, Zimmerly S. Insertion of group II intron retroelements after intrinsic transcriptional terminators. Proc Natl Acad Sci U S A. 2007;104:6620–5. doi: 10.1073/pnas.0700561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa M, Michel F, Westhof E. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 2000;19:5007–18. doi: 10.1093/emboj/19.18.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toor N, Robart AR, Christianson J, Zimmerly S. Self-splicing of a group IIC intron: 5′ exon recognition and alternative 5′ splicing events implicate the stem-loop motif of a transcriptional terminator. Nucleic Acids Res. 2006;34:6461–71. doi: 10.1093/nar/gkl820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhuang F, Karberg M, Perutka J, Lambowitz AM. EcI5, a group IIB intron with high retrohoming frequency: DNA target site recognition and use in gene targeting. RNA. 2009;15:432–49. doi: 10.1261/rna.1378909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y, McClane BA, Fisher DJ, Rood JI, Gupta P. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl Environ Microbiol. 2005;71:7542–7. doi: 10.1128/AEM.71.11.7542-7547.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA. 2006;12:1271–81. doi: 10.1261/rna.68706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zarscheler K, Janesch B, Zayni S, Schäffer C, Messner P. Construction of a gene knockout system for application in Paenibacillus alvei CCM 2051T exemplified with the initiation enzyme WsfP of S-layer glycan biosynthesis. Appl Environ Microbiol. 2009;75:3077–85. doi: 10.1128/AEM.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol. 2001;19:1162–7. doi: 10.1038/nbt1201-1162. [DOI] [PubMed] [Google Scholar]
- 27.Yao J, Lambowitz AM. Gene targeting in gram-negative bacteria by use of a mobile group II intron (“Targetron”) expressed from a broad-host-range vector. Appl Environ Microbiol. 2007;73:2735–43. doi: 10.1128/AEM.02829-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.García-Rodríguez FM, Barrientos-Durán A, Díaz-Prado V, Fernández-López M, Toro N. Use of RmInt1, a group IIB intron lacking the intron-encoded protein endonuclease domain, in gene targeting. Appl Environ Microbiol. 2011;77:854–61. doi: 10.1128/AEM.02319-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrientos-Durán A, Chillón I, Martínez-Abarca F, Toro N. Exon sequence requirements for excision in vivo of the bacterial group II intron RmInt1. BMC Mol Biol. 2011;12:24. doi: 10.1186/1471-2199-12-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perutka J, Wang W, Goerlitz D, Lambowitz AM. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol. 2004;336:421–39. doi: 10.1016/j.jmb.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 31.Durbin F, Eddy S, Krogh A, Mitchison G. Biological sequence analysis. Probabilistic models of proteins and nucleic acids. Cambridge, UK: Cambridge University Press; 1998. [Google Scholar]
- 32.Galibert F, Finan TM, Long SR, Pühler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–72. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 33.Martínez-Abarca F, Barrientos-Durán A, Fernández-López M, Toro N. The RmInt1 group II intron has two different retrohoming pathways for mobility using predominantly the nascent lagging strand at DNA replication forks for priming. Nucleic Acids Res. 2004;32:2880–8. doi: 10.1093/nar/gkh616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selbitschka W, Niemann S, Pühler A. Construction of gene replacement vectors for Gram- bacteria using a genetically modified sacRB gene as a positive selection marker. Appl Microbiol Biotechnol. 1993;38:615–8. doi: 10.1007/BF00182799. [DOI] [Google Scholar]
- 35.Zhong J, Karberg M, Lambowitz AM. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res. 2003;31:1656–64. doi: 10.1093/nar/gkg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duggin IG, Wake RG, Bell SD, Hill TM. The replication fork trap and termination of chromosome replication. Mol Microbiol. 2008;70:1323–33. doi: 10.1111/j.1365-2958.2008.06500.x. [DOI] [PubMed] [Google Scholar]
- 37.Mohr G, Smith D, Belfort M, Lambowitz AM. Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev. 2000;14:559–73. [PMC free article] [PubMed] [Google Scholar]
- 38.Mohr G, Hong W, Zhang J, Cui GZ, Yang Y, Cui Q, Liu YJ, Lambowitz AM. A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum. PLoS One. 2013;8:e69032. doi: 10.1371/journal.pone.0069032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohr G, Ghanem E, Lambowitz AM. Mechanisms used for genomic proliferation by thermophilic group II introns. PLoS Biol. 2010;8:e1000391. doi: 10.1371/journal.pbio.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, Lambowitz AM. A bacterial group II intron-encoded reverse transcriptase localizes to cellular poles. Proc Natl Acad Sci U S A. 2005;102:16133–40. doi: 10.1073/pnas.0507057102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nisa-Martínez R, Laporte P, Jiménez-Zurdo JI, Frugier F, Crespi M, Toro N. Localization of a bacterial group II intron-encoded protein in eukaryotic nuclear splicing-related cell compartments. PLoS ONE. 8:e84056. doi: 10.1371/journal.pone.0084056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertsen BK, Aiman P, Darvill AG, McNeil M, Alberstein P. The structure of acidic extracellular polysaccharides secreted by Rhizobium leguminosarum and Rhizobium trifolii. Plant Physiol. 1981;67:389–400. doi: 10.1104/pp.67.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1989. [Google Scholar]
- 44.Hofmann MA, Brian DA. Sequencing PCR DNA amplified directly from a bacterial colony. Biotechniques. 1991;11:30–1. [PubMed] [Google Scholar]
- 45.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nisa-Martínez R, Jiménez-Zurdo JI, Martínez-Abarca F, Muñoz-Adelantado E, Toro N. Dispersion of the RmInt1 group II intron in the Sinorhizobium meliloti upon acquisition by conjugative transfer. Nucleic Acids Res. 2007;35:214–22. doi: 10.1093/nar/gkl1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–6. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 48.Giacomini A, Ollero FJ, Squartini A, Nuti MP. Construction of multipurpose gene cartridges based on a novel synthetic promoter for high-level gene expression in gram-negative bacteria. Gene. 1994;144:17–24. doi: 10.1016/0378-1119(94)90197-X. [DOI] [PubMed] [Google Scholar]
- 49.Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.