Abstract
Protein crystallography is used to generate atomic resolution structures of protein molecules. These structures provide information about biological function, mechanism and interaction of a protein with substrates or effectors including DNA, RNA, cofactors or other small molecules, ions and other proteins. This technique can be applied to membrane proteins resident in the membranes of cells. To accomplish this, membrane proteins first need to be either heterologously expressed or purified from a native source. The protein has to be extracted from the lipid membrane with a mild detergent and purified to a stable, homogeneous population that may then be crystallized. Protein crystals are then used for X-ray diffraction to yield atomic resolution structures of the desired membrane protein target. Below, we present a general protocol for the growth of diffraction quality membrane protein crystals. The process of protein crystallization is highly variable, and obtaining diffraction quality crystals can require weeks to months or even years in some cases.
INTRODUCTION
Overview
Beginning with the structure of myoglobin 50 years ago1, X-ray crystallography has been used to generate atomic models of proteins that inform scientists about their structure and function. However, it was not until the photosynthetic reaction center was determined in 1985 that the structure of an integral membrane protein extracted from its natural source was determined at atomic resolution2. It was over a decade more before a membrane protein obtained from heterologous expression, the potassium channel KcsA3, was crystallized and its structure solved. The relative difficulty for obtaining membrane protein structures is largely a result of the difficulties associated with generating the milligram quantities of pure, monodisperse membrane protein generally required for crystallization4. Furthermore, it is imperative that the protein is stable in a discrete fold and oligomeric state over the lifetime of crystallization, which may be hours to over a year, in some cases. Overwhelmingly, we find that the greatest predictor of success in crystallization is the preparation of a pure, homogeneous and stable protein solution. Based empirically, we have found useful criteria to be > 98% pure, > 95% homogeneous and > 95% stable when stored unconcentrated at 4 °C for 2 weeks or when stored concentrated (i.e., the concentration used for crystallization experiments) at 4 °C for 1 week. Typically, 2 mg of protein meeting these criteria per preparation of protein is a useful starting point for crystallization screening, i.e., a final purified protein sample of 200 µl at 10mg ml−1 protein concentration. The protocol described here is aimed at meeting these criteria as the requisite for crystallization.
The protocol begins with heterologous expression of the target protein and then proceeds through cell membrane isolation, solubilization, purification, identification of conditions suitable for crystallization of the target membrane protein and the refinement of these conditions to improve crystal size and quality. Protocols for membrane protein crystallography are most often found accompanying the publication of specific targets. In these instances, the scope of a given protocol is generally limited to one particular protein. Even when extremely detailed, these protocols rarely give insight into the evolution of the protocol, failing to explain how the conditions for expression, solubilization, purification or crystallization were arrived at. Alternatively, there are general reviews that provide excellent overviews of the process of membrane protein crystallization. These works, however, do not usually serve as practical guides to direct crystallographers at the bench. The protocol described here is intended to provide enough experimental detail to serve as a practical guide. At the same time, we constructed the protocol to be adaptable to different protein systems by identifying potential impedances on the path to obtaining membrane protein crystals as well as emphasizing variables that should be optimized to overcome these obstacles.
Applications and limitations
This protocol focuses on the crystallization of membrane proteins heterologously expressed in Escherichia coli. However, bacterial expression systems may be unsuitable for expressing membrane proteins when the system cannot provide the folding machinery, post-translational modifications or specific lipid environment5 required for functional protein production. Although generally more expensive and more difficult to manipulate than E. coli, alternative expression systems, especially for producing eukaryotic membrane protein targets, may be better suited to meet these needs. Pichia pastoris yeast6–9, Saccharomyces cerevisiae yeast10,11, and Sf9 insect cells12 have all been used to produce membrane proteins for X-ray crystal structures. In addition, human embryonic kidney (HEK293S GnTI−) cells that can be grown in suspension cultures have great potential for expressing eukaryotic membrane proteins13,14. Regardless of what expression system is used, the same purification and crystallization principles apply. In that respect, the protocol presented here can be applied to membrane proteins expressed in systems other than E. coli. In these cases, the membrane fraction from the alternate expression system should be isolated and the protocol should be started from the Screening detergents procedure given in Box 1.
BOX 1 | SCREENING FOR AN APPROPRIATE DETERGENT FOR PROTEIN SOLUBILIZATION ●TIMING 2 D.
-
Thaw the frozen membranes obtained in Step 16 on ice.
▲ CRITICAL STEP All steps should be carried out on ice or at 4 °C. All buffers should be maintained at 4 °C.
To determine the appropriate concentration of membranes in suspension, remove a 10-µl sample of the resuspended membrane from Step 16 of PROCEDURE.
-
Make 10 twofold serial dilutions (i.e., 1×, (1/2) ×, (1/4)×… (1/512)×) of the resuspended membrane. Load the samples and molecular weight standards on a 4–20% (wt/vol) Tris-Glycine gel and run in SDS-PAGE gel running buffer.
▲ CRITICAL STEP Do not boil the samples before loading on the gel. All samples should be stored at 4 °C until just before running of the gel. At this point, the 2× SDS protein loading dye should be added.
Transfer the gels with a Trans-Blot SD semidry electrophoretic transfer cell according to the manufacturer’s instructions.
Blot the membranes with an anti-His antibody conjugated with horseradish peroxidase according to the manufacturer’s instructions and visualize with the SuperSignal West Pico Chemiluinescent Substrate.
Identify the most dilute sample that still produces a signal by western blotting. Dilute the membranes so that they are eight times more concentrated than the weakest signal on the western blot. This ensures that 25% of the target protein will be detectable by western blot if solubilized by a particular detergent.
On ice, aliquot 150 µl of the resuspended membrane from Step 22 fraction into six 1.5-ml ultracentrifuge tubes.
-
To each of the tubes, add 150 µl of one of the following solubilization buffers: OG, DDM, LDAO, CHAPS, FC-12 and SDS (see REAGENT SETUP).
▲ CRITICAL STEP To avoid the production of foam, do not vortex the solubilization mixtures at any time, mix by gently pipetting up and down.
Add a magnetic stir bar (7 mm × 2 mm) and agitate the mixtures using a magnetic stir plate for 12–18 h at 4 °C. The amount of time necessary for solubilization may be far less than this and can be optimized down to as little as 1 h once an appropriate detergent has been selected.
Remove the magnetic stir bar from the solubilization mixture. Take a 10-µl sample for analysis by SDS-PAGE and store at 4 °C.
To pellet the unsolubilized material, centrifuge the remainder of the sample for 30 min at 100,000g and 4 °C.
Remove the supernatant from each tube taking care not to disturb the pellet and transfer it to another clean, chilled 1.5-ml tube.
Mix the contents of each tube by gentle pipetting up and down. Take a 10-µl sample for analysis by SDS-PAGE.
-
Run a gel of each sample from Steps 10 and 13 and perform a western blot. Refer to Step 42 of the protocol for details on gel running and western blot conditions. Refer to the section on solubilization and Figure 2c to guide selection of the most suitable detergent.
? TROUBLESHOOTING
The expression and purification of a functional membrane protein as described here can be valuable for experiments other than crystallization. The purified membrane protein generated using this protocol can be used for structural analysis by other methods, such as electron microscopy15 or, with some modification of the protocol during expression, nuclearmagnetic resonance16–19. In addition, a variety of biochemical and functional studies use pure membrane protein reconstituted into proteoliposomes20,21 or planar lipid bilayer systems22. These types of analyses complement crystallography and provide an opportunity to study membrane proteins in isolated systems. Alternatively, the membrane fraction itself can be used in binding assays23,24. Finally, information about the oligomeric state, size, shape and fold of the protein can be obtained through biophysical methods, such as analytical ultracentrifugation25, small angle X-ray scattering26–28, size exclusion chromatography (SEC) with multiple detectors29 and circular dichroism30,31.
Experimental design
General considerations
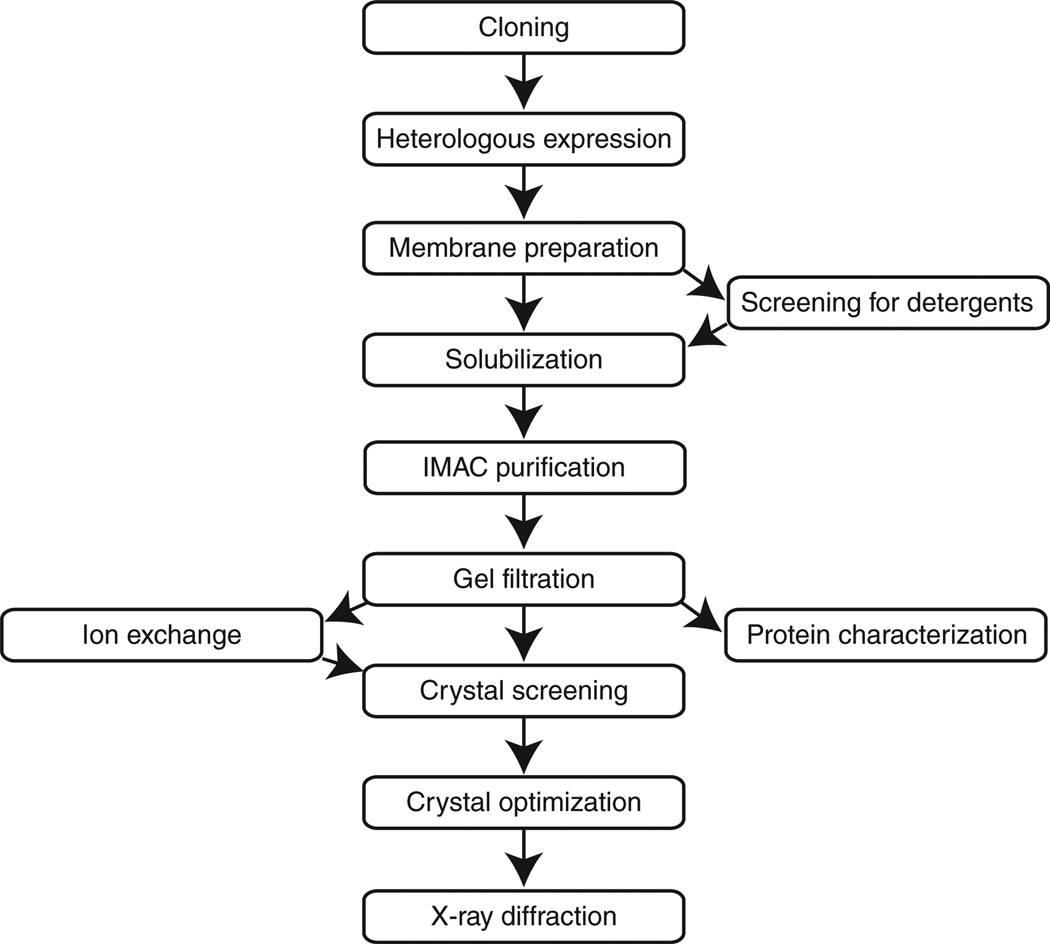
The pathway to high-resolution membrane protein crystals (Fig. 1) is inherently an iterative one. At every step in the process, information is obtained to improve the purity, stability and homogeneity of the subsequent preparations of the target protein, as well as to guide optimization of crystal growth conditions. The general protocol presented here has been used successfully by our group for the crystallization of several membrane proteins32–38. To make the process of generating such a protocol more transparent, some sections are accompanied by additional text in the Experimental design that highlights key parameters that require optimization. In some cases, alternatives to the methods presented are also provided. There are many excellent overviews of membrane protein crystallography that can be consulted for a comprehensive discussion of the method4,39–44.
Figure 1.
Workflow for generating membrane protein crystals. Ion-exchange chromatography can be incorporated into the process but is not absolutely necessary for purification. Screening for a suitable detergent for solubilization is not performed for every protein preparation but rather typically only once at the beginning of efforts to purify and crystallize a membrane protein.
Designing and cloning constructs
As immobilized metal-affinity chromatography (IMAC) is an effective first step in protein purification45,46, constructs are cloned with a cleavable N- or C-terminal poly-histidine tag. If a protein is predicted to have a signal sequence, N-terminal tags should be avoided; the tag may interfere with proper targeting of the protein or be cleaved during signal sequence processing. The pET E. coli expression vectors that are T7 RNA polymerase promoter driven47 and isopropyl-β-d-thiogalactopyranoside (IPTG) inducible are useful for the generation of expression constructs47. Alternatively, the pBAD vector system for E. coli expression uses arabinose induction48 and has been implemented successfully for the production of membrane proteins for X-ray studies49. These plasmids can encode either an N- or C-terminal thrombin-cleavable 6 × histidine tag. There is evidence that adding an N-terminal fusion protein can increase the amount of protein heterologously expressed in the membrane. Examples include maltose-binding protein, glutathione-S-transferase50, the PelB leader sequence51 and the membrane-integrating sequence for translation of integral membrane constructs (Mistic)52.
Expression system
Escherichia coli is traditionally the system of choice for structural biologists to heterologously produce protein for X-ray studies. It is inexpensive, easy to manipulate (e.g., transform, culture and harvest protein from) and is capable of producing milligram quantities of protein from culture sizes on the order of tens of liters53. Also, a few published structures indicate that E. coli can be used to produce not only prokaryotic membrane proteins for structural studies, but also eukaryotic membrane proteins38,54. To aid in membrane protein expression, a strain was selected by Miroux and Walker55 on the basis of its ability to overproduce membrane proteins. This is the strain predominantly used in our laboratory for expression of membrane proteins. In addition, E. coli is amenable to growth in a variety of temperatures and media conditions that can affect the quality and quantity of the protein being heterologously expressed56. A reduction in the growth temperature or change in media conditions, for example, can alter the expression product from being an improperly folded aggregate (inclusion bodies) to being correctly folded and inserted into the membrane.
Membrane preparations
We routinely isolate the membrane fraction of E. coli during protein preparations; although this is not absolutely necessary57, it is strongly recommended. Ideally, immediately after harvesting, the cells are lysed and the membranes are isolated by centrifugation. In addition to enriching for membrane proteins, this separates the membrane fraction from the soluble proteins that can degrade target proteins through proteolysis. As the target protein is in the membrane pellet, it does not need to be purified away from the soluble components, as they are discarded in the supernatant after the membrane fraction is harvested by centrifugation.
Solubilization
For purification and crystallization, membrane proteins need to be extracted from the lipid membrane in which they were expressed with a detergent. For most expression systems, this extraction is performed on the isolated membrane fraction. However, in HEK293S cells, membrane proteins can be extracted from whole cells using detergent14. Whether solubilizing from membranes or from whole cells, the goal is to yield a water-soluble protein–detergent–lipid complex (PDLC) (Fig. 2a). The identification of the detergent most suitable for a particular protein target is an empirical process. A variety of different classes of detergents have been used in membrane protein crystallography, although some, such as the maltosides and glucosides, have been used more often than others (Fig. 2b)58,59. Most of these detergents are available with a variety of hydrocarbon chain lengths, allowing fine-tuning of the size and stability of the PDLC41.
Figure 2.
Detergent solubilization of membrane proteins. (a) Schematic of the solubilization process. From left to right: free detergent monomers (a) associate to form detergent micelles (b) at concentrations above the CMC. When added to a membrane preparation (c), the micelles extract membrane proteins from the lipid bilayer yielding a solution containing PDLC complexes, free lipid-detergent micelles and detergent monomers (d). (b) Some common detergents used in solubilization, purification and crystallization of membrane proteins. (c) A western blot of E. coli proteins YiaA and YagU. The supernatant of a solubilization before (bs) and after (as) a high-speed spin that pellets unsolubilized material. YiaA is extracted nearly quantitatively, whereas only a portion of YagU is extracted from the lipid bilayer. Reprinted with permission from ref. 86.
The ability of a particular detergent to solubilize a membrane protein can be assessed by comparing the amount of the target protein present in the solubilized membrane fraction before the high-speed spin with the amount of protein present in the supernatant after the spin. Unsolubilized material will be pelleted during this spin. The material that remains in the supernatant is detergent-solubilized protein and can be taken through purification, characterization and crystallization. Therefore, a western blot of samples from the supernatant before and after the high-speed spin will give an indication of how much material was retained in the supernatant (i.e., solubilized). On comparison, the intensity of the band corresponding to the target protein in the before-spin sample will be the same as the corresponding band in the after-spin sample in the instance where the majority of a target is solubilized (Fig. 2c). The ideal detergent extracts all of the membrane protein target from the membrane, maintains the native fold of the protein and forms a PDC that is stable throughout purification and crystallization, as determined by analytical SEC. Less than complete extraction (Fig. 2c) is acceptable if the solubilized protein is stable throughout the purification and crystallization steps.
The concentration of detergent required for solubilization will depend, in part, on the critical micelle concentration (CMC) of the particular detergent, the concentration at which detergent monomers begin to self-associate to form micelles60. Concentrations well above the CMC are usually used for solubilization to ensure that a sufficient amount of detergent is present to saturate the bilayer, disrupt the bilayer and form micelles of lipid and detergent to extract the target protein from the membrane. The concentration of detergent will also depend on the protein system being studied. The ideal concentration of detergent in the solubilization, purification and/or crystallization buffers are empirically determined. Some suggestions for concentrations of commonly used detergents are listed in REAGENT SETUP.
It is not necessary to use the same detergent throughout the entirety of a crystallization experiment. The detergent used for solubilization does not necessarily need to be the same one used for purification and crystallization. A longer chain detergent used to extract the protein from the lipid membrane can be exchanged for a shorter chain detergent during the first step of affinity purification. The longer chain detergent may help to recover a greater percentage of the membrane protein from the lipid bilayer, whereas the shorter chain detergent may help to form a more compact PDC that is more amenable to crystallization. In addition, it may be beneficial to have a mixed detergent micelle composed of multiple detergents and/or lipids61. There are even examples of proteins in bicelles composed of detergent and lipids62. More in-depth discussions of detergents in solubilization, purification and crystallization can be found elsewhere40–44,60,63.
Immobilized metal affinity chromatography
Immobilized metal-affinity chromatography is an efficient, relatively inexpensive method for the first step in a purification protocol45,46. During the process of obtaining membrane protein crystals, IMAC will inevitably be performed multiple times. Slight adjustments in the number of imidazole wash steps and the concentration of imidazole in each wash step should be made to maximize the yield and purity of the target protein obtained after this first step of purification. Careful monitoring of each fraction by western blotting is necessary to ensure that the target protein is bound to the metal affinity resin and also that the protein is not being eluted prematurely in the imidazole wash steps. In the initial purifications performed on a membrane protein target, the protein purified by IMAC should be carried through the cleavage and size-exclusion steps of the protocol regardless of low purity or poor yield. Valuable information can be obtained from these subsequent steps despite the presence of contaminant proteins or limited amounts of material.
Cleavage of the affinity tag
There are a few added considerations for membrane proteins when cleaving an engineered affinity tag by proteolysis. First, the presence of the detergent micelle surrounding the target protein may occlude access to the cleavage site by the protease. Second, there is evidence that the presence of certain detergents may inhibit the activity of various proteases64. The former complication, if encountered, can be overcome by placing the tag at the opposite terminus of the protein or by adding a linker sequence to serve as a spacer between the cleavage site and the protein. The latter complication can sometimes be overcome by increasing the recommended amount of protease by as much as 5–10 times. Alternatively, the gene can be cloned into a vector that uses an alternate cleavage site specific for a protease that is not affected by the presence of the detergent being used.
Size-exclusion chromatography
Size-exclusion chromatography is useful for further purification of proteins after IMAC and cleavage of the affinity tag. Every membrane protein crystallized in our laboratory has been subject to SEC during purification32–38 (Table 1). SEC is also a powerful tool in assessing the homogeneity, stability and purity of a protein target. The retention time and shape of the chromatogram provides information about the oligomeric state and dispersion of the PDC (Fig. 3). A sharp, Gaussian peak with a retention time that corresponds to the mass of the correct biological oligomeric state is ideal. Although making a protein meet these criteria does not absolutely prove homogeneity, it is an excellent indicator65–68. The oligomeric state can be inferred by comparing the retention time of the target protein–detergent complex to those of size-exclusion molecular weight standards. Some caution needs to be exercised when performing this, as standards for size-exclusion columns are soluble proteins or small molecules, whereas membrane proteins are in complex with detergent molecules that may potentially expand the hydrodynamic radius60. As size-exclusion chromatography separates proteins on the basis of their hydrodynamic radius, membrane proteins can run at molecular weights higher than would be predicted.
TABLE 1.
Protein expression, purification and crystallization data.
| Protein | IMAC | Expression (mg liter −1) |
Size exclusion | Ion exchange | Detergent | Precipitant | PDB code | Resolution (Å) |
|---|---|---|---|---|---|---|---|---|
| GlpF | Nickel | 7 | TSK-GEL G3000SW | NA | OG | PEG 2000 | 1FX8 | 2.20 |
| AQPZ | Nickel | 10 | Superose 12 | NA | OG | PEGMME 2000 | 1RC2 | 2.50 |
| AQP0 | NA | NA | TSK-GEL G3000SW | Cation | OG | PEG 1000 | 1YMG | 2.24 |
| AmtB | Nickel | 3 | TSK-GEL G3000SW | NA | OG | PEG 400 | 1U7G | 1.35 |
| AQPM | Nickel | 0.2 | Superdex200 | NA | OG | PEG 4000 | 2F2B | 1.68 |
| AmtB/GlnK | Nickel | 3 | TSK-GEL G3000SW | NA | OG | PEGMME 550 | 2NS1 | 1.96 |
| PfAQP | Nickel | 0.2 | TSK-GEL G3000SW | NA | OG | PEGMME 2000 | 1C02 | 2.05 |
NA, not applicable; PEG, polyethylene glycol. PDB, reference code for protein structures deposited in the RCSB Protein Data Bank.
Figure 3.
Size-exclusion chromatography. (a) Protein eluted near the void limit of the size-exclusion column. This generally occurs when a protein is aggregated or misfolded. (b) Protein eluting from size exclusion in multiple peaks. This occurs when the PDC exists in multiple oligomeric states. (c) The human aquaporin channel, hAQP4, when initially purified on size exclusion (red) and after 6 d at 4 °C in concentrated form (blue). The broadening of the peak on the left-hand side (the appearance of ‘shoulders’) indicates that the protein forms higher oligomeric states in the concentrated form over time when stored at 4 °C. (d) hAQP4 when initially purified on size exclusion (red) and after 6 d at 4 °C in concentrated form (blue) once buffer conditions have been optimized as compared with c. The major peak containing the target protein detergent complex has the same shape and retention time in the initial purification and after 6 d.
A final consideration is that the matrix composing the size-exclusion column is not inert. The PDC may interact with the column matrix, altering the retention time of the protein on the column. The basis of this interaction can be either electrostatic or hydrophobic in nature and may occur between the matrix and either the protein or the detergent components of the PDC42. Although the presence of salt in the size-exclusion buffer helps to reduce potential electrostatic interactions between the PDC and the matrix, having different size-exclusion columns in hand that are composed of differing matrices can be helpful in overcoming these problems.
Ion-exchange chromatography
Ion-exchange chromatography is a purification method that separates molecules on the basis of their charge state at a given pH. Similar to SEC, IEC is a useful purification method after IMAC and cleavage of the affinity tag. A target protein that is stable in its native tertiary and quaternary structure can be expected to be present in a single, median overall charge state and therefore will ideally elute from the IEC column in a single Gaussian peak. IEC can be used as a final purification step after IMAC and SEC. Alternatively, it can replace size exclusion as the second and final step in the purification process. If the latter route is chosen, analytical amounts of the protein should still be injected onto a size-exclusion column as a further assessment of the quality of the protein.
Ion exchange is also useful for concentrating the protein sample. A sample containing protein at a low concentration in a large volume can be loaded onto the column and then eluted in a single, more concentrated fraction. In addition, detergents can be exchanged at this stage. While the protein is immobilized on the resin, it can be extensively washed with a buffer containing an alternative detergent.
Concentration and dialysis
To achieve the supersaturated solutions necessary for successful crystallization experiments, the protein solution needs to be concentrated; this should be carried out after the purification steps. The stability of the protein as a function of concentration should be monitored by analytical SEC. There are a few considerations when concentrating solutions of pure PDC. The most important is that during concentration of the protein, free detergent micelles that are large enough will be retained with the PDC. This accumulation of free micelles will increase the amount of phase separation that occurs in the crystallization trials, which can inhibit crystal formation.
Dialysis of the concentrated protein against a buffer that contains the minimal amount of detergent necessary to keep the protein in solution may also reduce the amount of excess detergent present in the protein sample used for crystallization. Dialysis is not always an absolute requirement for a membrane protein to crystallize. However, given the possibility of detergent micelles accumulating with the protein during concentration, it is advantageous to dialyze the protein to start with a known concentration of detergent before crystallization. It is important to note that detergents dialyze to equilibrium at different rates. Detergents that have an extremely low CMC (e.g., N-dodecyl-β-d-maltopyranoside) will not dialyze as easily as those with a higher CMC (e.g., octyl-β-d-glucopyranoside).
Phase separation in crystallization trials
The presence of detergent in the purification and crystallization buffers can complicate crystallization efforts. The tendency of detergent micelles to partition out of aqueous solution at high enough precipitant concentrations can introduce a separate detergent-rich phase in the crystallization drop60,63. This phenomenon, called phase separation, can inhibit crystal formation or cause variable results from trial to trial. This variability will hamper efforts to optimize or reproduce crystallization conditions. Efforts should be made to minimize significant phase separation observed in the drop during crystallization.
There are a few key areas where excess detergent can accumulate in the protein sample resulting in unnecessary phase separation during crystallization. The first is during size exclusion. Although not totally homogeneous, free detergent micelles in the purification buffer will elute in a single peak from the size-exclusion column69. If the peak containing the free detergent micelles overlaps with the peak containing the target PDC, there will be an enrichment of detergent in the protein sample leading to greater phase separation in crystal trials. It can be difficult to determine if this overlap of peaks is occurring, as most detergents do not absorb at 280 nm, the wavelength monitored during protein purification. Accumulation of free detergent micelles can also occur when concentrating the protein before crystallization. Regardless of the type of concentration device being used (centrifugal filter device or a stirred ultra-filtration cell), there are generally a variety of molecular weight cutoffs available to choose from. To avoid concentration of detergent micelles during this step, use a concentrator with the largest molecular weight cutoff possible that will retain the target protein. Frequently interrupting the concentration process to mix the protein solution by gentle pipetting can also help to minimize the accumulation of free micelles during protein concentration with a centrifugal filter device.
Sparse matrix screening
If there is no previous information about the crystallization of the target protein (or proteins closely related to the target protein), the hanging-drop vapor diffusion technique is used in a 96-well format on a 200–300 nanoliter scale. The goal of the initial screen is to identify a buffer condition from a sparse matrix screen that will yield crystals of the target PDC. Sparse matrix screens allow sampling of a broad range of conditions that have been selected on the basis of previous success in producing protein crystals suitable for X-ray diffraction studies70. The use of a robot to dispense crystal drops on a small scale makes screening less labor intensive and requires less protein sample than setting drops by hand on a microliter scale. In addition to sparse matrix screening, screens that are more systematic with respect to pH as well as precipitant type and concentration can be used.
We use a number of commercially available screens: MemStart, MemSys, MemGold, CP-Custom-IV and the Classics, PEGs, PEGsII, MbClass and MbClass II Suites. MemStart and MemSys are screens that contain 48 conditions each and can be combined to form a 96-condition screen. MemStart is a sparse matrix screen, whereas MemSys is a systematic exploration of pH, salt concentration/type and precipitant concentration/type. MemGold is a rationally designed 96-well screen based on the conditions that have successfully generated α-helical membrane protein crystals used to solve X-ray structures59. All of the screens besides Classics are heavily biased toward polyethylene glycol (PEG) precipitants because PEG is overwhelmingly the most successful precipitating agent in membrane protein crystallization experiments58. The effect of pH, salt and precipitants on crystallization have been covered extensively elsewhere43,70.
Grid screens
Although there are examples of crystals suitable for X-ray diffraction experiments being harvested from drops that are hundreds of nanoliters71, we still find it necessary in most cases to translate the initial crystal hits to microliter-sized drops. To reproduce the crystal hit identified in the sparse matrix screen, a broad grid screen is set around the crystallization hit to optimize for pH, precipitant concentration and salt (Fig. 4a). A broad grid screen is often necessary to reproduce the initial hit. This is due to the differences in the kinetics of mixing, vapor diffusion and crystallization that arise from the different drop sizes used in the sparse matrix screens and the grid screens.
Figure 4.
Example of grid screens. (a) A broad screen set around an initial crystallization condition identified through a nanoliter-scale screening of sparse matrix screens. The condition generating the initial hit is placed close to the center of the screen, and the concentration of the precipitant is screened against either pH (red) or salt (blue) if any salt was present in the hit. (b) An example of a more focused grid screen to further optimize the crystal hit that was reproduced in the broad grid screen.
Once the initial crystal hit has been reproduced, the grid screen focuses on a narrower range of pH, precipitant and salt concentrations (Fig. 4b). The breadth of the screen is designed to encompass a large area of crystallization space around the initial hit to increase the chances of reproducing the hit on the microliter scale. Figure 4 is intended to illustrate the concept of the grid screen. The actual increments and breadth for any parameter being optimized can be altered at the discretion of the crystallographer. The overall goal at this stage is to refine the conditions of the well solution to minimize nucleation and maximize crystal size and quality. Information gained from the broad grid screen should be used here to direct the focus of the screen. The variables having the largest impact on reducing nucleation and increasing crystal size should be varied (Fig. 5); those having little effect over the range of conditions explored in the broad grid screens should be held constant or eliminated from the well solution if not necessary for crystal formation. Although not always necessary, introduction of detergent or glycerol into the well solution at a concentration similar to that contained in the protein buffer condition can help to stabilize the protein in the crystallization drop and aid refinement of conditions. For example, inclusion of 40 mM OG into the well solution for a PDC purified in the same concentration of that detergent can help to reduce the amount of amorphous precipitation in the crystallization drop.
Figure 5.
Scoring membrane protein crystal trials. (a) Image of a crystal hit viewed with a light microscope. (b) The same image of the crystal hit in a viewed with the Korima PRS-1000 protein review station. (c) Image of amorphous precipitate formed during crystallization trial. (d,e) Two common types of phase separation observed in drops that contain excess detergent. (f) Crystals growing at the boundary of the aqueous phase and the detergent-rich phase of a drop containing detergent. (g) A crystallization drop containing a large amount of nucleation and crystals of small size (i.e., <40 µm). (h) A crystallization drop containing medium-sized crystals (i.e., 40–150 µm) and less nucleation than g. (i) A crystallization drop containing few nucleation sites and large crystals (i.e., > 150 µm).
Additives
When optimization with grid screens alone fails to generate crystals that diffract at high resolution, an additive screen can be helpful to improve the diffraction limit. To conserve sample, we generally perform this additive screen on the nanoliter scale using robotic dispensing of solutions. The following screens are available in 96-well format from Hampton Research: the Additive Screen, Detergent Screen and Silver Bullets screen72. There is also evidence in some cases8,73 that the addition of specific lipids to crystallization conditions helps to improve diffraction limits. Once a suitable additive is identified, a focused grid screen on the microliter scale is performed to find the optimal well condition in the presence of the additive.
Choosing a cryoprotectant
When data are collected under cryogenic conditions, choosing an appropriate cryoprotectant is imperative. In some cases, the crystallization drop itself may contain sufficient concentrations of precipitant (e.g., PEG400) or additive (e.g., glycerol) that can act as a cryoprotectant. If this is not the case, guidelines for selecting and assessing the quality of a cryoprotectant have been covered elsewhere and are not included here74.
Improving crystal quality
It may take multiple stages of optimization to obtain crystals that yield a diffraction pattern suitable for data collection and structure determination. Besides the steps detailed in the protocol and in Figure 6, we often treat crystals after crystallization to help in improving diffraction75. Gains in crystallization and diffraction limits can also be achieved through limited proteolysis76, co-crystallization with antibodies77,78, generating new constructs of truncated versions of the protein or pursuing species homologs. We generally only pursue these experiments when the options detailed in this protocol have been exhausted.
Figure 6.
Flowchart for crystal optimization. This diagram shows the process for generating and optimizing crystal hits for membrane proteins. The chart outlines the variables that should be optimized and suggests the order in which they should be addressed according to the problems encountered. This flowchart was adapted and reproduced with permission from ref. 87.
It is also important to note that there are a variety of different crystallization techniques besides hanging-drop vapor diffusion that can be used for screening or for optimization after an initial crystallization condition has been identified. Lipidic cubic phase79,80, sitting-drop vapor diffusion and crystallization under oil (e.g., microbatch) have all been used successfully to obtain membrane protein structures42. Capillary counterdiffusion81 and microfluidic chips82–84 have also attracted attention as a viable method for the crystallization of proteins.
MATERIALS
REAGENTS
Tris base (Fisher Scientific, cat. no. BP154-1)
Trizma HCl (Sigma, cat. no. T-3253)
HEPES (Fisher Scientific, cat. no. BP310-1)
NaCl (VWR, cat. no. BDH0286-500G)
Glycerol (Fisher Scientific, cat. no. BP229-4)
Imidazole (Sigma, cat. no. I2399-500G)
Dithiothreitol (DTT) (Fisher Scientific, cat. no. BP172-25)
β-Mercaptoethanol (Fisher Scientific, cat. no. 03446-100)
Bromophenol Blue (Fisher Scientific, cat. no. B392)
Methanol (Fisher Scientific, cat. no. A433P-4)
Acetic acid, glacial (Fisher Scientific, cat. no. A38-212) ! CAUTION Corrosive. Avoid exposure to inhalation, skin and eyes.
Phenylmethanesulfonyl fluoride (PMSF) (Sigma, cat. no. P7626-25G) ! CAUTION Corrosive. Avoid exposure to inhalation, skin and eyes.
Complete protease inhibitors, EDTA free (Roche, cat. no. 11 873 580 001)
Coomassie Brilliant Blue R (Sigma, cat. no. B7920)
Plasmid vectors: pET E. coli T7 expression vectors (Novagen)
E. coli XL1-blue cells for cloning (Stratagene, cat. no. 200228)
OverExpress C43 (DE3): E. coli cells for membrane protein expression (Lucigen, cat. no. 60345-1)
LB (Luria-Bertani Broth) Agar Miller (Fisher Scientific, cat. no. BP1425-2)
Antibiotics: e.g., kanamycin sulfate (Shelton Scientific, cat. no. IBO2120), Ampicillin sodium salt (Sigma, cat. no. A9518-100G)
LB Broth Miller (EMD, cat. no. 1.10258.5007)
IPTG (Anatrace, cat. no. I1003)
For SDS-PAGE: 4–20% (wt/vol) Tris-glycine gel, 1 mm × 15 well (Invitrogen, cat. no. EC60255BOX)
Invitrolon PVDF filter paper sandwich, 0.45-µm pore size (Invitrogen, cat. no. LC2005)
Anti-His antibody (Santa Cruz Biotechnology Inc., cat. no. SC-8036 HRP)
SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, cat. no. 34080)
Detergents (Anatrace): CHAPS (cat. no. C316), DDM (cat. no. D310), FC-12 (cat. no. F308), LDAO (cat. no. D360), OG (cat. no. O311)
Sodium dodecyl sulfate (Bio-Rad, cat. no. 161-0302)
Ni-NTA Agarose resin (Qiagen, cat. no. 30250)
Econo-Pac 10 DG disposable chromatography column (Bio-Rad, cat. no. 732-2010)
Thrombin (Novagen, cat. no. 69671)
Benzamidine sepharose 6B (GE Healthcare, cat. no. 17-0568-01)
Centrifugal filter devices: Ultra-4 or Ultra-15 series depending on what volume is being concentrated (Amicon)
Stirred ultrafiltration cells (Amicon)
Spectra/Por molecular porous membrane tubing (Spectrum Laboratories Inc.)
Spectra/Por Float-A-Lyzer apparatus for dialysis of milliliters of concentrated protein (Spectrum Laboratories Inc.)
Micro DispoDialyzer apparatus for dialysis of microliters of concentrated protein (Spectrum Laboratories Inc.)
Crystallization screens: MemStart (Molecular Dimensions, cat. no. MD1-21USA), MemSys (Molecular Dimensions, cat. no. MD1-25USA), MemGold (Molecular Dimensions, cat. no. MD1-41), CP-Custom-IV (Axygen Biosciences, cat. no. CP-CUSTOM-IV), Classics Suite (Qiagen, cat. no. 130901), PEGs Suite (Qiagen, cat. no. 130904), PEGs II Suite (Qiagen cat. no. 130916), MbClass Suite (Qiagen, cat. no. 130911), MbClass Suite II (Qiagen, cat. no. 130912), Additive Screen (Hampton Research, cat. no. HR2-138), Detergent Screen (Hampton Research, cat. no. HR2-406), Silver Bullets (Hampton Research, cat. no. HR2-096)
96-well flat-bottomed clear polypropylene plates (E&K Scientific, cat. no. EK-25201)
Protein crystallization covers for 96-well plates (Grace Bio-Labs, cat. no. 45232)
EasyXtal Tool: 24-well crystallization plate (Qiagen, cat. no. 132023)
EQUIPMENT
Gene Pulser Xcell total system (Bio-Rad 165-2660)
EmulsiFlex C3 or C5 (Avestin Inc.)
Trans-Blot SD semidry electrophoretic transfer cell (Bio-Rad, cat. no. 170-3940)
Trans-Blot SD system and Power-Pac HC power supply system (Bio-Rad, cat. no. 170-3848)
Innova 44R incubator shaker (New Brunswick Scientific, cat. no. M1282-014)
Spinbar, Teflon, Flea Micro (VWR, cat. no. 58948-976)
Superdex 200 10/300 GL (GE Healthcare Bio-Sciences, cat. no. 17-5175-01)
TSK-GEL G3000SW (Tosoh Bioscience, cat. no. 05103)
TSK-GEL G4000SW (Tosoh Bioscience, cat. no. 05104)
Guardcolumn SW (Tosoh Bioscience, cat. no. 05371) to be used with TSK-GEL columns
Shimadzu modular HPLC system (see EQUIPMENT SETUP)
Mosquito crystal nanoliter liquid handler (TTP Labtech)
Leica MZ16 stereomicroscope/KL 1500 LCD (Leica Microsystems)
PRS-1000 protein review station (Korima Inc.)
0.2-µm filter, Corning Disposable filter system (Fisher Scientific, cat. no. 09-761-40)
REAGENT SETUP
LB broth
Add 25 g of LB Broth Miller to 1 liter of water and autoclave for 30 min. Store at room temperature (25 °C) until use.
LB agar plates
Add 40 g of LB Agar Miller to 1 liter of water. Autoclave for 30 min. Allow LB agar to cool to 50 °C and add appropriate antibiotic. Dispense into sterile Petri dishes (10-cm diameter) and allow to set at room temperature. Store the plates at 4 °C.
Cell wash buffer
Mix 20 mM Tris base (pH 8) and 150 mM NaCl. The buffers are prepared at room temperature and therefore the pH of Tris is 7.4. When the experiment is performed at 4 °C, the pH is 8. Sterile-filter with a 0.2-µm filter and store buffer at 4 °C.
Cell lysis buffer
Mix 20 mM Tris base (pH 8), 250–500 mM NaCl, 4 mM β-Me, 1 mM PMSF and one complete protease inhibitor tablet. The buffers are prepared at room temperature and therefore the pH of Tris is 7.4 at this time. When the experiment is performed at 4 °C, the pH is 8. Sterile-filter with a 0.2-µm filter and store the buffer at 4 °C. The buffer is prepared without β-Me. This component is added before each use.
Membrane resuspension buffer
Mix 20 mM Tris base (pH 8), 100 mM NaCl, 20% (vol/vol) glycerol, 4 mM β-Me and 1 mM PMSF. The buffers are prepared at room temperature and therefore the pH of Tris is 7.4 at this time. When the experiment is performed at 4 °C, the pH is 8. Sterile-filter with a 0.2-µm filter and store the buffer at 4 °C. The buffer is prepared without β-Me. This component is added before each use.
SDS-PAGE gel running buffer
Dissolve 1 g of SDS, 3 g of Tris base and 14.4 g of glycine in 600 ml of water. Add water to a final volume of 1 liter. Store at room temperature.
2× SDS protein-loading dye
Mix 2.5 ml of 0.1 M Tris-HCl (pH 6.8), 1 g of SDS, 5 ml of 100% (vol/vol) glycerol and 5 ml of 1% (wt/vol) Bromophenol blue. Add water up to 25 ml. A quantity of 10 µl of β-Me is added to 240 µl of loading dye to make the 2× solution. Store at room temperature.
Coomassie stain
Combine 450 ml of methanol, 100 ml of glacial acetic acid, 400 ml of water. Dissolve 1 g of Coomassie Brilliant Blue R in the mixture. Stir with a magnetic stir bar extensively. Filter any particulate matter with filter paper. Store at room temperature.
Gel destain
Mix 120ml of methanol, 120 ml of glacial acetic acid and 460 ml of water. Store at room temperature.
Solubilization buffers
Buffer A: Mix 20 mM Tris base (pH 8), 100 mM NaCl, 10% (vol/vol) glycerol, 4 mM β-Me, 1 mM PMSF and detergent. Buffer B: Mix 20 mM Tris base (pH 7.4) at room temperature, 100 mM NaCl, 20% (vol/vol) glycerol, 4 mM β-Me, 1 mM PMSF and detergent. Either of the above buffers serves as the core of the solubilization buffer. The concentration of glycerol used is a matter of individual preference. Detergent is added to solubilization buffer A or B. The following are recommended concentrations for detergents commonly used to solubilize proteins: 270 mM OG, 20 mM DDM, 200 mM LDAO, 150 mM CHAPS, 20 mM FC-12 and 2% (wt/vol) SDS. SDS is used as a control for solubilization. For detergents other than those listed, a good starting point for a concentration of detergent in a solubilization buffer is at 10× the CMC for the detergent in the given buffer condition (i.e., ionic strength of the buffer). The buffers are prepared at room temperature and therefore pH of Tris is 7.4 at this time. When the experiment is performed at 4 °C, the pH is 8. Sterile-filter buffers with a 0.2-µm filter and store buffer at 4 °C. Buffers are prepared without β-Me. This component is added before each use.
Nickel zero buffer
Mix 20 mM Tris base (pH 8.0), 100 mM NaCl, 10% (vol/vol) glycerol, 4 mM β-Me, 1 mM PMSF and detergent. The need for an osmolite, such as glycerol, should be determined for each protein target. The following are recommended concentrations for detergents commonly used to solubilize proteins: 40 mM OG, 0.5 mM DDM, 12 mM LDAO, 16 mM CHAPS and 4 mM FC-12. For detergents other than those listed, a good starting point for a concentration of detergent in a chromatography buffer is at least 2× the CMC for the detergent in the given buffer condition (i.e., ionic strength of the buffer). The buffers are prepared at room temperature and therefore pH of Tris is 7.4 at this time. When the experiment is performed at 4 °C, the pH is 8. Sterile-filter the buffer with a 0.2-µm filter and store buffer at 4 °C. Buffer is prepared without β-Me. This component is added before each use.
Imidazole wash buffers
Mix 20 mM Tris base (pH 8), 300 mM NaCl, 10% (vol/vol) glycerol, 4 mM β-Me, 1 mM PMSF and detergent. The need for an osmolite, such as glycerol, should be determined for each protein target. See setup of nickel zero buffer for discussion of appropriate detergent concentrations. The buffers are prepared at room temperature and therefore the pH of Tris is 7.4 at this time. When the experiment is performed at 4 °C, the pH is 8. Sterile-filter the buffer with a 0.2-µm filter and store buffer at 4 °C. Buffer is prepared without β-Me. This component is added before each use.
Size-exclusion buffer
Mix 20 mM HEPES (pH 7.4), 150 mM NaCl, 10% (vol/vol) glycerol, 2 mM DTT, 1 mM PMSF and detergent. The need for an osmolite, such as glycerol, should be determined for each protein target. See setup of nickel zero buffer for discussion of appropriate detergent concentrations. Sterile-filter the buffer with a 0.2-µm filter and store buffer at 4 °C. The buffer is prepared without DTT. This component is added before each use.
EQUIPMENT SETUP
Shimadzu chromatography system
The modular chromatography system is set up with the SCL-10AVP system controller operating all of the fast protein liquid chromatography (FPLC) components. The other components are the LC-10AD solvent delivery system, the SPD-M20A PDA detector and FRC-10A fraction collector unit. The system is generally purged with water after the completion of purification. With the column in line, flow rates of 0.4–0.5 and 0.25–0.333 ml ml−1 are standard for the TSK-GEL and Superdex200 columns, respectively, when glycerol is present in the purification buffers. The flow rate can be increased in the absence of glycerol as dictated by the maximum pressure values for each column.
PROCEDURE
Expression in E. coli ● TIMING 2–3 d
-
1|
Transform E. coli OverExpress C43 (DE3) cells (by electroporation or heat shock according to the manufacturer’s protocol) with the vector containing the target gene. Plate the transformed cells onto LB agar plates with the appropriate antibiotic selection85. Grow at 37 °C overnight.
? TROUBLESHOOTING
-
2|
Pick a single colony and inoculate an LB culture of the appropriate size in a shaker flask. A range of 10–30 ml of inoculating culture should be grown for each liter of medium that will be used in Step 3. Place the culture in an incubator/shaker overnight at 37 °C and 225 r.p.m.
-
3|
Inoculate each liter of LB with 10–30 ml of the overnight growth culture. Add the appropriate antibiotic to the culture85.
-
4|
Grow the cultures in an incubator shaker at 37 °C and 175–225 r.p.m. until the optical density at 600 nm reaches 0.4–0.6 (see ref. 85).
-
5|
Induce protein expression by adding IPTG to a final concentration of 1 mM. Continue to grow the cultures for 3–5 h.
-
6|
Transfer the cells to centrifuge tubes and harvest the cells by centrifuging for 15 min at 5,000g and 4 °C.
-
7|
Discard the supernatant and resuspend the pellet in 50 ml of cell wash buffer for each liter worth of cell pellet.
-
8|
Harvest the cells by centrifuging for 15 min at 5,000g and 4 °C. Tare the tubes before centrifuging
-
9|
Discard the supernatant. Record the weight of the pellet.
■ PAUSE POINT The cells can be frozen at this point and stored at −80 °C, but continuing to Step 10 before freezing is recommended. Freezing at this point may negatively affect protein quality, and tolerance for storage should be determined empirically for each target protein.
Preparing membranes from E. coli ● TIMING 4 h
-
10|
If cell pellets were not frozen, proceed to Step 11. Thaw frozen cell pellets on ice.
▲ CRITICAL STEP All steps should be carried out on ice or at 4 °C. All buffers should be maintained at 4 °C.
-
11|
Resuspend the thawed cell pellet in 5 ml of cell lysis buffer for each gram of cell pellet. Agitate with a magnetic stir bar at 4 °C until the suspension is homogeneous.
-
12|
Lyse the cells with 3–5 passes through an EmulsiFlex-C5 or C3 microfluidizer at 10,000–15,000 psi. The apparatus should be cooled to 4 °C with a circulating water bath. Take a 10-µl sample of the lysate for analysis by SDS-PAGE and store at 4 °C.
-
13|
Centrifuge for 30 min at 15,000g and 4 °C to pellet unlysed cells and cellular debris.
-
14|
Remove the supernatant taking care not to disturb the pellet and transfer it to a clean, ultracentrifuge tube chilled on ice. Take a 10-µl sample of the supernatant for analysis by SDS-PAGE and store at 4 °C.
-
15|
To pellet the membrane fraction, centrifuge the supernatant from Step 14 for 2 h at 200,000 and 4 °C and discard the supernatant. Tare the tubes before centrifuging and record the weight of the pellet after the supernatant is discarded.
-
16|
Add 1 ml of membrane resuspension buffer for every gram of membrane pellet. A paintbrush is useful for helping to resuspend the membrane fraction. Subsequent agitation with a magnetic stir bar helps to yield a homogeneous suspension. If an appropriate detergent for solubilization of the membrane protein needs to be determined, refer to Box 1. If a suitable detergent has already been determined, proceed to Step 17.
■ PAUSE POINT The resuspended membrane fraction can be stored at −80 °C at this point or can be solubilized immediately. Freezing at this point may negatively affect protein quality and tolerance for storage should be determined empirically for each target protein.
Solubilization ● TIMING 4–24 h
-
17|
Thaw the frozen membranes on ice.
▲ CRITICAL STEP All steps should be carried out on ice or at 4 °C. All buffers should be at 4 °C.
-
18|
Resuspend the membrane pellet at the appropriate dilution in the solubilization buffer determined by following the procedure in Box 1. A paintbrush is useful for helping to resuspend the membrane fraction into solution. Subsequent agitation with a magnetic stir bar helps to yield a homogeneous suspension.
-
19|
Transfer the solubilization mixture to a clean, chilled beaker and agitate the resuspension with a magnetic stir bar in the cold room for 12–18 h at 4 °C. Take a 10-µl sample for analysis by SDS-PAGE at the end of this time. The amount of time necessary for solubilization may be far less than this and can be optimized down to as little as 1 h once an appropriate detergent has been selected.
-
20|
Transfer the solubilizaton mixture to a clean, chilled ultracentrifuge tube and pellet unsolubilized material by centrifugation for 2 h at 200,000g and 4 °C.
-
21|
Remove the supernatant from each tube taking care not to disturb the pellet and transfer it to a clean, chilled 50-ml Falcon tube. Take a 10-µl sample of the supernatant for analysis by SDS-PAGE and store at 4 °C.
Nickel affinity purification ● TIMING 3–4 h
-
22|
Prepare the appropriate amount of Ni-NTA agarose resin according to the manufacturer’s instructions. All affinity purification steps should be carried out at 4 °C with buffers chilled to 4 °C. Monitor the eluant at 280 nm.
-
23|
Load the supernatant collected in Step 20 onto a column containing the equilibrated Ni-NTA agarose resin. Collect and save the flow-through containing the unbound material. Take a 10-µl sample of the flow-through for analysis by SDS-PAGE and store at 4 °C. The flow-through should not contain the target protein but should be saved until this is confirmed by Coomassie gel and western blot of all the gel samples.
-
24|
Wash the column with nickel zero buffer until the signal of the eluant at 280 nm returns nearly to baseline.
-
25|
Wash the column with 10 mM imidazole wash buffer until the eluant at 280 nm returns nearly to baseline. Take a 10-µl sample of the elution for SDS-PAGE analysis and store at 4 °C.
-
26|
Optional: Wash the column with nickel zero buffer until the signal of the eluant at 280 nm returns to baseline.
-
27|
Wash the column with 25 mM imidazole wash buffer until the eluant at 280 nm returns nearly to baseline. Take a 10-µl sample of the elution for SDS-PAGE analysis and store at 4 °C.
-
28|
Optional: Wash the column with nickel zero buffer until the signal of the eluant at 280 nm returns to baseline.
-
29|
Wash the column with 40 mM imidazole wash buffer until the eluant at 280 nm returns nearly to baseline. Take a 10-µl sample of the elution for SDS-PAGE analysis and store at 4 °C.
-
30|
Optional: Wash the column with nickel zero buffer until the signal of the eluant at 280 nm returns to baseline.
-
31|
Reduce the flow rate of buffer through the column to minimize the volume in which the target protein is eluted.
-
32|
Elute the protein with 300 mM imidazole wash buffer and collect the peak on the chromatogram. Take a 10-µl sample for analysis by SDS-PAGE and store 4 °C.
Removal of imidazole ● TIMING 30 min
-
33|
Equilibrate an Econo-Pac 10 DG disposable chromatography column according to the manufacturer’s instructions. This can take quite some time and can be started while running the affinity purification column.
-
34|
Add 3 ml of eluate from Step 32 to the Econo-Pac column.
-
35|
Elute the protein with 4 ml of size exclusion buffer. Take a 10-µl sample for analysis by SDS-PAGE and store at 4 °C.
-
36|
Take an absorption spectrum reading and determine the protein yield by measuring the absorption value at 280 nm.
Cleavage of affinity tag ● TIMING 12–20 h
-
37|
Add four units of thrombin for every mg of protein obtained after Ni purification.
-
38|
Incubate for 12–18 h (or overnight) at 4 °C. The target protein may not require an overnight cleavage. This minimal time required for cleavage can be determined by running a gel of time points throughout the cleavage reaction.
-
39|
At the end of the cleavage incubation, take a 10-µl sample for analysis by SDS-PAGE and store the sample at 4 °C.
-
40|
Equilibrate the appropriate amount of benzamidine sepharose 6B resin according to the manufacturer’s instructions.
-
41|
Pass the Ni-purified, thrombin-cleaved protein and protease over the benzamidine sepharose 6B resin to remove the protease. Take a 10-µl sample of the flow-through for analysis by SDS-PAGE and store at 4 °C.
-
42|
Run two identical SDS-PAGE 4–20% (wt/vol) Tris-Glycine gels of the samples (collected in Steps 12, 14, 19, 21, 23, 25, 27, 29, 32, 35, 39 and 41). Use one gel for Coomassie staining85 and one for western blot.
? TROUBLESHOOTING
Size-exclusion chromatography ● TIMING 2–8 h
-
43|
Equilibrate a gel filtration column connected to the Shimadzu chromatography system (TSK-GEL G3000SW or Superdex200 10/300 GL) in size-exclusion buffer following the manufacturer’s instructions. The size-exclusion chromatography section contains additional information on selecting and running the column.
-
44|
Centrifuge a dilute sample (~250 µg ml−1) of the Ni-purified protein to be injected onto the gel filtration column for 10 min at > 10,000g and 4 °C. Remove the supernatant and discard any pellet.
▲ CRITICAL STEP Failure to centrifuge the protein sample before injection will leave aggregated or precipitated protein, if any, in the injection sample. This can clog the column, leading to increased backpressure in the purification system.
-
45|
Inject approximately 150–200 µg of the dilute sample onto the gel filtration column, collect 0.5- to 1-ml fractions of the eluate while monitoring at 280 nm.
? TROUBLESHOOTING
-
46|
To determine which peak on the chromatogram contains the target protein, run the fractions collected in Step 45 on a 4–20% (wt/vol) Tris-Glycine gel and stain with Coomassie.
-
47|
Inject the remainder of the Ni-purified protein. Monitor the eluate at 280 nm and collect 0.5- to 1-ml fractions. This may require multiple injections of the protein sample. Inject the maximum amount of protein allowable while still being able to resolve the target protein peak from contaminant protein peaks, if any are present.
-
48|
Pool the fractions from the size-exclusion purification step that contain the protein as determined by the Coomassiestained gel in Step 46. The majority of the purified protein will be taken to Step 49 for crystallization. A portion of the protein will be set aside for characterization as described in Box 2.
BOX 2 | CHARACTERIZING PROTEIN STABILITY, PURITY AND HOMOGENEITY ● TIMING 1–2 WEEKS.
-
Immediately after purification of the protein by size exclusion, inject 100 µg of the protein from the pooled fractions from Step 48 and repeat Step 45.
▲ CRITICAL STEP It is imperative that the injected sample appears as a single peak that maintains the same retention time on the column and shape as the parent peak on the original chromatogram (Fig. 3d).
? TROUBLESHOOTING
Store 500 µg of the protein at 4 °C and freeze a 250-µg aliquot at −80 °C.
-
The next day, repeat Step 1 of Box 2, injecting half of the protein (250 µg) that was stored at 4 °C.
▲ CRITICAL STEP Run the column under identical conditions as the original purification. The injected sample should appear as a single peak that maintains the same retention time on the column and shape as the parent peak obtained on the original chromatogram.
? TROUBLESHOOTING
-
One week later, repeat Step 1 of Box 2 injecting the remainder of the protein (250 µg) that was stored at 4 °C. Additionally, thaw the protein (250 µg) that was stored at −80 °C and repeat Step 1 of Box 2 with this protein sample.
? TROUBLESHOOTING
Sample preparation for crystallization ● TIMING 1 d
-
49|
Concentrate the membrane protein obtained from purification in Step 48 to 10 mg ml−1 using either an Amicon tangential-flow spin filter or an Amicon stirred cell concentrator following the manufacturer’s instructions.
▲ CRITICAL STEP For spin filters, frequently stop the concentration process and mix the sample by gentle pipetting to reduce the accumulation of free detergent micelles.
-
50|
Set aside 200 µg of the protein for characterization by size-exclusion chromatography (Box 2).
-
51|
Transfer the remainder of the protein from Step 49 to a dialysis apparatus (see EQUIPMENT) that has been extensively rinsed with water and size-exclusion buffer and is the appropriate molecular-weight cutoff. Place the dialysis bag into a beaker containing up to 1 liter of size-exclusion buffer at 4 °C. Place a stir bar at the bottom of the beaker and stir very gently for 24 h at 4 °C.
-
52|
Remove the protein from the dialysis apparatus and transfer to a clean, chilled Eppendorf tube.
-
53|
Determine the protein concentration by measuring the absorption of the sample at 280 nm.
-
54|
Immediately before crystallization, remove any aggregated protein or particulate matter by centrifugation in a micro-ultra centrifuge for 10 min at 75,000g and 4 °C.
-
55|
Remove the supernatant and transfer it to a clean, chilled Eppendorf tube.
Crystallization screen on nanoliter scale ● TIMING 1–2 h for setup; weeks/months to score results
-
56|
At room temperature, dispense 100 ml of a commercially available crystallization screen (conditions 1–96) into reservoirs 1–96 of a 96-well flat-bottomed polypropylene plate. See Experimental design for a discussion on how to choose an appropriate screen.
-
57|
Using the mosquito Crystal liquid handler, set hanging drops by mixing 150 µl of protein from Step 55 with 150 µl of a given screening condition.
? TROUBLESHOOTING
-
58|
Incubate the 96-well plates in a temperature-controlled environment at 25 °C.
-
59|Observe and score the drops immediately after they have been set as well as on days 1, 3, 7, 14, 30 and once a month after day 30 using a Leica stereomicroscope. Use the scale outlined in the table below and see Figure 5. Also use Figure 6 to guide setups if hits are not obtained.
-
60|
Image potential crystal hits with the Korima PRS-1000 protein review station to verify that crystals are protein.
Reproducing crystal hits on a microliter scale ● TIMING 1–4 h for setup; weeks/months to score results
-
61|
To optimize the crystal hits from Steps 56–60 for precipitant concentration, pH, salt concentration and drop volume ratios, use an EasyXtal Tool 24-well crystallization plate. Set up multiple drops per well with two different drop volume ratios: 1 µl of protein and 1 µl of well solution and 2 µl of protein and 1 ml of well solution. The total volume of the reservoir of the crystal tray should be between 500 and 1,000 µl. Compose the grid screen according to Figure 4a. In general, one will need to screen higher PEG or salt concentrations from the initial nanoliter-scale screen, to replicate the crystals in the larger 24-well plate format.
-
62|
Repeat Step 59.
? TROUBLESHOOTING
-
63|
Use the highest-ranking crystals from Step 61 in X-ray diffraction experiments. Refer to Experimental design for selection of a cryoprotectant. If the diffraction quality is sufficient for determining a structure, stop. If not, proceed to Step 64 and refer Figure 6.
Crystal optimization on a microliter scale ● TIMING 1–4 h for setup; weeks/months to score results
-
64|
Repeat Step 61 with parallel screens at 4, 18 and 25 °C. The total volume of the reservoir of the crystal tray should be between 500 and 1,000 µl, and the drop size and ratio should be the optimal size/ratio determined in Step 61. Compose the grid screen according to Figure 4b.
-
65|
Repeat Step 59 and then repeat Step 63. If the diffraction quality is sufficient for determining a structure, stop. If not, proceed to Step 66.
Additive screen ● TIMING 1–2 h for setup; weeks to score results
-
66|
Use the crystallization conditions optimized in Steps 64–65 to perform a 96-condition additive screen from Hampton Research. Place 100 µl of the optimized crystallization condition in each well of a 96-well flat-bottomed polypropylene plate. Use the mosquito Crystal nanoliter liquid handler to dispense 100 nl of protein, 100 nl of the optimized crystallization condition from the 96-well plate and 25 nl of each additive or detergent onto a protein crystallization cover for a 96-well plate at room temperature. For the Silver Bullets screen, dispense 100 nl of protein, 50 nl of well solution and 50 nl of each silver bullet onto a protein crystallization cover for a 96-well plate at room temperature.
-
67|
To find the optimal well condition in the presence of the additive, set up more grid screens in the large 24-well plate hanging-drop format with the best additives/detergents/silver bullets from Step 66 as determined with the scoring method described in Step 59. Screen different concentrations of additives/detergents/silver bullets in the crystallization drop.
-
68|
Use the highest-ranking crystals from Step 67 in X-ray diffraction experiments. If the diffraction quality is sufficient for determining a structure, stop. If not, refer Figure 6 for other options to pursue.
● TIMING
Steps 1–9, expression in E. coli: 2–3 d
Steps 10–16, preparing membranes from E. coli: 4 h
Steps 17–21, solubilization: 4–24 h
Steps 22–32, nickel affinity purification: 3–4 h
Steps 33–36, removal of imidazole: 30 min
Steps 37–42, cleavage of affinity tag: 12–20 h
Steps 43–48, size-exclusion chromatography: 2–8 h
Steps 49–55, sample preparation for crystallization: 1 d
Steps 56–60, crystallization screen on nanoliter scale: 1–2 h to set drops; weeks/months to score results
Steps 61–63, reproducing crystal hits on a microliter scale: 1–4 h to set drops; weeks/months to score results
Steps 64–65, crystal optimization on a microliter scale: 1–4 h to set drops; weeks/months to score results
Steps 66–68, additive screen: 1–2 h for setup; weeks to score results
Box 1, screening for an appropriate detergent for protein solubilization: 2 d
Box 2, characterizing protein stability, purity, and homogeneity: 1–2 weeks
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
TABLE 2.
Troubleshooting table
| Step | Problem | Possible reason | Possible reason |
|---|---|---|---|
| 1 | No colonies appear | Incorrect antibiotic was used for selection | Use the correct antibiotic for selection |
| The competency of the cells has been compromised | Confirm the competency of the cells with a control plasmid | ||
| Insufficient amount of plasmid was transformed | Transform a larger amount of plasmid | ||
| 42 | No protein expression was detectable by western blot against affinity tag | There was an error in cloning | Sequence gene and analyze for frame shifts, mutations, proper start and stop codons |
| Affinity tag is processed by proteintargeting machinery or is cleaved during undesired proteolysis | Use a protein specific antibody to probe for protein expression | ||
| Lower growth temperature to minimize proteolysis during expression | |||
| Insufficient expression time | Increase length of expression | ||
| Very low level of expression | Add an N-terminal fusion protein to the construct to enhance expression level. See Experimental design | ||
| Affinity tag interferes with expression | Clone affinity tag at the opposite terminus of target gene or use an alternate affinity tag | ||
| Codon usage of gene does not match with that of host expression system | Synthesize a gene to optimize the codon usage so that it matches with that of the host expression system | ||
| Heterologous expression system does not possess proper post-translational machinery and/or lipid membrane environment | Express protein in an alternative expression systems: Pichia pastoris, Saccharomyces cerevisiae, HEK293S or Sf9 cells | ||
| Protein pellets during 15,000g spin | The protein is expressed in inclusion bodies | During expression, lower culture temperature to 25 or 17 °C before induction, express in a minimal M9 media or combine both of the above | |
| Express protein in an alternative expression system: Pichia pastoris, Saccharomyces cerevisiae, HEK293S or Sf9 cells | |||
| Add an N-terminal fusion protein to the construct to improve solubility during expression. See Experimental design | |||
| Protein does not bind to Ni resin | Detergent used in solubilization is incompatible with IMAC, as concentration being used at PDC requires a long time to bind Ni resin | Refer to the manufacturer’s recommendations for the compatibility of IMAC resin detergents. Batch-bind protein by incubating the solubilization supernatant with Ni-NTA agarose beads that have been rinsed with Ni zero buffer | |
| Affinity tag is buried in protein structure or binding of the affinity tag is occluded by the detergent micelle | Add a linker between the histidine affinity tag and the protease cleavage site | ||
| Protein eluted from the Ni resin is heavily contaminated | Proteins from the heterologous expression system nonspecifically bind to the Ni resin | Add 10–20 mM imidazole to the solubilization buffer to reduce nonspecific binding to the Ni resin | |
| Increase the number of imidazole washes | |||
| Increase the concentration of the imidazole in the wash buffers | |||
| Protease does not cleave affinity tag completely | Affinity tag is buried in protein structure or binding of the affinity tag is occluded by the detergent micelle | Add a linker between the histidine affinity tag and the protease cleavage site | |
| Insufficient amount of protease was added | Add 2× the amount of protease | ||
| Precipitated protein appears after overnight cleavage | Protease, target protein or contaminant protein has crashed out of solution | Spin sample at 15,000g and 4 °C for 15 min to pellet precipitated protein. Remove the supernatant only and proceed with the protocol | |
| 42 and Box 1 | Protein pellets with unsolubilized material during 200,000g or 100,000g spin following detergent screen or solubilization | Protein is properly folded but not solubilized at all by detergents or only partially solubilized in screen | Refer to Figure 2 for alternative detergents to screen |
| Try solubilization at a different salt concentration, pH or higher temperature | |||
| 45 | Protein peaks are not resolved on the chromatogram | Protein exists in multiple oligomeric states or the protein sample is contaminated with proteins other than the target protein | Initially injecting a dilute sample of protein will allow resolving peaks of contaminant proteins on the chromatogram from the target protein. Injecting a concentrated sample can cause peaks to broaden and run together. This can obfuscate the interpretation of the chromatogram. The peak containing the target protein should ideally be a single Gaussian peak that elutes in the included volume of the column, i.e., not in or near the void volume (Fig. 3) |
| Protein does not elute from the size-exclusion column | The protein is an aggregate that gets retained in the size-exclusion column or pelleted in the spin in Step 56 | Try solubilizing in a detergent within the same family but with a longer hydrocarbon tail e.g., DDM instead of DM | |
| If other detergents were identified in the detergent screen, use a detergent from a different family for solubilization and purification | |||
| The protein detergent complex interacts with the size-exclusion matrix | Use a different size-exclusion column that is composed of a different matrix | ||
| Protein runs in the void volume of the size-exclusion column | Protein is aggregated or not stable in the buffer in which it is being solubilized and/or purified | Add an osmolite, such as glycerol or sucrose, to the buffer condition. If the buffer already contains an osmolite, increase the concentration in 2.5–5% increments up to 20% | |
| Vary the salt concentration and pH of the size-exclusion buffer | |||
| Try solubilizing in a detergent within the same family but with a longer hydrocarbon tail e.g., DDM instead of DM | |||
| If other detergents were identified in the detergent screen, use a detergent from a different family for solubilization and purification | |||
| Protein–detergent complex elutes in a non-Gaussian peak from the size exclusion | The protein–detergent complex does not exist in a single discrete fold | Try solubilizing in a detergent within the same family but with a longer hydrocarbon tail e.g., DDM instead of DM or NG instead of OG | |
| If other detergents were identified in the detergent screen, use a detergent from a different family for solubilization and purification | |||
| Vary the salt concentration and pH of the size-exclusion buffer | |||
| Protein detergent complex elutes in two or more peaks from the size-exclusion column | The protein-detergent exists in two or more oligomeric states that are in equilibrium with each other | Add an osmolite, such as glycerol or sucrose, to the buffer condition. If the buffer already contains an osmolite, increase the concentration in 2.5–5% increments up to 20% | |
| Try solubilizing in a detergent within the same family but with a longer hydrocarbon tail e.g., DDM instead of DM or NG instead of OG | |||
| If other detergents were identified in the detergent screen, use a detergent from a different family for solubilization and purification | |||
| Vary the salt concentration and pH of the size-exclusion buffer | |||
| The protein–detergent complex exists in two or more oligomeric states that are not in equilibrium with each other. This can be distinguished from the above possibility by reinjecting the protein collected from each peak and observing the retention time of the protein. If it is a single peak with the same retention time, the multiple oligomeric states are not in equilibrium with each other | Purify the peak that corresponds to the biological oligomeric state and proceed with purification | ||
| Add an osmolite, such as glycerol or sucrose, to the buffer condition. If the buffer already contains an osmolite, increase the concentration in 2.5–5% increments up to 20% to stabilize the biological oligomeric state | |||
| Vary the salt concentration and pH of the size-exclusion buffer until the biological oligomeric state is stable | |||
| 57 and 62 | Phase separation in crystallization drops | Coelution of the detergent micelle peak with the target PDC during size exclusion | Use an alternative size-exclusion column that may resolve the two peaks |
| Use a different detergent for purification and crystallization that will have a different micelle size and therefore not coelute with the target PDC | |||
| Perform ion-exchange chromatography on the purified sample. PDC will bind to the ion exchange column, and the micelles accumulated during size exclusion will flow through | |||
| Detergent micelles are being concentrated during the protein concentration step before crystallization | Use largest molecular weight cutoff concentration apparatus that will still retain the target PDC during concentration | ||
| Use different ratios of protein to well solution in the crystallization drop (e.g., 2:1 or 1:2) | |||
| Box 2 | Protein initially elutes in the included volume of the size-exclusion column but is eluted in the void volume if the protein is frozen or stored at 4 °C for any length of time | Protein is not stable in the buffer condition in which it is being solubilized and purified in | Add an osmolite, such as glycerol or sucrose, to the buffer condition. If the buffer already contains an osmolite, increase the concentration in 2.5–5% increments up to 20% |
| Try solubilizing in a detergent within the same family but with a longer hydrocarbon tail e.g., DDM instead of DM or NG instead of OG | |||
| If other detergents were identified in the detergent screen, use a detergent from a different family for solubilization and purification |
ANTICIPATED RESULTS
The core of this protocol has been used so far to crystallize a handful of membrane proteins in our laboratory (Table 1). As indicated by the reported quantities of expression, levels will vary greatly from target to target. Detergent solubilization can extract all, part or none of the target protein from the membrane fraction (Fig. 2c). All of the integral membrane protein structures published from our laboratory have been extracted in OG. This is, however, not necessarily to be expected, nor is it a prerequisite for crystallization, as membrane protein structures have been solved in other detergents59. Figure 3 contains examples and interpretations of size-exclusion traces for various protein targets. Deviations from ideal behavior (Fig. 3d) at this stage include aggregated protein that elutes in the void of a size-exclusion column (Fig. 3a) or a protein that yields multiple peaks due to the presence of multiple oligomeric states (Fig. 3b,c). In our experience, proteins showing ideal behavior shown in Figure 3d have a very high probability of success in crystallization trials. Owing to the presence of detergents in the purification buffers, formation of distinct phases in the crystallization drop during trials is not uncommon. This can inhibit the formation of crystals if severe (Fig. 5d,e). There are, however, examples of crystal nucleation and growth at phase boundaries (Fig. 5f). Optimization of a crystallization condition should limit nucleation events and maximize crystal size (Fig. 5g–i). The ultimate test of crystal quality, however, is the quality of the diffraction pattern (Supplementary Fig. 1 online).
It should be understood, however, that there is a tremendous amount of attrition that occurs along the pathway to membrane protein crystallization. That is to say, a particular target may fail at any stage, i.e., protein expression, solubilization, purification or crystallization. It is impossible to ask a priori if a particular target will make it through the entire pipeline successfully or at what stage it will fail during the process. Thus, if barriers to progress are encountered when implementing this protocol as written for a given protein target, the troubleshooting section (Table 2) should be used as a practical guide to direct variations in subsequent iterations of the protocol.
Supplementary Material
ACKNOWLEDGMENTS
Research was supported by NIH grant RO1 GM24485 to R.M.S., the NIH Roadmap center grant P50 GM073210 and the Specialized Center grant of the Protein Structure Initiative U54 GM074929-01. We thank Rebecca Robbins in the Stroud laboratory and Ryan R Atkinson for helpful discussions and help in preparing the manuscript. We also thank the reviewers and editors for their helpful suggestions.
Footnotes
Note: Supplementary information is available via the HTML version of this article.
References
- 1.Kendrew JC, et al. A three-dimensional model of the myoglobin molecule obtained by X-ray analysis. Nature. 1958;181:662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 2.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridis at 3Å resolution. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 3.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 4.Ostermeier C, Michel H. Crystallization of membrane proteins. Curr. Opin. Struct. Biol. 1997;7:697–701. doi: 10.1016/s0959-440x(97)80080-2. [DOI] [PubMed] [Google Scholar]
- 5.Opekarova M, Tanner W. Specific lipid requirements of membrane proteins—a putative bottleneck in heterologous expression. Biochim. Biophys. Acta. 2003;1610:11–22. doi: 10.1016/s0005-2736(02)00708-3. [DOI] [PubMed] [Google Scholar]
- 6.Cregg JM, Cereghino JL, Shi J, Higgins DR. Recombinant protein expression in Pichia pastoris. Mol. Biotechnol. 2000;16:23–52. doi: 10.1385/MB:16:1:23. [DOI] [PubMed] [Google Scholar]
- 7.Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 8.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 9.Tornroth-Horsefield S, et al. Structural mechanism of plant aquaporin gating. Nature. 2006;439:688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 10.Jidenko M, et al. Crystallization of a mammalian membrane protein overexpressed in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2005;102:11687–11691. doi: 10.1073/pnas.0503986102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- 12.Jasti J, Furukawa H, Gonzales EB, Gouaux E. Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature. 2007;449:316–323. doi: 10.1038/nature06163. [DOI] [PubMed] [Google Scholar]
- 13.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reeves PJ, Kim JM, Khorana HG. Structure and function in rhodopsin: a tetracycline-inducible system in stable mammalian cell lines for high-level expression of opsin mutants. Proc. Natl. Acad. Sci. USA. 2002;99:13413–13418. doi: 10.1073/pnas.212519199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonen T, et al. Lipid-protein interactions in double-layered two-dimensional AQP0 crystals. Nature. 2005;438:633–638. doi: 10.1038/nature04321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oxenoid K, Kim HJ, Jacob J, Sonnichsen FD, Sanders CR. NMR assignments for a helical 40 kDa membrane protein. J. Am. Chem. Soc. 2004;126:5048–5049. doi: 10.1021/ja049916m. [DOI] [PubMed] [Google Scholar]
- 17.Opella SJ, Marassi FM. Structure determination ofmembrane proteins by NMR spectroscopy. Chem. Rev. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howell SC, Mesleh MF, Opella SJ. NMR structure determination of a membrane protein with two transmembrane helices in micelles: MerF of the bacterial mercury detoxification system. Biochemistry. 2005;44:5196–5206. doi: 10.1021/bi048095v. [DOI] [PubMed] [Google Scholar]
- 19.Tian C, et al. Membrane protein preparation for TROSY NMR screening. Methods Enzymol. 2005;394:321–334. doi: 10.1016/S0076-6879(05)94012-3. [DOI] [PubMed] [Google Scholar]
- 20.Rigaud JL, Levy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 21.Morera FJ, Vargas G, Gonzalez C, Rosenmann E, Latorre R. Ion-channel reconstitution. Methods Mol. Biol. 2007;400:571–585. doi: 10.1007/978-1-59745-519-0_38. [DOI] [PubMed] [Google Scholar]
- 22.Andreoli TE. Planar lipid bilayer membranes. Methods Enzymol. 1974;32:513–539. [PubMed] [Google Scholar]
- 23.Bylund DB, Toews ML. Radioligand binding methods: practical guide and tips. Am. J. Physiol. 1993;265:L421–L429. doi: 10.1152/ajplung.1993.265.5.L421. [DOI] [PubMed] [Google Scholar]
- 24.Bylund DB, Deupree JD, Toews ML. Radioligand-binding methods for membrane preparations and intact cells. Methods Mol. Biol. 2004;259:1–28. doi: 10.1385/1-59259-754-8:001. [DOI] [PubMed] [Google Scholar]
- 25.Lebowitz J, Lewis MS, Schuck P. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 2002;11:2067–2079. doi: 10.1110/ps.0207702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong X, Weng YX, Li M. Determination of the topological shape of integral membrane protein light-harvesting complex LH2 from photosynthetic bacteria in the detergent solution by small-angle X-ray scattering. Biophys. J. 2004;86:1082–1088. doi: 10.1016/S0006-3495(04)74183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lipfert J, Doniach S. Small-angle X-ray scattering from RNA, proteins, and protein complexes. Annu. Rev. Biophys. Biomol. Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 28.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 29.Folta-Stogniew E, Williams KR. Determination of molecular masses of proteins in solution: implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Techniques. 1999;10:51–63. [PMC free article] [PubMed] [Google Scholar]
- 30.Swords NA, Wallace BA. Circular-dichroism analyses of membrane proteins: examination of environmental effects on bacteriorhodopsin spectra. Biochem. J. 1993;289(Part 1):215–219. doi: 10.1042/bj2890215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace BA, Lees JG, Orry AJ, Lobley A, Janes RW. Analyses of circular dichroism spectra of membrane proteins. Protein Sci. 2003;12:875–884. doi: 10.1110/ps.0229603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu D, Libson A, Stroud R. The structure of GlpF, a glycerol conducting channel. Novartis Found Symp. 2002;245:51–61. discussion 61–65, 165–168. [PubMed] [Google Scholar]
- 33.Savage DF, Egea PF, Robles-Colmenares Y, O’Connell JD, III, Stroud RM. Architecture and selectivity in aquaporins: 2.5 a X-ray structure of aquaporin Z. PLoS Biol. 2003;1:E72. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harries WE, Akhavan D, Miercke LJ, Khademi S, Stroud RM. The channel architecture of aquaporin 0 at a 2.2-A resolution. Proc. Natl. Acad. Sci. USA. 2004;101:14045–14050. doi: 10.1073/pnas.0405274101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khademi S, et al. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 36.Lee JK, et al. Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A. Proc. Natl. Acad. Sci. USA. 2005;102:18932–18937. doi: 10.1073/pnas.0509469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruswitz F, O’Connell J, III, Stroud RM. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. Proc. Natl. Acad. Sci. USA. 2007;104:42–47. doi: 10.1073/pnas.0609796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newby ZE, et al. Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum. Nat. Struct. Mol. Biol. 2008;15:619–625. doi: 10.1038/nsmb.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caffrey M. Membrane protein crystallization. J. Struct. Biol. 2003;142:108–132. doi: 10.1016/s1047-8477(03)00043-1. [DOI] [PubMed] [Google Scholar]
- 40.Garavito RM, Markovic-Housley Z, Jenkins JA. The growth and characterization of membrane protein crystals. J. Cryst. Growth. 1986;76:701–709. [Google Scholar]
- 41.Garavito RM, Picot D, Loll PJ. Strategies for crystallizing membrane proteins. J. Bioenerg. Biomembr. 1996;28:13–27. [PubMed] [Google Scholar]
- 42.Iwata S. Methods and Results in Crystallization of Membrane Proteins. Vol. 355. La Jolla, CA: International University Line; 2003. [Google Scholar]
- 43.Michel H. Crystallization of Membrane Proteins. Boca Raton, FL: CRC Press; 1991. [Google Scholar]
- 44.Wiener MC. A pedestrian guide to membrane protein crystallization. Methods. 2004;34:364–372. doi: 10.1016/j.ymeth.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 45.Hemdan ES, Porath J. Development of immobilized metal affinity chromatography II. Interaction of amino acids with immobilized nickel iminodiacetate. J. Chromatogr. 1985;323:255–264. [Google Scholar]
- 46.Hemdan ES, Porath J. Development of immobilized metal affinity chromatography III. Interaction of oligopeptides with immobilized nickel iminodiacetate. J. Chromatogr. 1985;323:255–264. [Google Scholar]
- 47.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 48.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 50.Junge F, et al. Large-scale production of functional membrane proteins. Cell Mol. Life Sci. 2008 doi: 10.1007/s00018-008-8067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- 52.Roosild TP, et al. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- 53.Makrides SC. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferguson AD, et al. Crystal structure of inhibitor-bound human 5-lipoxygenase-activating protein. Science. 2007;317:510–512. doi: 10.1126/science.1144346. [DOI] [PubMed] [Google Scholar]
- 55.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 56.Hannig G, Makrides SC. Strategies for optimizing heterologous protein expression in Escherichia coli. Trends Biotechnol. 1998;16:54–60. doi: 10.1016/s0167-7799(97)01155-4. [DOI] [PubMed] [Google Scholar]
- 57.Yeliseev AA, Wong KK, Soubias O, Gawrisch K. Expression of human peripheral cannabinoid receptor for structural studies. Protein Sci. 2005;14:2638–2653. doi: 10.1110/ps.051550305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raman P, Cherezov V, Caffrey M. The membrane protein data bank. Cell Mol. Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newstead S, Ferrandon S, Iwata S. Rationalizing alpha-helical membrane protein crystallization. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garavito RM, Ferguson-Miller S. Detergents as tools in membrane biochemistry. J. Biol. Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 61.Chiu ML, Tsang C, Grihalde N, MacWilliams MP. Over-expression, solubilization, and purification of G protein-coupled receptors for structural biology. Comb. Chem. High Throughput Screen. 2008;11:439–462. doi: 10.2174/138620708784911456. [DOI] [PubMed] [Google Scholar]
- 62.Faham S, Bowie JU. Bicelle crystallization: a new method for crystallizing membrane proteins yields a monomeric bacteriorhodopsin structure. J. Mol. Biol. 2002;316:1–6. doi: 10.1006/jmbi.2001.5295. [DOI] [PubMed] [Google Scholar]
- 63.Weckstrom K. Aqueous micellar systems in membrane protein crystallization. Partial miscibility of a nonionic surfactant in the presence of salt or polyethylene glycol. FEBS Lett. 1985;192:220–224. doi: 10.1016/0014-5793(85)80111-3. [DOI] [PubMed] [Google Scholar]
- 64.Mohanty AK, Simmons CR, Wiener MC. Inhibition of tobacco etch virus protease activity by detergents. Protein Expr. Purif. 2003;27:109–114. doi: 10.1016/s1046-5928(02)00589-2. [DOI] [PubMed] [Google Scholar]
- 65.Potschka M. Universal calibration of gel permeation chromatography and determination of molecular shape in solution. Anal. Biochem. 1987;162:47–64. doi: 10.1016/0003-2697(87)90009-1. [DOI] [PubMed] [Google Scholar]
- 66.Barth HG, Boyes BE, Jackson C. Size exclusion chromatography. Anal. Chem. 1994;66:595R–620R. doi: 10.1021/ac00084a022. [DOI] [PubMed] [Google Scholar]
- 67.Ricker RD, Sandoval LA. Fast, reproducible size-exclusion chromatography of biological macromolecules. J. Chromatogr. A. 1996;743:43–50. doi: 10.1016/0021-9673(96)00283-x. [DOI] [PubMed] [Google Scholar]
- 68.Trathnigg B. Size-exclusion chromatography of polymers. In: Meyers RA, editor. Encyclopedia of Analytical Chemistry. Chichester: John Wiley & Sons Ltd; 2000. pp. 8008–8034. [Google Scholar]
- 69.Strop P, Brunger AT. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 2005;14:2207–2211. doi: 10.1110/ps.051543805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bergfors TM. Protein Crystallization. Vol. 306. La Jolla, CA: International University Line; 1999. [Google Scholar]
- 71.Walter TS, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. I. Protocol design and validation. J. Appl. Cryst. 2003;36:308–314. [Google Scholar]
- 72.McPherson A, Cudney B. Searching for silver bullets: an alternative strategy for crystallizing macromolecules. J. Struct. Biol. 2006;156:387–406. doi: 10.1016/j.jsb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 73.Guan L, Smirnova IN, Verner G, Nagamori S, Kaback HR. Manipulating phospholipids for crystallization of a membrane transport protein. Proc. Natl. Acad. Sci. USA. 2006;103:1723–1726. doi: 10.1073/pnas.0510922103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McFerrin MB, Snell EH. The development and applicatioin of a method to quantify the quality of cryoprotectant solutions using standard area-detector X-ray images. J. Appl. Cryst. 2002;35:538–545. [Google Scholar]
- 75.Heras B, Martin JL. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr. D Biol. Crystallogr. 2005;61:1173–1180. doi: 10.1107/S0907444905019451. [DOI] [PubMed] [Google Scholar]
- 76.Cohen SL, Chait BT. Mass spectrometry as a tool for protein crystallography. Annu. Rev. Biophys. Biomol. Struct. 2001;30:67–85. doi: 10.1146/annurev.biophys.30.1.67. [DOI] [PubMed] [Google Scholar]
- 77.Hunte C, Michel H. Crystallisation of membrane proteins mediated by antibody fragments. Curr. Opin. Struct. Biol. 2002;12:503–508. doi: 10.1016/s0959-440x(02)00354-8. [DOI] [PubMed] [Google Scholar]
- 78.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 79.Landau EM, Rosenbusch JP. Lipidic cubic phases: a novel concept for the crystallization of membrane proteins. Proc. Natl. Acad. Sci. USA. 1996;93:14532–14535. doi: 10.1073/pnas.93.25.14532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nollert P. Lipidic cubic phases as matrices for membrane protein crystallization. Methods. 2004;34:348–353. doi: 10.1016/j.ymeth.2004.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ng JD, Gavira JA, Garcia-Ruiz JM. Protein crystallization by capillary counterdiffusion for applied crystallographic structure determination. J. Struct. Biol. 2003;142:218–231. doi: 10.1016/s1047-8477(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 82.Hansen CL, Skordalakes E, Berger JM, Quake SR. A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion. Proc. Natl. Acad. Sci. USA. 2002;99:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hansen C, Quake SR. Microfluidics in structural biology: smaller, faster em leader better. Curr. Opin. Struct. Biol. 2003;13:538–544. doi: 10.1016/j.sbi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 84.Hansen CL, Classen S, Berger JM, Quake SR. A microfluidic device for kinetic optimization of protein crystallization and in situ structure determination. J. Am. Chem. Soc. 2006;128:3142–3143. doi: 10.1021/ja0576637. [DOI] [PubMed] [Google Scholar]
- 85.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York, USA: Cold Spring Laboratory Press; 1989. [Google Scholar]
- 86.Savage DF, Anderson CL, Robles-Colmenares Y, Newby ZE, Stroud RM. Cell-free complements in vivo expression of the E. coli membrane proteome. Protein Sci. 2007;16:966–976. doi: 10.1110/ps.062696307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McPherson A. Current approaches to macromolecular crystallization. Eur. J. Biochem. 1990;189:1–23. doi: 10.1111/j.1432-1033.1990.tb15454.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.