Abstract
The reversible Y-O•/Y-OH redox properties of the α3Y model protein enable access to the electrochemical and thermodynamic properties of 3,5-difluorotyrosine. The unnatural amino acid has been incorporated at position 32, the dedicated radical site in α3Y, by in vivo nonsense codon suppression. Incorporation of 3,5-difluorotyrosine gives rise to very minor structural changes in the protein scaffold at pH below the apparent pK (8.0 ± 0.1) of the unnatural residue. Square-wave voltammetry on α3(3,5)F2Y provides an E°′(Y-O•/Y-OH) of 1026 ± 4 mV versus the NHE (pH 5.70 ± 0.02) and shows that the fluoro-substitutions lower E°′ by –30 ± 3 mV. These results illustrate the utility of combining the optimized α3Y tyrosine radical system with in vivo nonsense codon suppression to obtain the formal reduction potential of an unnatural aromatic residue residing within a well-structured protein. It is further observed that the protein E°′ values differ significantly from peak potentials derived from irreversible voltammograms of the corresponding aqueous species. This is significant since solution potentials have been the main thermodynamic data available for amino-acid radicals. These findings are discussed relative to recent mechanistic studies on the multistep radical-transfer process in E. coli ribonucleotide reductase site-specifically labeled with unnatural tyrosine residues.
Tyrosine serves as a one-electron redox cofactor in catalytic and multistep electron-transfer reactions (1-5). It has been challenging to obtain precise and accurate thermodynamic information for this high-potential protein redox species. Electrochemical characterization of the natural systems has not been feasible due to their size, complexity and sensitivity to oxidative damage. Mechanistic studies on redox proteins employing tyrosine radical (Y•) cofactors must thus partly rely on model systems to provide insights to the thermodynamics involved. Reduction potentials (E values) of aqueous Y and various analogues have been obtained by pulse radiolysis and voltammetry methods (6-11). Considerable uncertainty is associated with the reported values. This is in part due to varying experimental conditions, comparison of neutral and zwitterionic amino acids, and complicating issues such as solvent oxidation and the perturbation of solute/working electrode interactions. The most significant uncertainty, however, arises from the reactivity of the radical species themselves. Tyrosine radicals generated in solution will rapidly dimerize (~ 5 × 108 M–1 s–1; 12-16) and give rise to quasi/irreversible voltammograms (17, 18). Peak potentials (Epeak) derived from such data reflect the electrode process (electrode driven oxidation-reduction of Y) as well as the thermodynamics and kinetics of the coupled side reactions (chemical reduction of Y•). The observed potentials may thus differ by 10s even 100s of millivolts from the formal (true) reduction potential (E°′) of the Y redox system. Comparison between aqueous solution and protein Y redox chemistry is further complicated by the significant differences in the fundamental properties of these two media (19, 20).
The α3X family of well-structured proteins was developed to address these concerns (10, 21). This model protein system yields thermodynamic information that is uncompromised by electrochemical irreversibility and radical side reactions (22-24). The α3X proteins are based on a common de novo designed three.helix bundle scaffold: Scheme 1 displays helical and loop regions in bold and italic, respectively. The N-terminal GS of the 67-residue sequence form part of a thrombin cleavage site and are labeled as –2 and –1 to keep the amino-acid numbering consistent between the chemically synthesized (10) and recombinantly expressed (25, 26) α3X proteins. The buried redox site (position 32 in the middle of the central helix) is occupied by a tryptophan (to form the α3W protein), a tyrosine (α3Y), or a cysteine (α3C). C32 has been used to covalently attach phenol (24, 26) and quinone (27) molecules to the protein scaffold. All of the α3X proteins display very similar structural characteristics. They are stable and well-structured from pH ~4 to 10. Their single aromatic residue (W, Y, phenol or quinone) gives rise to UV-Vis, fluorescence and NMR spectra that are highly sensitive to the microenvironment of the redox site. Protein voltammetry has shown that the α3X system displays unique electrochemical properties. The protein scaffold is redox inert to at least +1.3 V vs. NHE (22, 24, 26). The system becomes redox active when a W (10), Y (22, 23), phenol (24, 26) or quinone (27) is introduced at position 32. Fully reversible voltammograms and E°′ values have thus far been reported for the Y and phenol-containing α3X proteins (22, 24).
Scheme 1.
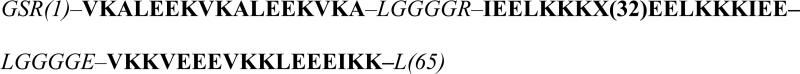
In this study we use the α3X system in combination with the in vivo nonsense codon suppression method (28) and square-wave voltammetry (SWV; 17, 29, 30) to determine E°′ for a protein 3,5-difluorotyrosine (3,5-F2Y) residue. We report ΔE°′ between 3,5-F2Y and Y residues obtained at highly comparable experimental conditions. The results are compared to published solution potentials and discussed relative to recent mechanistic studies on E. coli class Ia ribonucleotide reductase (RNR) site-specifically labeled with unnatural Y residues in an effort to understand the thermodynamic and kinetic landscape of the proton-coupled electron transfer (PCET) pathway in this system (28, 31, 32).
MATERIALS AND METHODS
Purification of tyrosine phenol lyase (TPL)
E. coli strain SVS370 harboring the plasmid pTZTPL was obtained as a gift from Dr. Robert Phillips (University of Georgia). pTZTPL encodes TPL under a constitutive promoter and the protein was expressed and purified (33, 34) using a slightly modified procedure. The elution fractions from the octyl-sepharose column were assayed using a coupled spectrophotometric assay where a small volume of the fraction was added to an assay mixture containing 2 mM L-tyrosine, 5 mM β-mercaptoethanol, 50 μM pyridoxyl-5′-phosphate, 0.3 mg/mL lactate dehydrogenase (L. leichmannii from Sigma-Aldrich) and 0.2 mM NADH in 50 mM potassium phosphate pH 8.0. The reaction was monitored at 340 nm for the disappearance of NADH. Fractions containing considerable activity were pooled and concentrated in an Amicon ultrafiltration cell with a YM 10 membrane and the protein was used with no additional purification steps. One unit (U) of activity is defined as 1 μmol product/min. A total of 62 U/g cell paste was obtained, in excellent agreement with the previously reported yield (33).
Enzymatic synthesis of 3,5-difluorotyrosine (3,5-F2Y)
TPL was used to enzymatically synthesize 3,5-F2Y from 2,6-difluorophenol (the numbering of the aromatic ring changes as the OH carbon is position 1 in the phenol and position 4 in the amino acid). Briefly, a one or two liter reaction mixture containing 10 mM 2,6-difluorophenol, 30 mM ammonium acetate pH 8.0, 60 mM sodium pyruvate, 5 mM β-mercaptoethanol, 40 μM pyridoxyl-5′-phosphate and 30 U of TPL at room temperature was stirred in the dark for a total of four days. The mixture was supplied with additional TPL, β-mercaptoethanol and pyridoxyl-5′-phosphate every other day. Once the reaction was complete (as assessed by TLC), the mixture was worked up as described in Ref. 34. The yield is typically > 85% for 3,5-F2Y and the final product was characterized by 1H and 19F NMR spectroscopy (11, 35).
Construction of α3TAG32 by site-directed mutagenesis
The amber stop codon (TAG) was introduced at position 32 of α3Y using a modified pET32b-α3Y plasmid (22, 25) as template and forward primer 5’- GGC GGC CGT ATT GAA GAA CTG AAA AAA AAA TAG GAA GAA CTG AAA AAA AAA ATT GAA GAA CTG GGC GGC GGC-3’ and reverse primer 5’- GCC GCC GCC CAG TTC TTC AAT TTT TTT TTT CAG TTC TTC CTA TTT TTT TTT CAG TTC TTC AAT ACG GCC GCC-3’. The mutation was performed using the Stratagene QuikChange kit and confirmed by sequencing at the MIT Biopolymers Laboratory.
Expression of α3(3,5)F2Y
pET32b-α3TAG32 was co-transformed with a plasmid (pEVOL-FnY-RS) encoding the fluorotyrosine tRNA and aminoacyl-tRNA synthetase (FnY-RS) genes (28) into BL21(DE3) competent cells and grown on media containing ampicillin (100 μg/mL) and chloramphenicol (35 μg/mL) at 37° C. Unnatural amino-acid concentration and protein expression were optimized on a small-scale resulting in the following protocol: 40 mL LB pre-cultures were started from single colonies, grown for ~ 16 h, and used to inoculate (50- fold dilution) 1 L 2x YT cultures containing 2.0 mM 3,5-F2Y. 3,5-F2Y additions were made from stock solutions freshly prepared in water, NaOH solubilized and sterile filtered. The expression of FnY-RS was induced at an OD600 of 0.2-0.3 (L-arabinose, final concentration 0.05% (w/v)). The expression of α3(3,5)F2Y was induced at an OD600 of 0.5 (IPTG , final concentration 1mM). Growth was continued for an additional 4 h and the cells were harvested by centrifugation (5000 × g, 15 min, 4° C). Protein expression was monitored by SDS-PAGE. No toxicity was seen due to the unnatural amino acid. A typical yield of 0.5 g cell paste/100 mL and 2.5 g cell paste/L media was obtained for the 0.1 and 1 L cultures, respectively.
Purification of α3(3,5)F2Y
Cell paste (~ 5-10 g) was resuspended (5 mL/g paste) in buffer A (20 mM Tris-HCl, 500 mM NaCl, 5 mM imidazole, pH 7.9), treated with lysozyme (300 μg/mL, 30 min, 30° C) and lysed by sonication. The lysate was clarified by centrifugation (12000 × g, 20 min, 4° C), passed over a nickel column (10 mL His•bind resin, EMD Millipore) equilibrated with buffer A, and the thioredoxin fusions eluted with a linear 0–40% buffer B (20 mM Tris-HCl, 500 mM NaCl, 1M imidazole, pH 7.9) gradient over 40 min (flow rate 1.5 mL/min). Fractions containing the thioredoxin fusions were identified by SDS-PAGE. Thrombin (T6634; Sigma-Aldrich) was added to the pooled fusion-protein fractions (thrombin/protein ratio 1:2000 (w/w)) and the resulting mixture dialyzed against 20 mM Tris-HCl, 500 mM NaCl, 2.5 mM CaCl2, pH 8.0 at RT for >16 h. The thrombin digestion produces three major products: His- tagged thioredoxin, the truncated α3(residue 1–31) peptide (MW 3579 Da) and the full-length α3(3,5)F2Y protein (MW 7557 Da). A major fraction of the truncated α3(residue 1–31) product was removed during the overnight dialysis step (Spectra/Por tubing MWCO 3500 Da). The digestion/dialysis mixture (~ 30 mL) was passed over a nickel column (10 mL His•bind resin equilibrated with buffer A) to remove the His-tagged thioredoxin and any remaining undigested fusion products. α3(3,5)F2Y (sample injection volume 5-10 mL) was isolated by reversed-phase HPLC (218TP C18 column, particle size 10 μm, column size 10 × 250 mm; Grace/VYDAC) using an linear water/acetonitrile/0.1% (w/v) trifluoroacetic acid gradient (30-60% acetonitrile over 45 min, flow rate 5 mL/min), and stored as lyophilized powder. The protein purification steps were monitored by SDS-PAGE. Purity was evaluated by reversed-phase HPLC (218TP C18 column, particle size 5 μm, column size 4.6 × 250 mm; Grace/VYDAC) using a linear water/acetonitrile/0.1% (w/v) trifluoroacetic acid gradient (20-70% acetonitrile over 50 min, flow rate 1 mL/min). Molecular weights were verified by matrix-assisted desorption/ionization-time of flight (MALDI-TOF) mass spectrometry using a Bruker Microflex 3.1. An average yield of ~3 mg pure α3(3,5)F2Y/L culture was observed.
Absorption spectroscopy
Absorption spectra were recorded on a Varian Cary 50 Bio UV-Vis spectrometer at RT. Protein concentrations were determined by the Bradford protein assay (Bio-Rad) with standard curves prepared using either bovine serum albumin (for thioredoxin fusion samples) or α3W (10) (for α3Y and α3(3,5)F2Y protein-characterization samples). An ε280 of 5690 M–1 cm–1 was used for α3W (10, 36, 37) when preparing the standard curves. The apparent tyrosinate/tyrosine pKa (pKapp) of (3,5)F2Y32 was measured by dissolving lyophilized α3(3,5)F2Y in 3.0 mL 20 mM sodium acetate, 20 mM potassium phosphate, 20 mM sodium borate (APB buffer), pH 12 to give an absorption of 0.2 at 277 nm (10 mm path). The protein stock solution was added to 20 mM APB buffer generating two samples with pH ~ 5 and 12, respectively. The pH was pre-adjusted in the APB buffer samples to generate the final pH upon addition of the protein stock. The protein dilution was 2-fold providing a final 277 nm absorption of 0.1 (10 mm path; pH 12). Equal-volume (1.6 mL) titration was performed manually by removing a 100-400 μL portion from the cuvette containing the high-pH sample and then adding the same amount of low-pH sample per pH adjustment. The pKapp was obtained by fitting the pH-induced increase in the (3,5)F2Y32-O– absorption to a single pKa.
Circular dichroism (CD) spectroscopy
CD data were collected on an Aviv 202 CD spectrometer at 25° C. The instrument was equipped with an automated titration system. For the α-helical measurements, CD spectra were collected from α3W, α3Y and α3(3,5)F2Y dissolved in 20 mM sodium acetate, pH 5.6. Protein concentrations were determined by the Bradford assay and using α3W as the standard. pH titrations were performed by dissolving lyophilized protein in 20 mM APB buffer, pH 4.8 to a 235 nm ellipticity of ~–175 mdegrees (10 mm path). The protein stock solution was added to 20 mM APB buffer generating two samples with pH ~ 4 and 10, respectively. The pH was pre-adjusted in the APB buffer samples to generate the final pH upon addition of the protein stock. The protein dilution was 4-fold generating a final 222 nm ellipticity ~–150 mdegrees (10 mm path; pH 4). Equal-volume (2.5 mL) titration was performed manually by removing a 40-1000 μL portion from the cuvette containing the high-pH sample and then adding the same amount of low-pH sample per pH adjustment. Protein stability measurements were conducted by dissolving lyophilized protein in 20 mM sodium acetate, 20 mM potassium phosphate (AP buffer), pH 5.0 or 5.5 buffer to a 230 nm ellipticity of –75 to –140 mdegrees (1 mm path). The protein stock solution was added to 20 mM AP buffer containing 0 and 9.5 M urea, respectively. The pH was 5.0 or 5.5 in the protein/AP buffer and protein/AP buffer/urea solutions. The protein dilution was 18-fold generating a final 222 nm ellipticity in the –110 to – 210 mdegrees range (10 mm path) at zero molar denaturant. The urea denaturation experiments were performed by automated equal-volume (2.0 mL) titration controlled from the Aviv software. Global stability values were determined by fitting the denaturation curves as described in Ref. 38.
Square-wave voltammetry (SWV)
Voltammetry measurements (17, 29, 30) were performed using an Autolab PGSTAT12 potentiostat equipped with a temperature-controlled, Faraday-cage protected three-electrode micro-cell (Princeton Applied Research). The Ag/AgCl reference electrode and the platinum wire counter electrode (Advanced Measurements Inc.) were stored dry and prepared by filling the former with a 3M KCl/saturated AgCl solution and the latter with sample buffer. All measurements were carried out using a 3 mm diameter pyrolytic graphite edge (PGE) working electrode (Bio-Logic, USA). The electrode surface was activated between measurements by manually polishing its surface for 60 s in a 1.0 μm diamond/water slurry on a diamond polishing pad (Bio-Logic, USA) followed by 60 s in a 0.05 μm alumina/water slurry on a microcloth pad (Bioanalytical systems Inc.). The electrode was rinsed with an excess of methanol followed by milli-Q water directed against the surface of the electrode. Measurements were performed immediately following the polishing procedures. Solution resistance was compensated for by using the positive feed-back iR compensation function of the Autolab system. Potentials are given versus the normal hydrogen electrode (NHE). All samples were prepared from ultra-pure chemicals and the measurements performed under an argon atmosphere. Preparation of the voltammetry protein samples is described in the main text. Protein concentration series were obtained by stepwise dilution of the protein samples in the electrochemical cell while keeping the sample volume constant at 2.5 mL. The pH was monitored directly in the electrochemical cell using a pH microelectrode (Microelectrodes Inc.) connected to a SevenMulti pH meter (Mettler Toledo). The pH electrode was disconnected from the pH meter during the active measurements to avoid the risk of introducing electric noise. Data processing and analyses were performed using the Autolab GPES software and PeakFit (Systat Software Inc.).
RESULTS
α3(3,5)F2Y expression and purification
3,5-difluorotyrosine was synthesized enzymatically (33, 34) as described in the Materials and Methods section. The amber stop codon (TAG) was introduced in place of the codon for residue 32 (Scheme 1) in a modified pET32b plasmid used for expression of the α3X proteins (22, 25). The α3TAG variant was co-expressed with an evolved polyspecific fluorotyrosine aminoacyl-tRNA synthetase (28) in E. coli BL21(DE3) cells. Protein expression was initially evaluated in 100 mL growth cultures by SDS-PAGE and mass spectrometry. Optimal expression was observed with 2.0 mM 3,5-F2Y in 2x YT media (see Fig. S1 in the Supporting Information). The growth cultures were scaled to 1 L to generate sufficient material for protein characterization and voltammetry studies. Protein expression and purification protocols are described in detail in Materials and Methods. The purity of the α3(3,5)F2Y preparations was evaluated by reversed-phase HPLC (Fig. S2A) and the correct protein molecular weight was verified by mass spectrometry (Fig. S2B). The average yield of pure α3(3,5)F2Y was ~3 mg/L culture, which represents a suppression efficiency of only ~10%. Efforts to improve the suppression efficiency are in progress.
Protein characterization
The aims of this study are to determine (i) the electrochemical reversibility of α3(3,5)F2Y, (ii) E°′ of (3,5)F2Y32, and (iii) ΔE°′ between (3,5)F2Y32 and Y32 when recorded at near identical conditions. Initially we compared the structural properties of α3Y and α3(3,5)F2Y. α3Y is highly helical and well-structured from pH 4.5 to 10 (10, 22). The apparent Y-O–/Y-OH pKa (pKapp) of Y32 is 11.3 ± 0.1 (Table 1). Exchanging Y32 to (3,5)F2Y32 was not expected to significantly perturb the protein scaffold (39, 40) at pH conditions where the buried phenol side chain is uncharged. Experiments were carried out to determine a suitable pH range for the SWV analysis, i.e., where (3,5)F2Y32 remains protonated and the structural properties of α3(3,5)F2Y closely resemble those of α3Y.
Table 1.
Properties of Y, 3,5-F2Y, α3Y and α3(3,5)F2Y
| System | λmax (nm) / ε (M–1 cm–1) | pKa [pKapp] | Potential vs. NHE (mV)e | Ref. |
|---|---|---|---|---|
| Y(Ac/NH2)a | 275/1400 (Y-OH) 293/2420 (Y-O-) |
10 | Epeak 830 (pH 7.0) | 31 |
| 3,5-F2Y(Ac/NH2)a | 263/560 (Y-OH) 275/1830 (Y-O-) |
7.0 ± 0.2 | Epeak 770 (pH 7.0) | 31 |
| α3Y | 277/1500 (Y-OH, pH 5.4) 294/2430 (Y-O-, pH 13.4) |
[11.3 ± 0.1] |
E°' 1059 (pH 5.7) E°' 980 (pH 7.0) |
10, 22, 23 this work |
| α3(3,5)F2Y | ~264/n.d. (Y-OH)a ~277/n.d. (Y-O-)a |
[8.0 ± 0.1] |
E°' 1026 (pH 5.7) E°' 952 (pH 7.0)f |
this work |
| System | [Θ]222 × 103 (deg cm2 dmol–1) | Helix (%)b | ΔG (kcal mol–1)g | Ref. |
|---|---|---|---|---|
| α3Y | −20.0 ± 0.8 (pH 5.6) | 75 ± 3 (pH 5.6)c 74 ± 1 (4.5-10)d |
−3.7 ± 0.1 (pH 5.0) −3.9 ± 0.1 (pH 5.5) |
10, 22, this work |
| α3(3,5)F2Y | −20.9 ± 0.6 (pH 5.6) | 79 ± 2 (pH 5.6)c | −3.7 ± 0.1 (pH 5.0) −3.7 ± 0.1 (pH 5.5) |
this work |
Acetyl-tyrosinamide (Ac/NH2), ε was not determined (n.d.).
[Protein] determined by the Bradford assay.
Anodic peak potentials from irreversible differential pulse voltammograms (Epeak; 10, 11, 31). The standard error in E°' is ± ± 4 mV.
Extrapolated from the pH 5.7 value and assuming the same pH dependence as observed for α3Y E°'(pH 5.7) and E°'(pH 7.0)(23).
From Fig. S5.
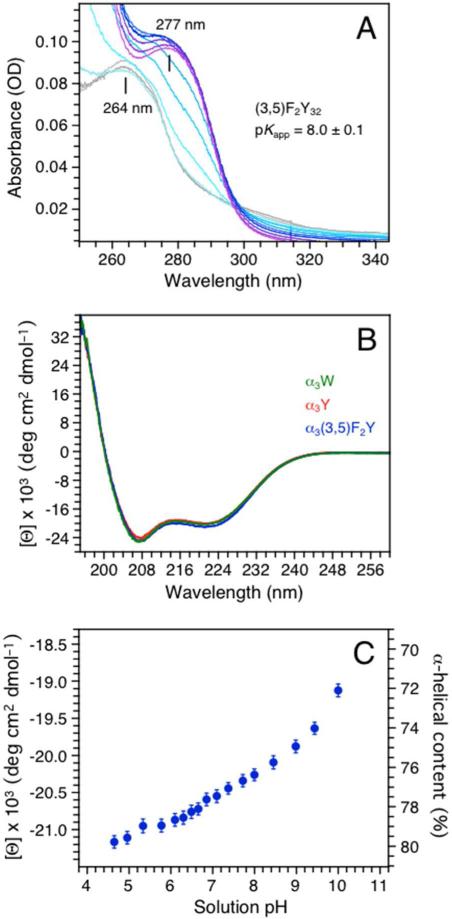
Figure 1A displays absorption spectra of α3(3,5)F2Y collected over a pH 5.7 (grey) to 10.4 (purple) range. The (3,5)F2Y32-O–/(3,5)F2Y32-OH transition titrates with a pKapp of 8.0 ± 0.1 (see Fig. S3 for details). The protein environment increases the pKapp by ~1 unit relative to aqueous 3,5-F2Y (pKa 7.0 ± 0.2; Table 1; 11, 35). The weak and blue shifted absorbance of aqueous 3,5-F2Y-OH (ε263 560 M–1 cm–1) relative to unmodified Y-OH (ε275 1490 M–1 cm–1; Table 1) was expected to translate into a rather poorly defined absorption spectrum of α3(3,5)F2Y. The blue shift increases the spectral overlap between the absorption of the aromatic side chain and the large absorption of the protein backbone (λmax 190-220 nm) making α3(3,5)F2Y concentration measurements based on UV absorbance unreliable. Thus, the Bradford method was used to determine protein concentrations with standard curves prepared from α3W. ε277 of 1500 ± 80 (pH 5.4) and ε294 of 2430 ± 130 (pH 13.4) M–1 cm–1 were obtained for Y32-OH and Y32-O– in α3Y, respectively. The ε277 value is consistent with the average ε280 of 1490 M–1 cm–1 typically used for protein Y-OH residues (36, 37) and close to the εmax values of aqueous Y-OH (Table 1; 11). We conclude that the Bradford assay determines the α3X protein concentration with good accuracy.
Figure 1.
(A) A pH titration of α3(3,5)F2Y monitored by UV-Vis absorption spectroscopy. Spectra were collected at pH 5.72 (gray), 6.38, 6.84, 7.49, 8.06, 8.53, 8.96, 9.31, 9.59, 10.02 and 10.42 (purple). (B) Far-UV CD spectra of α3(3,5)F2Y (blue), α3Y (red) and α3W (green) in 20 mM sodium acetate, pH 5.62 ± 0.01. The single aromatic residue in each protein is fully protonated at this pH. The CD spectra are displayed in units of mean residue molar ellipticity ([Θ]) obtained by: ([Θ]) = θobs(106/Cln) where θobs is the observed ellipticity in mdegrees (spectrometer raw signal), C the protein concentration in μM, l the cuvette path length in mm, and n the number of amino-acid residues (65). [Θ]222 equals –20.0 ± 0.8 × 103 deg cm2 dmol–1 at pH 5.6 for α3(3,5)F2Y (Table 1). (C) Changes in the mean residue molar ellipticity ([Θ]) and the corresponding % α-helical content of α3(3,5)F2Y between pH 4.7 and 10.
CD spectroscopy was used to measure the absolute α-helical content, the pH sensitivity of this parameter, and the global stability of α3(3,5)F2Y relative to α3Y (Table 1). Figure 1B displays far-UV CD spectra of α3(3,5)F2Y (blue) and α3Y (red) collected at pH 5.6. The reference spectrum of the structurally characterized α3W protein (green; 10, 25) is also shown. The protein concentration in the CD samples was determined by the Bradford method. The CD spectral line shapes are essentially identical and display the characteristic 208/222 nm double minima of a predominantly α-helical protein. The 222 nm mean residue molar ellipticity ([Θ]222) reflects the α-helical content, which was estimated to ~75-80% for α3(3,5)F2Y and α3Y at pH 5.6 (Table 1). This translates into 50 ± 1 residues with helical ψ and φ backbone angles in α3(3,5)F2Y and α3Y. This degree of helicity has consistently been observed for structurally characterized α3X proteins (α3W PDB ID 1LQ7 (25); 2-mercaptophenol-α3C 2LXY (24)). Figure 1C displays [Θ]222 and the corresponding % helix of α3(3,5)F2Y as a function of pH. The α-helical content of α3(3,5)F2Y decreases by only 2% from pH 4.7 to 6.7 and by 6% from pH 6.7 to 10. These small pH-induced changes in helical content are likely to be driven by the deprotonation of (3,5)F2Y32. The global stability of α3(3,5)F2Y and α3Y was thus compared at acidic pH > two pH units below the pKapp of (3,5)F2Y32 (Fig. S4). The stability of the two proteins was found to be identical at pH 5.0 and differ by only 0.2 kcal mol–1 at pH 5.5 (Table 1). The SWV analysis described below was conducted at low pH (5.6 ± 0.1) where the structural properties of α3(3,5)F2Y and α3Y are very similar and their phenols are protonated.
Preparation of SWV samples
The protein concentration is a key parameter to control when investigating the protein/working electrode interactions and optimizing the Faradaic response (23, 24). The high α-helical content of α3(3,5)F2Y makes CD spectroscopy a precise method to determine the protein concentration in a highly reproducible manner. Voltammetry samples were thus prepared by dissolving lyophilized protein in buffer until an appropriate θ222 and the protein concentration was calculated from the determined [Θ]222 value (see Fig. 1 legend and Table 1). The supporting electrolyte, KCl, was added to the samples after the CD measurements to avoid UV absorption of the chloride ion.
SWV analysis of α3(3,5)F2Y
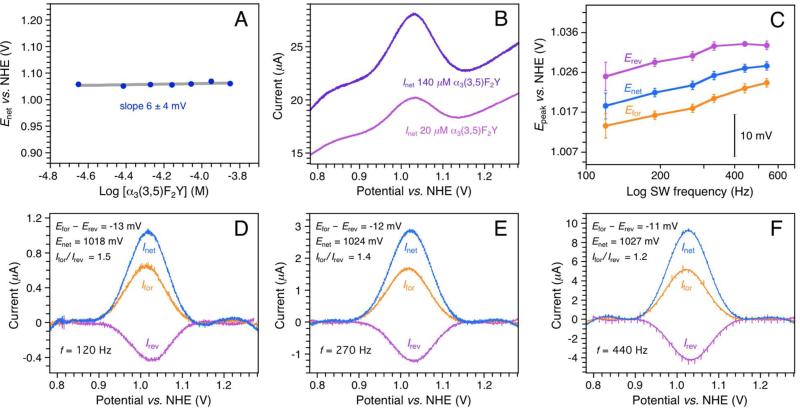
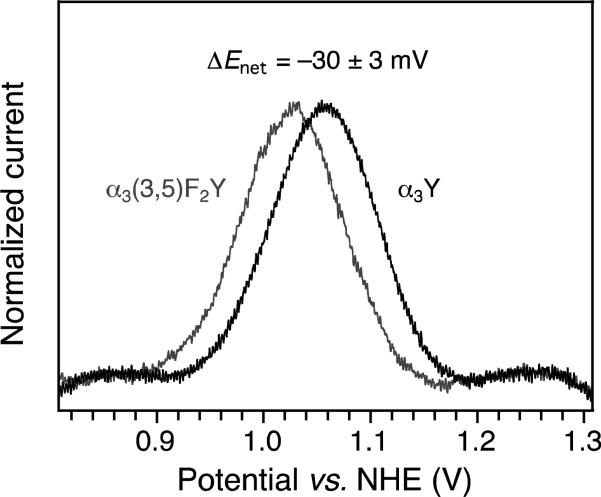
SWV voltammetry was conducted using a PGE working electrode. In SWV, the base potential (Estep) is changed incrementally in a series of forward and reverse pulses (17, 29, 30). The pulse height and length are set by the SW amplitude (ESW) and frequency (f), respectively. The current is sampled at the end of each alternating pulse and traced out as a function of Estep. The pulse train generates a forward (Ifor), a reverse (Irev), and a net (Inet = Ifor – Irev) voltammogram. Previous studies using the PGE electrode have shown that ~20–100 μM α3X in 20 mM APB buffer and 60–140 mM KCl yields Y and phenol voltammograms with optimal S/N at both acidic (5.5) and alkaline (8.5) pH (23, 24). At these conditions, the system is governed by diffusion-controlled kinetics, the protein does not unfold on the electrode surface, and the observed potential is independent of functional groups present at the electrode surface. The α3(3,5)F2Y SWV measurements were consequently conducted using 20 mM APB and 75 mM KCl. Overall, α3(3,5)F2Y displays electrochemical properties very similar to those of α3Y (23). The Faradaic response is consistent with diffusion-controlled kinetics (Fig. S5) and benign α3(3,5)F2Y/working electrode interactions (Figs. 2A and B). Figure 2C shows the change in the peak potential of the forward (Ifor), reverse (Irev) and net (Inet) currents as a function of the SW frequency. Background-corrected voltammograms from this data series are shown in Figs. 2D-F. Efor, Erev and Enet level off and come together as the SW frequency increases. This is consistent with a redox system that is shifting from the upper quasi-reversible region towards the fully reversible region as the SW frequency increases (29, 30). Enet of 1026 ± 3 mV, Efor of 1021 ± 3 mV, and Erev of 1032 ± 2 mV are observed for the 270-540 Hz range at pH 5.70 ± 0.02. The observed frequency insensitivity and small separation in peak maxima mean that Enet closely approximates E°′ (i.e. within a few mV; 23, 24, 30). With a pKapp of 8.0, we calculate E°′(pH 7.0) to be 952 mV (Table 1). Comparing these results with the earlier reported α3Y SWV study (23), provides an E°′ between Y32 and (3,5)F2Y32 of –28-33 mV. This result was reproduced in a control experiment where α3(3,5)F2Y and α3Y SW voltammograms were collected at identical experimental conditions, using the same electrode setup (Fig. 3, ΔEnet –30 ± 3 mV). We conclude that E°′ of the Y32-O•/Y32-OH redox pair is lowered by 30 ± 3 mV upon fluoro-substitution at ring positions 3 and 5 (carbon atoms next to the phenol oxygen). We further note that the difference between Epeak of aqueous Y and E°′ of α3Y is –150 mV, with the protein being the more oxidizing system (Table 1).
Figure 2.
SWV analysis of the Y-O•/Y-OH redox system in α3(3,5)F2Y. (A) Enet(270 Hz) as a function of [α3(3,5)F2Y], pH 5.53 ± 0.11. Enet is independent of the protein concentration, which is consistent with benign protein/working electrode interactions. (B) Representative net voltammograms (Inet) collected at the upper (140 μM, purple) and lower (20 μM, magenta) limit of the [α3(3,5)F2Y] data series shown in panel A. The Faradaic response is optimal in this protein concentration range. (C) Peak potential of the forward (Efor), reverse (Erev) and net (Enet) α3(3,5)F2Y voltammograms as a function of the SW frequency (f), pH 5.70 ± 0.02. (D, E and F) Background-corrected voltammograms from the data series shown in panel C. SWV settings: 20 mM APB, 75 mM KCl, PGE working electrode, temperature 25° C, step potential 0.15 mV, SW pulse amplitude 25 mV.
Figure 3.
Inet of α3(3,5)F2Y (grey) and α3Y (black) recorded at identical experimental conditions. SWV settings: 20 mM APB, 75 mM KCl, pH 5.62 ± 0.02, PGE working electrode, temperature 25° C, step potential 0.15 mV, SW frequency 190 Hz, SW pulse amplitude 25 mV.
DISCUSSION
Measuring tyrosine reduction potentials
There are three basic properties associated with tyrosine that make electrochemical characterization a major challenge. The measurements must be carried out at highly oxidizing conditions (~ 1.0 ± 0.3 V vs. NHE), the Y• state is reactive (e.g. 12-16), and both the reduced (ε280 1490 M–1 cm–1; 36, 37) and oxidized (ε408 of 2750 M–1 cm–1; 41) states have weak extinction coefficients. The combination of these three properties rule out redox titration as a viable method to study the thermodynamics of Y redox cofactors. This is significant since redox titration, using either chemical titrants and a redox cuvette or a potentiostat-controlled spectroelectrochemical cell, is the most common approach to measure reduction potentials of protein redox cofactors. The two former properties also make voltammetry a challenge. Measurements performed in the + 1.0 V range will generate background currents arising from the working electrode and from the water solvent itself. These background currents will compromise data analysis unless the Faradaic current reflecting the Y redox cofactor is prominent. Moreover, uncontrolled oxidation of surface residues and general oxidative damage to the host protein are likely events. In addition to these concerns are the inherent issues associated with protein voltammetry, i.e., to identify conditions for which the folded protein exhibits direct and reversible electron transfer between the working electrode and the redox site of interest (42-44). These are the main reasons that a direct voltammetry approach on a complex radical enzyme such as E. coli RNR is not practical or even possible.
The α3X system was developed to address these experimental barriers and allow rigorous electrochemical characterization of aromatic amino acids buried within a structured protein. In three recent studies (22-24), we have demonstrated that pulsed voltammetry methods (differential pulse voltammetry and SWV) generate reversible Y and phenol protein voltammograms of high quality. Control studies have shown that the characteristics of the α3X voltammograms are highly reproducible and that they uniquely reflect the aromatic residue at position 32 and its local environment. The more commonly used method of cyclic voltammetry (17) generates a poor Faradaic response from the α3X proteins (22) and this method was deemed too insensitive for this system. We note that the potential range probed in this study is very oxidizing for a protein system with measured Enet values well above +1.0 V. The high quality of the presented voltammograms is the result of combining the sensitivity provided by the pulsed SWV method (due to effective elimination of capacitative background currents) with carefully optimized protein/PGE working electrode conditions. The latter include electrode polishing procedures (described in the Materials and Methods sections), the sample composition (optimized for the α3X/PGE system in Refs. 23 and 24), and the protein concentration (Fig. 2A). Importantly, the electrochemical reversibility observed for the α3X proteins reflects the long halftimes (> 100s of ms) of the radicals generated in this system (23, 24). Thus, the protein environment efficiently blocks the deleterious Y• side reactions that typically compromise electrochemical characterization of the solvated species.
In this study the optimized α3X radical system was combined with in vivo nonsense codon suppression (28) to measure E°′ of a protein 3,5-F2Y residue. The use of unnatural amino acids to study protein electron-transfer (ET) and proton coupled electron-transfer (PCET) processes involving Y redox sites has emerged as an informative experimental approach (31, 45, 46). The Y analogues provide a means to introduce major changes in the pKa of the reduced state and in the E°′ values of the Y-O•/Y-OH and Y-O•/Y-O– redox couples. The pKa of the oxidized state (–2 for aqueous Y-OH•+; 47) is predicted to be non-accessible within the structural and catalytic pH ranges of proteins. Residue 32 resides in a structured, solvent-protected and low dielectric site typical of natural Y redox cofactors. The α3X radical system will be used to generate a consistent set of ΔE°′ values to provide a guide for interpreting mutation-induced changes in the free-energy profile of native ET/PCET chains involving aromatic amino-acid redox site(s). The availability of protein E°′ values removes the uncertainty associated with using solution potentials to interpret protein redox events. Table 1 shows that there are considerable differences in protein potentials obtained under reversible conditions relative to solution potentials obtained under irreversible conditions. We find a difference between ΔE°′ (protein Y32 and 3,5-F2Y32, –30 ± 3 mV) and ΔEpeak (aqueous Y and 3,5-F2Y, –60 mV) of 30 mV (Table 1). This difference most certainly arises from the irreversible characteristics of the small-molecule measurements. We also found that the absolute values of E°′ (protein Y32 and 3,5-F2Y32) vs. Epeak (aqueous Y and 3,5-F2Y) is rather large (–150-180 mV). This observation suggests that the influence by the protein matrix on the absolute potentials of radical cofactors may be substantial. We conclude that site-specific incorporation of an unnatural amino acid into the electrochemically reversible α3X redox system is a feasible and informative approach. It will be expanded to include other Y analogues in future studies, as described in more detail below.
Using the α3X system as a guide to interpret mechanistic studies of E. coli RNR
RNRs catalyze the formation of deoxynucleotides (dNDPs) from their corresponding nucleotides (1, 31). E. coli class Ia RNR is composed of two homodimeric subunits, α2 and β2. A stable diferric-Y122• cofactor in β2 generates a transient cysteine radical (C439•) in the active site of α2 located 35 Å away. The reversible long range radical-transfer process, triggered by substrate and effector binding to α2, is proposed to involve multiple PCET steps via a conserved pathway (Y122 ⇔ [W48] ⇔ Y356 in β2 to Y731 ⇔ Y730 ⇔ C439 in α2; 31). We have recently described the site-specific insertion of 3-nitrotyrosine (NO2Y) in place of Y122 inβ2 (48). The diferric-NO2Y122• cofactor was generated and studies with α2, substrate (CDP) and effector (ATP) allowed the first observation of a transient, kinetically competent Y• on pathway by electron paramagnetic resonance (EPR) spectroscopy (48). Pulsed electron-electron double resonance (PELDOR) spectroscopy was used to establish the primary location of the new radical as Y356•-β2 and suggested the formation of a small percentage of radical at either Y731 or Y730 in α2. To probe this further, the reaction of diferric-NO2Y122•-β2 with (3,5)F2Y731-α2 (or (3,5)F2Y730-α2), CDP and ATP was performed. Analysis of the reactions by X-band EPR spectroscopy demonstrated an equilibrium between Y356• in β2 and (3,5)F2Y731• or (3,5)F2Y730• in α2 with 85-90% of the spin localized at Y356 and 15-10% distributed over F2Y731/F2Y730 (32). To extrapolate the equilibrium observed with 3,5-F2Y to Y, the native pathway residue, requires a knowledge of E°′ values for 3,5-F2Y and Y. To date only solution Epeak values derived from irreversible voltammograms have been available for the fluorotyrosines (11, 31). The SWV measurements performed on α3Y and α3(3,5)F2Y provide the first E°′ values representing the radical species in a well-defined protein environment. The small ΔE°′ of –30 ± 3 mV between Y32 and 3,5-F2Y32 suggests that a thermodynamic landscape, formed by three transient tyrosyl radicals (Y356, Y731, Y730) of similar energy with one (Y356) being most prevalent, is a reasonable depiction of nature's design within this part of the pathway.
Future perspectives
We have shown that formal reduction potentials representing a reversible Y-O•/Y-OH redox system can be obtained for α3Y (23) and α3(3,5)F2Y (this work). Future studies will involve the incorporation of other modified amino acids at position 32 in the α3X scaffold to obtain a consistent set of E°′ values from Y and modified Y residues. To determine E°′ of 3-aminotyrosine (NH2Y) and 2,3,5-trifluorotyrosine (2,3,5F3Y) incorporated into the α3X protein is of particular interest. In the former case, studies with NH2Y in place of Y356, Y731 and Y730 show an active pathway and that dNDPs can be produced (49). This is an interesting observation since the solution potential of NH2Y (9) suggests that this species may act as a radical sink shutting down the pathway, as observed in experiments on 3,4-dihyroxyphenylalanine (DOPA)-labeled Y356-β2 (50). Obtaining a solid protein ΔE°′ value for NH2Y32 vs. Y32 would be valuable for interpreting the characteristics of the NH2Y-labeled RNR systems (see Ref. 31 for more details). In the latter case, experiments are in progress in an effort to measure the thermodynamics between Y122 and Y356 in β2. Toward that goal, diferric-FnY122• (n = 2 or 3)-β2s have been successfully generated and the reactions of (3,5)F2Y122•-β2 and (2,3,5)F3Y122•-β2 with α2, CDP and ATP have been analyzed by EPR spectroscopy (28, 31). These studies reveal that only (2,3,5)F3Y122• is capable of generating a new pathway radical that is postulated to be Y356•. Detailed investigations are ongoing with FnY-β2s and wt-α2 and Y731F-α2 to establish if there is an equilibrium between Y356• and FnY122. Thus, protein E°′ values of (2,3,5)F3Y and other FnY residues relative to Y may also play an important role in defining the thermodynamic landscape within this part of the pathway. Finally, work to complement the α3X protein E°′ data series with α3W is also in progress. Tryptophan is an important protein redox cofactor observed in a number of enzymes (e.g. 51-53), proposed to participate in the E. coli RNR radical-transfer pathway (54, 55), and engineered to study multistep electron tunneling in Pseudomonas aeruginosa azurin (56).
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH grants GM079190 (C.T.) and by GM29595 (J.S).
Abbreviations
- α2 and β2
subunits of E. coli RNR
- α3Y
α3W and α3C, de novo protein containing a single buried tyrosine, tryptophan or cysteine
- α3X
generic designation for this protein family
- α3(3,5)F2Y
α3Y containing 3,5-difluorotyrosine
- APB buffer
sodium acetate, potassium phosphate, sodium borate buffer
- ATP
adenosine 5′-triphosphate
- CDP
cytidine 5′-diphosphate
- E°′
formal reduction potential
- Epeak
voltammetry peak potential
- Efor
Erev and Enet, peak potential of the forward, reverse and net current in SWV
- Estep
staircase base potential in SWV
- ESW
square-wave amplitude
- EPR
electron paramagnetic resonance
- ET
electron transfer
- f
square-wave frequency
- FnY (n = 2, 3)
di- and tri-substituted fluorotyrosines
- FnY. RS
fluorotyrosine aminoacyl-tRNA synthetase
- pKapp
apparent pKa
- Ifor
Irev and Inet, forward, reverse and net current in SWV
- PCET
proton-coupled electron transfer
- PGE
pyrolytic graphite edge
- RNR
ribonucleotide reductase
- SWV
square-wave voltammetry
- TPL
tyrosine phenol lyase
- Y•
tyrosine radical
Footnotes
Supporting Information. Materials and Methods; α3(3,5)F2Y protein expression analysis (Fig. S1); HPLC and MALDI-TOF evaluation of purified α3(3,5)F2Y (Fig. S2); pKapp fitting analysis (Fig. S3); chemical denaturation plots of α3(3,5)F2Y and α3Y at pH 5.0 and 5.5 (Fig. S4); α3(3,5)F2Y Inet as a function of ESW (Fig S5). This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Stubbe J, van der Donk WA. Protein radicals in enzyme catalysis. Chem. Rev. 1998;98:705–762. doi: 10.1021/cr9400875. [DOI] [PubMed] [Google Scholar]
- 2.Tommos C, Babcock GT. Proton and hydrogen currents in photosynthetic water oxidation, Biochim. Biophys. Acta. 2000;1458:199–219. doi: 10.1016/s0005-2728(00)00069-4. [DOI] [PubMed] [Google Scholar]
- 3.Pesavento RP, van der Donk WA. Tyrosyl radical cofactors. Adv. Protein Chem. 2001;58:317–385. doi: 10.1016/s0065-3233(01)58008-0. [DOI] [PubMed] [Google Scholar]
- 4.Hoganson CW, Tommos C. The function and characteristics of tyrosyl radical cofactors. Biochim. Biophys. Acta. 2004;1655:116–122. doi: 10.1016/j.bbabio.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 5.Warren JJ, Winkler JR, Gray HB. Redox properties of tyrosine and related molecules. FEBS Lett. 2012;586:596–602. doi: 10.1016/j.febslet.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harriman A. Further comments on the redox potentials of tryptophan and tyrosine. J. Phys. Chem. 1987;91:6102–6104. [Google Scholar]
- 7.DeFelippis MR, Murthy CP, Faraggi M, Klapper MH. Pulse radiolytic measurement of redox potentials: The tyrosine and tryptophan radicals. Biochemistry. 1989;28:4847–4853. doi: 10.1021/bi00437a049. [DOI] [PubMed] [Google Scholar]
- 8.Lind J, Shen X, Eriksen TE, Merényi G. The one-electron reduction potential of 4-substituted phenoxyl radicals in water. J. Am. Chem. Soc. 1990;112:479–482. [Google Scholar]
- 9.DeFelippis MR, Murthy CP, Broitman F, Weinraub D, Faraggi M, Klapper MH. Electrochemical properties of tyrosine phenoxy and tryptophan indolyl radicals in peptides and amino acid analogues. J. Phys. Chem. 1991;95:3416–3419. [Google Scholar]
- 10.Tommos C, Skalicky JJ, Pilloud DL, Wand AJ, Dutton PL. De novo proteins as models of radical enzymes. Biochemistry. 1999;38:9495–9507. doi: 10.1021/bi990609g. [DOI] [PubMed] [Google Scholar]
- 11.Seyedsayamdost MR, Reece SY, Nocera DG, Stubbe J. Mono-, di-, tri-, and tetra-substituted fluorotyrosines: New probes for enzymes that use tyrosyl radicals in catalysis. J. Am. Chem. Soc. 2006;128:1569–1579. doi: 10.1021/ja055926r. [DOI] [PubMed] [Google Scholar]
- 12.Gross AJ, Sizer IW. The oxidation of tyramine, tyrosine, and related compounds by peroxide. J. Biol. Chem. 1959;234:1611–1614. [PubMed] [Google Scholar]
- 13.Lehrer SS, Fasman GD. Ultraviolet irradiation effects in poly-L-tyrosine and model compounds. Identification of bityrosine as a photoproduct. Biochemistry. 1967;6:757–767. doi: 10.1021/bi00855a017. [DOI] [PubMed] [Google Scholar]
- 14.Boguta G, Dancewicz AM. Radiation-induced dimerization of tyrosine and glycyltyrosine in aqueous solutions. Int. J. Radiat. Biol. 1981;39:163–174. doi: 10.1080/09553008114550181. [DOI] [PubMed] [Google Scholar]
- 15.Karam LR, Dizdaroglu M, Simic MG. OH radical-induced products of tyrosine peptides. Int. J. Radiat. Biol. 1984;46:715–724. doi: 10.1080/09553008414551951. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins CL, Davies MJ. Generation and propagation of radical reactions on proteins. Biochim. Biophys. Acta. 2001;1504:93–109. doi: 10.1016/s0005-2728(00)00252-8. [DOI] [PubMed] [Google Scholar]
- 17.Bard AJ, Faulkner LR. Electrochemical methods: Fundamentals and applications. 2nd Ed. John Wiley & Sons, Inc.; USA: 2001. [Google Scholar]
- 18.Savéant JM. Elements of molecular and biomolecular electrochemistry: An electrochemical approach to electron transfer chemistry. John Wiley & Sons, Inc.; USA: 2006. [Google Scholar]
- 19.Warshel A, Sharma PK, Kato M, Xiang Y, Hanbin Liu, Olsson MHM. Electrostatic basis for enzyme catalysis. Chem. Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 20.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 21.Westerlund K, Berry BW, Privett HK, Tommos C. Exploring amino-acid radical chemistry: protein engineering and de novo design. Biochim. Biophys. Acta. 2005;1707:103–116. doi: 10.1016/j.bbabio.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Martínez-Rivera MC, Berry BW, Valentine KG, Westerlund K, Hay S, Tommos C. Electrochemical and structural properties of a protein system designed to generate tyrosine Pourbaix diagrams. J. Am. Chem. Soc. 2011;133:17786–17795. doi: 10.1021/ja206876h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry BW, Martínez-Rivera MC, Tommos C. Reversible voltammograms and a Pourbaix diagram for a protein tyrosine radical. Proc. Nat. Acad. Sci. U.S.A. 2012;109:9739–9743. doi: 10.1073/pnas.1112057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tommos C, Valentine KG, Martínez-Rivera MC, Liang L, Moorman VR. Reversible phenol oxidation and reduction in the structurally well-defined 2-mercaptophenol-α3C protein. Biochemistry. 2013;52:1409–1418. doi: 10.1021/bi301613p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai Q-H, Tommos C, Fuentes EJ, Blomberg MRA, Dutton PL, Wand AJ. Structure of a de novo designed model protein of radical enzymes. J. Am. Chem. Soc. 2002;124:10952–10953. doi: 10.1021/ja0264201. [DOI] [PubMed] [Google Scholar]
- 26.Hay S, Westerlund K, Tommos C. Moving a phenol hydroxyl group from the surface to the interior of a protein: Effects on the phenol potential and pKA. Biochemistry. 2005;44:11891–11902. doi: 10.1021/bi050901q. [DOI] [PubMed] [Google Scholar]
- 27.Hay S, Westerlund K, Tommos C. Redox characteristics of a de novo quinone protein. J. Phys. Chem. B. 2007;111:3488–3495. doi: 10.1021/jp066654x. [DOI] [PubMed] [Google Scholar]
- 28.Minnihan EC, Young DD, Schultz PG, Stubbe J. Incorporation of fluorotyrosines into ribonucleotide reductase using an evolved, polyspecific aminoacyl-tRNA synthetase. J. Am. Chem. Soc. 2011;133:15942–15945. doi: 10.1021/ja207719f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osteryoung J, O'Dea JJ. Square-wave voltammetry. In: Bard AJ, editor. Electroanalytical chemistry. Vol. 5. Marcel Dekker; New York: 1986. pp. 209–308. [Google Scholar]
- 30.Mirčeski V, Komorsky-Lovrić Š, Lovrić M. Square-wave voltammetry: Theory and applications. In: F Scholz., editor. Monographs in electrochemistry. Springer-Verlag; Berlin, Germany: 2007. [Google Scholar]
- 31.Minnihan EC, Nocera DG, Stubbe J. Reversible, long-range radical transfer in E. coli class 1a ribonucleotide reductase. Acc. Chem. Res. 2013 doi: 10.1021/ar4000407. 10.1021/ar4000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yokoyama K, Smith AA, Corzilius B, Griffin RG, Stubbe J. Equilibration of tyrosyl radicals (Y356•, Y731•, Y730•) in the radical propagation pathway of the Escherichia coli Class 1a ribonucleotide reductase. J. Am. Chem. Soc. 2011;133:18420–18432. doi: 10.1021/ja207455k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H, Gollnick P, Phillips RS. Site-directed mutagenesis of His343 → Ala in Citrobacter freundii tyrosine phenol-lyase: Effects on the kinetic mechanism and rate-determining step. Eur. J. Biochem. 1995;229:540–549. [PubMed] [Google Scholar]
- 34.Seyedsayamdost MR, Yee CS, Stubbe J. Site-specific incorporation of fluorotyrosines into the R2 subunit of E. coli ribonucleotide reductase by expressed protein ligation. Nat. Protoc. 2007;2:1225–1235. doi: 10.1038/nprot.2007.159. [DOI] [PubMed] [Google Scholar]
- 35.Kim K, Cole PA. Kinetic analysis of a protein tyrosine kinase reaction transition state in the forward and reverse directions. J. Am. Chem. Soc. 1998;120:6851–6858. [Google Scholar]
- 36.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 37.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Prot. Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl α-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 39.Salwiczek M, Nyakatura EK, Gerling UIM, Ye S, Koksch B. Fluorinated amino acids: compatibility with native protein structures and effects on protein–protein interactions. Chem. Soc. Rev. 2012;41:2135–2171. doi: 10.1039/c1cs15241f. [DOI] [PubMed] [Google Scholar]
- 40.Buer BC, Marsh ENG. Fluorine: A new element in protein design. Prot. Sci. 2012;21:453–462. doi: 10.1002/pro.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feitelson J, Hayon E. Electron ejection and electron capture by phenolic compounds. J. Phys. Chem. 1973;77:10–15. [Google Scholar]
- 42.Rusling JF. Enzyme bioelectrochemistry in cast biomembrane-like films. Acc. Chem. Res. 1998;31:363–369. [Google Scholar]
- 43.Armstrong FA, Wilson GS. Recent developments in faradaic bioelectrochemistry. Electrochim. Acta. 2000;45:2623–2645. [Google Scholar]
- 44.Armstrong FA. Recent developments in dynamic electrochemical studies of adsorbed enzymes and their active sites. Curr. Opin. Chem. Biol. 2005;9:110–117. doi: 10.1016/j.cbpa.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 45.Rappaport F, Boussac A, Force DA, Peloquin J, Brynda M, Sugiura M, Un S, Britt RD, Diner BA. Probing the coupling between proton and electron transfer in photosystem II core complexes containing a 3-fluorotyrosine. J. Am. Chem. Soc. 2009;131:4425–4433. doi: 10.1021/ja808604h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren JJ, Herrera N, Hill MG, Winkler JR, Gray HB. Electron flow through nitrotyrosine in Pseudomonas aeruginosa azurin. J. Am. Chem. Soc. 2013;135:11151–11158. doi: 10.1021/ja403734n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixon WT, Murphy D. Determination of the acidity constants of some phenol radical cations by means of electron spin resonance. J. Chem. Soc. Faraday. Trans. II. 1976;72:1221–1230. [Google Scholar]
- 48.Yokoyama K, Uhlin U, Stubbe J. A hot oxidant, 3-NO2Y122 radical, unmasks conformational gating in ribonucleotide reductase. J. Am. Chem. Soc. 2010;132:15368–15379. doi: 10.1021/ja1069344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seyedsayamdost MR, Xie J, Chan CTY, Schultz PG, Stubbe J. Site-specific insertion of 3-aminotyrosine in subunit α2 of E. coli ribonucleotide reductase: Direct evidence for involvement of Y730 and Y731 in radical propagation. J. Am. Chem. Soc. 2007;129:15060–15071. doi: 10.1021/ja076043y. [DOI] [PubMed] [Google Scholar]
- 50.Seyedsayamdost MR, Stubbe J. Site-specific replacement of Y356 with 3,4-dihydroxyphenlalanine in the β2 subunit of E. coli ribonucleotide reductase. J. Am. Chem. Soc. 2006;128:2522–2523. doi: 10.1021/ja057776q. [DOI] [PubMed] [Google Scholar]
- 51.Tsai A-L, Kulmacz RJ. Prostaglandine H synthesis: Resolved and unresolved mechanistic issues. Arch. Biochem. Biophys. 2010;493:103–124. doi: 10.1016/j.abb.2009.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aubert C, Vos MH, Mathis P, Eker APM, Brettel K. Intraprotein radical transfer during photoactivation of DNA photolyase. Nature. 2000;405:586–590. doi: 10.1038/35014644. [DOI] [PubMed] [Google Scholar]
- 53.Tarboush NA, Jensen LMR, Yukl ET, Geng J, Liu A, Wilmot CM, Davidson VL. Mutagenesis of tryptophan 199 suggests that hopping is required for MauG-dependent tryptophan tryptophylquinone biosynthesis. Proc. Nat. Acad. Sci. U.S.A. 2011;108:16956–16961. doi: 10.1073/pnas.1109423108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 55.Ekberg M, Birgander P, Sjöberg B-M. In vivo assay for low-activity mutant forms of Escherichia coli ribonucleotide reductase. J. Bacteriol. 2003;185:1167–1173. doi: 10.1128/JB.185.4.1167-1173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shih C, Museth AK, Abrahamsson M, Blanco-Rodriguez AM, Di Bilio AJ, Sudhamsu J, Crane BR, Ronayne KL, Towrie M, Vlcek A, Jr., Richards JH, Winkler JR, Gray HB. Tryptophan-accelerated electron flow through proteins. Science. 2008;320:1760–1762. doi: 10.1126/science.1158241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.