Abstract
Tobacco smoke is the major environmental risk factor underlying lung carcinogenesis. However, approximately one-tenth smokers develop lung cancer in their lifetime indicating there is significant individual variation in susceptibility to lung cancer. And, the reasons for this are largely unknown. In particular, the genetic variants discovered in genome-wide association studies (GWAS) account for only a small fraction of the phenotypic variations for lung cancer, and gene–environment interactions are thought to explain the missing fraction of disease heritability. The ability to identify smokers at high risk of developing cancer has substantial preventive implications. Thus, we undertook a gene–smoking interaction analysis in a GWAS of lung cancer in Han Chinese population using a two-phase designed case–control study. In the discovery phase, we evaluated all pair-wise (591 370) gene–smoking interactions in 5408 subjects (2331 cases and 3077 controls) using a logistic regression model with covariate adjustment. In the replication phase, promising interactions were validated in an independent population of 3023 subjects (1534 cases and 1489 controls). We identified interactions between two single nucleotide polymorphisms and smoking. The interaction P values are 6.73 × 10− 6 and 3.84 × 10− 6 for rs1316298 and rs4589502, respectively, in the combined dataset from the two phases. An antagonistic interaction (rs1316298–smoking) and a synergetic interaction (rs4589502–smoking) were observed. The two interactions identified in our study may help explain some of the missing heritability in lung cancer susceptibility and present strong evidence for further study of these gene–smoking interactions, which are benefit to intensive screening and smoking cessation interventions.
Introduction
Lung cancer is the most commonly diagnosed cancer and the global leading cause of cancer death (1). In China, the incidence and mortality rates for lung cancer have increased over the last three decades, primarily due to tobacco use (2). Tobacco smoke contains numerous carcinogens that can induce various types of DNA damage, which is believed to be the major mechanism underlying lung carcinogenesis (3). Although tobacco smoke is the primary risk factor for lung cancer, approximately one-tenth smokers develops lung cancer in their lifetime indicating an individual variation in susceptibility to tobacco-induced lung cancer (4). It is most likely that multiple susceptibility factors must be accounted for to represent the true dimensions of gene–environment interactions (5). Take gene–smoking interaction as example. It means that the risk effect of smoking varies among subjects with different genetic background or the joint effect of gene and smoking deviates from the accumulated main effects of gene and smoking. Thus, the ability to identify smokers with the high risks of developing cancer has substantial preventive implications, such as the intensive screening and the smoking cessation interventions.
Lung cancer is a polygenic disease, many genetic factors seem to have an important role in disease development. During the past 3 years, several genome-wide association studies (GWAS) have successfully identified 16 genetic susceptibility loci with P ≤ 5.00 × 10− 8 that are associated with lung cancer risk (6–11). However, most of these studies were conducted in populations of European descent, and many identified risk alleles have not been adequately evaluated in Asian populations. We have recently identified five new susceptibility loci (5q32, 10p14, 13q12.12, 20q13.2 and 22q12.2) in the Han Chinese population (10,11). Even so, the genetic variants with a significant main effect account for only a small fraction of phenotypic variations (12,13).
Gene–environment interaction may account for the missing heritability of the complex diseases (14). However, few studies have investigated the gene–environment interactions on a genome-wide scale, following the initial findings of GWAS (not only lung cancer study). Because current GWAS is designed to detect the main effect of genetic variants, the ability to detect gene–environment interactions even in the single nucleotide polymorphism (SNP) analysis has been limited. Pilot studies have revealed that significant gene–smoking interactions may contribute to the risk of lung cancer based on candidate gene strategy in multiple ethnic populations (15–28). We also previously reported significant gene–smoking interactions, only focusing on those 13 most significant SNPs with main effects in a GWAS of lung cancer (10,11,29). In this study, we performed an extended gene–smoking interaction analysis on a genome-wide scale (591 370 SNPs) based on our GWAS data.
We adopted a two-phase designed case–control study. In the discovery phase, we screened all possible gene–smoking interactions using GWAS population of 5408 subjects (2331 cases and 3077 controls). We then validated the most promising interactions using an independent population of 3023 subjects (1534 cases and 1489 controls).
Materials and methods
Study participants
The populations used in the discovery phase (the Nanjing study and the Beijing study) have been described previously (10). The Nanjing study included 1473 cases and 1962 controls, and the Beijing study included 858 cases and 1115 controls (Supplementary Table S1, available at Carcinogenesis Online). Another independent population from Beijing that used in the replication phase has also been described previously and consisted of 1534 cases and 1489 controls (Supplementary Table S1, available at Carcinogenesis Online) (11). All controls were frequency-matched for age, gender and geographic regions to each set of the lung cancer cases. Smoking information was collected in a face-to-face interview. We used the same criteria to define the smokers as that in previous study (10). Individuals who had smoked an average of <1 cigarette per day and for <1 year in their lifetime were defined as non-smokers; otherwise, the subjects were defined as smokers.
Ethics statement
All subjects provided informed consent, and the institutional review boards of each participating institution approved this collaborative study.
Genotyping analysis and quality control
A total of 5543 subjects (2383 lung cancer cases and 3160 controls) were originally genotyped using Affymetrix Genome-Wide Human SNP Array 6.0 chips containing 906 703 SNPs, followed by a systematic quality control procedure before association analysis, as described elsewhere (10). SNPs were excluded when they fit the following criteria: (i) SNPs were not mapped on autosomal chromosomes (SNPs at chromosome X were only calculated for female participants), (ii) SNPs had a call rate <95% in all GWAS samples or in either the Nanjing Study or the Beijing Study samples, (iii) SNPs had minor allele frequency < 0.05, (iv) the genotype distributions of SNPs deviated from those expected by Hardy–Weinberg equilibrium (P < 1×10− 5 in all GWAS samples or P < 1×10− 4 in either the Nanjing Study or the Beijing Study samples) or (v) SNPs did not have clear genotyping clusters (Supplementary Figure S1, available at Carcinogenesis Online). The samples with overall genotype completion rates <95% were excluded from further analysis (13 subjects were excluded). Seven cases were excluded because they showed gender discrepancies. An additional 89 unexpected duplicates or probable relatives were excluded based on pair-wise identity by state according to their PI_HAT value in PLINK (all PI_HAT > 0.25). Heterozygosity rates were calculated, and >6 SD from the mean was used as the exclusion criteria (22 samples were excluded). In addition, four outliers in population stratification analysis were excluded (Supplementary Figure S2, available at Carcinogenesis Online). Finally, 2331 cases and 3077 controls with 591 370 SNPs were included in the discovery phase. We imputed the un-genotyped SNPs using Minimac software (30) based on linkage disequilibrium (LD) information from the 1000 Genomes database with Han Chinese in Beijing (CHB) and Japanese in Tokyo (JPT) as the reference set (released June 2010).
Genotyping analysis in the replication population was conducted using iPLEX Sequenom MassARRAY platform (Sequenom). The primers and probes used are available on request.
Statistical analysis
Genome-wide gene–smoking interaction association analysis was performed using a logistic regression model adjusted for age, gender and first four principal components generated by EIGENSTRAT 3.0 (31) (Equation 1). SNPs were coded in an additive genetic model (0, 1 and 2). Smoking was coded as smoker (1) or non-smoker (0).
| (1) |
We tested all possible (591 370) gene–smoking interactions in the Nanjing study, the Beijing study and the total population in the discovery phase. We used the following criteria for selecting SNPs for validation in the replication phase. (i) A P value of interaction <0.01 in both the Nanjing and the Beijing studies, and <5.0 × 10− 5 in the total discovery population. (ii) A consistent pattern of gene–smoking interaction between the Nanjing and the Beijing studies. (iii) A clear genotyping cluster for the SNP. (iv) Among multiple SNPs in LD (r 2 > 0.8) showing an interaction with smoking, we choose the one with the lowest missing rate.
We used PLINK 1.07 for the interaction analysis of the GWAS data (32). The quantile–quantile plot of interaction P values was generated using R 2.14.0 (The R Foundation for Statistical Computing). The chromosomal region related with to the interaction was plotted using LocusZoom 1.1 (33). The Manhattan plot of −log10 P of interaction was generated by using Haploview 4.1 (34). Differences in the expression levels of genes in smokers versus non-smokers were assessed by a Wilcoxon rank-sum test. The P values are two sided, and the 95% confidence intervals (95% CIs) of the odds ratios (ORs) are given.
The predictive ability of the newly identified gene–smoking interactions
The integrated discrimination improvement (IDI) and the net reclassification improvement (NRI) are two statistics proposed to evaluate the significance of novel predictors (35). The IDI measures the new model’s improvement in average sensitivity without sacrificing average specificity. The relative IDI is defined using Equation 2. Here, the baseline logistic regression model including age, gender, smoking, rs1316298 and rs4589502 was denoted as model 1. The new model added with rs1316298–smoking and rs4589502–smoking interactions was denoted as model 2. and are the mean of the predicted probability derived from logistic regression model for cases and controls, respectively.
| (2) |
The NRI measures the correctness of reclassification of subjects based on their predicted probabilities of events using the new model. The N case and N control are the number of cases and controls, respectively. P is the predicted probability for each subject, derived from logistic regression model. The category-free NRI is defined using Equation 3, which means the proportion for cases/controls with the predicted probability moving up/down in model 2 compared with model 1.
| (3) |
Quantitative reverse transcription–polymerase chain reaction
Quantitative reverse transcription–polymerase chain reaction was performed to determine the messenger RNA (mRNA) expressions of guanine nucleotide binding protein gamma 2 (GNG2), FERM-domain containing-6 (FRMD6) and nidogen-2 (NID2). RNAs from whole blood of 131 health individuals were isolated with the QIAamp RNA Blood Mini Kit (QIAGEN). We used TaqMan gene expression probes (Applied Biosystems) to perform quantitative reverse transcription–polymerase chain reaction assay. All real-time PCR reactions, including no-template controls and real-time minus controls, were run by using the ABI7900 Real-Time PCR System (Applied Biosystems) and performed in triplicate. β-Actin gene was used to normalize the expression levels. A relative expression was calculated using the equation 2− ΔCt (Ct, cycle threshold), in which ΔCt = Ctgene − Ctβ-actin.
Bioinformatics analysis
Functional annotation for SNPs was based on Regulome DB database(see ‘URLs’). Open chromatin regions, recognized by DNase I hypersensitive site sequencing (DNase-seq) are associated with gene regulatory elements, including promoters, enhancers, silencers, insulators and locus control region. Whether these SNPs located in the DNase-seq peaks were derived from the ENCODE database (see ‘URLs’). Putative exonic splicing enhancer motifs specific for human Ser/Arg-rich proteins (SR proteins) were predicted by ESEfinder3.0(see ‘URLs’).The extent by which the SNPs affect microRNAs binding was predicted by Patrocles (see ‘URLs’).
Results
Associations between genotypes, smoking and lung cancer risk
The logistic regression model in the total GWAS sample revealed 27 pairs of gene–smoking interactions with P < 5.0 × 10− 5 (Supplementary Figure S3, available at Carcinogenesis Online). The quantile–quantile plot for all P values of the gene–smoking interactions also revealed a good match between the distribution of observed interaction P values and those expected by chance (Supplementary Figure S4, available at Carcinogenesis Online). The small genomic control inflation factor (λ) of 1.021 indicates a low possibility of false positives due to population stratification. Among the 27 pairs of gene–smoking interactions screened out, 6 fit the criteria above were selected for further validation in the replication phase (Table I, Supplementary Table S2, available at Carcinogenesis Online). The other 21 were presented in Supplementary Table S3, available at Carcinogenesis Online.
Table I.
Gene–smoking interaction analysis using a logistic regression model
| Study | Gene | Smoking | Interaction | |||
|---|---|---|---|---|---|---|
| OR (95% CI)a | P a | OR (95% CI)a | P a | OR (95% CI)a | P a | |
| rs1316298 smokingb | ||||||
| GWAS (Nanjing) | 1.35 (1.12, 1.65) | 2.17×10−03 | 4.03 (3.12, 5.22) | 2.46×10−26 | 0.65 (0.50, 0.85) | 1.28×10−03 |
| GWAS (Beijing) | 1.20 (0.92, 1.55) | 1.78×10−01 | 4.04 (2.95, 5.52) | 2.73×10−18 | 0.61 (0.44, 0.84) | 3.09×10−03 |
| GWAS (Total) | 1.25 (1.08, 1.45) | 3.53×10−03 | 3.81 (3.15, 4.61) | 3.42×10−43 | 0.66 (0.55, 0.81) | 4.15×10−05 |
| Replication | 1.05 (0.88, 1.27) | 5.74×10−01 | 4.34 (3.45, 5.47) | 1.53×10−35 | 0.64 (0.50, 0.83) | 8.87×10−04 |
| Combined | 1.11 (1.00, 1.23) | 5.67×10−02 | 3.99 (3.48, 4.58) | 6.71×10−86 | 0.71 (0.62, 0.83) | 6.73×10−06 |
| rs4589502 smokingb | ||||||
| GWAS (Nanjing) | 0.68 (0.53, 0.88) | 3.39×10−03 | 2.80 (2.22, 3.55) | 6.24×10−18 | 1.69 (1.20, 2.36) | 2.33×10−03 |
| GWAS (Beijing) | 0.75 (0.54, 1.05) | 9.10×10−02 | 2.69 (2.04, 3.54) | 1.69×10−12 | 1.76 (1.16, 2.68) | 8.37×10−03 |
| GWAS (Total) | 0.70 (0.58, 0.85) | 3.95×10−04 | 2.66 (2.24, 3.16) | 2.40×10−29 | 1.72 (1.34, 2.22) | 2.61×10−05 |
| Replication | 0.78 (0.62, 0.98) | 2.93×10−02 | 3.26 (2.63, 4.03) | 1.17×10−27 | 1.39 (1.01, 1.91) | 4.40×10−02 |
| Combined | 0.74 (0.64, 0.84) | 1.14×10−05 | 3.01 (2.66, 3.42) | 4.07×10−66 | 1.55 (1.29, 1.87) | 3.84×10−06 |
aAdjusted for age, gender and principal components where appropriate.
brs1316298 (14q22.1), rs4589502 (15q22.32).
Replication and imputation analysis
We found that two SNPs (rs1316298 and rs4589502) had consistent interaction with smoking in both discovery and replication phase. The P values of gene–smoking interaction for the two SNPs in total GWAS samples were 4.15 × 10− 5 and 2.61 × 10− 5, respectively. In the replication phase, the P values of gene–smoking interaction for the two SNPs were 8.87 × 10− 4 and 4.40 × 10− 2, respectively. By combining the datasets from two phases, we observed that the P values of rs1316298–smoking and rs4589502–smoking interactions in 8431 subjects (3865 cases and 4566 controls) were 6.73 × 10− 6 and 3.84 × 10− 6, respectively, with estimates of interaction OR of 0.71 (95% CI: 0.62–0.83) and 1.55 (95% CI: 1.29–1.87).
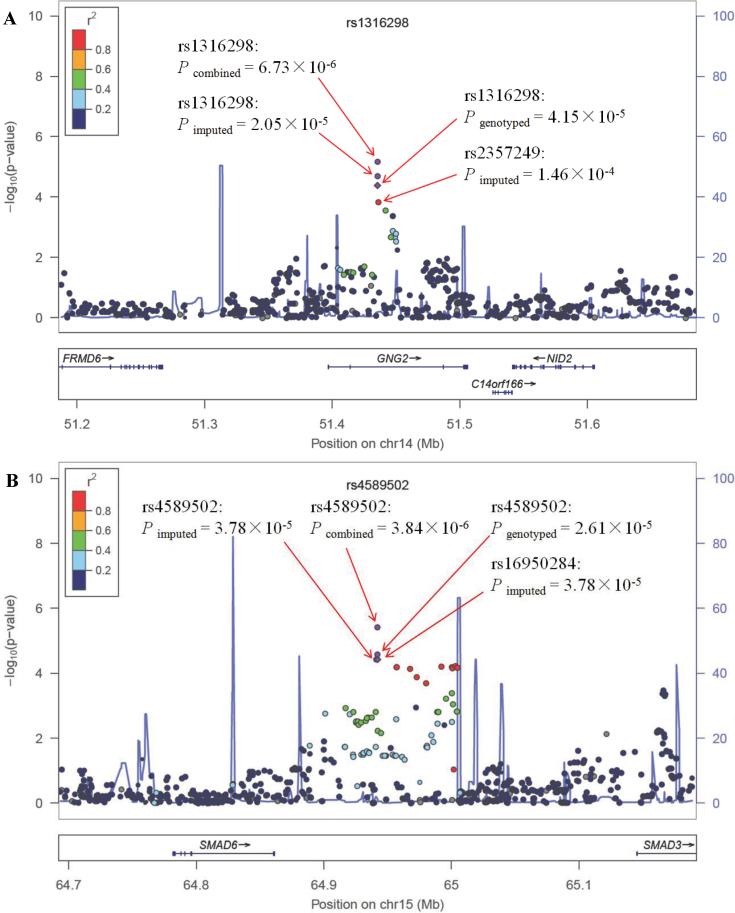
After imputation, we also tested gene–smoking interactions for the imputed SNPs located within 250kb of rs1316298 or rs4589502. The chromosome regional plots were presented in Figure 1. The P values of the imputed rs1316298–smoking and rs4589502–smoking interactions are 2.05 × 10− 5 and 3.78 × 10− 5, respectively. An imputed SNP (rs2357249), in high LD (r 2 = 0.81) with rs1316298, <1kb away, has an interaction P value = 1.46 × 10− 4. We observed that a series of nearby SNPs in high LD with rs4589502 (r 2 > 0.8) interacted with smoking (from P = 6.98 × 10− 5 to P = 3.78 × 10− 5).
Fig. 1.
The chromosome regional plot of the rs1316298 (A) and rs4589502 (B) using gene–smoking interaction P values derived from a logistic regression model adjusted for age, gender and principal components in the total GWAS samples. Results (−log10 P) are shown for SNPs in the 250kb flanking region on each side of the target SNP. The LD for each SNP is presented as colours representing r 2 values. The genes within the region of interest are annotated below each plot, with arrows indicating the direction of transcription.
Stratification analysis
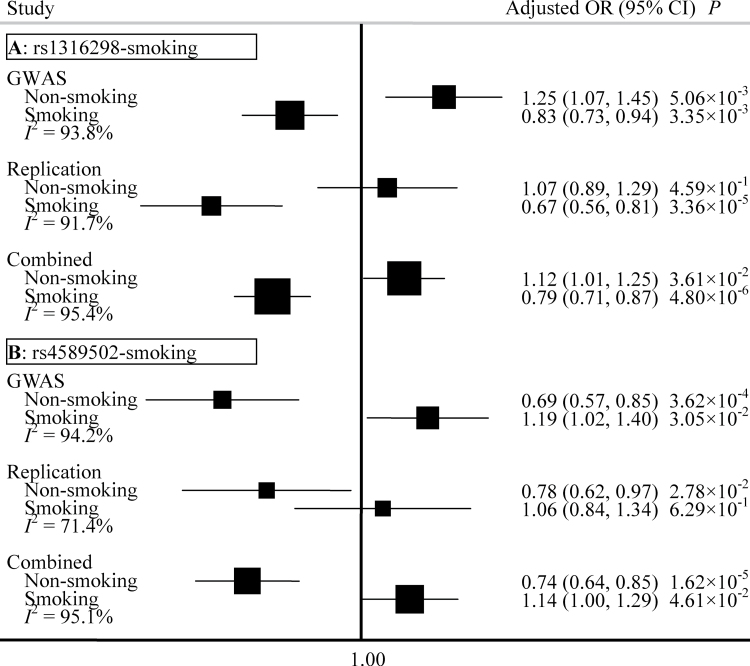
To further explore and understand the mechanism of these interactions, we completed a stratification analysis among non-smokers and smokers. We found the effect of rs1316298 to be different between non-smokers and smokers (Figure 2A). In the combined data from two phases, rs1316298 was found to be a risk factor for non-smokers (OR = 1.12, 95% CI: 1.01–1.25), whereas it had a protective effect for smokers (OR = 0.79, 95% CI: 0.71–0.87). In contrast, rs4589502 is a protective factor for non-smokers (OR = 0.74, 95% CI: 0.64–0.85) and a risk factor for smokers (OR = 1.14, 95% CI: 1.00–1.29) (Figure 2B). The I 2 (variation in OR attributed to heterogeneity) between the two groups is >95%, which also indicates that the two SNPs (rs1316298 and rs4589502) interact with smoking to convey susceptibility for lung cancer risk.
Fig. 2.
Forest plots represent the effect of rs1316298 among non-smokers and smokers (A), and the effect of rs4589502 among non-smokers and smokers (B). Each box represents the OR and the horizontal line represents the 95% CI, derived from the logistic regression model adjusted for age, gender and principal components where appropriate. I 2: the variation in OR attributed to heterogeneity.
We also found that the effect of smoking depends on the individual genetic background. The effect of smoking is OR = 4.21 (95% CI: 3.60–4.93) for subjects carrying the TT genotype of rs1316298, whereas it is smaller for those carrying the TC or CC genotype of rs1316298 (OR = 2.56, 95% CI: 2.15–3.04) (Supplementary Figure S5A, available at Carcinogenesis Online). The TC or CC genotype of rs1316298 weakens the effect of smoking. In contrast, the CT or TT genotype of rs4589502 enhances the effect of smoking (Supplementary Figure S5B, available at Carcinogenesis Online). The effect of smoking for subjects carrying the CC genotype or the CT or TT genotype is OR = 3.14 (95% CI: 2.74–3.59) and OR = 4.25 (95% CI: 3.37–5.37), respectively.
The effects of the identified variants on smoking behaviour and lung cancer risk
We also performed association analyses between rs1316298 and smoking behaviour in lung cancer subjects only. As presented in Table II, among the subjects carrying the TT genotype of rs1316298, 63.94% were smokers. Whereas, among those carrying the TC or CC genotype of rs1316298, only 60.58% were smokers (P = 3.39 × 10− 2). Thus, the TC or CC genotype of rs1316298 might be associated with abstain from smoking, which may explain both the results of our stratification analysis indicating that rs1316298 is a protective factor for smokers (Figure 2A), and our finding that the risk effect of smoking is smaller in subjects carrying the TC or CC genotype (Supplementary Figure S5A, available at Carcinogenesis Online). In contrast to rs1316298, 61.54% of subjects carrying the CC genotype and 65.86% of those carrying the CT or TT genotype of rs4589502 were smokers, which implies that the CT or TT genotype in this SNP is associated with addiction to smoking (P = 1.73 × 10− 2). Thus, rs4589502 is a risk factor for smokers (Figure 2B) and the risk effect of smoking is larger in subjects carrying the CT or TT genotype (Supplementary Figure S5B, available at Carcinogenesis Online).
Table II.
The association between two SNPs and smoking behaviour in all lung cancer subjects only
| SNP | Genotype | Sample size | Smoking (%) | P |
|---|---|---|---|---|
| rs1316298a | TT | 2224 | 1422 (63.94) | 3.39×10−2 |
| TC/CC | 1616 | 979 (60.58) | ||
| rs4589502b | CC | 2894 | 1781 (61.54) | 1.73×10−2 |
| CT/TT | 946 | 623 (65.86) |
aT/C: major/minor alleles.
bC/T: major/minor alleles.
The analysis of joint effect of smoking status and genetic mutation
In the light of these results, we evaluated the joint effects of genetic mutation of each SNP (No versus Yes) and the status of smoking (No versus Yes) on the risk of lung cancer (Table III). In the combined data from two phases, the effect of smoking is OR = 4.09 (95% CI: 3.55–4.71) and the effect of the TC or CC genotype of rs1316298 is OR = 1.13 (95% CI: 0.99–1.29). However, the joint effect of them is OR = 2.99 (95% CI: 2.58–3.46), which is less than the product of the two individual effects (4.09×1.13), indicating an antagonistic interaction between rs1316298 and smoking (OR = 0.65, 95% CI: 0.54–0.78, P = 2.23 × 10− 6).
Table III.
The joint effects of genetic mutation and smoking status on the risk of lung cancer
| Study | Mutationa | Smoking | rs1316298–smoking | rs4589502–smoking | ||
|---|---|---|---|---|---|---|
| OR (95% CI)b | P b | OR (95% CI)b | P b | |||
| GWAS | ||||||
| No | No | Reference | Reference | |||
| No | Yes | 3.95 (3.25, 4.80) | 2.95×10−43 | 2.68 (2.26, 3.18) | 2.09×10−29 | |
| Yes | No | 1.30 (1.08, 1.57) | 6.25×10−03 | 0.68 (0.55, 0.84) | 5.05×10−04 | |
| Yes | Yes | 2.96 (2.42, 3.62) | 4.21×10−26 | 3.20 (2.58, 3.96) | 1.69×10−26 | |
| P interaction b | 0.58 (0.45, 0.74) | 9.48×10 −06 | 1.76 (1.33, 2.33) | 8.94×10 −05 | ||
| Replication | ||||||
| No | No | Reference | Reference | |||
| No | Yes | 4.26 (3.36, 5.39) | 2.88×10−33 | 3.28 (2.65, 4.07) | 1.21×10−27 | |
| Yes | No | 1.02 (0.82, 1.27) | 8.83×10−01 | 0.76 (0.59, 0.97) | 2.83×10−02 | |
| Yes | Yes | 2.79 (2.19, 3.56) | 1.70×10−16 | 3.41 (2.58, 4.50) | 6.62×10−18 | |
| P interaction b | 0.65 (0.47, 0.88) | 5.66×10 −03 | 1.39 (0.98, 1.96) | 6.60×10 −02 | ||
| Combined | ||||||
| No | No | Reference | Reference | |||
| No | Yes | 4.09 (3.55, 4.71) | 1.31×10−84 | 3.03 (2.67, 3.44) | 4.48×10−66 | |
| Yes | No | 1.13 (0.99, 1,29) | 7.36×10−02 | 0.72 (0.62, 0.83) | 1.78×10−05 | |
| Yes | Yes | 2.99 (2.58, 3.46) | 1.87×10−48 | 3.39 (2.89, 3.99) | 1.13×10−49 | |
| P interaction b | 0.65 (0.54, 0.78) | 2.23×10 −06 | 1.56 (1.27, 1.92) | 2.64×10 −05 | ||
The results of gene–smoking interaction analysis is highlighted in boldface values.
aSubjects with C allele of rs1316298 or T allele of rs4589502 have genetic mutation.
bAdjusted for age, gender and principal components where appropriate.
For another interaction, in the combined data of two phase, the effect of smoking is OR = 3.03 (95% CI: 2.67–3.44) and the effect of the CT or TT genotype of rs4589502 is OR = 0.72 (95% CI: 0.62–0.83). However, the joint effect of them is OR = 3.39 (95% CI: 2.89–3.99), which is greater than the product of the two individual effects (3.03 × 0.72). The increase in risk demonstrates that rs4589502 and smoking have a synergistic interaction (OR = 1.56, 95% CI: 1.27–1.92, P = 2.64 × 10− 5).
The predictive ability of the newly identified gene–smoking interactions
We evaluated the predictive ability for the newly identified gene–smoking interactions using two logistic regression model (without or with gene–smoking interactions). In this study, the relative IDI for the model with the two interactions added is 8.35%, 95% CI: 5.69–10.86, P < 0.0001 (Supplementary Table S4, available at Carcinogenesis Online), whereas the category-free NRI is 24.77%, 95% CI: 20.47–29.07, P < 0.0001 (Supplementary Table S5, available at Carcinogenesis Online).
Discussion
Previous studies only evaluated gene–smoking interactions associated with lung cancer risk in a few of SNPs (15–28). In this study, we systematically evaluated all pair-wise gene–smoking interactions on a genome-wide scale in the discovery phase (two independent GWAS). These promising ones were again confirmed in the replication phase (another independent study) to identify the confident associations. The sample size of these studies of gene–smoking interaction in Asian populations is ranging from ~250 to ~3000 (16,17,19–23,25–27). We have a larger sample size (total 8431 subjects) in this study.
We also investigated the SNPs that have been reported in previous studies based on candidate gene strategy, in our GWAS dataset (15–28). Because these SNPs were not genotyped in the current study, we tested the SNPs that were in high LD (r 2 ≥ 0.80) with the previously identified SNPs according to the information from hg18/1000 Genomes database using SNAP 2.2 (36). One SNP rs2984915, which is in perfect LD (r 2 = 1.00) with rs2760501 (25), and ~3kb away, had a significant gene–smoking interaction (P = 1.04×10− 3). Another SNP rs4845882, which is also in perfect LD with rs1801131 (22), ~10 kb away, also had a significant gene–smoking interaction (P = 4.26×10− 2). The replication of interactions faces additional challenges (37). For example, the different definitions of smoking (20,26) and/or ethnic attributes (15,24) in previous studies may explain the missing of significant results in our study.
The effect of smoking varies for subjects carrying the different genotypes of a SNP. Based on our results of gene–smoking interaction analysis, smoking causes more damage (larger risk effect OR > 4.2) to subjects carrying the TT genotype of rs1316298 or the CT or TT genotype of rs4589502. It is necessary to persuade the carriers of those alleles to quit smoking. What we found helps for intensive screening and smoking cessation interventions of targeted populations.
Both the relative IDI and the category-free NRI are significant for these two identified gene–smoking interactions, indicating the improvement of the predicative ability with gene–smoking interactions added in the model. These results suggest that the interactions between these two SNPs and tobacco smoke may help explain some of the missing heritability of lung cancer susceptibility. However, this improvement still only explains a relatively small portion of the missing heritability of lung cancer, the remaining missing heritability may be due to rare variants, copy number variations or other unaccounted factors. Further studies of which are warranted to discover more missing heritability of lung cancer.
The SNP rs1316298, located at 14q22.1, lies in intron 3 of the GNG2, which is expressed in several foetal tissues as well as adult lung and malignant tissues (38,39). The gng2 gene has been shown to interact with the vegf pathway in the zebra fish model, and thereby to block pathologic angiogenesis (40). Thereby it is possible that GNG2 has a tumour suppressor function in humans. The nearby genes, FRMD6 (downstream 171.4kb) and NID2 (downstream 106.7kb) have also been shown to have tumour-related functions. FRMD6 may have tumour suppressor properties (41). FRMD6 loci have been associated with asthma (42) and Alzheimer’s disease (43) and the protein encoded by FRMD6 can activate the Hippo kinase pathway, which is an important regulator of cancer development in mammals (44–46). The methylation status of NID2, which is involved in basement membrane structure, is correlated with various types of cancer (47–49), including non-small cell lung cancer (50). The interaction with rs1316298 is perplexing since it is a protective factor in smokers and risk factor in non-smokers. In order to understand these findings, we then evaluated the expression levels of the three genes (GNG2, FRMD6 and NID2) surrounding rs1316298 in smokers versus non-smokers. We examined mRNA expression levels in total cellular RNA from whole blood of 131 healthy individuals using quantitative reverse transcription–polymerase chain reaction and observed that the relative expression of NID2 was significantly lower in smokers (n = 73) as compared with non-smokers (n = 58) (P = 0.0415) (Supplementary Figure S6, available at Carcinogenesis Online). However, non-significant results were observed for GNG2 (P = 0.3837) and FRMD6 (P = 0.2531). Considering smoking may induce methylation at many sites (51–53). It is possible that smoking induces methylation of the NID2 genes, and the expression of which may been down-regulated by methylation in smokers, hence may inhibit lung carcinogenesis.
The other SNP of interest, rs4589502, is located at 15q22.32, between SMAD family member 6 (SMAD6, 83kb downstream) and SMAD family member 3 (SMAD3, 206kb upstream). Tobacco smoke down-regulates SMAD6 in the airway epithelial cell line A549 (54). Cigarette smoking has also been shown to decrease Smad3 expression, and therefore promote lung cancer by increasing cell viability and decreasing apoptosis in the human lung adenocarcinoma cell line A549 (55). Furthermore, loss of Smad3 expression in cigarette smoke condensate-treated cells is associated with resistance to carboplatin and up-regulated expression of the anti-apoptotic Bcl2 in non-small cell lung cancer with associated poor survival (56). In addition, rs4589502 is located near a region that contains the DNase I hypersensitivity cluster. ENCODE ChIP-Seq data showed that rs4589502 is located in a region that may affect the binding of numerous transcription factors, including CTCF, ZNF263 and c-Myc. Among these factors, CTCF may decrease NY-ESO-1 transcription during lung carcinogenesis (57,58). The MYC oncogene is over-expressed in lung cancer cells and is associated with lung cancer metastasis (59–61).
We also performed functional annotation for the two marker SNPs (rs1316298 and rs4589502), as well as those are tagged by the two marker SNPs (r 2 > 0.8) based on public available datasets or tools (see Materials and methods; Supplementary Table S6, available at Carcinogenesis Online). Among the two marker SNPs and those 16 SNPs highly correlated with which, seven SNPs are located in motifs that may influence the binding of specified transcription factors. We then evaluated whether the SNPs modulate the mRNA expression levels through transcriptional or post-transcriptional mechanism. Based on the DNase-seq dataset, we found that three SNPs are within open chromatin regions associated with gene regulatory elements, whereas eight SNPs may influence pre-mRNA splicing as they may disrupt putative exonic splicing enhancer motifs. Furthermore, six SNPs may affect the microRNAs binding (Supplementary Table S6, available at Carcinogenesis Online). It is plausible that variations in the two SNPs (rs1316298 and rs4589502), or in SNPs in high LD with these two SNPs, collaboratively result in the aberrant activities of certain transcription or post-transcriptional factors. In turn, those factors and smoking may interactively regulate the expression of the same target genes nearby or throughout the genome, hence activating the crucial signalling pathways that drive lung carcinogenesis. Notably, since the functional annotation for SNPs was based on Regulome DB database, which was performed mainly in Caucasian. Just like most of the other reported GWAS, the variants identified in populations of European descent might not be applicable among Asians because of underlying genetic heterogeneity (both allelic and locus heterogeneity). Of course, we could not exclude the possibility of genetic heterogeneity from different populations. So, these functional annotation results are very preliminary and merit further investigations, especially in Han Chinese. Therefore, these results should be treated with caution. Further studies, especially in non-Han Chinese population, are warranted to confirm what we found.
In summary, this is the first attempt of a genome-wide gene–smoking interaction analysis in a GWAS of lung cancer, followed by an independent replication phase in Han Chinese population. The two interactions (rs1316298–smoking and rs4589502–smoking) identified in our study may help explain some of the missing heritability in lung cancer susceptibility and present strong evidence for further study of these gene–smoking interactions, which are benefit to intensive screening and smoking cessation interventions.
URLs
Minimac, http://genome.sph.umich.edu/wiki/Minimac/; The HapMap project, http://hapmap.ncbi.nlm.nih.gov/; PLINK 1.07, http://pngu.mgh.harvard.edu/~purcell/plink/; R 2.14.0, statistical environment, http://www.cran.r-project.org/; LocusZoom 1.1, http://csg.sph.umich.edu/locuszoom/; SNAP, http://www.broadinstitute.org/mpg/snap/ldsearch.php/; ENCODE, http://genome.ucsc.edu/ENCODE/; Regulomedb, http://regulomedb.org/; Patrocles, http://www.patrocles.org/; ESEfinder3.0, http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home.
Supplementary material
Supplementary Tables S1–S6 and Figures S1–S6 can be found at http://carcin.oxfordjournals.org/
Funding
National Key Basic Research Program Grant (2011CB503805, 2013CB911400); the National Natural Science Foundation of China (81230067, 81225020, 81270044, 81072389, 81373102, 81202283, 81302488); Jiangsu Natural Science Foundation (BK2012042); Natural Science Foundation of the Jiangsu Higher Education Institutions of China (11KJA330001, 10KJA33034, 10KJB330002); the Research Fund for the Doctoral Program of Higher Education of China (20113234110002); the US National Institutes of Health Grant (U19 CA148127); the New Century Excellent Talents in University (NCET-10–0178); the Research and Innovation Project for College Graduates of Jiangsu Province (CXZZ11_0733, CXZZ12_0591); the Science and Technology Innovation Foundation of Nanjing Medical University (2013NJMU016); the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Supplementary Material
Acknowledgements
We thank all of the study subjects, research staff and students who participated in this work.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations:
- CI
confidence interval
- DNase-seq
DNase I hypersensitive site sequencing
- FRMD6
FERM-domain containing-6
- GNG2
guanine nucleotide binding protein gamma 2
- GWAS
genome-wide association study
- IDI
integrated discrimination improvement
- LD
linkage disequilibrium
- mRNA
messenger RNA
- NID2
nidogen-2
- NRI
net reclassification improvement
- OR
odds ratio
- SNP
single nucleotide polymorphism.
References
- 1. Jemal A., et al. (2011). Global cancer statistics. Cancer J. Clin., 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. Zhang H., et al. (2003). The impact of tobacco on lung health in China. Respirology, 8, 17–21 [DOI] [PubMed] [Google Scholar]
- 3. Stavrides J.C. (2006). Lung carcinogenesis: pivotal role of metals in tobacco smoke. Free Radic. Biol. Med., 41, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 4. Doll R., et al. (1981). The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J. Natl Cancer Inst., 66, 1191–1308 [PubMed] [Google Scholar]
- 5. Spitz M.R., et al. (1999). Genetic susceptibility to tobacco carcinogenesis. Cancer Invest., 17, 645–659 [DOI] [PubMed] [Google Scholar]
- 6. Amos C.I., et al. (2008). Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat. Genet., 40, 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hung R.J., et al. (2008). A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature, 452, 633–637 [DOI] [PubMed] [Google Scholar]
- 8. McKay J.D., et al. ; EPIC Study. (2008). Lung cancer susceptibility locus at 5p15.33. Nat. Genet., 40, 1404–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang Y., et al. (2008). Common 5p15.33 and 6p21.33 variants influence lung cancer risk. Nat. Genet., 40, 1407–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu Z., et al. (2011). A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nat. Genet., 43, 792–796 [DOI] [PubMed] [Google Scholar]
- 11. Dong J., et al. (2012). Association analyses identify multiple new lung cancer susceptibility loci and their interactions with smoking in the Chinese population. Nat. Genet., 44, 895–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manolio T.A., et al. (2009). Finding the missing heritability of complex diseases. Nature, 461, 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moore J.H., et al. (2009). Epistasis and its implications for personal genetics. Am. J. Hum. Genet., 85, 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Maher B. (2008). Personal genomes: the case of the missing heritability. Nature, 456, 18–21 [DOI] [PubMed] [Google Scholar]
- 15. Zhou W., et al. (2002). Gene-environment interaction for the ERCC2 polymorphisms and cumulative cigarette smoking exposure in lung cancer. Cancer Res., 62, 1377–1381 [PubMed] [Google Scholar]
- 16. Ito H., et al. (2004). Gene-environment interactions between the smoking habit and polymorphisms in the DNA repair genes, APE1 Asp148Glu and XRCC1 Arg399Gln, in Japanese lung cancer risk. Carcinogenesis, 25, 1395–1401 [DOI] [PubMed] [Google Scholar]
- 17. Shah P.P., et al. (2008). Interaction of cytochrome P4501A1 genotypes with other risk factors and susceptibility to lung cancer. Mutat. Res., 639, 1–10 [DOI] [PubMed] [Google Scholar]
- 18. Spitz M.R., et al. (2008). The CHRNA5-A3 region on chromosome 15q24-25.1 is a risk factor both for nicotine dependence and for lung cancer. J. Natl Cancer Inst., 100, 1552–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Singh A.P., et al. (2010). Polymorphism in cytochrome P450 1A2 and their interaction with risk factors in determining risk of squamous cell lung carcinoma in men. Cancer Biomark., 8, 351–359 [DOI] [PubMed] [Google Scholar]
- 20. Lu J., et al. (2011). The polymorphism and haplotypes of PIN1 gene are associated with the risk of lung cancer in Southern and Eastern Chinese populations. Hum. Mutat., 32, 1299–1308 [DOI] [PubMed] [Google Scholar]
- 21. Hsia T.C., et al. (2011). Interaction of CCND1 genotype and smoking habit in Taiwan lung cancer patients. Anticancer Res., 31, 3601–3605 [PubMed] [Google Scholar]
- 22. Kiyohara C., et al. (2011). Methylenetetrahydrofolate reductase polymorphisms and interaction with smoking and alcohol consumption in lung cancer risk: a case-control study in a Japanese population. BMC Cancer, 11, 459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ihsan R., et al. (2011). Multiple analytical approaches reveal distinct gene-environment interactions in smokers and non smokers in lung cancer. PLoS One, 6, e29431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. VanderWeele T.J., et al. (2012). Genetic variants on 15q25.1, smoking, and lung cancer: an assessment of mediation and interaction. Am. J. Epidemiol., 175, 1013–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang B., et al. (2012). Functional genetic variants of c-Jun and their interaction with smoking and drinking increase the susceptibility to lung cancer in southern and eastern Chinese. Int. J. Cancer, 131, E744–E758 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Z., et al. (2012). Cigarette smoking strongly modifies the association of complement factor H variant and the risk of lung cancer. Cancer Epidemiol., 36, e111–e115 [DOI] [PubMed] [Google Scholar]
- 27. Cheng Z., et al. (2012). hOGG1, p53 genes, and smoking interactions are associated with the development of lung cancer. Asian Pac. J. Cancer Prev., 13, 1803–1808 [DOI] [PubMed] [Google Scholar]
- 28. Feng Z., et al. (2012). Association of ERCC2/XPD polymorphisms and interaction with tobacco smoking in lung cancer susceptibility: a systemic review and meta-analysis. Mol. Biol. Rep., 39, 57–69 [DOI] [PubMed] [Google Scholar]
- 29. Deng Q, et al. (2013). Imputation-based association analyses identify new lung cancer susceptibility variants in CDK6 and SH3RF1 and their interactions with smoking in Chinese populations. Carcinogenesis, 34, 2010–2016. [DOI] [PubMed] [Google Scholar]
- 30. Howie B., et al. (2012). Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat. Genet., 44, 955–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Price A.L., et al. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909 [DOI] [PubMed] [Google Scholar]
- 32. Purcell S., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pruim R.J., et al. (2010). LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barrett J.C., et al. (2005). Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265 [DOI] [PubMed] [Google Scholar]
- 35. Pencina M.J., et al. (2008). Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Stat. Med., 27, 157–72; discussion 207. [DOI] [PubMed] [Google Scholar]
- 36. Johnson A.D., et al. (2008). SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics, 24, 2938–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aschard H., et al. (2012). Challenges and opportunities in genome-wide environmental interaction (GWEI) studies. Hum. Genet., 131, 1591–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Modarressi M.H., et al. (2000). Cloning, characterization, and mapping of the gene encoding the human G protein gamma 2 subunit. Biochem. Biophys. Res. Commun., 272, 610–615 [DOI] [PubMed] [Google Scholar]
- 39. Yajima I., et al. (2012). Reduced GNG2 expression levels in mouse malignant melanomas and human melanoma cell lines. Am. J. Cancer Res., 2, 322–329 [PMC free article] [PubMed] [Google Scholar]
- 40. Leung T., et al. (2006). Zebrafish G protein gamma2 is required for VEGF signaling during angiogenesis. Blood, 108, 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Visser-Grieve S., et al. (2012). Human homolog of Drosophila expanded, hEx, functions as a putative tumor suppressor in human cancer cell lines independently of the Hippo pathway. Oncogene, 31, 1189–1195 [DOI] [PubMed] [Google Scholar]
- 42. Ungvári I., et al. (2012). Evaluation of a partial genome screening of two asthma susceptibility regions using bayesian network based bayesian multilevel analysis of relevance. PLoS One, 7, e33573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hong M.G., et al. (2012). Genome-wide and gene-based association implicates FRMD6 in Alzheimer disease. Hum. Mutat., 33, 521–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeng Q., et al. (2008). The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell, 13, 188–192 [DOI] [PubMed] [Google Scholar]
- 45. Pan D. (2010). The hippo signaling pathway in development and cancer. Dev. Cell, 19, 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Angus L., et al. (2012). Willin/FRMD6 expression activates the Hippo signaling pathway kinases in mammals and antagonizes oncogenic YAP. Oncogene, 31, 238–250 [DOI] [PubMed] [Google Scholar]
- 47. Ulazzi L., et al. (2007). Nidogen 1 and 2 gene promoters are aberrantly methylated in human gastrointestinal cancer. Mol. Cancer, 6, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Renard I., et al. (2010). Identification and validation of the methylated TWIST1 and NID2 genes through real-time methylation-specific polymerase chain reaction assays for the noninvasive detection of primary bladder cancer in urine samples. Eur. Urol., 58, 96–104 [DOI] [PubMed] [Google Scholar]
- 49. Guerrero-Preston R., et al. (2011). NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev. Res. (Phila)., 4, 1061–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Geng J., et al. (2012). Methylation status of NEUROG2 and NID2 improves the diagnosis of stage I NSCLC. Oncol. Lett., 3, 901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim J.S., et al. (2004). Aberrant methylation of the FHIT gene in chronic smokers with early stage squamous cell carcinoma of the lung. Carcinogenesis, 25, 2165–2171 [DOI] [PubMed] [Google Scholar]
- 52. Divine K.K., et al. (2005). Multiplicity of abnormal promoter methylation in lung adenocarcinomas from smokers and never smokers. Int. J. Cancer, 114, 400–405 [DOI] [PubMed] [Google Scholar]
- 53. Buro-Auriemma L.J., et al. (2013). Cigarette smoking induces small airway epithelial epigenetic changes with corresponding modulation of gene expression. Hum. Mol. Genet., 22, 4726–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Springer J., et al. (2004). SMAD-signaling in chronic obstructive pulmonary disease: transcriptional down-regulation of inhibitory SMAD 6 and 7 by cigarette smoke. Biol. Chem., 385, 649–653 [DOI] [PubMed] [Google Scholar]
- 55. Samanta D., et al. (2012). Smoking attenuates transforming growth factor-β-mediated tumor suppression function through downregulation of Smad3 in lung cancer. Cancer Prev. Res. (Phila)., 5, 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Samanta D., et al. (2012). Long-term smoking mediated down-regulation of Smad3 induces resistance to carboplatin in non-small cell lung cancer. Neoplasia, 14, 644–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hong J.A., et al. (2005). Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells. Cancer Res., 65, 7763–7774 [DOI] [PubMed] [Google Scholar]
- 58. Kang Y., et al. (2007). Dynamic transcriptional regulatory complexes including BORIS, CTCF and Sp1 modulate NY-ESO-1 expression in lung cancer cells. Oncogene, 26, 4394–4403 [DOI] [PubMed] [Google Scholar]
- 59. Zajac-Kaye M. (2001). Myc oncogene: a key component in cell cycle regulation and its implication for lung cancer. Lung Cancer, 34 (suppl. 2), S43–S46 [DOI] [PubMed] [Google Scholar]
- 60. Rapp U.R., et al. (2009). MYC is a metastasis gene for non-small-cell lung cancer. PLoS One, 4, e6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Allen T.D., et al. (2011). Interaction between MYC and MCL1 in the genesis and outcome of non-small-cell lung cancer. Cancer Res., 71, 2212–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.