Abstract
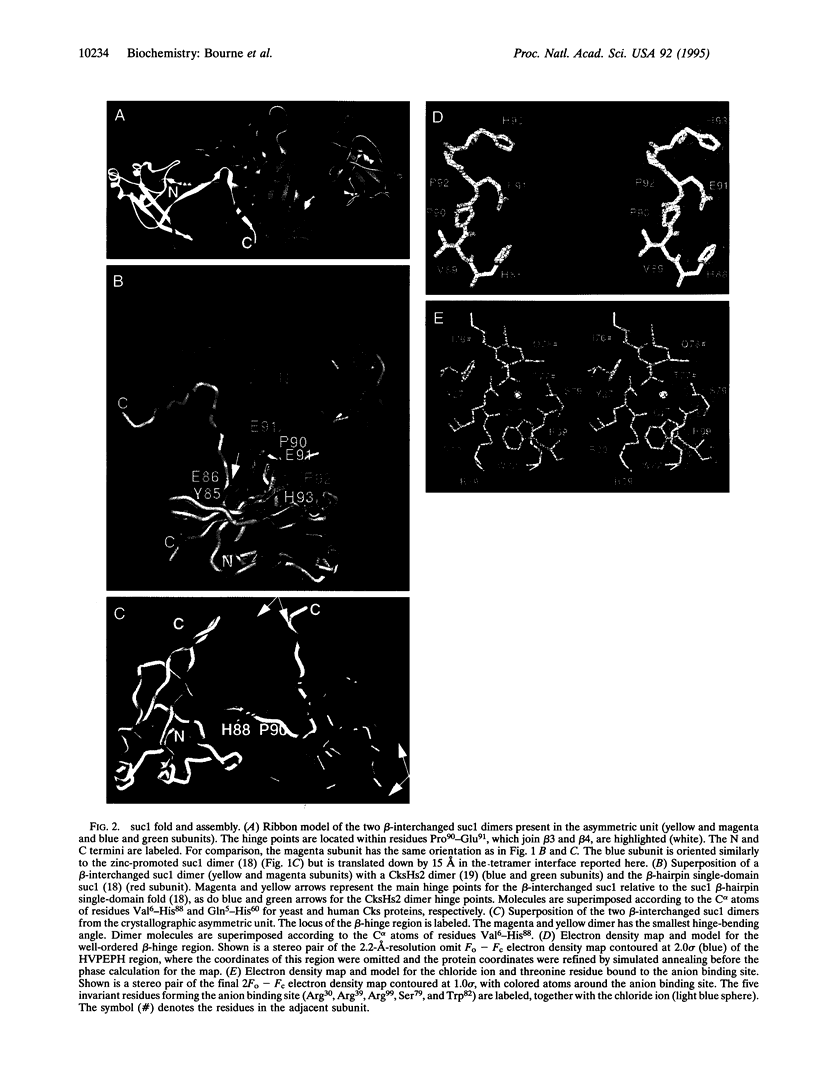
The Schizosaccharomyces pombe cell cycle-regulatory protein suc1, named as the suppressor of cdc2 temperature-sensitive mutations, is essential for cell cycle progression. To understand suc1 structure-function relationships and to help resolve conflicting interpretations of suc1 function based on genetic studies of suc1 and its functional homologs in both lower and higher eukaryotes, we have determined the crystal structure of the beta-interchanged suc1 dimer. Each domain consists of three alpha-helices and a four-stranded beta-sheet, completed by the interchange of terminal beta-strands between the two subunits. This beta-interchanged suc1 dimer, when compared with the beta-hairpin single-domain folds of suc1, reveals a beta-hinge motif formed by the conserved amino acid sequence HVPEPH. This beta-hinge mediates the subunit conformation and assembly of suc1: closing produces the intrasubunit beta-hairpin and single-domain fold, whereas opening leads to the intersubunit beta-strand interchange and interlocked dimer assembly reported here. This conformational switch markedly changes the surface accessibility of sequence-conserved residues available for recognition of cyclin-dependent kinase, suggesting a structural mechanism for beta-hinge-mediated regulation of suc1 biological function. Thus, suc1 belongs to the family of domain-swapping proteins, consisting of intertwined and dimeric protein structures in which the dual assembly modes regulate their function.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson B. F., Baker H. M., Norris G. E., Rumball S. V., Baker E. N. Apolactoferrin structure demonstrates ligand-induced conformational change in transferrins. Nature. 1990 Apr 19;344(6268):784–787. doi: 10.1038/344784a0. [DOI] [PubMed] [Google Scholar]
- Banzon J. A., Kelly J. W. Beta-sheet rearrangements: serpins and beyond. Protein Eng. 1992 Mar;5(2):113–115. doi: 10.1093/protein/5.2.113. [DOI] [PubMed] [Google Scholar]
- Bennett M. J., Choe S., Eisenberg D. Domain swapping: entangling alliances between proteins. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizuela L., Draetta G., Beach D. Activation of human CDC2 protein as a histone H1 kinase is associated with complex formation with the p62 subunit. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4362–4366. doi: 10.1073/pnas.86.12.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Cohen B. I., Presnell S. R., Cohen F. E. Origins of structural diversity within sequentially identical hexapeptides. Protein Sci. 1993 Dec;2(12):2134–2145. doi: 10.1002/pro.5560021213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colas P., Serras F., Van Loon A. E. Microinjection of suc1 transcripts delays the cell cycle clock in Patella vulgata embryos. Int J Dev Biol. 1993 Dec;37(4):589–594. [PubMed] [Google Scholar]
- Dunphy W. G., Newport J. W. Fission yeast p13 blocks mitotic activation and tyrosine dephosphorylation of the Xenopus cdc2 protein kinase. Cell. 1989 Jul 14;58(1):181–191. doi: 10.1016/0092-8674(89)90414-5. [DOI] [PubMed] [Google Scholar]
- Dunphy W. G., Newport J. W. Unraveling of mitotic control mechanisms. Cell. 1988 Dec 23;55(6):925–928. doi: 10.1016/0092-8674(88)90234-6. [DOI] [PubMed] [Google Scholar]
- Dunphy W. G. The decision to enter mitosis. Trends Cell Biol. 1994 Jun;4(6):202–207. doi: 10.1016/0962-8924(94)90142-2. [DOI] [PubMed] [Google Scholar]
- Endicott J. A., Noble M. E., Garman E. F., Brown N., Rasmussen B., Nurse P., Johnson L. N. The crystal structure of p13suc1, a p34cdc2-interacting cell cycle control protein. EMBO J. 1995 Mar 1;14(5):1004–1014. doi: 10.1002/j.1460-2075.1995.tb07081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber H. R., Matthews B. W. A mutant T4 lysozyme displays five different crystal conformations. Nature. 1990 Nov 15;348(6298):263–266. doi: 10.1038/348263a0. [DOI] [PubMed] [Google Scholar]
- Hadwiger J. A., Wittenberg C., Mendenhall M. D., Reed S. I. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol Cell Biol. 1989 May;9(5):2034–2041. doi: 10.1128/mcb.9.5.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamlin R. Multiwire area X-ray diffractometers. Methods Enzymol. 1985;114:416–452. doi: 10.1016/0076-6879(85)14029-2. [DOI] [PubMed] [Google Scholar]
- Harris L. J., Larson S. B., Hasel K. W., Day J., Greenwood A., McPherson A. The three-dimensional structure of an intact monoclonal antibody for canine lymphoma. Nature. 1992 Nov 26;360(6402):369–372. doi: 10.1038/360369a0. [DOI] [PubMed] [Google Scholar]
- Hayles J., Aves S., Nurse P. suc1 is an essential gene involved in both the cell cycle and growth in fission yeast. EMBO J. 1986 Dec 1;5(12):3373–3379. doi: 10.1002/j.1460-2075.1986.tb04653.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J., Beach D., Durkacz B., Nurse P. The fission yeast cell cycle control gene cdc2: isolation of a sequence suc1 that suppresses cdc2 mutant function. Mol Gen Genet. 1986 Feb;202(2):291–293. doi: 10.1007/BF00331653. [DOI] [PubMed] [Google Scholar]
- Hindley J., Phear G., Stein M., Beach D. Sucl+ encodes a predicted 13-kilodalton protein that is essential for cell viability and is directly involved in the division cycle of Schizosaccharomyces pombe. Mol Cell Biol. 1987 Jan;7(1):504–511. doi: 10.1128/mcb.7.1.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. Braking the cycle. Cell. 1993 Dec 3;75(5):839–841. doi: 10.1016/0092-8674(93)90528-x. [DOI] [PubMed] [Google Scholar]
- Johnson L. N., Acharya K. R., Jordan M. D., McLaughlin P. J. Refined crystal structure of the phosphorylase-heptulose 2-phosphate-oligosaccharide-AMP complex. J Mol Biol. 1990 Feb 5;211(3):645–661. doi: 10.1016/0022-2836(90)90271-M. [DOI] [PubMed] [Google Scholar]
- Kusubata M., Tokui T., Matsuoka Y., Okumura E., Tachibana K., Hisanaga S., Kishimoto T., Yasuda H., Kamijo M., Ohba Y. p13suc1 suppresses the catalytic function of p34cdc2 kinase for intermediate filament proteins, in vitro. J Biol Chem. 1992 Oct 15;267(29):20937–20942. [PubMed] [Google Scholar]
- Lohka M. J., Hayes M. K., Maller J. L. Purification of maturation-promoting factor, an intracellular regulator of early mitotic events. Proc Natl Acad Sci U S A. 1988 May;85(9):3009–3013. doi: 10.1073/pnas.85.9.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. J Mol Biol. 1968 Apr 28;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Meikrantz W., Suprynowicz F. A., Halleck M. S., Schlegel R. A. Identification of mitosis-specific p65 dimer as a component of human M phase-promoting factor. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9600–9604. doi: 10.1073/pnas.87.24.9600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Hayles J., Nurse P. Regulation of p34cdc2 protein kinase during mitosis. Cell. 1989 Jul 28;58(2):361–372. doi: 10.1016/0092-8674(89)90850-7. [DOI] [PubMed] [Google Scholar]
- Parge H. E., Arvai A. S., Murtari D. J., Reed S. I., Tainer J. A. Human CksHs2 atomic structure: a role for its hexameric assembly in cell cycle control. Science. 1993 Oct 15;262(5132):387–395. doi: 10.1126/science.8211159. [DOI] [PubMed] [Google Scholar]
- Piccoli R., Tamburrini M., Piccialli G., Di Donato A., Parente A., D'Alessio G. The dual-mode quaternary structure of seminal RNase. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1870–1874. doi: 10.1073/pnas.89.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J., Hunter T. Cyclin-dependent kinases: a new cell cycle motif? Trends Cell Biol. 1991 Nov;1(5):117–121. doi: 10.1016/0962-8924(91)90116-q. [DOI] [PubMed] [Google Scholar]
- Richardson H. E., Stueland C. S., Thomas J., Russell P., Reed S. I. Human cDNAs encoding homologs of the small p34Cdc28/Cdc2-associated protein of Saccharomyces cerevisiae and Schizosaccharomyces pombe. Genes Dev. 1990 Aug;4(8):1332–1344. doi: 10.1101/gad.4.8.1332. [DOI] [PubMed] [Google Scholar]
- Tang Y., Reed S. I. The Cdk-associated protein Cks1 functions both in G1 and G2 in Saccharomyces cerevisiae. Genes Dev. 1993 May;7(5):822–832. doi: 10.1101/gad.7.5.822. [DOI] [PubMed] [Google Scholar]