SUMMARY
In metazoa, the nuclear envelope (NE), together with the embedded nuclear pore complexes (NPCs), breaks down and reassembles during cell division. It is suggested that ELYS, a nucleoporin, binds to chromatin in an initial step of postmitotic NPC assembly and subsequently recruits the essential Y-subcomplex, the major scaffolding unit of the NPC. Here, we show that ELYS contains three domains: an N-terminal β-propeller domain, a central α-helical domain and a C-terminal disordered region. While the disordered region is responsible for the interactions with chromatin, the two preceding domains synergistically mediate tethering to the NPC. We present the crystal structure of the seven-bladed β-propeller domain at 1.9 Å resolution. Analysis of the β-propeller surface reveals the regions that are required for NPC anchorage. We discuss the possible roles of ELYS in the context of the NPC scaffold architecture.
INTRODUCTION
The compartmentalization of the genomic material into a membrane-enclosed organelle, the nucleus, is the central feature that distinguishes eukaryotes from prokaryotes. Nuclear pore complexes (NPCs) are huge macromolecular assemblies that span the double membrane of the nuclear envelope (NE) and control transport into and out of the nucleus (Wente and Rout, 2010). Despite an estimated total mass of 40–60 MDa, these macromolecular assemblies are composed of only about 30 distinct proteins, nucleoporins (Nups), that are present in multiple copies, as dictated by the eight-fold rotational symmetry of NPCs (Beck et al., 2007; Brohawn and Schwartz, 2009a).
In multicellular eukaryotes that undergo an open mitosis, NE breakdown (NEBD) is required during prophase for proper chromosome segregation (Güttinger et al., 2009; Asakawa et al., 2011). NEBD is accompanied by the disassembly of NPCs and by mixing of the nuclear compartment with the cytosol. Membrane-spanning nucleoporins are redistributed into the ER, which remains intact during mitosis, while the soluble nucleoporins are released into the cytosol as discrete subcomplexes (Hetzer et al., 2010). At the end of mitosis, NPCs reassemble from these subcomplexes in a defined order (Dultz and Ellenberg, 2010; Dultz et al., 2008). Whether NPCs reassemble before, during, or after the reformation of the NE is an ongoing debate (Antonin et al., 2005; Kiseleva et al., 2001; Lu et al., 2011). NPCs can also form during interphase by insertion into the intact double membrane of the NE. Many fungi undergo a closed mitosis without NEBD and thus rely exclusively on this type of NPC assembly. Recent data point to the mechanistic differences between post-mitotic and interphase NPC assembly (Rasala et al., 2008; Doucet et al., 2010; Doucet and Hetzer, 2010; Dultz and Ellenberg, 2010). Despite all efforts, though, the assembly of NPCs, which requires the coordination of ~500 proteins and the stabilization of highly curved openings in the NE, remains ill-defined (Bilokapic and Schwartz, 2011a).
Generally, postmitotic NPC assembly is better understood than interphase assembly, because it can be assayed in vitro through nuclear reconstitution assays using Xenopus egg extracts. Postmitotic NPC assembly presumably begins with the binding of ELYS (embryonic large molecule derived from yolk sac) to chromatin and the subsequent recruitment of the essential Nup107-160 complex (Rasala et al., 2006; Franz et al., 2007; Gillespie et al., 2007). This complex is composed of 7 universally conserved proteins (Nup160, Nup133, Nup107, Nup96, Nup85, Sec13, and Seh1) as well as three additional nucleoporins (Nup43, Nup37, ELYS), found in Homo sapiens and most metazoans (Cronshaw et al., 2002; Loïodice et al., 2004; Rasala et al., 2006). Negative stain electron microscopy data of the homologous Nup84 complex from S. cerevisiae revealed a Y-shaped structure with two short arms and one long stalk (Siniossoglou et al., 2000; Lutzmann et al., 2002). Thus, due to the characteristic shape, the Nup107-160 complex is also known as the Y-complex. It has a crucial role in NPC assembly and pore-free nuclei with a continuous NE form in depleted Xenopus egg extracts (Walther et al., 2003; Harel et al., 2003).
Time-course studies have revealed the sequential recruitment of nucleoporins to sites of postmitotic NPC reassembly (Dultz et al., 2008). ELYS is the earliest NPC component to be detected on the chromatin surface and on kinetochores, followed by the recruitment of the Y-complex (Franz et al. 2007). ELYS not only shows the same cellular localization as the Y-complex, but also interacts with it as shown by immunoprecipitation experiments in human cells and in Xenopus extracts (Rasala et al. 2006). The immunodepletion of ELYS from Xenopus nuclear reconstitution extracts abolished binding of the Y-complex to chromatin (Rasala et al., 2006; Franz et al., 2007). In the absence of ELYS in human cultured cells, NPC assembly occurred within ER membranes and triggered the formation of NPC studded annulate lamellae (Rasala et al., 2008). Thus, the association of ELYS with chromatin and the subsequent recruitment of the Y-complex are the earliest events in the post-mitotic assembly of nuclear pores. Next, the transmembrane nup Pom121 is recruited (Bodoor et al., 1999; Dultz et al., 2008), followed by the Nup93-containing subcomplex (Hase and Cordes, 2003; Dultz et al., 2008), which together with the Y-complex, builds the structural scaffold of the NPC (Bilokapic and Schwartz, 2011a). Finally, more peripheral nucleoporins are incorporated into newly forming pores.
Large-scale RNAi analyses in C. elegans (Sonnichsen et al., 2005) showed that components of the Ran-cycle including RAN-1 (RanGTPase), RAN-2 (RanGAP), NPP-9 (RANBP1), IMB-1 (importin β) and IMA-2 (importin α), were required for proper Mel-28 (ELYS homolog in C. elegans) localization (Fernandez and Piano, 2006). In agreement, nuclear reconstitution assays using Xenopus egg extract showed that importin β and transportin bind the ELYS-containing Y-complex and negatively regulate the first step of NPC assembly (Lau et al., 2009; Rotem et al., 2009). RanGTP, which is enriched at the surface of chromatin, plays a counteractive role to that of importin β, and positively regulates the recruitment of ELYS and the Y-complex to chromatin (Lau et al., 2009; Rotem et al., 2009). Furthermore, RNAi depletion of Mel-28 impairs not only NPC but also NE formation in C. elegans (Galy et al., 2006). Recent data also showed impaired NE formation in HeLa cells in the absence of ELYS (Clever et al., 2012).
ELYS was originally found in metazoa and available data suggested that the protein uses its C-terminal AT-hook domain to bind to the surface of the chromatin during mitosis (Rasala et al., 2008). Our previous data showed that the N-terminal half of protein, composed of a β-stranded domain and an α-helical domain, interacts with Nup160 (Bilokapic and Schwartz, 2012b). Additionally, a shorter ELYS homolog, composed of an α-helical domain, has been found in plants and some fungi (Amlacher et al., 2011; Liu et al., 2009; Tamura et al., 2010). Biochemical characterization of a single domain ELYS homolog from S. pombe showed that the α-helical domain interacts with Nup120 (Nup160 homolog from yeast), and thus with the Y-complex (Bilokapic and Schwartz, 2012b). Here we demonstrate that metazoan ELYS is constructed from three domains that have different functions. While the disordered C-terminal region is not involved in NPC binding, the central α-helical domain is effective in the binding only in conjunction with the N-terminal domain. To understand the functional role of the N-terminal β-stranded domain in metazoan ELYS we solved its crystal structure and, using structure-guided mutants, identified the binding determinants of ELYS in vivo in human cultured cells. Binding of ELYS to chromatin is the first known step in NPC reconstitution after cell division. Knowing how ELYS interacts with other Nups and how it guides their recruitment to chromatin is crucial for understanding nuclear pore formation. Therefore, our work provides insight into the earliest steps that govern NPC assembly.
RESULTS
Each domain of metazoan ELYS has a specific functional role
Secondary structure prediction and sequence conservation analysis suggest that the 252 kDa hELYS consists of three functionally distinct domains: an N-terminal, 55 kDa domain (NTD), which is mainly composed of β-strands, a central, 55 kDa α-helical domain (CHD) and a ~140 kDa C-terminal disordered region (CTR) (Figure 1). The disordered region of ELYS contains a putative nuclear localization signal (NLS), a putative AT-hook DNA-binding motif and an additional portion required for chromatin binding (Rasala et al., 2008) (Figure 1). Our previous work has shown that the CHD, the only domain conserved from S. pombe to H. sapiens, interacts with the NPC through Nup160 of the Y-complex (Figure 1) (Bilokapic and Schwartz, 2012b). However, there is no published data on the functional role of the N-terminal domain of metazoan ELYS.
Figure 1. Domain organization of ELYS.
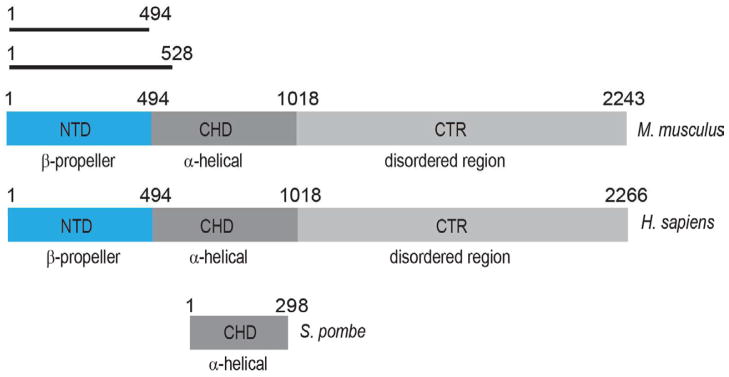
Secondary structure prediction methods suggest three domains in higher eukaryotes, with an N-terminal β-stranded domain (NTD), a central α-helical domain (CHD) and a disordered C-terminal region (CTR). In unicellular eukaryotes, i.e. S. pombe, only the CHD is conserved in ELYS homologs. The constructs used for crystallization are denoted with black bars. The residue numbering is based on UniProt entries Q8CJF7 (M. musculus), Q8WYP5 (H. sapiens), and O94384 (S. pombe).
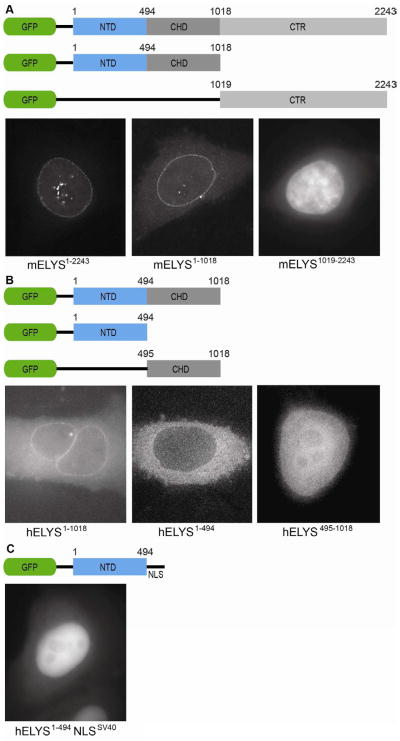
Thus, we first investigated potential roles of the ELYS NTD in vivo. We constructed GFP fusion of full-length mELYS (residues 1–2243) (Figure 2). Full-length mELYS was used instead of hELYS for technical reasons, but with 70 % sequence identity and 81 % sequence similarity we expected similar behavior. Moreover, major differences between metazoan ELYS proteins are in the disordered region, while two N-terminal structural domains are highly conserved (89 % identity, 95 % similarity). To study the function of each domain in vivo we created GFP fusion with the structured domains (residues 1–1018) or the disordered CTR (residues 1019–2243) (Figure 2A). As a control for the same behavior of H. sapiens and M. musculus homologs we included the GFP reporter protein containing both structured domains of hELYS (residues 1–1018) (Figure 2B).
Figure 2. In vivo localization of metazoan ELYS domains.
(A) Domain organization of the GFP-labeled mELYS constructs and their localization in HeLa cells. Full-length mELYS (residues 1–2243) shows nuclear rim staining and intra-nuclear puncta. The construct containing both structured domains (NTD and CHD, residues 1–1018) also localize to the nuclear rim, while the disordered region (CTR, residues 1019–2243) shows diffuse nuclear GFP-signal.
(B) Domain organization of the GFP-labeled hELYS constructs and their localization in HeLa cells. The constructs containing only the NTD (residues 1–494) or the CHD (residues 495–1018) fail to localize to the nuclear rim in HeLa cells, showing that in metazoa both domains (residues 1–1018) are required for interactions with the NPC.
(C) Fusion of the classical SV40 NLS to the C terminus of the hELYS NTD induces nuclear translocation but the domain does not concentrate at the nuclear rim.
The constructs were transfected into T98G and HeLa cells and cellular localization was followed using the fluorescent GFP signal. Consistent with published immunofluorescence data on human ELYS (Franz et al., 2007), full-length GFP-mELYS showed characteristic nuclear rim staining and also appeared as punctae within the nuclear interior during interphase (Figure 2A). The mELYS construct containing both the NTD and CHD, (residues 1–1018), showed similar localization as the full-length GFP-fusion (Figure 2A), consistent with the importance of the CHD for interaction with the Nup107-160 complex. As expected from the sequence conservation, GFP-hELYS (residues 1–1018) exhibited the same localization pattern in HeLa cells as the equivalent region from M. musculus homolog (Figure 2B). The disordered CTR construct, however, did not show nuclear rim staining, but was found throughout the nucleus (Figure 2A). Thus, CTR likely does not interact with the NPC directly, but due to an NLS close to its C terminus gets imported into the nucleus.
Metazoan ELYS contains two structured domains, the NTD and CHD, compared to single-cell eukaryotes that only contain a shortened α-helical domain (Figure 1) (Bilokapic and Schwartz, 2012b). To further investigate the NPC-targeting function separate GFP fusions of NTD and CHD were tested (Figure 2B). Interestingly, neither domain alone was able to incorporate into the NPC as shown by the absence of nuclear rim staining. While GFP-tagged ELYS NTD (residues 1–494) is dispersed throughout the cytosol, GFP-tagged ELYS CHD (residues 495–1018) showed both nuclear and cytosolic localization (Figure 2B). To exclude the possibility that ELYS NTD could not interact with the NPC due to its localization into the cytosol we introduced the SV40 NLS to the GFP-tagged construct. The N-terminal domain with the NLS signal localized to the nuclear interior, but nuclear rim staining was not detected (Figure 2C).
These results show that different roles can be attributed to individual ELYS domains. While, ELYS CTR governs chromatin binding, both structural domains are required for the efficient interactions with the NPC.
Structure Determination
To gain insight into the atomic details of ELYS interacting with the NPC, we sought to determine its crystal structure. We expressed the NTD recombinantly in E. coli and purified it to homogeneity using standard chromatography methods. Size exclusion chromatography indicated that the 55 kDa domain is a monomer in solution (Figure S1). Crystals of ELYS NTD appeared in the P21 space group with one monomer in the asymmetric unit. The structure was solved by SAD using a selenomethionine-labeled crystal (Table 1, Figure S2). The final structure of the derivatized protein was refined to 1.9 Å resolution and includes all residues except for three loops (residues 338–345, 362–373, 458–465) (Figure 3), which are probably flexible and therefore poorly resolved in the electron density.
Table 1.
Data collection and refinement statistics for selenomethionine labeled mELYS NTD. The highest resolution shell with 5 % of the data is shown in parentheses
See also Figure S2.
| Data Set | mELYS NTD SeMet |
|---|---|
| Data Collection | |
| Wavelength (Å) | 0.9792 |
| Space group | P21 |
| Cell dimensions | |
| a, b, c (Å) | 52.6, 78.1, 58.8 |
| β (°) | 106.6 |
| No. unique reflections | 69665 |
| Resolution (Å) | 50-1.90 (1.93-1.90) |
| Rsym (%) | 8.4 (71.4) |
| Completeness (%) | 98.9 (97.0) |
| Redundancy | 2.9 (2.7) |
| I/σ(I) | 11.0 (1.2) |
| Wilson B Factor (Å2) | 27.3 |
| Refinement | |
| Resolution (Å) | 26.5-1.90 |
| No. reflections | |
| Total | 69605 |
| Rfree | 3501 |
| Rwork/Rfree (%) | 18.0/21.4 |
| No. atoms | |
| Protein | 3629 |
| Water | 169 |
| B-factors (Å2) | |
| Protein | 35.7 |
| Water | 36.6 |
| R.m.s deviations | |
| Bond lengths (Å) | 0.012 |
| Bond angles (°) | 1.422 |
| Ramachandran Plot | |
| Favored/allowed (%) | 96.5/3.5 |
| MolProbity score | 2.01 (67th percentile) |
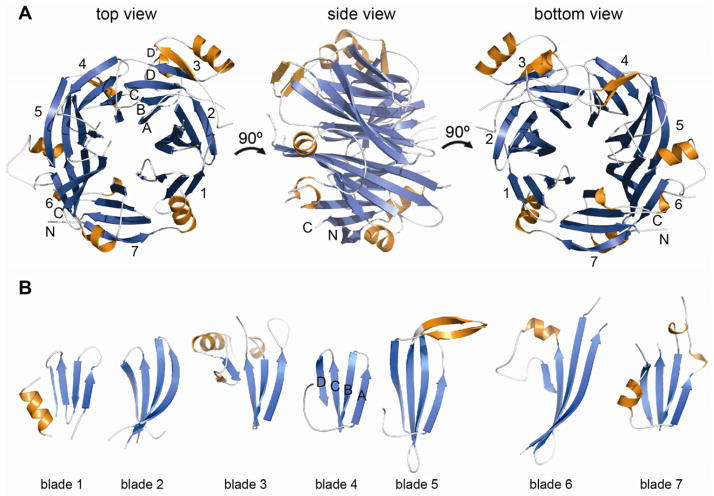
Figure 3. Structural analysis of mELYS NTD.
(A) Cartoon representation of the mELYS β–propeller domain with the blades labeled from 1 to 7. As a reference, the strands of blade 3 are labeled A–D, with the non-canonical 5th strand labeled D′. The β–propeller blades are colored in blue, loops are in white and the secondary structure insertions within the loops are shown in orange.
(B) Comparison of the β–propeller blades reveals long loops that decorate the structural core and contribute about 40% of the total mass of the 494 residue mELYS NTD.
See also Figure S1.
We also designed a series of expression constructs that encoded both the NTD and CHD of mELYS with the goal of crystallizing the entire structured domain of ELYS (residue 1–1018). All tested constructs were largely insoluble and heavily degraded in bacterial cells, likely indicating a compromised fold. Analysis of the main degradation products obtained after expression of the entire structured domain revealed that prominent degradation occurs in the loop following the second predicted α-helix of the central domain. We made a modified expression construct of mELYS that contained the NTD and two additional predicted α-helices from the CHD (residue 1–528), crystallized it and solved the structure with molecular replacement (Figure S1C and S1D). No additional electron density for the two expected α-helices could be observed, suggesting that NTD and CHD are not rigidly connected and possibly act as two rather flexibly tethered domains.
Structural Overview of the N-terminal domain of ELYS
ELYS NTD forms a seven-bladed β-propeller with dimensions 63 Å × 54 Å × 40 Å (Figure 3A). Each blade consists of four antiparallel β-strands labeled A to D, following the standard convention (Chaudhuri et al., 2008). The blades are arranged radially around a central water-filled pore, with β-strand A at the innermost position. Blade 7 is built from strands A–C at the C terminus of the protein, while strand D is formed by an N-terminal extension emanating from strand 1A. This arrangement for blade 7, known as a velcro-closure, is typical in β-propellers and contributes additionally to their structural stability (Chaudhuri et al., 2008; Xu and Min, 2011).
A regular β-propeller domain is constructed from ~300 residues, however the ELYS NTD is almost 500 residues in size. The β-propeller domain of ELYS is decorated with long loops, some of which contain additional α-helices and non-canonical β-strands. Together these elements make up for the ~200 extra residues (Figure 3A and 3B). The loops are named according to the canonical β-strands that they connect. DA loops (loops connecting β-strands D and A of two neighboring blades) together with BC loops define the top surface of the β-propeller, while the bottom surface is composed of AB and CD loops. Short BC loops are conserved in all blades, while long CD loops contribute with additional structural elements to the core scaffold of the NTD (Figure 3B). Most of the long loops contain α-helices that are pushed toward the perimeter of the β-propeller, giving it an asymmetric shape (Figure 3A). The 5CD loop, contains two anti-parallel β-strands, but they point away from the β-propeller rather than extending blade 5 (Figure 3B). β-propellers can have N-terminal extensions that add a fifth β-strand to the first blade, as, for example, observed in the β-propeller domain of S. pombe Nup120. However, in the ELYS β-propeller, blade 3 is five-stranded due to the long 3CD loop contributing, along with an α-helix, an additional β-strand (3D′) (Figure 3A).
Extensive loop insertions cause the length of the blade sequence repeat to vary between 44 and 95 residues (Figure 3B; Figure S3). In addition, blades 5 and 6 have significantly longer β-strands, leading to an asymmetry in the height of the β-propeller, while blade 3, with its fifth β-strand and α-helix located on the side of the β-propeller, distorts it radially (Figure 3A). Despite the overall asymmetry of the NTD, due to the non-canonical decorations, the rotational symmetry about the central axis of the core domain is preserved. Antiparallel β-strands within the core of the β-propeller align with a RMSD between 1.20–1.81 Å. The three inner β-strands of each blade superimpose very well, while β-strand D shows more structural divergence (Figure 3B). The connecting loops are, as expected, structurally very diverse.
3D structure comparison using the Dali server revealed different β-propellers as structural homologs of the ELYS NTD. Although all β-propellers have a very similar structural core, characteristic structural features projecting from the basic scaffold often define their specific functions. As the sequence conservation between the ELYS NTD and structurally similar proteins is low (<11 %), we conclude that they are functionally diverse.
Surface conservation reveals the interaction site of ELYS NTD with the NPC
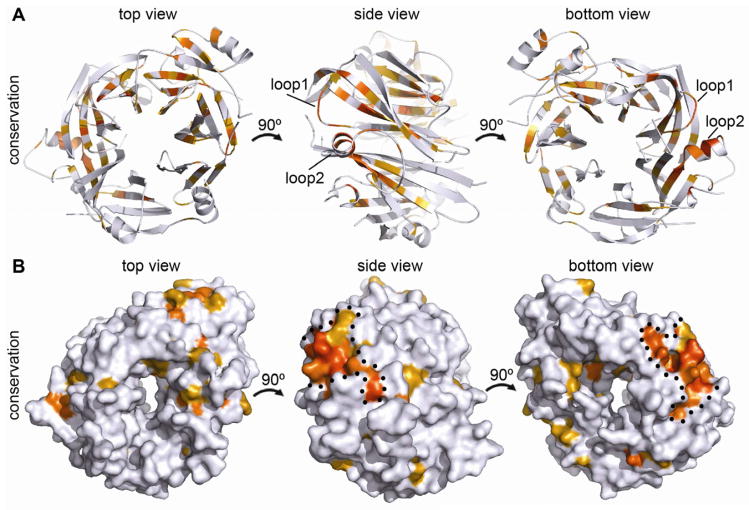
An alignment of ELYS NTD sequences from multiple organisms using T-coffee (Notredame et al., 2000) revealed that most conserved residues are buried in the hydrophobic core of the β-propeller and are important for its structural stability (Figure 4). Mapping the sequence conservation onto the surface of the protein identified a single, highly conserved region at the edge of the bottom surface (Figure 4B). This patch is formed by two loops, 5AB (loop1) and 6CD (loop2), and contains many charged and hydrophilic residues capable of forming salt bridges and H-bonds. Since sequence conservation typically indicates functional importance, we hypothesized that the conserved loops in the NTD might build binding sites for interacting nucleoporins, possibly guiding the incorporation of ELYS into the NPC. β-propeller domains are known to be mediators of diverse protein-protein interactions (Xu and Min, 2011).
Figure 4. Conserved features of the mELYS β–propeller.
(A) Cartoon representation of mELYS NTD from three different perspectives, related by rotation around the vertical axis as indicated. Conservation within the mELYS NTD is gradient-colored from gray (not conserved) to dark orange (most conserved) based on the alignment in Figure S3. The majority of conserved residues are structurally important as they build the core scaffold of the β–propeller.
(B) Surface representation showing conservation of residues on the β–propeller periphery. Color scheme as in A. The two conserved loops building the NPC binding interface are outlined with a black dotted line.
See also Figure S3.
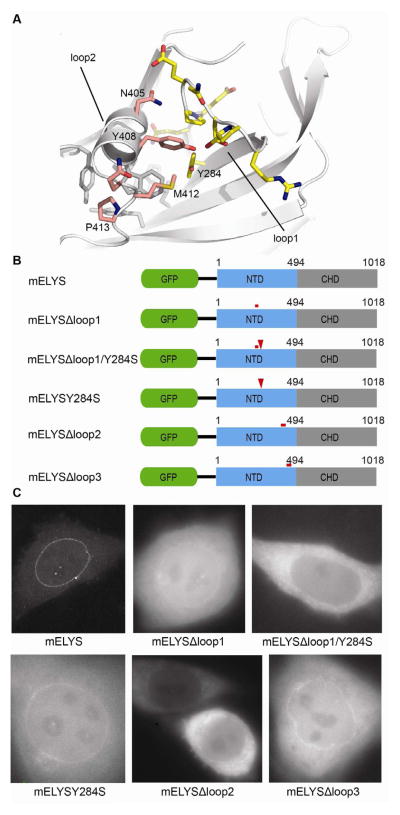
In vivo localization data showed that both structural domains (NTD and CHD) of metazoan ELYS contribute to proper NPC localization (Figure 2). To experimentally confirm the functional importance of the conserved surface loops in the ELYS NTD for NPC integration, we introduced loop deletions and point mutations into the GFP expression construct containing both structural domains of mELYS (residues 1–1018) and tested their localization in HeLa cells (Figure 5). Y284 from β-strand 5B is a strictly conserved residue and although its main chain atoms are part of the structural core of β-propeller, the side chain is exposed and accessible for interaction (Figure 5A; Figure S4). However, a Y284S mutation did not abolish NPC binding (Figure 5B and 5C). Replacement of the conserved 5AB loop (mELYSΔloop1, residue 272–280) with a glycine-serine rich sequence was also not sufficient to abrogate NPC-targeting (Figure 5B and 5C). Combining the Y284S mutation together with the loop1 deletion, however, did disrupt nuclear rim staining of mELYS (Figure 5C), indicating that this surface region is indeed involved in NPC targeting.
Figure 5. Mutations in the conserved β–propeller loops abolish proper NPC localization of mELYS.
(A) Cartoon representation of mELYS NTD with the residues in the conserved loops shown as sticks. Loop1 residues are highlighted in yellow and loop2 residues in pink. Residues forming a hydrophobic pocket beneath loop2 are shown in gray.
(B) Domain organization of the GFP-tagged mELYS mutants with the positions of the mutations indicated by red bars (loop deletions or multiple mutations) or red arrowheads (single mutations).
(C) Confocal fluorescence microscopy of HeLa cells expressing GFP-tagged mELYS proteins. Wild-type mELYS properly localizes to the nuclear envelope. Deletion of the disordered loop3 (control, residues 458–476), as well as milder mutations within the binding region show nuclear rim staining. The mELYSΔloop1/Y284S and mELYSloop2mut mutants abolish nuclear rim localization. See also Figure S4.
Replacement of the conserved 6CD loop (loop2, residues I404–Y427) with a glycine-serine rich sequence resulted in insoluble protein. Loop2 starts with an α-helix (residues I404–Q411) before it becomes less ordered. Two residues, I404 and W407, from the highly conserved α-helix form hydrophobic packing interactions with L416 and L422 of loop2, as well as with the hydrophobic core of the β-propeller (Figure 5A). Thus, replacing the entire loop2 with a flexible sequence might have caused folding problems. To maintain the structural integrity of the β-propeller fold, we then mutated just the conserved residues within loop2 that are not involved in building the structural core (mELYSloop2mut, mELYS N405S/Y408S/Q411A/M412S/P413G). While the expression of single mutants within loop2 had no affect on localization of the GFP reporter construct in HeLa cells, GFP-tagged mELYSloop2mut abolished nuclear rim staining suggesting that proper assembly into the NPC was affected (Figure 5B and 5C). Increased background levels can be noticed for mutants that show nuclear rim staining, indicating that their NPC binding affinity is somewhat decreased compared to wild-type.
To confirm the specificity of our mutants and their in vivo defects, we deleted the neighboring 7AB loop (mELYSΔloop3, residues 458–476) as a control. Most of the 7AB loop is disordered in our structure, however its terminal, confining residues are positioned on the same side of the β-propeller as the conserved region, suggesting the spatial proximity of all three loops. In contrast to the 5AB and 6CD loops, the sequence of the 7AB loop is not conserved. As expected, deletion of the 7AB loop did not influence the localization of GFP-reporter in HeLa cells (Figure 5B and 5C).
The tested mutants were designed based on the crystal structure, changing the protein surface without perturbing the protein fold. The mutants had the same expression levels in bacterial cells as the wild-type mELYS NTD, had identical solubility, and exhibited identical gelfiltration behavior (Figure S4B). Moreover, we obtained crystals of the mELYSΔloop1 mutant in similar growth conditions as used for the wild-type protein (Figure S4C), indicating that the overall protein fold of the mutants is not compromised. Therefore, the observed in vivo mislocalization of the mutants is the result of abolished critical interactions that tether ELYS to the NPC.
Both conserved loops are involved in crystal contacts with symmetry related molecules (Figure S4D, S4E and S4F), which likely influences the observed loop conformations. Thus, the conformation of loops 1 and 2 might be somewhat different in vivo. In addition, the conformation of the two loops may change upon interaction with binding partner.
Our previous data showed that ELYS homolog found in unicellular organisms consists of an short α-helical domain that binds Nup120. Since targeting of the metazoan ELYS to the NPC seems to be more complex, with both NTD and CHD contributing, this may well reflect structural differences within the core scaffold of the NPC.
DISCUSSION
Binding of ELYS to chromatin is an early step in NPC reassembly after cell division in metazoa. Biochemical pull-down data (Rasala et al., 2006; Franz et al., 2007) and in vivo time-course experiments (Franz et al., 2007) showed that ELYS subsequently recruits the Nup107-160 complex. Understanding how ELYS interacts with other nucleoporins is crucial for understanding of the NPC assembly process. Further, ELYS is a stable nucleoporin, and as a large architectural protein has substantial impact on the NPC structure.
To understand how this ~250 kDa protein fulfills its different functional roles, we studied the localization pattern of its domains. Transfection of HeLa cells with different truncations of GFP-tagged ELYS revealed that the combined structured domains, NTD and CHD, of mELYS (residues 1–1018) establish NPC anchorage, while the disordered CTR (residues 1019-end) localize to the nucleus and binds chromatin. Residues within the CTR have been shown to be phosphorylated in a cell cycle dependent manner (Nousiainen et al., 2006; Dephoure et al., 2008), perhaps indicating regulation and fine-tuning of its functional role. Interestingly, recent data show that ELYS interacts with the lamin B receptor in a phosphorylation-dependent manner in HeLa cells, thereby influencing NE formation (Clever et al., 2012).
Our previous data revealed that ELYS interacts directly with Nup160 of the nonameric Nup107-Nup160 complex (Bilokapic and Schwartz, 2012b). Based on the crystal structure of the ELYS NTD we were able to narrow down the interaction site with the NPC. In HeLa and T98G cells transfected with GFP-tagged mELYSΔloop1/Y284S or mELYSloop2mut, the fusion protein failed to localize to the NPC and was found in the cytoplasm instead (Figure 5C). Thus, we experimentally confirmed the functional significance of the conserved surface patch in the β-propeller domain of ELYS and establish that it is necessary for NPC interaction. Our data do not exclude the possibility that ELYS binds to other nups besides Nup160. In fact, already the small ELYS homolog in unicellular organisms binds to Nup37 as well, thus multiple binding partners are rather expected for metazoan ELYS.
In addition to its NPC localization, ELYS was found in distinct nucleoplasmic punctae during interphase (Figure 2 and Rasala et al., 2006), suggesting an additional function unrelated to NPC binding and nucleocytoplasmic transport. This is not uncommon for nucleoporins. In fact, several are thought to interact with chromatin away from the NPC (Liang and Hetzer, 2011). ELYS protein turnover has been studied by photobleaching experiments (Galy et al., 2006). It was shown that ELYS exists in two pools, one with high turnover inside the nucleus, and one with very low turnover and NPC-associated. Architectural nups have a very low protein turnover (D’Angelo et al, 2009; Savas et al., 2012), similar to the pore-associated ELYS pool. Thus, in addition to the structural role at the NPC, ELYS might also act away from the NPC, inside the nucleus. It will be interesting to determine what the function of this intranuclear pool of ELYS is.
Although several reports showed that ELYS binds to the surface of chromatin during mitosis (Rasala et al., 2006; Galy et al., 2006; Franz et al., 2007; Gillespie et al., 2007), it is not known whether this interaction remains intact after NPC assembly. DNA-binding is mediated via the ~150 kDa disordered CTR of ELYS (Rasala et al., 2008; Lau et al., 2009). Due to its flexible character the CTR can potentially stretch far into the nucleus, and thus maintain the connection with chromatin even after NPC formation. Alternatively, this domain could also be involved in targeting of genes to the NPC or in creating heterochromatin boundaries around the pores.
EM analysis of the purified and reconstituted heptameric S. cerevisiae Y-complex (homolog of the 107–160 complex) places Nup120 at one of the two short arms of the assembly structure. As ELYS and Nup37 interact with Nup160/Nup120 (Bilokapic and Schwartz, 2012b), but are absent in budding yeast, they contribute significantly to the molecular mass and shape of this short arm of the Y-complex. As a consequence the metazoan Y-complex is potentially substantially distorted from a regular Y-shape. The contribution of these additional proteins has to be taken into consideration when positioning the Y-complex into EM structure of whole intact NPCs of metazoa. It will be interesting to see whether ELYS is one of the main nucleoporins responsible for species-specific differences seen in EM reconstructions (Frenkiel-Krispin et al., 2010; Maimon et al., 2012).
EXPERIMENTAL PROCEDURES
Plasmid construction and protein purification
The N-terminal domain of Mus musculus ELYS (mELYS NTD, residues 1–494) was amplified from plasmid FFg-BOS-M (a generous gift from T. Taga) and cloned into an in-house modified version of the pET28a bacterial expression plasmid (Novagen) to yield an N-terminally 6xHis-tagged protein followed by a human rhinovirus 3C protease recognition site. The resulting vector was transformed into Escherichia coli BL21 (DE3) RIL strains (Stratagene).
Transformed cells were grown in LB medium containing the appropriate antibiotics and 0.5% (w/v) glucose. The bacterial culture was grown at 37 °C to an optical density at 600nm (OD600) of 0.6 before it was shifted to 18 °C for 30 min and induced by the addition of 0.2 mM isopropyl-thiogalactopyranoside. After overnight induction at 18 °C, cells were harvested by centrifugation, resuspended in lysis buffer (50 mM potassium-phosphate, pH 8.0, 300 mM KCl, 5 mM imidazole, 3 mM β-mercaptoethanol) and lysed using a french press. The crude lysate was supplemented with 1mM phenylmethanesulfonyl fluoride and Benzonase nuclease (Sigma) before it was clarified by centrifugation at 9,500 g for 25 min. The cleared supernatant was incubated with nickel-charged HisSelect resin (Sigma) for 30 min. After binding, the resin was washed three times with 10 bed volumes of lysis buffer in batch and loaded onto a disposable column (Pierce). The column was washed with 4 bed volumes of washing buffer (50 mM potassium-phosphate, pH 8.0, 300 mM KCl, 10 mM imidazole, 5 mM β-mercaptoethanol) and the bound protein was eluted with 4 bed volumes of elution buffer (50 mM potassium-phosphate, pH 8.0, 300 mM KCl, 150 mM imidazole, 5 mM β-mercaptoethanol). The protein was dialyzed overnight at 4 °C against 20 mM HEPES/KOH, pH 7.5, 100 mM KCl, 0.1 mM EDTA, 1 mM dithiothreitol (DTT). The N-terminal His tag was proteolytically removed during dialysis. The protein was further purified via cation-exchange chromatography by applying a linear 0.1–1 M NaCl gradient in elution buffer: 20 mM HEPES/KOH, pH 7.5, 1 mM DTT. Fractions with the protein were loaded onto size exclusion Superdex 200 column (GE Healthcare) equilibrated in crystallization buffer (10 mM HEPES/KOH pH 7.5, 150 mM KCl, 1 mM DTT). The elution profile of mELYS NTD indicated monomeric behaviour in solution. The protein was concentrated to 20 mg/ml.
Selenomethionine-labeled protein was prepared as described in (Brohawn et al., 2009b). Protein purification was carried out as for the native protein and incorporation of selenomethionine was confirmed by mass-spectometry.
Protein Crystallization
Initial crystals were detected in a sitting drop screen with a reservoir solution containing 0.1 M Bis-Tris/HCl pH 6.5, 20 % (w/v) PEG 3350 and 0.2 M ammonium acetate, using a protein concentration of 10 mg/ml. Diffraction quality crystals were obtained under oil in drops containing 1.5 μl of protein solution at 5 mg/ml and 1.5 μl of precipitant (0.1 M Bis-Tris/HCl pH 6.5, 25 % (w/v) PEG 3350, 0.2 M ammonium acetate, 0.5 % Octyl-β-D-glucopyranoside). Crystals appeared after two days. Prior to freezing, the crystals were gradually transferred into mother liquor solution containing 22 % (v/v) PEG 400. Selenomethionine-derivatized protein crystallized under identical conditions and the crystals diffracted significantly better than the native crystals.
Data Collection and Structure Determination
Diffraction data for a 1.9 Å single-anomalous dispersion (SAD) dataset was collected at 100 K at the NE-CAT beamlines 24-IDC/-E at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL). Data were processed with the HKL2000 package (Otwinowski and Minor, 1997). Four, out of six possible, selenium sites were found with SHELXC/D/E (Sheldrick, 2008) and further refined with SHARP (Bricogne et al., 2003). The seven-bladed β-propeller could initially be barely recognized in the resulting electron density map due to the low phasing power. To improve the initial map we performed density modification with DM. In the resulting electron density map protein-solvent boundaries were clearly visible and helped to position the basic β-propeller fold. We used the peptide backbone of the N-terminal domain of Homo sapiens Nup133 (PDB code 1XKS, Berke et al., 2004) as a starting model. The loops connecting β-strands as well as the D-strands from each blade were removed from the model and a Cα model was placed into the electron density using BrutePTF (Strokopytov et al., 2005). Combining model and SAD phases improved the electron density map; the connectivity of the main chain and the side chains started to become visible (Figure S2). ARP/wARP was used for automatic model building (Langer et al., 2008). More demanding protein loops were subsequently built manually in Coot (Emsley et al., 2010). The structure was refined using phenix.refine (Adams et al., 2010).
The refined structure of mELYS NTD consists of residues 3–494 with three loops (residues 338–345, 362–373, 458–465) absent from the final model, as they were not defined in the 2Fo-Fc electron density map. The statistics of the data collection and refinement are listed in Table 1.
Structure analysis
Sequence alignments were performed using T-Coffee (Notredame et al., 2000) and edited with JalView (Waterhouse et al., 2009). The model was validated using Molprobity (Chen et al., 2010). Structure figures were generated with PyMOL.
Transient expression in T98G cells
Human T98G and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium, supplemented with 10% heat-inactivated fetal calf serum (FCS), penicillin and streptomycin (100 IU/ml and 100 g/ml, respectively). The full-length mELYS, or domains thereof, were amplified from FFg-BOS-M plasmid and cloned into pIC242 (GFP-TEV-S-tag in pBABE retroviral vector) plasmid using XhoI/SacII restriction sites (Cheeseman et al., 2004). Homo sapiens ELYS (hELYS, residues 1–1018) and its individual domains were amplified from cMarathon DNA and cloned into pIC242 plasmid. For transient expression of GFP proteins in HeLa and T98G cells, the plasmids were transfected using Effectene (QIAGEN) according to the manufacturer’s instructions. Cells were imaged under the microscope 28–32 hours after transfection. Images were acquired on a DeltaVision Core deconvolution microscope (Applied Precision) equipped with a CoolSnap HQ2 CCD camera and deconvoluted using the DeltaVision software.
Supplementary Material
HIGHLIGHTS.
The vertebrate nucleoporin ELYS contains three structurally distinct domains
β-propeller and α-helical domain synergistically mediate integration into the NPC
Crystal structure of the N-terminal domain from M. musculus at 1.9 Å is presented
In vivo data using structure-based mutants reveal the determinants of NPC integration
Acknowledgments
We thank staff at beamlines 24-ID-C/-E at Argonne National Laboratory for assistance with data collection; I. Cheeseman for valuable advice and support during the work with human cell lines; N. Leksa and M. Halic for critically reading the manuscript. This work was supported by NIH Grant GM077537 (T.U.S.), a Pew Scholar Award (T.U.S.) and a Croatian Science Foundation fellowship (S.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with accession number 4I0O.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- Antonin W, Franz C, Haselmann U, Antony C, Mattaj IW. The integral membrane nucleoporin pom121 functionally links nuclear pore complex assembly and nuclear envelope formation. Mol Cell. 2005;17:83–92. doi: 10.1016/j.molcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Asakawa H, Hiraoka Y, Haraguchi T. Physical breakdown of the nuclear envelope is not necessary for breaking its barrier function. Nucleus. 2011;6:523–526. doi: 10.4161/nucl.2.6.16117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Lucić V, Förster F, Baumeister W, Medalia O. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- Berke IC, Boehmer T, Blobel G, Schwartz TU. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol. 2004;167:591–597. doi: 10.1083/jcb.200408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU. 3D ultrastructure of the nuclear pore complex. Curr Opin Cell Biol. 2012a;24:86–91. doi: 10.1016/j.ceb.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc Natl Acad Sci USA. 2012b;109:15241–15246. doi: 10.1073/pnas.1205151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor K, Shaikh S, Salina D, Raharjo WH, Bastos R, Lohka M, Burke B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J Cell Sci. 1999;112:2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Cryst D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JRR, Schwartz TU. The nuclear pore complex has entered the atomic age. Structure. 2009a;17:1156–1168. doi: 10.1016/j.str.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Schwartz TU. Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol. 2009b;16:1173–1177. doi: 10.1038/nsmb.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri I, Söding J, Lupas AN. Evolution of the β-propeller fold. Proteins. 2008;71:795–803. doi: 10.1002/prot.21764. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, 3rd, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clever M, Funakoshi T, Mimura Y, Takagi M, Imamoto N. The nucleoporin ELYS/Mel28 regulates nuclear envelope subdomain formation in HeLa cells. Nucleus. 2012;3:187–99. doi: 10.4161/nucl.19595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–95. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet CM, Talamas JA, Hetzer MW. Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell. 2010;141:1030–1041. doi: 10.1016/j.cell.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet C, Hetzer M. Nuclear pore biogenesis into an intact nuclear envelope. Chromosoma. 2010;19:469–477. doi: 10.1007/s00412-010-0289-2. [DOI] [PubMed] [Google Scholar]
- Dultz E, Zanin E, Wurzenberger C, Braun M, Rabut G, Sironi L, Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol. 2010;191:15–22. doi: 10.1083/jcb.201007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez AG, Piano F. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr Biol. 2006;16:1757–1763. doi: 10.1016/j.cub.2006.07.071. [DOI] [PubMed] [Google Scholar]
- Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkiel-Krispin D, Maco B, Aebi U, Medalia O. Structural analysis of a metazoan nuclear pore complex reveals a fused concentric ring architecture. J Mol Biol. 2010;395:578–586. doi: 10.1016/j.jmb.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Galy V, Askjaer P, Franz C, López-Iglesias C, Mattaj IW. MEL-28, a novel nuclear-envelope and kinetochore protein essential for zygotic nuclear-envelope assembly in C. elegans. Current Biology. 2006;16:1748–1756. doi: 10.1016/j.cub.2006.06.067. [DOI] [PubMed] [Google Scholar]
- Gillespie PJ, Khoudoli GA, Stewart G, Swedlow JR, Blow JJ. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Current Biology. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat Rev Mol Cell Biol. 2009;10:178–191. doi: 10.1038/nrm2641. [DOI] [PubMed] [Google Scholar]
- Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hase ME, Cordes VC. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol Biol Cell. 2003;14:1923–1940. doi: 10.1091/mbc.E02-09-0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol. 2010;2:a000539. doi: 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Rutherford S, Cotter LM, Allen TD, Goldberg MW. Steps of nuclear pore complex disassembly and reassembly during mitosis in early Drosophila embryos. J Cell Sci. 2001;114:3607–3618. doi: 10.1242/jcs.114.20.3607. [DOI] [PubMed] [Google Scholar]
- Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Delmar VA, Chan RC, Phung Q, Bernis C, Fichtman B, Rasala BA, Forbes DJ. Transportin regulates major mitotic assembly events: from spindle to nuclear pore assembly. Mol Biol Cell. 2009;20:4043–4058. doi: 10.1091/mbc.E09-02-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, De Souza CPC, Osmani AH, Osmani SA. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell. 2009;20:616–630. doi: 10.1091/mbc.E08-06-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loïodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Ladinsky MS, Kirchhausen T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol. 2011;194:425–40. doi: 10.1083/jcb.201012063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–397. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon T, Elad N, Dahan I, Medalia O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Nousiainen M, Silljé HHW, Sauer G, Nigg EA, Körner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;277:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Rasala BA, Ramos C, Harel A, Forbes DJ. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol Biol Cell. 2008;19:3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci USA. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotem A, Gruber R, Shorer H, Shaulov L, Klein E, Harel A. Importin beta regulates the seeding of chromatin with initiation sites for nuclear pore assembly. Mol Biol Cell. 2009;20:4031–4042. doi: 10.1091/mbc.E09-02-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, 3rd, Hetzer MW. Extremely long-lived nuclear pore proteins in the rat brain. Science. 2012;335:942. doi: 10.1126/science.1217421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM. A short history of SHELX. Acta Crystallogr, A, Found Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E. Structure and assembly of the Nup84p complex. J Cell Biol. 2000;149:41–54. doi: 10.1083/jcb.149.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, et al. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature. 2005;434:462–9. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]
- Strokopytov BV, Fedorov A, Mahoney NM, Kessels M, Drubin DG, Almo SC. Phased translation function revisited: structure solution of the cofilin-homology domain from yeast actin-binding protein 1 using six-dimensional searches. Acta Crystallogr D Biol Crystallogr. 2005;61:285–293. doi: 10.1107/S0907444904033037. [DOI] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–4097. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loïodice I, Hetzer M, Galy V, Hülsmann BB, Köcher T, Wilm M, Allen T, Mattaj IW, Doye V. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.