Abstract
The first link between sirtuins and longevity was made 15 years ago in yeast. These initial studies sparked efforts by many laboratories working in diverse model organisms to elucidate the relationships between sirtuins, lifespan, and age-associated dysfunction. Here we discuss the current understanding of how sirtuins relate to aging. We focus primarily on mammalian sirtuins SIRT1, SIRT3, and SIRT6, the three sirtuins for which the most relevant data are available. Strikingly, a large body of evidence now indicates that these and other mammalian sirtuins suppress a variety of age-related pathologies and promote healthspan. Moreover, increased expression of SIRT1 or SIRT6 extends mouse lifespan. Overall, these data point to important roles for sirtuins in promoting mammalian health, and perhaps in modulating the aging process.
Keywords: Aging, longevity, age-associated disease, NAD+, mitochondria, SIRT1, SIRT3, SIRT6
Phenotypes of aging: lifespan versus healthspan
Aging is a conserved phenomenon across all species, and imposes an everincreasing risk of dysfunction and death on older organisms. In humans, beginning in the 19th century, dramatic improvements in medicine, nutrition and hygiene significantly reduced early life mortality, while adult mortality in industrialized countries has decreased roughly 25-fold [1]. Collectively, these developments have led to a remarkable increase in average lifespan in industrialized societies, where life expectancy at birth has doubled in less than 10 generations, reaching 78.7 years in the United States in 2011. The estimated age-adjusted death rate, which accounts for changes in the age distribution of the population, reached a record low of 740.6 per 100,000 in 2011 in the US [2]. In the 75 years between 1935 and 2010, the overall risk of dying dropped by a remarkable 60%. Although the US population becomes overall older with each year, the rate of death continues to decline steadily across all races and sexes. Taken together, these data indicate that people living in advanced industrialized societies are living dramatically longer on average.
Although these changes represent a great boon for individuals fortunate enough to live in affluent societies – affording them the opportunity to live longer and healthier lives – they have also come with unintended consequences. As we live longer, and escape the (typically infectious) causes of early-life mortality that afflicted our forebears, we become more susceptible to age-associated pathologies, including cardiovascular disease, cancer, diabetes and neurodegenerative diseases. These chronic conditions exert an enormous toll, both in terms of human suffering and economic loss. For example, in the US the total cost of healthcare for people age 65 and older with dementia was estimated to be over $200 billion in 2013, and is expected to top $1 trillion yearly by 2050 [3]. This has led to intense interest in devising means to delay or even prevent this and other age-associated conditions.
Studies of the basic biology of aging may offer answers in this regard. A wealth of data from studies in model organisms has now shown conclusively that modulation of specific cellular signaling pathways – e.g. insulin/insulin-like growth factor-1 (IGF-1) signaling (IIS), mammalian target of rapamycin (mTOR), and sirtuins – can extend lifespan, in a manner that is conserved across distantly-related organisms [4]. Such interventions not only extend lifespan, but more importantly, extend the period of healthy living (i.e., healthspan) as well [5]. In mammals, interventions that slow the aging process concomitantly retard the onset and pace of an impressive array of degenerative, neoplastic, and metabolic diseases [6].
This review focuses on mammalian sirtuins, and addresses the key question of whether these proteins truly regulate aging in mammals. We focus our discussion on the three mammalian sirtuins currently thought to represent the most promising regulators of mammalian longevity: SIRT1, SIRT3, and SIRT6.
Initial observations of sirtuin-driven lifespan extension
In 1959, Mortimer and Johnston made the seminal observation that individual cells of the budding yeast Saccharomyces cerevisiae are not immortal. Instead, they found that a yeast mother cell divides only a finite number of times (replicative lifespan) [7]. Since that initial finding, numerous genes have been identified as regulators of longevity in this model organism [8]. The first evolutionarily conserved gene to be identified in such studies was the silent information regulator 2, or SIR2. Biochemically, Sir2p acts as an NAD+-dependent histone deacetylase (HDAC) [9]. Sir2p HDAC activity promotes increased chromatin compaction and decreased transcription at the silent mating-type loci, telomeric regions, and ribosomal DNA (rDNA). Inactivation of sir2 decreases lifespan by 50%; conversely a single extra copy of SIR2 extends lifespan by 30% [10]. Initial insights into mechanism focused on the finding that Sir2p protects yeast cells from a toxic accumulation of self-replicating extrachromosomal rDNA circles (ERCs) [11]. In this regard, deletion of the gene encoding the replication fork-blocking protein, Fob1p, enhances replicative lifespan by protecting against rDNA instability [10, 12], rescuing the short lifespan of sir2 mutant cells. Subsequent work has revealed additional, distinct mechanisms by which Sir2p promotes replicative lifespan. Sir2p facilitates retention of oxidized proteins, misfolded protein aggregates, and dysfunctional mitochondria in mother cells by maintaining cellular polarity [13, 14]. This allows nascent daughter cells to begin life with undamaged proteins and mitochondria, extending daughter cell lifespan at the expense of the mother. More recent studies have elucidated an additional epigenetic mechanism by which Sir2p promotes yeast replicative lifespan, involving the densely packaged (heterochromatic) regions adjacent to telomeres. Sas2p, a histone acetyltransferase, and Sir2p act in concert to regulate histone H4 lysine 16 acetylation (H4K16Ac) at the telomere-euchromatin boundary [15]. Levels of Sir2p decline over the yeast lifespan, concomitant with an increase in H4K16Ac. A mutation mimicking constitutive H4K16 acetylation reduces longevity, suggesting that derepression of subtelomeric silencing negatively affects yeast lifespan [16]. Altogether, these studies have revealed multiple distinct roles for Sir2p in promoting yeast replicative lifespan: first, in promoting rDNA array stability; second, in protecting daughter cells from inheriting damaged proteins and mitochondria; and third, in maintaining transcriptional silencing at subtelomeric regions.
SIR2 homologs have been identified in other species as well (BOX 1). Collectively, these Sir2p-like proteins are termed sirtuins and are present in eukaryotes and prokaryotes, implying an ancient origin [17]. Sirtuins possess NAD+-dependent deacetylase, deacylase, desuccinylase, demalonylase, deglutarylase, and ADP-ribosyltransferase activities, and regulate diverse cellular functions via modification of histones and many other proteins [9, 18–21]. Sirtuin-mediated deacetylation generates the feedback inhibitor nicotinamide (NAM), 2’-O-acetyl-ADP-ribose, and the deacetylated target protein [22]. In mammals, fluctuations in NAD+ levels due to varied dietary conditions, exercise, and circadian rhythm can all regulate sirtuin activities [23]. Owing to their requirement for oxidized NAD+, synthesized de novo or recycled via metabolic pathways, the enzymatic activity of sirtuins is, therefore, tightly linked to cellular nutritional milieu and metabolism [24]. Although mammalian sirtuins share a fairly conserved NAD+-binding catalytic domain, they have divergent amino and carboxy termini, and differ from one another with respect to catalytic activities, subcellular localization, protein targets, and biological functions [23].
Box 1: Sirtuin-driven lifespan extension in invertebrate models.
Much work has centered on delineating the genetic, molecular and biochemical roles sirtuins may play in promoting longevity in Caenorhabditis elegans and Drosophila melanogaster. The C. elegans genome encodes four sirtuin genes, SIR-2.1 though SIR-2.4, whose products are homologous to mammalian SIRT1 (SIR-2.1), SIRT4 (SIR-2.2/2.3) and SIRT6/SIRT7 (SIR-2.4) [17]. SIR-2.1 overexpression modestly extends mean lifespan in worms by 10–15%, via interaction with DAF-16 and the IIS pathway [161, 171–175]. C. elegans secrete dauer-inducing small molecules called ascarosides that control developmental timing and various social behaviors. Supplementation with pure synthetic ascarosides increases worm lifespan and stress resistance in a SIR-2.1-dependent manner, independent of IIS signaling through DAF-16. Inhibition of chemosensation in sensory neurons abolishes this phenotype, linking sensing of endogenous small molecules to sirtuin-dependent longevity and stress resistance [176]. In D. melanogaster, ubiquitous dSIR2 overexpression, or overexpression restricted to the nervous system or the fat body, also increases longevity [177–179]. Here it should be pointed out that some groups have been unable to reproduce these pro-longevity effects of invertebrate sirtuins [180]. Differences in genetic backgrounds or husbandry conditions may account for some of these discrepancies.
Impact of sirtuins on longevity in mouse models
Several studies in yeast, worms and flies have raised the important issue of whether sirtuins regulate aging in mammals (Box 1). Experimentally, this topic can be broken down into several distinct questions. First, does overexpression or hyperactivity of mammalian sirtuins increase lifespan in animal models, and/or suppress the appearance of age-sensitive traits? Conversely, does sirtuin loss-of-function shorten lifespan, and/or hasten the appearance of characteristics associated with normal aging? A closely related issue concerns how sirtuins interact with diseases of aging, i.e. what are their roles in regulating mammalian healthspan? We discuss below the fairly limited data on sirtuins and longevity in mammals, and then summarize the much larger body of literature concerning sirtuins and healthspan.
Sirtuins and mammalian aging
Sirt1
SIRT1 deacetylates a diverse array of cellular proteins – histones, transcription factors, DNA repair proteins, autophagy factors, and others – to modulate metabolism, stress responses, and many other cellular processes [25]. In vitro, SIRT1 possesses deacylase activity, though the functional significance of this activity in vivo remains unclear [26]. Though SIRT1 can localize to the cytoplasm in certain transformed cell lines [27, 28], adult cardiomyocytes [29] and neurons [30, 31], most studies have focused on nuclear SIRT1 functions.
Several studies address the question of whether increased SIRT1 levels can increase mammalian lifespan. In two Sirt1 transgenic mouse strains, overexpression of SIRT1 in many different tissues does not extend overall longevity [32]. For one of these strains, it was hypothesized that ectopically expressed SIRT1 did not increase overall longevity because it did not reduce the incidence of age-associated lymphoma, a major cause of death in many inbred mouse strains [33]. Sirt1 transgenic mice do show amelioration of several age-sensitive traits: improved maintenance of glucose homeostasis, wound healing, and neuromuscular function, as well as delayed bone loss, and a reduced incidence of carcinomas and sarcomas [33].
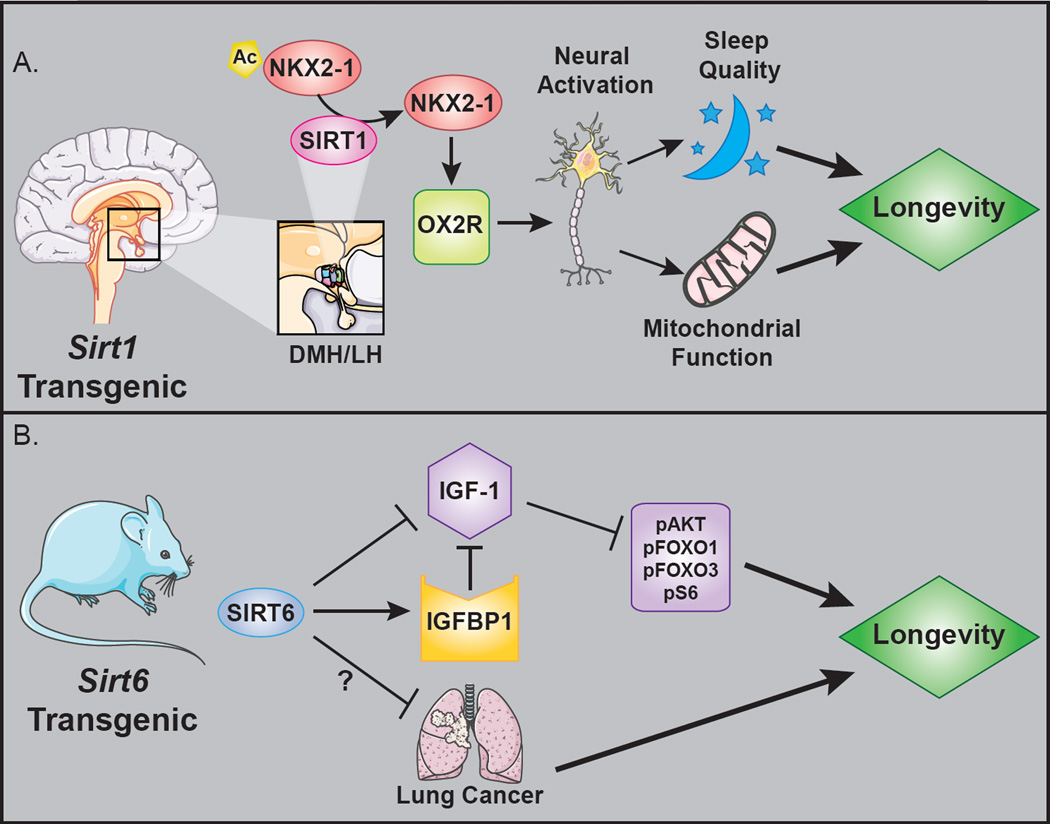
In contrast to results obtained with global SIRT1 overexpression, brain-specific SIRT1 overexpression in the BRASTO mouse line results in an ~11% increase in median lifespan, and delays cancer-related death in both sexes [34]. SIRT1 overexpression in the dorsomedial and lateral hypothalamic nuclei (DMH/LH) increases physical activity and whole-body oxygen consumption, preserves mitochondrial morphology in skeletal muscle, and promotes body temperature homeostasis in aged transgenic mice. These phenotypes are thought to result from an increase in neural activity due to SIRT1-mediated upregulation of brain-specific orexin type 2 receptor (OX2R) expression (Fig. 1A). Strikingly, in the DMH/LH of mice on a calorie-restricted diet, SIRT1 activity and OX2R expression mimics that of the BRASTO mouse [34]. Overexpression of SIRT1 has no lifespan effect in a second independently derived BRASTO transgenic line, possibly as a result of opposing longevity effects in different regions of the brain, and/or differences in SIRT1 overexpression levels. There is ample precedent for dose-dependent effects of sirtuin overexpression, both in invertebrates and in mammals. For example, in the mouse heart, modest SIRT1 overexpression (7.5-fold) protects against cardiac dysfunction, apoptosis and oxidative stress, whereas higher SIRT1 levels (12.5-fold overexpression) results in hypertrophy and impaired cardiac function [35]. Similar dose-dependent effects of SIR2 overexpression have been noted in invertebrate systems [36].
Figure 1. Lifespan extension in sirtuin transgenic mouse models.
A. SIRT1 overexpressed in the dorsomedial and lateral hypothalamic nuclei (DMH/LH) extends lifespan of both male and female mice by upregulating expression of the orexin type 2 receptor (OX2R), through deacetylation of the Nk2 homeobox 1 transcription factor (NKX2-1). B. Whole-body SIRT6 overexpression increases lifespan in male mice by inhibition of the insulin-like growth factor 1 (IGF-1) signaling cascade. Affected downstream effectors are indicated: phosphorylated AKT, (pAKT), phosphorylated forkhead box protein O1 and O3 (pFOXO1 and pFOXO3), and phosphorylated S6 kinase (pS6). IGFBP1, IGF-1 binding protein 1.
Germline Sirt1 deletion causes late prenatal or early postnatal lethality in an inbred 129/Sv strain background. Homozygous null embryos are developmentally delayed, with a subset showing exencephaly [37]. However, in a genetically outbred background, or on other inbred backgrounds [38], SIRT1 deficiency is compatible with adult viability [37]. SIRT1-deficient mice are runted, sterile and have a reduced lifespan under both normal feeding and dietary restricted conditions [31, 39, 40]. The pleiotropic nature of the phenotypes associated with SIRT1 deficiency renders it difficult to make firm conclusions about the appearance of age-sensitive traits in this strain.
Sirt3
The active form of SIRT3 is present in the mitochondrial matrix, where it deacetylates many proteins to regulate diverse mitochondrial functions such as ATP production, reactive oxygen species (ROS) management, β-oxidation, ketogenesis, cell death, and others [41]. Ectopically-expressed SIRT3 partially reverses the age-associated functional decline of murine hematopoietic stem cells [42]. Currently no pan- or tissue-specific transgenic animal models overexpressing SIRT3 have been described in the literature to determine whether SIRT3 overexpression confers lifespan extension or protects against age-associated pathologies. This is a critical question in clarifying current discrepancies regarding potential roles for SIRT3 in modulating human lifespan (BOX 2). SIRT3-deficient mice have no grossly apparent phenotype, but manifest a variety of defects associated with impaired mitochondrial functions, particularly under stress conditions or with increasing age (see below).
Box 2: SIRT3 Polymorphisms in Human Populations.
It is unknown whether increased sirtuin expression or activity would promote lifespan extension in humans. Studies testing links between sirtuins and human longevity have yielded conflicting results. Genetic linkage analysis in an Italian cohort identified an association between a synonymous single nucleotide polymorphism (SNP) in SIRT3 and increased male survival [181]. A follow-up study revealed this SNP, rs11555236, to be in linkage disequilibrium with a variable number tandem repeat found in a putative SIRT3 enhancer, and to be associated with male longevity [182]. However, a larger study was unable to validate this finding when samples from central and southern Italy were pooled with a German cohort [183].
A recent study of an Italian population, described in the prospective TRELONG study [184], analyzed SIRT3 genetic variants previously reported to be present in healthy elderly individuals of European ancestry [185]. Two SNPs, rs11555236 and rs4980329, were associated with longevity of the entire population, though this association remained significant only in females when the data were stratified by sex. Furthermore, this study confirmed the earlier finding that rs11555236 is in linkage disequilibrium with a putative SIRT3 enhancer. Individuals homozygous for this variant have increased SIRT3 expression in peripheral blood mononuclear cells [186], though it is unclear if genetic variation in this enhancer region directly affects SIRT3 mRNA expression. While current data are consistent the notion that SIRT3 genetic variation affects human longevity, and rs11555236 is a marker for longevity in some populations, further validation of the relationship between these SNPs and longevity in other human populations is necessary
SIRT3 may also impart protection from forms of metabolic dysfunction in humans, including obesity, insulin resistance and liver steatosis. Knockout of the Sirt3 gene in mice exacerbates the metabolic sequelae of a HFD. A nonsynonymous SNP in the catalytic domain of SIRT3 was found to be associated with metabolic syndrome in a cohort of human patients with nonalcoholic fatty liver disease. This correlation was further validated in a human population of ~8,000 males of Finnish decent who are part of the Metabolic Syndrome in Men cohort. This polymorphism reduces the catalytic activity of recombinant SIRT3 by ~34%. Thus, reduced SIRT3 activity may result in enhanced susceptibility to metabolic disease in humans [96]. Careful assessment of SIRT3 polymorphisms and expression in genetically diverse populations is essential to rigorously assess whether increased SIRT3 expression does indeed correlate with longer lifespan in either sex, protection against various age-related diseases, and/or cancer predisposition [127].
Sirt6
SIRT6 deacetylates specific cellular targets: H3K9Ac and H3K56Ac [43–45], the DNA repair factor CtIP [46], and the acetyltransferase GCN5 [47]. In vitro deacetylation of an H3K9Ac peptide by SIRT6 is stimulated by addition of physiologic concentrations of several long-chain fatty acids, representing a mode of endogenous catalytic regulation distinct from NAD+ concentration [26]. SIRT6 also activates poly[ADP-ribose] polymerase 1 (PARP1) via mono-ADP-ribosylation [48], and promotes secretion of the pro-inflammatory cytokine tumor necrosis factor alpha (TNFα) via deacylation [18]. Although nuclear SIRT6 functions are best characterized, SIRT6 and its invertebrate homolog SIR-2.4 promote formation of cytoplasmic stress granules, and associate with these structures [49–51]. Through these functions, SIRT6 plays essential roles in metabolic homeostasis, inflammation, stress responses, and genomic stability [52].
In two independent Sirt6 transgenic mouse strains, whole-body SIRT6 overexpression extends median lifespan of males by 14.5% and 9.9% respectively (Fig. 1B). In female transgenics, no significant increase in lifespan is observed. [53]. One possible rationale for this sexual dimorphism is that transgenic Sirt6 expression lowers serum IGF-1 levels and downstream signaling in male mice, but not in females. Reduced IIS in model organisms is strongly associated with increased longevity [54]. Male Sirt6 transgenics show some protection against lung tumors, and a trend towards preservation of glucose tolerance with age, but no protection against some other phenotypic characteristics of aging (e.g. osteoporosis and adrenal cortical hyperplasia) [53]. These data support a pro-longevity role for SIRT6 in male mice; however, a more comprehensive assessment of a variety of age-associated pathologies is necessary to characterize fully the effects of Sirt6 on healthspan.
Germline deletion of Sirt6 in inbred 129/SvJ-strain mice causes progressive, lethal hypoglycemia; SIRT6-deficient mice display a degenerative phenotype, lymphopenia, and genomic instability [55]. A subset of germline knockout mice on an outbred 129/Sv-C57BL/6 genetic background can survive up to 1 year if supplemented with glucose [56]. It is currently unknown whether these mice show accelerated appearance of aging characteristics. Germline or heart-specific deletion of Sirt6 results in cardiac pathologies (see below) [57].
Do sirtuins regulate aging?
Current data show that brain-specific (SIRT1) or ubiquitous (SIRT6) overexpression of specific sirtuin proteins can extend mammalian lifespan. Future studies must assess the robustness of these findings across different strain backgrounds and in different laboratories. These initial results are consistent with anti-aging functions for these proteins. However, disease-specific interventions can also extend lifespan, provided they mitigate a condition that is a common cause of mortality in the population. To prove that SIRT1 and SIRT6 slow aging in mice, many more studies are required to comprehensively elucidate the impact of increased levels of these proteins on diverse age-sensitive traits in different tissues, as has been done in the context of dietary restriction (BOX 3) and reduced growth hormone signaling [6]. Recent controversies in the literature regarding whether the mTOR inhibitor rapamycin truly delays aging emphasize the difficulty of establishing a primary effect of a singular intervention on aging [58–60].
Box 3: Dietary Restriction and Sirtuins.
In many diverse organisms, dietary restriction (DR) is a robust means to extend lifespan. In rodents, DR elicits lifespan extension up to 50% and protection against a variety of age-associated conditions [187]. Given that sirtuin catalytic activity is NAD+-dependent, and NAD+ levels rise upon nutrient stress in certain tissues, sirtuins have been proposed to link reduced intake with the pro-longevity response [188].
In S. cerevisiae, reduced glucose extends replicative lifespan in a strain-specific manner. There are conflicting reports as to whether this requires Sir2p, discrepancies which may result from strain differences and the use of varying degrees of glucose restriction [189–192]. Likewise, inconsistent results have been reported regarding the requirement for Sir2p homologs for DR-induced longevity in C. elegans and D. melanogaster. In C. elegans, most investigators find that SIR-2.1 is dispensable for lifespan extension in the eat-2 DR model [193–198]. In D. melanogaster, dSIR2 is required for DR-induced longevity, a finding that was not replicated by a subsequent study, but later supported using tissue-specific and global dSIR2 RNAi [68, 178, 180, 199]. In the fat body, DR induces gene expression changes distinct from those provoked by dSIR2 overexpression, indicating that DR and SIR-2.1 act, at least in part, through separate pathways in this tissue in flies [179].
Roles for mammalian sirtuins during DR are under active investigation (reviewed in [188]). SIRT1 is required for certain physiological changes in response to DR, in a tissue-specific manner. SIRT1 levels increase in WAT, skeletal muscle, and hypothalamus in response to DR in mice, but hepatic SIRT1 and NAD+ levels decline; thus SIRT1 activity is modulated in a tissue-specific manner during DR [38, 200]. Consistent with this, Sirt1 ablation in skeletal muscle abrogates the increased insulin sensitivity associated with DR; in contrast no metabolic defect in response to DR is observed when Sirt1 is deleted in the liver [200, 201]. Sirt1 ablation in the CNS and peripheral neurons also impairs insulin sensitivity in response to DR [202]; however, deletion in CNS neurons alone enhances insulin sensitivity and glucose tolerance compared to controls [203]. SIRT1 supports maintenance of body temperature and neuronal activity in hypothalamic nuclei during DR [38]. During DR, SIRT1 protects against age-related decline in renal function [204]. DR does not extend lifespan in Sirt1 global knockout mice [39, 205], though these mice are too short-lived to draw definitive conclusions regarding the DR response in this strain.
Increased SIRT3 expression in liver, skeletal muscle, and adipose tissue occurs during DR [92, 206–211]. Hepatic SIRT3 deacetylates and activates SOD2 and IDH2, reducing ROS levels during DR, [210, 212–214]. SIRT3 is required for protection against age-associated hearing loss induced by DR, presumably by suppressing ROS levels [210]. In wild-type mice, DR decreases hepatic acetyl-CoA, serum insulin and triglyceride levels, changes that require SIRT3 [210, 214]. SIRT3 is clearly necessary for multiple metabolic responses to DR, but whether or not it is required for longevity in response to this intervention remains unclear. To date, no role for SIRT6 in response to DR has been identified.
Sirtuin effects on diseases
In contrast to the modest amount of data regarding mammalian sirtuins and longevity per se, a very large literature exists substantiating roles for these proteins as guarantors of mammalian healthspan (Fig. 2). The following section focuses on the salient findings from current research concerning sirtuins in neurodegenerative disease, metabolic dysfunction, inflammation, cancer, and cardiovascular disease. Due to space requirements, only recent work will be highlighted here; the reader is referred to any of a number of excellent recent reviews in this area for a more comprehensive discussion (e.g. [61]).
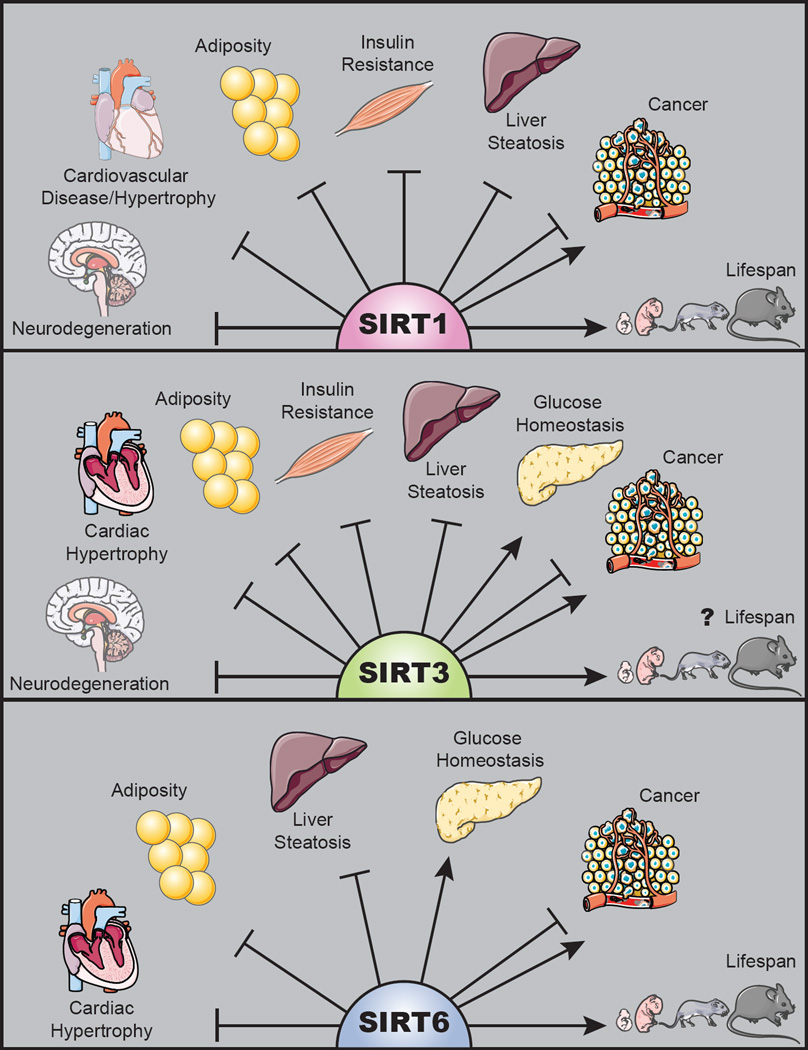
Figure 2. Sirtuins as guardians of mammalian healthspan.
SIRT1, 3, and 6 attenuate multiple age-associated diseases as indicated. Tumor suppressor and oncogenic properties have been reported for each of these proteins. SIRT1 and SIRT6 overexpression extend longevity in mice.
Neurodegenerative disease
The risk of neurodegenerative disease – e.g. Alzheimer’s disease (AD), Huntington’s disease (HD), Parkinson’s disease (PD), and others – increases sharply with age [62]. Several studies have revealed important roles for sirtuins in neuronal development and neurodegenerative disease [63].
Sirt1
AD is characterized by an accumulation of extracellular β-amyloid plaques and intracellular neurofibrillary tau protein tangles. In an AD mouse model, SIRT1 overexpression diminishes toxic β-amyloid plaque accumulation, mitigates loss of learning and memory, and increases survival [64]. Conversely, brain-specific Sirt1 knockout increases β-amyloid aggregates and worsens the phenotypes observed in this strain [64]. Mechanistically, SIRT1-mediated deacetylation of the retinoic acid receptor RAR β activates expression of the α-secretase ADAM10, which directs amyloid precursor proteins away from processing by β-secretase, thereby attenuating production of β-amyloid [64]. A separate study in AD mice deficient for SIRT1 demonstrated that SIRT1 deacetylates tau, promoting its ubiquitin-mediated degradation to suppress its pathogenic accumulation [65, 66].
Parkinson’s disease (PD) is associated with aggregation of presynaptic α-synuclein, and death of dopamine-secreting neurons in the substantia nigra. In transgenic mice harboring a mutant human α-synuclein gene associated with familial PD, brain-specific SIRT1 overexpression suppresses α-synuclein aggregation and extends lifespan. Conversely, compared to controls, PD mice lacking SIRT1 in the brain have a truncated lifespan, and elevated levels of α-synuclein aggregates [67]. Mechanistically, SIRT1 deacetylates the transcription factor heat shock factor 1, upregulating expression of the molecular chaperone HSP70, and reducing pathogenic α -synuclein aggregation [67].
HD is an autosomal dominant disease caused by polyglutamine repeat expansion in the huntingtin protein, resulting in neuronal dysfunction and death, primarily in the basal ganglia. Although conflicting roles have been described for SIRT1 orthologs in C. elegans and D. melanogaster HD models [68, 69], mammalian SIRT1 is clearly neuroprotective in mouse models of HD [32, 70]. SIRT1 overexpression in the brain of HD mice results in delayed onset and slower progression of disease, whereas Sirt1 deletion in the brain exacerbates HD symptomatology. SIRT1-dependent activation of p53, target of rapamycin complex 1 (TORC1), forkhead box O3a (FOXO3A), and peroxisome proliferator-activated receptor (PPAR)γ coactivator 1α (PGC-1α) pathways in the brain have been hypothesized as the molecular mechanisms by which SIRT1 promotes cell survival and confers neuroprotection in HD [70].
It is clear that increased SIRT1 activity can ameliorate disease effects in animal models of neurodegeneration. Whether or not modulating SIRT1 activity in human patients might prove therapeutically useful in treating these diseases awaits the results of clinical trials using SIRT1 activators (BOX 4).
Box 4: Pharmacologic approaches to activate sirtuin activities.
Screens have been conducted to identify small molecule sirtuin-activating compounds (STACs), work that has primarily focused on SIRT1 activation. The most widely-used STAC is the polyphenol resveratrol (RSV). RSV has been reported to extend longevity in yeast, worms, flies, and short-lived fish [215]. In mammals, RSV supplementation rescues the shortened lifespan of mice on a HFD [216], but does not extend longevity in mice fed a standard diet [58, 217]. RSV supplementation shifts the gene expression pattern observed in mice on a HFD towards a profile associated with a standard diet. This shift occurs in parallel with improved overall health, insulin sensitivity, increased mitochondrial content and maintenance of motor function.
Whether RSV and other STACs actually activate mammalian SIRT1 or its paralogs in vivo, and if so whether they function via direct mechanisms, or through upstream mediators such as AMPK, has been a hotly contested topic in the literature [218, 219]. In mice, RSV treatment stimulates mitochondrial function, activates AMPK and increases cellular NAD+ levels. These effects of RSV are SIRT1-dependent, though at higher doses RSV dose not require functional SIRT1 to activate AMPK [219]. In human patients with non-alcoholic fatty liver disease, RSV treatment appeared to be toxic to hepatocytes, and did not ameliorate liver steatosis or insulin resistance [220]. This study was limited, however, by including only male participants, and no assessment of RSV metabolites was performed.
To obviate potential confounding off-target effects of RSV, structurally unrelated synthetic STACs have been generated, one of which, SRT1720, activates SIRT1 in vitro and extends mean mouse lifespan by 18% in response to a HFD [221] and by 8.8% in mice fed a standard diet [222]. In this regard, a specific amino acid in SIRT1 targeted by STACs has recently been identified. Cells bearing a SIRT1 allele mutated at this site do not show the increased mitochondrial copy number and ATP content normally induced by STAC treatment [223]. Pharmacologic approaches to increase cellular NAD+ levels via administration of NR and NMN have recently provided alternative means of promoting increased sirtuin function.
Sirt3
SIRT3 is neuroprotective in an ex vivo cell culture system expressing nonpathogenic and mutant forms of huntingtin. Expression of mutant huntingtin results in decreased SIRT3 activity, reduced cellular NAD+ levels, and inhibition of mitochondrial biogenesis. Conversely, treatment with viniferin, a dimer of resveratrol (BOX 4), activates AMP-activated kinase (AMPK), promotes mitochondrial biogenesis, and increases SIRT3 expression. Sirt3 knockdown inhibits viniferin-mediated AMPK activation and attenuates its neuroprotective effects [71]. Given that mitochondrial dysfunction is thought to contribute to HD pathogenesis [72], it will be of interest to assess potential protective effects of SIRT3 in HD in vivo.
Metabolic dysfunction
Body composition changes with age, resulting in progressive loss of skeletal muscle mass and accumulation of white adipose tissue (WAT) [73, 74]. Increased visceral WAT is associated with an elevated risk of cardiovascular disease, type 2 diabetes (T2D), insulin resistance, hypertension and cancer [75, 76]. In this regard, obesity is associated with increased risk for cancers at diverse sites, including pancreas, breast, thyroid, kidney, colon and rectum. The 2007 NCI Surveillance, Epidemiology, and End Results (SEER) study estimated that about 4% of new cases of cancer in men and 7% in women are due to obesity in the US [77]. Sirtuin-related pathways, such as inflammatory responses, IIS, mTOR and AMPK signaling have all been hypothesized to mediate this obesity-cancer connection [78]. A large body of evidence now supports physiological roles for SIRT1, SIRT3, and SIRT6 in regulating metabolic homeostasis, with the potential to affect other age-related diseases such as cancer.
Sirt1
SIRT1 is a primary regulator of metabolism, an area reviewed extensively by others [79, 80]. For example, in the liver, SIRT1 regulates gluconeogenesis, fatty acid oxidation, cholesterol efflux, bile acid synthesis, and lipogenesis. Deletion of Sirt1 in hepatocytes sensitizes mice to liver steatosis on a high-fat diet (HFD); in humans this condition is associated with T2D, and represents a common cause of liver dysfunction. Aged mice with hepatic Sirt1 deletion also present with hepatosteatosis under normal dietary conditions [81, 82]. In the rat hypothalamus, pharmacological or siRNA-mediated inhibition of SIRT1 results in increased activity of anorexinergic pro-opiomelanocortin (POMC) neurons and decreased activity of orexinergic agouti-related protein (NPY/AgRP) neurons. Subsequently, these rats eat less and gain less weight [83]. However, another study found that Cre-mediated Sirt1 deletion specifically in POMC neurons predisposes mice to diet-induced obesity [84]. These discrepant results could be explained by differences in model organisms, neuronal populations affected, acute versus sustained inhibition of SIRT1 activity, and/or degrees of SIRT1 inhibition.
Under fasting conditions, SIRT1 expression is elevated in skeletal muscle, where activated PGC-1α supports mitochondrial biogenesis [85]. SIRT1 deletion in skeletal muscle results in reduced mitochondrial gene (mtDNA) expression, while mitochondrial copy number and expression of nuclear-encoded mitochondrial genes remain unchanged [86]. SIRT1 ablation in skeletal muscle reduces promoter activity of the nuclear mitochondrial transcription factor A (TFAM). These results suggest mtDNA gene expression can be regulated by a PGC-1α-independent pathway to maintain mitochondrial homeostaisis. Also, via PPARγ deacetylation, SIRT1 induces “browning” of WAT, suggesting a potential for SIRT1-dependent inhibition of adiposity [87, 88].
Pancreatic β cell-specific SIRT1 overexpression promotes insulin secretion and improves glucose tolerance in younger mice [89, 90]. Despite sustained SIRT1 overexpression, insulin secretion and glucose tolerance declines as this cohort ages, due to an age-associated decrease in plasma nicotinamide mononucleotide (NMN) [89]. NMN is an NAD+ precursor that ameliorates glucose intolerance by elevating NAD+ levels in a mouse T2D model [91]. NMN supplementation restores the age-related glucose-stimulated insulin secretion defect in aged β cell-specific Sirt1 transgenic females [89]. Taken together, these studies suggest that in addition to SIRT1 protein levels, maintenance of proper cellular NAD+ concentrations is critical for guarding against metabolic dysfunction. Indeed, an age-related reduction in the skeletal muscle NAD+ pool negatively affects mitochondrial homeostasis and ATP production by impairment of SIRT1-dependent TFAM expression [86]. Strikingly, this defect is prevented by chronic calorie restriction, or reversed by NMN supplementation. A more careful investigation of cell type-specific effects of SIRT1 modulation will be necessary to fully elucidate the numerous mechanisms by which SIRT1 affects energy expenditure, food intake and diet-related obesity.
Sirt3
SIRT3 governs diverse mitochondrial functions [41], and plays important roles in regulating fuel utilization. In this regard, SIRT3 deacetylates metabolic enzymes to promote fatty acid β-oxidation [92], glucose oxidation [93], and ketogenesis [94]. Sirt3 knockout mice have increased amounts of WAT and are glucose intolerant [95]. In response to a long-term HFD challenge, Sirt3 germline knockout mice develop obesity, insulin resistance, and hepatic steatosis at an accelerated rate [96]. Interestingly, Sirt3 gene ablation in either skeletal muscle or the liver alone does not apparently recapitulate the metabolic dysfunction observed in the whole-body knockout strains, suggesting that SIRT3 may exert metabolic control in part through non-tissue autonomous means, via effects in other tissues (e.g. the CNS) [41, 97]. SIRT3 plays important roles in response to dietary restriction (Box 3).
Sirt6
SIRT6 plays significant functions in glucose and lipid homeostasis. Sirt6 global knockout mice typically die from severe hypoglycemia by 4 weeks of age [55]. This phenotype has been attributed to cell-autonomous roles for SIRT6 in promoting mitochondrial respiration via suppression of hypoxia-inducible factor 1 α (HIF-1 α) function, and in reducing cellular insulin signaling through mechanisms that remain obscure [56, 98]. In contrast to the phenotype observed in whole-body SIRT6-deficient mice, hepatic Sirt6 knockdown increases both blood glucose levels and gluconeogenic gene expression [47, 99]. SIRT6 inhibits gluconeogenesis by indirectly inactivating a key transcription factor of gluconeogenic genes, PGC-1α [47]. Liver-specific Sirt6 deletion also results in liver steatosis, reduced fatty acid β-oxidation, and increased expression of lipogenic genes [99]. The role of SIRT6 in regulating lipid homeostasis has recently been investigated; SIRT6 negatively regulates activities of the lipogenic transcription factors SREBP1 and SREBP2, via multiple mechanisms [100, 101].
These diverse metabolic functions of SIRT6 may eventually render it a therapeutic target in the context of metabolic dysfunction. In this regard, SIRT6 overexpression ameliorates aspects of age-associated metabolic decline, such as insulin insensitivity and glucose intolerance. Sirt6 transgenic mice fed a HFD are protected against WAT accumulation, elevation of serum triglyceride and LDL cholesterol levels, and impaired glucose tolerance relative to non-transgenic controls [102]. In an obese/diabetic mouse model, SIRT6 expression in the liver is reduced, and gluconeogenesis is hyperactive. Adenoviral-mediated hepatic SIRT6 expression decreases blood glucose levels and suppresses expression of gluconeogenic genes [47].
Inflammation
Aging is associated with sub-clinical, chronic inflammation that has been linked to a number of age-related diseases including cancer, T2D, and cardiovascular disease [103]. The nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) family of transcription factors coordinates cellular responses to diverse genotoxic, inflammatory, and oxidative stresses. NF-κB is critical in regulating cellular processes such as apoptosis, proliferation, inflammation, immunity, and has been directly implicated in the aging process [104]. Several cellular signaling pathways involved in lifespan extension, including IIS, forkhead box O (FOXO) transcription factors, mTOR, and sirtuins, overlap with NF-κB signaling [104].
SIRT1 suppresses inflammation in multiple tissues by deacetylation of regulatory factors, including the NF-κB p65 subunit and FOXO proteins [105–107]. Consequently, liver-specific Sirt1 knockout mice fed a HFD show increased inflammation and elevated NF-κB signaling [81]. SIRT1 has also been shown to modulate NF-κB responses in macrophages [79]. Cartilage breakdown is increased in Sirt1 knockout mice, and increased levels of matrix metalloproteases that are typically elevated in osteoarthritis are present, suggesting an anti-inflammatory role for SIRT1 in osteoarthritis [108]. In humans, a mutation in the SIRT1 coding sequence was identified in a family with individuals suffering from the autoimmune disorders type 1 diabetes and ulcerative colitis [109]. This finding provides compelling evidence that SIRT1 suppresses inflammation and autoimmunity in humans.
Evidence that SIRT6 plays an anti-inflammatory role comes from studies in Sirt6-null mice. SIRT6 interacts with the NF-κB RELA/p65 subunit and localizes to the promoter regions of NF-κB targets, where it deacetylates H3K9Ac to silence expression of these genes. Haploinsufficiency of the RELA/p65 subunit in SIRT6-deficient mice rescues some of these mice from early demise, suggesting that elevated inflammation mediated through NF-κB plays an important role in the pathologies associated with SIRT6 deficiency [106]. Recent in vitro analyses have demonstrated that SIRT6 stimulates pro-inflammatory TNF α secretion by removal of long-chain fatty acyl groups. More detailed analysis in vivo is required to elucidate the role that SIRT6 may play in enhancing inflammation [18].
Cancer
Accumulated genetic and epigenetic alterations, as well as metabolic reprogramming, are among the hallmarks of cancer [110]. Sirtuins regulate many aspects of chromatin biology, genome stability, and metabolism. Perhaps not surprisingly, then, a large body of evidence now links sirtuins to cancer in a complex, cell- and context-specific manner.
Sirt1
Both oncogenic and tumor suppressor roles have been ascribed to SIRT1. SIRT1 regulates more than 50 distinct substrates relevant to cancer proliferation and survival, notably including the c-MYC oncoprotein and the tumor suppressor p53 [111]. Increased SIRT1 expression has been detected in many human cancers including: breast, prostate, lung, colon, liver, pancreatic, lymphoma, leukemia and some ovarian and cervical cancers, suggesting an oncogenic role for SIRT1 [111].
Conversely, evidence primarily from mouse models supports a role for SIRT1 as a tumor suppressor, where SIRT1 promotes the maintenance of genome stability [112], and inhibition of cell growth via survivin (an inhibitor of apoptosis), β-catenin, and other pathways [113, 114]. Consequently, the incidence of spontaneous carcinomas and sarcomas is reduced in Sirt1 transgenic mice [33]. Also, ectopic expression of SIRT1 in the APC+/min mouse model of intestinal cancer decreases tumorigenesis [115]. Paradoxically however, global or enterocyte-specific deletion of Sirt1 reduces the size and number of intestinal polyps formed in this model [40, 116]. Sirt1 deletion results in increased apoptosis in polyps arising in APC+/min mice, implying SIRT1 can play a pro-survival role during tumor progression [116].
SIRT1 directly binds to and deacetylates HIF-1α, a transcription factor that mediates gene expression in response to increased ROS and metabolic reprogramming in cancer cells [113]. Whether SIRT1-dependent deacetylation promotes HIF-1α activity, or attenuates its ability to promote expression of its target genes, is currently a matter of debate. One study demonstrated SIRT1 knockdown in vivo impairs transcriptional output of HIF target genes in hepatocellular carcinoma cells [117]. Others have shown that SIRT1-dependent deacetylation of HIF-1α inhibits transcriptional co-activator binding to promoters of HIF-1α target genes, and negatively regulates tumor growth and angiogenesis in xenograft tumor models [113]. SIRT1 also interacts with the oncoprotein c-MYC, though the functional outcome of this interaction is also controversial [112, 118, 119]. Overexpression of c-MYC and SIRT1 is present in several cancer types, and is often associated with higher tumor grade [111]. Although SIRT1 expression is stimulated by c-MYC, deacetylated c-MYC is more susceptible to degradation; therefore SIRT1 indirectly inhibits c-MYC function [112, 119]. However, others have found that deacetylated c-MYC is stabilized, and enhances SIRT1 activity by promoting NAD+ synthesis, which establishes a positive feedback loop to enhance cellular transformation [118]. Thus, while SIRT1 expression may inhibit oncogenesis in some contexts (e.g. primary cells), increased SIRT1 expression may confer a growth advantage in cells that have already acquired oncogenic mutations. Future studies focusing on the relationship between SIRT1 and its substrates, in particular HIF-1 α and c-MYC, in various tissue- and cell-type specific contexts will clarify the multiple ways in which SIRT1 interacts with carcinongenesis.
Sirt3
As with SIRT1, conflicting data exist regarding SIRT3’s roles in cancer. One study found that the human SIRT3 gene is deleted in up to 20% of all human cancers analyzed overall, and up to 40% of breast and ovarian cancers [120]. Expression of SIRT3 is reduced in a number of other human malignancies including testicular, prostate, and hepatocellular cancers [99]. SIRT3-deficient mice show an increased incidence of mammary cancer [120, 121]. Mechanistically, elevated ROS levels promote tumorigenesis via increased mutation rate and genomic instability, and activation of pathways regulating metabolism, cell survival, and proliferation [122]. Therefore, suppression of ROS levels is likely a principal means by which this sirtuin inhibits tumorigenesis. In this regard, SIRT3 deacetylates and activates mitochondrial antioxidant proteins, superoxide dismutase 2 (SOD2) and isocitrate dehydrogenase 2 (IDH2); deacetylation of IDH2 in turn allows regeneration of the antioxidant glutathione [123]. Sirt3 deletion increases ROS and stabilizes HIF-1 α, which results in protumorigenic metabolic reprogramming [120, 121, 124]. Deletion of Sirt3 in mouse embryonic fibroblasts increases intracellular superoxide levels and aneuploidy in response to genotoxic stressors, suggesting that SIRT3 guards against chromosomal instability and tumorigenesis by maintaining genomic integrity [121]. A novel function of SIRT3 in tumor suppression was recently revealed in studies demonstrating a role for SIRT3 in stimulating activity of the pyruvate dehydrogenase complex (PDC) to promote mitochondrial respiration [93]. Suppression of PDC activity is an important component of metabolic reprogramming in many tumors [125].
However, data also exist supporting an oncogenic role for SIRT3. SIRT3 has been reported to deacetylate p53 and counteract p53-induced growth arrest in human bladder cancer cells [126]. Li-Fraumeni Syndrome (LFS) is characterized by a predisposition to a wide spectrum of cancers, and frequently results from mutations in the TP53 gene. In a study of a family with LFS without a detectable TP53 mutation, a genomic duplication of the entire SIRT3 gene was identified, leading to increased SIRT3 mRNA expression [127]. To evaluate the role of SIRT3 as a cause of tumorigenesis in these patients, the authors of this study demonstrated that SIRT3 overexpression in a glioma cell line prevents apoptosis, deregulates cell cycle progression and results in hypermethylation of CpG islands, features typically associated with cancer [110]. Furthermore, elevated levels of SIRT3 have been detected in node-positive breast cancer [128] and oral squamous cell cancer (OSCC). In OSCC, SIRT3 knockdown inhibits proliferation and sensitizes cells to radiation and chemotherapeutic treatments in vitro [129]. Thus, the oncogenic and tumor suppressor activities of SIRT3 are likely highly context specific.
Sirt6
As with SIRT1 and SIRT3, evidence exists supporting both oncogenic and tumor suppressor properties for SIRT6. Elevated SIRT6 expression is associated with reduced overall survival in breast cancer patients, where SIRT6 promotes chemoresistance [130]. Similarly, knockdown of SIRT6 expression in prostate cancer cells sensitizes cells to chemotherapeutics and decreases cancer cell survival [131].
However, supporting a tumor suppressor role for SIRT6, a survey of ~1,000 human cancer cell lines revealed that the SIRT6 locus was deleted in 35% of samples [132]. In a mouse intestinal polyposis model, Sirt6 deletion resulted in a dramatic exacerbation of adenoma number and severity [132]. Mechanistically, SIRT6 inhibits the transcriptional activities of HIF-1α and c-MYC, resulting in repression of glycolytic metabolic reprogramming [98, 132]. In a mouse liver cancer model, increased SIRT6 expression reduces survivin expression by deacetylating H3K9Ac at the survivin promoter, and induces apoptosis during cancer initiation [133]. In this context, SIRT6 expression averts tumorigenesis by inhibiting tumor cell survival. In human cancer cell lines, ectopic SIRT6 overexpression results in increased p53/p73-dependent apoptosis. Cell death occurs through the ataxia telangiectasia mutated (ATM) kinase, and requires SIRT6 mono-ADP-ribosyltransferase activity [134]. The opposing actions of SIRT6 to suppress or facilitate tumorigenesis may be specific to tumor type and stage. Modulation of SIRT6 activity or expression in pre-malignant and advanced tumors in vivo would help further delineate these roles of SIRT6.
Although we focus here on SIRT1, SIRT3, and SIRT6, other sirtuins have important roles in cancer, reviewed in depth elsewhere [111]. For example, SIRT2 can also function as an oncogene or a tumor suppressor [111]. SIRT4 functions as tumor suppressor by suppressing glutamine metabolism and promoting genomic stability [135–137]. SIRT7 has recently been shown to sustain malignant properties of cancer cells by deacetylating H3K18Ac [138].
Cardiovascular Disease
Cardiovascular disease (CVD) accounts for roughly one-third of all deaths in the United States, [139] and the risk of developing clinically evident CVD increases dramatically with advancing age [140]. Sirtuins have been implicated in cardiac and vascular protection by regulating mechanisms associated with CVD risk factors, such as vasorelaxation [141], vasoprotection [142], endothelial ischemic recovery, and cholesterol metabolism [143]. For example, SIRT1 knockdown inhibits endothelial cell migration and angiogenesis in human endothelial cells [144], and endothelial cell-specific Sirt1 knockout mice are unable to revascularize tissue in response to ischemic injury [145]. Mechanistically, SIRT1 deacetylates and inactivates the FOXO family of transcription factors and components of the NOTCH signaling pathway that have anti-angiogeneic properties [146–150].
Sirt1
SIRT1 promotes endothelial homeostasis in response to atheroprotective vascular flow and pulsatile shear (PS) stress. PS stress induces expression of anti-inflammatory and antioxidant genes such as Sod1 and Sod2, an effect that is lost upon knockdown of the gene encoding Ca2+/calmodulin-dependent protein kinase kinase (CaMKK)β. CaMKKβ, an AMP-activated protein kinase, phosphorylates and subsequently stabilizes SIRT1, and together are able to suppress the oxidative stress and inflammation pathways in an atheroprotective PS stress model. When crossed onto an apolipoprotein E-deficient background and fed an atherogenic diet, increased atherosclerotic lesions are observed in mice lacking either CaMKKβ or SIRT1, indicating that this pathway may be a clinically relevant target for intervention to treat age-associated CVD [151].
Pathological cardiac hypertrophy (CH) is a response to chronic hypertension or other sources of cardiac injury that manifests as increased cardiomyocyte size and re-induction of developmental genes whose expression is typically restricted to the fetal heart. Cardiac metabolism of fatty acids shifts to favor glucose in CH, likely due to repression of fatty acid oxidation and oxidative phosphorylation transcriptional networks in the adult heart [152]. SIRT1 interacts with and activates peroxisome proliferator-activated receptor-α (PPARα) to prevent this metabolic switch and the development of hypertrophy, by promoting fatty acid oxidation gene expression, and inactivates the NF-κB pro-inflammatory pathway. Treating mice with resveratrol (BOX 4) attenuates markers of induced CH in wild-type, but not PPARα-null mice [153]. Thus, SIRT1 is able to orchestrate metabolic reprogramming and inflammatory responses to protect against development of CH. Overexpressing both PPARα and SIRT1 in mice impairs mitochondrial function and promotes heart failure during CH, by downregulating genes involved in mitochondrial respiration, oxidative stress and cardiac contractility [154]. These data demonstrate that the activation of mechanisms that coordinate a predisposition to CH by SIRT1 is dose-dependent. Correspondingly, Sirt1-null mice are resistant to the development of exercise- or agonist-induced CH, phenotypes associated with reduced AKT activation in the heart [155].
Sirt3 and Sirt6
AKT is a serine-threonine kinase that is central in a network of diverse cellular processes, such as cell proliferation, apoptosis, glucose metabolism, and angiogenesis [156]. The end result of persistent hyperactive IGF-AKT signaling in myocardium is hypertrophy and eventual heart failure [157, 158]. Both SIRT3 and SIRT6 negatively regulate CH in part by inhibiting the IGF-AKT signaling cascade. Overexpressing Sirt3 in cultured cardiomyocytes and transgenic mouse lines, or supplementing mice with exogenous NAD+ precursors, represses the hallmarks of agonist-induced CH [159, 160]. SIRT3 protects against CH by inducing expression of the antioxidant proteins, SOD2 and catalase. SIRT3-mediated reduction in ROS levels inhibits AKT signaling and downstream gene expression associated with CH induction [159].
In failing human and hypertrophic mouse hearts, SIRT6 protein expression is reduced compared to controls. Sirt6 deletion in the mouse heart or globally results in CH, whereas cardiac-specific SIRT6 overexpression protects mice from induction of CH. Mechanistically, SIRT6 functions as a negative regulator of CH by co-repressing c-Jun-dependent transcription at the chromatin level, thereby inhibiting downstream IGF-AKT signaling. Importantly, in vivo inhibition of IGF signaling in whole-body and cardiac-specific Sirt6 knockout mice abrogates the hypertrophic response [57]. Overall, SIRT1, SIRT3, and SIRT6 all protect against CH in cell culture and mouse models, through overlapping and distinct mechanisms. It will be of great interest to assess the therapeutic potential of sirtuin activation in patients with CH.
Concluding remarks
The finding that sirtuin overexpression in yeast, worms and flies can extend longevity in these organisms has led to intensive efforts to test whether mammalian sirtuins might act similarly. With the recent observations that SIRT1 overexpression in the brain, and SIRT6 overexpression globally, increases lifespan in mice, this question has now been settled decidedly in the affirmative [34, 53]. Moreover, a large body of literature in mouse models demonstrates that sirtuins play powerful roles as guardians of mammalian healthspan in many diverse disease contexts by interacting with a variety of cellular factors (Fig. 3).
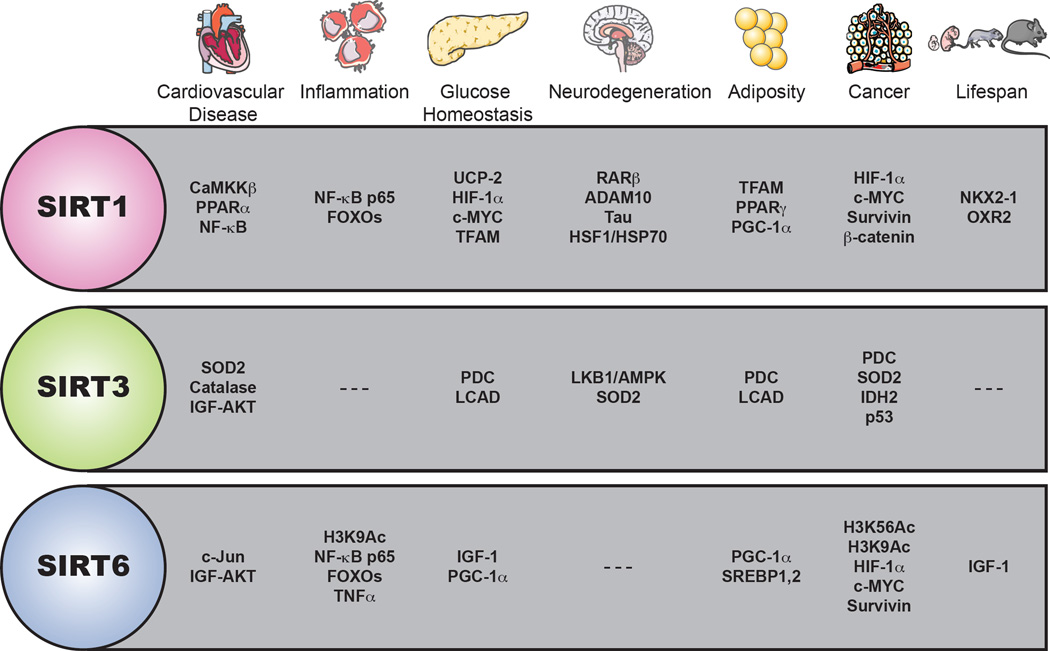
Figure 3. Representative major sirtuin interactors/substrates.
Examples of sirtuin protein interacting partners or substrates relevant to pathways outlined in the text.
However, these findings lead to a series of important downstream questions (Box 5). From the perspective of aging biology, it is vital to determine whether SIRT1 and SIRT6 extend lifespan by modulating the pace of aging per se, by suppressing major age-associated diseases, or through a combination of both activities. Since the onset and course of aging pathologies is itself tied to the aging rate, this distinction can be difficult to tease out, and will require careful assessment of numerous age-associated phenotypes in many tissue types in sirtuin overexpressors and controls [6]. In this regard, suppression of lung cancer may represent an important element of the pro-longevity effects of SIRT6 in male mice [53].
Box 5: Outstanding Questions.
Over the past decade, intense research interest in mammalian sirtuins has revealed pleiotropic roles for these proteins in blunting a remarkable range of age-associated diseases. Key important question remain, however, that need to be addressed before the full therapeutic potential of sirtuin modulation can be realized clinically:
First, many different sirtuins affect common pathogenic states, and even shared molecular targets. The extent to which mammalian sirtuins possess unique versus redundant functions, and the degree to which cross-talk exists between these proteins, are key questions that need to be systematically tackled.
Second, to what extent does activation of all sirtuins occur as a consequence of increased cellular NAD+ levels? In disease states, where different sirtuins may have opposing roles (e.g., neoplasia), pan-sirtuin activation may not be a viable therapeutic strategy.
Third, what are the consequences of long-term increases in sirtuin activity in vivo? Sustained overexpression of SIRT1 and SIRT6 in specific contexts confers health benefits and a modest increase in longevity. However, as a general matter, it is unknown whether long-term increased sirtuin activity might have deleterious consequences.
At a basic level, what does the extension of mammalian longevity by sirtuins imply? One unifying feature of sirtuins is they require NAD+ for catalytic function. NAD+ levels decrease in the liver, muscle and pancreas of aged mice [89, 161], in several tissues of aged rats [162, 163], in aged C. elegans [161] and in skin of elderly humans [164]. This age-related decline in NAD+ would be predicted to reduce sirtuin activity, potentially predisposing organisms to phenotypes associated with sirtuin loss-of-function. In this model, elevated sirtuin levels could compensate for reduced NAD+ levels through simple mass-action; that is, the elevated numbers of sirtuin protein molecules in the cell would compete more effectively for the increasingly-limited pool of NAD+. Importantly, several studies have now demonstrated that supplementation with the NAD+ precursors nicotinamide riboside (NR) or NMN can restore cellular NAD+ levels and protect against diet-induced obesity [165] and the decline in mitochondrial function in aged mice [86]. It will be of great interest to test sirtuin function and longevity in mouse strains with chronically elevated NAD+ levels. In this regard, the CD38 protein represents the major NADase in many mouse tissues; mice lacking CD38 show elevated NAD+ levels in vivo and increased SIRT1 activity, coupled with protection from diet-induced obesity [166].
There are also hints that sirtuin expression may decline in some tissues with aging. SIRT3 expression is decreased in hematopoietic stem/progenitor cells isolated from aged mice (18–24 months) compared to 3 month-olds [42]. SIRT6 protein levels are significantly lower in human dermal fibroblasts derived from males >50 years old compared with males <18 years old [167]. This may represent an evolutionarily conserved phenomenon; in yeast, levels of Sir2p decline with age, resulting in chromatin dysregulation [16]. It will be of interest to determine systematically whether levels of SIRT1 and other sirtuins decline in aged tissues. Increased longevity and disease suppression conferred by sirtuin overexpression would then represent a means to bypass a hypothetical reduction in sirtuin expression in aged animals.
The striking fact that sirtuins can mitigate so many diverse mammalian diseases – involving both proliferative and non-proliferative tissues – likely indicates that these proteins as a group modulate process(es) that are central to many pathologies. In this regard, one current hypothesis concerning the role of mTOR signaling in mammalian disease and aging postulates that mTOR signaling increases with physiologic aging and in many different disease states; hence the mTOR inhibitor rapamycin suppresses diverse pathologies and extends mammalian lifespan [168]. Analogously, perhaps a decline in cellular NAD+ levels occurs as a final common event in many different pathologic states, in turn reducing sirtuin activities and contributing to disease severity. Restoration of sirtuin activity via overexpression or NAD+ precursor supplementation could mitigate downstream pathologies in any disease state where such reduced NAD+ occurs. Unfortunately, little is known about what happens to cellular NAD+ levels in pathological states, apart from obesity and diabetes, where the NAD+/NADH ratio declines due to dysregulated energy metabolism, an increase in the activity of the NAD+-consuming DNA-repair enzyme, PARP1, and a decrease in hepatic nicotinamide phosphoribosyltransferase expression, the enzyme that catalyzes the rate-limiting step of NAD+ biosynthesis [91, 169, 170].
Intensive pharmaceutical research has centered on developing specific activators of SIRT1 as a means to stave off age-related disease (BOX 4); little is known about specific activators of other sirtuins. As sirtuin functions are further elucidated using systematic tissue-specific approaches, additional target pathways for potential sirtuin-directed pharmacological intervention will certainly be identified. Approaches to maintain NAD+ concentrations at a youthful level may provide a means to maintain sirtuin function and promote health during aging.
Highlights.
Overexpression of the mammalian sirtuins SIRT1 and SIRT6 increases lifespan in mouse models.
Sirtuin activity is protective against a variety of age-associated diseases.
Sirtuins affect tumor initiation and maintenance via diverse mechanisms.
Small molecule interventions are emerging to modulate sirtuin functions in vivo.
Acknowledgements
We thank B. Zwaans for helpful discussions, and apologize to those whose work is not cited due to space limitations. Work in our laboratory is supported by NIH (R01GM101171 and R21CA177925). W.G. is supported by NIH Training Grant T32-AG000114. Some graphics in the figures were obtained and modified from Servier Medical Art from Servier (http://www.servier.com/Powerpoint-image-bank).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Finch CE. Evolution in health and medicine Sackler colloquium: Evolution of the human lifespan and diseases of aging: roles of infection, inflammation, and nutrition. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1718–1724. doi: 10.1073/pnas.0909606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoyert DL, Xu Jiaquan. Deaths: Preliminary data for 2011. National vital statistics reports. 2012;61 [PubMed] [Google Scholar]
- 3.Thies W, et al. 2013 Alzheimer's disease facts and figures. Alzheimer's & dementia : the journal of the Alzheimer's Association. 2013;9:208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Otin C, et al. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gems D. Tragedy and delight: the ethics of decelerated ageing. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2011;366:108–112. doi: 10.1098/rstb.2010.0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombard DB, Miller RA. Aging, Disease, and Longevity in Mice. Annual Review of Gerontology and Geriatrics. 2014;34:93–138. [Google Scholar]
- 7.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 8.Kaeberlein M, Kennedy BK. Large-scale identification in yeast of conserved ageing genes. Mech Ageing Dev. 2005;126:17–21. doi: 10.1016/j.mad.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 9.Imai S, et al. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 10.Kaeberlein M, et al. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 12.Defossez PA, et al. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 13.Liu B, et al. The polarisome is required for segregation and retrograde transport of protein aggregates. Cell. 2010;140:257–267. doi: 10.1016/j.cell.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Higuchi R, et al. Actin dynamics affect mitochondrial quality control and aging in budding yeast. Curr Biol. 2013;23:2417–2422. doi: 10.1016/j.cub.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suka N, et al. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- 16.Dang W, et al. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- 18.Jiang H, et al. SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du J, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tan M, et al. Lysine Glutarylation Is a Protein Posttranslational Modification Regulated by SIRT5. Cell Metabolism. 2014;19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng C, et al. The first identification of lysine malonylation substrates and its regulatory enzyme. Mol Cell Proteomics. 2011;10:M111 012658. doi: 10.1074/mcp.M111.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanner KG, et al. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product: 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canto C, et al. Crosstalk between poly(ADP-ribose) polymerase and sirtuin enzymes. Mol Aspects Med. 2013 doi: 10.1016/j.mam.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi: 10.1038/nrm1616. [DOI] [PubMed] [Google Scholar]
- 25.Rahman S, Islam R. Mammalian Sirt1: insights on its biological functions. Cell Commun Signal. 2011;9:11. doi: 10.1186/1478-811X-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feldman JL, et al. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem. 2013;288:31350–31356. doi: 10.1074/jbc.C113.511261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin Q, et al. Cytoplasm-localized SIRT1 enhances apoptosis. Journal of cellular physiology. 2007;213:88–97. doi: 10.1002/jcp.21091. [DOI] [PubMed] [Google Scholar]
- 28.Byles V, et al. Aberrant cytoplasm localization and protein stability of SIRT1 is regulated by PI3K/IGF-1R signaling in human cancer cells. Int J Biol Sci. 2010;6:599–612. doi: 10.7150/ijbs.6.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanno M, et al. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 30.Hisahara S, et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci U S A. 2008;105:15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8:38–48. doi: 10.1016/j.cmet.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong H, et al. Sirt1 mediates neuroprotection from mutant huntingtin by activation of the TORC1 and CREB transcriptional pathway. Nat Med. 2012;18:159–165. doi: 10.1038/nm.2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herranz D, et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satoh A, et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab. 2013;18:416–430. doi: 10.1016/j.cmet.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alcendor RR, et al. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 36.Whitaker R, et al. Increased expression of Drosophila Sir2 extends life span in a dose-dependent manner. Aging (Albany NY) 2013;5:682–691. doi: 10.18632/aging.100599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McBurney MW, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh A, et al. SIRT1 promotes the central adaptive response to diet restriction through activation of the dorsomedial and lateral nuclei of the hypothalamus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:10220–10232. doi: 10.1523/JNEUROSCI.1385-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mercken EM, et al. SIRT1 but not its increased expression is essential for lifespan extension in caloric-restricted mice. Aging Cell. 2014;13:193–196. doi: 10.1111/acel.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boily G, et al. SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene. 2009;28:2882–2893. doi: 10.1038/onc.2009.147. [DOI] [PubMed] [Google Scholar]
- 41.Lombard DB, Zwaans BM. SIRT3: As Simple As It Seems? Gerontology. 2014;60:56–64. doi: 10.1159/000354382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown K, et al. SIRT3 reverses aging-associated degeneration. Cell reports. 2013;3:319–327. doi: 10.1016/j.celrep.2013.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang B, et al. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michishita E, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaidi A, et al. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.Dominy JE, Jr, et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol Cell. 2012;48:900–913. doi: 10.1016/j.molcel.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mao Z, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jedrusik-Bode M, et al. The sirtuin SIRT6 regulates stress granule formation in C. elegans and mammals. J Cell Sci. 2013;126:5166–5177. doi: 10.1242/jcs.130708. [DOI] [PubMed] [Google Scholar]
- 50.Michishita E, et al. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simeoni F, et al. Proteomic analysis of the SIRT6 interactome: novel links to genome maintenance and cellular stress signaling. Sci Rep. 2013;3:3085. doi: 10.1038/srep03085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beauharnois JM, et al. Sirtuin 6: a review of biological effects and potential therapeutic properties. Mol Biosyst. 2013;9:1789–1806. doi: 10.1039/c3mb00001j. [DOI] [PubMed] [Google Scholar]
- 53.Kanfi Y, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- 54.Rincon M, et al. The paradox of the insulin/IGF-1 signaling pathway in longevity. Mech Ageing Dev. 2004;125:397–403. doi: 10.1016/j.mad.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 55.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 56.Xiao C, et al. SIRT6 deficiency results in severe hypoglycemia by enhancing both basal and insulin-stimulated glucose uptake in mice. J Biol Chem. 2010;285:36776–36784. doi: 10.1074/jbc.M110.168039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sundaresan NR, et al. The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med. 2012;18:1643–1650. doi: 10.1038/nm.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller RA, et al. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci. 2011;66:191–201. doi: 10.1093/gerona/glq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neff F, et al. Rapamycin extends murine lifespan but has limited effects on aging. J Clin Invest. 2013;123:3272–3291. doi: 10.1172/JCI67674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Richardson A. Rapamycin, anti-aging, and avoiding the fate of Tithonus. J Clin Invest. 2013;123:3204–3206. doi: 10.1172/JCI70800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hall JA, et al. The sirtuin family's role in aging and age-associated pathologies. J Clin Invest. 2013;123:973–979. doi: 10.1172/JCI64094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown RC, et al. Neurodegenerative diseases: an overview of environmental risk factors. Environmental health perspectives. 2005;113:1250–1256. doi: 10.1289/ehp.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Herskovits AZ, Guarente L. SIRT1 in Neurodevelopment and Brain Senescence. Neuron. 2014;81:471–483. doi: 10.1016/j.neuron.2014.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donmez G, et al. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 65.Min SW, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen TJ, et al. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat Commun. 2011;2:252. doi: 10.1038/ncomms1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donmez G, et al. SIRT1 protects against alpha-synuclein aggregation by activating molecular chaperones. J Neurosci. 2012;32:124–132. doi: 10.1523/JNEUROSCI.3442-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Pallos J, et al. Inhibition of specific HDACs and sirtuins suppresses pathogenesis in a Drosophila model of Huntington's disease. Hum Mol Genet. 2008;17:3767–3775. doi: 10.1093/hmg/ddn273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parker JA, et al. Integration of beta-catenin, sirtuin, and FOXO signaling protects from mutant huntingtin toxicity. J Neurosci. 2012;32:12630–12640. doi: 10.1523/JNEUROSCI.0277-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jiang M, et al. Neuroprotective role of Sirt1 in mammalian models of Huntington's disease through activation of multiple Sirt1 targets. Nat Med. 2012;18:153–158. doi: 10.1038/nm.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fu J, et al. trans-(-)-epsilon-Viniferin increases mitochondrial sirtuin 3 (SIRT3), activates AMP-activated protein kinase (AMPK), and protects cells in models of Huntington Disease. J Biol Chem. 2012;287:24460–24472. doi: 10.1074/jbc.M112.382226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song W, et al. Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nat Med. 2011;17:377–382. doi: 10.1038/nm.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fukagawa NK, et al. Effect of age on body composition and resting metabolic rate. The American journal of physiology. 1990;259:E233–E238. doi: 10.1152/ajpendo.1990.259.2.E233. [DOI] [PubMed] [Google Scholar]
- 74.St-Onge MP, Gallagher D. Body composition changes with aging: the cause or the result of alterations in metabolic rate and macronutrient oxidation? Nutrition. 2010;26:152–155. doi: 10.1016/j.nut.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Basen-Engquist K, Chang M. Obesity and cancer risk: recent review and evidence. Curr Oncol Rep. 2011;13:71–76. doi: 10.1007/s11912-010-0139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morley JE. The metabolic syndrome and aging. J Gerontol A Biol Sci Med Sci. 2004;59:139–142. doi: 10.1093/gerona/59.2.m139. [DOI] [PubMed] [Google Scholar]
- 77.Polednak AP. Estimating the number of U.S. incident cancers attributable to obesity and the impact on temporal trends in incidence rates for obesity-related cancers. Cancer detection and prevention. 2008;32:190–199. doi: 10.1016/j.cdp.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 78.Roberts DL, et al. Biological mechanisms linking obesity and cancer risk: new perspectives. Annual review of medicine. 2010;61:301–316. doi: 10.1146/annurev.med.080708.082713. [DOI] [PubMed] [Google Scholar]
- 79.Li X. SIRT1 and energy metabolism. Acta Biochim Biophys Sin (Shanghai) 2013;45:51–60. doi: 10.1093/abbs/gms108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2013 doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Purushotham A, et al. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang RH, et al. Liver steatosis and increased ChREBP expression in mice carrying a liver specific SIRT1 null mutation under a normal feeding condition. Int J Biol Sci. 2010;6:682–690. doi: 10.7150/ijbs.6.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cakir I, et al. Hypothalamic Sirt1 regulates food intake in a rodent model system. PLoS One. 2009;4:e8322. doi: 10.1371/journal.pone.0008322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramadori G, et al. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metab. 2010;12:78–87. doi: 10.1016/j.cmet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gerhart-Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gomes AP, et al. Declining NAD(+) Induces a Pseudohypoxic State Disrupting Nuclear-Mitochondrial Communication during Aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qiang L, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ramsey KM, et al. Age-associated loss of Sirt1-mediated enhancement of glucose-stimulated insulin secretion in beta cell-specific Sirt1-overexpressing (BESTO) mice. Aging Cell. 2008;7:78–88. doi: 10.1111/j.1474-9726.2007.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Moynihan KA, et al. Increased dosage of mammalian Sir2 in pancreatic beta cells enhances glucose-stimulated insulin secretion in mice. Cell Metab. 2005;2:105–117. doi: 10.1016/j.cmet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 91.Yoshino J, et al. Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 2011;14:528–536. doi: 10.1016/j.cmet.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hirschey MD, et al. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–125. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fan J, et al. Tyr Phosphorylation of PDP1 Toggles Recruitment between ACAT1 and SIRT3 to Regulate the Pyruvate Dehydrogenase Complex. Mol Cell. 2014;53:534–548. doi: 10.1016/j.molcel.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shimazu T, et al. SIRT3 deacetylates mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2 and regulates ketone body production. Cell Metab. 2010;12:654–661. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jing E, et al. Sirtuin-3 (Sirt3) regulates skeletal muscle metabolism and insulin signaling via altered mitochondrial oxidation and reactive oxygen species production. Proc Natl Acad Sci U S A. 2011;108:14608–14613. doi: 10.1073/pnas.1111308108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hirschey MD, et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Molecular cell. 2011;44:177–190. doi: 10.1016/j.molcel.2011.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fernandez-Marcos PJ, et al. Muscle or liver-specific Sirt3 deficiency induces hyperacetylation of mitochondrial proteins without affecting global metabolic homeostasis. Scientific reports. 2012;2:425. doi: 10.1038/srep00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhong L, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kim HS, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Elhanati S, et al. Multiple Regulatory Layers of SREBP1/2 by SIRT6. Cell Reports. 2013 doi: 10.1016/j.celrep.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 101.Tao R, et al. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. Journal of lipid research. 2013;54:2745–2753. doi: 10.1194/jlr.M039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanfi Y, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 103.Barbaresko J, et al. Dietary pattern analysis and biomarkers of low-grade inflammation: a systematic literature review. Nutr Rev. 2013;71:511–527. doi: 10.1111/nure.12035. [DOI] [PubMed] [Google Scholar]
- 104.Tilstra JS, et al. NF-kappaB in Aging and Disease. Aging and disease. 2011;2:449–465. [PMC free article] [PubMed] [Google Scholar]
- 105.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jung KJ, et al. Effect of short term calorie restriction on pro-inflammatory NF-kB and AP-1 in aged rat kidney. Inflammation research : official journal of the European Histamine Research Society ... [et al.] 2009;58:143–150. doi: 10.1007/s00011-008-7227-2. [DOI] [PubMed] [Google Scholar]
- 108.Gabay O, Gabay C. Hand osteoarthritis: new insights. Joint, bone, spine : revue du rhumatisme. 2013;80:130–134. doi: 10.1016/j.jbspin.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 109.Biason-Lauber A, et al. Identification of a SIRT1 mutation in a family with type 1 diabetes. Cell Metab. 2013;17:448–455. doi: 10.1016/j.cmet.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 111.Yuan H, et al. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. OncoTargets and therapy. 2013;6:1399–1416. doi: 10.2147/OTT.S37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yuan J, et al. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lim JH, et al. Sirtuin 1 modulates cellular responses to hypoxia by deacetylating hypoxia-inducible factor 1alpha. Mol Cell. 2010;38:864–878. doi: 10.1016/j.molcel.2010.05.023. [DOI] [PubMed] [Google Scholar]
- 114.Srisuttee R, et al. Hepatitis B virus X (HBX) protein upregulates beta-catenin in a human hepatic cell line by sequestering SIRT1 deacetylase. Oncol Rep. 2012;28:276–282. doi: 10.3892/or.2012.1798. [DOI] [PubMed] [Google Scholar]
- 115.Firestein R, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Leko V, et al. Enterocyte-specific inactivation of SIRT1 reduces tumor load in the APC(+/min) mouse model. PLoS One. 2013;8:e66283. doi: 10.1371/journal.pone.0066283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Laemmle A, et al. Inhibition of SIRT1 impairs the accumulation and transcriptional activity of HIF-1alpha protein under hypoxic conditions. PLoS One. 2012;7:e33433. doi: 10.1371/journal.pone.0033433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Menssen A, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci U S A. 2012;10:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mao B, et al. Sirt1 deacetylates c-Myc and promotes c-Myc/Max association. Int J Biochem Cell Biol. 2011;43:1573–1581. doi: 10.1016/j.biocel.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 120.Finley LW, et al. SIRT3 Opposes Reprogramming of Cancer Cell Metabolism through HIF1alpha Destabilization. Cancer Cell. 2011;19:416–428. doi: 10.1016/j.ccr.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim HS, et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liou GY, Storz P. Reactive oxygen species in cancer. Free radical research. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Finley LW, Haigis MC. Metabolic regulation by SIRT3: implications for tumorigenesis. Trends Mol Med. 2012;18:516–523. doi: 10.1016/j.molmed.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bell EL, et al. SirT3 suppresses hypoxia inducible factor 1alpha and tumor growth by inhibiting mitochondrial ROS production. Oncogene. 2011;30:2986–2996. doi: 10.1038/onc.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kaplon J, et al. A key role for mitochondrial gatekeeper pyruvate dehydrogenase in oncogene-induced senescence. Nature. 2013;498:109–112. doi: 10.1038/nature12154. [DOI] [PubMed] [Google Scholar]
- 126.Li S, et al. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS One. 2010;5:e10486. doi: 10.1371/journal.pone.0010486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aury-Landas J, et al. Germline copy number variation of genes involved in chromatin remodelling in families suggestive of Li-Fraumeni syndrome with brain tumours. Eur J Hum Genet. 2013 doi: 10.1038/ejhg.2013.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ashraf N, et al. Altered sirtuin expression is associated with node-positive breast cancer. British journal of cancer. 2006;95:1056–1061. doi: 10.1038/sj.bjc.6603384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Alhazzazi TY, et al. Sirtuin-3 (SIRT3), a novel potential therapeutic target for oral cancer. Cancer. 2011;117:1670–1678. doi: 10.1002/cncr.25676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khongkow M, et al. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis. 2013;34:1476–1486. doi: 10.1093/carcin/bgt098. [DOI] [PubMed] [Google Scholar]
- 131.Liu Y, et al. Inhibition of SIRT6 in prostate cancer reduces cell viability and increases sensitivity to chemotherapeutics. Protein & cell. 2013 doi: 10.1007/s13238-013-3054-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sebastian C, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Min L, et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol. 2012;14:1203–1211. doi: 10.1038/ncb2590. [DOI] [PubMed] [Google Scholar]
- 134.Van Meter M, et al. SIRT6 overexpression induces massive apoptosis in cancer cells but not in normal cells. Cell Cycle. 2011;10:3153–3158. doi: 10.4161/cc.10.18.17435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jeong Seung M, et al. SIRT4 Has Tumor-Suppressive Activity and Regulates the Cellular Metabolic Response to DNA Damage by Inhibiting Mitochondrial Glutamine Metabolism. Cancer Cell. 2013;23:450–463. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Csibi A, et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jeong SM, et al. SIRT4 protein suppresses tumor formation in genetic models of Myc-induced B cell lymphoma. J Biol Chem. 2014;289:4135–4144. doi: 10.1074/jbc.M113.525949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Barber MF, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Go AS, et al. Heart Disease and Stroke Statistics--2014 Update: A Report From the American Heart Association. Circulation. 2013 doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lakatta EG, Levy D. Arterial and cardiac aging: major shareholders in cardiovascular disease enterprises: Part II: the aging heart in health: links to heart disease. Circulation. 2003;107:346–354. doi: 10.1161/01.cir.0000048893.62841.f7. [DOI] [PubMed] [Google Scholar]
- 141.Mattagajasingh I, et al. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc Natl Acad Sci U S A. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Csiszar A, et al. Vasoprotective effects of resveratrol and SIRT1: attenuation of cigarette smoke-induced oxidative stress and proinflammatory phenotypic alterations. Am J Physiol Heart Circ Physiol. 2008;294:H2721–H2735. doi: 10.1152/ajpheart.00235.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Potente M, Dimmeler S. Emerging roles of SIRT1 in vascular endothelial homeostasis. Cell Cycle. 2008;7:2117–2122. doi: 10.4161/cc.7.14.6267. [DOI] [PubMed] [Google Scholar]
- 144.Potente M, et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Guarani V, et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473:234–238. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Oellerich MF, Potente M. FOXOs and sirtuins in vascular growth, maintenance, and aging. Circ Res. 2012;110:1238–1251. doi: 10.1161/CIRCRESAHA.111.246488. [DOI] [PubMed] [Google Scholar]
- 147.Paik JH. FOXOs in the maintenance of vascular homoeostasis. Biochem Soc Trans. 2006;34:731–734. doi: 10.1042/BST0340731. [DOI] [PubMed] [Google Scholar]
- 148.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Potente M, et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by notch. Dev Cell. 2009;16:196–208. doi: 10.1016/j.devcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 151.Wen L, et al. Ca2+/calmodulin-dependent protein kinase kinase beta phosphorylation of Sirtuin 1 in endothelium is atheroprotective. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1309354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kolwicz SC, Jr, Tian R. Glucose metabolism and cardiac hypertrophy. Cardiovasc Res. 2011;90:194–201. doi: 10.1093/cvr/cvr071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Planavila A, et al. Sirt1 acts in association with PPARalpha to protect the heart from hypertrophy, metabolic dysregulation, and inflammation. Cardiovasc Res. 2011;90:276–284. doi: 10.1093/cvr/cvq376. [DOI] [PubMed] [Google Scholar]
- 154.Oka S, et al. PPARalpha-Sirt1 complex mediates cardiac hypertrophy and failure through suppression of the ERR transcriptional pathway. Cell Metab. 2011;14:598–611. doi: 10.1016/j.cmet.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sundaresan NR, et al. The deacetylase SIRT1 promotes membrane localization and activation of Akt and PDK1 during tumorigenesis and cardiac hypertrophy. Sci Signal. 2011;4:ra46. doi: 10.1126/scisignal.2001465. [DOI] [PubMed] [Google Scholar]
- 156.Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Condorelli G, et al. Akt induces enhanced myocardial contractility and cell size in vivo in transgenic mice. Proc Natl Acad Sci U S A. 2002;99:12333–12338. doi: 10.1073/pnas.172376399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Shiojima I, et al. Akt signaling mediates postnatal heart growth in response to insulin and nutritional status. J Biol Chem. 2002;277:37670–37677. doi: 10.1074/jbc.M204572200. [DOI] [PubMed] [Google Scholar]
- 159.Sundaresan NR, et al. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–2771. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Pillai VB, et al. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem. 2010;285:3133–3144. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mouchiroud L, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Braidy N, et al. Age related changes in NAD+ metabolism oxidative stress and Sirt1 activity in wistar rats. PLoS One. 2011;6:e19194. doi: 10.1371/journal.pone.0019194. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 163.Braidy N, et al. Mapping NAD(+) metabolism in the brain of ageing Wistar rats: potential targets for influencing brain senescence. Biogerontology. 2014;15:177–198. doi: 10.1007/s10522-013-9489-5. [DOI] [PubMed] [Google Scholar]
- 164.Massudi H, et al. Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS One. 2012;7:e42357. doi: 10.1371/journal.pone.0042357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Canto C, et al. The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab. 2012;15:838–847. doi: 10.1016/j.cmet.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Barbosa MT, et al. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J. 2007;21:3629–3639. doi: 10.1096/fj.07-8290com. [DOI] [PubMed] [Google Scholar]
- 167.Sharma A, et al. The role of SIRT6 protein in aging and reprogramming of human induced pluripotent stem cells. J Biol Chem. 2013;288:18439–18447. doi: 10.1074/jbc.M112.405928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kennedy BK, Pennypacker JK. Drugs that modulate aging: the promising yet difficult path ahead. Transl Res. 2013 doi: 10.1016/j.trsl.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kim HJ, et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. Journal of proteome research. 2011;10:722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- 170.Bai P, Canto C. The role of PARP-1 and PARP-2 enzymes in metabolic regulation and disease. Cell Metab. 2012;16:290–295. doi: 10.1016/j.cmet.2012.06.016. [DOI] [PubMed] [Google Scholar]
- 171.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 172.Tissenbaum HA, Guarente L. Model organisms as a guide to mammalian aging. Dev Cell. 2002;2:9–19. doi: 10.1016/s1534-5807(01)00098-3. [DOI] [PubMed] [Google Scholar]
- 173.Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–E2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- 174.Rizki G, et al. The evolutionarily conserved longevity determinants HCF-1 and SIR-2.1/SIRT1 collaborate to regulate DAF-16/FOXO. PLoS Genet. 2011;7:e1002235. doi: 10.1371/journal.pgen.1002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Berdichevsky A, et al. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 176.Ludewig AH, et al. Pheromone sensing regulates Caenorhabditis elegans lifespan and stress resistance via the deacetylase SIR-2.1. Proc Natl Acad Sci U S A. 2013;110:5522–5527. doi: 10.1073/pnas.1214467110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Banerjee KK, et al. dSir2 in the adult fat body, but not in muscles, regulates life span in a diet-dependent manner. Cell Rep. 2012;2:1485–1491. doi: 10.1016/j.celrep.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 179.Hoffmann J, et al. Overexpression of Sir2 in the adult fat body is sufficient to extend lifespan of male and female Drosophila. Aging (Albany NY) 2013;5:315–327. doi: 10.18632/aging.100553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Burnett C, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rose G, et al. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–1070. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- 182.Bellizzi D, et al. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–263. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 183.Lescai F, et al. Human longevity and 11p15.5: a study in 1321 centenarians. Eur J Hum Genet. 2009;17:1515–1519. doi: 10.1038/ejhg.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gallucci M, et al. The Treviso Longeva (Trelong) study: a biomedical, demographic, economic and social investigation on people 70 years and over in a typical town of North-East of Italy. Archives of gerontology and geriatrics. 2007;44(Suppl 1):173–192. doi: 10.1016/j.archger.2007.01.025. [DOI] [PubMed] [Google Scholar]
- 185.Halaschek-Wiener J, et al. Genetic variation in healthy oldest-old. PLoS One. 2009;4:e6641. doi: 10.1371/journal.pone.0006641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Albani D, et al. Modulation of human longevity by SIRT3 single nucleotide polymorphisms in the prospective study "Treviso Longeva (TRELONG)". Age (Dordr) 2014;36:469–478. doi: 10.1007/s11357-013-9559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Speakman JR, Mitchell SE. Caloric restriction. Mol Aspects Med. 2011;32:159–221. doi: 10.1016/j.mam.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 188.Guarente L. Calorie restriction and sirtuins revisited. Genes Dev. 2013;27:2072–2085. doi: 10.1101/gad.227439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lamming DW, et al. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- 190.Lin SJ, et al. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 191.Kaeberlein M, Powers RW., 3rd Sir2 and calorie restriction in yeast: a skeptical perspective. Ageing Res Rev. 2007;6:128–140. doi: 10.1016/j.arr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 192.Longo VD, et al. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Hansen M, et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 194.Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 195.Mair W, et al. Optimizing dietary restriction for genetic epistasis analysis and gene discovery in C. elegans. PLoS ONE. 2009;4:e4535. doi: 10.1371/journal.pone.0004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lee GD, et al. Dietary deprivation extends lifespan in Caenorhabditis elegans. Aging Cell. 2006;5:515–524. doi: 10.1111/j.1474-9726.2006.00241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kaeberlein TL, et al. Lifespan extension in Caenorhabditis elegans by complete removal of food. Aging Cell. 2006;5:487–494. doi: 10.1111/j.1474-9726.2006.00238.x. [DOI] [PubMed] [Google Scholar]
- 198.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bauer JH, et al. dSir2 and Dmp53 interact to mediate aspects of CR-dependent lifespan extension in D. melanogaster. Aging (Albany NY) 2009;1:38–48. doi: 10.18632/aging.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chen D, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Schenk S, et al. Sirt1 enhances skeletal muscle insulin sensitivity in mice during caloric restriction. The Journal of clinical investigation. 2011;121:4281–4288. doi: 10.1172/JCI58554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cohen DE, et al. Neuronal SIRT1 regulates endocrine and behavioral responses to calorie restriction. Genes Dev. 2009;23:2812–2817. doi: 10.1101/gad.1839209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lu M, et al. Neuronal Sirt1 deficiency increases insulin sensitivity in both brain and peripheral tissues. The Journal of biological chemistry. 2013;288:10722–10735. doi: 10.1074/jbc.M112.443606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kume S, et al. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. The Journal of clinical investigation. 2010;120:1043–1055. doi: 10.1172/JCI41376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Palacios OM, et al. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging (Albany NY) 2009;1:771–783. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Shi T, et al. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–13567. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- 208.Schwer B, et al. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–606. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hallows WC, et al. Sirt3 promotes the urea cycle and fatty acid oxidation during dietary restriction. Mol Cell. 2011;41:139–149. doi: 10.1016/j.molcel.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Someya S, et al. Sirt3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–812. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Nakagawa T, et al. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–570. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Qiu X, et al. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–667. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 213.Tao R, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hebert AS, et al. Calorie Restriction and SIRT3 Trigger Global Reprogramming of the Mitochondrial Protein Acetylome. Molecular cell. 2012 doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Baur JA. Resveratrol, sirtuins, and the promise of a DR mimetic. Mech Ageing Dev. 2010;131:261–269. doi: 10.1016/j.mad.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Baur JA, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Pearson KJ, et al. Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab. 2008;8:157–168. doi: 10.1016/j.cmet.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Pacholec M, et al. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J Biol Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Price NL, et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab. 2012;15:675–690. doi: 10.1016/j.cmet.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Chachay VS, et al. Resveratrol Does Not Benefit Patients with Non-alcoholic Fatty Liver Disease. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2014 doi: 10.1016/j.cgh.2014.02.024. [DOI] [PubMed] [Google Scholar]
- 221.Minor RK, et al. SRT1720 improves survival and healthspan of obese mice. Sci Rep. 2011;1:70. doi: 10.1038/srep00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Mitchell SJ, et al. The SIRT1 Activator SRT1720 Extends Lifespan and Improves Health of Mice Fed a Standard Diet. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hubbard BP, et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]