Abstract
The methyltransferase, M.EcoKI, recognizes the DNA sequence 5′-AACNNNNNNGTGC-3′ and methylates adenine at the underlined positions. DNA methylation has been shown by crystallography to occur via a base flipping mechanism and is believed to be a general mechanism for all methyltransferases. If no structure is available, the fluorescence of 2-aminopurine is often used as a signal for base flipping as it shows enhanced fluorescence when its environment is perturbed. We find that 2-aminopurine gives enhanced fluorescence emission not only when it is placed at the M.EcoKI methylation sites but also at a location adjacent to the target adenine. Thus it appears that 2-aminopurine fluorescence intensity is not a clear indicator of base flipping but is a more general measure of DNA distortion. Upon addition of the cofactor S-adenosyl-methionine to the M.EcoKI:DNA complex, the 2-aminopurine fluorescence changes to that of a new species showing excitation at 345 nm and emission at 450 nm. This change requires a fully active enzyme, the correct cofactor and the 2-aminopurine located at the methylation site. However, the new fluorescent species is not a covalently modified form of 2-aminopurine and we suggest that it represents a hitherto undetected physicochemical form of 2-aminopurine.
INTRODUCTION
2-Aminopurine (2-AP), an analogue of adenine, emits fluorescence when excited with radiation between 310 and 320 nm (1–4), a region which largely avoids absorbance by proteins and DNA and excitation of protein fluorescence. 2-AP has a high quantum yield in aqueous solution but the fluorescence is highly quenched when it is incorporated into DNA (1,4). The quenching mechanisms depend upon the surrounding bases and involve electron transfer to nearby guanine and adenine, base stacking, hydrogen bonding to other bases and collisional quenching with neighbouring bases due to DNA dynamics (4–9). Electron transfer and base stacking (6,8,9) are the most quantitative mechanisms proposed so far to explain the quenching of 2-AP in DNA and we believe these are the dominant mechanisms at work (unpublished results). 2-AP can base pair with thymine and the stability of this 2-AP:T base pair is only slightly lower than the normal A:T base pair (5,10–12). Perturbations of the 2-AP within a DNA structure can change its fluorescence behaviour. Fluorescence intensity changes caused by the binding of a protein to DNA is indicative of a change in the environment of the 2-AP. This sensitivity means that 2-AP has frequently been incorporated into DNA to study interactions with proteins such as polymerases, helicases, methyltransferases and repair enzymes (13–30). Fluorescence enhancement of 2-AP located at specific sites within DNA has been caused by several DNA methyltransferases and repair enzymes (13,14,16–25,27,28). These enzymes use or are postulated to use a nucleotide base flipping mechanism (31,32). In this mechanism, the specific nucleotide base targeted for either methylation or repair is rotated by approximately 180° around the phosphate backbone of the DNA strand into an extrahelical position which places the base in the catalytic site of the enzyme. The base flipping mechanism suggests that if 2-AP were located at the appropriate position, its fluorescence would be dramatically enhanced as it was swung out of the strongly-quenched DNA double helical environment into the enzyme catalytic site. Such enhancements in fluorescence emission intensity have been observed for enzymes known to use base flipping from the availability of crystal structures such as HhaI and TaqI methyltransferases and Uracil glycosylase (33–36). Fluores cence enhancements have been observed for 2-AP placed at methylation sites in DNA sequences recognized by other methyltransferases. These enzymes, although structures are either not available or are not bound specifically to their DNA target, are also highly likely to use base flipping (13,18,22,24). However, some methyltransferases which almost certainly use the base-flipping mechanism have induced different fluorescent behaviour of 2-AP (21,23,28). M.EcoRV gave no enhancement of 2-AP fluorescence when it was incorporated at the base flipping site whilst both M.EcoRV and M.EcoP15I induced enhanced 2-AP fluorescence when the 2-AP was placed at a location other than the base flipping site. Thus it appears, as one may have expected from many studies predating the discovery of base flipping, that 2-AP is not only a probe for base flipping but also probes other aspects of DNA structure and dynamics that can also be altered by protein binding.
In this paper, we present results obtained using the EcoKI DNA methyltransferase, M.EcoKI, binding to DNA containing 2-AP at various locations including the methylation sites proposed to undergo base flipping. M.EcoKI is a part of the EcoKI type I DNA restriction and modification system and as such recognizes a bipartite DNA target sequence, 5′-AACNNNNNNGTGC-3′ with methylation occurring at the underlined adenine and at the adenine complementary to the underlined thymine (37–39). The enzyme comprises three subunits, one HsdS specificity subunit for recognizing the DNA sequence and two HsdM modification subunits for determining the methylation state of the target and carrying out methyl group transfer from the cofactor S-adenosyl methionine (AdoMet) to the adenine. The enzyme displays a very strong preference for methylating target sequences which already contain one methyl group on either adenine, i.e. for methylating hemi-methylated DNA. The methylation of unmodified targets is extremely slow as these substrates are the preferred target for restriction by the R.EcoKI endonuclease. The ability to recognize the methylation state of the target lies within the HsdM subunits as mutations have been isolated which reduce this ability and allow effective methylation of unmodified DNA (40). Each HsdM subunit contains a catalytic domain which has been successfully modelled upon the known structures of the HhaI and TaqI type II DNA methyltransferases (41). Mutations which reduce catalytic activity have been identified (42) and support the structural correlation between type I methyltransferases and the type II methyltransferases. Thus it is highly probable that type I methyltransferases use a base flipping mechanism during catalysis and experimental support for this, obtained via DNA footprinting of the EcoR124I methyltransferase, is available (43,44). The use of the spectroscopic properties of 2-AP to investigate the type I methyltransferases has not hitherto been investigated. In this paper, we have found that 2-AP fluorescence increases both at the base targeted for methylation but also at an adjacent base. This suggests, as found by others, that 2-AP is not a definitive probe for base flipping. The addition of the AdoMet cofactor causes a further novel change in the fluorescence of 2-AP located at the methylation site indicative of the formation of a new fluorescent species. In this paper, we describe our efforts to identify the cause of this novel fluorescence and we conclude that it is due to some physicochemical affect on 2-AP rather than a covalent modification.
MATERIALS AND METHODS
M.EcoKI and mutant versions were prepared as previously described (45,46). These methyltransferase preparations do not contain bound AdoMet. DNA oligodeoxynucleotides were synthesized using standard phosphoramidite chemistry and purified by reversed-phase HPLC. Annealing of duplexes was performed by mixing appropriate amounts of each oligodeoxynucleotide, heating to 95°C and allowing the sample to cool slowly over several hours to room temperature. A 2-fold excess of oligodeoxynucleotide lacking 2-AP was annealed to oligodeoxynucleotides containing 2-AP to ensure that there was no single-stranded DNA containing 2-AP left free in solution. The concentration of 2-AP labelled strands was 2 µM after annealing. The oligodeoxynucleotide sequences are given in Table 1. AdoMet was from New England Biolabs (32 mM stock solution) and [3H-methyl]AdoMet (2.74 TBq/mmol, 13.5 µM stock) was from Amersham Pharmacia Biotech. The buffer used in all experiments was 20 mM Tris–HCl, 50 mM NaCl, 0.1 mM EDTA, 6 mM MgCl2 buffer, pH 8 at 25°C.
Table 1. The DNA duplexes used to investigate M.EcoKI.
| Duplex number | DNA duplexa | Fluorescence emission maxima (nm) and intensity increase | ||
|---|---|---|---|---|
| DNA alone | DNA + M.EcoKI | DNA + M.EcoKI + AdoMet | ||
| 1 | 5′(N)nA(AmP)C(N)6GTGC(N)n3′ | 380 | 370 (10-fold) | 430 |
| 3′(N)nT T G(N)6CACG(N)n5′ | ||||
| 2 | 5′(N)nAAC(N)6G T GC(N)n3′ | 380 | 370 (10-fold) | 370b |
| 3′(N)nTTG(N)6C(AmP)CG(N)n5′ | ||||
| 3 | 5′(N)nA(AmP)C(N)6G T GC(N)n3′ | 380 | 365 | 365b |
| 3′(N)nT T G(N)6C(AmP)CG(N)n5′ | ||||
| 4 | 5′(N)nA(AmP)C(N)6G T GC(N)n3′ | 380 | 370 (11-fold) | 430 |
| 3′(N)nT T G(N)6C(MeA)CG(N)n5′ | ||||
| 5 | 5′(N)n(AmP)AC(N)6GTGC(N)n3′ | 380 | 370 (12-fold) | 365 |
| 3′(N)nT TG(N)6CACG(N)n5′ | ||||
aAll oligodeoxynucleotides are based on the ‘parent’ duplex (M.EcoKI recognition site in bold, bases that are the target for CH3 group addition underlined): 5′ CACGGGCCTAACGATATCGTGCGTACGAGC 3′; 3′ GTGCCCGGATTGCTATAGCACGCATGCTCG 5′. Only the bases that comprise the M.EcoKI recognition sequence are shown in full in the table, other bases being designated by the letter N. AmP is 2-AP, MeA is 6-methyl-adenine. Underlined bases are at the position of methyl group addition. The parent duplex was used in either the fully un-methylated form or a hemi-methylated form with the lower oligodeoxynucleotide methylated.
bThe 2-AP* spectrum was a weak shoulder on the side of the normal 2-AP spectrum for these duplexes.
Fluorescence spectra were recorded using an Edinburgh Instruments FS900 T-geometry single photon counting fluorimeter with magic angle polarization and with samples thermostatted at 25°C. Excitation and emission bandwidths were 6 nm and the sample cuvette excitation and emission pathlengths were 3 mm. Samples used contained 2-AP labelled DNA duplexes at a concentration of 1 µM in 2-AP to which small volumes of concentrated M.EcoKI (60 µM stock) and AdoMet (32 mM stock) were added to final concentrations of 1.92 and 307 µM, respectively. Weak baseline fluorescence and Raman scattering from the buffer was subtracted from the excitation and emission spectra.
A Gilson HPLC equipped with absorption detection at 254 nm and fluorescence emission detection was used for analysis of DNA. The reverse phase column (C18 Jones Chromatography) was thermostatted at 65°C. Two sets of fluorescence excitation/emission wavelengths, 310/380 nm and 350/450 nm, and absorption at 254 nm were used to monitor the elution of the samples. A linear acetonitrile gradient was generated by mixing 0.1 M acetic acid, 5% acetonitrile and 0.1 M acetic acid, 65% acetonitrile. The pH of the two solvents was adjusted to 6.5 with triethylamine.
Methyltransferase activity was measured by the transfer of tritiated methyl groups from [3H-methyl]AdoMet to DNA oligodeoxynucleotide duplexes labelled on one strand at the 5′ end with biotin (Table 1). These duplexes were used in assays based upon the method of Roth and Jeltsch (47) as modified by O’Neill et al. (48). Sixty microlitre samples containing 1 µM of biotin-labelled oligodeoxynucleotide annealed to a complementary strand (2 µM concentration) and 2 µM [3H]AdoMet in 20 mM Tris, 50 mM NaCl, 0.1 mM EDTA, pH 8 were prepared. Then 2 µl of M.EcoKI was added to give an enzyme concentration of 2 µM. Samples were incubated for 30 min at 25°C. For duplexes containing 2-AP, fluorescence spectra were recorded to confirm the fluorescence enhancement of 2-AP by M.EcoKI and the formation of M-AP upon addition of [3H-methyl]AdoMet. After the 30 min incubation, non-radioactive AdoMet and NaCl were added to concentrations of 267 µM and 0.2 M, respectively to stop further incorporation of 3H-methyl groups into the DNA. Biotinylated DNA was then bound to streptavidin-coated microtitre plates (Cat. no. 95029273, Labsystems) and washed to remove all molecules except the DNA. Scintillation fluid (Microscint™ PS, Packard) was added and the amount of radioactivity bound to the plates determined by scintillation counting in a scintillation counter (Top-Count Microplate Scintillation Counter, Canberra-Packard).
RESULTS
Steady-state spectra of 2-AP substituted DNA and DNA-M.EcoKI complexes
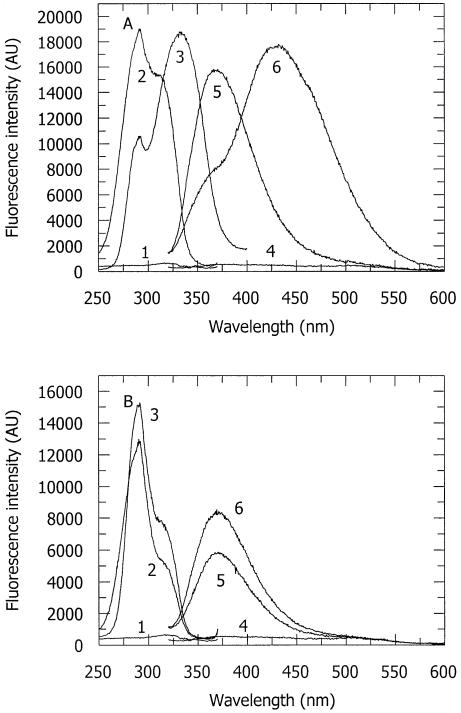
A set of 2-AP-containing oligodeoxynucleotides, derived from a ‘parent’ duplex that contains an unmodified M.EcoKI recognition site (Table 1), have been synthesized and used to probe the interaction of the methyltransferase with DNA. The steady-state fluorescence excitation and emission spectra of a duplex, containing a single 2-AP at the target site for methylation (Table 1, duplex 1) are shown in Figure 1A (spectra 1 and 4). A broad fluorescence emission was observed around 380 nm with excitation at 310 nm. On addition of M.EcoKI, the fluorescence emission intensity at 380 nm increased by nearly 10-fold, and the emission peak was blue shifted by about 10 nm (Fig. 1A, spectrum 5). The large fluorescence enhancement of 2-AP after the addition of M.EcoKI suggests that the 2-AP, which is located at the methylation target position, was exposed to an environment different from the base stacking in the DNA double helix. Furthermore, the blue shift of the emission peak indicates that the 2-AP was placed in an environment with a lower dielectric constant than inside the helix. The excitation spectrum (Fig. 1A, spectrum 2) shows enhanced excitation of 2-AP around 310 nm and excitation of both 2-AP and protein fluorescence with a maximum around 280 nm as expected. The addition of AdoMet to the M.EcoKI–DNA complex gives rise, after a 15 min incubation, to the unusual spectra, numbers 3 and 6, in Figure 1A, a phenomenon further elaborated below. (Note that the drop in excitation intensity at 280 nm is due to the loss of the contribution from 2-AP as it is converted into the excitation peak around 330 nm.) The fluorescence spectra of a number of related duplexes, all containing 2-AP at the methylation target site, were essentially identical (spectra not shown, results summarized in Table 1). As shown in the table, the oligodeoxynucleotides comprised: duplex 2 (2-AP in ‘bottom’ strand), duplex 3 (2-AP in both strands) and duplex 4 (2-AP purine in ‘top’ strand, 6-methyl-adenine in ‘bottom’ strand).
Figure 1.
Fluorescence excitation and emission spectra of 2-AP incorporated in various DNA duplexes. Curves 1, 2 and 3 are excitation spectra and 4, 5 and 6 are emission spectra. (A) Spectra of 2-AP in duplex 1 (i.e. having 2-AP at the position of CH3 addition, Table 1). Spectra 1 and 4, DNA duplex 1 only; spectra 2 and 5, DNA duplex 1 with M.EcoKI; spectra 3 and 6, complex of DNA duplex 1 with M.EcoKI and AdoMet taken after a 15 min incubation period with AdoMet. In these spectra, the weak buffer background and Raman scattering have been subtracted. (B) Spectra of 2-AP in duplex 5 (i.e. having 2-AP adjacent to the position of CH3 addition, Table 1). Spectra 1 and 4, DNA duplex 5 only; spectra 2 and 5, DNA duplex 5 with M.EcoKI; spectra 3 and 6, complex of DNA duplex 5 with M.EcoKI and AdoMet taken after a 15 min incubation period with AdoMet. In these spectra, the weak buffer background and Raman scattering have been subtracted.
Experiments have also been carried out with a duplex containing 2-AP in the position adjacent to the target adenine for methylation (Table 1, duplex 5). As shown in Figure 1B, the addition of M.EcoKI caused a significant increase in the fluorescence intensity of both excitation and emission spectra (spectra 2 and 5 compared to spectra 1 and 4). This is unexpected as the 2-AP in this location is not expected to undergo base flipping upon binding of M.EcoKI. The addition of AdoMet gives rise to a further small enhancement in fluorescence intensity (spectra 3 and 6) and we believe this is due to AdoMet enhancing binding of M.EcoKI to the duplex and shifting the equilibrium between free and bound DNA towards a greater proportion of bound DNA.
Effect of AdoMet on steady-state spectra of 2-AP-containing DNA duplexes
The addition of a saturating amount of the cofactor AdoMet to the DNA–M.EcoKI complex induces a major change in the fluorescence of 2-AP, providing this base is located at the methylation target site (Fig. 1A, Table 1, duplexes 1 to 4). Immediately after addition of AdoMet, the 2-AP fluorescence intensity increases slightly (not shown). This effect is of the same magnitude as observed in Figure 1B for duplex 5 containing 2-AP adjacent to the target site and is attributed to enhanced formation of protein–DNA complexes, as before. However, this enhancement lasts for only a few seconds as the 2-AP fluorescence undergoes a subsequent change over the next few minutes. Figure 1A (spectra 3 and 6) shows the dramatic alteration in both excitation and emission spectra produced on incubating the 2-AP duplex/M.EcoKI complex (Table 1, duplex 1) with AdoMet for 15 min. Spectral changes (not shown) are observed for other duplexes containing 2-AP at the methylation position (Table 1, duplexes 2, 3 and 4). Duplex 4 gave spectra identical to those obtained with duplex 1 whereas the spectral change was much weaker for duplexes 2 and 3. In all cases, the fluorescence emission maximum at 370 nm, generated by the 2-AP in the binary complex, decreases and a new emission peak is formed to a greater or lesser extent at 430 nm. The excitation spectrum shows a peak at 280 nm (due to protein fluorescence) but the shoulder around 310 nm, arising from 2-AP is replaced completely by a new maximum at 340 nm. Since the excitation spectrum of the 2-AP, after addition of the cofactor, is different from the excitation spectrum in the absence of AdoMet, the 430 nm fluorescence emission must arise from a new ground state species, which we term 2-AP*. Although the appearance of 2-AP* is rapid upon addition of AdoMet to binary complexes, the 2-AP* fluorescence also appears in the absence of cofactor; however, formation is very slow and does not go to completion (data not shown). This may be due to the presence of some AdoMet in the preparation of the enzyme, as it has been found that cofactor can remain tightly bound to other type I methyltransferases (49). Alternatively, the formation of 2-AP* may be an intrinsic property of M.EcoKI, that is only enhanced by the addition of AdoMet.
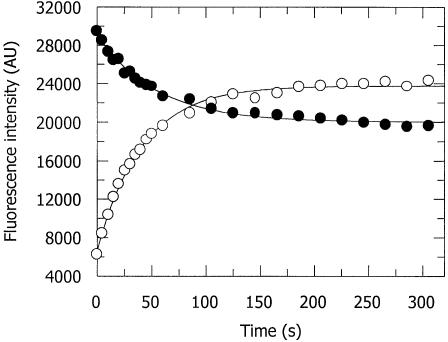
The time course for the formation of the 2-AP*, after the addition of AdoMet to a binary complex formed from M.EcoKI and a duplex containing 2-AP in a hemi-methylated environment (Table 1, duplex 4), is shown in Figure 2. The fluorescence emission of 2-AP at 370 nm decayed gradually, and was accompanied by a corresponding increase in the fluorescence of 2-AP* at 430 nm; the variation in the fluorescence intensities at the two wavelengths ceased within 5 min. The time courses were fitted using a first-order rate equation with a rate constant for the formation of 2-AP* of 0.027 ± 0.003 s–1 and a rate constant of 0.016 ± 0.004 s–1 for the fluorescence intensity decrease of 2-AP. The relatively small difference in these two rate constants may be attributed to the fact that we have not accounted for the initial rapid rise in 2-AP emission immediately after the addition of AdoMet. A rate constant of 0.005 ± 0.0001 s–1 was obtained for the appearance of 2-AP*, upon the addition of AdoMet to a complex of M.EcoKI with 2-AP in an un-methylated context (Table 1, duplex 1; data not shown); a roughly 5-fold slower appearance of 2-AP* than was observed with the hemi-methylated duplex.
Figure 2.
Variation of fluorescence emission intensity as a function of time for duplex 4 (i.e. having 2-AP at the position of CH3 addition and MeA on the complementary oligodeoxynucleotide, Table 1) in the presence of M.EcoKI. The reaction is initiated by the addition of AdoMet and the formation of 2-AP* (open circle, emission at 430 nm) and the decay of 2-AP (filled circle, emission at 370 nm) observed. A first-order rate equation was used to fit the data and the reaction was essentially complete after 5 min although the spectra shown in other figures were obtained after 15 min of reaction.
It is worth recapitulating the effect of AdoMet on the duplex containing 2-AP at a position adjacent to one of the target adenine bases (Table 1, duplex 5). As shown in Figure 1B, no formation of 2-AP* was observed on the addition of cofactor to the complex of this DNA duplex and M.EcoKI.
Requirements for appearance of 2-AP*
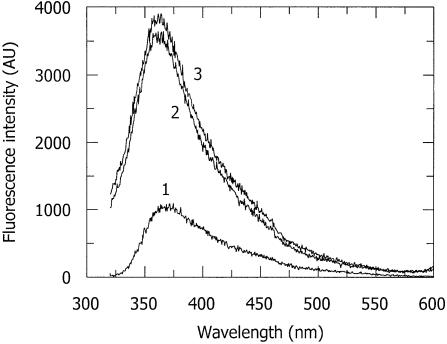
The influence of specific amino acids in M.EcoKI on the formation of 2-AP* was examined by measuring the fluorescence spectra of complexes containing 2-AP (Table 1, duplexes 1 and 4) with various mutated forms of M.EcoKI in the presence of AdoMet. The amino acid changes G177D, N266D, F269G, F269C, F269Y and F269W (located in the M subunit of the enzyme) give rise to catalytic mutants (42), all of which can bind normally to the EcoKI DNA recognition sequence. The alteration G177D abolishes AdoMet binding and activity; proteins containing the substitutions N266D, F269G and F269C bind AdoMet, but completely lack methyltransferase activity. The variants F269Y and F269W are capable of binding AdoMet and retain partial enzymatic activity. A typical example, obtained with the mutant F269Y, of the fluorescence spectra of a bound oligodeoxynucleotide containing 2-AP (duplex 1) in the absence and presence of AdoMet is given in Figure 3. The addition of protein to the 2-AP duplex DNA, enhanced the fluorescence of 2-AP in the same way as observed for the wild-type protein. However, no spectral changes, characteristic of the formation of 2-AP*, were observed following cofactor addition. Similar results were obtained with all the other catalytic mutants, irrespective of whether they were inactive or retained partial activity (data not shown).
Figure 3.
Fluorescence emission spectra (excitation at 310 nm) of 2-AP incorporated in duplex 1 (i.e. having 2-AP at the position of CH3 addition, Table 1) with a mutant of M.EcoKI. Spectrum 1, DNA duplex 1 alone; spectrum 2, DNA duplex 1 with M.EcoKI F269Y mutant; spectrum 3, complex of DNA duplex 1, M.EcoKI F269Y mutant and AdoMet taken after a 15 min incubation period with AdoMet. In these spectra, the buffer background has been subtracted. Although the mutant protein can bind AdoMet and retains some methyltransferase activity, no 2-AP* is formed.
A further series of amino acid substitutions in the M subunit of M.EcoKI, namely L85Q, S144Y, E48K and L113Q, give rise to m* mutants (40). These substitutions produce variants which have lost the preference of wild-type M.EcoKI to add CH3 groups to hemi-methylated DNA recognition sequences rather than unmodified DNA; reaction with both hemi-methylated and un-methylated target sequences occurring effectively. The mutation sites are located in the N-terminal region of M.EcoKI. Addition of any of these mutant proteins to 2-AP-containing duplex (Table 1, duplex 4) enhanced the fluorescence of 2-AP in the same way as observed for the wild-type protein (data not shown). On addition of AdoMet to the complexes of a 2-AP-containing duplex (Table 1, duplex 4) with the m* mutants, the formation of the 2-AP* fluorescence spectra was observed. The amount of 2-AP* produced by the mutants L85Q and S144Y was smaller than observed with the wild-type enzyme and the other m* proteins and gave rise to just a broad shoulder around 430 nm next to the residual 2-AP fluorescence emission (results not shown). The kinetics of this process, although not investigated in detail, was similar to that observed using the wild-type EcoKI methyltransferase.
S-Adenosyl homocysteine (AdoHcy) is the product produced from AdoMet, following methyl group transfer to DNA. The effect of AdoHcy on the DNA–M.EcoKI complexes formed using the oligodeoxynucleotides that constitute duplexes 1 and 4 in Table 1 (i.e. containing 2-AP at the methylation site in either an un- or hemi-methylated context) was investigated. Measurement of both excitation and emission fluorescence spectra gave no sign of the formation of 2-AP* (result not shown).
DNA methylation assay
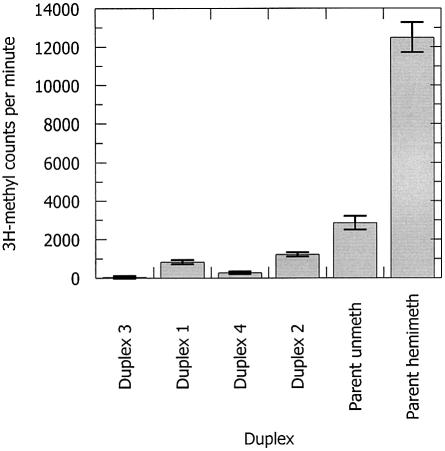
The base 2-AP lacks a 6-amino group, the site of CH3 group addition when M.EcoKI acts on ‘parent’ sequences (Table 1), containing adenine at the target site. Although the catalytic sites of M.EcoKI are considered unlikely to methylate 2-AP, this possibility was formally eliminated using biotin-labelled oligodeoxynucleotide duplexes. The addition of the biotin probe did not affect the conversion of 2-AP into 2-AP* upon addition of the [3H-methyl]AdoMet, as determined by fluorescence spectroscopy, in cases were the 2-AP was located at the methylation target site (duplexes 1 to 4, data not shown). Control experiments using duplexes containing the parent sequence indicated extensive CH3 addition to hemi- methylated duplexes containing adenine and significant methylation of un-methylated duplexes with adenine (Fig. 4, lanes 6 and 5, respectively). The difference in extent of methylation between the two duplexes is expected from the known preference of M.EcoKI for hemi-methylated target sequences. For un-methylated duplexes containing one adenine and one 2-AP at the methylation sites (Fig. 4, lanes 2 and 4, duplexes 1 and 2), the extent of reaction was half that observed for the un-methylated duplex containing two adenines (Fig. 4, lane 5). Critically, no CH3 group addition was observed when both methylation sites were substituted with 2-AP, or when one site contained 2-AP and the other adenine was already methylated (Fig. 4, lanes 1 and 3, duplexes 3 and 4). We conclude that M.EcoKI is unable to add CH3 groups to 2-AP and, therefore, that 2-AP* is not a methylated derivative of 2-AP.
Figure 4.
Tritium assay of methylation of different 2-AP-containing duplexes by the wild-type M.EcoKI showing the amount of radioactivity incorporated into the DNA during a 30 min incubation with [3H-methyl]AdoMet. Lanes 1 to 4 are duplexes 3, 1, 4 and 2, respectively. Lanes 5 and 6 are the parent duplex (Table 1) in fully un-methylated and hemi-methylated forms, respectively.
Attempted HPLC isolation of 2-AP*
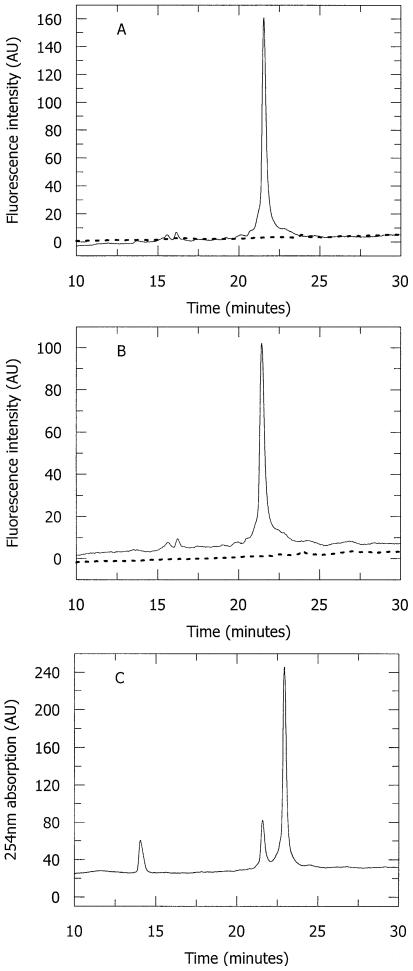
The appearance of a new excitation spectrum associated with 2-AP* requires a new ground state species which may arise from an unsuspected chemical transformation, differing from the previously eliminated methylation reaction. To investigate whether 2-AP* is a stable chemical entity, we attempted its isolation after its formation in a 2-AP-containing duplex (Table 1, duplex 4) by M.EcoKI and AdoMet. Prior to running the chromatograms (which denature M.EcoKI and result in dissociation of DNA and cofactor), the formation of 2-AP* was confirmed by fluorescence spectroscopy (data not shown). Figure 5A shows the elution of the mixture of the duplex, M.EcoKI and AdoMet after formation of 2-AP*, monitored using fluorescence detection at two wavelengths corresponding to excitation/emission of 2-AP (310/380 nm) and 2-AP* (350/450 nm). Only a single peak, eluting at 21 min, is observed with fluorescence properties corresponding to material containing only 2-AP and lacking any 2-AP*. Confirmation that this peak is the starting oligodeoxynucleotide containing 2-AP comes from its co-elution with an authentic standard, on co-injection (Fig. 5B). Repeating the HPLC, but using absorbance at 254 nm for detection (Fig. 5C) reveals three peaks. As before the peak at 21 min corresponds to, and co-elutes with, the starting 2-AP-containing duplex (the peaks at 14 and 23 min are due to AdoMet and the excess of the complementary DNA strand used to prepare the duplex). It is clear that there is no extra peak which might suggest the existence of a DNA strand containing 2-AP*. These results convincingly demonstrate that while 2-AP* can be formed from 2-AP, on incubation of appropriate DNA in the presence of M.EcoKI and AdoMet, this material is only stable when bound to the enzyme. Attempts to isolate free DNA containing 2-AP*, result in its reversion to 2-AP starting material. It appears that 2-AP* does not arise from a chemical conversion of 2-AP and is not a new species which can be isolated by chromatography.
Figure 5.
Attempted isolation of 2-AP* using reversed-phase HPLC chromatography. Duplex 4 (i.e. containing 2-AP at the CH3 target site in a hemi-methylated context) was incubated with M.EcoKI and AdoMet to form 2-AP* and the mixture denatured and analysed by HPLC. (A) HPLC eluate monitored by fluorescence excitation and emission of 310/380 nm (solid line) and 350/450 nm (dashed line). (B) As (A) but with the addition of the starting oligodeoxynucleotide containing 2-AP (i.e. the top strand of duplex 1). The added standard co-elutes with the fluorescent product obtained from the mixture of DNA duplex 4 with M.EcoKI and AdoMet. (C) As (A) but eluate monitored by absorption at 254 nm.
DISCUSSION
The base-flipping mechanism has been proposed as a general means used by methyltransferases for AdoMet-dependent methylation of DNA (31,32) and 2-AP has been broadly used as a fluorescence probe for detection of the existence of base flipping (13,14,16–20,22,25,27). An increase in 2-AP fluorescence intensity induced by the binding of M.EcoKI to DNA containing 2-AP at the methylation site, i.e. duplexes 1–4 in Table 1, has been observed in our study. However, a similar fluorescence increase after the addition of M.EcoKI to DNA with 2-AP in the position adjacent to the target adenine base, i.e. duplex 5 in Table 1, has also been observed. Therefore, these results suggest that, although the base-flipping mechanism may be used by M.EcoKI for DNA methylation, the enhancement in 2-AP fluorescence caused by the binding of M.EcoKI to its recognition sequence is not a definitive signal of base-flipping at the methylation site. This is consistent with the conclusions drawn by Reddy and Rao (23) and Jeltsch and colleagues (21,28) and would be expected given that 2-AP has been used for diverse assays of other DNA binding enzymes. Changes in 2-AP fluorescence emission intensity appear to be only indicative of a general change in the environment of the probe upon perturbation by protein binding.
However, the addition of AdoMet to complexes of M.EcoKI with DNA containing 2-AP at a methylation site produced a most unusual effect. A new and significant fluorescence emission peak appeared at 430 nm and this new peak was maximally excited by irradiation around 345 nm, a region far removed from the normal excitation spectrum of 2-AP. These excitation and emission characteristics indicate that the fluorescence is derived from a new ground electronic state species which we refer to as 2-AP*. The requirements to produce 2-AP* are (i) an active M.EcoKI enzyme with no amino acid substitutions in the two known AdoMet-binding and catalytic motifs (42), (ii) the cofactor substrate AdoMet rather than the cofactor product AdoHcy, and (iii) the 2-AP located at either of the two methylation sites within the recognition sequence. The 2-AP* product appears rapidly when using hemi-methylated DNA with 2-AP on one DNA strand and N6-methyl adenine (Table 1, duplex 4). The rate of appearance is similar to the rates of methylation of hemi-methylated DNA observed in assays of M.EcoKI (45,48). 2-AP* appears more slowly when using un- methylated substrates with 2-AP on one strand and adenine on the other (duplexes 1 and 2, Table 1) but its rate of appearance is much faster than the addition of CH3 groups to un-methylated DNA by M.EcoKI. This would suggest that 2-AP* appearance is actually related to a step in the overall reaction prior to the actual methyl group transfer from AdoMet to the DNA. We have found that the 2-AP* species is not simply a methylated form of 2-AP and indeed does not survive the removal of protein and AdoMet from the DNA duplex nor the separation of the two DNA strands. From our results, we conclude that 2-AP* is formed by the specific three-dimensional arrangement of protein, AdoMet and DNA around the 2-AP itself. This conformation is adopted over the course of several minutes, a time scale similar to the reaction cycle of the enzyme. Despite possessing enzyme activity, the two mutant proteins containing the substitutions F269Y and F269W do not trigger formation of 2-AP* suggesting that the location and chemical character of Phe269 are as important for the formation of 2-AP* as the cofactor AdoMet and the correct location of the 2-AP in the DNA sequence. It is regrettable that a high resolution structure of M.EcoKI is not available and structural models, based on homology with other methyltransferases (41,50), are not accurate enough to define how 2-AP* is formed. However, there is evidence that type I methyltransferases do carry out base flipping in the same manner as structurally characterized methyltransferases (43,44), so we assume that 2-AP* requires base flipping of the 2-AP into the active site of M.EcoKI, where it comes into contact with AdoMet and Phe269 amongst other unspecified interactions. There are no literature reports of a species having the spectral properties of 2-AP* but its formation could arise as a consequence of alterations to hydrogen bonding, protonation and van der Waals interactions, following flipping of 2-AP into the active site. Stacking of aromatic groups on either side of the 2-AP base is likely, as observed for M.TaqI, the only DNA dA methyltransferase for which co-crystal structures are available (36). However, stacking would be expected to have a large quenching effect on fluorescence so it would seem likely, given the large amount of emission intensity observed for 2-AP*, that this is not a major contributor. Hydrogen bonding or pH dependent protonation could occur at the N1, N3 or N7 positions of the flipped 2-AP. Position N1 hydrogen bonds to thymine in duplex DNA in a normal Watson–Crick base pair so it is unlikely that simple hydrogen bonding, to this position, following base flipping is going to cause a dramatic change in fluorescence. Intriguingly, protonation and deprotonation of the base analogue formycin at positions N1 and N6 (equivalent to N7 and N1, respectively in 2-AP) give rise to the formation of fluorescence emission at wavelengths longer than observed for formycin in neutral solution (1) so perhaps when 2-AP is flipped into the M.EcoKI active site a protonation/deprotonation event occurs. One could also consider hydrogen bonding or protonation of the N3 position. In addition, tautomerization of free 2-AP between positions N7 and N9 has been proposed from theoretical studies (51) and, although the N9 position is not available for such a process for 2-AP in DNA, it has also been calculated that the N1 position can also take part in tautomerization with 2-AP existing in the imino form. The calculations indicated that such a form would be very rare but perhaps the environment in the M.EcoKI–DNA–AdoMet complex can influence the nature of the N1 position. Clearly, there is a need for better understanding of the physical chemistry of 2-AP as our HPLC results appear to indicate that 2-AP* is actually 2-AP existing in some unusual state.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge discussions with Professor N. E. Murray (Institute of Cell and Molecular Biology, Edinburgh), Dr A. C. Jones and Mr R. Neely (School of Chemistry, Edinburgh) and assistance in early experiments by Richard Mathison and Malcolm Watson. This work was supported by grant GR/N19748 to D.T.F.D. from the Engineering and Physical Sciences Research Council and a Royal Society University Research Fellowship to D.T.F.D. Research in the laboratory of B.A.C. is supported by the Biotechnology and Biological Sciences Research Council, Wellcome Trust and the European Union.
REFERENCES
- 1.Ward D.C., Reich,E. and Stryer,L. (1969) Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside and their derivatives. J. Biol. Chem., 244, 1228–1237. [PubMed] [Google Scholar]
- 2.Holmen A., Norden,B. and Albinsson,B. (1997) Electronic transition moments of 2-aminopurine. J. Am. Chem. Soc., 119, 3114–3121. [Google Scholar]
- 3.Jean J.M. and Hall,K.B. (2000) Theoretical study of the excited state properties and transitions of 2-aminopurine in the gas phase and in solution. J. Phys. Chem. A, 104, 1930–1937. [Google Scholar]
- 4.Rachofsky E.L., Osman,R. and Ross,J.B. (2001) Probing structure and dynamics of DNA with 2-aminopurine: effects of local environment on fluorescence. Biochemistry, 40, 946–956. [DOI] [PubMed] [Google Scholar]
- 5.Guest C.R., Hochstrasser,R.A., Sowers,L.C. and Millar,D.P. (1991) Dynamics of mismatched base pairs in DNA. Biochemistry, 30, 3271–3279. [DOI] [PubMed] [Google Scholar]
- 6.Kelley S.O. and Barton,J.K. (1999) Electron transfer between bases in double helical DNA. Science, 283, 375–381. [DOI] [PubMed] [Google Scholar]
- 7.Wan C., Fiebig,T., Schiemann,O., Barton,J.K. and Zewail,A.H. (2000) Femtosecond direct observation of charge transfer between bases in DNA. Proc. Natl Acad. Sci. USA, 97, 14052–14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jean J.M. and Hall,K.B. (2001) 2-Aminopurine fluorescence quenching and lifetimes: Role of base stacking. Proc. Natl Acad. Sci. USA, 98, 37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neill M.A. and Barton,J.K. (2002) Effects of strand and directional asymmetry on base–base coupling and charge transfer in double-helical DNA. Proc. Natl Acad. Sci. USA, 99, 16543–16550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lycksell P.O., Graslund,A., Claesens,F., McLaughlin,L.W., Larsson,U. and Rigler,R. (1987) Base pair opening dynamics of a 2-aminopurine substituted EcoRI restriction sequence and its unsubstituted counterpart in oligonucleotides. Nucleic Acids Res., 15, 9011–9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu D., Evans,K.O. and Nordlund,T.M. (1994) Melting and premelting transitions of an oligomer measured by DNA base fluorescence and absorption. Biochemistry, 33, 9592–9599. [DOI] [PubMed] [Google Scholar]
- 12.Law S.M., Eritja,R., Goodman,M.F. and Breslauer,K.J. (1996) Spectroscopic and calorimetric characterizations of DNA duplexes containing 2-aminopurine. Biochemistry, 35, 12329–12337. [DOI] [PubMed] [Google Scholar]
- 13.Allan B.W. and Reich,N.O. (1996) Targeted base stacking disruption by the EcoRI DNA methyltransferase. Biochemistry, 35, 14757–14762. [DOI] [PubMed] [Google Scholar]
- 14.McCullough A.K., Dodson,M.L., Scharer,O.D. and Lloyd,R.S. (1997) The role of base flipping in damage recognition and catalysis by T4 endonuclease V. J. Biol. Chem., 272, 27210–27217. [DOI] [PubMed] [Google Scholar]
- 15.Beechem J.M., Otto,M.R., Bloom,L.B., Eritja,R., Reha-Krantz,L.J. and Goodman,M.F. (1998) Exonuclease-polymerase active site partitioning of primer-template DNA strands and equilibrium Mg2+ binding properties of bacteriophage T4 DNA polymerase. Biochemistry, 37, 10144–10155. [DOI] [PubMed] [Google Scholar]
- 16.Holz B., Klimasauskas,S., Serva,S. and Weinhold,E. (1998) 2-Aminopurine as a fluorescent probe for DNA base flipping by methyltransferases. Nucleic Acids Res., 26, 1076–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stivers J.T. (1998) 2-Aminopurine fluorescence studies of base stacking interactions at abasic sites in DNA: metal-ion and base sequence effects. Nucleic Acids Res., 26, 3837–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allan B.W., Reich,N.O. and Beechem,J.M. (1999) Measurement of the absolute temporal coupling between DNA binding and base flipping. Biochemistry, 38, 5308–5314. [DOI] [PubMed] [Google Scholar]
- 19.Pues H., Bleimling,N., Holz,B., Wolcke,J. and Weinhold,E. (1999) Functional roles of the conserved aromatic amino acid residues at position 108 (Motif IV) and position 196 (Motif VIII) in base flipping and catalysis by the N6-adenine DNA methyltransferase from Thermus aquaticus. Biochemistry, 38, 1426–1434. [DOI] [PubMed] [Google Scholar]
- 20.Stivers J.T., Pankiewicz,K.W. and Watanabe,K.A. (1999) Kinetic mechanism of damage site recognition and uracil flipping by Escherichia coli uracil DNA glycosylase. Biochemistry, 38, 952–963. [DOI] [PubMed] [Google Scholar]
- 21.Gowher H. and Jeltsch,A. (2000) Molecular enzymology of the EcoRV DNA-(Adenine-N (6))-methyltransferase: kinetics of DNA binding and bending, kinetic mechanism and linear diffusion of the enzyme on DNA. J. Mol. Biol., 303, 93–110. [DOI] [PubMed] [Google Scholar]
- 22.Malygin E.G., Lindstrom,W.M.,Jr, Schlagman,S.L., Hattman,S. and Reich,N.O. (2000) Pre-steady state kinetics of bacteriophage T4 dam DNA-[N(6)-adenine] methyltransferase: interaction with native (GATC) or modified sites. Nucleic Acids Res., 28, 4207–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy Y.V. and Rao,D.N. (2000) Binding of EcoP15I DNA methyltransferase to DNA reveals a large structural distortion within the recognition sequence. J. Mol. Biol., 298, 597–610. [DOI] [PubMed] [Google Scholar]
- 24.Szegedi S.S., Reich,N.O. and Gumport,R.I. (2000) Substrate binding in vitro and kinetics of RsrI [N6-adenine] DNA methyltransferase. Nucleic Acids Res., 28, 3962–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vilkaitis G., Dong,A., Weinhold,E., Cheng,X. and Klimasauskas,S. (2000) Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem., 275, 38722–38730. [DOI] [PubMed] [Google Scholar]
- 26.Bandwar R.P. and Patel,S.S. (2001) Peculiar 2-aminopurine fluorescence monitors the dynamics of open complex formation by bacteriophage T7 RNA polymerase. J. Biol. Chem., 276, 14075–14082. [DOI] [PubMed] [Google Scholar]
- 27.Rachofsky E.L., Seibert,E., Stivers,J.T., Osman,R. and Ross,J.B. (2001) Conformation and dynamics of abasic sites in DNA investigated by time-resolved fluorescence of 2-aminopurine. Biochemistry, 40, 957–967. [DOI] [PubMed] [Google Scholar]
- 28.Beck C. and Jeltsch,A. (2002) Probing the DNA interface of the EcoRV DNA-(adenine-N6)-methyltransferase by site-directed mutagenesis, fluorescence spectroscopy and UV cross-linking. Biochemistry, 41, 14103–14110. [DOI] [PubMed] [Google Scholar]
- 29.Dillingham M.S., Wigley,D.B. and Webb,M.R. (2002) Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry, 41, 643–651. [DOI] [PubMed] [Google Scholar]
- 30.Mandal S.S., Fidalgo da Silva,E. and Reha-Krantz,L.J. (2002) Using 2-aminopurine fluorescence to detect base unstacking in the template strand during nucleotide incorporation by the bacteriophage T4 DNA polymerase. Biochemistry, 41, 4399–4406. [DOI] [PubMed] [Google Scholar]
- 31.Dryden D.T.F. (1999) Bacterial DNA methyltransferases. In Cheng,X. and Blumenthal,R.M. (eds), S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions. World Scientific Publishing, Singapore, pp. 283–340. [Google Scholar]
- 32.Cheng X. and Roberts,R.J. (2001) AdoMet-dependent methylation, DNA methyltransferases and base flipping. Nucleic Acids Res., 29, 3784–3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klimasauskas S., Kumar,S., Roberts,R.J. and Cheng,X. (1994) Hhal methyltransferase flips its target base out of the DNA helix. Cell, 76, 357–369. [DOI] [PubMed] [Google Scholar]
- 34.Savva R., McAuley-Hecht,K., Brown,T. and Pearl,L. (1995) The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature, 373, 487–493. [DOI] [PubMed] [Google Scholar]
- 35.Slupphaug G., Mol,C.D., Kavli,B., Arvai,A.S., Krokan,H.E. and Tainer,J.A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- 36.Goedecke K., Pignot,M., Goody,R.S., Scheidig,A.J. and Weinhold,E. (2001) Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat. Struct. Biol., 8, 121–125. [DOI] [PubMed] [Google Scholar]
- 37.Murray N.E. (2000) Type I restriction systems: sophisticated molecular machines. Microbiol. Mol. Biol. Rev., 64, 412–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dryden D.T., Murray,N.E. and Rao,D.N. (2001) Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res., 29, 3728–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Loenen W.A. (2003) Tracking EcoKI and DNA fifty years on: a golden story full of surprises. Nucleic Acids Res., 31, 7059–7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelleher J.E., Daniel,A.S. and Murray,N.E. (1991) Mutations that confer de novo activity upon a maintenance methyltransferase. J. Mol. Biol., 221, 431–440. [DOI] [PubMed] [Google Scholar]
- 41.Dryden D.T.F., Sturrock,S.S. and Winter,M. (1995) Structural modelling of a type I DNA methyltransferase. Nat. Struct. Biol., 2, 632–635. [DOI] [PubMed] [Google Scholar]
- 42.Willcock D.F., Dryden,D.T.F. and Murray,N.E. (1994) A mutational analysis of the two motifs common to adenine methyltransferases. EMBO J., 13, 3902–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mernagh D.R. and Kneale,G.G. (1996) High resolution footprinting of a type I methyltransferase reveals a large structural distortion within the DNA recognition site. Nucleic Acids Res., 24, 4853–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mernagh D.R., Taylor,I.A. and Kneale,G.G. (1998) Interaction of the type I methyltransferase M.EcoR124I with modified DNA substrates: sequence discrimination and base flipping. Biochem. J., 336, 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dryden D.T.F., Cooper,L.P. and Murray,N.E. (1993) Purification and characterization of the methyltransferase from the type 1 restriction and modification system of Escherichia coli K12. J. Biol. Chem., 268, 13228–13236. [PubMed] [Google Scholar]
- 46.Dryden D.T.F., Cooper,L.P., Thorpe,P.H. and Byron,O. (1997) The in vitro assembly of the EcoKI type I DNA restriction/modification enzyme and its in vivo implications. Biochemistry, 36, 1065–1076. [DOI] [PubMed] [Google Scholar]
- 47.Roth M. and Jeltsch,A. (2000) Biotin-avidin microplate assay for the quantitative analysis of enzymatic methylation of DNA by DNA methyltransferases. Biol. Chem., 381, 269–272. [DOI] [PubMed] [Google Scholar]
- 48.O’Neill M., Powell,L.M. and Murray,N.E. (2001) Target recognition by EcoKI: the recognition domain is robust and restriction-deficiency commonly results from the proteolytic control of enzyme activity. J. Mol. Biol., 307, 951–963. [DOI] [PubMed] [Google Scholar]
- 49.Janscak P., Abadjieva,A. and Firman,K. (1996) The type I restriction endonuclease R.EcoR124I: over-production and biochemical properties. J. Mol. Biol., 257, 977–991. [DOI] [PubMed] [Google Scholar]
- 50.Sturrock S.S. and Dryden,D.T.F. (1997) A prediction of the amino acids and structures involved in DNA recognition by type I DNA restriction and modification enzymes. Nucleic Acids Res., 25, 3408–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Broo A. and Holmén,A. (1996) Ab initio MP2 and DFT calculations of geometry and solution tautomerism of purine and some purine derivatives. Chem. Phys. 211, 147–161. [Google Scholar]