Abstract
The Mid-South Clinical Data Research Network (CDRN) encompasses three large health systems: (1) Vanderbilt Health System (VU) with electronic medical records for over 2 million patients, (2) the Vanderbilt Healthcare Affiliated Network (VHAN) which currently includes over 40 hospitals, hundreds of ambulatory practices, and over 3 million patients in the Mid-South, and (3) Greenway Medical Technologies, with access to 24 million patients nationally. Initial goals of the Mid-South CDRN include: (1) expansion of our VU data network to include the VHAN and Greenway systems, (2) developing data integration/interoperability across the three systems, (3) improving our current tools for extracting clinical data, (4) optimization of tools for collection of patient-reported data, and (5) expansion of clinical decision support. By 18 months, we anticipate our CDRN will robustly support projects in comparative effectiveness research, pragmatic clinical trials, and other key research areas and have the capacity to share data and health information technology tools nationally.
Keywords: Research Network, Interoperability, Clinical Research, Health Information Exchange, Data Standards
Introduction
The Mid-South Clinical Data Research Network (CDRN) will create a large research network to support pragmatic trials and comparative effectiveness research. The Mid-South CDRN will connect three major health system networks: (1) the Vanderbilt University Health System (VU), which currently includes electronic medical records for over 2 million patients, (2) a growing Vanderbilt Healthcare Affiliated Network (VHAN), which currently includes over 40 hospitals and hundreds of ambulatory practices, and will cover over 3 million patients in the Mid-South region, and (3) ambulatory practices served by Greenway Medical Technologies, covering over 24 million patients across the country. The Mid-South CDRN will leverage current infrastructure, health information technologies, and data standards to connect the three health systems (see figure 1). The Mid-South CDRN will have a broad reach that includes a diverse population of patients across a large geographic region (see figure 2).
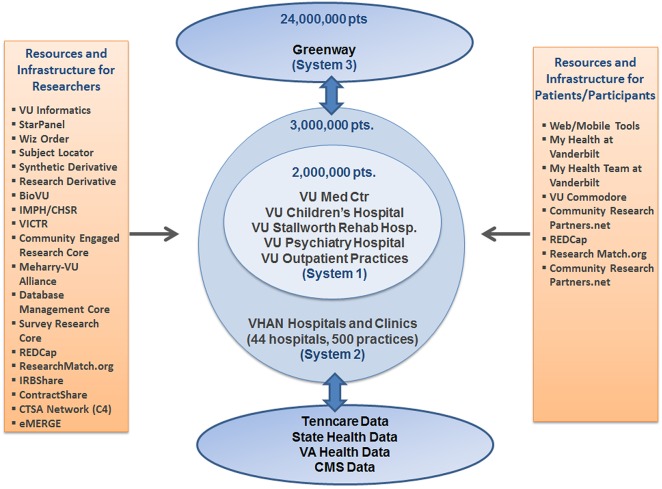
Figure 1.
Mid-South Clinical Data Research Network (CDRN) systems and resources. VU, Vanderbilt University; IMPH, Vanderbilt Institute for Medicine and Public Health; CHSSR, Vanderbilt Center for Health Services Research; VICTR, Vanderbilt Institute for Clinical and Translational Research; REDCap, Research Electronic Data Capture; IRB, Institutional Research Board; CTSA, Clinical and Translational Science Award Program; eMERGE, Electronic Medical Records and Genomics Network; VHAN, Vanderbilt Health Affiliate Network; VA, Veterans Administration; CMS, Centers for Medicare & Medicaid Services.
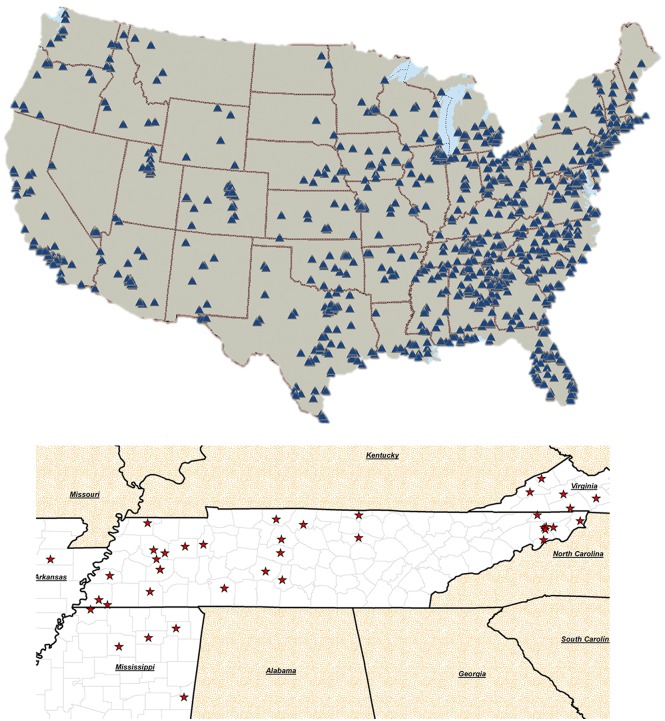
Figure 2.
Mid-South Clinical Data Research Network (CDRN) reach. The top map shows the location of Greenway PrimeRESEARCH sites participating in their research network. The bottom map shows Vanderbilt Health Affiliate Network (VHAN) partner hospital sites.
Overview of existing clinical systems
The primary objective of creating the Mid-South CDRN is to permit research across numerous sites of healthcare delivery through the Southeast USA. To accomplish this, the Mid-South CDRN will accommodate the diverse health information technologies installed at participating sites, and accept the different data formats they produce. We will develop connections among the installed technologies in the three major health systems, described below.
The Vanderbilt Health System (VU) network utilizes a comprehensive electronic health record (EHR) system called StarPanel.1 Developed at Vanderbilt, StarPanel is an integrated web-based user interface to a number of clinically facing tools, such as clinical documentation systems, communication tools supporting secure provider-to-provider and patient-to-provider messaging, reminders, alerts, management of work queues, and notification of new results. StarPanel is used throughout Vanderbilt and has been in place for over 15 years, with peak usage routinely exceeding 8000 concurrent sessions. Because StarPanel is the only system used across the VU network, all clinical data generated during patient care are immediately available to any other Vanderbilt provider. Underlying StarPanel are a data abstraction, a data aggregation, and a data storage layer, collectively called StarChart. StarChart additionally connects and can integrate data from numerous disparate health information technologies, such as commercial EHR systems installed elsewhere.2 3 StarChart accepts patient data in diverse forms and integrates them through a common header format that identifies the patient, the nature of the report, who generated the report at what time, and demographic data. Data in StarChart are encoded to external standards where feasible (see table 1).
Table 1.
StarChart data standards
| Component | Technical approach/data standard |
|---|---|
| Business transactions | HL7 (Health Level 7), ASC (Accredited Standards Committee) X12 |
| Diagnostic imaging | DiCOM (Digital Imaging and Communications in Medicine) |
| Laboratory | LOINC (Logical Observations: Identifiers, Names, Codes) |
| Medications | RX Norm, FDB (First Data Bank) |
| Pharmacy | CPDP (National Council for Prescription Drug Programs), NDC (National Drug Code) |
| Providers | UPIN (Universal Physician Identifier Number) |
| Concept terminology | SNOMED-CT (Systematized Nomenclature of Medicine-Clinical Terms), UMLS (Unified Medical Language System) |
| Procedures | CPT (Current Procedural Terminology), HCPCS (Healthcare Common Procedure Coding System) |
| Diagnoses | ICD (International Classification of Diseases, versions 9 and 10), ICD-O (International Classification of Diseases for Oncology), SNOMED-CT |
| Billing and claims | UB (Uniform Bill) 92, CMS (Center for Medicare and Medicaid Services) 1500 |
The VHAN is a clinically integrated network, chartered by the state of Tennessee and managed through a formal board structure and sub-committees. VHAN is currently composed of seven health systems that include over 40 hospitals and 400 ambulatory practices, covering an estimated 3000 clinicians ranging from ambulatory care to advanced subspecialties, with an estimated reach of over 3 million patients in the Mid-South area. The VHAN is a collection of different healthcare sites, each with its own installed EHR and health information technology (HIT) tools. Installed EHR systems across VHAN will not be standardized to one technology, and therefore VHAN represents the full spectrum of EHR adoption and utilization diversity. Given multiple EHR platforms, VHAN will encourage the use of standards and integration technologies to allow affiliate healthcare providers access to information across the network, with each provider still using a locally installed EHR system. As the VHAN Affiliate Exchange Infrastructure becomes more established and network sites become more sophisticated, VHAN will provide information exchange across the network as a whole. Data exchange will start with administrative and structured clinical data and progress to include clinical narratives and summaries.
Greenway Medical Technologies provides integrated EHR and practice management software and services to over 2000 ambulatory care practices across the country. The PrimeSuite EHR includes the patient chart, numerous clinical tools, and content management pages, as well as practice management functionality. PrimeSuite's integrated platform supports a wide variety of primary care and specialty practices including Federally Qualified Health Centers. The platform also supports a patient-centered medical home for facilities that utilize specific modules and reporting tools. Greenway uses defined syntactic, semantic, and vocabulary standards for standardizing and normalizing data from other systems. Throughout the data build and exchange process of translation, protocol configuration, formatting, and end-point arrivals, Greenway technology supports CCD/CDA, XDS.b, PIX, PDQ, HL7v2, Direct XDR, and custom clinical content needs. Greenway is fostering Consolidated CDA (clinical document architecture), an emerging patient data standard that consolidates and accommodates existing CCD/CDA clinical data for transferring summary of care records from within a single source. A small number of VHAN practices use Greenway PrimeSuite as their EHR solution, and so may become part of PrimeRESEARCH. Any decision about whether to target them first will be made during the infrastructure-building and governance-development process of rolling out the Mid-South CDRN.
Research systems overview
The planned technical infrastructure for the Mid-South CDRN is designed to maximize usage of agreed-upon standards where possible, and apply methods that have been well-established at Vanderbilt3 to exchange data where standards adoption is not immediately feasible. Currently accepted standards include both document-level standards (ie, ‘syntactic standards’ such as CDA) and data formatting standards (ie, ‘semantic standards’). Standards in use include CDIS (The Clinical Data Interchange Standards),4 MedDRA (Medical Dictionary for Regulatory Activities), HL7 (Health Level 7), LOINC (Logical Observations: Identifiers, Names, Codes), and SNOMED CT (Systematized Nomenclature of Medicine Clinical Terms). Where structured and standardized content is not available, the technical infrastructure will leverage existing tools to extract information from unstructured documents. For example, text processing services will identify clinical attributes and phenotypes from corpora of narrative texts in support of clinical research.5 This infrastructural approach has been used successfully in VU and in a health information exchange program in West Tennessee. In addition to this basic infrastructure, VU has developed, operationalized, and disseminated a number of technologies used around the country to support clinical research. These technologies will support data integration, streamline research activities, and standardize administrative processes across the Mid-South CDRN.
The Vanderbilt Research Data Warehouse5 mirrors all clinical information contained in the VU EHR system, administrative systems, and local research databases, and applies novel informatics tools to analyze these data (see figure 3). The Research Data Warehouse contains structured data and applies text processing algorithms to support research.6–14 The Research Data Warehouse includes the Synthetic Derivative (SD), a fully de-identified database containing longitudinal clinical information derived from 2.2 million patients represented in Vanderbilt's EHR. The SD is routinely used as a stand-alone resource, and has been employed to capture data on disease status, disease onset and progression, drug utilization, drug responses, instances of polypharmacy and multimorbidity, medical procedures, hospital and health system utilization, longitudinal laboratory measures, vital signs, social characteristics, and health-related behaviors. The SD can also be used in conjunction with Vanderbilt's 175 000-sample DNA biologic repository called the BioVU biobank, to identify patient sets for genome–phenome analysis.15–17 The Research Data Warehouse also includes the Research Derivative (RD), an identified database used for cohort identification and data extraction for research purposes. The infrastructure framework for the SD and RD databases, and the other related investigator self-service tools, will be expanded or replicated across health systems within the Mid-South CDRN.
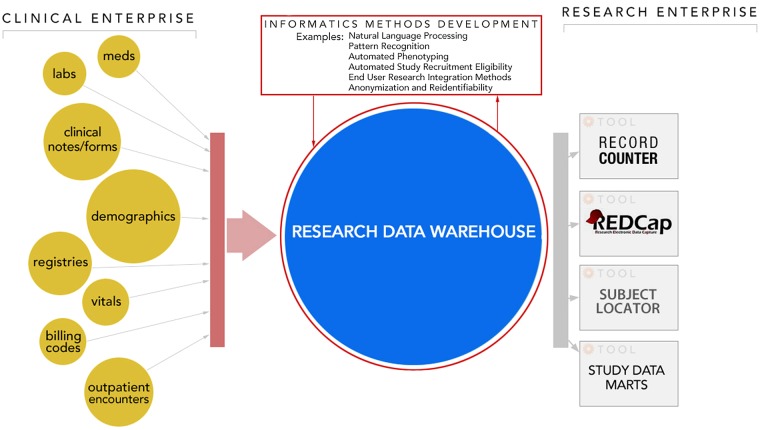
Figure 3.
The Mid-South CDRN's research data warehousing model includes integrating data from multiple clinical systems into a common architecture supporting both de-identified and identified use cases. Beneficiaries include informatics methods development research teams (e.g. natural language processing, de-id and re-id) whose work translates into improving both research and clinical practice. Scientific investigators across the research enterprise also benefit through tools and services built on data warehousing platforms.
REDCap (Research Electronic Data Capture) is a Vanderbilt-developed, secure, web-based platform for building and managing online surveys and research databases. Since its original creation in 2004, REDCap has become a de facto standard for clinical and translational research around the world, with use in over 98 000 studies engaging nearly 1000 academic and non-profit partner organizations across 75 countries.18 19 REDCap supports standardization and shared data management for networked clinical research, including comprehensive data management workflow and capacity. It also provides a user-friendly interface for data entry and validation, audit trails for tracking data manipulation, export procedures for common statistical packages, data quality checks and (if desired) data query functionality, and resolution workflow. In addition, REDCap contains preliminary procedures for importing data from external sources such as EHRs, allowing clinical data to directly feed case report forms.20 REDCap also offers a robust, flexible platform for straightforward definition of survey instruments and administration to patients or family members for detailed data collection. Over the past several years, we have recorded over 691 000 surveys completed in our Vanderbilt-specific installation of REDCap alone.
Subject Locator is a researcher-facing tool that supports patient enrollment into clinical and translational research studies. By mapping study inclusion and exclusion criteria to computable rules, Subject Locator signals whenever a patient is deemed ‘close to appropriate’ for a study. Once a particular patient is flagged, triggering notifications (based on study requirements) are followed by participant contact and consent. A related tool, Record Counter, uses the Research Data Warehouse to support efficient feasibility testing based on counts of the number of possible subjects within the VU system. Record Counter applies a sophisticated search mechanism that allows for complex system queries and returns counts of records stratified by race, sex, and age.
Research Match21 is a disease-neutral, geographic-neutral research registry developed by our team that currently connects 51 000 patient and family volunteers with 1900 researchers at 87 participating medical centers across the country. The platform has proven effective for volunteer/researcher connectivity aimed at study recruitment.22 We have also found Research Match volunteers to be receptive and eager to participate in research prioritization focus groups and patient/family surveys.23 The Research Match consortium of diverse patients and research teams from around the country can play an important role in the planning and executing of operations and individual CDRN studies.
IRBshare is a new shared institutional review board (IRB) review model for multi-site studies consisting of participating institutions utilizing shared review documents and a shared review process, supported by a centralized, secure web portal and the IRBshare Master Agreement. IRBshare is a national project, designed and managed at Vanderbilt, and is comprised of 35 participating institutions to date. ContractShare is an evolving initiative patterned after IRBShare that will streamline contracting processes for multi-site studies. The master contract has been drafted (collaboratively by ∼25 Clinical and Translational Science Awards (CTSA) sites) and is under review with industry sponsors and other stakeholders. The model can be readily expanded to support the CDRNs in future work that requires multiple subcontracts across organizations.
A clinical decision support (CDS) service will be a central component of our CDRN to embed research activities within the healthcare systems without disrupting the business of providing healthcare. The CDS service will enable interventional studies, including by providing randomization of interventions in the form of best practice formats of standard CDS types (eg, alerts, reminders, ordering support, guidelines, forms, templates).24
In addition to the tools described above, our CDRN plans to incorporate significant resources to support comparative effectiveness research and stakeholder engagement. The planned CDRN will benefit from engagement with the Vanderbilt Center for Health Services Research, which currently has over 120 faculty funded by over $50 million for annual funded research in comparative effectiveness research, pragmatic clinical trials, health economics and decision sciences, health communication research, health disparities research, community-based participatory research, and implementation sciences. Authentic stakeholder involvement is critical to the successful implementation of patient-centered outcomes research and our CDRN. Our overarching goals for stakeholder engagement are to develop the infrastructure to facilitate the meaningful involvement of patients, families, and clinicians in all aspects of our CDRN, and cultivate a research environment that values input from stakeholders and respects stakeholders’ perceptions of the relevance and acceptability of research generally, and of specific research studies.
During the first 18 months, we will demonstrate CDRN capacity through identification, recruitment, and data collection from three cohorts: (1) sickle cell disease, (2) coronary heart disease, and (3) a cohort focused on weight status (see table 2). Establishing these cohorts will demonstrate our ability to identify and extract data from our EHR, reach patients through our web portal, collect patient-reported data, and link to genomic and other clinical data.
Table 2.
Proposed initial cohorts
| Cohort name | Target population size | Proposed membership sources | Proposed data elements |
|---|---|---|---|
| Rare disease cohort | Over 400 patients with sickle cell disease (adult and pediatric) | Identification of patients in the VU and VHAN (systems 1 and 2), and patients cared for at Vanderbilt Meharry Matthew Walker Center of Excellence in Sickle Cell Disease | Patient reported willingness to participate in future clinical studies, emergency department hospitalizations, and current medications, solicitation of attitudes to decrease hospitalization, ED visits, and readmissions |
| Weight status cohort | 10 000 adult patients | Identification of patients in the VU EHR (system 1), VHAN EHRs (system 2), and Greenway PrimeRESEARCH network (system 3) | Body mass index (BMI), weight, height, blood pressure, presence of comorbidities (ICD codes), current medications, select laboratory measures (A1C, BG, liver tests) patient reported characteristics, and attitudes related to study participation. EMA of health behaviors in a subset of 1000 patients |
| Network cohort of choice (coronary heart disease) | 10 000 adult patients | Identification of patients in the VU EHR (system 1), and if needed, VHAN EHRs (system 2) | Sociodemographic characteristics, health literacy, medication adherence, diet, exercise, tobacco use status, BMI, weight, height, blood pressure, presence of comorbidities (ICD codes), current medications, select laboratory measures (A1C, creatinine), attitudes related to study participation |
A1C, hemoglobin A1C; BG, blood glucose; ED, emergency department; EHR, electronic health record; EMA, ecological momentary assessment; ICD, International Classification of Diseases; VHAN, Vanderbilt Healthcare Affiliated Network; VU, Vanderbilt Health System.
Summary
Developing the Mid-South CDRN and the larger PCORNet research network represents an exciting and innovative opportunity to collaboratively support comparative effectiveness research and pragmatic clinical trials, and to improve health for patients with both common and rare conditions. By the end of 18 months, we anticipate the Mid-South CDRN will be positioned to take on projects in comparative effectiveness research, pragmatic clinical trials, and other key research areas, and have the capacity to share data and HIT nationally.
Footnotes
Contributors: All authors have contributed substantively to this work.
Funding: The project was supported with funding from the Patient-Centered Outcomes Research Institute (PCORI) #CDRN-1306-04869.
Competing interests: None.
Provenance and peer review: Commissioned; internally peer reviewed.
References
- 1.Guise JM, O'Haire C, McPheeters M, et al. A practice-based tool for engaging stakeholders in future research: a synthesis of current practices. J Clin Epidemiol 2013;66:666–74 [DOI] [PubMed] [Google Scholar]
- 2.Frisse ME, King JK, Rice WB, et al. A regional health information exchange: architecture and implementation. AMIA Annu Symp Proc 2008:212–16 [PMC free article] [PubMed] [Google Scholar]
- 3.Stead WW, Miller RA, Musen MA, et al. Integration and beyond: linking information from disparate sources and into workflow. J Am Med Inform Assoc 2000;7:135–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vadakin AKR. CDISC standards and innovations. Clin Eval 2012; 40(Suppl XXXI):217–28 [Google Scholar]
- 5.Danciu I, Cowan JD, Basford M, et al. Secondary use of clinical data: the Vanderbilt approach. J Biomed Inform 2014. pii: S1532-0464(14)00039-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delaney JT, Ramirez AH, Bowton E, et al. Predicting clopidogrel response using DNA samples linked to an electronic health record. Clin Pharmacol Ther 2012;91:257–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu H, Jiang M, Oetjens M, et al. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J Am Med Inform Assoc 2011;18:387–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denny JC, Ritchie MD, Crawford DC, et al. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation 2010;122:2016–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010;26:1205–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denny JC, Choma NN, Peterson JF, et al. Natural language processing improves identification of colorectal cancer testing in the electronic medical record. Med Decis Making 2012;32:188–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denny JC, Miller RA, Waitman LR, et al. Identifying QT prolongation from ECG impressions using a general-purpose Natural Language Processor. Int J Med Inform 2009;78(Suppl 1)S34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny JC, Peterson JF, Choma NN, et al. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. J Am Med Inform Assoc 2010;17:383–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu H, Doan S, Birdwell KA, et al. An automated approach to calculating the daily dose of tacrolimus in electronic health records. AMIA Summits on Translational Science Proceedings; 2010;vol. 2010:71–5 [PMC free article] [PubMed] [Google Scholar]
- 14.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clin Pharmacol Ther 2011;89:379–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pulley J, Clayton E, Bernard GR, et al. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci 2010;3:42–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010;86:560–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obeid JS, McGraw CA, Minor BL, et al. Procurement of shared data instruments for Research Electronic Data Capture (REDCap). J Biomed Inform 2013;46:259–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antman EM, Harrington RA. Transforming clinical trials in cardiovascular disease: mission critical for health and economic well-being. JAMA 2012;308:1743–4 [DOI] [PubMed] [Google Scholar]
- 21.Research Match. https://www.researchmatch.org/
- 22.Harris PA, Scott KW, Lebo L, et al. ResearchMatch: a national registry to recruit volunteers for clinical research. Acad Med 2012;87:66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pulley J, Hassan NN, Bernard GR, et al. Identifying unpredicted drug benefit through query of patient experiential knowledge: a proof of concept web-based system. Clin Transl Sci 2010;3:98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horsky J, Schiff GD, Johnston D, et al. Interface design principles for usable decision support: a targeted review of best practices for clinical prescribing interventions. J Biomed Inform 2012;45:1202–16 [DOI] [PubMed] [Google Scholar]