Abstract
Fluorescence resonance energy transfer (FRET) is one of the most powerful and promising tools for single nucleotide polymorphism (SNP) genotyping. However, the present methods using FRET require expensive reagents such as fluorescently labeled oligonucleotides. Here, we describe a novel and cost-effective method for SNP genotyping using FRET. The technique is based on allele-specific primer extension using mononucleotides labeled with a green dye and a red dye. When the target DNA contains the sequence complementary to the primer, extension of the primer incorporates the green and red dye-labeled nucleotides into the strand, and red fluorescence is emitted by FRET. In contrast, when the 3′ end nucleotide of the primer is not complementary to the target DNA, there is no extension of the primer, or FRET signal. Therefore, discrimination among genotypes is achieved by measuring the intensity of red fluorescence after the extension reaction. We have validated this method with 11 SNPs, which were successfully determined by end-point measurements of fluorescence intensity. The new strategy is simple and cost-effective, because all steps of the preparation consist of simple additions of solutions and incubation, and the dye-labeled mononucleotides are applicable to all SNP analyses. This method will be suitable for large-scale genotyping.
INTRODUCTION
Over the past few years, it has become apparent that single nucleotide polymorphisms (SNPs) will represent the next generation of markers. Use of SNPs is expected to lead to a better understanding of the genetic basis for complex diseases, and to realize the potential of pharmacogenetics (1). More than 1.4 million SNPs have been identified in the human genome (2), and approximately 300 000 SNPs would be required for whole-genome association studies (3). To meet this demand, high-throughput genotyping is required. However, such high-throughput assays must be straightforward. The original SNP genotyping methods of DNA sequencing, single strand conformation polymorphism (SSCP) (4) and restriction fragment length polymorphisms (RFLPs) (5), and the methods based on mass spectrometry (6–8) require multiple steps, including size separation, that limit throughput and the potential for automation. A recent approach to a simple procedure for genotyping is the use of fluorescence resonance energy transfer (FRET), which is observed when two fluorescent dyes are in close proximity (9) and one fluorophore’s emission overlaps the other’s excitation spectrum. FRET is one of the most promising tools for SNP genotyping and has provided novel and unique detection systems, such as the 5′ nuclease or TaqMan assay (10), molecular beacons (11,12), template-directed dye-terminator incorporation (TDI) (13), allele-specific hybridization (14), hybridization probe assay (15), flap endonuclease discrimination (invader assay) (16) and allele-specific PCR with universal energy-transfer-labeled primer (17). These methods do not require size separation of products or removal of surplus fluorescence substances. In these methods, all steps of the preparation can be performed in the same vessel by simple additions of solutions and incubations, and then genotypes are discriminated by measuring fluorescence intensity during or after incubations. Therefore, these methods are suitable for automation. However, the cost of analysis is high because they require fluorescently labeled oligonucleotides.
Here, we describe the use of FRET in a cost-effective novel method to identify SNPs. The key feature of this method is to use fluorescently labeled mononucleotides as universal FRET reagents instead of the fluorescently labeled oligonucleotides that are used by current methods. This approach avoids the time and cost of synthesis of fluorescently labeled oligonucleotides. In this method, fluorescently labeled mononucleotides are inserted into the strand by the allele-specific primer extension reaction and emit FRET signals. We validated this novel method with 11 SNPs in the genomes of volunteers. The SNPs were successfully discriminated by measuring the fluorescence intensity after the extension reaction.
Principle
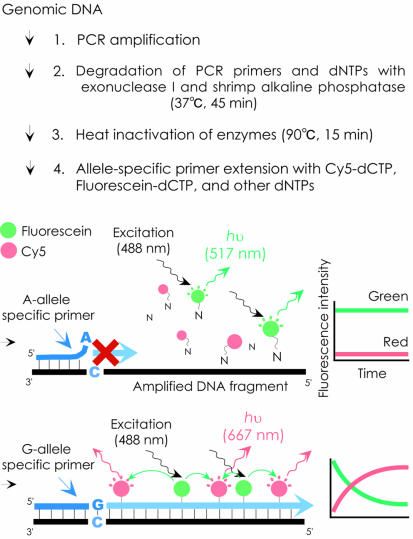
A scheme of the proposed method is shown in Figure 1. A green dye, fluorescein (excitation/emission = 494/517 nm), and a red dye, Cy5 (650/667 nm), are employed as the energy donor and acceptor, respectively. The green dye is directly excited and red fluorescence is measured as a FRET signal. The amplified genomic DNA containing a polymorphic site is incubated with an allele-specific primer (designed to contain a single allele-specific nucleotide at the 3′ terminus) in the presence of a green dye-labeled mononucleotide, a red dye-labeled mononucleotide and DNA polymerase. When the target DNA fragment contains the sequence complementary to the allele-specific primer, extension of the primer with green and red dye-labeled nucleotides occurs, and red fluorescence is emitted by FRET. In contrast, there is no primer extension with target DNA containing a nucleotide that is not complementary to the 3′ end of the primer; the dye-labeled nucleotides remain free in solution, and no FRET occurs. Therefore, the SNP can be discriminated by measuring the intensity of red fluorescence after the extension reaction. To enhance the extension, the temperature of the reaction mixture is held at 94°C for 2 min, after which there are 30 cycles of denaturation at 94°C for 20 s, annealing at 52–60°C for 60 s, and extension at 72°C for 20 s. To enhance discrimination between the two alleles, an artificial mismatch was introduced into the third position from the 3′ terminus (18,19).
Figure 1.
Schematic presentation of the new method. The SNP shown here as an example is a G/A substitution. Allele-specific primers with perfectly matching terminal bases can be extended to generate a FRET signal that would result in an increase of red fluorescence intensity, corresponding to Cy5 dye emission (667 nm), whereas the fluorescence intensity remains unchanged with the mismatched primer.
MATERIALS AND METHODS
Chemicals
A dCTP labeled with fluorescein-12 or Cy5 and a dUTP labeled with fluorescein-12 were obtained from PerkinElmer. Vent (exo-) DNA polymerase was purchased from New England Biolabs. Accuprime Supermix II was purchased from Invitrogen. Shrimp alkaline phosphatase and Escherichia coli exonuclease I were purchased from USB.
Oligonucleotides
All the oligonucleotides were synthesized and purified by Espec Oligo Service. The allele-specific primers and PCR primers used in this study are described in Tables 1 and 2, respectively.
Table 1. Allele-specific primers used in this study.
| Experiment | Locus name | Reference | Allele-specific primer sequence (5′–3′) | Annealing temp. (°C) |
|---|---|---|---|---|
| A | ALDH2 | (20) | tcccacactcacagttttcacAtC/T | 58 |
| B | GNB3 | (21) | atcatctgcggcatcacgAcC/T | 60 |
| C | HTR2A (T102C) | (22) | ggctctacagtaatgactttaacAcC/T | 52 |
| D | HTR2A (intron 3) | dbSNP ID; rs2296973 | ctggaaattgtcataggatagggTtA/C | 59 |
| E | PSEN2 | dbSNP ID; rs1800681 | gtcaggctgatatcccaaatcTaG/C | 58 |
| F | DRD1 | dbSNP ID; rs4532 | actgacccctattccctgcAtA/G | 60 |
| G | CYP2C19 | (23) | ttcccactatcattgattatttcAcG/A | 54 |
| H | TAP2 | dbSNP ID; rs241442 | actgtcccctgccctctcTcG/A | 60 |
| I | P53 codon 273 | (24) | acggaacagctttgaggAgC/T | 57 |
| J | P53 codon 282 | (25) | gtgcctgtcctgggagagTcC/T | 59 |
| K | AGTR1 | (26) | ccttcaattctgaaaagtagctGaT/G | 55 |
| L | AGTR1 | ccttcaattctgaaaagtagctaaT/G | 55 |
The two upper case letters at the 3′ terminus of the primer are the specific bases of the SNP type, and the third upper case letter is the artificially mismatched base. The primer used in experiment L did not contain an artificially mismatched base.
Table 2. PCR primers, annealing temperature, product size and restriction enzymes for RFLP.
| No. | PCR primer 1 (5′–3′) | PCR primer 2 (5′–3′) | Annealing temp. (°C) | Product size (bp) | Restriction enzyme |
|---|---|---|---|---|---|
| A | GTGTAACCCATAACCCCCAAGA | CACCAGCAGACCCTCAAGC | 60 | 226 | Eco57I |
| B | CTGATCTGCTTCTCCCACGAG | GCAGTAAGAGAGTCCGAAATGG | 60 | 243 | BtgI |
| C | TCAACTACGAACTCCCTAATGCAA | GATGACGAGTATGTTTCCAGCAA | 60 | 252 | MspI |
| C(1)a | TCAACTACGAACTCCCTAATGCAA | GAGGCACCCTTCACAGGAAA | 60 | 147 | – |
| C(2)a | TCAACTACGAACTCCCTAATGCAA | TTATAGTTTGTTTGCCCCCTGA | 60 | 477 | – |
| D | TGAAATATGTTGCCACTTACCTACTG | CCCAGAGCTGGAAATTGTCATA | 60 | 309 | BamHI |
| E | GGGATGCTTGGAACAGATCAC | CTTGACACAGGGGAGTGGAAG | 60 | 343 | BslI |
| F | GACTGACCCCTATTCCCTGCTT | ACACAGCCAAGGAGATGACAAAG | 60 | 275 | DdeI |
| G | CAGAGCTTGGCATATTGTATC | GTAAACACACAACTAGTCAATG | 49 | 321 | SmaI |
| H | TACCTGCTGTGCACTTGTCC | CCCAATTCTGCACAGTCTGA | 60 | 295 | ScaI |
| I/J | ACCTGATTTCCTTACTGCCTCTTGC | GTCCTGCTTGCTTACCTCGCTTAGT | 60 | 200 | BcgIb/Cfr101 |
| K/L | CAAAAATGAGCACGCTTTCCT | GGCTTTGCTTTGTCTTGTTGC | 60 | 284 | DdeI |
aPCR products of experiments C(1) and C(2) were used in the validation of the method.
bFor RFLP analysis in experiment I, the amplified 200 bp DNA fragment was again amplified with primer 2 and a mutagenesis primer (5′- TACTGGGACGGAACACGATTGAGGTG-3′) to generate a BcgI site in the wild-type fragment.
Examination of the optimal distance between fluorescein and Cy5 for the FRET signal with synthetic templates
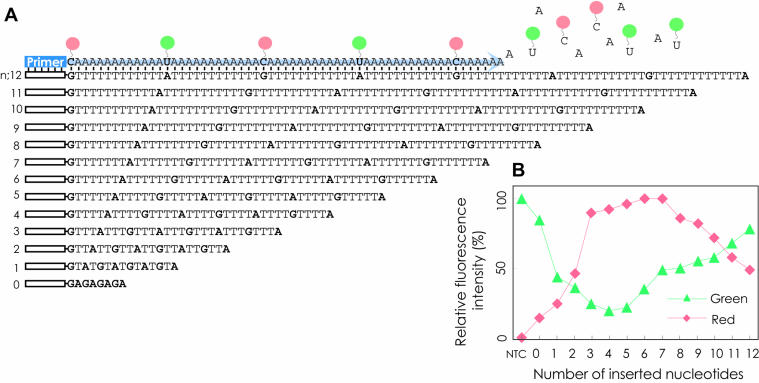
A dCTP labeled with Cy5 and a dUTP labeled with fluorescein-12 were used for primer extension reactions with 13 synthetic oligonucleotide templates that contained a sequence complementary to a primer (5′-CAGACT CGACAGTGTAGACCCG-3′) at the 5′ terminus and subsequently (GTnATn)3GTnA (n = 0–12) (Fig. 2A). The reactions were carried out in a total volume of 25 µl containing 0.4 µM dUTP labeled with fluorescein-12, 0.4 µM dCTP labeled with Cy5, 2 µM dATP, 200 nM primer, 20 nM synthetic oligonucleotide template, 1× polymerase buffer provided by the supplier and 0.25 U of vent (exo-) DNA polymerase. Thermal cycling was performed in a SmartCycler (Takara) under the following conditions: 94°C for 2 min, followed by 30 cycles of 94°C for 20 s, 62°C for 60 s and 72°C for 20 s. The no-template control (NTC) contained water instead of the template.
Figure 2.
Examination of the optimal distance between fluorescein and Cy5 for the FRET signal. (A) The 13 synthetic oligonucleotide templates prepared for this study. (B) Fluorescence intensities in the final extension cycle. The data were normalized to a maximum intensity of 100 in a set of 13 experiments. NTC, no-template control.
Real sample preparation by PCR
Genomic DNA was extracted with ISOHAIR (Nippon Gene) from black hair samples of 18 volunteers according to the manufacturer’s manual. PCR was performed with 20 ng of genomic DNA template, 200 nM of each primer shown in Table 2 and Accuprime Supermix II (Invitrogen) in a final reaction volume of 25 µl in an I-Cycler (Bio-Rad Laboratories). The reaction mixture was held at 94°C for 2 min followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 49 or 60°C for 30 s, and extension at 68°C for 40 s, and then kept at 68°C for 5 min. The quantity of genomic DNA extracted from the hair samples was not large enough to amplify by PCR all SNP sites shown in Table 1 for all 18 extracts, and therefore six SNP sites were amplified from fewer than 18 extracts.
Pre-treatment of PCR product for allele-specific primer extension reaction
The dNTPs and PCR primers left over from the PCR were considered to disturb subsequent allele-specific extension, and therefore they were removed by enzyme treatment or purification of the PCR product by the following procedures. In the experiments to determine the optimal ratio of labeled to unlabeled nucleotides in the reaction mixture and to genotype SNPs in the genomes of volunteers, the PCR products were treated with 2 U of shrimp alkaline phosphatase and 10 U of E.coli exonuclease I for 45 min at 37°C and then heat treated at 90°C for 15 min for enzyme inactivation. A 4 µl sample of the treated PCR product was used as the DNA template for the allele-specific primer extension reaction. In the experiments to validate the method and to examine the effect of template size, the PCR products were purified with a QIA PCR Purification kit (QIAGEN) and quantified by measuring the OD260. Each DNA fragment was used in the primer extension reaction at a concentration of 5 nM.
Allele-specific primer extension reaction
The allele-specific primer sequences and annealing temperatures are shown in Table 1. The reaction mixture for allele-specific extension contained 20 mM Tris–HCl pH 8.8, 10 mM KCl, 10 mM (NH4)2SO4, 2.5 mM MgCl2, 0.2% Triton X-100, 7% dimethylsulfoxide (DMSO), 0.25 U of vent (exo-) DNA polymerase, 2 µM dATP, 2 µM dGTP, 2 µM dTTP, 1.2 µM dCTP, 0.4 µM fluorescein-labeled dCTP, 0.4 µM Cy5-labeled dCTP, 200 nM allele-specific primer and template DNA in a total volume of 25 µl. In each experiment, we included two controls that contained water instead of the template DNA. Thermal cycling was performed in a SmartCycler under the following conditions: 94°C for 2 min, followed by 30 cycles of denaturation at 94°C for 20 s, annealing at 52–60°C for 60 s, and extension at 72°C for 20 s.
Fluorescence measurement
Fluorescence intensities were measured with a SmartCycler during the annealing phase of the allele-specific primer extension reaction. Excitation was at 450–495 nm, with a 505–537 nm emission filter for green fluorescence and a 605–800 nm emission filter for red fluorescence. The fluorescence was measured again after the end of the reaction at 40°C.
Allelic ratio calculation
Red fluorescence intensity obtained with the major allele-specific primer was divided by the intensity obtained with the minor allele-specific primer after the intensity of the NTC had been subtracted from both.
RESULTS AND DISCUSSION
Determination of the optimal ratio of labeled to unlabeled nucleotides in the reaction mixture for FRET
First, the optimal distance between fluorescein and Cy5 in a DNA strand for the greatest FRET signal was examined with 13 synthetic oligonucleotide templates containing (GTnATn)3GTnA (n = 0–12) (Fig. 2A). Primer extension by vent (exo-) polymerase was performed with fluorescein-labeled dUTP, Cy5-labeled dCTP and non-labeled dATP. As shown in Figure 2B, the templates with n = 6–7 showed the strongest red fluorescence. This suggests that the optimal ratio of labeled to unlabeled nucleotides in the reaction mixture for FRET would be 1:6–7.
Next, the optimal ratio of labeled to unlabeled nucleotides was examined with a 252 bp PCR product containing a polymorphic site (T102C) of the HTR2A gene prepared from genomic DNA. The PCR product of homozygous C/C was incubated with a C allele-specific primer, vent (exo-) DNA polymerase and 2 µM of each of the four dNTPs. Some of the dCTP was replaced with Cy5- and fluorescein-labeled dCTP. The ratio of Cy5-dCTP, fluorescein-dCTP and unlabeled dCTP was adjusted to 1:1:a (a = 1, 2, 3, 4 and 5) with a final concentration of dCTP of 2 µM. Consequently, the ratio of dye-labeled nucleotides to total unlabeled nucleotides in the reaction mixture was 1:a′ (a′ = 5, 7, 9, 11 and 13, respectively). The relative increase in red fluorescence with these mixtures was 89, 100, 77, 74 and 53, respectively, indicating that the optimal ratio of labeled to unlabeled nucleotides is 1:7. This result was consistent with that of the first experiment using synthetic oligonucleotides. The range of ratios giving strong fluorescence intensity (>70% of the maximum) was 1:5–11, and we selected a ratio of 1:9 for the subsequent detection of SNPs. We selected this ratio in an attempt to lower costs by reducing the amount of labeled nucleotide.
The first experiments were repeated with Taq polymerase instead of vent (exo-) polymerase, but only weak emissions of red fluorescence were observed (data not shown), showing that Taq polymerase is not suitable for this method.
Validation of the method
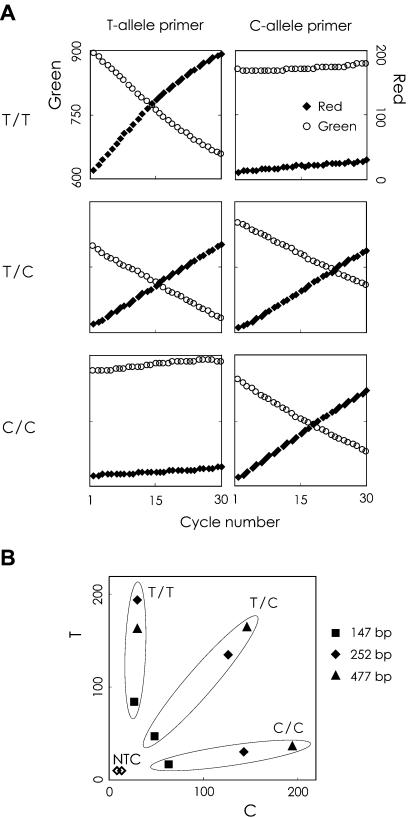
The new method was examined with the 252 bp DNA fragment of the HTR2A gene, which contains a T102C polymorphism (T/T, T/C and C/C). The DNA fragment was prepared from genomic DNA by PCR, and then subjected to the primer extension reaction with the C- or T-allele-specific primers. Figure 3A shows the real-time fluorescence intensity profiles of the three known genotypes in the 252 bp fragment. The three genotypes gave clearly different profiles, with the two specific primers recognizing the C and T alleles. For example, the T/T homozygous sample showed a progressive drop in green fluorescence and a corresponding rise in red fluorescence with the T allele-specific primer, whereas no fluorescence changes were observed with the C allele-specific primer. This indicates that the three genotypes can be clearly distinguished from one another by measuring the intensity of the red fluorescence at the end-point of the reaction, and that measurement of the intensity of green fluorescence is useful to confirm the result.
Figure 3.
Validation of the method and effect of template size on genotyping. (A) Real-time fluorescence intensity profiles for the three known genotypes (T/T, C/C and T/C) of the HTR2A gene (252 bp). (B) Scatter plot of the end-point red fluorescence intensity obtained with 147, 252 and 477 bp PCR fragments as templates.
Effect of template size
The effect of template size on the identification of SNPs was examined with 147, 252 and 477 bp fragments of the HTR2A gene. The allele-specific primer could extend 81, 186 and 411 bases, respectively. The final red fluorescence intensities from the primer extension reactions with the C and T allele-specific primers were plotted on the x-axis and the y-axis, respectively (Fig. 3B). The three genotypes were discriminated as clusters on the graph, and the two negative controls were separated from them. The genotypes were then distinguished by comparing their allelic ratios. The T per C allelic ratios for the 147, 252 and 477 bp fragments were: T/T-homozygous, 4.45, 9.46, 7.92; T/C-heterozygous, 0.97, 1.08, 1.15; C/C-homozygous, 0.12, 0.15, 0.15, respectively. From these data, the genotypes were easily distinguishable with all template sizes. In subsequent experiments, we prepared 200–343 bp template DNAs for SNP genotyping.
SNP genotyping
The new method was used to type 11 different SNPs (Table 1), which were selected to represent various types of single nucleotide substitutions, i.e. A/G, C/T, G/C, G/T and A/C. The SNPs were also analyzed in parallel by a PCR-RFLP assay.
The genotypes were determined by our method with the two allele-specific primers containing the artificial mismatch base shown in Table 1. The allelic ratios of end-point red fluorescence intensity were calculated, and are summarized in Table 3. The allelic ratios for the minor homozygous samples, the heterozygous samples and the major homozygous samples were 0.00–0.30, 0.6–1.5 and 3.6–147, respectively, except for one major homozygous sample of the AGTR1 SNP (K in Table 3), which showed an allelic ratio of 2.1. Thus, except for the AGTR1 SNP, the three genotypes were clearly distinguishable. The allele-specific extension method for genotyping the AGTR1 SNP was repeated for all 16 samples with a primer without an artificial mismatch base at the third position from the 3′ terminus. As a result, the allelic ratio of the major homozygous sample was improved to more than 5.5 (L in Table 3). It has been reported that an artificial mismatch introduced into the third position from the 3′ end of primers will enhance discrimination of the two alleles (18,19), but this is a case where a perfectly matching allele-specific primer is more effective.
Table 3. The allelic ratio for 11 SNPs.
| No. | Allele | Allelic ratio (number of samples) | |||
|---|---|---|---|---|---|
| Major | Minor | Genotype | |||
| A1 | A2 | A1/A1 | A1/A2 | A2/A2 | |
| A | C | T | 5.0–147 (11) | 1.2–1.4 (5) | 0.16–0.19 (2) |
| B | C | T | 7.4–8.2 (3) | 1.1–1.3 (9) | 0.26–0.30 (2) |
| C | C | T | 11.9–18.3 (6) | 1.0–1.5 (9) | 0.00–0.08 (3) |
| D | A | C | 6.8–21.9 (5) | 0.8–1.6 (10) | 0.06–0.07 (3) |
| E | G | C | 8.0–126 (12) | 1.0–1.5 (3) | – (0) |
| F | A | G | 3.8–11.4 (10) | 0.6–0.9 (4) | – (0) |
| G | G | A | 7.4–97.5 (12) | 1.1–1.4 (4) | – (0) |
| H | G | A | 9.8–14.2 (2) | – (0) | – (0) |
| I | C | T | 3.6–10.8 (18) | – (0) | – (0) |
| J | C | T | 5.4–16.8 (18) | – (0) | – (0) |
| K | T | G | 2.1–19.0 (15) | 0.7 (1) | – (0) |
| L | T | G | 5.5–146 (15) | 1.1 (1) | – (0) |
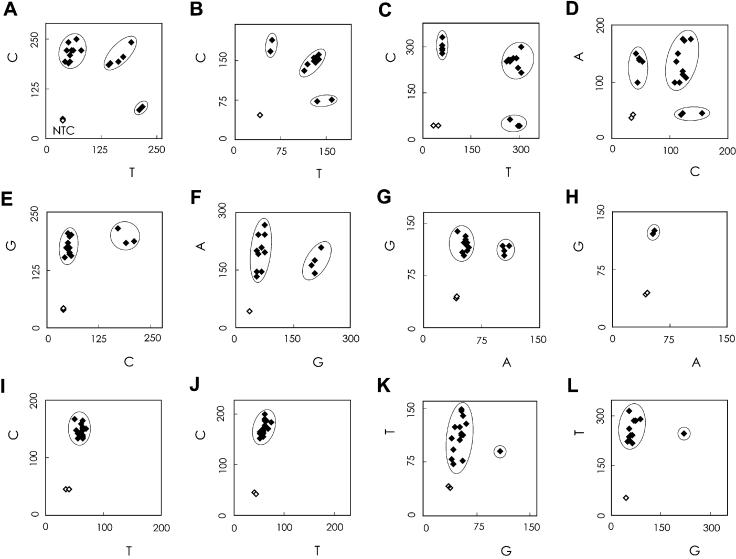
A scatter plot is shown in Figure 4. In this figure, the red fluorescence intensities generated by one (major) allele-specific primer are plotted on the y-axis, and the red intensities from the second (minor) allele-specific primer are plotted on the x-axis. For each SNP, homozygous and heterozygous SNP genotypes were discriminated as clusters on the graph, and the two negative controls were separated from them. All genotypes were determined independently by PCR-RFLP to verify the results of our method.
Figure 4.
Genotyping results for 11 SNPs. A scatter plot of the end-point red fluorescence intensity is shown. The genotypes were also determined by RFLP, and samples with the same genotype are circled in the figure. The genotype assignment is for the (A) ALDH2, (B) GNB3, (C) HTR2A (T102C), (D) HTR2A (intron 3), (E) PSEN2, (F) DRD1, (G) CYP2C19, (H) TAP2, (I) p53 codon 273 (J) p53 codon 282 and (K) AGTR1 gene polymorphisms determined with primers containing an artificial mismatch base at the third position from the 3′ end, and (L) the AGTR1 gene polymorphism determined with primers containing no artificial mismatch base.
Thermal cycling for allele-specific primer extension was performed for 30 cycles in our study. We used the end-point fluorescence intensity to discriminate SNPs, but we also measured the intensity at every cycle. The allelic ratio of the major homozygote was greater than 3 after 13, 29, 8, 22, 28, 16, 16, 15, 11, 15 and 16 cycles in experiments A–L (excluding K), respectively, so the genotypes were clearly distinguishable at these cycle numbers. Thus, for many of the SNPs we examined, a cycle number of less than 30 would be adequate to determine the genotype.
With this method, the target SNP region is first amplified by PCR from genomic DNA. Multiplex PCR, which amplifies many target regions simultaneously in one tube with many sets of primers, reduces time, reagent cost and requirement for genomic DNA. We amplified the ALDH2 gene and the p53 gene (A and I/J in Table 2) by multiplex PCR, and subjected the product to genotyping. As shown in Figure 4A, I and J, the genotypes were clearly discriminated from one another.
Advantages of the method
The method has several useful features, as follows. (i) The method uses fluorescently labeled mononucleotides instead of labeled oligonucleotides for the FRET reagent, and therefore the cost is low. Fluorescently labeled mononucleotides can be purchased from chemical suppliers, whereas fluorescently labeled oligonucleotides are tailor-made. The optimal sequences of oligonucleotides needed for geno typing new SNPs are often determined by trial and error. In many of the present methods, this requires fluorescently labeled oligonucleotides, with the attendant high costs, whereas our method uses cost-effective unlabeled oligonucleotides. (ii) Genotypes can be determined by end-point fluorescence measurement using simple equipment with a simple optical system. This simple procedure can significantly increase throughput. (iii) All reactions, including PCR, enzyme treatment and allele-specific primer extension, can be performed simply in the same vessel by additions of solutions and incubations, and then the fluorescence of the resulting product can be measured directly in the same vessel. Hence, this approach has the potential for automation with a robotic workstation for high throughput. (iv) This homogeneous assay is not restricted to a particular format, making it possible to envisage different high-throughput engineering strategies, such as DNA microarrays with many kinds of allele-specific primers immobilized on one chip to type many SNPs at one time.
In summary, we showed that a novel method using an allele-specific primer extension reaction with fluorescently labeled mononucleotides was useful for genotyping SNPs. This novel method will contribute to large-scale genotyping studies.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Masakazu Inazuka and Dr Yoshihiro Takatsu for advice and for constructive criticism of this manuscript.
REFERENCES
- 1.Risch N. and Merikangas,K. (1996) The future of genetic studies of complex human diseases. Science, 273, 1516–1517. [DOI] [PubMed] [Google Scholar]
- 2.Sachidanandam R., Weissman,D., Schmidt,S.C., Kakol,J.M., Stein,L.D., Marth,G., Sherry,S., Mullikin,J.C., Mortimore,B.J., Willey,D.L. et al. (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature, 409, 928–933. [DOI] [PubMed] [Google Scholar]
- 3.Patil N., Berno,A.J., Hinds,D.A., Barrett,W.A., Doshi,J.M., Hacker,C.R., Kautzer,C.R., Lee,D.H., Marjoribanks,C., McDonough,D.P. et al. (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science, 294, 1719–1723. [DOI] [PubMed] [Google Scholar]
- 4.Orita M., Iwahana,H., Kanazawa,H., Hayashi,K. and Sekiya,T. (1989) Detection of polymorphisms of human DNA by gel electrophoresis as single-strand conformation polymorphisms. Proc. Natl Acad. Sci. USA, 86, 2766–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kan Y.W. and Dozy,A.M. (1978) Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc. Natl Acad. Sci. USA, 75, 5631–5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun A., Little,D.P. and Koster,H. (1997) Detecting CFTR gene mutations by using primer oligo base extension and mass spectrometry. Clin. Chem., 43, 1151–1158. [PubMed] [Google Scholar]
- 7.Sauer S., Lechner,D., Berlin,K., Lehrach,H., Escary,J.L., Fox,N. and Gut,I.G. (2000) A novel procedure for efficient genotyping of single nucleotide polymorphisms. Nucleic Acids Res., 28, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haff L.A. and Smirnov,I.P. (1997) Single-nucleotide polymorphism identification assays using a thermostable DNA polymerase and delayed extraction MALDI-TOF mass spectrometry. Genome Res., 7, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Livak K.J., Flood,S.J., Marmaro,J., Giusti,W. and Deetz,K. (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl., 4, 357–362. [DOI] [PubMed] [Google Scholar]
- 10.Livak K.J., Marmaro,J. and Todd,J.A. (1995) Towards fully automated genome-wide polymorphism screening. Nature Genet., 9, 341–342. [DOI] [PubMed] [Google Scholar]
- 11.Piatek A.S., Tyagi,S., Pol,A.C., Telenti,A., Miller,L.P., Kramer,F.R. and Alland,D. (1998) Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat. Biotechnol., 16, 359–363. [DOI] [PubMed] [Google Scholar]
- 12.Tyagi S., Bratu,D.P. and Kramer,F.R. (1998) Multicolor molecular beacons for allele discrimination. Nat. Biotechnol., 16, 49–53. [DOI] [PubMed] [Google Scholar]
- 13.Chen X., Zehnbauer,B., Gnirke,A. and Kwok,P.Y. (1997) Fluorescence energy transfer detection as a homogeneous DNA diagnostic method. Proc. Natl Acad. Sci. USA, 94, 10756–10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howell W.M., Jobs,M. and Brookes,A.J. (2002) iFRET: an improved fluorescence system for DNA-melting analysis. Genome Res., 12, 1401–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schutz E., von Ahsen,N. and Oellerich,M. (2000) Genotyping of eight thiopurine methyltransferase mutations: three-color multiplexing, ‘two-color/shared’ anchor and fluorescence-quenching hybridization probe assays based on thermodynamic nearest-neighbor probe design. Clin. Chem., 46, 1728–1737. [PubMed] [Google Scholar]
- 16.Hall J.G., Eis,P.S., Law,S.M., Reynaldo,L.P., Prudent,J.R., Marshall,D.J., Allawi,H.T., Mast,A.L., Dahlberg,J.E., Kwiatkowski,R.W. et al. (2000) Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc. Natl Acad. Sci. USA, 97, 8272–8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myakishev M.V., Khripin,Y., Hu,S. and Hamer,D.H. (2001) High-throughput SNP genotyping by allele-specific PCR with universal energy-transfer-labeled primers. Genome Res., 11, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okimoto R. and Dodgson,J.B. (1996) Improved PCR amplification of multiple specific alleles (PAMSA) using internally mismatched primers. Biotechniques, 21, 20–22, 24,, 26. [DOI] [PubMed] [Google Scholar]
- 19.Zhou G., Kamahori,M., Okano,K., Chuan,G., Harada,K. and Kambara,H. (2001) Quantitative detection of single nucleotide polymorphisms for a pooled sample by a bioluminometric assay coupled with modified primer extension reactions (BAMPER). Nucleic Acids Res., 29, e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomasson H.R., Edenberg,H.J., Crabb,D.W., Mai,X.L., Jerome,R.E., Li,T.K., Wang,S.P., Lin,Y.T., Lu,R.B. and Yin,S.J. (1991) Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am. J. Hum. Genet., 48, 677–681. [PMC free article] [PubMed] [Google Scholar]
- 21.Siffert W., Rosskopf,D., Siffert,G., Busch,S., Moritz,A., Erbel,R., Sharma,A.M., Ritz,E., Wichmann,H.E., Jakobs,K.H. et al. (1998) Association of a human G-protein beta3 subunit variant with hypertension. Nature Genet., 18, 45–48. [DOI] [PubMed] [Google Scholar]
- 22.Lohmueller K.E., Pearce,C.L., Pike,M., Lander,E.S. and Hirschhorn,J.N. (2003) Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nature Genet., 33, 177–182. [DOI] [PubMed] [Google Scholar]
- 23.deMorais S.M., Wilkinson,G.R., Blaisdell,J., Nakamura,K., Meyer,U.A. and Goldstein,J.A. (1994) The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J. Biol. Chem., 269, 15419–15422. [PubMed] [Google Scholar]
- 24.Levine A.J., Momand,J. and Finlay,C.A. (1991) The p53 tumour suppressor gene. Nature, 351, 453–456. [DOI] [PubMed] [Google Scholar]
- 25.Toguchida J., Yamaguchi,T., Dayton,S.H., Beauchamp,R.L., Herrera,G.E., Ishizaki,K., Yamamuro,T., Meyers,P.A., Little,J.B., Sasaki,M.S. et al. (1992) Prevalence and spectrum of germline mutations of the p53 gene among patients with sarcoma. N. Engl. J. Med., 326, 1301–1308. [DOI] [PubMed] [Google Scholar]
- 26.Bonnardeaux A., Davies,E., Jeunemaitre,X., Fery,I., Charru,A., Clauser,E., Tiret,L., Cambien,F., Corvol,P. and Soubrier,F. (1994) Angiotensin II type 1 receptor gene polymorphisms in human essential hypertension. Hypertension, 24, 63–69. [DOI] [PubMed] [Google Scholar]