Abstract
Despite a regain of interest recently in ERK3 kinase signaling, the molecular regulations of both ERK3 gene expression and protein kinase activity are still largely unknown. While it is shown that disruption of ERK3 gene causes neonatal lethality, cell type-specific functions of ERK3 signaling remain to be explored. In this study, we report that ERK3 gene expression is upregulated by cytokines through c-Jun in endothelial cells; c-Jun binds to the ERK3 gene and regulates its transcription. We further reveal a new role for ERK3 in regulating endothelial cell migration, proliferation and tube formation by upregulating SRC-3/SP-1-mediated VEGFR2 expression. The underlying molecular mechanism involves ERK3-stimulated formation of a transcriptional complex involving coactivator SRC-3, transcription factor SP-1 and the secondary coactivator CBP. Taken together, our study identified a molecular regulatory mechanism of ERK3 gene expression and revealed a previously unknown role of ERK3 in regulating endothelial cell functions.
Keywords: ERK3, Cytokines, c-Jun, VEGFR2, SRC-3, SP-1, Endothelial cell functions
Introduction
Angiogenesis is a fundamental process in physiology and pathology, through which new blood vessels form from pre-existing vessels (Risau, 1997). Vascular endothelial growth factors (VEGFs) and their receptors (VEGFRs) play essential roles in angiogenesis (Lohela et al., 2009). There are three VEGFRs, including VEGFR1, VEGFR2, and VEGFR3, and multiple VEGFs, such as VEGFs-A, -B, -C, and –D. VEGFR2 is the primary transducer of VEGF signals in endothelial cells during angiogenesis (Olsson et al., 2006). VEGF/VEGFR signals activate multiple downstream protein kinases such as MAPKs and Akt, subsequently leading to the activation of transcriptional factors (e.g. AP-1 and SP-1) and the expression of target genes including VEGFR2 itself, and eventually promoting endothelial cell proliferation, migration and permeability (Shibuya and Claesson-Welsh, 2006).
Steroid receptor coactivator 3 (SRC-3) is a transcriptional coactivator for nuclear receptors and other types of transcriptional factors including AP-1 and SP-1 (Liao et al., 2002). The transcriptional activity of SRC-3 is regulated by a variety of signals such as hormones, growth factors, and cytokines (Xu et al., 2009). SRC-3 integrates multiple signaling pathways to regulate cell proliferation, migration and invasion (Long et al., 2010; Zhou et al., 2005), and has been characterized as a bona fide oncogene with amplification and/or overexpression in multiple human cancers (Anzick et al., 1997; Tien and Xu, 2012; Torres-Arzayus et al., 2004). In addition to its functions in epithelial cells, SRC-3 was also shown to be important for the functions of endothelial cells including in vivo angiogenesis (Al-Otaiby et al., 2012; Fereshteh et al., 2008; Ying et al., 2008). The molecular mechanisms underlying SRC-3's function in endothelial cells, however, are largely unknown.
ERK3 belongs to the atypical MAPK subfamily (Coulombe and Meloche, 2007). While the signaling pathways of the conventional MAPKs (such as ERK1 and ERK2) are well characterized, little is known about the constitution and regulation of ERK3 signaling cascade (Cargnello and Roux, 2011). As to the downstream targets, ERK3 was shown to phosphorylate MAPK-activated protein kinase 5 (MK5) (Schumacher et al., 2004; Seternes et al., 2004) and SRC-3 (Long et al., 2012), both of which are important downstream mediators of ERK3 signaling in regulating cell migration and invasion (Gerits et al., 2007; Long et al., 2012; Tak et al., 2007). In addition, several studies have demonstrated ERK3's function in cellular differentiation (Coulombe et al., 2003; Hansen et al., 2008; Julien et al., 2003). While these in vitro studies suggest a role for ERK3 in suppressing cell cycle progression, ERK3-deficient mice display intrauterine growth restriction and neonatal lethality mainly due to pulmonary immaturity (Klinger et al., 2009) with loss of differentiation of Type II pneumocytes. The role of ERK3 signaling in angiogenesis, however, remains to be explored. Importantly, both of the two known EKR3 substrates, SRC-3 and MK5, are known to positively regulate angiogenesis (Al-Otaiby et al., 2012; Yoshizuka et al., 2012), suggesting that ERK3 may play a similar role in this process. The functional activity of a protein kinase is regulated by both its gene expression level and its protein enzymatic activation. Similar to the lack of knowledge regarding the molecular regulation of ERK3 kinase activation, it is virtually unknown how ERK3 gene expression is regulated.
In the current study, we have found that ERK3 expression is upregulated by cytokine signals via c-Jun-mediated transcription in primary human umbilical vein endothelial cell (HUVEC); ERK3 stimulates the interactions of SRC-3 and CBP coactivators with SP-1 transcriptional factor, upregulates SRC-3/SP-1-mediated VEGFR2 gene expression, and promotes HUVEC migration, proliferation and tuber formation.
Materials and Methods
Cell culture, transfection and cytokine or growth factor stimulation
Primary human umbilical vein endothelial cells (HUVECs) at passage 2 were purchased from Lonza Inc., USA and were cultured in EGM-2 medium (Lonza) containing 2% FBS along with growth supplements. HeLa cells from ATCC (American Type Culture Collection) were maintained in DMEM (Dulbecco's modified Eagle medium, Invitrogen) supplemented with 10% FBS. Plasmids and siRNAs were transfected into cells using Fugene HD reagent (Roche) and TransIT-TKO Transfection Reagent (Mirus Bio Corporation), respectively, following the manufacturer's instructions. The silencer select siRNAs targeting human ERK3, c-Jun, c-Fos, p65, or p50, and the silencer negative control #1 were purchased from Ambion. SRC-3 siRNA on-target plus SMART pool and non-targeting siRNA pool were purchased from Dharmacon.
For cytokine or growth factor stimulation, cells were starved in EGM-2 basic medium overnight, and then treated with 10 ng/ml of TNF-α, IL1-beta, EGF, bFGF, or vehicle for 6 hours. Cells were then harvested for either RNA or protein extraction.
Real time quantitative reverse transcriptase-PCR (RT-qPCR)
Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer's instruction. cDNA was synthesized from total RNA using SuperScript® III First-Strand Synthesis System (Invitrogen). Real time PCR was performed using TaqMan Probe system (Roche Diagnostics) on the ABI Prism 7700 sequence detector (Applied Biosystems, CA). GAPDH was used as the internal control.
ChIP assay
Chromatin-immunoprecipitation (ChIP) assay was performed using the EZ-Magna ChIP Kit (Upstate/Millipore) following the manufacturer's protocol. The antibodies used for ChIP assay were anti–c-Jun (Cell Signaling), anti-phospho-c-Jun (Cell Signaling), or the normal rabbit IgG (Santa Cruz Biotechnology). The occupancy of c-Jun on ERK3 gene promoter was analyzed by quantitative real-time PCR using SYBR Green technology (Roche Applied Science) with the following primers: the forward primer (5′-TACTTTGCTGAAGGGGATGG-3′) and the reverse primer (5′-AAAAAGCCACGTAGCAGTCC-3′). Primers for non-relevant binding site were as follows: the forward primer (5′-CAAAATAATGCAACGCAGGA-3′) and the reverse primer (5′-TCAAGGCAAGGTTTGTTTC-3′). Results were presented as the percentage of sheared chromatin input.
Luciferase reporter assay
To determine the regulation of ERK3 gene promoter activity by c-Jun, an ERK3 gene fragment (500 bp in length) containing a canonic c-Jun binding site was amplified by PCR using the forward primer [5′-GGCGCTAGCGATCACCTAGGTATTACTGTTGAG-3′] and the reverse primer [5′-GGCGCTAGCGAGAGAGAATTTCAATGTGGACAG-3′] and subcloned into pGL3-promoter luciferase vector (pGL3-promoter-Luc, Promega) via Nhe I site. pGL3-promoter-Luc construct contains a SV40 promoter driving luciferase gene expression. The resulted construct is designated as pGL3-promoter-Luc-BS. pGL3-promoter-Luc or pGL3-promoter-Luc-BS was transfected together with pcDNA3-c-Jun or pcDNA3 empty vector into HUVECs using Fugene 6 Reagent (Promega). Luciferase activity was determined using the Promega luciferase assay kit. Similarly, VEGFR2 gene promoter-driven luciferase reporter assays were performed by expressing pGL3-VEGFR2-225-Luc (Addgene plasmid # 21306) and pEGFP-SP1 (Addgene, plasmid #39325), pSG5ERK3, or pSG5SRC3 construct in HUVECs.
Tube formation/in vitro angiogenesis assay
HUVECs were transfected with the silencer negative control siRNA, c-Jun siRNA, or ERK3 siRNA. 48 hours post-transfection, 1 × 105 cells were plated in each well of the 24-well plate coated with Matrigel (BD Biosciences). 16 hours later, endothelial tube structures were stained with Calcein AM fluorescent dye (BD Biosiences) and tube formation was analyzed using a Nikon inverted microscope at 20X magnification. Tube area was quantified using Image J software.
Two-chamber transwell cell migration assay
Cell migration was analyzed using a modified two-chamber transwell system (BD Biosciences) following the manufacturer's instructions. HUVECs were detached by trypsin-EDTA, washed once with 1X PBS, and then resuspended in serum-free medium. 0.5 ml of complete culture media was added to each bottom well. 1 × 105 cells were added in each transwell insert and allowed to migrate for 12 hrs in a 37°C cell incubator. Cells in the upper surface of the transwell were removed using cotton swabs. Migrated cells attached on the undersurface were fixed with 4% paraformaldehyde for 10 min and stained with crystal violet solution (0.5% in water) for 10 min. Cells were photographed and counted under microscopy at 100 x magnification.
Cell proliferation assay
Cell proliferation was determined using CellTiter 96® AQueous one solution cell proliferation assay kit (Promega) by following the manufacturer's instructions.
Immunoprecipitation and Western blotting
HeLa cells were co-transfected with pSG5SRC-3Flag and pSG5ERK3 or its corresponding empty vector, or together with ERK3 siRNA or the silencer negative control siRNA. 48 hours post-transfection, cells were lysed with EBC lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.5% NP-40, 1 mM PMSF, 1 mM Complete protease inhibitors (Roche Diagnostics), 10 mM NaF, 1 mM sodium orthovanadate, and 1 mM Phosphatase Inhibitor Cocktail III (Sigma)). 1mg of total protein lysate was used for each IP using a Flag antibody or a mouse IgG control. The supernatant was precleared with 40 μl protein A/G agarose beads for 1 hr at 4°C with constant rotation. The samples were then incubated with desired antibody for 2 hrs, followed by the addition of 40 μl protein A/G agarose beads for additional 1 hr. The beads were washed three times (5 min/wash) with lysis buffer. Proteins were boiled off the beads in 2× Laemmili sample buffer and resolved on 4–15% SDS-PAGE gels (BioRad). 2% of the amount of protein supernatant for IP was loaded as the input control. Western blotting was performed by first blocking nitrocellulose membranes with 5% nonfat milk in PBS-T buffer for 30min, followed by overnight incubation with primary antibody at 4°C and 1 hr incubation with appropriate secondary antibody at room temperature. The Western blot was visualized by chemiluminescence (Amersham) and quantified by Image J software (NIH, US). Primary antibodies used in western blotting were: anti-Flag (Sigma), anti-ERK3 (Epitomics), anti-CBP (Cell Signaling), anti-SP1 (Cell Signaling), anti–β-actin (Millipore), anti-ERK1/2 (Cell Signaling), anti-phospho-ERK1/2 (Santa Cruz), and anti-VEGFR2 (Cell Signaling).
Statistics
Results are expressed as mean ± s.e. of three separate experiments unless otherwise indicated. Statistical significance was determined by a 2-sided Student's t test. A P-value less than 0.05 is considered statistically significant. * indicates P <0.05, ** P <0.01, and *** P <0.001.
Results
ERK3 expression is upregulated in HUVECs by TNF-α through c-Jun
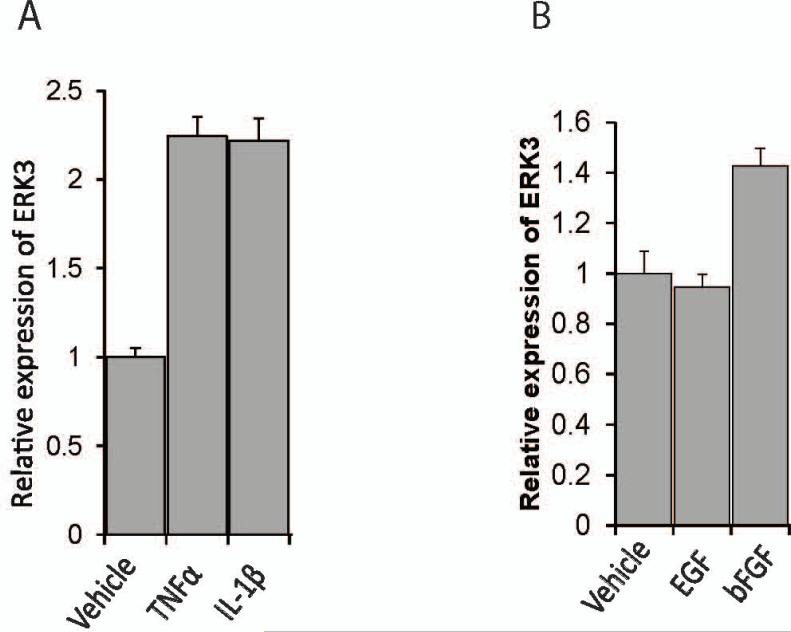
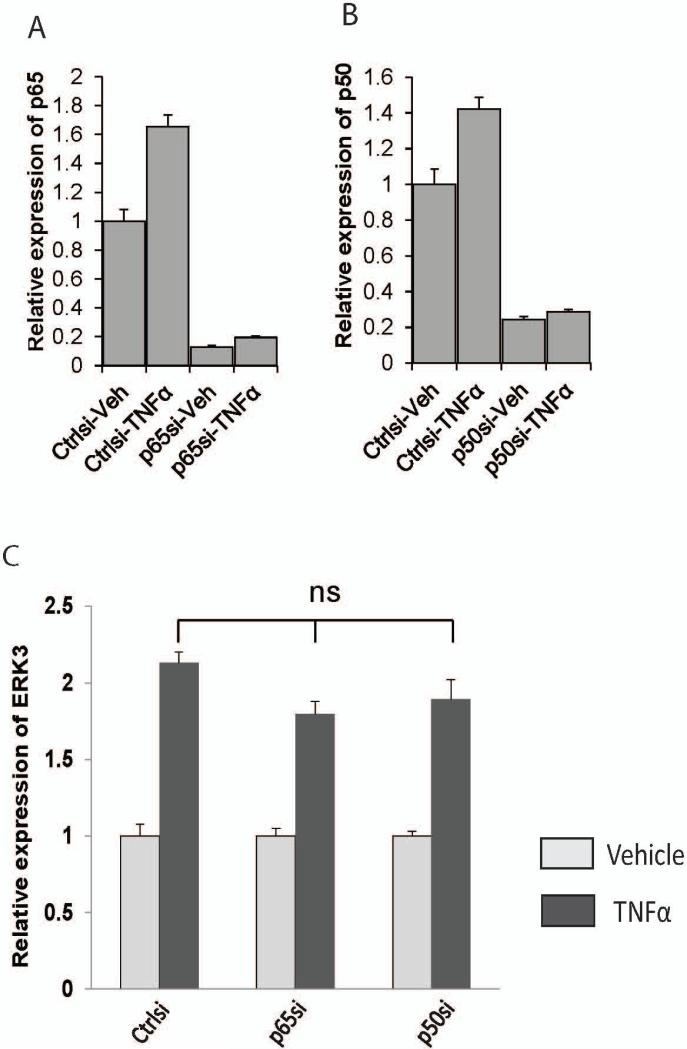
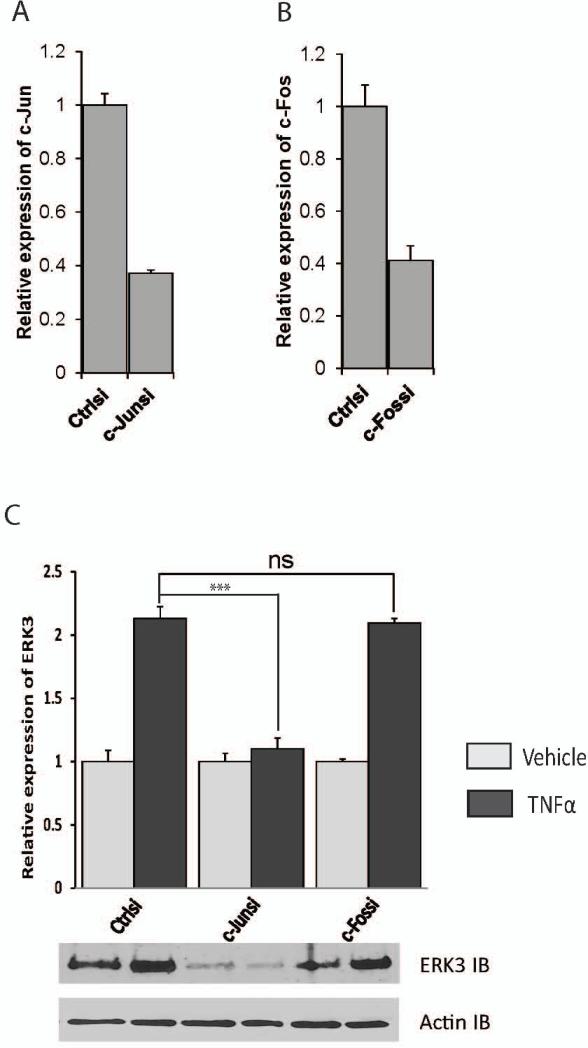
To explore the role of ERK3 in endothelial cells, we first investigated the regulation of ERK3 gene expression in endothelial cells using primary HUVECs. We treated primary HUVECs with a variety of growth factors or cytokines and analyzed the changes of ERK3 gene expression. Both TNF-α and IL-1β cytokines greatly stimulated ERK3 expression (over two-fold, Fig. 1A), whereas bFGF had a modest effect and EGF had little effect (Fig. 1B). TNF-α/IL-1β cytokine signals activate two major families of transcriptional factors (Baud and Karin, 2001; Dunne and O'Neill, 2003): nuclear factor-kappa B (NF-κB) family, including p65, p50 and p52, and the activation protein-1 (AP-1) family consisting of primarily c-Jun and c-Fos. To identify which transcription factor (s) mediates the upregulation of ERK3 in response to the cytokine signal, we compared ERK3 gene expression in HUVECs with or without TNF-α stimulation following transient knockdown of each essential component of these two transcription factor families. Knockdown of either p65 (Fig. 2A) or p50 (Fig. 2B) had no significant effect on TNF-α-stimulated upregulation of ERK3 (Fig. 2C), indicating that the NF-κB pathway is not involved in this regulation. Importantly, knockdown of c-Jun (Fig.3A) nearly abolished the stimulatory effect of TNF-α on ERK3 gene expression both at mRNA and at protein levels (Fig. 3C), whereas knockdown of c-Fos (Fig. 3B) had little effect (Fig. 3C). These results suggest that TNF-α -induced ERK3 gene expression is regulated by c-Jun.
Figure 1. ERK3 expression is stimulated by cytokines.
Serum-starved HUVECs cells were exposed to either cytokines (A), such as TNF-α and IL1-beta, or growth factors (B), such as EGF and bFGF, or the vehicles for 6 hours. ERK3 gene expression was measured by RT-qPCR.
Figure 2. TNFα-induced ERK3 expression is not regulated by NF-κB pathway.
HUVECs were transiently transfected with p65 siRNA (p65si), p50 siRNA (p50si), or the silencer negative control siRNA (Ctrlsi), then serum-starved overnight, followed by the treatment with TNFα or vehicle. Gene expression was determined by RT-qPCR. ns: no significance.
Figure 3. ERK3 expression is regulated by TNFα through c-Jun.
HUVECs were transiently transfected with c-Jun siRNA (c-Junsi), c-Fos siRNA (c-Fossi), or the silencer negative control siRNA (Ctrlsi). Cells were then treated with TNFα (10 ng/ml) or vehicle for 6 hours. Knockdown of c-Jun (A) and c-Fos (B) was verified by RT-qPCR. ERK3 gene expression was determined by both RT-qPCR (Bar-graph) at mRNA level and by Western blotting at protein level (C). ERK3 IB: ERK3 immunoblot.
c-Jun binds to ERK3 gene and regulates its transcriptional activity
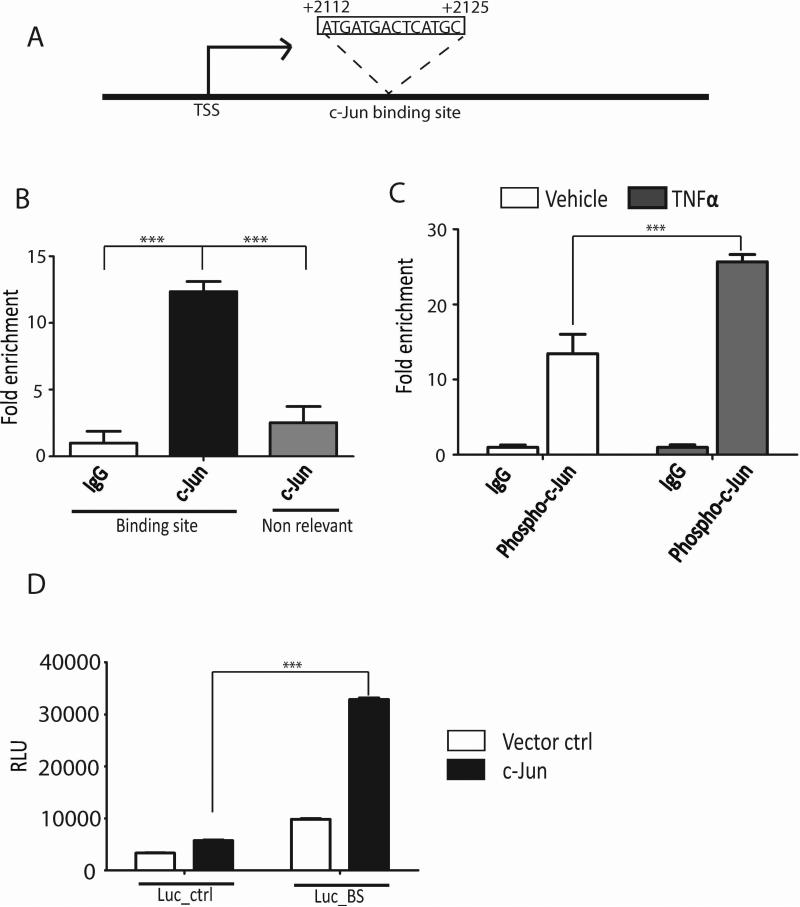
c-Jun is activated by phosphorylation upon the stimulation of TNF-α (Karin et al., 1997). Phosphorylated c-Jun then binds to the promoters of its target genes and regulates transcription. To investigate whether ERK3 gene transcription is regulated directly by c-Jun, we scanned for c-Jun binding site (s) within the ERK3 gene promoter region (from −3 kb to +3 kb of the transcription start site; TSS). A canonic c-Jun binding site (Halazonetis et al., 1988) was found within the fragment spanning +2112 bp to +2125 bp downstream of the TSS (Fig. 4A) and would function as a transcription enhancer as it resides in intron 1 of ERK3 gene. As demonstrated by the chromatin-IP (ChIP) assays using either a c-Jun antibody (Fig. 4B) or a phospho-c-Jun antibody (Fig. 4C), c-Jun binds to the region of ERK3 gene containing this canonic c-Jun binding element. Importantly, TNF-α significantly stimulated the binding of phosphorylated c-Jun to the ERK3 gene (Fig. 4C). To further demonstrate the regulation of ERK3 gene transcription by c-Jun, we cloned a fragment (500 bp in length) containing the c-Jun binding site from ERK3 gene into the pGL3-promoter luciferase vector that contains a SV40 promoter driving luciferase gene expression. Presumably, this ERK3 gene fragment would function as a transcriptional enhancer in this luciferase reporter system. Indeed, expression of c-Jun greatly increased the luciferase activity of pGL3-promoter luciferase plasmid incorporated with the c-Jun binding site from ERK3 gene promoter (Luc-BS, Fig. 4D), but only had a modest effect on the activity of the control reporter vector (Luc-Ctrl, Fig. 4D). Taken together, these results suggest that c-Jun binds to ERK3 gene and regulates its transcriptional activity.
Figure 4. c-Jun binds to ERK3 gene promoter and regulates its transcriptional activity.
(A). A canonic c-Jun binding site exists in intron 1 of ERK3 gene, which presumably functions as a transcription enhancer. TSS: transcription start site. (B). c-Jun is bound to ERK3 gene promoter in HUVECs. ChIP assays were performed using either a c-Jun Ab or goat IgG. The occupancy of c-Jun protein on ERK3 gene promoter region containing the canonic c-Jun binding site or on a non-relevant region was analyzed by quantitative real-time PCR and presented as the percentage of sheared chromatin input. (C). TNF-α enhances the binding of phosphorylated c-Jun to ERK3 gene promoter. HUVECs were stimulated with TNF-α or vehicle control. ChIP assays were performed as shown in (B) except that a phospho-c-Jun Ab was used to analyze the occupancy of phosphorylated c-Jun on ERK3 gene promoter. (D). Luciferase assays in HUVECs transfected with either the pGL3-promoter luciferase vector control (Luc-ctrl) or the pGL3-promoter luciferase plasmid incorporated with the c-Jun binding site from ERK3 gene promoter (Luc-BS), together with c-Jun plasmid or the empty vector control (Vector ctrl). Luciferase activity is represented as relative luciferase units (RLU) on the Y-axis.
ERK3 regulates in vitro angiogenesis of HUVECs by promoting cell migration and proliferation
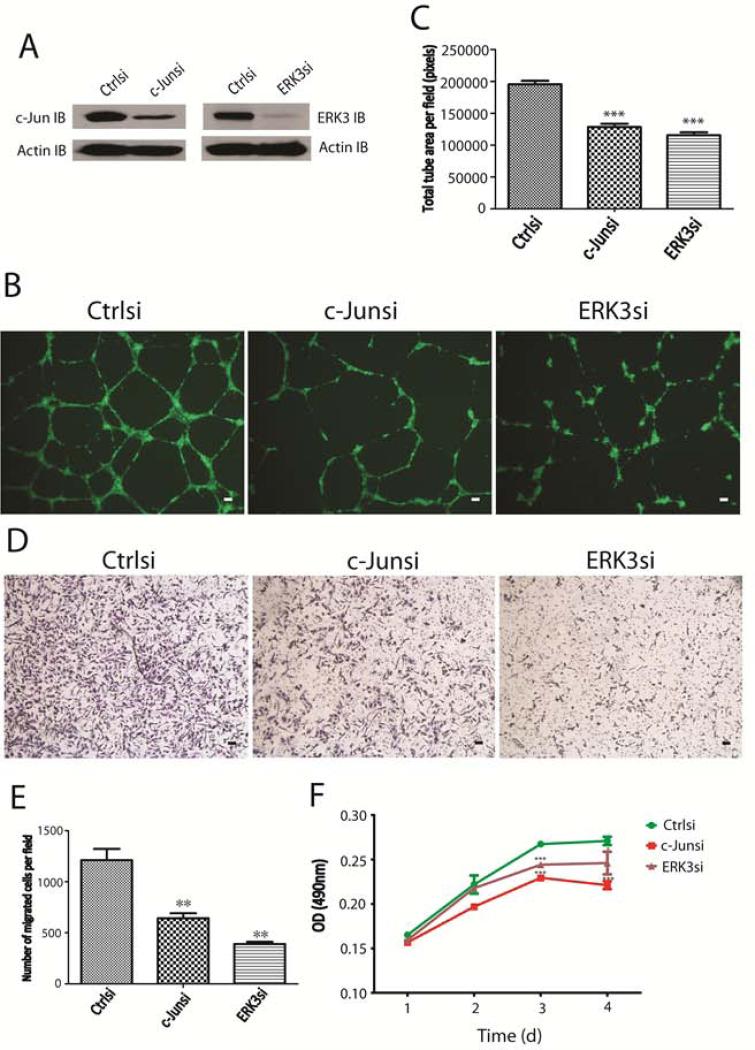
c-Jun functions as a critical factor for angiogenesis (Fahmy et al., 2006). Having identified ERK3 as a target gene of c-Jun, we went further to determine the cellular functions of ERK3 in HUVECs by knocking down either ERK3 or c-Jun (Fig. 5A). As expected, knockdown of c-Jun (c-Junsi) inhibited tube formation (in vitro angiogenesis) of HUVECs in matrigel (Fig. 5B and 5C). Importantly, knockdown of ERK3 (ERK3si) also greatly impaired tube formation of HUVECs (Fig. 5B and 5C). Tube formation is a process involving both cell migration and proliferation (Beck and D'Amore, 1997). Notably, knockdown of either c-Jun or ERK3 significantly inhibited both cell migration (Fig. 5D and 5E) and proliferation (Fig. 5F) of HUVECs, suggesting that ERK3 may regulate tube formation by promoting endothelial cell migration and proliferation.
Figure 5. Knockdown of c-Jun or ERK3 inhibits tube formation of HUVECs by diminishing cell migration and proliferation.
(A). HUVEC cells were transiently transfected with c-Jun siRNA (c-Junsi), ERK3 siRNA (ERK3si), or the silencer negative control siRNA (Ctrlsi). Knockdown of c-Jun and ERK3 was verified by Western blotting using a c-Jun Ab and an ERK3 Ab, respectively. (B) and (C). Knockdown of either c-Jun or ERK3 significantly inhibited tube formation of HUVECs in matrigel. (B): representative images of tube formation of HUVECs treated with different siRNAs. The scale bar represents 10μm. (C): Quantitation of tube formation by measuring the average tube area per field using Image J software. (D) and (E). Knockdown of either c-Jun or ERK3 significantly inhibited migration of HUVECs. Representative images of migration of HUVECs treated with different siRNAs were shown in (D) and quantitative migration ability under each condition was presented as number of migrated cells per filed (E). (F). MTS cell proliferation assay of HUVECs treated with different siRNAs.
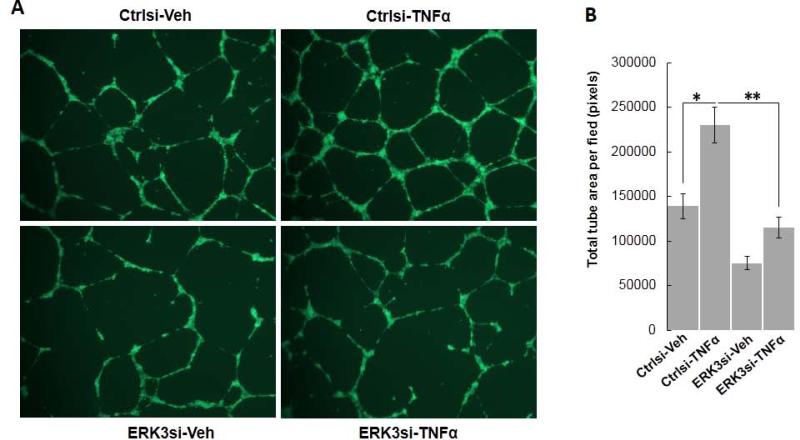
The action of TNFα in endothelial cell functions is complex. Both pro-angiogenic and anti-angiogenic roles of TNFα have been shown (Balkwill, 2009; Leek et al., 1994). Interestingly, a study by (Sainson et al., 2008) elegantly revealed that short-term TNFα treatment (“priming”) stimulated cell migration and angiogenesis, whereas long-term TNFα treatment triggered endothelial cell death and inhibited angiogenesis. Consistent with this report, we found that short-term (3 hours) TNFα treatment significantly stimulated capillary tube formation of HUVECs (Ctrlsi-TNFα versus Ctrlsi-Veh, Fig. 6). Importantly, knock-down of ERK3 greatly inhibited the stimulatory effect of TNFα (Fig. 6), suggesting ERK3 is important for TNFα to stimulate endothelial cell functions.
Figure 6. Knockdown of ERK3 inhibits TNFα-stimulated tube formation of HUVECs.
HUVEC cells were transiently transfected with ERK3 siRNA (ERK3si) or the silencer negative control siRNA (Ctrlsi). Two days later, cells were trypsinized, resuspended in medium containing either 10 ng/ml of TNF-α or vehicle (1 x PBS), then transferred into 24-well plate coated with matrigel. Three hours later, medium was replaced with regular EGM-2 culture medium to stop TNF-α treatment and tube formation by HUVECs resumed in the incubator for another 13 hours. (A). Representative images of tube formation of HUVECs with different treatments. (B). Quantitation of tube formation by measuring the average tube area per field using Image J software.
ERK3 regulates VEGFR2 expression and its signaling
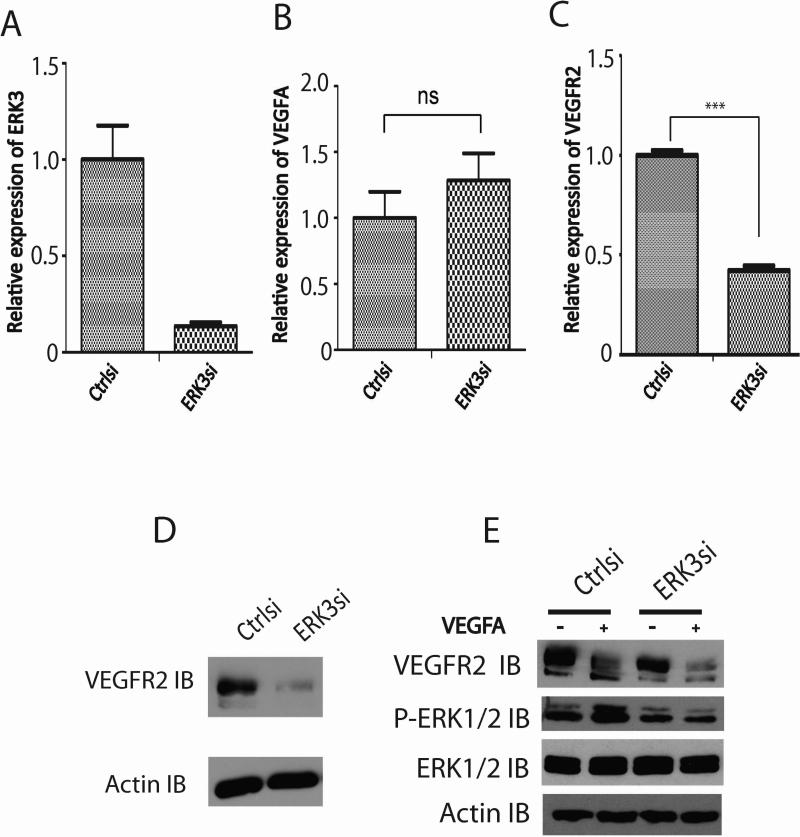
VEGF/VEGFR2 signaling plays a dominant role in endothelial cell function (Olsson et al., 2006). To elucidate the molecular mechanism of ERK3-regulated endothelial cell functions, we determined the role of ERK3 on the expression of VEGFA and VEGFR2. While knockdown of ERK3 (Fig. 7A) had no significant effect on VEGFA expression (Fig. 7B), it caused a significant decrease in VEGFR2 expression at both mRNA level (Fig. 7C) and protein level (Fig. 7D). Accordingly, VEGFR signaling was reduced by the depletion of ERK3 (Fig. 7E), which was demonstrated by the remarkable inhibition of VEGFA-stimulated ERK1/2 phosphorylation (PERK1/2 IB, Fig. 7E) in the cells treated with ERK3 siRNA (ERK3si) in comparison with that in the cells treated with non-targeting control siRNA (Ctrlsi). In conclusion, ERK3 regulates VEGFR2 expression and signaling, which attributes to ERK3's function in regulating endothelial cell proliferation, migration and tube formation.
Figure 7. Knockdown of ERK3 downregulates VEGFR2 expression and signaling.
HUVEC cells were transiently transfected with ERK3 siRNA (ERK3si) or the silencer negative control siRNA (Ctrlsi). (A). Verification of ERK3 knockdown by RT-qPCR. (B). RT-qPCR analysis of VEGFA mRNA expression. ns: no significance. (C). RT-qPCR analysis of VEGFR2 mRNA expression. (D). Western blot analysis of VEGFR2 protein expression. Actin was probed as a loading control. (E). HUVEC cells were transfected with ERK3 siRNA (ERK3si) or the silencer negative control siRNA (Ctrlsi), starved in serum-free medium, and then treated with or without VEGFA (25 ng/ml) for 10 min. Protein expression of VEGFR and ERK1/2 and phosphorylation of ERK1/2 (P- ERK1/2) were analyzed by Western blotting.
ERK3 and SRC3 synergistically regulate SP-1- mediated VEGFR2 gene promoter activity
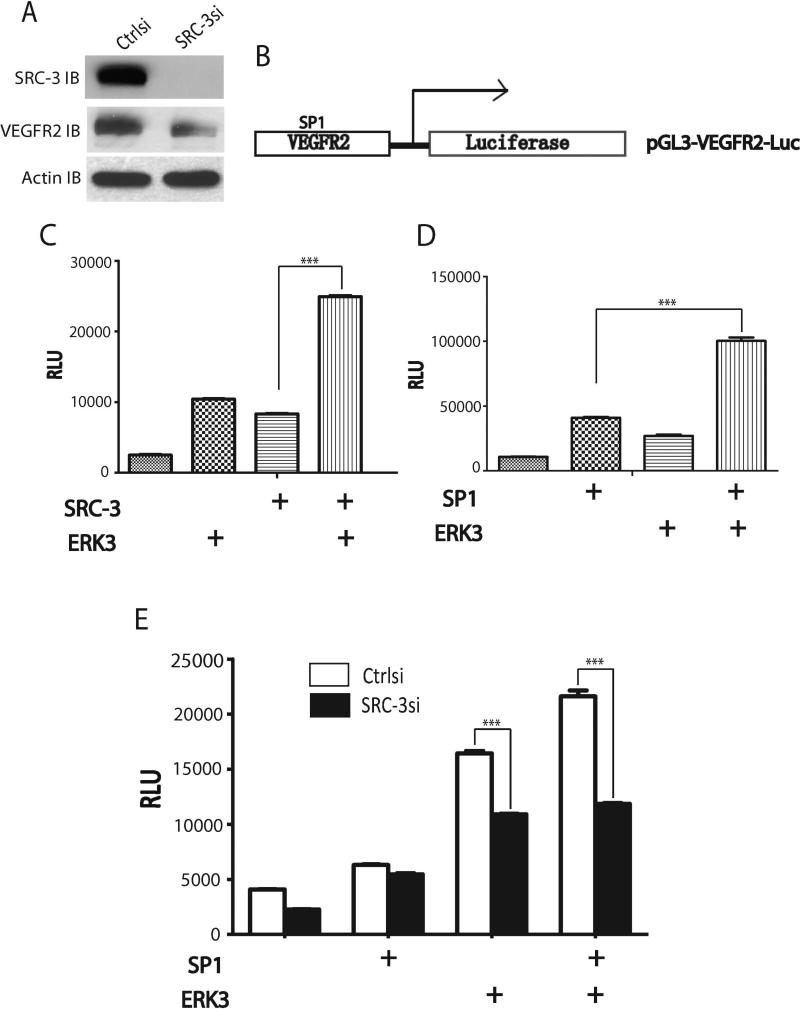
SP1 is a major transcription factor for the regulation of VEGFR2 expression (Meissner et al., 2008; Meissner et al., 2004). SRC-3 plays an important role in regulating endothelial cell functions and was shown to be a coactivator of SP1 (Mussi et al., 2006). It is not known, however, whether SRC-3's function in endothelial cells is through SP1 and whether SRC-3 regulates VEGFR2 expression. Given that ERK3 was shown to phosphorylate SRC-3 and regulate its transcriptional activity (Long et al., 2012), we postulated that the ERK3/SRC-3 signaling axis might regulate SP1-mediated VEGFR2 gene expression. Indeed, depletion of SRC-3 greatly reduced VEGFR2 expression (Fig. 8A). We then performed a VEGFR2 promoter-driven luciferase reporter assay (Fig. 8B) to test whether ERK3 can synergistically work with SRC-3 and/or SP1 to regulate VEGFR2 gene transcription. The VEGFR2 promoter fragment in pGL3-VEGFR2-Luc construct was shown to contain a functional SP1 binding site (Meissner et al., 2004). As shown in Fig. 8C and Fig. 8D, overexpression of ERK3, SRC-3, or SP1 alone greatly increased VEGFR2 gene promoter-driven luciferase activity. More importantly, cooverexpression of ERK3 with either SRC-3 (Fig. 8C) or SP1 (Fig. 8D) further significantly increased luciferase activity. Knockdown of SRC-3 significantly diminished the stimulatory effect of ERK3 and SP1 on VEGFR2 gene promoter-driven luciferase activity (Fig. 8E). Taken together, these results demonstrate that ERK3 and SRC3 synergistically regulate SP1-mediated VEGFR2 gene promoter activity.
Figure 8. ERK3 and SRC-3 synergistically regulate SP1-mediated VEGFR2 gene promoter activity.
(A). HUVEC cells were transfected with SRC-3 siRNA (SRC-3si) or the non-targeting negative control siRNA (Ctrlsi). Expression levels of SRC-3 and VEGFR2 were analyzed by Western blotting. (B). A schematic illustration of a VEGFR2 gene promoter-driven luciferase reporter construct (pGL3-VEGFR2-Luc). A SP1 binding element resides in the VEGFR2 gene promoter. (C) and (D). ERK3, SRC-3 and SP1 synergistically promote VEGFR2 promoter-driven luciferase activity. HUVECs were transfected with pGL3-VEGFR2-Luc and SRC-3, ERK3, or SP1 construct as indicated. Luciferase activity is represented as relative luciferase units (RLU). (E). Knockdown of SRC-3 diminished the stimulatory effect of ERK3 and SP1 on VEGFR2 gene promoter-driven luciferase activity. HUVECs were transfected with pGL3-VEGFR2-Luc and ERK3 or SP1 construct, together with either SRC-3 siRNA (SRC-3si) or non-targeting control siRNA (Ctrlsi) as indicated.
ERK3 promotes the interactions of SRC3 with CBP and SP1
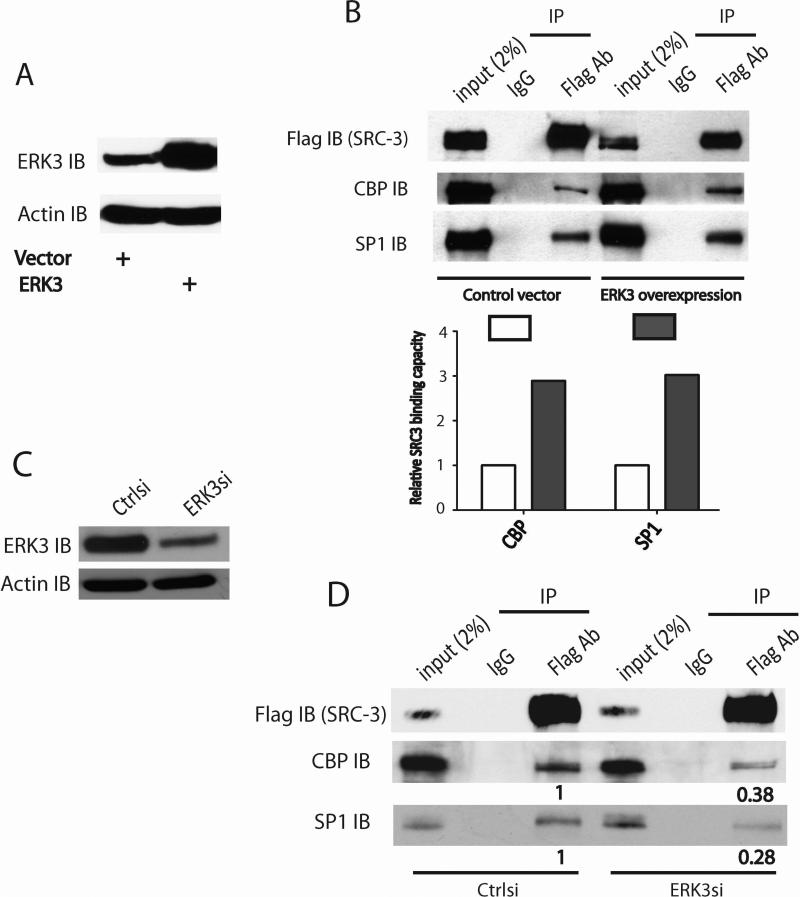
As a scaffolding transcriptional coactivator, SRC-3 binds to DNA-bound transcriptional factors such as SP1 and recruits secondary coactivators such as CREB binding protein (CBP, a histone acetyltransferase) to facilitate gene transcription (Liao et al., 2002). Thus, it is likely that ERK3 regulates SP1-mediated VEGFR2 gene expression by affecting the association of SRC-3 with SP1 and/or CBP. Indeed, overexpression of ERK3 (Fig. 9A) enhanced the interactions of SRC-3 with CBP and SP1 (Fig. 9B). On the contrary, knockdown of ERK3 (Fig. 9C) decreased these interactions (Fig. 9D).
Figure 9. ERK3 enhances the interactions of SRC3 with CBP and SP1.
(A) and (B). Cells were co-transfected with SRC-3Flag and ERK3 or the corresponding empty vector. The interactions of SRC-3 with CBP and SP1 were analyzed by immunoprecipitation (IP) using a Flag Ab or a mouse IgG, followed by Western blotting. The relative SRC-3 binding capacity with CBP or SP1 was determined by measuring the CBP or SP1 immunoblot (IB) band intensity in the IP samples. The band intensity in control vector group was arbitrarily set as “1.0”. Overexpression of ERK3 (A) enhanced the interactions of SRC-3 with CBP and SP1 shown in (B). (C) and (D). Cells were transfected with ERK3 siRNA (ERK3si) or the silencer negative control siRNA (Ctrlsi), together with SRC-3Flag plasmid. The interactions of SRC-3 with CBP and SP1 were analyzed by IP using a Flag Ab, followed by Western blotting. Numbers below the immunoblots of CBP and SP1 in Flag Ab/IP samples represent the relative SRC-3 binding capacity with these proteins, which is determined by the ratio of the band intensity in Flag Ab/IP over that in the corresponding input. For the purpose of comparison, the relative binding capacity (the ratio) in Ctrlsi group was arbitrarily set as “1.0”. Knockdown of ERK3 (C) greatly decreased the association of SRC3 with CBP and SP1 (D).
Discussion
In contrast to the well-studied ERK1 and ERK2, much less is known about ERK3 in regard to the molecular regulation of its signaling (both expression and activation). Although it has been shown that the ERK3 knockout mice display neonatal lethality mainly due to pulmonary immaturity (Klinger et al., 2009), the functions of ERK3 in specific tissues and physiological processes are still largely unknown. In the current study we found that ERK3 gene expression is regulated by cytokines through c-Jun transcription factor and we revealed a previously unknown function for ERK3 in regulating endothelial cell migration, proliferation and angiogenesis. It is worth noting that cytokines induce ERK3 expression not only in endothelial cells but also in macrophages and cancer cells (data not shown), suggesting the regulation of ERK3 by cytokines is probably a general event in mammalian cells. While both IKKs/NFκB and MAPK/AP-1 are the major signaling pathways that are activated by cytokine signals (Baud and Karin, 2001), the induction of ERK3 expression by TNFα is mainly regulated by AP-1-mediated transcription. Interestingly, we found that c-Jun, but not c-Fos, is required for this regulation, demonstrating another level of regulatory specificity in cytokine-induced ERK3 gene expression. In line with c-Jun's function as an angiogenesis-promoting factor (Zhang et al., 2004; Zhang et al., 2006), ERK3, as a c-Jun target, also plays an important role in endothelial cell function including in vitro angiogenesis. To our knowledge, our study is the first revealing a molecular regulatory mechanism of ERK3 gene transcription in response to extracellular signals. In addition, it is of note that while ERK1/2 signaling is highly regulated by growth factors such as EGF, neither activation (Deleris et al., 2008) nor expression of ERK3 (Fig. 1B) is affected by this growth factor.
SRC-3 was identified as a target of ERK3 kinase in our recent study (Long et al., 2012). SRC-3, as a coactivator for nuclear receptors and a variety of other transcriptional factors, plays an important role in endothelial cell function and neoangiogenesis (Al-Otaiby et al., 2012; Fereshteh et al., 2008). The molecular mechanisms by which SRC-3 regulates endothelial cell function, however, remain to be explored. SRC-3 acts as a scaffolding factor by recruiting secondary factors such as CBP/p300 to DNA-bound transcriptional factors and facilitating gene transcription. Here we found that ERK3 promotes the interactions of SRC-3 with CBP and the transcription factor SP-1 and facilitates SRC-3/SP-1-mediated transcription of VEGFR2, a primary VEGF receptor essential for endothelial cell function. The enhanced functional interaction of this transcriptional complex by ERK3 can be explained by our previous findings that ERK3 phosphorylates and activates SRC-3 (Long et al., 2012) so that SRC-3 can interact with CBP and other coregulators and transcription factors (Wu et al., 2004).
To date, SRC-3 and MK5 are the only two identified substrates for ERK3 kinase (Long et al., 2012; Seternes et al., 2004). As both MK5 and SRC-3 now have been shown to promote tumor angiogenesis (Al-Otaiby et al., 2012; Yoshizuka et al., 2012), it would be interesting to know whether ERK3 has a similar angiogenesis-promoting function during tumor progression. ERK3 has been shown to be upregulated in multiple cancers and its upregulation is frequently associated with tumor metastasis (Nambiar et al., 2005; Pitulescu et al., 2010). Consistent with these clinical findings, we have recently found that ERK3 promotes cancer cell invasiveness (Long et al., 2012). Taken together, our current and previous findings raise an intriguing possibility that ERK3 functions in multiple compartments of the tumor microenvironment: in the tumor compartment, it promotes cancer cell migration and invasion; and in the stromal compartment, it promotes tumor-associated endothelial cell function and neoangiogenesis, both of which might synergistically contribute to tumor progression and metastasis.
Acknowledgements
We would like to thank the laboratory's colleagues, including Dr. Subhamoy Dasgupta, Dr. Ping Yi and Dr. Amber Johnson, for providing reagents and critical discussion. This work was supported by the grants NIH 5R01HD-8188, HD07857, and Texas CPRIT to B.W.O.
Literature Cited
- Al-Otaiby M, Tassi E, Schmidt MO, Chien CD, Baker T, Salas AG, Xu J, Furlong M, Schlegel R, Riegel AT, Wellstein A. Role of the nuclear receptor coactivator AIB1/SRC-3 in angiogenesis and wound healing. Am J Pathol. 2012;180(4):1474–1484. doi: 10.1016/j.ajpath.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277(5328):965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- Balkwill F. Tumour necrosis factor and cancer. Nat Rev Cancer. 2009;9(5):361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- Baud V, Karin M. Signal transduction by tumor necrosis factor and its relatives. Trends Cell Biol. 2001;11(9):372–377. doi: 10.1016/s0962-8924(01)02064-5. [DOI] [PubMed] [Google Scholar]
- Beck L, Jr., D'Amore PA. Vascular development: cellular and molecular regulation. FASEB J. 1997;11(5):365–373. [PubMed] [Google Scholar]
- Cargnello M, Roux PP. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol Mol Biol Rev. 2011;75(1):50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe P, Meloche S. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim Biophys Acta. 2007;1773(8):1376–1387. doi: 10.1016/j.bbamcr.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Coulombe P, Rodier G, Pelletier S, Pellerin J, Meloche S. Rapid turnover of extracellular signal-regulated kinase 3 by the ubiquitin-proteasome pathway defines a novel paradigm of mitogen-activated protein kinase regulation during cellular differentiation. Mol Cell Biol. 2003;23(13):4542–4558. doi: 10.1128/MCB.23.13.4542-4558.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris P, Rousseau J, Coulombe P, Rodier G, Tanguay PL, Meloche S. Activation loop phosphorylation of the atypical MAP kinases ERK3 and ERK4 is required for binding, activation and cytoplasmic relocalization of MK5. J Cell Physiol. 2008;217(3):778–788. doi: 10.1002/jcp.21560. [DOI] [PubMed] [Google Scholar]
- Dunne A, O'Neill LA. The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE. 2003;2003(171):re3. doi: 10.1126/stke.2003.171.re3. [DOI] [PubMed] [Google Scholar]
- Fahmy RG, Waldman A, Zhang G, Mitchell A, Tedla N, Cai H, Geczy CR, Chesterman CN, Perry M, Khachigian LM. Suppression of vascular permeability and inflammation by targeting of the transcription factor c-Jun. Nat Biotechnol. 2006;24(7):856–863. doi: 10.1038/nbt1225. [DOI] [PubMed] [Google Scholar]
- Fereshteh MP, Tilli MT, Kim SE, Xu J, O'Malley BW, Wellstein A, Furth PA, Riegel AT. The nuclear receptor coactivator amplified in breast cancer-1 is required for Neu (ErbB2/HER2) activation, signaling, and mammary tumorigenesis in mice. Cancer Res. 2008;68(10):3697–3706. doi: 10.1158/0008-5472.CAN-07-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits N, Mikalsen T, Kostenko S, Shiryaev A, Johannessen M, Moens U. Modulation of F-actin rearrangement by the cyclic AMP/cAMP-dependent protein kinase (PKA) pathway is mediated by MAPK-activated protein kinase 5 and requires PKA-induced nuclear export of MK5. J Biol Chem. 2007;282(51):37232–37243. doi: 10.1074/jbc.M704873200. [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Georgopoulos K, Greenberg ME, Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988;55(5):917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- Hansen CA, Bartek J, Jensen S. A functional link between the human cell cycle-regulatory phosphatase Cdc14A and the atypical mitogen-activated kinase Erk3. Cell Cycle. 2008;7(3):325–334. doi: 10.4161/cc.7.3.5354. [DOI] [PubMed] [Google Scholar]
- Julien C, Coulombe P, Meloche S. Nuclear export of ERK3 by a CRM1-dependent mechanism regulates its inhibitory action on cell cycle progression. J Biol Chem. 2003;278(43):42615–42624. doi: 10.1074/jbc.M302724200. [DOI] [PubMed] [Google Scholar]
- Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9(2):240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- Klinger S, Turgeon B, Levesque K, Wood GA, Aagaard-Tillery KM, Meloche S. Loss of Erk3 function in mice leads to intrauterine growth restriction, pulmonary immaturity, and neonatal lethality. Proc Natl Acad Sci U S A. 2009;106(39):16710–16715. doi: 10.1073/pnas.0900919106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek RD, Harris AL, Lewis CE. Cytokine networks in solid human tumors: regulation of angiogenesis. J Leukoc Biol. 1994;56(4):423–435. doi: 10.1002/jlb.56.4.423. [DOI] [PubMed] [Google Scholar]
- Liao L, Kuang SQ, Yuan Y, Gonzalez SM, O'Malley BW, Xu J. Molecular structure and biological function of the cancer-amplified nuclear receptor coactivator SRC-3/AIB1. J Steroid Biochem Mol Biol. 2002;83(1-5):3–14. doi: 10.1016/s0960-0760(02)00254-6. [DOI] [PubMed] [Google Scholar]
- Lohela M, Bry M, Tammela T, Alitalo K. VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr Opin Cell Biol. 2009;21(2):154–165. doi: 10.1016/j.ceb.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Long W, Foulds CE, Qin J, Liu J, Ding C, Lonard DM, Solis LM, Wistuba, Tsai SY, Tsai MJ, O'Malley BW. ERK3 signals through SRC-3 coactivator to promote human lung cancer cell invasion. J Clin Invest. 2012;122(5):1869–1880. doi: 10.1172/JCI61492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long W, Yi P, Amazit L, LaMarca HL, Ashcroft F, Kumar R, Mancini MA, Tsai SY, Tsai MJ, O'Malley BW. SRC-3Delta4 mediates the interaction of EGFR with FAK to promote cell migration. Mol Cell. 2010;37(3):321–332. doi: 10.1016/j.molcel.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner M, Pinter A, Michailidou D, Hrgovic I, Kaprolat N, Stein M, Holtmeier W, Kaufmann R, Gille J. Microtubule-targeted drugs inhibit VEGF receptor-2 expression by both transcriptional and post-transcriptional mechanisms. J Invest Dermatol. 2008;128(8):2084–2091. doi: 10.1038/jid.2008.37. [DOI] [PubMed] [Google Scholar]
- Meissner M, Stein M, Urbich C, Reisinger K, Suske G, Staels B, Kaufmann R, Gille J. PPARalpha activators inhibit vascular endothelial growth factor receptor-2 expression by repressing Sp1-dependent DNA binding and transactivation. Circ Res. 2004;94(3):324–332. doi: 10.1161/01.RES.0000113781.08139.81. [DOI] [PubMed] [Google Scholar]
- Mussi P, Yu C, O'Malley BW, Xu J. Stimulation of steroid receptor coactivator-3 (SRC-3) gene overexpression by a positive regulatory loop of E2F1 and SRC-3. Mol Endocrinol. 2006;20(12):3105–3119. doi: 10.1210/me.2005-0522. [DOI] [PubMed] [Google Scholar]
- Nambiar S, Mirmohammadsadegh A, Doroudi R, Gustrau A, Marini A, Roeder G, Ruzicka T, Hengge UR. Signaling networks in cutaneous melanoma metastasis identified by complementary DNA microarrays. Arch Dermatol. 2005;141(2):165–173. doi: 10.1001/archderm.141.2.165. [DOI] [PubMed] [Google Scholar]
- Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat Rev Mol Cell Biol. 2006;7(5):359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- Pitulescu ME, Schmidt I, Benedito R, Adams RH. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat Protoc. 2010;5(9):1518–1534. doi: 10.1038/nprot.2010.113. [DOI] [PubMed] [Google Scholar]
- Risau W. Mechanisms of angiogenesis. Nature. 1997;386(6626):671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- Sainson RC, Johnston DA, Chu HC, Holderfield MT, Nakatsu MN, Crampton SP, Davis J, Conn E, Hughes CC. TNF primes endothelial cells for angiogenic sprouting by inducing a tip cell phenotype. Blood. 2008;111(10):4997–5007. doi: 10.1182/blood-2007-08-108597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher S, Laass K, Kant S, Shi Y, Visel A, Gruber AD, Kotlyarov A, Gaestel M. Scaffolding by ERK3 regulates MK5 in development. EMBO J. 2004;23(24):4770–4779. doi: 10.1038/sj.emboj.7600467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seternes OM, Mikalsen T, Johansen B, Michaelsen E, Armstrong CG, Morrice NA, Turgeon B, Meloche S, Moens U, Keyse SM. Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J. 2004;23(24):4780–4791. doi: 10.1038/sj.emboj.7600489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya M, Claesson-Welsh L. Signal transduction by VEGF receptors in regulation of angiogenesis and lymphangiogenesis. Exp Cell Res. 2006;312(5):549–560. doi: 10.1016/j.yexcr.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Tak H, Jang E, Kim SB, Park J, Suk J, Yoon YS, Ahn JK, Lee JH, Joe CO. 14-3-3epsilon inhibits MK5-mediated cell migration by disrupting F-actin polymerization. Cell Signal. 2007;19(11):2379–2387. doi: 10.1016/j.cellsig.2007.07.016. [DOI] [PubMed] [Google Scholar]
- Tien JC, Xu J. Steroid receptor coactivator-3 as a potential molecular target for cancer therapy. Expert Opin Ther Targets. 2012;16(11):1085–1096. doi: 10.1517/14728222.2012.718330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Arzayus MI, Font de Mora J, Yuan J, Vazquez F, Bronson R, Rue M, Sellers WR, Brown M. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6(3):263–274. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O'Malley BW. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol Cell. 2004;15(6):937–949. doi: 10.1016/j.molcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Xu J, Wu RC, O'Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9(9):615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Willingham MC, Cheng SY. The steroid receptor coactivator-3 is a tumor promoter in a mouse model of thyroid cancer. Oncogene. 2008;27(6):823–830. doi: 10.1038/sj.onc.1210680. [DOI] [PubMed] [Google Scholar]
- Yoshizuka N, Chen RM, Xu Z, Liao R, Hong L, Hu WY, Yu G, Han J, Chen L, Sun P. A novel function of p38-regulated/activated kinase in endothelial cell migration and tumor angiogenesis. Mol Cell Biol. 2012;32(3):606–618. doi: 10.1128/MCB.06301-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Dass CR, Sumithran E, Di Girolamo N, Sun LQ, Khachigian LM. Effect of deoxyribozymes targeting c-Jun on solid tumor growth and angiogenesis in rodents. J Natl Cancer Inst. 2004;96(9):683–696. doi: 10.1093/jnci/djh120. [DOI] [PubMed] [Google Scholar]
- Zhang G, Fahmy RG, diGirolamo N, Khachigian LM. JUN siRNA regulates matrix metalloproteinase-2 expression, microvascular endothelial growth and retinal neovascularisation. J Cell Sci. 2006;119(Pt 15):3219–3226. doi: 10.1242/jcs.03059. [DOI] [PubMed] [Google Scholar]
- Zhou HJ, Yan J, Luo W, Ayala G, Lin SH, Erdem H, Ittmann M, Tsai SY, Tsai MJ. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65(17):7976–7983. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]