Abstract
Patients with resistance to thyroid hormone (RTH) exhibit elevated thyroid hormone levels and inappropriate thyrotropin (thyroid-stimulating hormone, or TSH) production. The molecular basis of this disorder resides in the dominant inhibition of endogenous thyroid hormone receptors (TRs) by a mutant receptor. To determine the relative contributions of pituitary versus hypothalamic resistance to the dysregulated production of thyroid hormone in these patients, we developed a transgenic mouse model with pituitary-specific expression of a mutant TR (Δ337T). The equivalent mutation in humans is associated with severe generalized RTH. Transgenic mice developed profound pituitary resistance to thyroid hormone, as demonstrated by markedly elevated baseline and non–triodothyronine (T3)-suppressible serum TSH and pituitary TSH-β mRNA. Serum thyroxine (T4) levels were only marginally elevated in transgenic mice and thyrotropin-releasing hormone (TRH) gene expression in the paraventricular hypothalamus was downregulated. After TRH administration, T4 concentrations increased markedly in transgenic, but not in wild-type mice. Transgenic mice rendered hypothyroid exhibited a TSH response that was only 30% of the response observed in wild-type animals. These findings indicate that pituitary expression of this mutant TR impairs both T3-mediated suppression and T3-independent activation of TSH production in vivo. The discordance between basal TSH and T4 levels and the reversal with TRH administration demonstrates that resistance at the level of both the thyrotroph and the hypothalamic TRH neurons are required to elevate thyroid hormone levels in patients with RTH.
Introduction
Resistance to thyroid hormone (RTH) is a dominantly inherited condition with affected individuals harboring mutations in the β isoforms of the thyroid hormone receptor (TR) (1). Nearly all affected individuals are heterozygous for the mutant allele, and the molecular basis of the syndrome is presumed to be interference of wild-type TR function by the mutant TR (2, 3). Under normal circumstances, thyroid hormone concentrations are regulated by a feedback loop in which thyroid hormones inhibit the production of thyrotropin-releasing hormone (TRH) by the hypothalamus and thyrotropin (TSH) production by the thyrotrophs in the anterior pituitary (4–9). This pathway is critically dependent upon the presence of normal TRs, which bind to the promoters of TRH and TSH subunit genes and regulate their expression (10–14). In the presence of its ligand, triodothyronine (T3), TRs mediate ligand-dependent repression of the transcription of these genes, and in the absence of T3, the transcription rate is not simply returned to baseline, but ligand-independent activation is observed (15–17).
Patients with RTH have varying degrees of insensitivity to T3-mediated inhibition of TSH production (1). The impact of RTH on TSH regulation in the absence of thyroid hormone is unknown. Although it has been presumed that the basis for the endocrine abnormalities is the presence of mutant TRs in the pituitary and the hypothalamic TRH neurons, the relative roles of the pituitary versus the hypothalamus in the pathogenesis of RTH remains unknown. Most of the insight into the molecular basis of RTH has been obtained from experiments using transfected mutant TRs in various cell lines. It is not always possible to generalize or apply the observations derived from these experiments to thyroid hormone–responsive cells in the in vivo milieu. Thus, targeted expression of mutant thyroid hormone receptors to the hypothalamus or to the thyrotrophs of transgenic mice should give valuable insight into the roles of these compartments in the genesis of the resistance phenotype.
A few in vivo models of RTH currently exist. Adenoviral gene transfer of a mutant TR (G345R) into mice has recapitulated some of the hepatic abnormalities observed in RTH (18). The same group also expressed this receptor transgenically in the pituitaries of mice using a fragment of the TSH-β promoter to drive transgene expression (19). Mild increases in thyroxine (T4) concentrations with a tendency toward lower TSH concentrations were reported in the F1 transgenic mice. Unfortunately, the elevated T4 phenotype was lost in subsequent generations. Hence, it is difficult to know whether or not pituitary resistance truly existed in this model. Transgenic mice with ubiquitous overexpression of the frameshift mutant PV were reported to develop hyperactivity and mild increases in T4 concentrations (20). The pituitary expression of the transgene was variable and the extent of hypothalamic expression unknown. Finally, mice with targeted ablation of the β isoform of the TR exhibit impaired T3-mediated suppression of TSH, which underscores the important role of TR-β in the regulation of thyrotropin (21). TR-β knockout mice, however, are a model of recessive resistance and are therefore pathophysiologically distinct from the human syndrome (1, 21).
Hence, to gain further insight into the impact of mutant TR expression on the hypothalamic-pituitary-thyroid axis, we targeted a mutant TR associated with generalized RTH selectively to the pituitary in transgenic mice. Extensive characterization of the hypothalamic-pituitary-thyroid axis has been performed, and insights into the relative roles of the pituitary and the hypothalamus in the RTH phenotype have been obtained. These mice have also provided the opportunity to assess in vivo the impact of the mutant TR on ligand-dependent, as well as ligand-independent, functions of the endogenous receptor.
Methods
Animals.
All aspects of animal care and experimentation performed in this study were approved by the Institutional Animal Care and Use Committee of the Beth Israel Deaconess Medical Center. Animals were maintained on a 12-h light/12-h dark schedule (light on at 0600) and fed laboratory chow and water ad libitum. Blood was obtained for total thyroxine (T4) and thyrotropin (TSH) from the tail vein. Experimental hypothyroidism was induced by administering 150 μCi of iodine-131 by intraperitoneal injection to mice that had been placed on a low-iodine diet (Harlan Teklad, Madison, Wisconsin, USA) for 8 days. Experiments on hypothyroid animals were performed three weeks after the administration of iodine-131, because all mice develop undetectable T4 concentrations by the third week. Suppression of pituitary TSH production was attempted by daily intraperitoneal injections of T3 (1 μg/ml) in buffered HEPES for 3 weeks at doses of 0.2 μg/100 g mouse body weight during the first week, 0.5 μg/100 g mouse body weight during the second week, and 1.0 μg/100 g mouse body weight during the third week. All comparisons were made between transgenic mice and littermate controls to minimize the effect of genetic variability.
Generation of transgenic mice.
The naturally occurring mutation Δ337T (22, 23) was introduced into the human TR-β1 cDNA by site-directed mutagenesis (CLONTECH Laboratories Inc., Palo Alto, California, USA), and the presence of the mutant was confirmed by direct sequencing. The ability of this cDNA to produce an intact protein was confirmed by in vitro translation and by the ability of the proteins to bind to thyroid hormone response elements (TREs) in gel mobility shift assays (data not shown). The cDNA was initially subcloned as an EcoRI fragment into the expression vector pGEX4T2 (Pharmacia Biotech, Piscataway, New Jersey, USA). The transgene was made by cloning a 4.6-kb promoter fragment of the mouse α-subunit gene (24) (gift of University of Colorado Health Sciences Center, Denver, Colorado, USA) into the KpnI and HindIII sites of the expression plasmid pREP7 (Invitrogen Corp., San Diego, California, USA), which contains the SV40 polyadenylation sequence downstream of its multiple cloning site. The mutant TR cDNA was removed from pGEX4T2 as a BamHI–NotI fragment and then cloned downstream of the α-promoter fragment, immediately proximal to the SV40 polyA. The 6.2-kb transgene was released from this plasmid by KpnI and SalI restriction digests and gel purified by electroelution (Fig. 1a). In the Beth Israel Transgenic Facility, approximately 4 ng of the purified transgenic vector was microinjected into the male pronuclei of 1-day-old mouse zygotes of the inbred strain FVB/N and reimplanted into the uteri of pseudopregnant foster mothers. Litters were obtained after 21 days. Between the age of 14 and 18 days, preweaned mice were identified by sex and marked by earlobe punching. Then, 15 mm of tail was removed for genomic DNA extraction. Genomic DNA was extracted by overnight digestion with proteinase K and SDS. After high-salt precipitation of the SDS, phenol/chloroform extraction was performed and the DNA obtained by ethanol precipitation. Genomic DNA (10 μg) was digested with BamHI, subjected to agarose gel electrophoresis, and transferred to nylon membranes (Micron Separations Inc., Westboro, Massachusetts, USA). Southern blotting was performed using the entire human TR-β1 cDNA as a probe. Transgenic mice were identified by the presence of a specific 1.6-kb fragment.
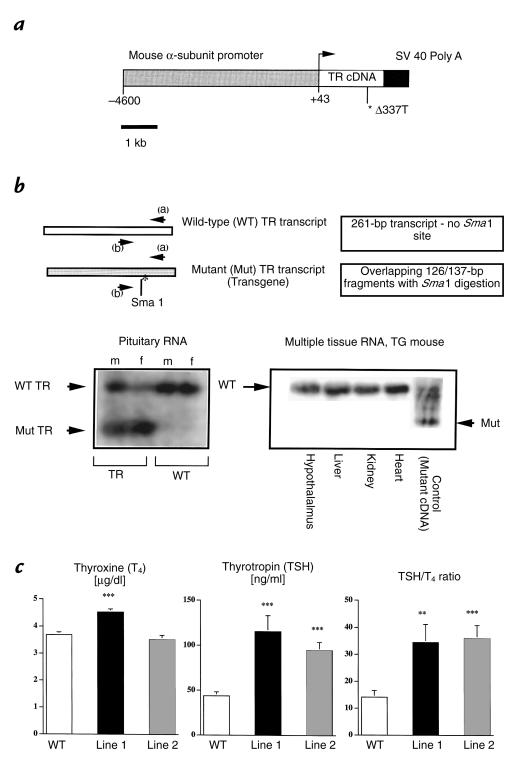
Figure 1.
Transgenic construct, pituitary expression of the transgene, and impact on basal thyroid hormone concentrations. (a) Schematic representation of the transgene. The vector contains 4.6 kb of the mouse glycoprotein subunit α gene cloned upstream of the human TR-β1 cDNA harboring the mutation Δ337T and ligated to a 400-bp fragment containing the SV40 polyadenylation signal. (b) RT-PCR scheme for detecting transgene expression and expression of the mutant and wild-type TR transcripts in mouse pituitaries and other tissues. RNA was obtained from pituitary, hypothalamus, liver, kidney, and heart, and analyzed by RT-PCR using primers (denoted a and b) that are homologous both to the human and mouse TR sequences. These primers amplify 261 bp of the carboxyl-terminus of the human and mouse TR-β gene. A polymorphism in the human gene results in the presence of a Sma1 restriction site that is not present in the mouse gene. Restriction digestion with this enzyme results in the appearance of 126/137-bp overlapping fragments indicating transgene expression. Gels shown were performed after Sma1 digestion of the RT-PCR products. Note that the smaller transcript appears only in the pituitaries of TG mice but is absent from WT mouse pituitaries and from multiple other tissues, including the hypothalamus in TG mice. Data is shown for line 1 mice; similar results were obtained for line 2 mice, which had 50% lower pituitary transgene expression vs. line 1. (c) Basal total T4, TSH concentrations, and TSH/T4 ratios (means ± SEM) in the WT and TG mice from two independent transgenic founder lines (1 and 2). The number of animals used for T4 determinations are WT: n = 89; line 1: n = 84; and line 2: n = 39, respectively. The number of animals used for TSH determinations are WT: n = 51; line 1: n = 40; and line 2: n = 46. The number of samples on which T4 and TSH were simultaneously measured for the purpose of calculating the TSH/T4 ratio are WT: n = 24; line 1: n = 26; and line 2: n = 26. Note the small increase in T4 concentration in line 1 and the lack of T4 elevation in line 2, despite large increases in TSH concentration. **P < 0.01 vs. WT; ***P < 0.0001 vs. WT. RT, reverse transcription; T4, thyroxine; TG, transgenic; TSH, thyroid-stimulating hormone; WT, wild-type.
RNA analysis.
RNA was extracted from pooled pituitaries (5–8 animals), using guanidinium thiocyanate, selective precipitation, and isopycnic centrifugation with lithium chloride and cesium trifluoroacetate (Pharmacia Biotech). Wild-type TR versus transgene expression was established by performing semiquantitative reverse transcription (RT)-PCR on DNAseI-treated samples using the following primers: 5′-GGCCAGCTGAAAAATGGGGG-3′ and 5′-TGTCACCTTCATCAGGAGTTT-3′, which amplify 261 bp of the COOH-terminus of TR-β. The size of the wild-type and transgenic PCR products are identical. However, the mutant human transcript contains a SmaI restriction site that is absent in the wild-type murine transcript. The appearance of overlapping 126- and 137-bp fragments upon SmaI restriction digestion of the PCR product is indicative of transgene expression. The PCR products were resolved on a 3% agarose gel, transferred to nylon membranes, and hybridized with the human and mouse PCR products. TSH-β gene expression was assayed in pituitary total RNA by Northern blotting. In hypothyroidism studies, serial dilutions of total RNA was performed, and 5, 2.5, 1.25, and 0.625 μg of RNA were resolved on a 1.2% formaldehyde agarose gel and transferred to a nylon membrane. The membrane was hybridized to the entire cDNA of the β subunit of murine TSH under high-stringency conditions. In studies of hyperthyroid mice, 15 μg of RNA was used.
Localization and quantification of preprothyrotropin (prepro)-TRH mRNA by in situ hybridization histochemistry.
Male wild-type and transgenic mice (n = 3 per group) were anesthetized with an intraperitoneal injection of sodium pentobarbital. They were perfused transcardially with PBS prepared with diethylpyrocarbonate (DEPC)-treated water, followed by 10% neutral buffered formalin. Brains were removed, immersed in the same fixative overnight, and then cryoprotected in 20% sucrose in PBS-DEPC at 4°C. Five series of 20-μm coronal free-floating sections were cut on a tabletop cryotome and mounted on Superfrost Plus glass slides (Fisher Scientific, Pittsburgh, Pennsylvania, USA). The protocol for in situ hybridization histochemistry has been described previously (25). Prepro-TRH mRNA was localized using a sulfur-35 UTP-labeled complementary RNA probe synthesized from a plasmid containing the cDNA for the mouse prepro-TRH gene (26) (kind gift of Masatomo Mori, Gunma University, Maebashi, Japan). Control sections were hybridized with a sense cRNA probe. The slides were exposed to Kodak Biomax MR film (Eastman Kodak Co. Scientific Imaging Systems, Rochester, New York, USA) for 2 days, after which they were dipped in Kodak NTB2 photographic emulsion and developed after 4 days of exposure; then they were counterstained with cresyl violet acetate. Sections were analyzed with a Zeiss Axioplan light microscope using bright and dark-field optics. Dark-field photomicrographs were taken with Kodak daylight slide film and the images printed in black and white using a dye sublimation printer (Kodak 8600). Film autoradiograms of the hypothalamic paraventricular nucleus (PVN) and thalamic reticular nucleus, corresponding to bregma –0.58 mm, –0.82 mm, and –1.22 mm, respectively (Figures 36, 38, and 41 of ref. 27), from three wild-type and three transgenic mice were selected, coded, and analyzed blindly by laser densitometry (Molecular Dynamics, Sunnyvale, California, USA). Integrated densities of prepro-TRH mRNA in each half of the brain were summed, and mean densities in the PVN and reticular nucleus were determined in each animal.
Radioimmunoassay.
Total T4 levels were measured in 10-μl serum samples in duplicate determinations by a mouse-specific RIA (ImmuChem coated tube-T4 iodine-125 RIA Kit; ICN Pharmaceuticals, Costa Mesa, California, USA). TSH was measured in 25-μl serum samples in triplicate determinations by a specific mouse TSH RIA using a mouse TSH/luteinizing hormone (LH) reference preparation (AFP51718mp), a mouse TSH antiserum (AFP98991), and rat TSH antigen for radioiodination (NIDDK-rTSH-I-9). All reagents were obtained from A.F. Parlow (Harbor University of California at Los Angeles Medical Center, Torrance, California, USA). The standard curve was performed in hyperthyroid mouse serum, and the limit of sensitivity was <20 ng/ml. The inter- and intra-assay variations were less than 6%.
TRH stimulation.
TRH stimulation was performed by implanting pellets impregnated with 1 mg of TRH (Innovative Research of America, Sarasota, Florida, USA) into the interscapular space. Blood was subsequently withdrawn from the tail vein for determination of total T4 at 0, 6, 24, 48, and 72 h after pellet insertion. TSH was assayed on the 0-, 6-, and 24-h samples.
Statistical analysis.
Differences were assessed by one-tailed Student's t test (two sample unequal variance).
Results
Generation of transgenic mice and expression level of transgene.
Because of the negligible effect of the transgene on basal T4 concentrations, founder lines were initially screened by measuring total T4 levels after three weeks of T3 suppression. Three lines exhibited incompletely suppressed T4 levels. Two of these lines, designated 1 and 2, respectively, are analyzed in detail and reported in this paper. Transgene expression was documented by RT-PCR (Fig. 1b). Endogenous TR expression was downregulated in transgenic mice when compared with wild-type controls. The relative abundance of mutant TR to wild-type TR transcripts determined from multiple assays was approximately 1.5:1. Transgene expression was approximately twofold higher in line 1 versus line 2. Mutant TR expression was assayed by RT-PCR in the hypothalamus and in peripheral tissues (heart, liver, kidney). Expression of the transgene outside of the pituitary was not seen (Fig. 1b).
Basal T4 and TSH concentrations.
Mean total T4 concentrations were increased by approximately 20% in mice from line 1 but were not increased in mice from line 2 (Fig. 1c). In contrast to the small effect of the transgene on T4 concentrations, TSH concentrations were increased by greater than twofold in transgenic mice from both lines (Fig. 1c). The disparity between TSH and T4 concentrations suggests an impairment in the bioactivity of the TSH produced by transgenic versus wild-type mice and is evidenced by the significant increase in the TSH/T4 ratio observed (Fig. 1c).
Analysis of in vivo regulation of thyrotroph function by T3.
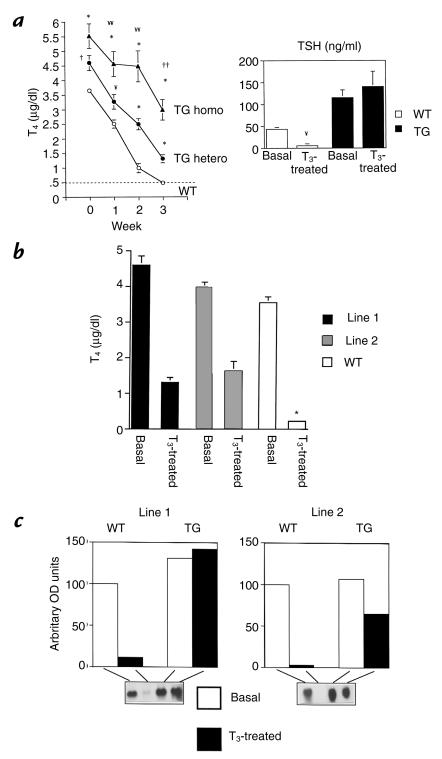
Administration of T3 suppresses endogenous TSH secretion and ultimately T4 production by the thyroid gland. We therefore sought to determine the sensitivity of the hypothalamic-pituitary-thyroid axis in transgenic mice by measuring total T4 concentrations at weekly intervals during three weeks of intraperitoneal administration of pharmacological doses of T3. TSH concentrations were measured at the end of the T3 treatment period. In wild-type mice, T3 administration resulted in a progressive decline in T4 concentrations, and serum T4 levels were not detectable by the end of three weeks. In contrast, thyroxine concentrations were only partially suppressible in transgenic mice. Compared with wild-type mice, T4 concentrations were significantly higher in mice (line 1) harboring one transgenic allele at all time points (Fig. 2a). Line 2 mice demonstrated a similar pattern of impaired T4 suppressibility, despite the fact that they did not have any increase in baseline thyroxine concentration (Fig 2b). To assess whether increasing transgene expression resulted in increased resistance, mice from line 1 were bred to homozygosity for the transgenic allele. Homozygous animals developed more severe T3 resistance. The increase in basal T4 concentrations was not significantly higher than the values seen in heterozygotes. However, in response to exogenously administered T3, the degree of T4 suppression was significantly blunted when compared with heterozygous and wild-type mice (Fig. 2a). To confirm that the nonsuppressibilty of T4 was due to impaired suppression of TSH, serum TSH concentrations and pituitary TSH-β mRNA were measured before and after T3 suppression. In wild-type mice, TSH concentrations were suppressed below the detection limit of the TSH RIA after T3 administration. This contrasts with line 1 transgenic mice in which no measurable reduction in TSH concentrations could be discerned (Fig. 2a). In line 2 mice, TSH concentrations fell by 50% but were not suppressible. TSH-β gene expression followed the pattern of serum TSH. Line 1 mice revealed complete absence of TSH-β mRNA suppression, and line 2 mice revealed a suppression of 40% (Fig 2c). In contrast, TSH-β mRNA was nearly completely obliterated by T3 administration in wild-type mice. These data indicate that transgenic mice from both lines exhibited profound pituitary resistance to thyroid hormone.
Figure 2.
Effect of T3 administration on serum T4, TSH concentrations, and pituitary TSH-β mRNA. (a) Total T4 concentrations (line 1) obtained at weekly intervals during the administration of pharmacological doses of T3 for 3 weeks. The hatched line represents the limit of detection of the T4 RIA. Mice with homozygous allelic expression of the transgene (TG homo; n = 5) were compared with TG mice harboring one transgenic allele (TG hetero; n = 13) and WT mice (n = 21). TSH concentrations were determined at baseline and at the end of 3 weeks in 11 heterozygous TG mice and 8 WT mice. Note that T4 and TSH are undetectable in WT mice after 3 weeks. In contrast, TG mice reveal only partial suppression of T4 and lack of suppression of TSH, indicating pituitary resistance. For the T4 data: ¥¥P < 0.01 and ††P< 0.0001, TG homo vs. TG hetero; ¥P < 0.01, †P< 0.001, *P < 0.0001, TG homo and hetero vs. WT. For TSH data: ¥P < 0.01, basal vs. treated WT. (b) Comparison of T4 concentrations before and after the administration of T3 in line 1 and line 2 TG mice. Note that despite basal thyroid hormone concentrations in line 2 that were similar to those of WT mice, T4 concentrations in line 2 are only partially suppressed to a level that is similar to that observed in line 1. TSH concentrations in line 2 mice were also partially suppressed to levels that were equivalent to untreated WT mice (44 ng/ml) after 3 weeks of T3 administration (data not shown). *P < 0.0001 compared with T3 treated values in lines 1 and 2. (c) TSH-β mRNA expression in pituitaries of T3 treated mice from both lines. Total RNA from pooled pituitaries (six mice per group) was analyzed by Northern blotting. The blots were initially hybridized with the mouse TSH-β cDNA and then with cyclophillin. Densitometry was used to determine mRNA abundance and cyclophyllin expression used to adjust the TSH-β message for differences in loading. TSH mRNA abundance is expressed in arbitrary OD units with WT basal expression normalized to 100. Note complete suppression of TSH-β message in WT mice, lack of suppression in line 1 mice, and partial suppression in line 2 mice. T3, triiodothyronine.
Analysis of the role of TRH.
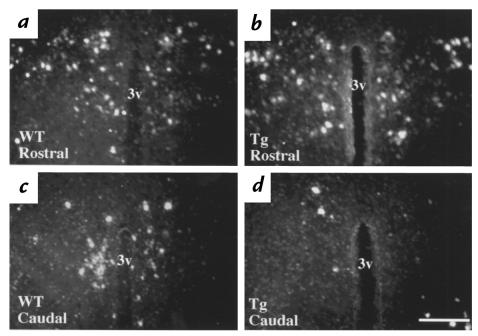
The mutant TR in our transgenic model was targeted only to the pituitary, and given the modest effect of the transgene on T4 concentrations, we speculated that increased TRH production may play a critical role in elevating thyroid hormone concentrations in RTH. To assess the impact of pituitary transgene expression on hypothalamic TRH gene expression, in situ hybridization histochemistry was performed on the brains of line 1 and line 2 transgenic mice and littermate controls. A comparison of the distribution of neurons expressing prepro-TRH mRNA in the rostral and caudal PVN of wild-type (Fig. 3, a and c, respectively) and transgenic mice (Fig. 3, b and d, respectively) reveal evidence of downregulated TRH gene expression in distinct subsets of TRH neurons in transgenic mice. The level of prepro-TRH mRNA in the rostral PVN of transgenic mice was similar to wild-type mice (compare Fig. 3, a and b). In contrast, prepro-TRH mRNA was markedly decreased in the caudal PVN of transgenic mice from both lines (compare Fig. 3, c and d). Densitometric analysis of film autoradiograms showed an overall decrease in PVN prepro-TRH mRNA content of 35% in transgenic mice from line 1 and 55% in transgenic mice from line 2 (P < 0.001). We also examined prepro-TRH mRNA content in the reticular thalamic nuclei of transgenic mice, which contain a population of TRH neurons previously demonstrated to lack T3 regulation in rats, and found no change in TRH gene expression (data not shown).
Figure 3.
(a–d) Representative dark-field photomicrographs showing prepro-TRH mRNA at two rostral and caudal levels in the PVN of mice. (a and c) WT mice. (b and d) TG mice. The rostral PVN is represented by a and b, and c and d represent the caudal PVN. Note the paucity of labeled cells in the caudal PVN of the TG mouse. As shown in this data from line 1, a similar degree of downregulation of prepro-TRH expression in the caudal PVN was observed in line 2 mice. 3v (third ventricle): scale bar = 300 μm. PVN, paraventricular hypothalamic nuclei; TRH, thyrotropin-releasing hormone.
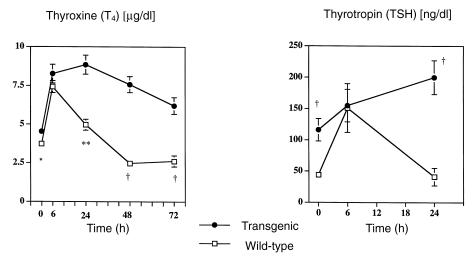
To test the hypothesis that TRH downregulation was minimizing the phenotype in transgenic mice, TSH and T4 responses to TRH administration were examined in transgenic and wild-type mice. Six hours after the insertion of the TRH-containing pellet, both transgenic and wild-type mice exhibited increased concentrations of T4 and TSH (Fig. 4). This increase was transient in wild-type mice, and their concentrations returned to baseline within 24 hours. In contrast, TSH concentrations of transgenic mice were still rising at 24 hours, and this was mirrored by a sustained increase in T4 concentrations that persisted beyond 72 hours (Fig. 4).
Figure 4.
Effect of TRH administration. Serum T4 and TSH concentrations, before and after the implantation of a 1-mg sustained release TRH pellet. Nineteen TG and 18 WT mice were studied. Basal T4 and TSH concentrations were obtained in all mice. To minimize the stress of repeated bleeding, and given the volume of serum required, TSH and T4 concentrations were obtained in a subset of mice (12 TG and 12 WT) at 6 h and 24 h. The remaining 7 TG and 6 WT mice were bled daily starting at 24 h for T4 only. Mice from both lines were studied, and combined data are shown. Note that TG and WT mice both show an initial T4 and TSH response to TRH, but the response is sustained beyond 24 h in TG mice at a time when values have returned to normal in WT animals. For T4 data: *P < 0.05, **P < 0.001, †P < 0.0001, TG vs WT. For TSH data: †P < 0.0001, TG vs. WT.
TSH response to hypothyroidism.
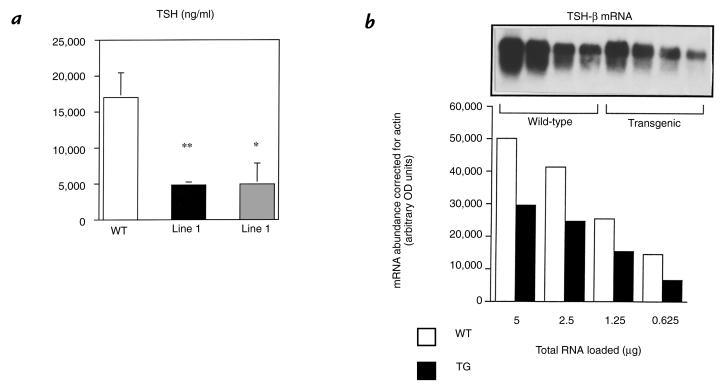
Three weeks after radioactive iodine ablation, all animals studied had thyroxine levels that were at or below the level of detection of the T4 RIA and had an associated robust increase in serum TSH concentrations. Wild-type mice increased their TSH more than 400-fold, but the response in transgenic mice was blunted, with only a 40-fold increase in TSH concentration (Fig. 5a). To determine whether these changes reflected changes at the level of TSH subunit gene expression, a Northern blot of pituitary total RNA obtained from hypothyroid animals was performed and probed with the TSH-β cDNA. In concordance with the changes in serum TSH, the TSH-β mRNA response to hypothyroidism was also blunted in transgenic mice (Fig. 5b).
Figure 5.
Effect of transgene expression on ligand-independent activation of TSH. (a) TSH concentrations were measured 3–4 weeks after the mice were rendered hypothyroid by iodine-131 administration. TSH levels rose markedly in TG (40-fold) and WT (>400-fold) when compared with their respective baseline values. However, the rise in TSH was 10-fold greater in WT mice. Number of animals in each group are WT: n = 13; line 1: n = 9; and line 2: n = 5. **P < 0.01, *P < 0.05 vs. WT. (b) TSH-β mRNA in WT and TG hypothyroid mice (pooled pituitaries). A Northern blot was performed using serial dilutions of pituitary total RNA. Data from line 1 are shown. Similar data (not shown) exist for line 2 mice. The blots were initially hybridized with the mouse TSH-β cDNA, and then with actin. Densitometry determined mRNA abundance, and actin expression was used to adjust the TSH-β message for differences in loading. TSH-β mRNA abundance was 40% lower in TG vs. WT mice.
Discussion
We have successfully generated a transgenic mouse model of selective pituitary resistance to thyroid hormone. The mutant TR Δ337T does not bind thyroid hormone and is associated in humans with a severe phenotype of generalized resistance to thyroid hormone (22). The promoter fragment used to drive pituitary transgene expression was previously shown to direct expression selectively in thyrotrophs and gonadotrophs (24). The expression of the transgene in the gonadotrophs did not appear to have any effect on fertility in these mice. In two lines with independent sites of transgene integration, the phenotype observed was one of severe in vivo thyrotroph resistance to T3, but with a small effect on baseline T4 concentration. Analysis of these mice provide novel insight into the relative contributions of the pituitary and hypothalamus to the neuroendocrine abnormalities associated with RTH.
Basal concentrations of thyroxine are determined by TSH stimulation of thyroid follicular cells. As TSH concentration rises, there is an associated increase in thyroid hormone synthesis and production (28). TSH production in the thyrotroph is regulated by feedback inhibition by circulating thyroid hormone and via stimulation by hypothalamic TRH (4). There is some evidence that TRH enhances the biologic activity of TSH by modifying its glycosylation pattern (29–31). This was recently confirmed in mice with targeted ablation of the TRH gene who exhibit T4 levels that were 50% of those in wild-type mice, despite TSH concentrations that were elevated twofold (32). In our mice, TSH concentrations were significantly elevated, which confirms the important role of pituitary TRs in regulating basal TSH output. The TSH elevation likely reflects impairment of endogenous TR-mediated regulation of TSH gene expression by the mutant TR transgene at ambient thyroid hormone concentrations. Paradoxically, T4 levels were not proportionately increased. The discrepancy between TSH levels and T4 production (as evidenced by the increased TSH/T4 ratio in transgenic mice) could reflect a primary abnormality in the thyroids of transgenic mice. This is extremely unlikely given the robust T4 response to TRH administration observed in transgenic mice. A more plausible explanation is that the bioactivity of the TSH produced in transgenic animals is reduced. This contrasts with patients with RTH who have been shown to produce a TSH with enhanced bioactivity (33). It is likely, then, that the difference between our mice and patients with RTH reside in differences in TRH production.
Since the mutant TR was selectively targeted to the pituitary, but not to the hypothalamus, basal TRH production in our transgenic mice should not be increased. In fact, when we directly assayed for hypothalamic TRH expression, we found marked downregulation of prepro-TRH mRNA in the neurons of the caudal paraventricular hypothalamus. Mapping of the distribution of T3-responsive TRH neurons in the PVN has been performed in the rat (7, 34, 35). In rats, the distribution of the T3 responsive neurons are restricted to the medial parvocellular neurons of the PVN (6, 7). Mice do not possess the same discreet hypothalamic nuclear architecture as rats, and as such, the exact location of T3 responsive neurons in the mouse are unknown. In this study, we have been able to demonstrate regional differences in TRH gene expression within the PVN of mice and show that the overall prepro-TRH mRNA content in the PVN is diminished in transgenic mice. The basis for the downregulation of TRH gene expression is likely due to the small increase in thyroxine concentrations observed in transgenic mice. This observation also suggests that the hypothalamic TRH neurons possess exquisite sensitivity to small changes in thyroid hormone concentrations, as evidenced by the downregulation of prepro-TRH gene expression in both lines of transgenic mice.
If TRH overproduction is an important component of the RTH phenotype, we hypothesized that chronic administration of TRH would result in further elevations in T4 concentrations in transgenic mice. Persistent elevations in T4 in response to TRH was indeed observed in transgenic mice. This suggests that resistance at the level of the hypothalamus plays an important role in sustaining the elevated thyroid hormone concentrations that are seen in RTH. There are several possible explanations for the augmented T4 response of the transgenic mice. Because of impaired T3-mediated gene regulation, the thyrotroph in the transgenic mice might respond to TRH administration as would a hypothyroid pituitary with an exaggerated TSH response. In addition, TRH could be differentially changing the glycosylation pattern of TSH in a way that could prolong the half-life of TSH. Because of the small amounts of TSH that can be extracted from mouse serum, we have not directly assayed for TSH glycosylation. However, previous published data in rats and humans render this hypothesis plausible (29–31). Changes in the glycosylation of TSH could also impact its bioactivity. Hence, increased bioactivity of TSH could also be playing a role in the elevated T4 concentrations observed.
The fall in T4 concentration observed in wild-type mice after T3 administration reflects, in large part, decreased thyroid gland T4 synthesis occurring in response to TSH inhibition. The possibility exists, however, that accelerated catabolism of T4 in response to T3-induced hyperthyroidism could account, in part, for its decline (36, 37). Transgenic mice displayed incomplete suppression of T4, lack of TSH inhibition in line 1, and partial TSH inhibition in line 2. The presence of the mutant TR in the pituitary clearly prevents the inhibition of TSH production in the face of T3 administration. It is intriguing that the partial suppression in T4 levels seen in transgenic mice appears greater than the corresponding change in TSH. Two possible explanations for this can be entertained. First, it is possible that T3 administration further impairs the bioactivity of TSH through its effect on TRH neurons, which retain their sensitivity to thyroid hormone. Second, the decline observed in transgenic mice may reflect the effect of increased clearance of T4 occurring in response to T3-induced hyperthyroidism; the fact that we were unable to completely suppress T4 levels reflects the persistent effect of the nonsuppressed TSH.
The molecular mechanisms governing negative regulation of thyrotroph TSH production are unclear but are thought to involve interactions between the TR and other nuclear proteins in a ligand-dependent manner (38). In this study we observed a direct relationship between transgene expression and resistance. Line 2 mice had lower transgene expression than line 1. This is the likely reason for the milder phenotype as evidenced by partial suppression of serum TSH and pituitary TSH-β mRNA by T3, which contrasts with complete lack of suppression in line 1. Furthermore, doubling of the transgene dose in homozygote line 1 mice produced a greater degree of resistance than that seen in heterozygotes. There are a number of possible explanations for the relationship between increasing transgene expression and pituitary resistance. There could simply be increasing amounts of the mutant TR bound to the promoters of the TSH subunit genes. This non–T3-binding mutant would then be incapable of mediating T3-mediated inhibition of TSH subunit gene transcription. The Δ337T mutant TR constitutively binds to the nuclear corepressor protein NCoR, and the binding is not relieved by T3 (39). Hence, one mechanism for resistance is that the aberrant interaction between the mutant TR and nuclear corepressors is not relieved by thyroid-hormone binding, and this prevents the recruitment of other cofactors responsible for mediating inhibition. Downregulation of endogenous TR expression was observed in heterozygous transgenic mice. Additional downregulation of the endogenous TR by the mutant TR in homozygous transgenic mice could also contribute to the dose-related increase in pituitary resistance.
Our mice also exhibited impaired ligand-independent activation of TSH-β gene expression in vivo. This finding is the first demonstration that ligand-independent activation of TSH production can be modulated in vivo and raises interesting questions about potential mechanisms. Impaired ligand-independent activation was demonstrated both at the level of measured serum TSH and at the level of TSH-β gene expression. The apparent greater differences in serum TSH, as compared with TSH-β gene expression, likely represents differences in the kinetics and half-life of TSH protein in the serum versus that of TSH-β mRNA in the pituitary. A simple explanation for the deficient TSH response to hypothyroidism may be diminished thyrotroph reserve. Transgenic mice under basal conditions have TSH concentrations that are two- to threefold higher than wild-type mice. This chronic overproduction of TSH could render the thyrotroph incapable of mounting a vigorous TSH response to hypothyroidism. Transgenic mice are still able, however, to increase their TSH some 40-fold in the face of hypothyroidism. Although this is 10-fold less than the wild-type response, our data indicate that they retain a significant degree of responsiveness to the withdrawal of ligand. Hence, we feel that thyrotroph failure might not be the sole explanation for this phenomenon. Another contributing factor could be differential degrees of TRH induction by hypothyroidism in transgenic versus wild-type mice. We know from the TRH administration experiments that the transgenic mice exhibit an augmented response to TRH administration; thus, any deficiency in TRH rise in transgenic mice resulting from the lower baseline expression would tend to be offset by the enhanced responsiveness of the thyrotrophs of the transgenic mice to TRH.
Potential molecular mechanisms for the impaired ligand-independent activation of the TSH subunit genes observed in this study are also worthy of discussion. Recent reports have attempted to address the role of nuclear corepressors in the ligand-independent activation of the TRH and TSH subunit genes, but the results are controversial (17, 40). Hollenberg et al. cloned a truncated NCoR molecule (NCoRI) that retains the interacting domains but lacks the repressing domains of full-length NCoR. In transfection experiments, NCoRI acts as a dominant inhibitor of endogenous NCoR. They reported that transfection of this NCoR inhibitor results in enhanced ligand-independent activation of the TRH gene by TR-β1 and TR-α, but not TR-β2 (40). Their data imply that by competitively inhibiting NCoR, ligand-independent activation by the TR-β1 and TR-α isoforms could be enhanced, and suggest that the TR-β2 isoform may mediate ligand-independent activation via an NCoR-independent mechanism. If this is true in the pituitary, then two potential explanations for our in vivo observations become apparent. First, the mutant TR used in our studies was expressed as TR-β1. If NCoR binding limits ligand-independent activation, then the degree of activation that could be mediated by this mutant that constitutively binds to NCoR would be expected to be less than that of the wild-type TR-β2. In addition, the downregulation of the endogenous TR by the mutant transgene could further limit the degree of ligand-independent activation mediated by the wild- type TRs. It will, therefore, be important to determine whether expression of this mutation as TR-β2 will produce a similar effect on ligand-independent activation of the TSH subunit genes.
In transfection experiments with full-length NCoR, Tagami et al. reported that cotransfection of NcoR augmented ligand-independent activation of the TSH-α, TSH-β, and the TRH genes by TR-β1. In addition, a mutant TR with defective NCoR binding was defective in ligand-independent activation (17). In contrast to the studies of Hollenberg et al., they postulated that nuclear corepressor proteins may act to enhance ligand-independent TSH-β gene transcription and that TR binding to DNA may not be required. If the Tagami hypothesis is correct, we would expect that the Δ337T mutant used in our transgenic mice should mediate enhanced ligand-independent activation, because it constitutively binds to NCoR (39). Our observation of impaired ligand-independent activation of TSH transcription does not support their hypothesis. One potential reason for this could be differences in the content of additional regulatory cofactors between the embryonic kidney cell lines in which their transfections were performed and the pituitary in vivo. The possibility also remains that the Δ337T mutant TR interferes with additional pathways that govern ligand-independent activation of the TSH subunit genes by TR. Hence, it will be important to perform similar in vivo expression experiments using other RTH mutant TRs with different transcriptional defects than that described for Δ337T (41–46). These studies should shed light on whether the defective ligand-independent activation of the TSH subunit genes observed with the Δ337T mutant in vivo is can be generalized to other mutant TRs associated with RTH.
In summary, we have successfully demonstrated that a transgenic approach can be used to study the effect of mutant TR expression on pituitary function in vivo. The model can therefore be used for the in vivo analysis of other mutant TRs that may possess alternative transcriptional defects or are associated with varying clinical phenotypes. We have demonstrated that pituitary expression of the TR Δ337T mutant results in impaired T3-mediated repression, as well as impaired T3-independent activation of TSH production. Furthermore, the discordance between basal TSH and thyroxine levels and the reversal with TRH administration, demonstrate that resistance at the level of both the pituitary and the hypothalamus is required to elevate thyroid hormone levels in patients with RTH. We suggest that central resistance, rather than pituitary resistance, may be a more appropriate term to describe the etiology of elevated thyroid hormone levels in patients with RTH.
Acknowledgments
We thank W.W. Wood and E.C. Ridgway for providing the mouse α-subunit promoter, M. Mori for providing the prepro-TRH cDNA plasmid, and Joseph Kelley for technical assistance. The work was supported by National Institutes of Health (NIH) grants DK02485 (to E.D. Abel), DC 03441 (to D. Forrest), and DK49126 and DK50564 (to F.E. Wondisford). E.D. Abel was a recipient of a Faculty Development Award from the Robert Wood Johnson Foundation, the Eleanor and Miles Shore 50th Anniversary Scholars in Medicine Fellowship (Harvard Medical School), and a Thyroid Research Advisory Council Award. H.C. Kaulbach was supported by NIH training grant T32 DK07516. This work was also supported, in part, by the Human Frontiers Science Program and a Sinsheimer Scholarship to D. Forrest. A. Campos-Barros was supported by grant PF 97 00679951 of the Spanish Ministry of Education and Culture.
References
- 1.Refetoff S, Weiss RE, Usala SJ. The syndromes of resistance to thyroid hormone. Endocr Rev. 1993;14:348–399. doi: 10.1210/edrv-14-3-348. [DOI] [PubMed] [Google Scholar]
- 2.Yen PM, Chin WW. Molecular mechanisms of dominant negative activity by nuclear hormone receptors. Mol Endocrinol. 1994;8:1450–1454. doi: 10.1210/mend.8.11.7877614. [DOI] [PubMed] [Google Scholar]
- 3.Jameson JL. Thyroid hormone resistance: pathophysiology at the molecular level. J Clin Endocrinol Metab. 1992;74:708–711. doi: 10.1210/jcem.74.4.1548331. [DOI] [PubMed] [Google Scholar]
- 4.Larsen PR. Thyroid-pituitary interaction: feedback regulation of thyrotropin secretion by thyroid hormones. N Engl J Med. 1982;306:23–32. doi: 10.1056/NEJM198201073060107. [DOI] [PubMed] [Google Scholar]
- 5.Segerson TP, et al. Thyroid hormone regulates TRH biosynthesis in the paraventricular nucleus of the rat hypothalamus. Science. 1987;238:78–80. doi: 10.1126/science.3116669. [DOI] [PubMed] [Google Scholar]
- 6.Kakucska I, Rand W, Lechan RM. Thyrotropin-releasing hormone gene expression in the hypothalamic paraventricular nucleus is dependent upon feedback regulation by both triiodothyronine and thyroxine. Endocrinology. 1992;130:2845–2850. doi: 10.1210/endo.130.5.1572297. [DOI] [PubMed] [Google Scholar]
- 7.Dyess EM, et al. Triiodothyronine exerts direct cell-specific regulation of thyrotropin-releasing hormone gene expression in the hypothalamic paraventricular nucleus. Endocrinology. 1988;123:2291–2297. doi: 10.1210/endo-123-5-2291. [DOI] [PubMed] [Google Scholar]
- 8.Wood WM, Kao MY, Gordon DF, Ridgway EC. Thyroid hormone regulates the mouse thyrotropin β-subunit promoter in transfected primary thyrotrophs. J Biol Chem. 1989;61:497–504. [PubMed] [Google Scholar]
- 9.Chatterjee VK, Lee JK, Rentoumis A, Jameson JL. Negative regulation of the thyroid-stimulating hormone alpha gene by thyroid hormone: receptor interaction adjacent to the TATA box. Proc Natl Acad Sci USA. 1989;86:9114–9118. doi: 10.1073/pnas.86.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wondisford FE, et al. Thyroid hormone inhibition of human thyrotropin beta-subunit gene expression is mediated by a cis-acting element located in the first exon. J Biol Chem. 1989;264:14601–14604. [PubMed] [Google Scholar]
- 11.Hollenberg AN, et al. The human thyrotropin-releasing hormone gene is regulated by thyroid hormone through two distinct classes of negative thyroid hormone response elements. Mol Endocrinol. 1995;9:540–550. doi: 10.1210/mend.9.5.7565802. [DOI] [PubMed] [Google Scholar]
- 12.Madison LD, Ahlquist JA, Rogers SD, Jameson J. Negative regulation of the glycoprotein alpha gene promoter by thyroid hormone: mutagenesis of a proximal receptor binding site preserves transcriptional repression. Mol Cell Endocrinol. 1993;94:129–136. doi: 10.1016/0303-7207(93)90060-w. [DOI] [PubMed] [Google Scholar]
- 13.Pennathur S, Madison LD, Kay TW, Jameson JL. Localization of promoter sequences required for thyrotropin-releasing hormone and thyroid hormone responsiveness of the glycoprotein hormone alpha-gene in primary cultures of rat pituitary cells. Mol Endocrinol. 1993;7:797–805. doi: 10.1210/mend.7.6.7689696. [DOI] [PubMed] [Google Scholar]
- 14.Lechan RM, Yanping Q, Jackson I, Mahdavi V. Identification of thyroid hormone receptor isoforms in thyrotropin-releasing hormone neurons of the hypothalamic paraventricular nucleus. Endocrinology. 1994;135:92–100. doi: 10.1210/endo.135.1.7516871. [DOI] [PubMed] [Google Scholar]
- 15.Wondisford FE, Steinfelder HJ, Nations M, Radovick S. AP-1 antagonizes thyroid hormone receptor action on the thyrotropin beta-subunit gene. J Biol Chem. 1993;268:2749–2754. [PubMed] [Google Scholar]
- 16.Sjoberg M, Vennstrom B. Ligand-dependent and -independent transactivation by thyroid hormone receptor beta 2 is determined by the structure of the hormone response element. Mol Cell Biol. 1995;15:4718–4726. doi: 10.1128/mcb.15.9.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tagami T, Madison LD, Nagaya T, Jameson JL. Nuclear receptor corepressors activate rather than suppress basal transcription of genes that are negatively regulated by thyroid hormone. Mol Cell Biol. 1997;17:2642–2648. doi: 10.1128/mcb.17.5.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi Y, Mangoura D, Refetoff S. A mouse model of resistance to thyroid hormone produced by somatic gene transfer of a mutant thyroid hormone receptor. Mol Endocrinol. 1996;10:100–106. doi: 10.1210/mend.10.1.8838149. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi Y, Xie J, Weiss RE, Pohlenz J, Refetoff S. Selective pituitary resistance to thyroid hormone produced by expression of a mutant thyroid hormone receptor β gene in the pituitary gland of transgenic mice. Biochem Biophys Res Comm. 1998;245:204–210. doi: 10.1006/bbrc.1998.8396. [DOI] [PubMed] [Google Scholar]
- 20.Wong R, et al. Transgenic mice bearing a mutant thyroid hormone beta 1 receptor manifest thyroid function anomalies, weight reduction, and hyperactivity. Mol Med. 1997;3:303–314. [PMC free article] [PubMed] [Google Scholar]
- 21.Forrest D, et al. Recessive resistance to thyroid hormone in mice lacking thyroid hormone receptor beta: evidence for tissue-specific modulation of receptor function. EMBO J. 1996;15:3006–3015. [PMC free article] [PubMed] [Google Scholar]
- 22.Usala SJ, et al. A homozygous deletion in the c-erbA β thyroid hormone receptor gene in a patient with generalized thyroid hormone resistance: isolation and characterization of the mutant receptor. Mol Endocrinol. 1991;5:327–335. doi: 10.1210/mend-5-3-327. [DOI] [PubMed] [Google Scholar]
- 23.Baniahmad A, Tsai SY, O’Malley BW, Tsai MJ. Kindred S thyroid hormone receptor is an active and constitutive silencer and a repressor for thyroid hormone and retinoic acid responses. Proc Natl Acad Sci USA. 1992;89:10633–10637. doi: 10.1073/pnas.89.22.10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kendall SK, et al. Enhancer-mediated high level expression of mouse pituitary glycoprotein hormone a-subunit transgene in thyrotropes, gonadotropes and developing pituitary gland. Mol Endocrinol. 1994;8:1420–1433. doi: 10.1210/mend.8.10.7531821. [DOI] [PubMed] [Google Scholar]
- 25.Elmquist JK, Bjorbaek C, Ahima RS, Flier JS, Saper CB. Distribution of leptin receptor mRNA isoforms in the rat brain. J Comp Neurol. 1998;395:535–547. [PubMed] [Google Scholar]
- 26.Satoh T, Yamada M, Monden T, Iizuka M, Mori M. Cloning of the mouse hypothalamic preprothyrotropin-releasing hormone (TRH) cDNA and tissue distribution of its mRNA. Mol Brain Res. 1992;14:131–135. doi: 10.1016/0169-328x(92)90020-c. [DOI] [PubMed] [Google Scholar]
- 27.Franklin, K.B., and Paxinos, G. 1997. In The mouse brain in stereotaxic coordinates. Academic Press. New York, NY. 210 pp.
- 28.Vassart G, Dumont JE. The thyrotropin receptor and the regulation of thyrocyte function and growth. Endocr Rev. 1992;13:596–611. doi: 10.1210/edrv-13-3-596. [DOI] [PubMed] [Google Scholar]
- 29.Taylor T, Gesundheit N, Gyves PW, Jakobowitz DM, Weintraub BD. Hypothalamic hypothyroidism caused by lesions in rat paraventricular nuclei alters the carbohydrate structure of secreted thyrotropin. Endocrinology. 1988;122:283–290. doi: 10.1210/endo-122-1-283. [DOI] [PubMed] [Google Scholar]
- 30.Taylor T, Weintraub BD. Altered thyrotropin (TSH) carbohydrate structures in hypothalamic hypothyroidism created by paraventricular nuclear lesions are corrected by in vivo TSH-releasing hormone administration. Endocrinology. 1989;125:2198–2203. doi: 10.1210/endo-125-4-2198. [DOI] [PubMed] [Google Scholar]
- 31.Beck-Peccoz P, Amr S, Menezes-Ferreira MM, Faglia G, Weintraub BD. Decreased receptor binding of biologically inactive thyrotropin in central hypothyroidism Effect of treatment with thyrotropin releasing hormone. N Engl J Med. 1985;312:1085–1090. doi: 10.1056/NEJM198504253121703. [DOI] [PubMed] [Google Scholar]
- 32.Yamada M, et al. Tertiary hypothyroidism and hyperglycemia in mice with targeted disruption of the thyrotropin-releasing hormone gene. Proc Natl Acad Sci USA. 1997;94:10862–10867. doi: 10.1073/pnas.94.20.10862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Persani L, et al. Evidence for the secretion of thyrotropin with enhanced bioactivity in syndromes of thyroid hormone resistance. J Clin Endocrinol Metab. 1994;78:1034–1039. doi: 10.1210/jcem.78.5.8175956. [DOI] [PubMed] [Google Scholar]
- 34.Lechan RM, Jackson IMD. Immunohistochemical localization of thyrotropin releasing hormone in the rat hypothalamus and pituitary. Endocrinology. 1982;111:56–65. doi: 10.1210/endo-111-1-55. [DOI] [PubMed] [Google Scholar]
- 35.Lechan RM, et al. Thyrotropin-releasing hormone precursor: characterization in rat brain. Science. 1986;231:159–161. doi: 10.1126/science.3079917. [DOI] [PubMed] [Google Scholar]
- 36.Cullen MJ, Doherty GF, Ingbar SH. The effect of hypothyroidism and thyrotoxicosis on thyroxine metabolism in the rat. Endocrinology. 1973;92:1028–1033. doi: 10.1210/endo-92-4-1028. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan MM, Utiger RD. Iodothyronine metabolism in liver and kidney homogenates from hyperthyroid and hypothyroid rats. Endocrinology. 1978;103:156–161. doi: 10.1210/endo-103-1-156. [DOI] [PubMed] [Google Scholar]
- 38.Cohen O, Flynn TR, Wondisford FE. Ligand-dependent antagonism by retinoid X receptors of inhibitory thyroid hormone response elements. J Biol Chem. 1995;270:13899–13905. doi: 10.1074/jbc.270.23.13899. [DOI] [PubMed] [Google Scholar]
- 39.Safer JD, Cohen R, Hollenberg AN, Wondisford FE. Defective release of corepressor by hinge mutants of the thyroid hormone receptor found in patients with resistance to thyroid hormone. J Biol Chem. 1998;273:30175–30182. doi: 10.1074/jbc.273.46.30175. [DOI] [PubMed] [Google Scholar]
- 40.Hollenberg AN, Monden T, Madura JP, Lee K, Wondisford FE. Function of nuclear co-repressor protein on thyroid hormone response elements is regulated by the receptor A/B domain. J Biol Chem. 1996;271:28516–28520. doi: 10.1074/jbc.271.45.28516. [DOI] [PubMed] [Google Scholar]
- 41.Safer JD, et al. Isoform variable action among thyroid hormone receptor mutants provides insight into pituitary resistance to thyroid hormone. Mol Endocrinol. 1997;11:16–26. doi: 10.1210/mend.11.1.9867. [DOI] [PubMed] [Google Scholar]
- 42.Yoh SM, Chatterjee VKK, Privalsky ML. Thyroid hormone resistance syndrome manifests as an aberrant interaction between mutant T3 receptors and transcriptional corepressors. Mol Endocrinol. 1997;11:470–480. doi: 10.1210/mend.11.4.9914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collingwood TN, et al. A natural transactivation mutation in the thyroid hormone β receptor: impaired interaction with putative transcriptional mediators. Proc Natl Acad Sci USA. 1997;94:248–253. doi: 10.1073/pnas.94.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Collingwood TN, Adams M, Tone Y, Chatterjee VKK. Spectrum of transcriptional dimerization and dominant negative properties of twenty different mutant thyroid hormone beta-receptors in thyroid hormone resistance syndrome. Mol Endocrinol. 1994;8:1262–1277. doi: 10.1210/mend.8.9.7838159. [DOI] [PubMed] [Google Scholar]
- 45.Adams M, et al. Genetic analysis of 29 kindreds with generalized and pituitary resistance to thyroid hormone. Identification of thirteen novel mutations in the thyroid hormone receptor β gene. J Clin Invest. 1994;94:506–515. doi: 10.1172/JCI117362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tagami T, Jameson JL. Nuclear corepressors enhance the dominant negative activity of mutant receptors that cause resistance to thyroid hormone. Endocrinology. 1998;139:640–650. doi: 10.1210/endo.139.2.5742. [DOI] [PubMed] [Google Scholar]