Abstract
Monoclonal antibody-conjugated sphere-shaped gold nano particles were combined with single-walled carbon nanotubes (SWCNTs) to create a nanohybrid system to selectively detect and eradicate multiple drug resistant Salmonella (MDRS) typhimurium DT104 bacteria. The Raman signal intensity from Rhodamine 6G (Rh6G) modified monoclonal AC04 antibody SWCNTs-Gold nanoparticle (SWCNT-GNPs) hybrid provided a SERS enhancement by several orders of magnitude to detect the MDRS bacteria over the GNP system. A targeted photothermolysis experiment using 670 nm light at 2 W/cm2 for 15 min, resulted in selective and irreparable damage to more than 99% Salmonella DT104 at the concentration of 105 CFU/mL. In comparison to solely SWCNTs or GNPs, our SWCNT-GNPs nanohybrids have also shown a better photo thermal efficiency.
Salmonella enteric serover Typhimurium definitive type 104 presents a great challenge in today’s public health care.1–7 Every year, approximately 42,000 cases of salmonellosis are reported in the United States.8 Due to the fact that many milder cases are not diagnosed or reported, the actual number of infections may be twenty-nine or more times greater. Salmonella typhimurium is an emerging food borne pathogen detected from different food samples in USA and Europe. It is now estimated that multiple drug resistant Salmonella (MDRS) cause approximately 60% of nosocomial infections.9
Due to antibiotic resistance, most of the currently available antibiotics cannot effectively kill MDR Salmonella inside of the human body.10 As a result, the development of physiologically low toxic, highly sensitive methods for the detection/killing of MDRS are in high demand.1–5,9 Recently published articles have demonstrated that gold nanoparticles (GNPs) of different sizes and shapes can be used for the hyperthermic destruction of bacteria.10–18 It has also been shown that single walled carbon nanotubes (SWCNTs) can be used as an agent in photo-thermal therapy due to its unique structure.19–23 Furthermore, when exposed to a laser, hybrid nanomaterials will generate a high temperature environment which enables the photo-thermal process to be both effective and rapid24–26, and SWCNTs-GNPs hybrids are expected to have better biocompatibility and low toxicity than GNPs formed through toxic surfactants such as cetyl trimethyl ammonium bromide (CTAB).9 As a result, bacteria such as MDRS are an excellent target for the application of these SWCNTs-GNPs nano-hybrids toward the selective photo thermally induced eradication of the aforementioned bacteria. This communication reports the first utilization of SWCNTs attached with GNPs for highly sensitive detection and destruction of Salmonella DT104 (ATCC700408).
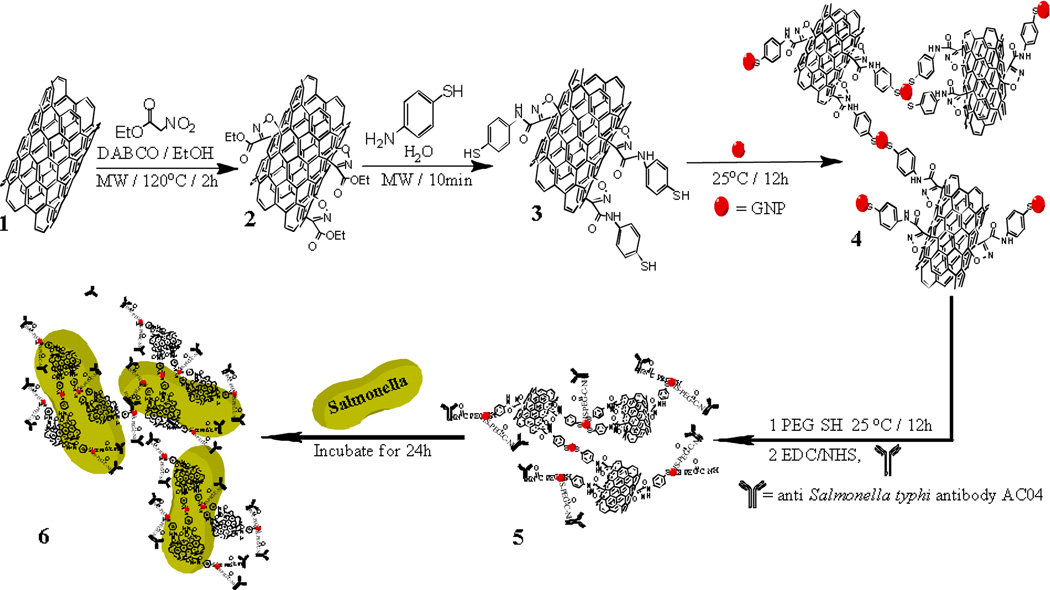
Due to the presence of strong Vander Waals interactions that tightly hold them together, SWCNTs are insoluble in a variety of solvents.18 As a result, modification of SWCNTs using chemical functionalization to enhance their solubility is necessary. In this context, our continuing research in 1,3-dipolar cycloaddition enabled us to functionalize SWCNTs through a 1,4-diazabicyclo[2.2.2]octane (DABCO) mediated 1,3-dipolar cycloaddition reaction with ethyl nitroacetate under microwave reaction conditions (Scheme 1).27 As a result of the cycloaddition, the ester group on the isoxazoline rings on the SWCNTs were then further functionalized to an amide through a quick reaction with para-aminothiophenol which offers a thiol group for linking with GNPs.12 After synthesizing 20 nm, sphere-shaped GNPs through a one-step process by using sodium citrate to reduce gold ions at 100 °C,18 the GNPs were attached to the SWCNTs through the Au-S bond via amino-thiophenol. After the GNPs were attached to the SWCNTs, the free GNPs were separated by centrifuging the mixture at 4000 rpm for 30 min.
Scheme 1.
Schematic representation shows the synthetic protocol for the functionalization of SWCNTs, the formation of GNP attached SWCNTs, and SWCNTs-GNPs-antibody-Salmonella.
For selective binding of the SWCNTs with MDR Salmonella, we modified the GNPs surface with monoclonal antibody AC04 which is specific for MDR Salmonella (Scheme 1). First, we modified the SWCNTs hybrid GNPs with carboxyl groups by using terminal thiol polyethylene glycol (PEG-SH). Then, covalent immobilization of the amine group of the antibody onto the carboxyl groups of PEG-SH was performed by ethyl-(dimethylaminopropyl)-carbodiimide (EDC)/N-hydroxysuccinimide (NHS) coupling catalysis.19
MDR Salmonella Typhimurium DT104 was obtained from the ATCC. For culturing, we followed the ATCC protocol as instructed. The bacteria were incubated at 37 °C with 5% CO2 for 24 h to attach the bacteria to the SWCNTs-GNPs-antibody (Scheme 1), and the nano-hybrid-antibody-MDRS were washed three times to remove any residual unbound materials.
For photo-thermal destruction of MDRS, we used a continuous wavelength 60 mW laser operating at 670 nm, as an excitation light source for 15 min. We then utilized the bacterial coating method to determine the percentage of dead bacteria.
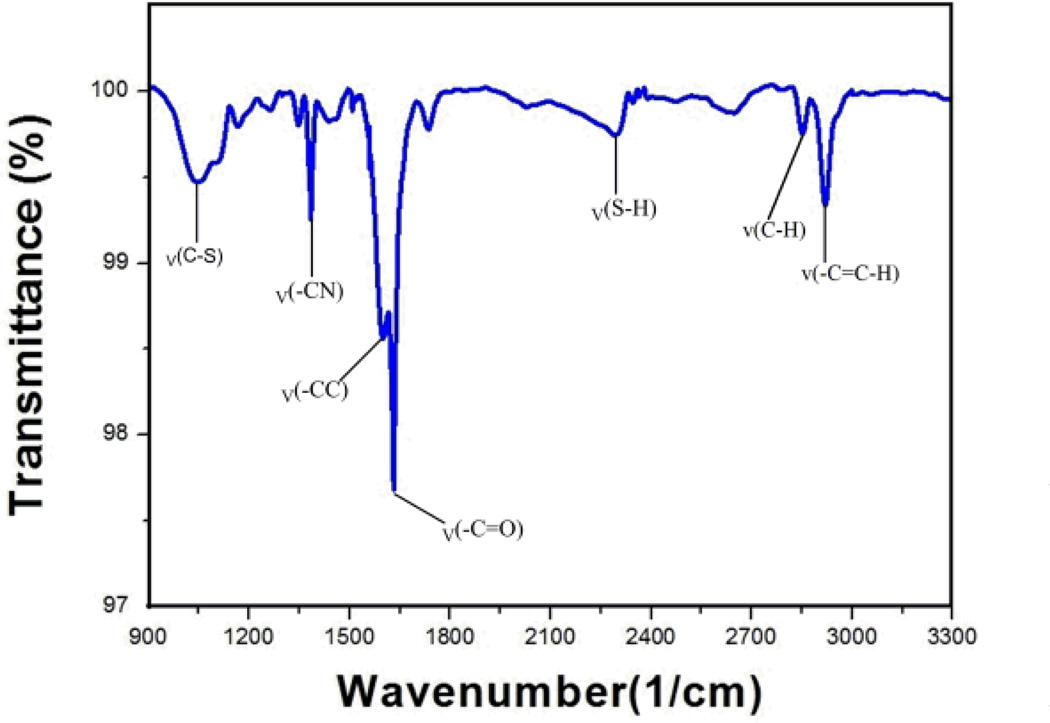
In order to confirm the formation of the amide bond for 3 from the reaction of aminothiophenol with 2, the functionalized SWCNT 3, was analyzed through FTIR spectroscopy. The FTIR spectrum in Figure 1, clearly shows the characteristic peaks associated with the expected amide formation from aminothiophenol that include the -SH and -CS stretching vibrations at 2292 and 1045 cm−1 respectively. The absorption peak at 1347 cm−1 is a -CN stretching vibration, and the 1598 and 2919 cm−1 peaks are the amide -C=O and -CH asymmetrical stretching vibrations respectively. The remaining peaks are mainly due to the -C=C- stretching vibration bands of the newly formed aromatic ring from aminothiophenol and the SWCNTs.
Fig. 1.
FTIR spectra of aminothiophenol attached SWCNTs.
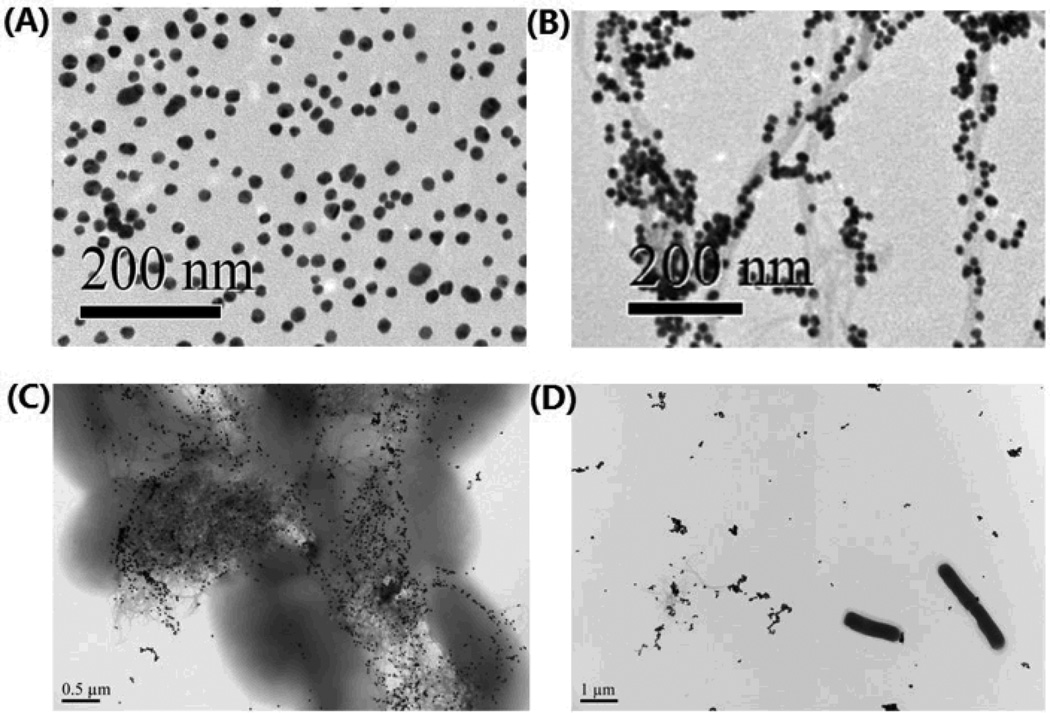
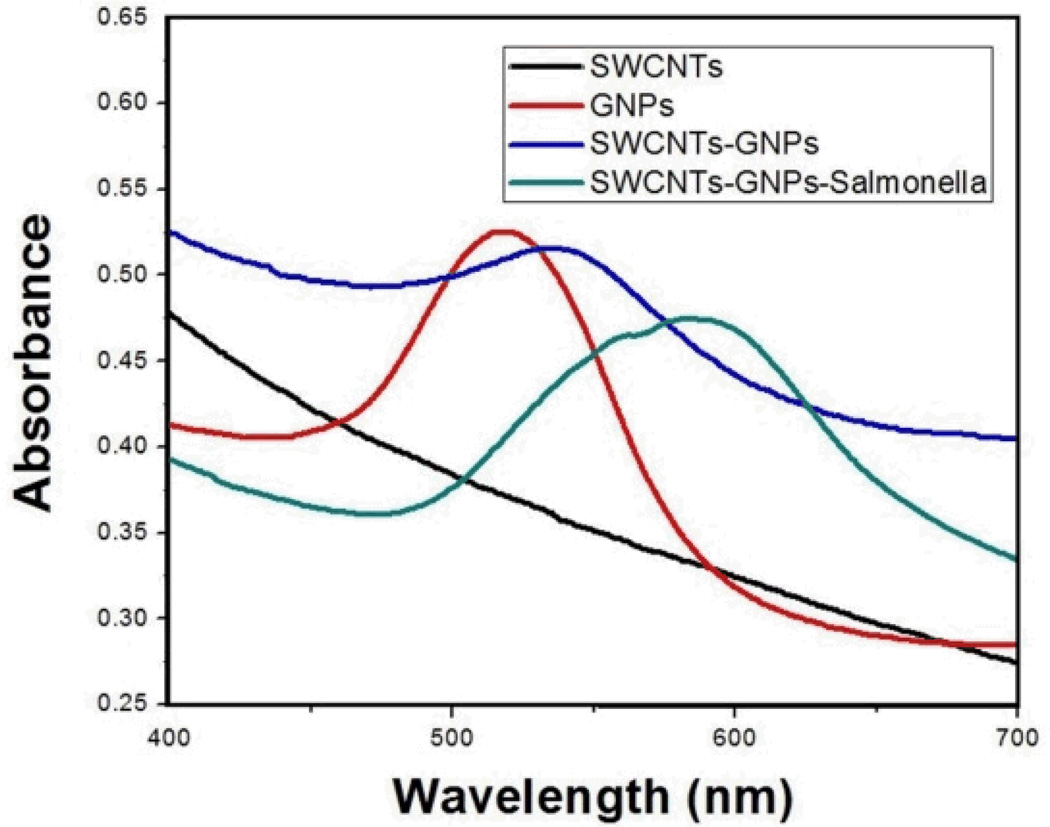
The shape of GNPs and its binding with SWCNTs were confirmed through the transmission electron microscope (TEM) and ultraviolet-visible spectroscopy (UV). Figures 2A and 2B show the TEM images of GNPs and SWCNTs-GNPs. Based upon the images in Figure 2B, the GNPs are nicely decorated onto the SWCNTs. The absorption spectra of only SWCNTs, 0.3 nM GNPs and 0.3 nM GNPs attached SWCNTs are depicted in Figure 4. Broad absorption spectrum from near-infrared and visible region from 400 to 700 nm is mainly due to the E11 and E22 transitions of nanotubes. As shown in Figure 4, in the case of GNPs, we observed a strong long wavelength plasmon band around 520 nm that is due to the oscillation of the conduction band electrons. The absorption spectra of SWCNTs-GNPs shift toward higher wavelength and become broadened. This broadening is mainly due to the fact that as we add GNPs, the GNPs become very closely packed on the SWCNTs as shown in Figure 2B. As a result, the absorption band shifts to higher wavelength, because of the interparticle interactions and change in the local refractive index on the nanoparticle surface.
Fig. 2.
TEM images illustrating: (A) GNPs. (B) GNPs-decorated SWCNTs. (C) Salmonella bound by AC04 antibody conjugated GNPs-decorated SWCNTs. (D) Lack of E.Coli binding by AC04 antibody conjugated GNPs-decorated SWCNTs.
Fig. 4.
UV Absorption spectra for only SWCNTs, 0.3 nM GNPs, 0.3 nM GNPs attached SWCNTs, and SWCNTs-GNPs-Salmonella
Salmonella bacteria are classified into over 2500 serotypes based on antigenic differences relating to lipopolysaccharide (O-antigen) and the flagellar structures (H-antigen).1–5 Our Salmonella typhimurium DT104 identification is based on the fact that anti-Salmonella typhi antibody AC04 conjugated GNPs can readily and specifically identify its bacterium O-antigen, through antibody-antigen recognition. To understand whether the identification and attachment is highly selective, we performed UV, TEM and surface-enhanced Raman scattering (SERS). As a control, we also tested if the SWCNTs attached AC04 Salmonella antibody conjugated GNPs would respond to E. coli O157:H7 bacteria. TEMs clearly confirm that our antibody conjugated SWCNT- GNP nanohybrids bind well with MDRS (Figure 2C) when compared to E. Coli O157:H7 bacteria (Figure 2D).
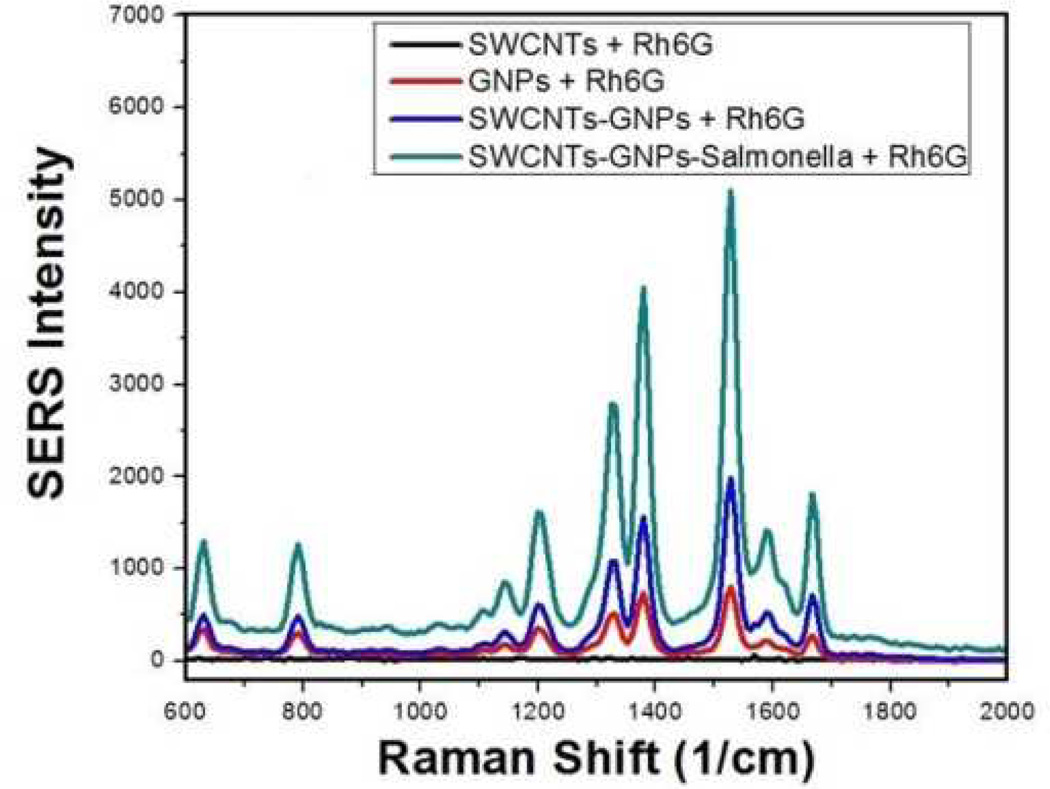
For our SERS experiment, we used a continuous wavelength diode-pumped solid-state laser operating at 670 nm as an excitation light source. As shown in Figure 3, the AC04 antibody-conjugated GNPs-decorated SWCNTs aggregate in the presence of MDRS. As a result, the aggregate formed several hot spots and provided a significant enhancement of the Raman signal intensity from the Rhodamine 6G modified monoclonal AC04 antibody by several orders of magnitude, while the SERS intensity change was negligible in the presence of only GNPs or SWCNTs.
Fig. 3.
SERS enhancement (SERS intensity change before and after addition of bacteria) due to the addition of Salmonella to monoclonal AC04 antibody-conjugated, GNPs-decorated SWCNTs.
Since Salmonella DT104 bacteria is more than an order of magnitude larger in size (1–3 µm) than the antibody-conjugated, GNPs-decorated SWCNTs, in the presence of Salmonella DT104 bacteria, several antibody-conjugated GNPs-decorated SWCNTs bind with one Salmonella bacteria, which leads to the change in the UV spectrum. As shown in Figure 4, the absorption maximum for the plasmon absorption band of antibody conjugated SWCNTs-GNPs at 539 nm decreased in the presence of MDR Salmonella DT104, whereas a new broad band appears at around 596 nm. This red shift is mainly due to the aggregation of nanoparticles on the Salmonella surface.9
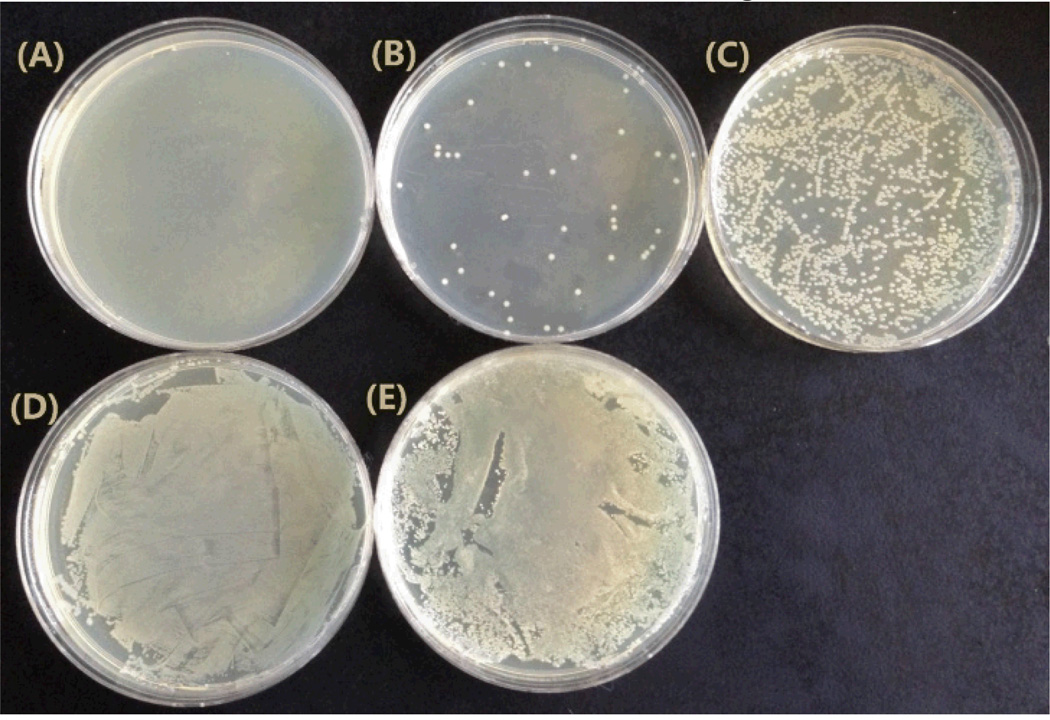
To verify the role of the laser and the hybrid SWCNTs-GNPs toward killing Salmonella, we performed 5 comparative photo-thermal experiments. Based on the results shown in Figure 5A, after laser irradiation (2 W/cm2 power of 670 nm light, 15 min exposure) of the MDR Salmonella in the presence of the AC04 Salmonlella antibody hybrid nanomaterial, more than 99% of the MDR Salmonella at the concentration of 105 CFU/mL were eradicated, while the hybrid nanomaterial without laser irradiation (Figure 5D) and MDR Salmonella with laser irradiation (no hybrid nanomaterial, Figure 5E) killed a negligible amount of MDR Salmonella. The data from Figure 5 confirmed that both hybrid SWCNTs-GNPs and laser irradiation play a vital role in the photo-thermal destruction of Salmonella DT104, which validates the mechanism that laser irradiated hybrid nanomaterials composed of SWCNTs-GNPs to produce enough heat to destruct the MDRS.
Fig. 5.
Photo thermal killing of Salmonella. (A) SWCNTs-GNPs-antibody-Salmonella under laser irradiation for 15 min. (B) GNPs-antibody-Salmonella under laser irradiation for 15 min. (C) SWCNTs-antibody-Salmonella under laser irradiation for 15 min. (D) SWCNTs-GNPs-antibody-Salmonella before laser irradiation. (E) Only Salmonella under laser irradiation for 15 min
To understand the advantages of using hybrid SWCNTs-GNPs instead of only GNPs or only SWCNTs, we have also performed photo-thermal experiments using antibody-conjugated GNPs and antibody-conjugated SWCNTs separately. As shown in Figure 5B and 5C, at the same power and exposure time, in the case of only GNPs or SWCNTs, they kill 10 to 40 percent less of MDRS than using hybrid SWCNTs-GNPs. On the other hand, the laser power had to be doubled (4W/cm2) and 20 minutes were required to eradicate the same amount of MDRS when SWCNTs and GNPs were used separately. Our experimental results depicted that the photo-thermal effect with the SWCNTs-GNPs hybrid is much more than either SWCNTs or GNPs alone. This effect is mainly because of two reasons: (1) In the case of the hybrid SWCNTs-GNPs, both SWCNTs and GNPs can generate heat when exposed to laser irradiation, and the amount of heat generation is expected to be higher than only SWCNTs or GNPs alone, and (2) after the attachment of SWCNTs-GNPs hybrids with Salmonella, the hybrids has a very strong absorption peak at around 590 nm, near to the 670 nm laser source compared with GNPs attached MDRS and SWCNTs-attached MDRS, as shown in Figure 4.
In conclusion, in this communication, we have reported for the first time a method of selected photo-thermal destruction of MDR Salmonella DT104 by using anti-Salmonella typhi antibody AC04-conjugated GNPs-decorated SWCNTs. We have shown that in the presence of targeted Salmonella DT104, antibody-conjugated hybrid nanomaterials provide a significant enhancement of the Raman signal intensity by 5 orders of magnitude. Our TEM image with Salmonella and E.Coli clearly demonstrate that our SERS assay is highly sensitive in targeting Salmonella DT104 and have the ability to distinguish between other non-targeted bacteria through antibody–antigen recognition. We have also shown that during photo-thermal experiments of antibody conjugated hybrid SWCNTs-GNPs attached Salmonella, the localized heating that occurs due to the absorption of 670 nm continuous NIR irradiation, gives rise to irreparable bacterial damage and selectively eradicates 99% of the MDRS Salmonella within 15 min at 2 W/cm2 power. Our data show that photo thermal response of the hybrid nanomaterial is far better than the examined solitary SWCNT and GNP nanomaterials. After optimization of some parameters, we believe that our method could have enormous potential application in rapid detection and photo-thermal therapy of clinical samples.
Supplementary Material
Acknowledgments
The project described was supported by the National Science Foundation (HBCU-RISE: HRD-1137763 and PREM: DMR-633156). The TEM CORE Facilities were supported by the National Institutes of Health/ National Center for Research Resources (Award Number: G12RR013459) and the National Institutes of Health /National Institute on Minority Health and Health Disparities (Award Number: G12MD007581).
Footnotes
Electronic supplemantary information (ESI) available: See DOI: 10.139/
Notes and references
- 1.Pignato S, Coniglio MA, Faro G, Lefevre M, Weill F-X, Giammanco G. Foodborne Pathog. Dis. 2010;7:945–951. doi: 10.1089/fpd.2009.0504. [DOI] [PubMed] [Google Scholar]
- 2.Wolter A, Niessner R, Seidel M. Anal. Chem. 2008;80:5854–5863. doi: 10.1021/ac800318b. [DOI] [PubMed] [Google Scholar]
- 3.Parashar V, Pandey SK, Pandey AC. Chem. Commun. 2010;46:3143–3145. doi: 10.1039/c000553c. [DOI] [PubMed] [Google Scholar]
- 4.Wang C, Irudayaraj J. Small. 2008;4:2204–2208. doi: 10.1002/smll.200800309. [DOI] [PubMed] [Google Scholar]
- 5.Singh AK, Senapati D, Wang S, Griffin J, Neely A, Candice P, Naylor KM, Varisli B, Kalluri JR, Ray PC. ACS Nano. 2009;3:1906–1912. doi: 10.1021/nn9005494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delibato E, Volpe G, Romanazzo D, Palleschi G. J. Agric. Food Chem. 2009;57:7200–7204. doi: 10.1021/jf901181m. [DOI] [PubMed] [Google Scholar]
- 7.Wen CY, Hu J, Zhang ZL, Tian ZQ, Pang DW. Anal. Chem. 2013;85:1223–1230. doi: 10.1021/ac303204q. [DOI] [PubMed] [Google Scholar]
- 8. http://www.cdc.gov/salmonella/general/
- 9.Ray PC, Khan SA, Singh AK, Senapati D, Fan Z. Chem Soc Rev. 2012;41:3193–3209. doi: 10.1039/c2cs15340h. [DOI] [PubMed] [Google Scholar]; Khan SA, Singh AK, Senapati D, Fan Z, Ray PC. Chem. Commun. 2011;47:9444–9446. doi: 10.1039/c1cc13199k. [DOI] [PubMed] [Google Scholar]
- 10.Gao J, Huang XY, Liu H, Zan F, Ren J. Langmuir. 2012;28:4464–4471. doi: 10.1021/la204289k. [DOI] [PubMed] [Google Scholar]
- 11.Jain PK, Huang X, El-Sayed IH, El-Sayed MA. Acc Chem. Res. 2008;41:1578–1586. doi: 10.1021/ar7002804. [DOI] [PubMed] [Google Scholar]
- 12.Dasary SSR, Senapati D, Singh AK, Anjaneyulu Y, Yu HT, Ray PC. Appl. Mater. Interfaces. 2010;2:3455–3460. doi: 10.1021/am1005139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang X, Tan W. Acc.Chem.Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lal S, Clare S, Halas N. J. Acc. Chem. Res. 2008;41:1842–1851. doi: 10.1021/ar800150g. [DOI] [PubMed] [Google Scholar]
- 15.Peer D, Karp J, Hong S, Farokhzad O, Margalit R, Langer R. Nat. Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 16.Giljohann D, Mirkin C. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yong KT, Hu R, Roy I, Ding H, Vathy LA, Bergey EJ, Mizuma M, Maitra A, Prasad PN. ACS Appl. Mater. Interfaces. 2009;1:710–719. doi: 10.1021/am8002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji X, Song XN, Li J, Bai YB, Yang WS, Peng XG. J. Am. Chem. Soc. 2007;129:13939–13948. doi: 10.1021/ja074447k. [DOI] [PubMed] [Google Scholar]
- 19.Beqa L, Fan Z, Senapati D, Ray PC. ACS Appl. Mater. Interfaces. 2011;3:3316–3324. doi: 10.1021/am2004366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kostarelos K, Bianco A, Prato MP. Nat. Nanotechnol. 2009;4:627–633. doi: 10.1038/nnano.2009.241. [DOI] [PubMed] [Google Scholar]
- 21.Xiang J, Drzal LT. Appl. Mater. Interfaces. 2011;3:1325–1332. doi: 10.1021/am200126x. [DOI] [PubMed] [Google Scholar]
- 22.Poon L, Zandberg W, Hsiao D, Erno Z, Sen D, Gates BD, Branda NR. ACS Nano. 2010;4:6395–6403. doi: 10.1021/nn1016346. [DOI] [PubMed] [Google Scholar]
- 23.Kolosnjaj TJ, Hartman KB, Boudjemaa S, Ananta JS, Wilson LJ, Moussa F. ACS Nano. 2010;4:1481–1492. doi: 10.1021/nn901573w. [DOI] [PubMed] [Google Scholar]
- 24.Huang XH, El-Sayed MA. Journal of Advanced Research. 2010;1:13–28. [Google Scholar]
- 25.Govorov AO, Richardson H. Nanotoday. 2007;2:30–38. [Google Scholar]
- 26.Jeyamohan P, Hasumura T, Nagaoka Y, Yoshida Y, Maekawa T, Kumar DS. International Journal of Nanomedicine. 2013;8:2653–2667. doi: 10.2147/IJN.S46054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Machetti F, Cecchi L, Trogu E, De Sarlo F. Eur. J. Org. Chem. 2007:4352–4359. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.