Abstract
The continuous assembly and disassembly of actin filament networks is vital for cellular processes including division, growth, and motility. Network remodeling is facilitated by cofilins, a family of essential regulatory proteins that fragment actin filaments. Cofilin induces net structural changes in filaments that render them more compliant in bending and twisting. A model in which local stress accumulation at mechanical discontinuities, such as boundaries of bare and cofilin-decorated filament segments, accounts for the cofilin concentration dependence of severing, including maximal activity at sub-stoichiometric binding densities. Real-time imaging of cofilin-mediated filament severing supports the boundary-fracture model. The severing model predicts that fragmentation is promoted by factors modulating filament mechanics (e.g. tethering, cross-linking, or deformation), possibly explaining enhanced in vivo severing activities.
Keywords: biopolymer mechanics, cytoskeleton, fracture, flexural rigidity, Ising model
Self-association (i.e. polymerization) of the protein actin into linear double-stranded helical filaments provides forces that drive eukaryotic cell motility (1; 2). Central to developing predictive models of motility is quantitative knowledge of actin behavior and how it is influenced by the vast number of actin regulatory proteins in cells. The majority of actin-based motility models are derived from biochemical and biophysical studies carried out in solution with purified protein components.
Assembled actin filaments elongate (e.g. grow and shrink) only from their ends (3). Sustained motility requires the regeneration of assembled subunits and replenishment of the actin monomer pool. Accordingly, factors that increase the filament end concentration provide a greater number of sites from which subunits can add or dissociate, thereby accelerating the overall subunit flux of filament networks.
Filament subunit interactions are non-covalent, yet filaments in solution assembled from purified actin can reach several microns in length (~365 subunits per micron) because of the stabilizing lateral and longitudinal contacts associated with the double-stranded filament geometry (4; 5). The mechanical properties of filaments are comparable to some familiar commercial plastics, enabling filaments and filament networks to sustain forces associated with cell motility as well as provide cells with mechanical strength and integrity. However, filaments in solution at their assembled microscopic length scales behave as semi-flexible polymers that readily undergo thermally driven bending and twisting shape fluctuations.
The actin regulatory protein cofilin binds and severs actin filaments in vitro and in vivo (6–9). Cofilin is essential (10; 11) and dysfunction is associated with human pathologies (12; 13). The recognition that filaments are mechanically stiff, yet readily fragmented by regulatory proteins, has attracted the utility of physics and engineering formalisms, specifically those developed in polymer mechanics. Severing is driven by cofilin binding interactions and linked equilibria; no external energy sources (e.g. ATP hydrolysis) are required. Elucidation of the cofilin severing mechanism has therefore benefited from interpretive power of physical chemistry, specifically thermodynamics and kinetics.
The contents of this review article focus on the later two aspects - physics and chemistry - of cofilin severing activity. Given space limitations, we focus on recent developments with the understanding that considerable bodies of work preceded and facilitated these advances. We encouragingly direct readers to these earlier works (cited within papers cited in this review) to fully grasp current understanding of the cofilin severing mechanism. We note that an extensive body of literature exists evaluating the interaction of cofilin with actin monomers, but these works are not discussed here.
Cofilin binding thermodynamics and kinetics
Cofilin binds actin stoichiometrically - one cofilin molecule per actin filament subunit (14–17) - and, in some cases, with positive cooperativity. Vertebrate cofilin binds vertebrate actin filaments cooperatively and equilibrium binding isotherms display a characteristic sigmoidal shape, independent of the detection method used to assay binding (e.g. cosedimentation (18; 19), fluorescence (17; 20), or phosphorescence (21)). Some non-vertebrate cofilins bind their native, species compatible actin non-cooperatively (22), while binding of some cofilins (e.g. S. cerevisae) is so strong that it precludes reliable detection of cooperative interactions should they exist (23).
Studies assessing cooperative binding commonly employ the familiar Hill Equation and yield Hill coefficients ranging from 4–40, indicating (relatively) weak positive cooperativity (14–17). This is a surprising behavior because actin-bound cofilins do not directly interact (24). Cooperative interactions must therefore arise from linked changes in (average) filament structure (25–28) and/or dynamics (21; 29; 30; 23).
Despite being an assuring indicator of cooperative interactions, the Hill formalism is limited in its ability to translate observed binding behaviors into meaningful thermodynamic binding parameters, as experimentally measured apparent binding constants represent weighted average composites of individual binding events (31). A one dimensional Ising lattice model with nearest neighbor cooperative interactions (32) has proven to be more informative than the more conventional Hill analysis, despite it not explicitly accounting for actin filament subunit geometry.
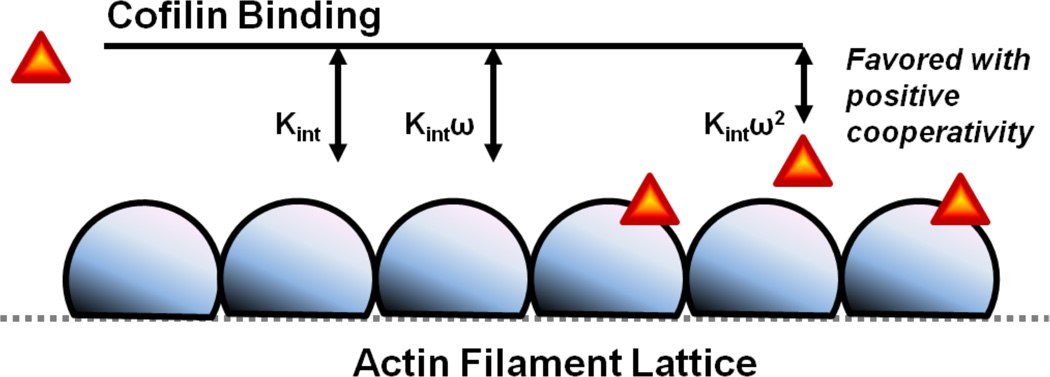
A two-state Ising model in which each lattice site (i.e. an individual actin subunit) exists as vacant or occupied by a cofilin molecule (Figure 1) reliably describes both the equilibrium (17) and kinetic (33) binding of vertebrate cofilin and vertebrate actin, and provides intrinsic binding constants (Kint) and cooperativity parameters (ω). According to this model, bound cofilin can exist in one of three distinct binding modes: a) isolated, with neighboring sites vacant and overall binding affinity Kint, b) singly contiguous, with one of the two neighboring sites occupied and overall binding affinity Kintω, and c) doubly contiguous, with both neighboring sites occupied with overall affinity Kintω2.
Figure 1.
One-dimensional Ising lattice model with nearest-neighbor cooperative interactions.
The body of equilibrium and kinetic data collected to date suggest that vertebrate cofilin binds with an intrinsic binding affinity of ~10–20 µM and a cooperativity parameter of ~8–20 (20). Cofilin binding is slow when compared to most other actin binding proteins (34) and on the timescale of ligands that bind cryptic filament sites (e.g. phalloidin (35; 36)). Time courses of cooperative association identified a slow isomerization subsequent to cofilin encounter with actin, indicating that bound cofilactin adopts (at least) two distinct conformations (33).
The nearest-neighbor Ising model lacks structural information and considers only nearest neighbor cooperative interactions. Although evidence for non-nearest neighbor cofilactin interactions exists (37; 38), they are not required to account for cooperative equilibrium (17; 8; 31) and kinetic binding data (33). More sensitive assays are needed to determine if non-nearest neighbor effects contribute to cofilin binding cooperativity. We note that although a two-state Ising model accounts for cooperative vertebrate cofilactin interactions, more reliable fits to non-vertebrate cofilin equilibrium binding data are obtained with a four-state conformational ensemble model that considers intrinsic equilibria among two distinct cofilin-occupied conformations as well as two distinct vacant states (31).
When binding is positively cooperative, clusters of bound cofilin will form along filaments. Knowledge of the various cofilin binding mode affinities (defined above) allows for prediction of the average (and distribution)(39) of cofilin cluster sizes at a given concentration (17). The predicted cofilin cluster sizes are small – with an average size of near unity (17)– at cofilin concentrations yielding efficient severing (10; 40), indicating that only one or few cofilin proteins are sufficient to sever filaments.
Cofilin binding is surprisingly weakling dependent on temperature over the 4–37°C range, indicative of a modest interaction enthalpy and a binding equilibrium that is driven primarily by positive entropy changes (20). The entropy changes driving cofilactin interactions must originate from reorganization of solvent, salts and/or protein (cofilin and actin) since these are the only components present in experimental samples. Solvent contributions appear to be small, as assessed from the dependence of binding free energy and cooperativity on inert crowding agents (20), see Ref. (41) for additional discussions.
Cofilin binding is salt-dependent, as one might expect for binding to a charged polymer (42). The intrinsic cofilin binding free energy is affected by salt, but the cooperativity is not (20). Salt-dependent cofilin binding has been interpreted in terms of a competition between cofilin and filament-associated cations (20) that likely bind at specific and discrete sites on filaments (43). Isolated cofilin binding is linked to release of a single Mg2+ (or approximately two K+) (20). Cooperative interactions have no apparent linkage to ion release, suggesting that actin conformational changes associated with cofilin binding and ion release are not propagated cooperatively beyond sites with bound cofilin.
Actin filament conformational dynamics
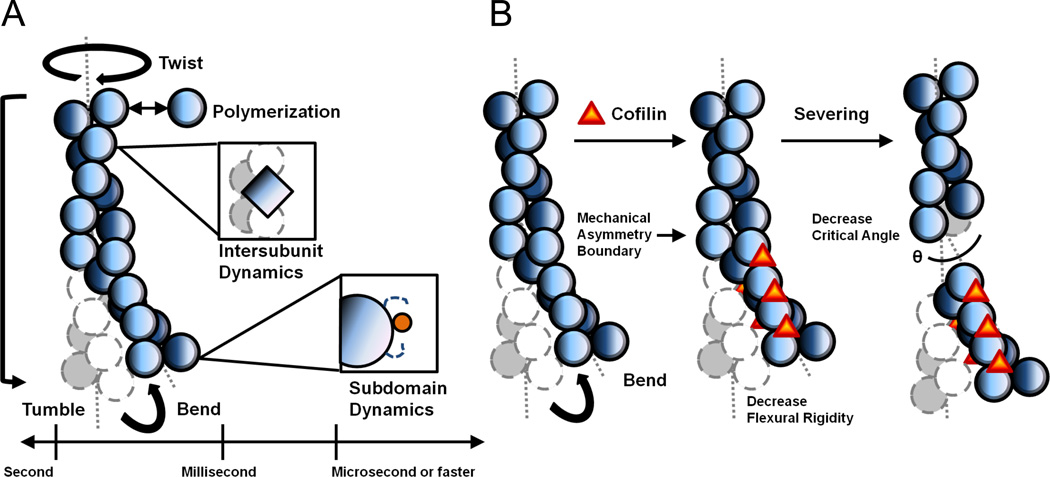
Ion release contributes partially to the entropic changes driving cofilin binding. The remaining entropy changes originate from enhanced conformational fluctuations of the cofilactin complex. The molecular origins of these changes manifest as increases in spontaneous, thermally driven actin filament shape fluctuations upon cofilin binding. Such filament conformational dynamics concern both large-scale filament shape changes as well as small amplitude fluctuations of individual subunits, and thus span a broad range of timescales (Figure 2A). Experimental and computational studies indicate that cofilin alter actin conformational dynamics on all of these length and time scales.
Figure 2.
(A) Actin filament conformational fluctuations span broad timescales. Filaments display large-scale tumbling, bending, and twisting motions. Individual actin subunits undergo sub-microsecond timescale dynamics that modulate subunit interfaces. (B) Cofilin binding introduces mechanical discontinuity that promotes filament severing.
Filament bending and twisting deformations occur on the microseconds to seconds time scale, depending on the amplitude of the motion. Single filaments in solution display diffusive tumbling, though these motions are very slow and negligible on the seconds time scale associated with cofilin-mediated severing events. Analysis of actin filament shape fluctuations generally employ conditions that eliminate or reduce tumbling motions.
Conformational dynamics of individual actin subunits span the same breadth of timescales as a typical globular protein, with functional motions ranging from millisecond to picoseconds timescales. Domain rotational motions or local folding/unfolding reactions on millisecond to microsecond timescales modulate subunit stability, the interface between neighboring subunits, and potentially regulate accessibility to binding partners (44–47). Additionally, loop fluctuations and side chain rearrangements on nanosecond or picoseconds timescales may regulate solvent or ion interactions that modulate filament stability (43).
Cofilin modulates actin subunit structure and filament mechanics
Cofilin binding yields net structural changes at both the subunit and filament scale. On the subunit scale, a conformational change occurs in the DNAase I binding loop (48–51) as well as the actin C-terminus (52). Otherwise cryptic sites within subunits become accessible to protease cleavage (44). On the filament scale, a change in twist and subunit tilt is observed (28; 27; 24), which could destabilize both the longitudinal (48) and lateral (53) filament subunit interfaces. These filament structural changes presumably contribute to cooperative binding since no direct contacts exist between bound cofilin molecules (24).
Computational modeling has provided valuable insight concerning the atomic level changes in the actin subunits and filament mechanical properties that result from cofilin binding (30; 54). Coarse-grain and molecular dynamics simulations of actin filaments (30; 54) reveal an outward radial movement of the DNAase I binding loop in actin subdomain 2. This conformational change alters the long-axis subunit interface, potentially destabilizing filament contacts, as inferred from biochemical solution studies (48).
The cofilin-mediated reorganization of actin subdomain 2 presumably narrows the overall filament radial contact distribution between actin subunits (29) yielding cofilactin filaments that bend and twist more readily than their bare counterparts. Direct visualization of filaments undergoing thermally-driven fluctuations in shape allows for measurement of filament flexural rigidity (55–57). The apparent cofilactin filament Young’s modulus is ~20-fold lower (after accounting for the predicted radial mass distribution change) than that of bare filaments (29). As a results, cofilin binding increases the thermally-driven filament bending amplitudes and modes (23)(Figure 2B). Cofilin also makes actin filaments ~18-fold more compliant in twisting (21).
Cofilin promotes severing at mechanical discontinuities
It has been proposed that discontinuities in filament mechanics and dynamics generate local accumulation of stress at boundaries of bare and cofilin-decorated segments (17; 58; 29; 38; 23; 59; 19). Such a model predicts that bare and fully decorated filaments sever less readily than partially decorated, which has been observed (17; 58; 29; 59). Direct visualization of cofilin, actin, and subsequent filament severing events demonstrate that the severing probability is higher at boundaries than within bare or cofilin-decorated segments (38). The model also predicts that cofilins that do not alter filament mechanics will not sever. Consistent with this prediction, vertebrate cofilin minimally alters yeast actin filament flexibility and weakly severs them (23).
Severing at mechanical discontinuities may also contribute to severing of filament bundles and networks. Cofilin severs fascin cross-linked bundles more readily than single filaments, despite reduced access to filament binding sites (60). These results suggest that both the stiffening effect of fascin (61), as well as the non-uniform distribution of cofilin on the bundles due to competitive fascin binding, enhances cofilin severing activity by promoting more pronounced mechanical discontinuity.
In contrast, Arabidopsis thaliana villin 1-crosslinked filament bundles are resistant to ADF/cofilin-mediated depolymerization (62). The factors contributing to the distinct severing of fascin and villin bundles are not understood. Competition and limited site accessibility is likely to influence severing efficiency towards bundles, and may fully account for the inhibiting effect of villin (62). However, the enhanced severing of fascin bundles (60) favors a mechanism that overcomes reduced site accessibility. Differences in crosslinker compliance or effects on filament mechanics could modulate cofilin severing efficiency, as can the crosslinker binding mode (e.g. contacting multiple filament subunits could accumulate stress at the subunit interfaces more effectively than contacts made predominantly with an individual subunit, as in the case of villin (63)).
The critical bending angle (i.e. angle at which bending deformations become irreversible) is smaller for fragmentation at bare and cofilin-decorated segment boundaries compared to bending sites within the segments (23). This behavior suggests that boundaries (either between cofilin-decorated segments and bare or potentially certain actin-binding protein decorated segments) are more susceptible to fracture than non-boundary segments, analogous to the adhesive joint failure of non-protein materials(64) . Such behavior could result from the altered actin subunit interfaces resulting from the filament twist and subunit tilt that occurs upon cofilin binding.
Although the proposed severing mechanism described here applies to vertebrate cofilin severing, not all cofilins may function in the same way. In contrast to vertebrate cofilin, yeast cofilin that is competent to sever yeast actin may act through a different mechanism (23). Other severing proteins such as gelsolin require other cofactors such as calcium and bind to other sites on the actin filament (65; 66).
Acknowledgement
Cofilactin research activities are supported by grants from the NIH (RO1-GM097348 awarded to EMDLC) and an American Heart Association Established Investigator Award (0655849T awarded to EMDLC).
Footnotes
The authors declare no conflict of interest.
References
- 1.Pollard T, Borisy G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 2.Fass J, Gehler S, Sarmiere P, Letourneau P, Bamburg J. Regulating filopodial dynamics through actin-depolymerizing factor/cofilin. Anatomical Science International. 2004;79:173–183. doi: 10.1111/j.1447-073x.2004.00087.x. [DOI] [PubMed] [Google Scholar]
- 3.Oosawa F, Asakura S. Thermodynamics of the polymerization of protein. Academic Press; 1975. [Google Scholar]
- 4.Howard J. Mechanics of motor proteins and the cytoskeleton. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- 5.De La Cruz E, Roland J, McCullough B, Blanchoin L, Martiel J. Origin of twist-bend coupling in actin filaments. Biophysical Journal. 2010;99:1852–1860. doi: 10.1016/j.bpj.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bamburg J. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annual Review of Cellular and Developmental Biology. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- 7.Wang W, Eddy R, Condeelis J. The cofilin pathway in breast cancer invasion and metastasis. Nature Reviews Cancer. 2007;7:429–440. doi: 10.1038/nrc2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De La Cruz E. How cofilin severs an actin filament. Biophysical Reviews. 2009;1:51–59. doi: 10.1007/s12551-009-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein B, Bamburg J. ADF/cofilin: A functional node in cell biology. Trends in Cell Biology. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moriyama K, Yahara I. The actin-severing activity of cofilin is exerted by the interplay of three distinct sites on cofilin and essential for cell viability. Biochemical Journal. 2002;365:147–155. doi: 10.1042/bj20020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurniak C, Perlas E, Witke W. The actin depolymerizing factor n-cofilin is essential for neural tube morphogenesis and neural crest cell migration. Developmental Biology. 2005;278:231–241. doi: 10.1016/j.ydbio.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 12.Agrawal P, Greenleaf R, Tomczak K, Lehtokari V, Wallgren-Pettersson C, Wallefeld W, Laing N, Darras B, Maciver S, Dormitzer P, Beggs A. Nemaline myopathy with minicores caused by mutation of the CFL2 gene encoding the skeletal muscle actin-binding protein, cofilin-2. American Journal of Human Genetics. 2007;80:162–167. doi: 10.1086/510402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ockeloen C, Gilhuis H, Pfundt R, Kamsteeg E, Agrawal P, Beggs A, Hama-Amin A, Diekstra A, Knoers N, Lammens M, van Alfen N. Congenital myopathy caused by a novel missense mutation in the CFL2 gene. Neuromuscular Disorders. 2012;22:632–639. doi: 10.1016/j.nmd.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawkins M, Pope B, Maciver S, Weeds A. Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry. 1993;32:9985–9993. doi: 10.1021/bi00089a014. [DOI] [PubMed] [Google Scholar]
- 15.Hayden S, Miller P, Brauweiler A, Bamburg J. Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry. 1993;32:9994–10004. doi: 10.1021/bi00089a015. [DOI] [PubMed] [Google Scholar]
- 16.Ressad F, Didry D, Xia G, Hong Y, Chua N, Pantaloni D, Carlier M. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins: Comparison of plant and human ADFs and effect of phosphorylation. Journal of Biological Chemistry. 1998;273:20894–20902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- 17.De La Cruz E. Cofilin binding to muscle and non-muscle actin filaments: Isoform-dependent cooperative interactions. Journal of Molecular Biology. 2005;346:557–564. doi: 10.1016/j.jmb.2004.11.065. [DOI] [PubMed] [Google Scholar]
- 18.Pope B, Gonsior S, Yeoh S, McGough A, Weeds A. Uncoupling actin filament fragmentation by cofilin from increased subunit turnover. Journal of Molecular Biology. 2000;298:649–661. doi: 10.1006/jmbi.2000.3688. [DOI] [PubMed] [Google Scholar]
- 19.Yeoh S, Pope B, Mannherz H, Weeds A. Determining the differences in actin binding by human ADF and cofilin. Journal of Molecular Biology. 2002;315:911–925. doi: 10.1006/jmbi.2001.5280. [DOI] [PubMed] [Google Scholar]
- 20.Cao W, Goodarzi J, De La Cruz E. Energetics and kinetics of cooperative cofilin-actin filament interactions. Journal of Molecular Biology. 2006;361:257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 21.Prochniewicz E, Janson N, Thomas D, De La Cruz E. Cofilin increases the torsional flexibility and dynamics of actin filaments. Journal of Molecular Biology. 2005;353:990–1000. doi: 10.1016/j.jmb.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 22.Blanchoin L, Pollard T. Mechanicsm of interaction of Acanthamoeba actophorin (ADF/cofilin) with actin filaments. Journal of Biological Chemistry. 1999;274:15538–15546. doi: 10.1074/jbc.274.22.15538. [DOI] [PubMed] [Google Scholar]
- 23.McCullough B, Grintsevich E, Chen C, Kang H, Hutchison A, Henn A, Cao W, Suarez C, Martiel J, Blanchion L, Reisler E, De La Cruz E. Cofilin-linked changes in actin filament flexibility promote severing. Biophysical Journal. 2011;101:151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGough A, Pope B, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. Journal of Cell Biology. 1997;138:771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galkin V, Orlova A, Van Loock M, Shvetsov A, Reisler E, Egelman E. ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. Journal of Cell Biology. 2003;163:1057–1066. doi: 10.1083/jcb.200308144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orlova A, Egelman E. A conformational change in the actin subunit can change the flexibility of the actin filament. Journal of Molecular Biology. 1993;232:334–341. doi: 10.1006/jmbi.1993.1393. [DOI] [PubMed] [Google Scholar]
- 27.Galkin V, Orlova A, Lukoyanova N, Wriggers W, Egelman E. Actin depolymerizing factor stabilizes and existing state of F-actin and can change the tilt of F-actin subunits. Journal of Cell Biology. 2001;153:75–86. doi: 10.1083/jcb.153.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galkin V, Orlova A, Kudryashov D, Solodukhin A, Reisler E, Schroder G, Egelman E. Remodeling of actin filaments by ADF/cofilin proteins. PNAS. 2011;108:20568–20572. doi: 10.1073/pnas.1110109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCullough B, Blanchion L, Martiel J, De La Cruz E. Cofilin increases the bending flexibility of actin filaments: Implications for severing and cell mechanics. Journal of Molecular Biology. 2008;381:550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfaendtner J, De La Cruz E, Voth G. Actin filament remodeling by actin depolymerization factor/cofilin. PNAS. 2010;107:7299–7304. doi: 10.1073/pnas.0911675107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mochrie S, Mack A, Regan L. Allosteric conformational spread: Exact results using a simple transfer matrix method. Physical Review E, statistical, nonlinear , and soft matter physics. 2010;82:031913. doi: 10.1103/PhysRevE.82.031913. [DOI] [PubMed] [Google Scholar]
- 32.McGhee J, Von Hippel P. Theoretical aspects of DNA-protein interactions: Co-operative and non-co-operative binding of large ligands to a one-dimensional homogeneous lattice. Journal of Molecular Biology. 1974;86:469–489. doi: 10.1016/0022-2836(74)90031-x. [DOI] [PubMed] [Google Scholar]
- 33.De La Cruz E, Sept D. The kinetics of cooperative cofilin binding reveals two states of the cofilin-actin filament. Biophysical Journal. 2010;98:1893–1901. doi: 10.1016/j.bpj.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hannemann D, Cao W, Olivares A, Robblee J, De La Cruz E. Magnesium, ADP, and actin binding linkage of myosin V: Evidence for multiple myosin V - ADP and actomyosin V - ADP states. Biochemistry. 2005;44:8826–8840. doi: 10.1021/bi0473509. [DOI] [PubMed] [Google Scholar]
- 35.De La Cruz E, Pollard T. Transient kinetic analysis of rhodamine phalloidin binding to actin filaments. Biochemistry. 1994;33:14387–14392. doi: 10.1021/bi00252a003. [DOI] [PubMed] [Google Scholar]
- 36.De La Cruz E, Pollard T. Kinetics and thermodynamics of phalloidin binding to actin filaments from three divergent species. Biochemistry. 1996;35:14054–14061. doi: 10.1021/bi961047t. [DOI] [PubMed] [Google Scholar]
- 37.Bobkov A, Muhlrad A, Pavlov D, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. Journal of Molecular Biology. 2006;356:325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- 38.Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough B, Reymann A, Guerin C, Martiel J, De La Cruz E, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Current Biology. 2011;21:862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bujalowski W, Lohman T, Anderson C. On the cooperative binding of large ligands to a one-dimensional homogeneous lattice: The generalized three-state lattice model. Biopolymers. 1989;28:1637–1643. doi: 10.1002/bip.360280912. [DOI] [PubMed] [Google Scholar]
- 40.Moriyama K, Yahara I. Two activities of cofilin, severing and accelerating directional depolymerization of actin filaments, are effected differentially by mutations around the actin-binding helix. EMBO Journal. 1999;18:6752–6761. doi: 10.1093/emboj/18.23.6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frederick K, Sept D, De La Cruz E. Effects of solution crowding on actin polymerization reveal the energetic basis for nucleotide-dependent filament stability. Journal of Molecular Biology. 2008;378:540–550. doi: 10.1016/j.jmb.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang J, Janmey P. The polyelectrolyte nature of F-actin and the mechanism of actin bundle formation. Journal of Biological Chemistry. 1996;271:8556–8563. doi: 10.1074/jbc.271.15.8556. [DOI] [PubMed] [Google Scholar]
- 43.Kang H, Bradley M, McCullough B, Pierre A, Grintsevich E, Reisler E, De La Cruz E. Identification of cation-binding sites on actin that drive polymerization and modulate bending stiffness. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:16923–16927. doi: 10.1073/pnas.1211078109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muhlrad A, Kudryashov D, Peyser Y, Bobkov A, Almo S, Reisler E. Cofilin induced conformational changes in F-actin expose subdomain 2 to proteolysis. Journal of Molecular Biology. 2004;342:1559–1567. doi: 10.1016/j.jmb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Chu J, Voth G. Allostery of actin filaments: Molecular dynamics simulations and coarse-grained analysis. PNAS. 2005;102:13111–13116. doi: 10.1073/pnas.0503732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chu J, Voth G. Coarse-grained modeling of the actin filament derived from atomistic-scale simulations. Biophysical Journal. 2006;90:1572–1582. doi: 10.1529/biophysj.105.073924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prochniewicz E, Chin H, Henn A, Hannemann D, Olivares A, Thomas D, De La Cruz E. Myosin isoform determines the conformational dynamics and cooperativity of actin filaments in the strongly bound actomyosin complex. Journal of Molecular Biology. 2010;396:501–509. doi: 10.1016/j.jmb.2009.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bobkov A, Muhlrad A, Kokabi K, Vorobiev S, Almo S, Reisler E. Structural effects of cofilin on longitudinal contacts in F-actin. Journal of Molecular Biology. 2002;323:739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- 49.Dedova I, Dedov V, Nosworthy N, Hambly B, Dos Remedios C. Cofilin and DNase 1 affect the conformation of the small domain of actin. Biophysical Journal. 2002;82:3134–3143. doi: 10.1016/S0006-3495(02)75655-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paavilainen V, Oksanen E, Goldman A, Lappalainen P. Structure of the actin-depolymerizing factors homology domain in complex with actin. Journal of Cell Biology. 2008;182:51–59. doi: 10.1083/jcb.200803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scoville D, Stamm J, Altenbach C, Shvetsov A, Kokabi K, Rubenstein P, Hubbell W, Reisler E. Effects of binding factors on structural elements in F-actin. Biochemistry. 2009;48:370–378. doi: 10.1021/bi801649j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim E, Reisler E. Intermolecular coupling between loop 38–52 and the C-terminus in actin filaments. Biophysical Journal. 1996;71:1914–1919. doi: 10.1016/S0006-3495(96)79390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bobkov A, Muhlrad A, Shvetsov A, Benchaar S, Scoville D, Almo S, Reisler E. Cofilin (ADF) affects lateral contacts in F-actin. Journal of Molecular Biology. 2004;337:93–104. doi: 10.1016/j.jmb.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Yogurtcu O, Kim J, Sun S. A mechanochemical model of actin filaments. Biophysical Journal. 2012;103:719–727. doi: 10.1016/j.bpj.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. Journal of Cell Biology. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isambert H, Venier P, Maggs A, Fattoum A, Kassab R, Pantaloni D, Carlier M. Flexibility of actin filaments derived from thermal fluctuations: Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. Journal of Biological Chemistry. 1995;270:11437–11444. doi: 10.1074/jbc.270.19.11437. [DOI] [PubMed] [Google Scholar]
- 57.Le Goff L, Hallatschek O, Frey E, Amblard F. Tracer studies on F-actin fluctuations. Physical Review Letters. 2002;89:258101. doi: 10.1103/PhysRevLett.89.258101. [DOI] [PubMed] [Google Scholar]
- 58.Andrianantoandro E, Pollard T. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Molecular Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Pavlov D, Muhlrad A, Cooper J, Wear M, Reisler E. Actin filament severing by cofilin. Journal of Molecular Biology. 2007;365:1350–1358. doi: 10.1016/j.jmb.2006.10.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Breitsprecher D, Koestler S, Chizhov I, Nemethova M, Mueller J, Goode B, Small J, Rottner K, Faix J. Cofilin cooperates with fascin to disassemble filopodial actin filaments. Journal of Cell Science. 2011;124:3305–3318. doi: 10.1242/jcs.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lieleg O, Claessens M, Heussinger C, Frey E, Bausch A. Mechanics of bundled semi-flexible polymer networks. Physics Review Letters. 2007;99:088102. doi: 10.1103/PhysRevLett.99.088102. [DOI] [PubMed] [Google Scholar]
- 62.Huang S, Robinson R, Gao L, Matsumoto T, Brunet A, Blanchoin L, Staiger C. Arabidopsis VILLIN1 generates actin filament cables that are resistant to depolymerization. The Plant Cell. 2005;17:486–501. doi: 10.1105/tpc.104.028555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hampton C, Liu J, Taylor D, DeRosier D, Taylor K. The 3D structure of villin as an unusual F-actin crosslinker. Structure. 2008;16:1882–1891. doi: 10.1016/j.str.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith W, Hashemi J. Foundations of materials science and engineering. New York, NY: McGraw-Hill; 2004. [Google Scholar]
- 65.Yin H, Zaner K, Stossel T. Ca2+ control of actin gelation. Journal of Biological Chemistry. 1980;255:9494–9500. [PubMed] [Google Scholar]
- 66.Janmey P, Stossel T. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature. 1987;325:362–364. doi: 10.1038/325362a0. [DOI] [PubMed] [Google Scholar]