Abstract
Background
In forensic science there are many types of crime that involve animals. Therefore, the identification of the species has become an essential investigative tool. The exhibits obtained from such offences are very often a challenge for forensic experts. Indeed, most biological materials are traces, hair or tanned fur. With hair samples, a common forensic approach should proceed from morphological and structural microscopic examination to DNA analysis. However, the microscopy of hair requires a lot of experience and a suitable comparative database to be able to recognize with a high degree of accuracy that a sample comes from a particular species and then to determine whether it is a protected one. DNA analysis offers the best opportunity to answer the question, ‘What species is this?’ In our work, we analyzed different samples of fur coming from China used to make hats and collars. Initially, the samples were examined under a microscope, then the mitochondrial DNA was tested for species identification. For this purpose, the genetic markers used were the 12S and 16S ribosomal RNA, while the hypervariable segment I of the control region was analyzed afterwards, to determine whether samples belonged to the same individual.
Results
Microscopic examination showed that the fibres were of animal origin, although it was difficult to determine with a high degree of confidence which species they belonged to and if they came from a protected species. Therefore, DNA analysis was essential to try to clarify the species of these fur samples.
Conclusions
Macroscopic and microscopic analysis confirmed the hypothesis regarding the analyzed hair belonging to real animals, although it failed to prove with any kind of certainty which actual family it came from, therefore, the species remains unknown. Sequence data analysis and comparisons with the samples available in GenBank showed that the hair, in most cases, belonged to the Canidae family, and in one case only to Felidae.
Keywords: Fur samples, High degraded samples, Microscopic analysis, mtDNA, Species identification
Background
Forensic analysis tends to focus mainly on human DNA typing, but this is not the only source of DNA that can be found at a crime scene. Many types of crime involve nonhuman DNA samples, such as those of animals, plants, pollen, bacterial and viruses [1-4]. Despite being a relatively new subdiscipline of forensic science, the interest of the forensic community regarding nonhuman DNA samples is considerable, as evidenced by [5]. The identification of a species and the linking of a sample to a specific individual are becoming important tools in forensic investigation, owing to an increase in wildlife crime. One of the main reasons for the increase of wildlife crime probably comes from the fact that it is highly profitable and the penalties for guilty people are often light. Most of the time, the potential profit far outweighs the maximum penalty for the alleged crime [6]. Interpol estimates that illegal wildlife trade is the largest black market worldwide, second only to the traffic in drugs [7-9].
Many mammalian species, such as tigers, elephants, bears, rhinoceroses, and, mainly, reptiles and amphibians are trafficked every year around the world [10-19]. Crimes range from the abuse of illegal trade in protected species, or parts of them, such as horns, eggs [20] and tusks used to make ornaments or jewellery [21-26] or fur used to make gloves, hats and collars, to poaching and hunting out of season and many others. Italy was the first in Europe, and second in the world after the USA, to have imposed a ban on imports and the domestic trade of skins and furs from protected animals, firstly, through the ordinances of Dr Sirchia, the Minister of Health, issued in 2001 and renewed within two years, and then, in 2004, with the inclusion of the prohibition into the law against the mistreatment of animals [27]. The Italian law (article 2) prohibits the use of skins and furs for commercial purposes: ‘Dogs (Canis familiaris) and cats (Felis catus) may not be used in the production or packaging of skins, furs, clothing and leather goods made or produced, in whole or in part, from the their skins or fur, as well as their skins or fur, cannot be marked or introduced into the national territory. Violation shall be punishable by imprisonment for periods of three months to one year or by a fine from 5,000 to 100,000 euros’. Therefore, the ability to identify a particular species and the capacity to determine whether the biological sample belongs to a specific individual of that species are an important tool in investigations.
Morphologic and structural microscopic analyses are the first approach in investigating the nature of the samples, excluding synthetic origins of the fibres, distinguishing between human and animal hair and trying to identify the species. These techniques are also important in comparing the features of the questioned hair with those collected from a known individual. In the case of animal hair, it is important to focus on shapes and sizes, colour banding, cuticular patterns, medulla organization, root structure and scales for its identification. A table presented in the Encyclopedia of Forensic Science[28] shows in detail all animal hair characteristics and traits that have to be considered during microscopic examination. Moreover, an adequate reference collection is essential for accurate identification of samples. Indeed, the wide variation in characteristics of hair taken from different body areas on the same animal and the large differences among animals that can occur within the same species do not allow the identification of the species of animal hair if a complete and accurate reference sample of hair is not available. Furthermore, much experience in microscopic hair analysis is required before one can confirm with a high degree of confidence whether or not a sample belongs to a protected species. However, even with experience, sometimes only the taxonomic rank of family may be identified.
When species identification using these methods is inconclusive, the only possible solution is to resort to more expensive DNA analysis. The application of DNA-based technologies to species identification has made the investigation of wildlife crimes possible. For several reasons, the DNA markers used in forensic species identification are located in the mitochondrial genome and derived from taxonomic and phylogenetic studies [29]. One of the main reasons for choosing mitochondrial DNA (mtDNA) as a forensic marker in identification studies is that there is no recombination. With the exception of mutations, all the mtDNA molecules of an individual are equal and inherited through the maternal line [30,31]. Moreover, the mtDNA molecules are present in multiple copies per cell [32] and the mitochondrion has a protein coat that helps to protect it from degradation: in highly degraded samples, such as bones, teeth and, obviously, the hair shaft [33-39], these characteristics make this approach more suitable than the nuclear DNA approach. Additionally, enzymes in the mitochondria are not able to read and correct DNA bases added incorrectly during replication [40], so that mutations can be accumulated in mtDNA up to five times more quickly than those accumulated during nuclear DNA replication.
There are many studies of species identification and many different markers can be utilized. The choice of marker for forensic species identification shows little intraspecific variability, that is, variability between members of the same species, but demonstrates sufficient interspecific variability, that is, between members of different species, to discriminate between individuals of different species. One of these loci is cytochrome b[41-43]. Over the years, several forensic and taxonomic studies have utilized cytochrome b as a marker for species identification [44-56]. However, more recently, there has been an increase in the use of another marker called cytochrome c oxidase I (COI). This is used in DNA barcoding; it has been adopted as a marker by the Barcode for Life Consortium [57-60]. Initially, it was used in the identification of invertebrate species [61-65]; for example, it has been exploited as a marker in forensic entomology to identify the beetle larvae on a corpse [66,67]. Since then, COI has been proposed for the identification of many organisms [68-82]. Other mitochondrial genes can be utilized as markers in forensic species identification: 12S ribosomal RNA [83-85], 16S ribosomal RNA [86-88] and the NDH family [89,90]. While all these genes have been investigated for species identification, the D-loop analysis has mostly been exploited for intraspecies identification [91-96], though sometimes it can be applied to species identification [23,43,85,97-99]. Usually, the standard analysis of species identification requires amplification of part of these genes, followed by sequencing and comparison of the obtained sequence with reference sequences kept in a database, such as GenBank.
Here, we report a case of the alleged illegal trade of a protected species of fur. Our work consisted of ascertaining whether the furs had a synthetic origin or belonged to Canis lupus familiaris, Felis silvestris catus or other species. Furthermore, we had to assess whether the different furs belonged to the same individual. Initially, we focused on ruling out a synthetic origin for the fur and on trying to assess which species the hair belonged to by microscopic analysis. Finally, the species was identified by DNA analysis. The genetic markers used were 12S and 16S rRNA and the control region (hypervariable segment I, HVS-I) for individual identification. The advantages of this assay were: (1) the abundance of the mtDNA genome, (2) the broad reactivity of the primers that permitted the amplification of the DNA from unknown samples, (3) the small size of the amplicons, particularly the 12S rRNA, which was ≈ 150 base pairs (bp) and (4) the amplification of seven overlapping HVS-I fragments with sizes ranging from 130 to 250 bp.
Methods
Samples
Twelve fur samples (named A1, A2, A3, A4, A5, B1, B2, B3, B4, B5, B6 and B7) of alleged animal origin from protected species, in Italy, coming from China and used to make hats and collars, were seized just outside Genoa. In Italy, since the law 189/2004 has prohibited use of the skins and furs of domestic dogs (Canis lupus familiaris) and cats (Felis silvestris catus) for commercial purposes, microscopic and DNA evidence was necessary to ascertain the origin of these fibres and permit identification of the species. The seized fur was of different colours: black, brown, grey and red. Despite being subjected to thermal, chemical and mechanical stress during the processing cycle of tanning, the furs showed an apparently good state of preservation. However the amount of DNA in hair [100] is low compared with other tissues. Indeed, hair cells undergo dehydration and catabolic breakdown of nucleic acids and organelles during keratinization. Nevertheless, forensic scientists have successfully amplified DNA from modern human hair [101] and analyzed mtDNA from degraded and old hair samples [102-105].
Microscopic analysis
Stereomicroscopy
First, the samples were carefully observed using a Leica M 651 stereomicroscope (Leica Microsystems Srl, Milano, Italy). This allowed the observation of samples in three dimensions, thereby avoiding the flattening effect typical of other microscopes. At low magnifications of approximately 6.5× to 40×, this analysis permitted the reproduction a three-dimensional image in direct vision and the examination of colour, texture, structure and treatment effects of samples.
Light microscopy
In forensic laboratories, the examination of the internal structures of this type of sample is typically carried out using a light microscope. Therefore, the hair samples were prepared for microscopic examination by placing them on glass slides with a colourless mounting medium in the refractive index range of 1.5 to 1.60. The mounting medium utilized was Hi-MO (Bio Optica, Milano S.p.A., Italy) with a refractive index of 1.5. To see a cuticular pattern more clearly, it was necessary to make a scale cast of the hair specimens. A thin coating of clear nail polish was set on a microscope slide. While still wet, each hair was positioned in the nail polish, ensuring that the entire length of the hair was included. Then, when the nail polish was completely dry, the hair was gently removed to obtain the scale cast [106]. To prevent contamination of each fur sample, the microscope slides were prepared under a hood. The light microscope utilized was a Leica DM LB2 (Leica Microsystems Srl, Milano, Italy).
Genetic analysis
Highly degraded DNA analysis
To obtain reliable results, owing to the stress caused by the tanning process and the characteristics of the hair, genetic analysis for these findings was conducted following the most stringent criteria proposed for ancient DNA studies [107-109]. Accepted authentication methods must be employed to assess the authenticity of the results. Different precautions were taken to avoid contamination of samples with extraneous DNA: (1) all DNA extraction and PCR involving the samples was carried out in a laboratory physically separated from the laboratory in which PCR cycling and post-PCR analyses were performed; (2) disposable masks, gloves and laboratory coats were worn throughout and were changed frequently; and (3) all DNA extractions and PCR reactions included negative controls. Repeated amplification from the same or different extracts from the same specimens are necessary: (1) to identify contamination of a particular extraction or amplification; (2) to allow DNA analysis when the extracts only sporadically contain DNA template molecules; and (3) to detect nucleotide misincorporations that produce consistent changes in the DNA molecule. In highly degraded samples, one of the most common modifications of (or damage caused to) the DNA molecule is the hydrolytic loss of amino groups from the nitrous bases [110]. The deamination products of cytosine (uracil), 5-methylcytosine (thymine) and adenine (hypoxanthine) have particular relevance to the amplification of ancient DNA, since they cause the incorrect bases (A instead of G and C instead of T) to be inserted when new DNA strands are synthesized by a DNA polymerase [111,112]. In addition, ‘jumping PCR’ , that is, the occurrence of template switching during PCR, may contribute to these substitutions. To distinguish between these two types of miscoding lesion, multiple amplifications of DNA extracts were performed and the PCR products were cloned. By comparing different clones from multiple amplifications, it will be possible to highlight the nucleotide differences that occur in all clones of one amplification from those that occur only sporadically [113]. A consensus sequence can be constructed from the selected clone sequences once data from possible jumping PCR events has been identified and removed. Bower et al.[114] demonstrated that 20 clones must be sampled from a single forensic or ancient DNA sample in order to determine a reliable consensus sequence.
DNA fragments derived from genomes of organelles, such as mitochondria [115], are often present in the nuclear genome [116] and, as reported by Jan den Tex et al.[117], they cause another problem for ancient DNA analysis. Since mitochondrial DNA is the molecule of interest in most ancient DNA projects, such nuclear integrations may occasionally be amplified by PCR and be mistaken for the organellar DNA sequences. This is particularly likely to happen if the primers used differ from the organellar DNA sequence in the individual specimen but not from the version of the same sequence that exists as a nuclear insertion. Erroneous conclusions regarding intraspecific variation [118], as well as species phylogenies [119], will then result. To prevent this problem, different primer sets can be used to amplify different overlapping fragments covering a target region, since it is very unlikely that two or more primer sets would preferentially amplify a particular nuclear insertion [119,120]. Furthermore, the use of multiple genes improves the chances of correctly identifying the species of the sample [121,122].
Sample preparation and DNA extraction
Sample preparation, DNA extraction and PCR reaction setup were carried out in a laboratory dedicated to ancient DNA analysis. The pre-PCR area was physically separated from the area in which PCR cycling and post-PCR analyses were conducted. The reagents, bench surface and nondisposable equipment were sterilized under ultraviolet light. Four or five hairs from each fur sample were decontaminated by soaking in a 5% bleach solution for 30 s, followed by a rinse carried out initially in absolute ethanol and then in sterile distilled water. The samples were left to air-dry and then cut into small pieces, about 2 to 3 mm, and collected in a sterile 1.5 ml vial. The DNA extraction was performed according to the protocol of Gilbert et al.[105]. The fragmented samples were digested overnight at 55°C in a 0.5 ml extraction buffer (modified, as to the protocol of Barnes et al.[123]) containing 0.01 M Tris buffer, 0.01 M NaCl solution, 1% SDS, 0.5 mg/ml proteinase K, 10 mg/ml dithiothreitol and 0.001 M N-phenacylthiazolium bromide. The DNA was extracted using a phenol-chloroform DNA extraction protocol, purified and concentrated by Amicon ultra 0.5 ml centrifugal filters (Millipore), according to the user’s manual, to yield a final volume of approximately 50 μl. At least two independent extracts were obtained from each fur sample. Negative controls were included in each extraction set.
PCR amplification of 12S and 16S
Two microlitres of DNA from each extract were amplified as follows: 94°C for 10 min (Taq polymerase activation), followed by 40 cycles of PCR (denaturation, 94°C for 45 s, annealing, 53°C for 1 min and extension, 72°C for 1 min) and then a final step at 72°C for 10 min. The 50 μl reaction mix contained 2 U of AmpliTaq Gold (Applied Biosystems), 200 μM of each deoxyribonucleotide triphosphate (dNTP), 1.5 mM MgCl2, 1× PCR buffer and 1 μM of each primer. According to Melton et al.[84], the 12S primer sequences amplified a 150 bp fragment. According to Kitano et al.[85], the 16S primer sequences amplified a 244 bp fragment. Each extract was amplified in duplicate. The extraction reagent control, a known positive control (human DNA) and a negative PCR control were amplified in parallel. Thirty-five microlitres of each PCR product were analyzed by electrophoresis, on a 1.5% agarose gel for 45 min at 80 V and visualized with ethidium bromide. To identify the molecular-weight size of PCR products we used a 50 bp ladder (New England BioLab Inc.). The PCR products were purified using a Montage™ DNA gel extraction kit (Millipore), according to the user’s manual.
PCR amplification of dog HVS-I
The control region of dog mtDNA is 1270 bp long and contains two hypervariable segments (HVS-I and HVS-II) and a 10 nt variable number tandem repeat (VNTR) between HVS-I and HVS-II [95]. We analyzed the HVS-I region and, as recommended for analysis of ancient DNA samples, the hypervariable region was divided into overlapping fragments, to prevent the amplification of nuclear insertion and increase the probability of amplification success if the DNA is degraded and fragmented [119,120]. The 672-bp-long HVS-I was subdivided into seven overlapping fragments, using the primer pairs reported in Table 1. Some primer sequences, such as L15422/H15555 and L15511/H15692, utilized for analysis, were obtained from Verginelli et al.[124,125]. The other primers were designed using the Primer3 program [126]. The nomenclature of the primers corresponds with the 5′-end of the reference sequence, according to [127]. The size of PCR products ranged from 130 to 250 bp. The HVS-I was amplified in a final volume of 50 μl in seven different assays including 2 U of AmpliTaq Gold (Applied Biosystems), 200 μM of each dNTP, 1.5 mM MgCl2, 1× PCR buffer and 1 μM of each primer. Each fragment was amplified in duplicate. The extraction reagent control, a known positive control (dog DNA) and a negative PCR control were amplified in parallel. The PCR products were analyzed by electrophoresis on a 1.5% agarose gel, as described in the previous paragraph.
Table 1.
Primer pairs used to amplify the seven overlapping fragments of HVS-I with molecular details
| Amplicon size (bp) | Primer name | Primer range | Primer sequence 5′-3′ | Sequence information range (bp) | Reference |
|---|---|---|---|---|---|
|
170 |
L15422 H15555 |
15404-15422 15555-15574 |
CTCTTGCTCCACCATCAGC TTATATGCATGGGGCAAACC |
133 |
[124] |
|
219 |
L15511 H15692 |
15490-15511 15692-15709 |
ACTGTGCTATGTCAGTATCTCC TTGATGGTTTCTCGAGGC |
181 |
[124] |
|
246 |
L15574 H15780 |
15555-15574 15780-15801 |
GGTTTGCCCCATGCATATAA AAGTAAGAACCAGATGCCAGGT |
206 |
[126] |
|
148 |
L15764 H15873 |
15744-15764 15873-15892 |
CCCATACTAACGTGGGGGTTA TGTGTGATCATGGGCTGATT |
109 |
[126] |
|
129 |
L15857 H15945 |
15836-15857 15945-15965 |
ATTCTCGCAAATGGGACATC GCGGTCGTAGGTGAGTGATAG |
88 |
[126] |
|
137 |
L15892 H15991 |
15873-15892 15991-16010 |
AATCAGCCCATGATCACACA GGCATATTCCCTGAGAGCAG |
99 |
[126] |
| 154 | L15982 H16097 | 15963-15982 16097-16117 | CGCAACGGCACTAACTCTAA TGTCCTGAAACCATTGACTGA | 115 | [126] |
Cloning and sequencing
The PCR products were cloned using a TOPO TA Cloning Kit (Invitrogen), according to the manufacturer’s instructions. Screening of 20 white recombinant colonies was accomplished by PCR, and the colonies were transferred into a 30 ml reaction mix (67 mM Tris HCl (pH 8.8), 2 mM MgCl2, 1 mM of each primer, 0.125 mM of each dNTP, 0.75 units of Taq polymerase) containing M13 forward and reverse universal primers. After 5 min at 92°C, 30 cycles of PCR (30 s at 90°C, 1 min at 50°C, 1 min at 72°C) were carried out and clones with samples of the expected size were identified by agarose gel electrophoresis. After the purification of all PCR products, using a PureLink™ PCR Purification kit (Invitrogen™), a volume of 1 μl was cycle-sequenced using a BigDye® Terminator v3.1 cycle sequencing kit™ (Applied Biosystems), following the supplier’s instructions. Sequencing involves initial denaturation at 96°C for 1 min followed by 25 cycles at 96°C for 10 s, 50°C for 5 s and 60°C for 4 min. Sequencing reaction products were purified from residual dye terminators using Agencourt® CleanSEQ® (Beckman Coulter), according to the manufacturer’s manual. Sequencing was carried out on Applied BioSystems 3130 × l Genetic Analyzer (Applied Biosystems) using POP7, 36 cm capillary arrays and the default instrument settings, as recommended by the manufacturer. The data were analyzed using SeqScape® Software Version 2.5 (Applied Biosystems) and default software settings, as recommended by the manufacturer. Twenty clones for each sample were sequenced, and the sequences obtained were aligned using the Clustal X program [128] and compared across clones. Then, for each sample, the consensus sequence of 12S, 16S and HVS-I was copied and pasted into the nucleotide-nucleotide BLAST function at the National Centre for Biotechnology Information (NCBI) website [129]. The BLAST portal generated a report showing the degree of sequence similarity with organisms in the NCBI database. If no NCBI reference presented a similarity of 100% with all the query sequences, further phylogenetic analyses were performed. Species identifications were made based on the percentage similarity of the query sequence to the reference sequence (>98%) as described in previous papers [20,130].
Phylogenetic analysis
A comparison dataset of sequences of the mitochondrial HVS-I was compiled, in order to represent the known intra- and interspecific variation between the species and subspecies for which almost 98% of similarity was detected by the nucleotide-nucleotide BLAST search with the results obtained from the fur samples. All the sequences (from the dataset and the fur samples) were analyzed using MEGA version 6 [131]; they were aligned using the MUSCLE algorithm [132], and the best substitution model was assessed and used to develop a neighbour-joining tree [133], including a bootstrap analysis with 1,000 replications, to determine which species or subspecies the fur samples clustered with.
Results and discussion
Macroscopic and microscopic analysis
The analysis of hair samples began with a macroscopic approach and then moved to a microscopic one. The visible characteristics of the samples were initially observed with the naked eye and described, and then studied using a stereomicroscope (macroscopic analysis) to determine colour, texture, structure and treatment effects of the samples. The seized samples consisted of fur of different colours (Table 2).Some of the fur samples, such as A1, A2, A3, A4, A5, B5 and B6, were a natural colour, with the colour changing over short distances (banding) (Figure 1). Others, such as B1, B2, B3, B4 and B7 were of artificial colours, roughly homogeneous (Figure 2).
Table 2.
Different colours of the seized furs
| Samples | Colour of fur |
|---|---|
| A1 |
Banding: yellowish and black |
| A2 |
Banding: yellowish and black |
| A3 |
Banding: white and dark brown |
| A4 |
Banding: white and dark brown |
| A5 |
Banding: yellowish and black |
| B1 |
Red with dark shades |
| B2 |
Dark brown |
| B3 |
Brown |
| B4 |
Dark brown |
| B5 |
Banding: white and dark brown |
| B6 |
Banding: white and dark brown |
| B7 | Dark brown |
Figure 1.

Sample A2. Example of colour changing over relatively short sections of the shaft of the sample.
Figure 2.

Sample B7. Example of the artificial, roughly homogeneous colour of the sample.
The natural colour banding, only present in some samples, is a typical characteristic of several animal species, such as the mink, muskrat, raccoon, otter and many others, but is generally not typical of the cat or the dog. The homogeneous colour of the other samples was unnatural and led us to believe that the furs were dyed. Careful examination also revealed the presence of both longer, thicker hair and shorter, thinner hair. The observed differences in length and texture were compatible with animal furs: in mammalian species, with the exclusion of human beings and cetaceans, the longest and thickest hairs are known as guard hairs, while the shortest and thinnest are known as wool hairs. The guard hair protects the animal from rain and snow, and determines the colour of the coat, while the wool hair represents the bottom layer of the coat and its main function is to keep the body temperature stable. Both guard and wool hair appeared heterogeneous in length, thickness and consistency. Moreover, a thorough inspection revealed that some of the hair seemed to originate from a single structure: this was probably due to complex hair follicles. The complex hair follicle, a typical structure of, for example, dog and cat hair, is an epidermal invagination in the dermis, whereby different hairs can grow from the same follicle. The internal structure of the hair was then investigated using a light microscope. Microscopic analysis showed the morphostructural characteristics of the hair. In particular, it confirmed the presence of three distinct layers from the outside inwards: the cuticle, the cortex and the medulla. The morphostructural characteristics observed for each hair are shown in Table 3.
Table 3.
Morphostructural characteristics of the seized furs[134]
| Samples | Cuticle pattern | Cortex | Medulla pattern | Individual characteristics |
|---|---|---|---|---|
|
A1 |
Double chevron |
Uniform |
Opaque unbroken cellular |
Cortex: more dense towards medulla |
|
A2 |
Double chevron |
Uniform |
Opaque unbroken cellular |
Root shape: spade-like |
|
A3 |
Double chevron |
Uniform |
Opaque unbroken cellular |
Cortex: more dense towards medulla |
|
A4 |
Double chevron |
Uniform |
Opaque unbroken |
Cortex: more dense towards medulla |
|
A5 |
Double chevron |
Uniform |
Vacuolated |
Cortex: more dense towards medulla |
|
B1 |
Double chevron |
Uniform |
Vacuolated |
|
|
B2 |
Regular petal |
Uniform |
Opaque unbroken |
Medulla pattern: wide |
|
B3 |
Double chevron |
Uniform |
Vacuolated |
|
|
B4 |
Regular petal |
Uniform |
Opaque unbroken or uniserial ladder |
Medulla pattern not recognizable from cortex |
|
B5 |
Double chevron |
Uniform |
Vacuolated |
Cortex: more dense towards medulla |
|
B6 |
Double chevron |
Uniform |
Vacuolated |
Root shape: spade-like |
| Cortex: more dense towards medulla | ||||
| B7 | Regular petal | Uniform | Vacuolated or uniserial ladder | Medulla pattern: wide |
The cuticle is a translucent layer of the hair shaft, consisting of scales covering the shaft. The observed cuticle scales ran from the root to the tip of the hair. In most samples, the structure of the scales was generally not prominent and showed an imbricate scale patterns (the double chevron of Table 3, shown in Figure 3); in others (B2, B4 -Figure 4- and B7) the scales were prominent and exhibited a spinous scale pattern (regular petal) that stuck out of the hair shaft and covered its entire length.
Figure 3.

Sample A3. Example of the double-chevron scale pattern.
Figure 4.
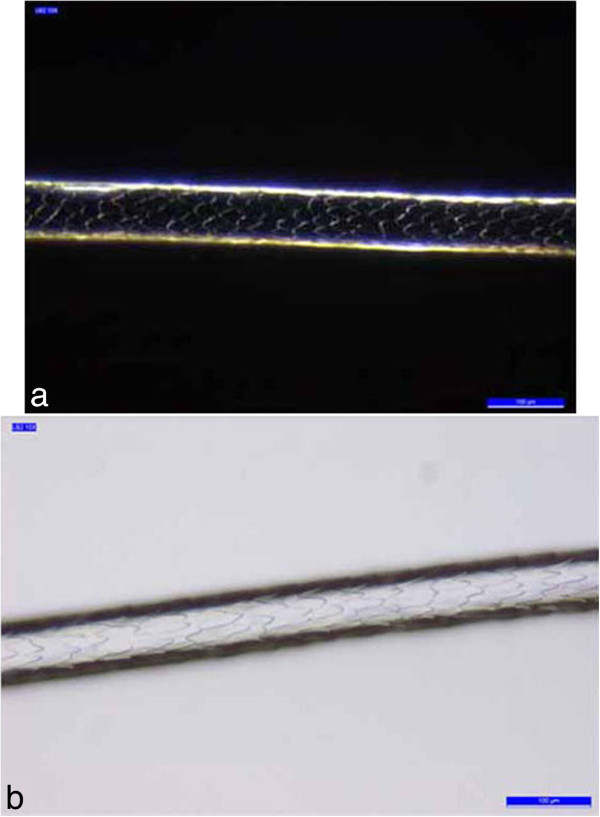
Example of the regular petal scale pattern of the samples. (a) B2 (b) B4.
The nonprominent scale pattern is typical of dog hair, while the prominent scale pattern is typical of several animals, such as the sable, lynx, seal and cat. The cortex is the main body of the hair and is highly structured and organized. It is composed of elongated and fusiform cells with a pyknotic nucleus and fibrils that are the primary source of mechanical strength and water uptake. The cortical cells constitute the bulk of hair and the cortex provides the hair fibre with its eventual shape, resilience, elasticity and curl. In every sample, the cortex appeared uniform. However, in the cortex of some hair samples (A1, A3, A4, A5, B5 and B6, Table 3), an inhomogeneous pigmentation was noted along the length of the hair, mainly spread over the centre of the medullary region (Figure 5). The medulla is the innermost region of the hair, and is not always present. As with the other morphological characteristics, even the medulla can be a useful parameter to identify the species. Indeed both the diameter of the medulla and its structure are typical features of particular species. The medulla appeared continuous along the structure of the hair in every sample examined. In most samples, the medulla was cellular (Figure 6) or vacuolated (Figure 7) (the typical structure of a dog’s medulla) but in samples B2, B4 and B7 its structure was opaque unbroken and a uniserial ladder (Figure 8) (the representative configuration of a cat’s medulla).
Figure 5.
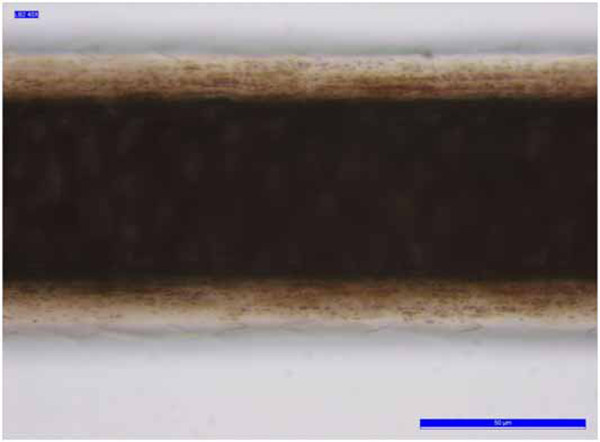
Sample A3. The cortex is more dense towards the medulla.
Figure 6.

Sample A2. Example of cellular medulla pattern.
Figure 7.
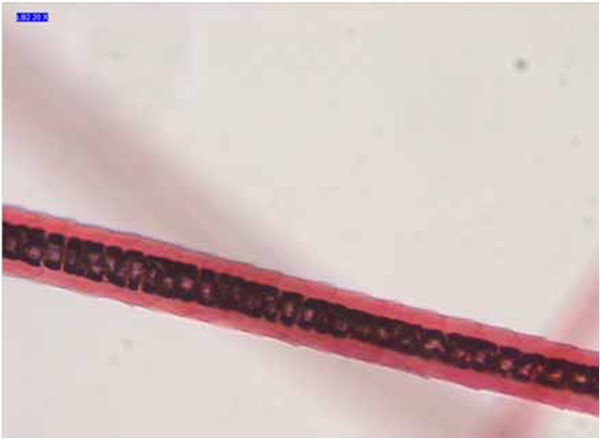
Sample B1. Example of vacuolated medulla pattern.
Figure 8.

Sample B7. Example of uniserial ladder medulla pattern.
In these samples, the medulla appeared wide. Generally, in animal hair, the medulla occupies an area greater than one-third of the overall diameter of the hair shaft and its value can change slightly among different species. Table 4 shows the medullary index of each sample (guard hair).
Table 4.
Medullary diameter, overall diameter of hair shaft and medullary index of hair samples
| Samples | Medullary diameter (μm) | Overall diameter of hair shaft (μm) | Medullary index |
|---|---|---|---|
| A1 |
94 |
122 |
0.77 |
| A2 |
81 |
103 |
0.78 |
| A3 |
56 |
85 |
0.66 |
| A4 |
73 |
113 |
0.65 |
| A5 |
38 |
54 |
0.70 |
| B1 |
45 |
66 |
0.68 |
| B2 |
42 |
64 |
0.68 |
| B3 |
43 |
70 |
0.61 |
| B4 |
Not detected |
95 |
Not detected |
| B5 |
91 |
147 |
0.61 |
| B6 |
51 |
85 |
0.60 |
| B7 | 30 | 40 | 0.75 |
As can be deduced by the values reported in Table 4, the hair samples presented a fairly uniform distribution of the medullary index value of about 0.6 to 0.7; however, the index was never higher than 0.8 for any sample. These values were compatible with those of animal hair; indeed animals have a medullary index ranging from 0.5 to 0.9. However, despite the medullary index values being compatible with the Canidae and Felidae families, they were not indicative of any species in particular, since they ranged from half to three-quarters of the width, as is the case with the majority of species. In samples A2 and B6, microscopic analysis also showed the presence of spade-shaped roots. This is a typical characteristic of dog hair roots (Figure 9).
Figure 9.
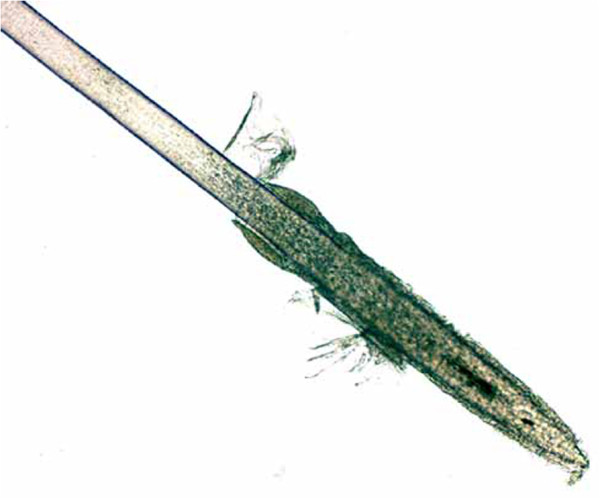
Sample A2. Example of spade-shaped root.
In summary, the furs appeared to be in good condition despite signs of physical and chemical stress due to tanning. These signs were particularly clear in the state of conservation of the underlying substrate and in the strong artificial dyeing of some hair. Microscopic inspection and evaluation of the medullary index [135] allowed us to establish the animal nature of the samples examined. The morphostructural data also provided an indication of the species to which the hair belonged, although they did not provide any kind of certainty about the specific species. Samples A1, A2, A3, A4, A5, B1, B3, B5 and B6 appeared as a homogeneous group with similar features, both macroscopic and microscopic, although macroscopically B1 and B3 showed intense artificial colouring different from the other fur samples. Therefore, on the basis of macro- and microscopic analysis, it was plausible to hypothesize that they shared a common origin, which could be ascribed to both the Canidae and the Procyonidae family. Yet, while the cuticle patterns (double chevron) and medulla patterns (vacuolated) are features of the Canidae family and the spade-like root shape of samples A2 and B6 is a typical structure in the hair roots of dogs, the medulla patterns (cellular), medullary indices and colour banding of the fur were more similar to a laboratory comparison sample from a raccoon [136]. Moreover, samples B2, B4 and B7 could be considered a separate group, with the same artificial colour of fur and comparable morphostructural characteristics consistent with a feline origin.
On the basis of the macroscopic and microscopic characteristics, it could be assumed that we were in the presence of hair belonging to the Canidae, Procyonidae or Felidae families. However, the identification of the family (Canidae or Procyonidae) and the species of these samples could not be confirmed by microscopic analysis, owing to the great variability of hair, the small amount of hair analyzed and the lack of a suitable comparative database or reference sample to compare the assay samples.
Genetic and phylogenetic analysis
Owing to the chemical and physical damage [109,137-140] of the samples’ DNA during the processing cycle of tanning, PCR amplification of the 12S and 16S markers did not produce results in every sample analyzed. With regard to the 12S marker, 10 samples out of 12 were successfully amplified, as shown in Table 5. For the 16S marker, only 4 samples out of 12 were successfully amplified, probably as a result of the larger fragment size (244 bp, in contrast with the 150 bp of the 12S), as shown in Table 6. The PCR products were cloned and 20 colonies were sequenced for each of them, to detect possible nucleotide misincorporations or contaminations. To identify the species, the consensus sequence produced for each sample was compared with the NCBI database on GenBank. A great many sequences are held on the GenBank database and even if a complete match cannot be found, a BLAST search allows identification of possibly closely related species, providing information at the level of the genus or family. The results obtained from this search and the percentage of similarity between the fur samples and the NCBI references for each marker analyzed are summarized in Table 5 and Table 6.
Table 5.
Results of 12S marker analysis
| Sample name | 12S consensus sequence | Identified species | Percentage similarity |
|---|---|---|---|
| A1 |
✓ |
Canis lupus laniger, Canis lupus chanco and Canis lupus familiaris |
98% match |
| A2 |
✓ |
Canis lupus laniger, Canis lupus chanco and Canis lupus familiaris |
98% match |
| A3 |
✓ |
Canis lupus laniger, Canis lupus chanco and Canis lupus familiaris |
98% match |
| A4 |
✓ |
Canis lupus laniger, Canis lupus chanco and Canis lupus familiaris |
98% match |
| A5 |
✓ |
Homo sapiens |
93% match |
| B1 |
✓ |
Homo sapiens |
93% match |
| B2 |
✓ |
Homo sapiens |
93% match |
| B3 |
✓ |
Homo sapiens |
93% match |
| B4 |
= |
Not available |
|
| B5 |
= |
Not available |
|
| B6 |
✓ |
Canis lupus laniger, Canis lupus chanco and Canis lupus familiaris |
98% match |
| B7 | ✓ | Felis silvestris catus | 100% match |
Species identification of fur samples and percentage of similarity after research conducted using the BLAST tool. ✓ success in obtaining the consensus sequence; = failure in obtaining the consensus sequence.
Table 6.
Results of 16S marker analysis
| Sample name | 16S consensus sequence | Identified species | Percentage of similarity |
|---|---|---|---|
| A1 |
✓ |
Canis lupus familiaris |
99% match |
| A2 |
✓ |
Canis lupus familiaris |
99% match |
| A3 |
✓ |
Homo sapiens |
95% match |
| A4 |
= |
Not available |
|
| A5 |
= |
Not available |
|
| B1 |
= |
Not available |
|
| B2 |
✓ |
Homo sapiens |
95% match |
| B3 |
= |
Not available |
|
| B4 |
= |
Not available |
|
| B5 |
= |
Not available |
|
| B6 |
= |
Not available |
|
| B7 | = | Not available |
Species identification of fur samples and percentage of similarity after research conducted using the BLAST tool. ✓ success in obtaining the consensus sequence; = failure in obtaining the consensus sequence.
Probably as a result of contamination by human genetic material from those who treated or handled the furs, which was not entirely eliminated during the cleaning of the samples, the consensus sequence obtained from samples A5, B1, B2 and B3 (12S marker) and A3 and B2 (16S marker) showed a 93 to 95% degree of similarity to the reference sequence for Homo sapiens. A match with 100% of homology between the short 12S fragment (150 bp) from the B7 sequence (Figure 10) and the NCBI reference of Felis silvestris catus was observed. This complete degree of homology could be explained as follows: either the sample comes from the species with which it matches or, because of the shortness of the sequence fragment, the sample matches that NCBI reference by chance and comes from an unknown species in GenBank. Furthermore, a search on GenBank for a comparison with sequences belonging to different Felis species or subspecies returned results only for the domestic species concerning this genetic marker. Owing to the availability of this short DNA sequence and the lack of knowledge about the variability between different species or subspecies for the 12S fragment, no phylogenetic analysis was performed for this sample and the genus Felis. For these reasons, it was not possible to attribute the hair solely to the domestic subspecies.
Figure 10.

12S consensus sequences of fur samples. Alignments of the 12S consensus sequences of fur samples (A1, A2, A3, A4, B6 and B7). Nucleotide positions are numbered according to GenBank GU256221.1 [141].
The 12S consensus sequences of samples A1, A3, A4 and B6 (Figure 10) were identical to each other, as was the sequence for the 16S in samples A1 and A2 (Figure 11), presumably because the hair analyzed belonged to the same species. Only the 12S consensus sequence of sample A2 showed a transversion (T instead of A) when compared with the other samples analyzed (Figure 10). The 98% homology between these sequences and NCBI references of Canis lupus laniger, Canis lupus chanco and Canis lupus familiaris was observed and could be explained in two ways. Either the unknown samples came from one of these species and the differences were due to intraspecific variation, or they came from an unknown but closely related species that was not present on the database.
Figure 11.

16S consensus sequence of fur samples. Alignments of the 16S consensus sequences of fur samples (A1 and A2). Nucleotide positions are numbered according to GenBank GU256221.1 [141].
To obtain an unambiguous attribution of the hair to the subspecies listed, and distinguish the fur samples from potential different individuals, the analysis focused on the study of the HVS-I of the canine D-loop. The amplification of HVS-I using seven overlapping fragments (Figure 12) led to a complete consensus sequence for samples A1 and A2. For samples A3, A4 and B6, the amplification of the IV fragment (150 bp) failed, maybe because of degradation phenomena with possible modification in the annealing site of the primers. However, it is most likely that the failure to obtain a result could be explained by the presence of mutations in the DNA template that prevented the annealing of one or both of the two primers. The complete consensus sequences of A1 and A2 samples were aligned with each other and with the partial consensus sequences of samples A3, A4 and B6. All consensus sequences are available at the National Center for Biotechnology [GeneBank Accession Numbers: KJ828711-KJ828715]. The sequences were similar but not the same (Figure 13). Owing to the large variability of the genetic region analyzed, we presumed that the hair could have belonged to different individuals. Both the complete consensus sequences of samples A1 and A2 and the partial ones of A3, A4 and B6 were compared with those held on GenBank. The results obtained showed a homology of 97% to 99% with Nyctereutes procyonoides. The degree of match was not 100% and this could be explained in the following ways: (1) different individuals of the same species could have different genetic profiles because the marker analyzed was a highly variable region; (2) some of the differences observed between unknown and reference samples were the result of post mortem damage [112,142,143], that is, the modifications in DNA sequence arose subsequent to cell death or as a result of the tanning process. The apparent inconsistency found when analyzing the results of mtDNA (12S, 16S and HVS-I) can be explained by the small amount of data available in the literature on the genome of Nyctereutes procyonoides and, at the time of the realization of this work, by the absence of the 12S sequence of this species in the NCBI database. Therefore, the most likely diagnosis of the species was that of the Nyctereutes procyonoides, the raccoon dog. This conclusion was confirmed when the 12S sequence of the Nyctereutes procyonoides genome was published in GenBank.
Figure 12.
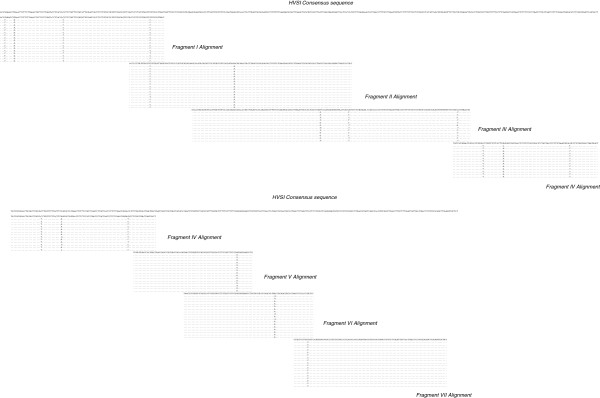
Alignment of HVS-I amplicons from the cloning of sample A2. DNA sequences from the clones analyzed for the A2 sample are reported as an example. Nucleotides identical to the A2 sequence are indicated by dots. Some misincorporations can be observed in some clones.
Figure 13.

HVS-I consensus sequences of fur samples. Alignments of the HVS-I consensus sequences of fur samples (A1, A2, A3, A4 and B6). The numbering of the nucleotide positions is at the top, according to GenBank GU256221.1 [141]. Nucleotides identical to the A1 sequence are indicated by dots.
The next comparison of the 12S consensus sequence of samples A1, A2, A3, A4 and B6 with those held on GenBank showed the highest homology (100%) with Nyctereutes procyonoides for samples A1, A3, A4 and B6 and 99% homology with the same species for sample A2. The next comparison of the consensus sequence for 16S of samples A1 and A2 showed the highest homology (100%) with Nyctereutes procyonoides. These data confirmed the information obtained from the HVS-I. Several sequences (280 for the 12S marker and 1,186 for the HVS-I) from the NCBI database relative to all species or subspecies that showed a homology of almost 98% with fur samples were collected in a dataset (Table 7). To better investigate how the fur samples fell into the genetic variability of the comparison dataset, phylogenetic analyses were performed. The neighbour-joining tree for the 12S marker (Figure 14) was calculated using the Kimura two-parameter model [144]: this described the substitution pattern in the dataset of sequences better than other available models. Genera Canis and Nyctereutes were separated into two groups with a bootstrap value of 79%. The three available sequences of Canis lupus chanco were distributed as follows: one in the subgroup of Canis lupus and Canis lupus familiaris, and the others in that of Canis lupus laniger. All fur samples fell into the same group of Nyctereutes procyonoides but the three NCBI reference sequences differed from ours. The neighbour-joining tree obtained from the alignment between the HVS-I consensus sequences of the fur samples and the comparison dataset is shown in Figure 15. The tree was calculated using the Tamura three-parameter model of pairwise base substitution [145]. This was the best model to describe sequence divergence in the dataset. Overall groupings of the sequences in the dataset were as expected: genera Canis and Nyctereutes were separated into two groups, with the maximum value of statistical support. A 100% bootstrap value at the node connecting these genera indicated that the genetic marker (HVS-I) we used had enough informative power to make the two groups distinguishable.
Table 7.
Number of sequences and their origin for the dataset used in the phylogenetic analysis
| Species | Number of sequences | GenBank accession numbers |
|---|---|---|
|
Canis lupus |
37 |
AF005296-005314, |
| AF008135-008142, | ||
| AF008160-008167 [146] | ||
|
AY240073, AY240155 [125] | ||
|
Canis lupus familiaris |
1,111 |
NC002008, U96639* [127] |
| EU223385-223811 [147] | ||
| AY656703-656710 [148] | ||
| AY656737-656755* [149] | ||
| EU816456-816557 [150] | ||
| AF531654-531741, | ||
|
HQ261490 [151] | ||
| AF005280-005295, | ||
| AF008143-008157, | ||
| AF008168-008182 [146] | ||
| JF342807-342906 * [152] | ||
| AY240030-240072, | ||
| AY240074-240093, | ||
| AY240095-240154, | ||
| AY2400156-2400157 [125] | ||
| AF064569-064586 [153] | ||
| EF122413-122428 [94] | ||
| AB622516-622568, | ||
| AB700664-700665 [154] | ||
| EU789667-789786** [150] | ||
|
Canis lupus laniger |
3 |
KF573616* [155] |
| NC011218, FJ032363* [156] | ||
|
Canis lupus chanco |
42 |
AB007375-007379 [157] |
| AB480743-480744 [158] | ||
| AY333738-333742 [159] | ||
|
EU442884* [160] | ||
|
GQ374438* [141] | ||
| JX415343-415379 [161] | ||
| NC010340* [156] | ||
| Nyctereutes procyonoides | 113 |
AB292740 [162] |
| D83614-83615 [163] | ||
| EU642411-642457 [164] | ||
| FJ888513-888521 [165] | ||
| JF809819-809848 [166] | ||
| KC344215-344235, | ||
|
KC509604 [167] | ||
|
GU256221* [141] | ||
| NC013700* [168] | ||
| KF781340** [169] |
All sequences reported in the table were used for HVS-I comparison. Those marked with * were used both for HVS-I and 12S and those marked with ** were used only for 12S comparison.
Figure 14.
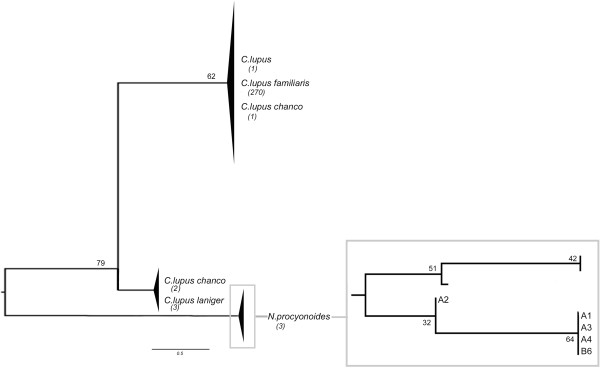
Neighbour-joining tree built for 12S marker. The bootstrap neighbour-joining tree inferred from 1,000 replicates represents the evolutionary history of 285 sequences (280 NCBI reference sequences and five fur samples) of the 12S marker. The reference sequences were retrieved from the species that presented almost 98% of similarity to the sequences obtained from fur samples A1, A2, A3, A4 and B6. The bootstrap percentage values are indicated at the nodes. In each clade, the number of comparison sequences is indicated below the name of the species. The Nyctereutes procyonoides clade is enlarged for better visibility within the grey box.
Figure 15.
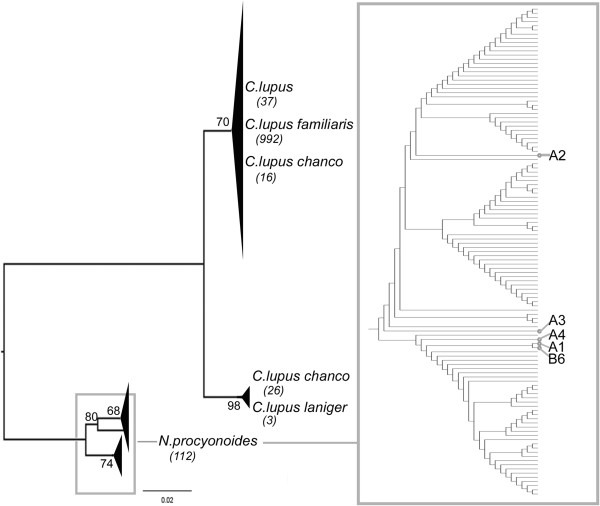
Neighbour-joining tree built for HVS-I. The bootstrap neighbour-joining tree inferred from 1,000 replicates represents the evolutionary history of 1,191 sequences (1,186 NCBI reference sequences and five fur samples) of the HVS-I marker. The reference sequences were retrieved from the species that presented almost 98% of similarity to the sequences obtained from fur samples A1, A2, A3, A4 and B6. The bootstrap percentage values are indicated at the nodes whenever larger than 50. In each clade, the number of comparison sequences is indicated below the name of the species. The Nyctereutes procyonoides clade is enlarged for better visibility within the grey box. This subtree is condensed for bootstrap values lower than 50 and only topology is shown. The position of the fur samples is indicated by grey dots linked to the name of the sample.
In the genus Canis, some sequences of Canis lupus chanco cluster together with Canis lupus and Canis lupus familiaris, while others belong to the same group of Canis lupus laniger. Consensus sequences of the fur samples fell clearly into the variability of the Nyctereutes procyonoides group. The phylogenetic analysis confirmed the indications given by the BLAST search, showing samples A1, A2, A3, A4 and B6 as belonging to the Nyctereutes procyonoides species. Although Nyctereutes procyonoides is often confused with raccoons and badgers, it does not belong to the Procyonidae family but to the Canidae, as does the domestic dog. Nyctereutes procyonoides and the domestic dog present the genus they belong to: the raccoon dog actually belongs to the genus Nyctereutes, while the domestic dog belongs to the genus Canis. The physical characteristics of the raccoon dog are substantially similar to those of a medium-sized domestic dog with the indicative feature of a raccoon snout, although it is distinguished taxonomically from the raccoon itself. Nyctereutes comes from Japan, Siberia and Manchuria.
Conclusions
Macroscopic and microscopic analysis allowed confirmation of the hypothesis regarding the analyzed hair belonging to real animals, although it failed to provide any kind of certainty regarding the actual family or species. The sequence data and the comparisons with the samples held on GenBank showed that, in most cases, the hair belonged to the Canidae family, and in only one case to Felidae. The genetic data related to the diagnosis of the genus and the species of Canidae hair was heterogeneous: the sequence results of samples A1, A2, A3, A4 and B6 (12S, 16S and HVS-I) showed a high homology with the genus Canis and the genus Nyctereutes (raccoon dog). This initial uncertainty led obviously to a likely ambiguous diagnosis of the species that was impossible to predict. However, the genetic results confirmed a unique diagnosis of nonattribution for the protected subspecies Canis lupus familiaris. Despite previous reservations advanced, the species that appeared more consistent with the data obtained from the BLAST search was Nyctereutes procyonoides, and phylogenetic analysis confirmed this observation.
The data of the diagnosis of the genus and the species of the hair belonging to the Felidae family appeared homogeneous. The study revealed a high similarity between our sequences and the genus Felis, species Felis silvestris. However, the failure to diagnose a single domestic subspecies (Felis silvestris catus) was mainly due to the limited amount of genetic data for comparative purposes in the literature. In fact, no information on the genetic differences between the various subspecies and on the process of cat domestication [170,171] are known and no genetic data are available on GenBank for different subspecies of Felis silvestris regarding the 12S marker. For these reasons, an unambiguous attribution of one of the fur samples to the subspecies Felis silvestris catus was not possible. Finally, we would like to add that beyond scientific classification, prohibition of the use for commercial purposes should not be just confined to the subspecies Canis lupus familiaris or Felis silvestris catus but should be extended to other Canidae and Felidae, particularly when taking into account sociocultural practices and geographical regions far from the West.
Abbreviations
BLAST: Basic Local Alignment Search Tool; bp: base pair; COI: cytochrome c oxidase subunit 1; dNTP: deoxy-nucleotide-triphosphate; HVS: hypervariable segment; HVS-I: hypervariable segment I; HVS-II: hypervariable segment II; mtDNA: mitochondrial DNA; NDH: nicotinamide adenine dinucleotide; nt: nucleotide; PCR: polymerase chain reaction; SDS: sodium dodecyl sulphate; VNTR: variable number tandem repeat.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
EP conceived and designed the experiments. EP, RC, AV and GD performed the experiments. EP, RC and SV analyzed the data. FB, AB, GL and DC contributed reagents, materials and analysis tools. EP wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Elena Pilli, Email: elena.pilli@unifi.it.
Rosario Casamassima, Email: rosario.casamassima@carabinieri.it.
Stefania Vai, Email: stefania.vai@unifi.it.
Antonino Virgili, Email: antonino.virgili@carabinieri.it.
Filippo Barni, Email: filippo.barni@carabinieri.it.
Giancarlo D’Errico, Email: giancarlo.derrico@carabinieri.it.
Andrea Berti, Email: andrea.berti@carabinieri.it.
Giampietro Lago, Email: giampietro.lago@carabinieri.it.
David Caramelli, Email: david.caramelli@unifi.it.
Acknowledgements
We are very grateful to Jacopo Marconcini for providing language support during the drafting of our paper and Claire Stypulkowski for invaluable advice in proofreading the article.
References
- Butler JM. In: Advanced Topics in Forensic DNA Typing Methodology. Butler JM, editor. London: Academic; 2011. Non-human DNA; pp. 473–487. [Google Scholar]
- Barbanera F, Guerrini M, Beccani C, Forcina G, Anayiotos P, Panayides P. Conservation of endemic and threatened wildlife: molecular forensic DNA against poaching of the Cypriot mouflon (Ovis orientalis ophion, Bovidae) Forensic Sci Int Genet. 2012;6:671–675. doi: 10.1016/j.fsigen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Berger C, Berger B, Parson W. Sequence analysis of the canine mitochondrial DNA control region from shed hair samples in criminal investigations. Methods Mol Biol. 2012;830:331–348. doi: 10.1007/978-1-61779-461-2_23. [DOI] [PubMed] [Google Scholar]
- Allgeier L, Hemenway J, Shirley N, LaNier T, Coyle HM. Field testing of collection cards for Cannabis sativa samples with a single hexanucleotide DNA marker. J Forensic Sci. 2011;56:1245–1249. doi: 10.1111/j.1556-4029.2011.01818.x. [DOI] [PubMed] [Google Scholar]
- Butler JM, Schneider PM, Carracedo A. Journal update. Forensic Sci Int Genet. 2010;4:143–144. doi: 10.1016/j.fsigen.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Yi-Ming L, Zenxiang G, Xinhai L, Sung W, Niemelä J. Illegal wildlife trade in the Himalayan region of China. Biodivers Conserv. 2000;9:901–918. [Google Scholar]
- Wilson-Wilde L. Combating wildlife crime. Forensic Sci Med Pathol. 2010;6:149–150. doi: 10.1007/s12024-010-9179-4. [DOI] [PubMed] [Google Scholar]
- Wilson-Wilde L. Wildlife crime: a global problem. Forensic Sci Med Pathol. 2010;6:221–222. doi: 10.1007/s12024-010-9167-8. [DOI] [PubMed] [Google Scholar]
- Alacs EA, Georges A, FitzSimmons NN, Robertson J. DNA detective: a review of molecular approaches to wildlife forensics. Forensic Sci Med Pathol. 2010;6:180–194. doi: 10.1007/s12024-009-9131-7. [DOI] [PubMed] [Google Scholar]
- World Wildlife Fund. Tiger. [ https://worldwildlife.org/species/tiger]
- World Wildlife Fund. Elephant. [ https://worldwildlife.org/species/elephant]
- Wozney KM, Wilson PJ. Real-time PCR detection and quantification of elephantid DNA: species identification for highly processed samples associated with the ivory trade. Forensic Sci Int. 2012;219:106–112. doi: 10.1016/j.forsciint.2011.12.006. [DOI] [PubMed] [Google Scholar]
- Peppin L, McEwing R, Carvalho GR, Ogden R. A DNA-based approach for the forensic identification of Asiatic black bear (Ursus thibetanus) in a traditional Asian medicine. J Forensic Sci. 2008;53:1358–1362. doi: 10.1111/j.1556-4029.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- Dubey B, Meganathan PR, Haque I. DNA mini-barcoding: an approach for forensic identification of some endangered Indian snake species. Forensic Sci Med Pathol. 2011;5:181–184. doi: 10.1016/j.fsigen.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Meganathan PR, Dubey B, Jogayya KN, Haque I. Validation of a multiplex PCR assay for the forensic identification of Indian crocodiles. J Forensic Sci. 2011;56:1241–1244. doi: 10.1111/j.1556-4029.2011.01812.x. [DOI] [PubMed] [Google Scholar]
- Reid BN, LE M, McCord WP, Iverson JB, Georges A, Bergmann T, Amato G, Desalle R, Naro-Maciel E. Comparing and combining distance-based and character-based approaches for barcoding turtles. Mol Ecol Resour. 2011;11:956–967. doi: 10.1111/j.1755-0998.2011.03032.x. [DOI] [PubMed] [Google Scholar]
- Rosen GE, Smith KF. Summarizing the evidence on the international trade in illegal wildlife. Ecohealth. 2010;7:24–32. doi: 10.1007/s10393-010-0317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman V. An overview of international wildlife trade from Southeast Asia. Biodivers Conserv. 2009;19:1101–1114. [Google Scholar]
- Alacs E, Georges A. Wildlife across our borders: a review of the illegal trade in Australia. Aust J Forensic Sci. 2008;40:147–160. [Google Scholar]
- Coghlan ML, White NE, Parkinson L, Haile J, Spencer PBS, Bunce M. Egg forensics: an appraisal of DNA sequencing to assist in species identification of illegally smuggled eggs. Forensic Sci Int Genet. 2012;6:268–273. doi: 10.1016/j.fsigen.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC-I, Hsieh H-M, Huang L-H, Kuo Y-C, Wu J-H, Chin S-C, Lee A-H, Linacre A, Tsai L-C. Ivory identification by DNA profiling of cytochrome b gene. Int J Legal Med. 2009;123:117–121. doi: 10.1007/s00414-008-0264-0. [DOI] [PubMed] [Google Scholar]
- Comstock KE, Ostrander EA, Wasser SK. Amplifying nuclear and mitochondrial DNA from African elephant ivory: a tool for monitoring the ivory trade. Conserv Biol. 2003;17:1840–1843. [Google Scholar]
- Gupta S, Thangaraj K, Singh L. A simple and inexpensive molecular method for sexing and identification of the forensic samples of elephant origin. J Forensic Sci. 2006;51:805–807. doi: 10.1111/j.1556-4029.2006.00154.x. [DOI] [PubMed] [Google Scholar]
- Wasser SK, Clark B, Laurie C. The ivory trail. Sci Am. 2009;301:68–74. doi: 10.1038/scientificamerican0709-68. 76. [DOI] [PubMed] [Google Scholar]
- Wasser SK, Joseph Clark W, Drori O, Stephen Kisamo E, Mailand C, Mutayoba B, Stephens M. Combating the illegal trade in African elephant ivory with DNA forensics. Conserv Biol. 2008;22:1065–1071. doi: 10.1111/j.1523-1739.2008.01012.x. [DOI] [PubMed] [Google Scholar]
- Biscardi B, Welsh W, Kennedy A. Discrimination of the hard keratins animal horn and chelonian shell using attenuated total reflection-infrared spectroscopy. Appl Spectrosc. 2012;66:606–608. doi: 10.1366/11-06565. [DOI] [PubMed] [Google Scholar]
- Parlamento Italiano. Disposizioni concernenti il divieto di maltrattamento degli animali, nonché di impiego degli stessi in combattimenti clandestini o competizioni non autorizzate. [ http://www.camera.it/parlam/leggi/04189l.htm]
- Bisbing RE. In: Encyclopedia of Forensic Science, Volume 3. Siegel J, Knupfer G, Saukko P, editor. London: Academic; 2000. Hair comparison: microscopic; pp. 1002–1016. [Google Scholar]
- Simon C, Buckley TR, Frati F, Stewart JB, Beckenbach AT. Incorporating molecular evolution into phylogenetic analysis, and a new compilation of conserved polymerase chain reaction primers for animal mitochondrial DNA. Annu Rev Ecol Evol Syst. 2006;37:545–579. [Google Scholar]
- Clayton DA. Replication of animal mitochondrial DNA. Cell. 1982;28:693–705. doi: 10.1016/0092-8674(82)90049-6. [DOI] [PubMed] [Google Scholar]
- Hayashi J, Tagashira Y, Yoshida MC. Absence of extensive recombination between inter- and intraspecies mitochondrial DNA in mammalian cells. Exp Cell Res. 1985;160:387–395. doi: 10.1016/0014-4827(85)90185-5. [DOI] [PubMed] [Google Scholar]
- Robin ED, Wong R. Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J Cell Physiol. 1988;136:507–513. doi: 10.1002/jcp.1041360316. [DOI] [PubMed] [Google Scholar]
- Pfeiffer H, Benthaus S, Rolf B, Brinkmann B. The Kaiser’s tooth. Int J Legal Med. 2003;117:118–120. doi: 10.1007/s00414-002-0336-5. [DOI] [PubMed] [Google Scholar]
- Ginther C, Issel-Tarver L, King MC. Identifying individuals by sequencing mitochondrial DNA from teeth. Nat Genet. 1992;2:135–138. doi: 10.1038/ng1092-135. [DOI] [PubMed] [Google Scholar]
- Loreille OM, Diegoli TM, Irwin JA, Coble MD, Parsons TJ. High efficiency DNA extraction from bone by total demineralization. Forensic Sci Int Genet. 2007;1:191–195. doi: 10.1016/j.fsigen.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Anslinger K, Weichhold G, Keil W, Bayer B, Eisenmenger W. Identification of the skeletal remains of Martin Bormann by mtDNA analysis. Int J Legal Med. 2001;114:194–196. doi: 10.1007/s004140000176. [DOI] [PubMed] [Google Scholar]
- Budowle B, van Daal A. Forensically relevant SNP classes. Biotechniques. 2008;44:603–608. doi: 10.2144/000112806. 610. [DOI] [PubMed] [Google Scholar]
- Parson W. Relevance of mtDNA analysis for forensic applications. Rechtmedizin. 2009;19:183–192. [Google Scholar]
- Allen M, Engström AS, Meyers S, Handt O, Saldeen T, von Haeseler A, Pääbo S, Gyllensten U. Mitochondrial DNA sequencing of shed hairs and saliva on robbery caps: sensitivity and matching probabilities. J Forensic Sci. 1998;43:453–464. [PubMed] [Google Scholar]
- Brown WM, George M, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979;76:1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwayama R, Ozawa T. Phylogenetic relationships among European red deer, wapiti, and sika deer inferred from mitochondrial DNA sequences. Mol Phylogenet Evol. 2000;15:115–123. doi: 10.1006/mpev.1999.0731. [DOI] [PubMed] [Google Scholar]
- Irwin DM, Kocher TD, Wilson AC. Evolution of the cytochrome b gene of mammals. J Mol Evol. 1991;32:128–144. doi: 10.1007/BF02515385. [DOI] [PubMed] [Google Scholar]
- Kocher TD, Thomas WK, Meyer A, Edwards SV, Pääbo S, Villablanca FX, Wilson AC. Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A. 1989;86:6196–6200. doi: 10.1073/pnas.86.16.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobe SS, Linacre A. A method to identify a large number of mammalian species in the UK from trace samples and mixtures without the use of sequencing. Forensic Sci Int Genet Suppl Ser. 2008;1:625–627. [Google Scholar]
- An J, Lee M-Y, Min M-S, Lee M-H, Lee H. A molecular genetic approach for species identification of mammals and sex determination of birds in a forensic case of poaching from South Korea. Forensic Sci Int. 2007;167:59–61. doi: 10.1016/j.forsciint.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Tobe S, Linacre A. Species identification of human and deer from mixed biological material. Forensic Sci Int. 2007;169:278–279. doi: 10.1016/j.forsciint.2006.07.016. [DOI] [PubMed] [Google Scholar]
- De Pancorbo MM, Castro A, Fernandez-Fernandez I, Gonzalez-Fernandez M, Martnez-Bouzas C, Cuevas N. Cytochrome b and HVI sequences of mitochondrial DNA to identify domestic animal hair in forensic casework. Int Congr Ser. 2003;1239:841–845. [Google Scholar]
- Wan Q-H, Fang S-G. Application of species-specific polymerase chain reaction in the forensic identification of tiger species. Forensic Sci Int. 2003;131:75–78. doi: 10.1016/s0379-0738(02)00398-5. [DOI] [PubMed] [Google Scholar]
- Wetton JH, Tsang CSF, Roney CA, Spriggs AC. An extremely sensitive species-specific ARMs PCR test for the presence of tiger bone DNA. Forensic Sci Int. 2004;140:139–145. doi: 10.1016/j.forsciint.2003.11.018. [DOI] [PubMed] [Google Scholar]
- Tobe SS, Linacre A. Identifying endangered species from degraded mixtures at low levels. Forensic Sci Int Genet Suppl Ser. 2009;2:304–305. [Google Scholar]
- Moore MK, Bemiss JA, Rice SM, Quattro JM, Woodley CM. Use of restriction fragment length polymorphisms to identify sea turtle eggs and cooked meats to species. Conserv Genet. 2003;4:95–103. [Google Scholar]
- Hsieh H-M, Huang L-H, Tsai L-C, Liu C-L, Kuo Y-C, Hsiao C-T, Linacre A, Lee JC-I. Species identification of Kachuga tecta using the cytochrome b gene. J Forensic Sci. 2006;51:52–56. doi: 10.1111/j.1556-4029.2005.00004.x. [DOI] [PubMed] [Google Scholar]
- Rohilla MS, Tiwari PK. Restriction fragment length polymorphism of mitochondrial DNA and phylogenetic relationships among five species of Indian freshwater turtles. J Appl Genet. 2008;49:167–182. doi: 10.1007/BF03195610. [DOI] [PubMed] [Google Scholar]
- Meganathan PR, Dubey B, Haque I. Molecular identification of crocodile species using novel primers for forensic analysis. Conserv Genet. 2008;10:767–770. [Google Scholar]
- Hsieh H-M, Huang L-H, Tsai L-C, Kuo Y-C, Meng H-H, Linacre A, Lee JC-I. Species identification of rhinoceros horns using the cytochrome b gene. Forensic Sci Int. 2003;136:1–11. doi: 10.1016/s0379-0738(03)00251-2. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Verma SK, Singh L. Molecular insight into a wildlife crime: the case of a peafowl slaughter. Forensic Sci Int. 2005;154:214–217. doi: 10.1016/j.forsciint.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Barcode of Life Data Systems. [ http:www.boldsystems.org]
- Hebert PDN, Cywinska A, Ball SL, de Waard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisenko AV, Lim BK, Ivanova NV, Hanner RH, Hebert PDN. DNA barcoding in surveys of small mammal communities: a field study in Suriname. Mol Ecol Resour. 2008;8:471–479. doi: 10.1111/j.1471-8286.2007.01998.x. [DOI] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, De Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc Biol Sci. 2003;270(Suppl 1):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier R, Shiyang K, Vaidya G, Ng PKL. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst Biol. 2006;55:715–728. doi: 10.1080/10635150600969864. [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. DNA barcodes distinguish species of tropical Lepidoptera. Proc Natl Acad Sci U S A. 2006;103:968–971. doi: 10.1073/pnas.0510466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Rojo Velasco GS. Testing molecular barcodes: invariant mitochondrial DNA sequences vs the larval and adult morphology of West Palaearctic Pandasyopthalmus species (Diptera: Syrphidae: Paragini) Eur J Entomol. 2006;103:443–458. [Google Scholar]
- Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN. DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae) Proc Natl Acad Sci U S A. 2006;103:3657–3662. doi: 10.1073/pnas.0511318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cywinska A, Hunter FF, Hebert PDN. Identifying Canadian mosquito species through DNA barcodes. Med Vet Entomol. 2006;20:413–424. doi: 10.1111/j.1365-2915.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- Nelson LA, Wallman JF, Dowton M. Using COI barcodes to identify forensically and medically important blowflies. Med Vet Entomol. 2007;21:44–52. doi: 10.1111/j.1365-2915.2007.00664.x. [DOI] [PubMed] [Google Scholar]
- Mitchell A. DNA barcoding demystified. Aust J Entomol. 2008;47:169–173. [Google Scholar]
- Smith MA, Poyarkov NA, Hebert PDN. DNA barcoding: CO1 DNA barcoding amphibians: take the chance, meet the challenge. Mol Ecol Resour. 2008;8:235–246. doi: 10.1111/j.1471-8286.2007.01964.x. [DOI] [PubMed] [Google Scholar]
- Holmes BH, Steinke D, Ward RD. Identification of shark and ray fins using DNA barcoding. Fish Res. 2009;95:280–288. [Google Scholar]
- Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HS, Eah J-Y, Kim JS, Kim Y-J, Min M-S, Paek WK, Lee H, Kim C-B. DNA barcoding Korean birds. Mol Cells. 2006;22:323–327. [PubMed] [Google Scholar]
- Tavares ES, Baker AJ. Single mitochondrial gene barcodes reliably identify sister-species in diverse clades of birds. BMC Evol Biol. 2008;8:81. doi: 10.1186/1471-2148-8-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman DJ, Prawiradilaga DM, Meier R. Improved COI barcoding primers for Southeast Asian perching birds (Aves: Passeriformes) Mol Ecol Resour. 2009;9:37–40. doi: 10.1111/j.1755-0998.2008.02221.x. [DOI] [PubMed] [Google Scholar]
- Wong EH-K, Shivji MS, Hanner RH. Identifying sharks with DNA barcodes: assessing the utility of a nucleotide diagnostic approach. Mol Ecol Resour. 2009;9(Suppl s1):243–256. doi: 10.1111/j.1755-0998.2009.02653.x. [DOI] [PubMed] [Google Scholar]
- Huang J, Xu Q, Sun ZJ, Tang GL, Su ZY. Identifying earthworms through DNA barcodes. Pedobiologia (Jena) 2007;51:301–309. [Google Scholar]
- Dalton DL, Kotze A. DNA barcoding as a tool for species identification in three forensic wildlife cases in South Africa. Forensic Sci Int. 2011;207:e51–e54. doi: 10.1016/j.forsciint.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Guo H, Wang W, Yang N, Guo B, Zhang S, Yang R, Yuan Y, Yu J, Hu S, Sun Q, Yu J. DNA barcoding provides distinction between Radix Astragali and its adulterants. Sci China Life Sci. 2010;53:992–999. doi: 10.1007/s11427-010-4044-y. [DOI] [PubMed] [Google Scholar]
- Tyagi A, Bag SK, Shukla V, Roy S, Tuli R. Oligonucleotide frequencies of barcoding loci can discriminate species across kingdoms. PLoS One. 2010;5:e12330. doi: 10.1371/journal.pone.0012330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botti S, Giuffra E. Oligonucleotide indexing of DNA barcodes: identification of tuna and other scombrid species in food products. BMC Biotechnol. 2010;10:60. doi: 10.1186/1472-6750-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni I, De Mattia F, Galimberti A, Galasso G, Banfi E, Casiraghi M, Labra M. Identification of poisonous plants by DNA barcoding approach. Int J Legal Med. 2010;124:595–603. doi: 10.1007/s00414-010-0447-3. [DOI] [PubMed] [Google Scholar]
- Boehme P, Amendt J, Disney RHL, Zehner R. Molecular identification of carrion-breeding scuttle flies (Diptera: Phoridae) using COI barcodes. Int J Legal Med. 2010;124:577–581. doi: 10.1007/s00414-010-0429-5. [DOI] [PubMed] [Google Scholar]
- Ferri G, Alù M, Corradini B, Beduschi G. Forensic botany: species identification of botanical trace evidence using a multigene barcoding approach. Int J Legal Med. 2009;123:395–401. doi: 10.1007/s00414-009-0356-5. [DOI] [PubMed] [Google Scholar]
- Balitzki-Korte B, Anslinger K, Bartsch C, Rolf B. Species identification by means of pyrosequencing the mitochondrial 12S rRNA gene. Int J Legal Med. 2005;119:291–294. doi: 10.1007/s00414-005-0537-9. [DOI] [PubMed] [Google Scholar]
- Melton T, Holland C. Routine forensic use of the mitochondrial 12S ribosomal RNA gene for species identification. J Forensic Sci. 2007;52:1305–1307. doi: 10.1111/j.1556-4029.2007.00553.x. [DOI] [PubMed] [Google Scholar]
- Kitano T, Umetsu K, Tian W, Osawa M. Two universal primer sets for species identification among vertebrates. Int J Legal Med. 2007;121:423–427. doi: 10.1007/s00414-006-0113-y. [DOI] [PubMed] [Google Scholar]
- Mitani T, Akane A, Tokiyasu T, Yoshimura S, Okii Y, Yoshida M. Identification of animal species using the partial sequences in the mitochondrial 16S rRNA gene. Leg Med (Tokyo) 2009;11(Suppl 1):S449–S450. doi: 10.1016/j.legalmed.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Imaizumi K, Akutsu T, Miyasaka S, Yoshino M. Development of species identification tests targeting the 16S ribosomal RNA coding region in mitochondrial DNA. Int J Legal Med. 2007;121:184–191. doi: 10.1007/s00414-006-0127-5. [DOI] [PubMed] [Google Scholar]
- Rastogi G, Dharne MS, Walujkar S, Kumar A, Patole MS, Shouche YS. Species identification and authentication of tissues of animal origin using mitochondrial and nuclear markers. Meat Sci. 2007;76:666–674. doi: 10.1016/j.meatsci.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Schwenke PL, Rhydderch JG, Ford MJ, Marshall AR, Park LK. Forensic identification of endangered Chinook salmon (Oncorhynchus tshawytscha) using a multilocus SNP assay. Conserv Genet. 2006;7:983–989. [Google Scholar]
- Junqueira ACM, Lessinger AC, Torres TT, da Silva FR, Vettore AL, Arruda P, Azeredo Espin AML. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae) Gene. 2004;339:7–15. doi: 10.1016/j.gene.2004.06.031. [DOI] [PubMed] [Google Scholar]
- Mayer F, Dietz C, Kiefer A. Molecular species identification boosts bat diversity. Front Zool. 2007;4:4. doi: 10.1186/1742-9994-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford SL, Anthony NM, Bawe-Johnson M, Abernethy KA, Tutin CEG, White LJT, Bermejo M, Goldsmith ML, McFarland K, Jeffery KJ, Bruford MW, Wickings EJ. Mitochondrial DNA phylogeography of western lowland gorillas (Gorilla gorilla gorilla) Mol Ecol. 2004;13:1551–1565,1567. doi: 10.1111/j.1365-294X.2004.02140.x. [DOI] [PubMed] [Google Scholar]
- Zhang W, Zhang Z, Shen F, Hou R, Lv X, Yue B. Highly conserved D-loop-like nuclear mitochondrial sequences (Numts) in tiger (Panthera tigris) J Genet. 2006;85:107–116. doi: 10.1007/BF02729016. [DOI] [PubMed] [Google Scholar]
- Himmelberger AL, Spear TF, Satkoski JA, George DA, Garnica WT, Malladi VS, Smith DG, Webb KM, Allard MW, Kanthaswamy S. Forensic utility of the mitochondrial hypervariable region 1 of domestic dogs, in conjunction with breed and geographic information. J Forensic Sci. 2008;53:81–89. doi: 10.1111/j.1556-4029.2007.00615.x. [DOI] [PubMed] [Google Scholar]
- Eichmann C, Parson W. Molecular characterization of the canine mitochondrial DNA control region for forensic applications. Int J Legal Med. 2007;121:411–416. doi: 10.1007/s00414-006-0143-5. [DOI] [PubMed] [Google Scholar]
- Schneider PM, Seo Y, Rittner C. Forensic mtDNA hair analysis excludes a dog from having caused a traffic accident. Int J Legal Med. 1999;112:315–316. doi: 10.1007/s004140050257. [DOI] [PubMed] [Google Scholar]
- Pun K-M, Albrecht C, Castella V, Fumagalli L. Species identification in mammals from mixed biological samples based on mitochondrial DNA control region length polymorphism. Electrophoresis. 2009;30:1008–1014. doi: 10.1002/elps.200800365. [DOI] [PubMed] [Google Scholar]
- Nussbaumer C, Korschineck I. Non-human mtDNA helps to exculpate a suspect in a homicide case. Int Congr Ser. 2006;1288:136–138. [Google Scholar]
- Fumagalli L, Cabrita CJ, Castella V. Simultaneous identification of multiple mammalian species from mixed forensic samples based on mtDNA control region length polymorphism. Forensic Sci Int Genet Suppl Ser. 2009;2:302–303. [Google Scholar]
- Gilbert MTP, Wilson AS, Bunce M, Hansen AJ, Willerslev E, Shapiro B, Higham TFG, Richards MP, O’Connell TC, Tobin DJ, Janaway RC, Cooper A. Ancient mitochondrial DNA from hair. Curr Biol. 2004;14:R463–R464. doi: 10.1016/j.cub.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Bengtsson CF, Olsen ME, Brandt LØ, Bertelsen MF, Willerslev E, Tobin DJ, Wilson AS, Gilbert MTP. DNA from keratinous tissue: part I: hair and nail. Ann Anat. 2012;194:17–25. doi: 10.1016/j.aanat.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Rasmussen M, Li Y, Lindgreen S, Pedersen JS, Albrechtsen A, Moltke I, Metspalu M, Metspalu E, Kivisild T, Gupta R, Bertalan M, Nielsen K, Gilbert MTP, Wang Y, Raghavan M, Campos PF, Kamp HM, Wilson AS, Gledhill A, Tridico S, Bunce M, Lorenzen ED, Binladen J, Guo X, Zhao J, Zhang X, Zhang H, Li Z, Chen M, Orlando L. et al. Ancient human genome sequence of an extinct Palaeo-Eskimo. Nature. 2010;463:757–762. doi: 10.1038/nature08835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton T, Dimick G, Higgins B, Yon M, Holland C. Mitochondrial DNA analysis of 114 hairs measuring less than 1 cm from a 19-year-old homicide. Investig Genet. 2012;3:12. doi: 10.1186/2041-2223-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MTP, Janaway RC, Tobin DJ, Cooper A, Wilson AS. Histological correlates of post mortem mitochondrial DNA damage in degraded hair. Forensic Sci Int. 2006;156:201–207. doi: 10.1016/j.forsciint.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Menez L, Janaway RC, Tobin DJ, Cooper A, Wilson AS. Resistance of degraded hair shafts to contaminant DNA. Forensic Sci Int. 2006;156:208–212. doi: 10.1016/j.forsciint.2005.02.021. [DOI] [PubMed] [Google Scholar]
- Ogle RR, Mitosinka GT. A rapid technique for preparing hair cuticular scale casts. J Forensic Sci. 1973;18:82–83. [PubMed] [Google Scholar]
- Cooper A, Poinar HN. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M. Genetic analyses from ancient DNA. Annu Rev Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- Willerslev E, Cooper A. Ancient DNA. Proc Biol Sci. 2005;272:3–16. doi: 10.1098/rspb.2004.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedber EC, Walker GC, Siede W, Wood RD, Schulttz RA, Ellenberger T. DNA Repair and Mutagenesis. 2. Washington DC: ASM Press; 2005. [Google Scholar]
- Hansen A, Willerslev E, Wiuf C, Mourier T, Arctander P. Statistical evidence for miscoding lesions in ancient DNA templates. Mol Biol Evol. 2001;18:262–265. doi: 10.1093/oxfordjournals.molbev.a003800. [DOI] [PubMed] [Google Scholar]
- Gilbert MTP, Hansen AJ, Willerslev E, Rudbeck L, Barnes I, Lynnerup N, Cooper A. Characterization of genetic miscoding lesions caused by postmortem damage. Am J Hum Genet. 2003;72:48–61. doi: 10.1086/345379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Pääbo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower MA, Spencer M, Matsumura S, Nisbet RER, Howe CJ. How many clones need to be sequenced from a single forensic or ancient DNA sample in order to determine a reliable consensus sequence? Nucleic Acids Res. 2005;33:2549–2556. doi: 10.1093/nar/gki550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat Rev Genet. 2004;5:123–135. doi: 10.1038/nrg1271. [DOI] [PubMed] [Google Scholar]
- Thalmann O, Hebler J, Poinar HN, Pääbo S, Vigilant L. Unreliable mtDNA data due to nuclear insertions: a cautionary tale from analysis of humans and other great apes. Mol Ecol. 2004;13:321–335. doi: 10.1046/j.1365-294x.2003.02070.x. [DOI] [PubMed] [Google Scholar]
- den Tex R-J, Maldonado JE, Thorington R, Leonard JA. Nuclear copies of mitochondrial genes: another problem for ancient DNA. Genetica. 2010;138:979–984. doi: 10.1007/s10709-010-9481-9. [DOI] [PubMed] [Google Scholar]
- Van der Kuyl AC, Kuiken CL, Dekker JT, Perizonius WR, Goudsmit J. Nuclear counterparts of the cytoplasmic mitochondrial 12S rRNA gene: a problem of ancient DNA and molecular phylogenies. J Mol Evol. 1995;40:652–657. doi: 10.1007/BF00160513. [DOI] [PubMed] [Google Scholar]
- Greenwood AD, Pääbo S. Nuclear insertion sequences of mitochondrial DNA predominate in hair but not in blood of elephants. Mol Ecol. 1999;8:133–137. doi: 10.1046/j.1365-294x.1999.00507.x. [DOI] [PubMed] [Google Scholar]
- Greenwood AD, Capelli C, Possnert G, Pääbo S. Nuclear DNA sequences from late Pleistocene megafauna. Mol Biol Evol. 1999;16:1466–1473. doi: 10.1093/oxfordjournals.molbev.a026058. [DOI] [PubMed] [Google Scholar]
- Oliveira R, Castro D, Godinho R, Luikart G, Alves PC. Species identification using a small nuclear gene fragment: application to sympatric wild carnivores from South-western Europe. Conserv Genet. 2009;11:1023–1032. [Google Scholar]
- Song H, Buhay JE, Whiting MF, Crandall KA. Many species in one: DNA barcoding overestimates the number of species when nuclear mitochondrial pseudogenes are coamplified. Proc Natl Acad Sci USA. 2008;105:13486–13491. doi: 10.1073/pnas.0803076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. Dynamics of Pleistocene population extinctions in Beringian brown bears. Science. 2002;295:2267–2270. doi: 10.1126/science.1067814. [DOI] [PubMed] [Google Scholar]
- Verginelli F, Capelli C, Coia V, Musiani M, Falchetti M, Ottini L, Palmirotta R, Tagliacozzo A, De Grossi Mazzorin I, Mariani-Costantini R. Mitochondrial DNA from prehistoric canids highlights relationships between dogs and south-east European wolves. Mol Biol Evol. 2005;22:2541–2551. doi: 10.1093/molbev/msi248. [DOI] [PubMed] [Google Scholar]
- Gundry RL, Allard MW, Moretti TR, Honeycutt RL, Wilson MR, Monson KL, Foran DR. Mitochondrial DNA analysis of the domestic dog: control region variation within and among breeds. J Forensic Sci. 2007;52:562–572. doi: 10.1111/j.1556-4029.2007.00425.x. [DOI] [PubMed] [Google Scholar]
- Primer3. [ http://frodo.wi.mit.edu/primer3/input.htm]
- Kim KS, Lee SE, Jeong HW, Ha JH. The complete nucleotide sequence of the domestic dog (Canis familiaris) mitochondrial genome. Mol Phylogenet Evol. 1998;10:210–220. doi: 10.1006/mpev.1998.0513. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Basic Local Alignment Search Tool. [blast.ncbi.nlm.nih.gov] [DOI] [PubMed]
- Coghlan ML, White NE, Murray DC, Houston J, Rutherford W, Bellgard MI, Haile J, Bunce M. Metabarcoding avian diets at airports: implications for birdstrike hazard management planning. Investig Genet. 2013;4:27. doi: 10.1186/2041-2223-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Wheeler B, Wilson LJ. Practical Forensic Microscopy: A Laboratory Manual. Chichester: Wiley; 2008. Animal hair examinations; pp. 160–169. [Google Scholar]
- Deedrick DW, Koch SL. Microscopy of hair Part II: a practical guide and manual for animal hairs. Forensic Sci Commun. 2004;6 [ http://www.fbi.gov/about-us/lab/forensic-science-communications/fsc/july2004/research/2004_03_research02.htm] [Google Scholar]
- Arbidar Fiber and Hair Collections. [ http://www.microtracellc.com/service/arbidar-reference-collections]
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- Pääbo S. Ancient DNA: extraction, characterization, molecular cloning, and enzymatic amplification. Proc Natl Acad Sci U S A. 1989;86:1939–1943. doi: 10.1073/pnas.86.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höss M, Jaruga P, Zastawny TH, Dizdaroglu M, Pääbo S. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D, Willerslev E, Hansen A. Damage and repair of ancient DNA. Mutat Res. 2005;571:265–276. doi: 10.1016/j.mrfmmm.2004.06.060. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhang HH. Nyctereutes procyonoides Mitochondrion, Complete Genome. GenBank: GU256221.1. [ http://www.ncbi.nlm.nih.gov/nuccore/GU256221.1]
- Bär W, Kratzer A, Mächler M, Schmid W. Postmortem stability of DNA. Forensic Sci Int. 1988;39:59–70. doi: 10.1016/0379-0738(88)90118-1. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972;11:3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G + C-content biases. Mol Biol Evol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- Vilà C, Savolainen P, Maldonado JE, Amorim IR, Rice JE, Honeycutt RL, Crandall KA, Lundeberg J, Wayne RK. Multiple and ancient origins of the domestic dog. Science. 1997;276:1687–1689. doi: 10.1126/science.276.5319.1687. [DOI] [PubMed] [Google Scholar]
- Webb KM, Allard MW. Identification of forensically informative SNPs in the domestic dog mitochondrial control region. J Forensic Sci. 2009;54:289–304. doi: 10.1111/j.1556-4029.2008.00953.x. [DOI] [PubMed] [Google Scholar]
- Angleby H, Savolainen P. Forensic informativity of domestic dog mtDNA control region sequences. Forensic Sci Int. 2005;154:99–110. doi: 10.1016/j.forsciint.2004.09.132. [DOI] [PubMed] [Google Scholar]
- Shahid SA, Xiao Y, Khan S, Feng D, Johnson GS, Ha J. Canis lupus familiaris Isolate 1 Breed Welsh Springer Spaniel Mitochondrion, Complete Genome. GenBank: AY656747.1. [ http://www.ncbi.nlm.nih.gov/nuccore/AY656747]
- Pang J-F, Kluetsch C, Zou X-J, Zhang A, Luo L-Y, Angleby H, Ardalan A, Ekström C, Sköllermo A, Lundeberg J, Matsumura S, Leitner T, Zhang Y-P, Savolainen P. mtDNA data indicate a single origin for dogs south of Yangtze River, less than 16,300 years ago, from numerous wolves. Mol Biol Evol. 2009;26:2849–2864. doi: 10.1093/molbev/msp195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savolainen P, Zhang Y, Luo J, Lundeberg J, Leitner T. Genetic evidence for an East Asian origin of domestic dogs. Science. 2002;298:1610–1613. doi: 10.1126/science.1073906. [DOI] [PubMed] [Google Scholar]
- Imes DL, Wictum EJ, Allard MW, Sacks BN. Identification of single nucleotide polymorphisms within the mtDNA genome of the domestic dog to discriminate individuals with common HVI haplotypes. Forensic Sci Int Genet. 2012;6:630–639. doi: 10.1016/j.fsigen.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Kim K, Jeong H, Sohn H, Ha J. Canis familiaris Breed Jindo Mitochondrial D-Loop, Partial Sequence. Genbank: AF064569.1. [ http://www.ncbi.nlm.nih.gov/nuccore/AF064569]
- Kasuga M, Sugiyama S, Tsuchida S, Omi T. Canis lupus familiaris DNA, Hypervariable Region 1 (Control Region), Partial Sequence, Haplotype: NVLU057 Genbank: AB700664.1. [ http://www.ncbi.nlm.nih.gov/nuccore/AB700664]
- Zhang J, Zhao C, Liu GS, Chen L, Zhang HH. Canis lupus laniger Mitochondrion, Complete Genome. Genbank: KF573616.1. [ http://www.ncbi.nlm.nih.gov/nuccore/KF573616]
- Meng C, Zhang H, Meng Q. Canis lupus laniger Mitochondrion, Complete Genome. GenBank: FJ032363.2. [ http://www.ncbi.nlm.nih.gov/nuccore/FJ032363]
- Tsuda K, Kikkawa Y, Yonekawa H, Tanabe Y. Extensive interbreeding occurred among multiple matriarchal ancestors during the domestication of dogs: evidence from inter- and intraspecies polymorphisms in the D-loop region of mitochondrial DNA between dogs and wolves. Genes Genet Syst. 1997;72:229–238. doi: 10.1266/ggs.72.229. [DOI] [PubMed] [Google Scholar]
- Ishiguro N, Inoshima Y, Shigehara N. Mitochondrial DNA analysis of the Japanese wolf (Canis lupus hodophilax Temminck, 1839) and comparison with representative wolf and domestic dog haplotypes. Zoolog Sci. 2009;26:765–770. doi: 10.2108/zsj.26.765. [DOI] [PubMed] [Google Scholar]
- Sharma DK, Maldonado JE, Jhala YV, Fleischer RC. Ancient wolf lineages in India. Proc Biol Sci. 2004;271(Suppl 3):S1–S4. doi: 10.1098/rsbl.2003.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng C, Zhang H. Canis lupus chanco Mitochondrion, Complete Genome. GenBank: EU442884.2. [ http://www.ncbi.nlm.nih.gov/nuccore/EU442884]
- Dou H. Canis lupus chanco Haplotype 1 D-Loop, Partial Sequence; Mitochondrial. GenBank: JX415343.1. [ http://www.ncbi.nlm.nih.gov/nuccore/JX415343]
- Nonaka N, Sano T, Inoue T, Fukui D, Katakura K, Oku Y. Nyctereutes procyonoides Mitochondrial DNA, D-Loop Region, Partial Sequence, isolate:R1. GenBank: AB292740.1. [ http://www.ncbi.nlm.nih.gov/nuccore/AB292740]
- Okumura N, Ishiguro N, Nakano M, Matsui A, Sahara M. Intra- and interbreed genetic variations of mitochondrial DNA major non-coding regions in Japanese native dog breeds (Canis familiaris) Anim Genet. 1996;27:397–405. doi: 10.1111/j.1365-2052.1996.tb00506.x. [DOI] [PubMed] [Google Scholar]
- Park SK, Lee M-Y, Min MS, Lee H. Nyctereutes procyonoides Haplotype L_179 D-Loop, Partial Sequence; Mitochondrial. GenBank: EU642411.1. [ http://www.ncbi.nlm.nih.gov/nuccore/EU642411]
- Pitra C, Schwarz S, Fickel J. Going west - invasion genetics of the alien raccoon dog Nyctereutes procynoides in Europe. Eur J Wildl Res. 2009;56:117–129. [Google Scholar]
- Korablev NP, Korablev MP, Rozhnov VV, Korablev PN. Polymorphism of the mitochondrial DNA control region in the population of raccoon dog (Nyctereutes procyonoides Gray, 1834) introduced into the Upper Volga basin. Russ J Genet. 2011;47:1227–1233. [PubMed] [Google Scholar]
- Paulauskas A, Griciuviene L, Radzijevskaja J, Gedminas V. Nyctereutes procyonoides Isolate LV1 D-Loop, Partial Sequence; Mitochondrial. GenBank: KC509604.1. [ http://www.ncbi.nlm.nih.gov/nuccore/KC509604]
- Chen L, Zhang HH. Nyctereutes procyonoides Mitochondrion, Complete Genome. NCBI Reference Sequence: NC_013700.1. [ http://www.ncbi.nlm.nih.gov/nuccore/NC_013700]
- Wodecka B, Rymaszewska A, Skotarczak B. Host and pathogen DNA identification in blood meals of nymphal Ixodes ricinus ticks from forest parks and rural forests of Poland. Exp Appl Acarol. 2014;62:543–555. doi: 10.1007/s10493-013-9763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll CA, Macdonald DW, O’Brien SJ. From wild animals to domestic pets, an evolutionary view of domestication. Proc Natl Acad Sci U S A. 2009;106(Suppl 1):9971–9978. doi: 10.1073/pnas.0901586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll CA, Menotti-Raymond M, Roca AL, Hupe K, Johnson WE, Geffen E, Harley EH, Delibes M, Pontier D, Kitchener AC, Yamaguchi N, O’Brien SJ, Macdonald DW. The Near Eastern origin of cat domestication. Science. 2007;317:519–523. doi: 10.1126/science.1139518. [DOI] [PMC free article] [PubMed] [Google Scholar]


