We performed transcriptomic and metabolomic analysis of rosette leaves of Arabidopsis autophagy mutants to elucidate the housekeeping process of autophagy, which plays a key role in immunity, senescence, nutrient recycling, and environmental adaptation. We deciphered the wide range of effects generated by autophagy defects in plants and uncovered links between autophagy, metabolism, and signaling.
Abstract
Autophagy is a fundamental process in the plant life story, playing a key role in immunity, senescence, nutrient recycling, and adaptation to the environment. Transcriptomics and metabolomics of the rosette leaves of Arabidopsis thaliana autophagy mutants (atg) show that autophagy is essential for cell homeostasis and stress responses and that several metabolic pathways are affected. Depletion of hexoses, quercetins, and anthocyanins parallel the overaccumulation of several amino acids and related compounds, such as glutamate, methionine, glutathione, pipecolate, and 2-aminoadipate. Transcriptomic data show that the pathways for glutathione, methionine, raffinose, galacturonate, and anthocyanin are perturbed. Anthocyanin depletion in atg mutants, which was previously reported as a possible defect in flavonoid trafficking to the vacuole, appears due to the downregulation of the master genes encoding the enzymes and regulatory proteins involved in flavonoid biosynthesis. Overexpression of the PRODUCTION OF ANTHOCYANIN PIGMENT1 transcription factor restores anthocyanin accumulation in vacuoles of atg mutants. Transcriptome analyses reveal connections between autophagy and (1) salicylic acid biosynthesis and response, (2) cytokinin perception, (3) oxidative stress and plant defense, and possible interactions between autophagy and the COP9 signalosome machinery. The metabolic and transcriptomic signatures identified for the autophagy mutants are discussed and show consistencies with the observed phenotypes.
INTRODUCTION
Autophagy (namely, macro-autophagy) is an evolutionarily conserved process in eukaryotic cells. It participates in catabolic processes and the recycling of cytoplasmic and organelle constituents through the sequestration of cell material into vesicles. These vesicles are then delivered for breakdown to the central vacuole in yeast and plants or to the lysosome in animals. The autophagy system, discovered in yeast and extensively studied in mammals, has also now been well described in Arabidopsis thaliana (Liu and Bassham, 2012). Many ATG (autophagy) genes play a role in the formation of cup-shaped double membrane preautophagosomal structures. These expand and curve to engulf malfunctioning or unneeded organelles or proteins and to remove cellular debris. Under normal conditions, autophagy is a housekeeping mechanism managing cell waste, recycling cell constituents, and controlling organelle quality. Its function in remobilizing nutrients becomes essential when cells are faced with nutrient deprivation or stress.
In plants, ATG genes are upregulated during leaf senescence and in response to a large range of stresses, such as pathogen attack, drought, salt stress, and starvation (Liu et al., 2009; Breeze et al., 2011). Phenotypic analyses of Arabidopsis atg mutants showed that autophagy is essential for plant survival upon carbon, nitrogen, or water deprivation and is involved in plant immunity (Hayward and Dinesh-Kumar, 2011). As autophagy mutants displayed early senescence symptoms under nitrogen limitation, autophagy-dependent nitrogen recycling and remobilization efficiency appeared critical. The use of 15N tracing at the whole-plant level demonstrated that autophagy has an important role in the remobilization of organic nitrogen from the leaves to the seeds. It was also shown that despite higher protease activities in rosette leaves, atg mutants accumulate soluble proteins and other nitrogen compounds in their vegetative tissues (Guiboileau et al., 2012, 2013).
Although it is now widely accepted that autophagy participates in recycling cellular components and, thus, in nutrient recycling and mobilization, it is likely that autophagy activity is also essential for numerous other cellular processes related to cell longevity, cell reprogramming, or organelle repair and maintenance. Thus, defects in autophagy housekeeping functions would generate a large set of intracellular changes and defects that would differ depending on age and environmental constraints. Indeed, recent reports showed that autophagy can be selective and that this selectivity might depend on the environmental stress conditions sensed by the plants and on the specific organelles damaged or cellular pathways impaired (Floyd et al., 2012). Specific autophagy occurring in response to sulfur stress was documented in tobacco (Nicotiana tabacum) plants by the discovery of interactions between the two sulfur-responsive proteins UPC9 and Joka2 and between Joka2 and ATG8f (Zientara-Rytter et al., 2011). The Joka2 protein sequence was revealed to have high homology with the NBR1 adapter involved in autophagy selectivity in mammals. A specific involvement of autophagy in chloroplast degradation (chlorophagy) was also previously documented. Ono et al. (2013) reported that in individual darkened leaves, vacuolar transfer and processing of chloroplastic Rubisco–green fluorescent protein (GFP) fusion is not observed in the atg5 mutant. More recently, we showed that atg mutants accumulate peptides identified as Rubisco and glutamine synthetase GS2 fragments, which might be due to incomplete degradation of stromal proteins (Guiboileau et al., 2013). In addition to specificities associated with substrates and stress conditions, some reports suggest that the core autophagy machinery might also be involved in trafficking cellular components other than unwanted organelles or proteins and as such might participate in metabolic pathways such as anthocyanin maturation and trafficking to the vacuole (Floyd et al., 2012).
Beside its role in cellular component degradation, autophagy also plays a role in plant immunity (Hayward et al., 2009). Several reports have shown that autophagy is required for resistance to necrotrophic pathogens and constitutes a prosurvival mechanism (Lai et al., 2011; Lenz et al., 2011). Responses of atg mutants to biotrophic pathogens, such as Pseudomonas, appear more complex. Autophagy could restrict or conversely promote cell death responses under incompatible plant pathogen interaction (Liu et al., 2005; Hofius et al., 2009). Autophagy activity appears to be involved in the regulation of the salicylic acid (SA) pathway, and elevated SA accumulation in autophagy mutants is responsible for their early leaf senescence and the extensive cell death symptoms when inoculated with incompatible Pseudomonas strains (Yoshimoto et al., 2009). However, the exact process by which the SA/autophagy feedback loop is working remains to be elucidated.
Our aim is to obtain an overview of the pleiotropic effects of autophagy activity on the regulation of plant metabolism and cell signaling in Arabidopsis. Since autophagy activity is enhanced in leaves during ageing and when nitrate is limiting, atg mutants grown under sufficient or limiting nitrate conditions were used for metabolomics and transcriptomics analyses. The results allowed us to decipher the large range of effects generated by autophagy defects in plants and to reveal links between autophagy, metabolism, and signaling.
RESULTS
Metabolite Profiles of atg Mutants and RNAi18 Reveal Prominent Changes in Amino Acid, Sterol, Sugar, and Flavonoid Metabolism
Our first analyses were performed on atg5, atg9, ATG18a RNA interference lines (RNAi18), and Columbia (Col) wild-type rosette samples from plants grown under low and high nitrate conditions for 30, 60, and 75 d according to Guiboileau et al. (2013). As shown in our previous reports, no senescing phenotype can be observed 30 d after sowing (DAS) for any of the genotypes under low or high nitrate conditions. At 60 DAS, senescing leaves were observed under low nitrate conditions on atg5, atg9, and RNAi18 rosettes but not on the wild type. At 75 DAS, leaf senescence was observed for all genotypes under low nitrate conditions but not the wild type under high nitrate conditions (Guiboileau et al., 2013).
Metabolic changes (raw data available online as Supplemental Data Set 1) were estimated by computing the fold changes in metabolite concentrations in atg5, atg9, and RNAi18 relative to the wild type at each time point and for each nitrate condition and each planting (three separate plantings, each containing three plant samples; see Methods). Only significant and reproducible differences between the mutant/RNAi and wild type (based on t test, P < 0.05) are presented in Supplemental Data Set 2. Metabolites were classified as amino acids, amines, secondary metabolites, cyano compounds, tricarboxylic acid cycle compounds, and sugars compounds to simply presentation of results.
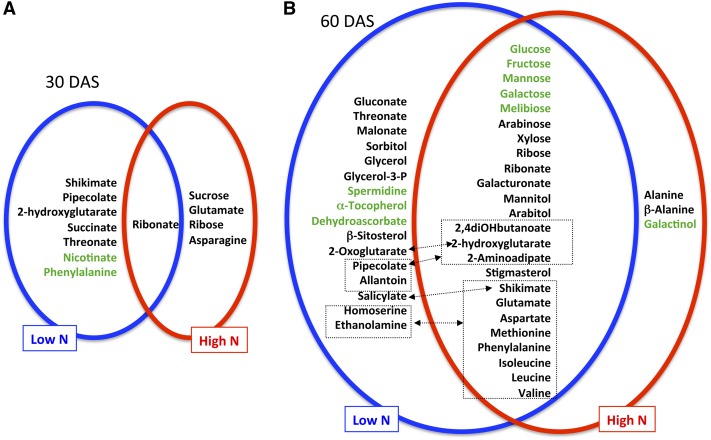
Under both low and high nitrate conditions, only a few changes were observed in atg5, atg9, and RNAi18 at 30 DAS (Figure 1A). With the exception of the ribonate concentration, which increased under both low and high nitrate conditions and in at least two of the three mutant/RNAi lines, no other metabolite changed in a similar way under both conditions (Supplemental Data Set 2; Figure 1A). Under low nitrate conditions, the accumulation of pipecolate, 2-hydroxyglutarate, shikimate, succinate, ribonate, and threonate was observed as well as a decrease in phenylalanine and nicotinate. After 30 DAS under high nitrate conditions, glutamate, asparagine, ribose, ribonate, and sucrose accumulated (Supplemental Data Set 2; Figure 1A).
Figure 1.
Changes in the Metabolite Contents of atg Mutants Compared with the Wild Type.
The metabolites that increased (black font) or decreased (gray or green font) significantly in the rosettes of atg mutant/RNAi lines, grown under low nitrate or high nitrate for 30 (A) or 60/75 (B) DAS are shown. Amino acids and related compounds are in dashed boxes, and arrows indicate metabolic links between compounds.
[See online article for color version of this figure.]
At 60 and 75 DAS, further differences between the wild type and the mutants were observed, and these were more pronounced under low nitrate conditions (Supplemental Data Set 2; Figure 1B). Under both low and high nitrate conditions, the mutant/RNAi lines accumulated a large set of amino acids among which glutamate and aspartate are the known precursors of the others. The mutant/RNAi lines also accumulated minor amino acids such as methionine, phenylalanine, and branched-chain amino acids such as valine, leucine, and isoleucine as well as shikimate, which is the precursor of aromatic amino acids and the starting point of the biosynthesis of phenolic compounds and SA (Figure 1B).
Changes in carbohydrates were more contrasted than those of the amino acids. While hexoses (glucose, fructose, mannose, and galactose) were lower in the mutant/RNAi lines than in the wild type, their corresponding sugar alcohols (mannitol and sorbitol) and aldonic acids (gluconate, galacturonate, ribonate, threonate, and malonate) were higher (Figure 1B). Depletion in hexose forms was consistent with previous results (Guiboileau et al., 2013) and the concomitant accumulation of sugar alcohols and acids suggests a defect in the redox management of sugar molecules in the autophagy defective lines. The accumulation of cell wall sugars such as xylose, arabinose, and ribonate also suggests that cell wall edification is affected in the mutants.
2-Hydroxyglutarate, 2-aminoadipate, and 2,4-dihydroxybutanoate were found to accumulate in the atg mutant/RNAi lines, suggesting that degradation of lysine and other aromatic and branched-chain amino acids is different in the mutant/RNAi lines compared with the wild type (Araújo et al., 2011; Figure 1B). 2-Hydroxyglutarate is known to participate in butanoate metabolism and is involved in lysine and aromatic amino acid catabolic pathways that can be used under certain circumstances to support mitochondrial respiration (Engqvist et al., 2011). 2-Aminoadipate is also involved in the lysine degradation pathway as a pipecolate catabolite (Goyer et al., 2004). Interestingly, pipecolate also accumulated in the mutant/RNAi lines, but only under low nitrate supply. Allantoin overaccumulation, which is symptomatic of purine degradation, was also observed. Accumulation of all these compounds suggests then that amino acid and purine catabolism is modified in autophagy mutants, and this feature could explain why autophagy mutants accumulate nitrogen and especially amino acids in their rosette leaves, as shown previously by Guiboileau et al. (2013). Indeed, it is difficult to explain why amino acids accumulate in the rosettes leaves of autophagy mutants when protein degradation is impaired (Guiboileau et al., 2013). The interconversion of all the amino acids released from protein degradation to glutamine and asparagine, which are considered as the two major amino acids dedicated to phloem uploading in plants, might be a bottleneck for amino acid translocation (reviewed in Masclaux-Daubresse et al., 2008). The accumulation of glutamate and aspartate, but not asparagine or glutamine, in the atg mutant/RNAi lines is consistent with this hypothesis.
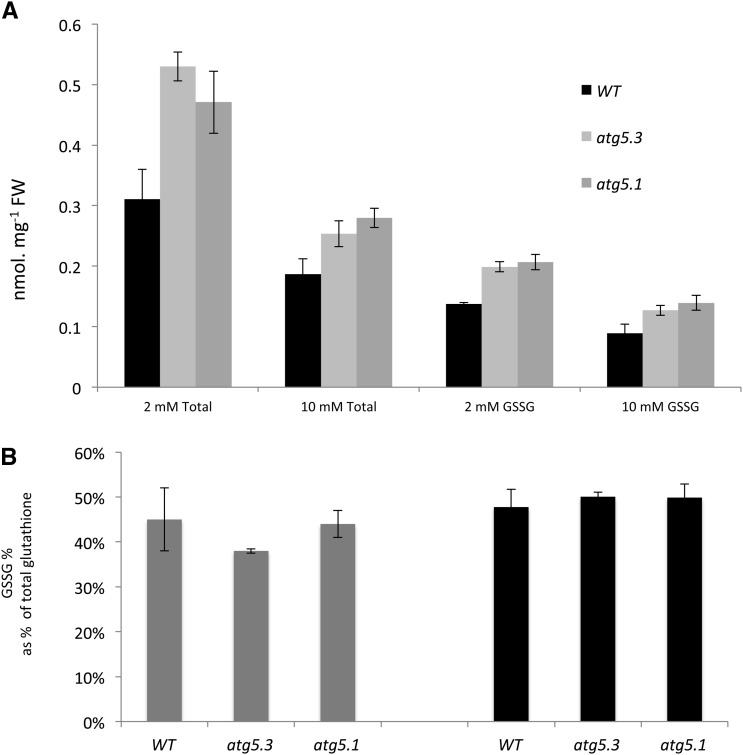
From these observations, the question of whether these metabolic defects could be related to symptoms of early senescence was raised. The main reason why autophagy mutants senesce earlier than the wild type was partially answered previously and linked to the overaccumulation of reactive oxygen species (ROS) and SA (Yoshimoto et al., 2009). Our results on sugar forms (hexoses, acids, and alcohol) show that autophagy mutants are suffering from some kind of disturbed redox (Figure 1B). In addition, we observed a depletion of antioxidant molecules such as α-tocopherol and spermidine in mutant/RNAi lines. Thus, to examine redox further, we then measured glutathione (GSSG and GSH) concentrations using enzymatic assays since glutathione cannot be determined using gas chromatography–mass spectrometry (GC-MS). Both total glutathione and oxidized glutathione (Figure 2A) were higher in atg mutants, while the percentage of glutathione oxidation was not modified (Figure 2B). These results suggest that atg mutant/RNAi lines do modulate their redox status at least through the glutathione pathway. However, the pronounced overaccumulation of glutathione that parallels the increases in glutamate and methionine suggests that amino acid metabolism is modified due to a more intensive activity of the glutathione pathway in these lines.
Figure 2.
Glutathione Concentrations Are Higher in atg Mutants.
(A) Total glutathione (GSH + GSSG) and oxidized glutathione (GSSG) concentrations are significantly higher in wild-type rosettes (black bars) than in atg5 mutant rosettes (gray bars; atg5.1 allele, dark gray; atg5.3 allele light gray; see Methods; mean and sd are shown; n = 3 plant replicates), grown under low (2 mM nitrate) or high (10 mM nitrate) for 60 DAS.
(B) The percentage of glutathione oxidation is not different in atg5 mutant compared with wild-type rosettes grown under low (2 mM nitrate; gray bars) or high (10 mM nitrate; black bars) for 60 DAS (mean and sd are shown; n = 3 plant replicates).
In addition to SA, the autophagy mutants also overaccumulate its two precursors, shikimate and phenylalanine. Interestingly, we observed that the autophagy defective lines did not turn red under nitrate limitation, as is usually observed in the wild type and under high nitrate conditions (Figure 3). This phenotype was especially striking on the reverse side of the rosettes and suggested differences in anthocyanin contents. We then used liquid chromatography–mass spectrometry (LC-MS) to measure the concentrations of flavonoid compounds that might be important as protectants against oxidative stress and are also produced from shikimate and phenylalanine. Overall, under high nitrate conditions, there was no significant difference in flavonoid contents between atg5 and Col wild type except for the cyanidins A1342 and A1182, which were less abundant in atg5 (Table 1; Supplemental Table 1). However, when grown under nitrate limitation, we found that the levels of all the anthocyanin molecules were 2 to 6 times lower in atg5 than in the wild type at 60 DAS, and 3 to 60 times less at 75 DAS. Quercetin was 2 times less abundant in atg5 at both 60 and 75 DAS, while no difference was observed for kampferol. The increase in SA that paralleled the decrease in flavonoids suggested that phenylalanine products are being preferentially rerouted to the SA pathway, thus depleting the biosynthesis of other flavonoid compounds.
Figure 3.
Unlike the Wild Type, the Rosettes of the atg Mutant Do Not Accumulate Anthocyanin or Turn Dark Red.
Photos of the reverse side of Col, atg5, and atg9 rosettes grown under low or high nitrate conditions for 60 DAS.
[See online article for color version of this figure.]
Table 1. Anthocyanin, Quercetin, and Kampferol in the atg5 Mutant Compared to the Wild Type.
| 30 DAS | 60 DAS | 75 DAS | ||||||
|---|---|---|---|---|---|---|---|---|
| Low N 2 mM | atg5 | Col | atg5 | Col | atg5 | Col | ||
| ng⋅g−1 FW | Cyanidin 3-O-[2’’-O-(xylosyl)-6’’… | Mean | 47.1 | 221.2 | 349.7 | 2373.2 | 7.9 | 253.5 |
| A974 | sd | 80.9 | 282.7 | 275.1 | 327.4 | 17.8 | 158.7 | |
| Cyanidin 3-O-[2’’-O-(2’’’-O-(sinapoyl) … | Mean | 194.4 | 266.1 | 470.5 | 1642.8 | 25.5 | 255.3 | |
| A1182 | sd | 181.1 | 153.4 | 170.8 | 191.0 | 42.7 | 62.2 | |
| Cyanidin 3-O-[2’’-O-(xylosyl) 6’’… | Mean | 0.0 | 0.0 | 1309.2 | 5535.5 | 42.0 | 2498.0 | |
| A1136 | sd | 0.0 | 0.0 | 233.4 | 651.5 | 73.4 | 922.1 | |
| Cyanidin 3-O-[2’’-O-(6’’’-O-(sinapoyl)… | Mean | 465.3 | 376.9 | 3783.8 | 7878.6 | 714.7 | 3138.0 | |
| A1342 | sd | 83.3 | 131.9 | 722.3 | 514.9 | 190.9 | 1109.8 | |
| Isomer Cyanidin 3-O-[2’’-O-(6’’’-… | Mean | 167.5 | 151.9 | 933.6 | 2171.5 | 463.5 | 1151.7 | |
| A1342 2 | sd | 99.9 | 117.0 | 93.8 | 108.7 | 77.7 | 316.9 | |
| Cyanidin 3-O-[2’’-O-(2’’’-O-(… | Mean | 33.1 | 36.8 | 602.7 | 1288.5 | 136.5 | 659.3 | |
| A1256 | sd | 22.2 | 29.3 | 81.4 | 114.2 | 28.6 | 258.0 | |
| μg⋅g−1 FW | K-3,7-di-O-Rha | Mean | 153.9 | 153.7 | 337.6 | 340.1 | 265.5 | 262.3 |
| sd | 8.1 | 14.5 | 11.2 | 32.6 | 43.5 | 22.7 | ||
| K-3-O-Glc-7-O-Rha | Mean | 96.4 | 95.2 | 147.8 | 135.9 | 140.1 | 124.7 | |
| sd | 6.5 | 9.9 | 12.8 | 22.1 | 26.5 | 19.1 | ||
| K-3-O-Rha(1-2)Glc-7-O-Rha | Mean | 245.0 | 231.9 | 694.5 | 574.6 | 532.5 | 481.2 | |
| sd | 36.6 | 38.5 | 84.2 | 62.4 | 109.4 | 52.0 | ||
| Q-3-O-Glc-7-O-Rha | Mean | 4.1 | 5.1 | 28.4 | 58.9 | 12.5 | 31.7 | |
| sd | 1.6 | 1.8 | 5.0 | 10.0 | 3.3 | 8.5 | ||
| Q-3-O-Rha(1-2)Glc-7-O-Rha | Mean | 8.6 | 8.0 | 52.3 | 90.1 | 16.9 | 46.9 | |
| sd | 1.8 | 2.2 | 5.6 | 8.4 | 7.5 | 13.6 | ||
| I-Glc-Rha | Mean | 1.2 | 1.5 | 3.9 | 5.4 | 3.3 | 3.8 | |
| sd | 0.9 | 1.4 | 1.0 | 1.5 | 0.9 | 0.6 | ||
| Q-3,7-di-O-Rha 60V | Mean | 5.6 | 5.0 | 15.5 | 41.6 | 6.5 | 21.4 | |
| sd | 0.6 | 0.8 | 2.8 | 7.6 | 2.3 | 7.4 | ||
| 30 DAS | 60 DAS | 75 DAS | ||||||
|---|---|---|---|---|---|---|---|---|
| High N 10 mM | atg5 | Col | atg5 | Col | atg5 | Col | ||
| ng⋅g−1 FW | Cyanidin 3-O-[2’’-O-(xylosyl)-6’’… | Mean | 0.0 | 0.0 | 12.3 | 0.0 | 15.6 | 7.2 |
| A974 | sd | 0.0 | 0.0 | 21.3 | 0.0 | 27.0 | 12.4 | |
| Cyanidin 3-O-[2’’-O-(2’’’-O-(sinapoyl) … | Mean | 136.6 | 274.5 | 0.0 | 102.3 | 0.0 | 0.0 | |
| A1182 | sd | 118.4 | 115.3 | 0.0 | 6.2 | 0.0 | 0.0 | |
| Cyanidin 3-O-[2’’-O-(xylosyl) 6’’… | Mean | 0.0 | 0.0 | 0.0 | 0.0 | 73.0 | 0.0 | |
| A1136 | sd | 0.0 | 0.0 | 0.0 | 0.0 | 126.5 | 0.0 | |
| Cyanidin 3-O-[2’’-O-(6’’’-O-(sinapoyl)… | Mean | 351.1 | 352.7 | 90.1 | 368.8 | 77.4 | 609.9 | |
| A1342 | sd | 52.4 | 118.6 | 41.2 | 105.8 | 134.1 | 200.7 | |
| Isomer Cyanidin 3-O-[2’’-O-(6’’’-… | Mean | 145.0 | 200.8 | 36.2 | 143.0 | 41.2 | 48.1 | |
| A1342 2 | sd | 6.0 | 92.2 | 62.7 | 46.5 | 71.3 | 83.3 | |
| Cyanidin 3-O-[2’’-O-(2’’’-O-(… | Mean | 32.0 | 35.8 | 12.8 | 51.2 | 0.0 | 0.0 | |
| A1256 | sd | 27.7 | 7.9 | 11.2 | 22.5 | 0.0 | 0.0 | |
| μg⋅g−1 FW | K-3,7-di-O-Rha | Mean | 179.3 | 168.8 | 199.8 | 194.4 | 150.3 | 198.3 |
| sd | 6.6 | 18.2 | 1.5 | 21.4 | 34.3 | 31.3 | ||
| K-3-O-Glc-7-O-Rha | Mean | 107.5 | 100.2 | 119.4 | 116.8 | 82.7 | 109.4 | |
| sd | 6.6 | 8.7 | 0.8 | 11.9 | 24.3 | 20.1 | ||
| K-3-O-Rha(1-2)Glc-7-O-Rha | Mean | 262.1 | 271.0 | 266.1 | 295.2 | 203.0 | 238.7 | |
| sd | 15.7 | 44.0 | 28.4 | 3.9 | 88.8 | 50.4 | ||
| Q-3-O-Glc-7-O-Rha | Mean | 6.1 | 5.3 | 6.8 | 6.3 | 9.4 | 3.8 | |
| sd | 0.6 | 0.4 | 0.4 | 0.4 | 2.1 | 4.6 | ||
| Q-3-O-Rha(1-2)Glc-7-O-Rha | Mean | 10.7 | 8.7 | 8.7 | 11.7 | 6.0 | 10.8 | |
| sd | 0.8 | 0.9 | 2.0 | 0.4 | 3.1 | 2.6 | ||
| I-Glc-Rha | Mean | 1.1 | 2.1 | 2.4 | 1.6 | 0.9 | 0.0 | |
| sd | 0.3 | 1.3 | 0.6 | 1.3 | 1.5 | 0.0 | ||
| Q-3,7-di-O-Rha 60V | Mean | 7.3 | 5.5 | 5.2 | 6.6 | 2.9 | 4.9 | |
| sd | 0.6 | 0.2 | 0.8 | 0.2 | 1.2 | 0.4 | ||
Anthocyanin, quercetin, and kampferol concentrations (mean ± se; n = 6 plants) in atg5 and Col wild type grown under low (Low N 2 mM) and high (High N 10 mM) nitrate conditions for 30, 60, and 75 DAS. Significant differences between atg5 and the wild type are indicated by bold and italic numbers. Three culture rounds were tested giving similar results. Complete names of cyanidin compounds can be found in Supplemental Table 1. Anthocyanin concentrations are in ng·g−1 FW. Quercetin and kampferol concentrations are in μg·g−1 FW. FW, fresh weight.
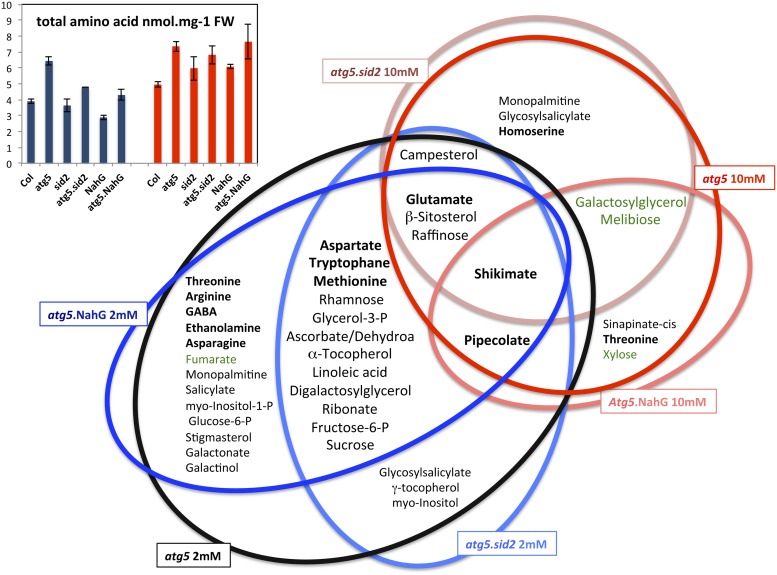
The question of whether the changes in metabolite profiles in the atg mutants are due to their early leaf senescence phenotype was also addressed using the SA-defective atg mutants previously described by Yoshimoto et al. (2009). As those SA-defective atg mutants senesce later, similarly to the wild type, these were used with the aim of detecting autophagy-related and senescence-independent metabolic changes. Thus, metabolites were determined in the Col wild type, two SA defective lines (the sid2 mutant and NahG overexpressor), the SA-deficient atg mutants (atg5-3.sid2 and atg5-1.NahG), and the two different atg5-1 and atg5-3 mutants used previously by Yoshimoto et al. (2009) and Guiboileau et al. (2012) (see Methods). Supplemental Figure 1 shows that there was no significant difference between the metabolomes of mutant alleles atg5-1 and atg5-3, so these were not distinguished further in this report. In order to determine SA-dependent and SA-independent autophagy-related metabolome modifications, significant metabolic differences were determined between atg5 and Col, between atg5.sid2 and sid2, and between atg5.NahG and NahG (Supplemental Table 2). These comparisons allowed us to determine which metabolites were significantly different in atg mutants independent of senescence symptoms, SA production, and genetic backgrounds. In the atg5/Col, atg5sid2/sid2, and atg5.NahG/NahG comparisons, we found that the most robust fingerprint specific to the autophagy defect was shikimate accumulation in atg5 mutants, which was observed regardless of genetic background and nitrate conditions (Supplemental Table 2; Figure 4). The other characteristics that were confirmed in five of the six comparisons performed were the significant accumulation of glutamate, pipecolate, β-sitosterol, and raffinose. The accumulation of other amino acids such as aspartate, methionine, and tryptophan were also significant in all the atg5 backgrounds under low nitrate conditions. Measurement of the total amino acid concentration also confirmed their increase in the autophagy defective lines compared with their respective controls (Figure 4). As a result, we concluded that the increases in glutamate, pipecolate, β-sitosterol, raffinose, aspartate, and methionine are robust fingerprints of defects in autophagy.
Figure 4.
SA-Dependent and -Independent Metabolic Changes in atg Mutants.
Venn diagram representing the metabolites that increased or decreased in the rosettes of atg5, atg5 sid2, and atg5 NahG relative to Col, sid2, and NahG controls, respectively, grown under low nitrate or under high nitrate for 60 DAS. The histogram (top left) shows that total amino acid concentrations are significantly different (Student’s t test *P < 0.05) in atg5, atg5 sid2, and atg5 NahG relative to Col, sid2, and NahG controls, respectively (mean and sd are shown; n = 3 plant replicates).
[See online article for color version of this figure.]
Gene Ontology Analysis of Transcriptomic Data for atg5 and atg9 Relative to the Wild Type Highlights That the atg Mutants Are Hypersensitive to a Large Set of Stresses
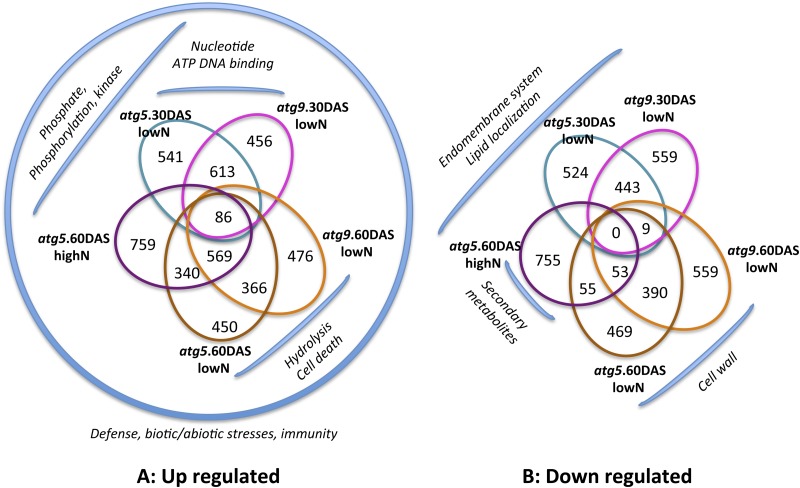
Genes showing significant changes (based on false discovery rate [FDR] percentage values; see Methods) in expression between (1) atg5 or atg9 mutants and the wild type grown under low nitrate conditions for 30 or 60 DAS, (2) the two atg mutants (atg5 and atg9) and the wild type grown under low nitrate conditions for 30 or 60 DAS, and (3) atg5 and the wild type grown under high nitrate for 60 DAS were identified. The full data sets are available online as supplemental Excel files (Supplemental Data Sets 3 to 9). The lists of genes that were significantly up- or downregulated in each condition, genotype, and harvest time are shown in Supplemental Data Sets 10 and 11. These lists were analyzed to determine intersections and Gene Ontologies (GOs) (Figure 5).
Figure 5.
The Number of Genes Up- or Downregulated in atg Mutants Relative to the Wild Type Fluctuates Depending on Nitrate Conditions and Plant Age.
Venn diagrams represent the number of genes upregulated (A) and downregulated (B) in atg5 or atg9 mutants grown under low (lowN) or high (highN) nitrate conditions for 30 or 60 DAS. The significantly up- or downregulated genes relative to the wild type (based on FDR% values; see Methods) were recorded (Supplemental Data Set 3 and 4) and analyzed using VirtualPlant1.3 (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/) and AGRIGO (http://bioinfo.cau.edu.cn/agriGO/index.php) bioinformatics tools to extract intersection lists for Venn diagrams and analyze GO classes. The most significant GO term classes found in the different lists are indicated by the brackets surrounding the Venn diagrams.
[See online article for color version of this figure.]
Eighty-six genes were significantly upregulated in both atg5 and atg9 under both low and high nitrate conditions at 30 and 60 DAS (Figure 5A; Supplemental Table 3 for more details). The frequency of the GO terms of these 86 upregulated genes shows that they are mainly involved in response to stimuli such as biotic stress (e.g., PATHOGENESIS-RELATED GENE1 [PR1], PLANT NATRIURETIC PEPTIDE-A, UPI serine protease inhibitor, the transcription factor genes WRKY38 and WRKY18, the resistance factor gene HR4, ARABINOGALACTAN-PROTEIN5, and the putative cytochrome 450 gene CYP71B25), chemicals and abiotic stresses (e.g., the transcription factor genes ANAC061, ANAC036, MYB15, MYB2, and MC8) and SA (e.g., PHYTOALEXIN DEFICIENT3 [PAD3], PAD4, and the GH3 gene PBS3) (Supplemental Table 4). No genes were downregulated in both atg5 and atg9 and in all nitrate conditions and at all time points (Figure 5B). To facilitate GO term analysis of all the genes listed in Supplemental Data Sets 10 and 11, we considered the genes up- or downregulated separately in both atg5 and atg9 grown for (1) 30 DAS under low nitrate conditions or (2) 60 DAS under low nitrate conditions and in atg5 grown for 60 DAS under high nitrate conditions (Supplemental Table 5). As shown in Supplemental Table 5, the most significant classes of genes upregulated in the mutants relative to the wild type were related to defense, biotic and abiotic stress responses, and immunity. Classes of GO terms related to phosphorus, such as phosphate/phosphorus metabolic processes, protein amino acid phosphorylation, phosphorylation and kinase activities, transferase activities transferring phosphorus, and posttranslational protein modifications, were also found in both atg5 and atg9 grown under low nitrate conditions for 30 DAS and in atg5 grown under high nitrate conditions for 60 DAS. The few classes of genes upregulated in both atg5 and atg9 under low nitrate conditions at 60 DAS were related to hydrolysis (GO:0016798, GO:0004553, and GO:0016787), cell death (GO:0016265 and GO:0008219), and regulation of defense or response to stimulus (GO:0048583 and GO:0031347). Overrepresentation of hydrolase and cell death–related genes in atg5 and atg9 under low nitrate conditions at 60 DAS is consistent with the fact that under nitrate limitation, mutants present severe senescence symptoms that certainly involve hydrolysis processes linked to cell component degradation (Breeze et al., 2011). Indeed, protease activities are also much higher in the mutants than in the wild type (Guiboileau et al., 2013).
The number of genes significantly downregulated in atg5 or atg9 under the various conditions and at the different time points was quite similar to the number of genes upregulated under the same conditions. However, their intersections and the significance (FDR%) of their GO terms were much weaker (Figure 5B; Supplemental Table 6). The few GO term classes detected were also all different depending on time points and nitrate conditions. We can note that they are related to the endomembrane system, lipid localization, and cell wall and secondary metabolism (phenylpropanoid, flavonoid, and isoprenoid), which makes sense regarding our metabolome results.
Transcript Profiles of atg Mutants Are Consistent with Metabolite Profiles and a Deficiency in Flavonoid Biosynthesis
In order to relate the transcriptome data to our previous metabolite profiles, we specifically examined the expression of genes related to the different metabolic pathways affected in atg mutant/RNAi lines.
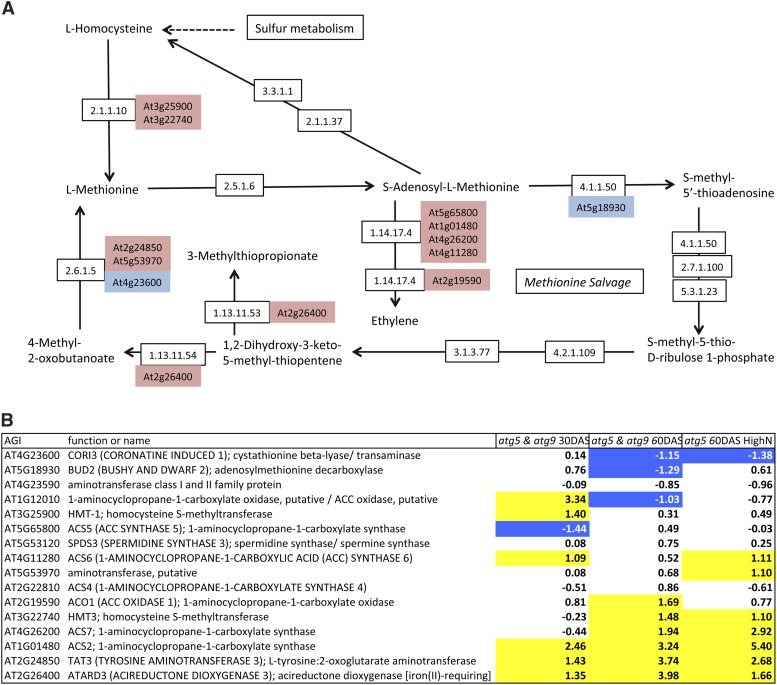
Since amino acid concentrations, especially glutamate, aspartate, and methionine, were shown to increase in the atg mutant/RNAi lines and in the SA-deficient atg mutants compared with the wild type, we first examined nitrogen and amino acid metabolism (Supplemental Table 7). Only a few genes involved in nitrate and ammonium assimilation and in glutamine, glutamate, aspartate, and asparagine metabolism were differentially expressed in atg5 and atg9, suggesting that amino acid accumulation in the mutants is not regulated at the transcriptional level by modification of the expression of the corresponding biosynthetic enzymes. Indeed, the upregulation of genes involved in nitrogen metabolism, such as GDH2 and ASN1, which are dark-induced genes, cannot explain the differences in amino acid composition, especially in the case of aspartate, asparagine, glutamate, and glutamine. In the case of methionine, which was significantly higher in all the autophagy-defective genotypes, examination of its biosynthesis pathway on KEGG (http://www.genome.jp/kegg/pathway.html) showed that expression of many genes involved in methionine synthesis and salvage pathways are significantly, and for some highly, upregulated (Figure 6). Thus, methionine metabolism and salvage pathways are certainly very active in the atg mutants. The link between methionine and ethylene also suggests that ethylene biosynthesis could be higher in the mutants, despite the fact that Yoshimoto et al. (2009) did not find any phenotype recovery in the atg5 ein2 double mutant. In addition to the methionine pathway, we observed that several genes involved in the glutathione pathway and encoding the glutathione S-transferase and γ-glutamyl transpeptidase were upregulated in the atg mutants. In good agreement with our metabolomic data, the active conversion of glutathione to glutamate suggested by these transcriptomic data might explain the higher concentrations of glutamate in the atg mutants (Figure 7). Although pipecolate accumulation was very high in the atg mutants compared with the wild type, the transcriptomic data did not reveal any significant difference in the expression levels of the few genes known in Arabidopsis to be involved in lysine or branched-chain amino acid catabolism.
Figure 6.
Genes Involved in the Methionine Pathway Are Upregulated in atg5 and atg9.
The l-methionine pathway was adapted from the KEGG methionine pathway (A) available online (http://www.genome.jp/kegg/pathway.html). Several genes identified on KEGG as encoding enzymes connected to methionine biosynthesis and methionine salvage pathways and ethylene biosynthesis are upregulated (pink box) or downregulated (blue box) in atg5 and atg9 relative to the wild type as shown in (B). Relative expression is presented as the log2(fold change) Significant up- or downregulation (FDR% <10−5) is indicated by yellow- or blue-colored cells, respectively.
Figure 7.
The Glutathione-to-Glutamate Conversion Pathway Is Upregulated in atg5 and atg9 Mutants.
The glutathione-to-glutamate pathway was adapted from the KEGG glutathione metabolism pathway (A) available online (http://www.genome.jp/kegg/pathway.html). Many genes identified on KEGG as encoding the glutathione S-transferase (EC.2.5.1.18) and γ-glutamyl-transpeptidase (EC.2.3.2.2) are upregulated in atg5 and atg9 relative to the wild type as shown in (B). Relative gene expression in atg5 and atg9 compared with the wild type is presented as the log2(fold change). Significant up- or downregulation (FDR% <10−5) is indicated by yellow-colored (light) and blue-colored (dark) cells, respectively.
[See online article for color version of this figure.]
Regarding other metabolites, such as some phytosterols and galacturonate, the transcriptome data showed that upregulation of the genes involved in their biosynthesis was reliably well correlated with the increase in their concentrations in the atg mutants (Supplemental Figures 2 and 3). Similarly, the increase in raffinose in the mutants was consistently well correlated with the upregulation of its biosynthetic genes DARK INDUCIBLE10 and RAFFINOSE SYNTHASE2.
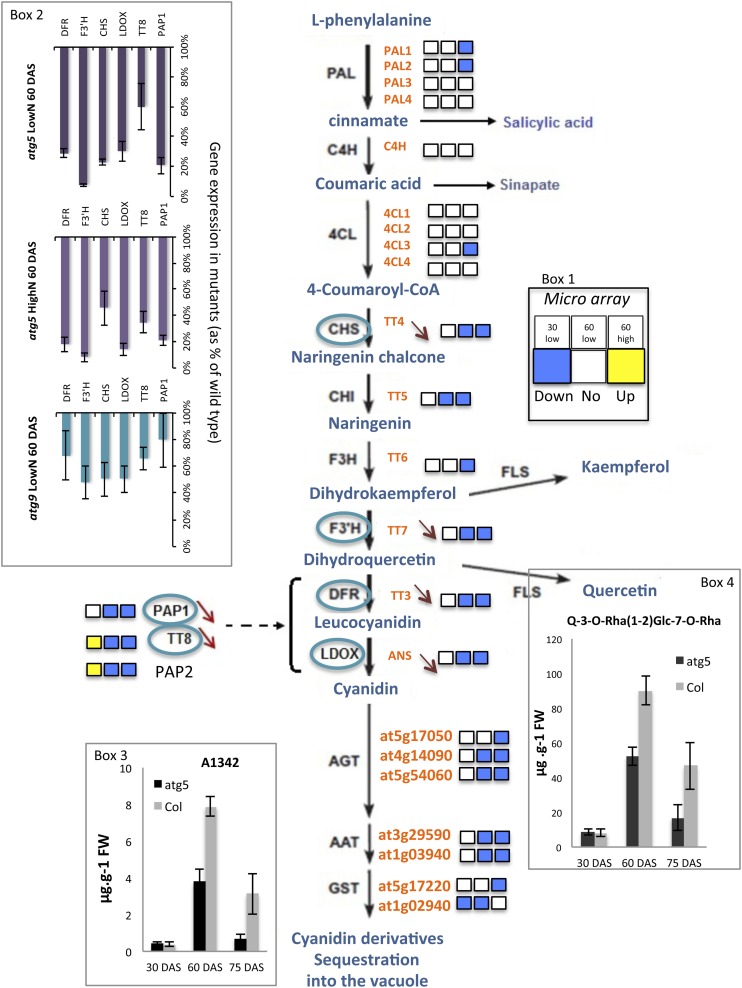
Among the other compounds found to overaccumulate in atg mutants, shikimate is a building block for many secondary metabolites, such as phenolics, anthocyanin, and SA. Each of these pathways could thus influence shikimate accumulation or depletion. When each was examined, we only found differences between the atg mutants and the wild type for the genes involved in the anthocyanin pathway. Different levels of anthocyanin and quercetin (Table 1, Figure 8, Boxes 3 and 4) were observed in the atg mutants relative to the wild type as previously reported by Pourcel et al. (2010). In their report, the authors suggested that autophagy is needed for trafficking and loading anthocyanin in the vacuole, and this conclusion was further reported in a review (Floyd et al., 2012). When expression of genes involved in phenylpropanoid pathways (KEGG and Lillo et al., 2008) was examined, we found that only a few genes involved in the shikimate pathway (e.g., the alanine glyoxylate aminotransferase gene AGT3) are upregulated in atg5 and atg9 (Supplemental Table 8). In contrast, many genes involved in the biosynthetic and regulatory flavonoid pathway, among which the master regulatory genes PRODUCTION OF ANTHOCYANIN PIGMENT 1 PAP1, PRODUCTION OF ANTHOCYANIN PIGMENT 2 PAP2 and TRANSPARENT TESTA 8 TT8 and the key enzymes CHALCONE SYNTHASE CHS, LEUCOANTHOCYANIDIN DIOXYGENASE LDOX, DIHYDROFLAVONOL 4-REDUCTASE DFR and FLAVANONE 3-HYDROXYLASE F3′H, were significantly downregulated in both atg5 (Supplemental Table 8) and atg9 (Supplemental Data Set 11) grown under low or high nitrate conditions at 60 DAS. Quantitative real-time PCR confirmed this downregulation of the major regulatory (PAP1 and TT8) and biosynthetic (CHS, DFR, F3′H, and LDOX) genes (2- to 5-fold in atg9 and atg5, respectively) (Figure 8, Box 2).
Figure 8.
Genes Involved in the Flavonoid Biosynthesis and Regulatory Pathways Are Downregulated in atg5.
A diagram of the flavonoid pathway was adapted from Lillo et al. (2008). The steps are catalyzed by phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonol synthase (FLS), dihydroflavonol 4-reductase (DFR), anthocyanidin synthase (ANS or LDOX), anthocyani(di)n glycosyltransferase (AGT), anthocyanin acyl transferase (AAT), and glutathione S-transferase (GST). PAP1, TT8, and PAP2, the main regulators of DFR and LDOX steps, are also shown. Genes encoding all the enzymes involved in the flavonoid pathway are in green typeface. Microarray data (Box 1) are presented as three squares for the relative gene expressions in atg5 grown under low nitrate for 30 d (first square), under low nitrate for 60 d (second square), and under high nitrate for 60 d (third square). Square colors in yellow, white, or blue indicate significant upregulation in atg5, no difference between atg5 and the wild type, or downregulation in atg5, respectively. Real-time quantitative RT-PCR performed on transcripts of atg5 and atg9 confirmed downregulation (red arrows) of the genes surrounded in blue and histograms (Box 2; mean and sd are shown on three plant replicates; two technical replicates showed similar results) of the relative gene expressions are presented in the top left box. The decreases observed in quercetin and anthocyanin compounds are illustrated by the representation of the histograms of A1342 and Q-3-Rha(1-2)Glc7-O-Rha concentrations in atg5 (black bars) and the wild type (gray bars) (Boxes 3 and 4, respectively; mean and sd are shown; n = 3 plant replicates).
Inhibition of Flavonoid Biosynthetic and Regulatory Pathways Reduces Anthocyanin Accumulation in atg Mutants
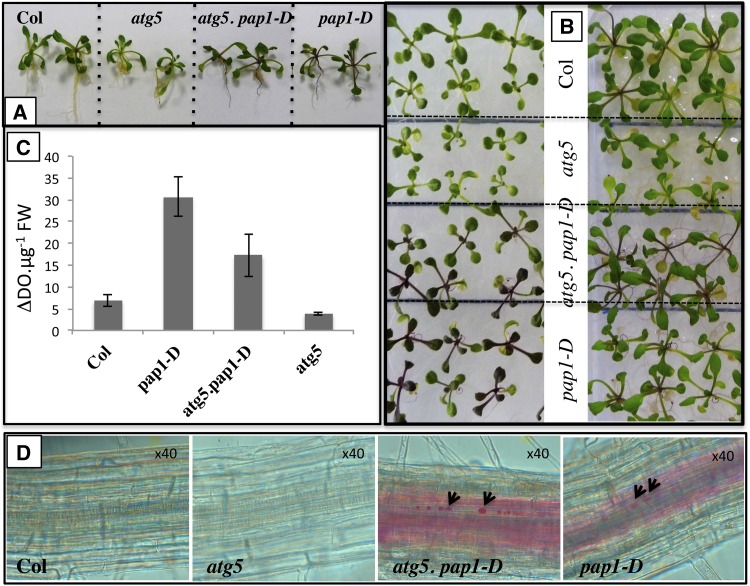
Although Pourcel et al. (2010) suggested that autophagy participates in trafficking and loading anthocyanin in the vacuole for storage, our transcriptomic data suggest that anthocyanin levels are down in atg mutants due to the suppression of their biosynthesis through downregulation of the master genes of the pathways. To verify this hypothesis, atg5 was crossed with the bright purple production of anthocyanin pigment 1-dominant (pap1-D) line in which the PAP1 gene is overexpressed (Borevitz et al., 2000). A double mutant was obtained (Supplemental Figure 4), and the red phenotype was recorded visually and by measuring the total anthocyanin concentration with a spectrometer. Interestingly, the double atg5 pap1-D plants grown in vitro turned red very soon after sowing like the pap1-D single mutant (Figures 9A and 9B); they were slightly bigger than atg5 but smaller than pap1-D. Measurement of absorbance at 525 nm of acidic methanolic extracts confirmed that atg5 pap1-D were accumulating more anthocyanin than the wild type and atg5, although slightly less than pap1-D (Figure 9C). The fact that in atg mutants both PAP2 and TT8 are downregulated in addition to PAP1 might explain why the pap1-D dominant allele introduced in atg5 did not fully restore anthocyanin production to the same level as measured in the pap1-D single mutant. Observations of seedling roots (Figure 9D) showed that in atg5 pap1-D, large amounts of anthocyanin accumulate in the vacuoles and are more concentrated in some intense red subvacuolar inclusions, described as anthocyanic vacuolar inclusions (AVIs) by Poustka et al. (2007) and Pourcel et al. (2010) (Figure 9). It seems then that the main reason why autophagy mutants do not accumulate anthocyanin is the downregulation of the anthocyanin biosynthesis pathway. As anthocyanins are antioxidants that could be involved in the early senescence phenotype of atg mutants, we compared atg mutants and wild-type phenotypes in order to determine whether the recovery of anthocyanin biosynthetic capacity has effects on other autophagy mutant phenotypes. We found that the phenotypes of atg5 pap1-D and pap1-D were very similar when grown under low or high nitrate conditions, even though atg5 pap1-D rosettes were slightly smaller than pap1-D (Figures 10A and 10B). The atg5 pap1-D rosettes were significantly bigger than the atg5 rosettes. However, both atg5 pap1-D and atg5 plants senesced earlier than the wild type and pap1-D (Figure 10C). Rosette biomass of atg5 was partially recovered by PAP1 overexpression, but their early senescence phenotype did not change.
Figure 9.
Overexpression of PAP1 in atg5 Enhanced Anthocyanin Accumulation and the Production of AVIs.
(A) and (B) The pap1-D dominant allele, introduced in the atg5 mutant, confers red phenotype to atg5 pap1-D similar to that of pap1-D single mutant.
(C) Anthocyanin concentration is increased in atg5 pap1-D compared with atg5 (mean and sd are shown; n = 4).
(D) Anthocyanin accumulation and AVI (arrows) can be observed in the roots of atg5 pap1-D and pap1-D but not in the roots of atg5 and the wild type.
Figure 10.
The Overexpression of PAP1 in atg5 Confers Red Phenotype but Does Not Modify Early Senescence Phenotype.
(A) Comparison of redness between atg5, atg5 pap1-D, pap1-D, and wild-type plants grown under low nitrate conditions and long days.
(B) Comparison of redness between atg5, atg5 pap1-D, pap1-D, and wild-type plants grown under high nitrate conditions and long days.
(C) Comparison of leaf senescence in atg5, atg5 pap1-D, pap1-D, and wild-type plants grown under high nitrate conditions and long days.
Transcript Profiling of atg Mutants Confirmed Hormonal Deregulation and Stress Response
As described previously, we found that autophagy mutants are early senescing and accumulate SA. Consistent with that, we also found that the SA biosynthetic genes are upregulated in atg5 (Supplemental Figure 5). Early senescence of atg mutants, observed as early yellowing symptoms, was positively correlated with upregulation of several senescence-associated genes, such as SAG12, SAG13, SAG21, SRG1, and SRG3 (Supplemental Table 9). However, while the differential expression of SAGs in the atg mutants compared with the wild type was significant from 30 DAS, surprisingly, the fold changes did not increase with age (Supplemental Table 9). In addition, the expression of many known senescence-related genes was not different in the atg mutants relative to the wild type. This suggests that the early leaf senescence of atg mutants is not typical developmental senescence but appears to be related to other stresses, possibly excess SA production.
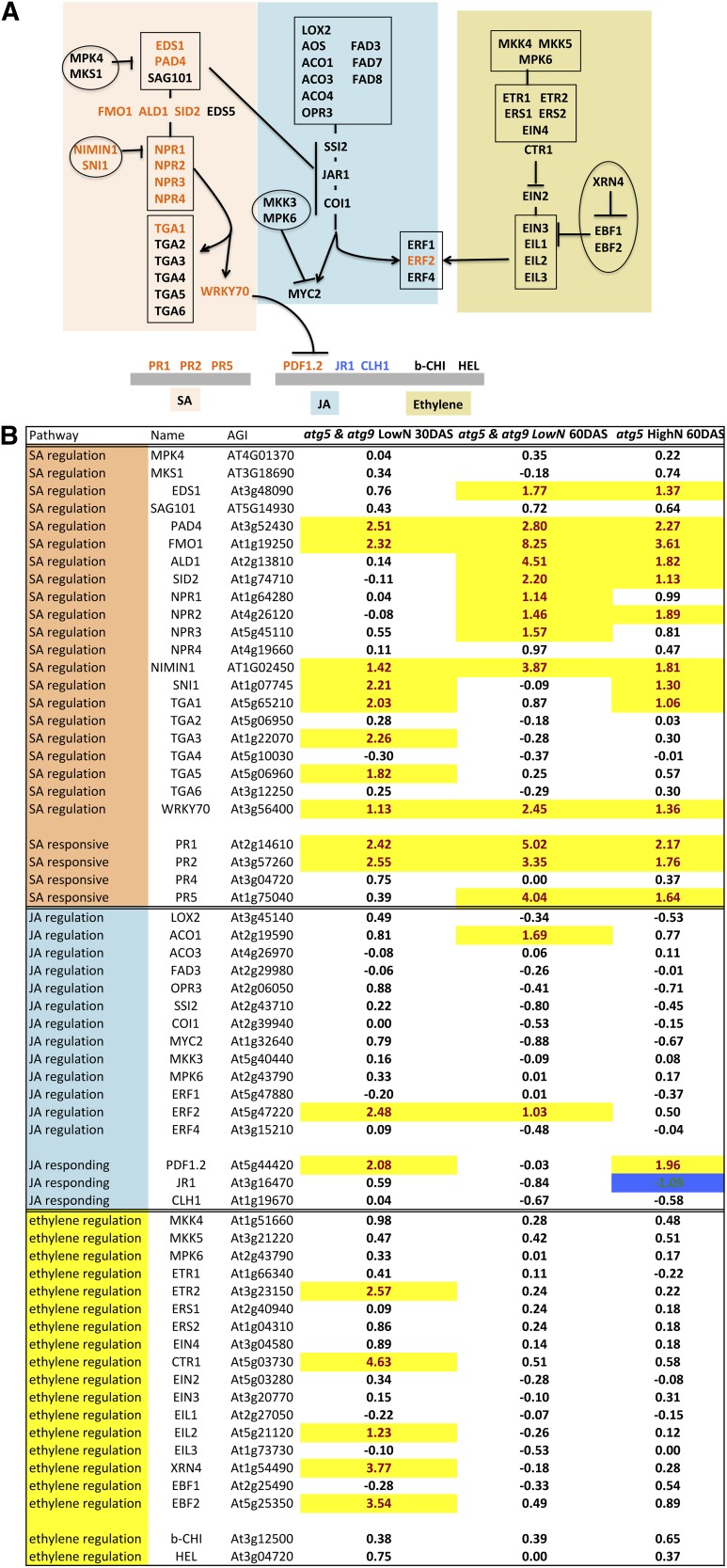
To confirm that autophagy mutants were suffering from stress and that stress hormone signaling pathways are affected, the relative expression of marker genes responding to pathogens, SA, jasmonic acid (JA), and ethylene was examined (Figure 11A). Many of the SA-related genes involved in the SA signaling pathway (ENHANCED DISEASE SUSCEPTIBILITY1, PAD4, SALICYLIC ACID INDUCTION DEFICIENT2, and NONEXPRESSER OF PR GENES [NPR1-4]) or in SA responses (PR1 and PR2) were upregulated in atg5 and atg9 relative to the wild type (Figure 11B; Supplemental Table 10). Consistent with Yoshimoto et al. (2009), who did not detect any difference in JA contents between atg mutants and the wild type, very few genes involved in the JA signaling pathway (like ETHYLENE RESPONSE FACTOR2) or responding to JA (like PLANT DEFENSIN 1.2) were differentially expressed in atg5 and atg9 mutants compared with the wild type. The very few ethylene signaling related genes (e.g., ETHYLENE RESPONSE2 and CONSTITUTIVE TRIPLE RESPONSE1) that were upregulated in atg5 and atg9 at 30 DAS could suggest that ethylene is overproduced in the atg mutants, at least in young rosettes (Figure 11B). However, the differential expression of ethylene-related genes was only observed at 30 DAS but not at 60 and 75 DAS. Accordingly, many WRKY transcription factors known to be involved in stress responses and response to pathogen attack were also upregulated in atg5 and atg9 relative to the wild type (Supplemental Table 11). Some of these, but not all, are also related to developmental leaf senescence (Breeze et al., 2011). In addition to WRKY, several NAC transcription factors were also upregulated in atg5 and atg9, among which some are SA or senescence related (Supplemental Table 12). However, the ANAC055 gene described recently as a master negative regulator of SA biosynthesis (Hickman et al., 2013) was highly upregulated in atg5 and atg9, compared with ANAC019, which is a positive regulator of SA biosynthesis. This suggests that the defects in the regulation of the SA pathway are downstream of the ANAC055 regulation node.
Figure 11.
SA Signaling Pathway Is Upregulated in atg5 and atg9.
(A) The model of interaction of the key genes involved in SA (orange), JA (blue), and ethylene (yellow) was adapted from Ascencio-Ibáñez et al. (2008). Genes up- or downregulated in atg5 and atg9 are in orange or blue typefaces, respectively.
(B) Genes involved in SA, JA, or ethylene regulation and response are listed. Their relative expressions in atg5 compared with the wild type are reported as log2(fold change) values. Significant up- or downregulations (FDR% <10−5) are indicated by yellow- or blue-colored cells, respectively.
Comparison of Transcriptome Profiles
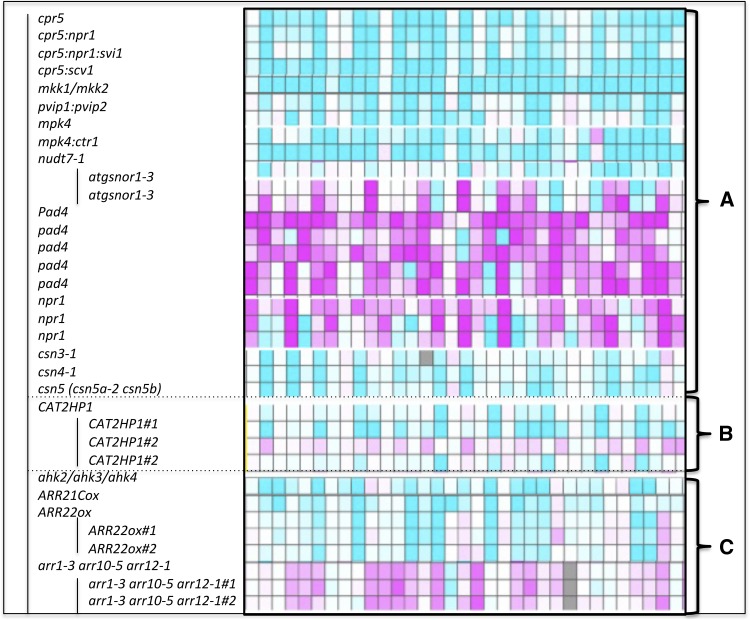
The transcriptome profiles of the 100 genes showing the highest overexpression levels in the atg5 and atg9 mutants relative to the wild type, under low nitrate at 60 DAS, were compared using the Genevestigator analysis tools (https://www.genevestigator.com/gv/). Figure 12 summarizes the most significant profile similarities found in other mutants. It was not a surprise to find that mutants affected in plant immunity and systemic acquired resistance, such as cpr5, mkk1/mkk2, nudt7-1, pvip1 pvip2, and mpk4, showed similar patterns to the atg mutants. In contrast, we observed that the top genes upregulated in the atg mutants were downregulated in the pad4 and npr1 mutants and affected in plant immunity and SA signaling. Similarities with the CAT2HP lines (mutated in the catalase 2 gene) confirmed that atg mutants are defective at detoxifying ROS, as described by Yoshimoto et al. (2009).
Figure 12.
Transcriptome Profile Comparisons.
Genes upregulated in atg5 and atg9 shows similarities with mutants affected in plant immunity (A), oxidative stress (B), and cytokinin signaling (C). Using the 100 genes most upregulated in atg5 and atg9, transcriptome comparison was performed using the Genevestigator Perturbation tool that displays the response of genes to a wide variety of genotypes. Each vertical line displays the expression data for one gene. Each horizontal line displays gene relative expressions in the genetic background indicated. Blue and pink indicate up- and downregulation, respectively. Intensity of the colors is proportional to the absolute values of the fold changes. Only representative similarities/differences are shown, and only few gene patterns are shown (pattern was similar for the 100 genes).
The atg expression profiles were also similar to those of the ARR2 overexpressor and qhk2 ahk3 ahk4 triple mutants. In contrast, genes that were upregulated in the atg mutants were downregulated in the arr1 arr10 arr12 triple mutants, suggesting defects in the perception and response of atg mutants to cytokinins.
DISCUSSION
Why Do Autophagy Mutants Accumulate Amino Acids?
We previously reported that atg mutants accumulate many forms of nitrogen in their rosette leaves (soluble proteins, peptides, ammonium, and amino acids, but not nitrate) and have depleted carbon in the form of hexose, sucrose, and starch (Guiboileau et al., 2013). This report confirmed that amino acids accumulate in atg rosettes, especially when grown under low nitrate conditions. The accumulation of glutamate was detected early, as soon as 30 DAS, when no yellowing phenotype can be observed. The accumulation of glutamate, which was confirmed at 60 and 75 DAS, was also found in the SA-deficient backgrounds, thus showing that glutamate accumulation in atg mutants was senescence independent. In addition to increased glutamate levels, the other most striking metabolomic differences found between atg mutants and the wild type, such as the accumulation of shikimate, aspartate, pipecolate, and methionine, were also found in the SA-deficient backgrounds.
An accumulation of glutamate and aspartate, while the glutamine, asparagine, γ-aminobutyric acid, and proline concentrations are maintained, is not an usual feature in plants. Glutamate is usually considered as a highly buffered metabolite (Scheible et al., 2000), while glutamine, proline, or γ-aminobutyric acid concentrations are more flexible. Glutamate and aspartate accumulation in the atg mutants suggests that asparagine synthetase and glutamine synthetase, which is known to be very efficient for ammonium assimilation (Masclaux-Daubresse et al., 2006), were the bottleneck of amino acid conversion in atg mutants. However, our transcriptomics data did not reveal downregulation of asparagine synthetase or glutamine synthetase (GS) transcript levels. No upregulation of any aspartate amino transferase or glutamate oxoglutarate amino transferase (GOGAT) transcripts was found either. In addition, previously, we did not find any difference between the wild type and atg mutant GS activities (Guiboileau et al., 2013). Therefore, the accumulation of glutamate and aspartate does not appear to be due to changes in GOGAT, GS, asparagine synthetase, or aspartate amino transferase expression levels or activities.
Amino acid biosynthesis occurs in both mesophyll and phloem cells. Thus, amino acids could accumulate in atg mutants due to some defects in phloem uploading or cell-to-cell amino acid translocation. In atg mutants, GS1 activity in the veins was previously found to be higher than that of GS2 (see Guiboileau et al., 2013). The GOGAT isoenzymes, especially the ferredoxin-GOGAT, are preferentially located in the chloroplasts of mesophyll cells; the translocation of glutamate or aspartate from the mesophyll to the veins could be a bottleneck in amino acid export. However, at present, this hypothesis is not supported by any evidence and no genes encoding amino acid or peptide transporters were downregulated.
The increase in shikimate, glutamate, methionine, pipecolate, and glutathione levels in the atg mutants strongly suggests that the major metabolic changes could be linked to oxidative stress management. Glutamate, methionine, and cysteine are essential for glutathione GSH and GSSG synthesis. The upregulation of several glutathione S-transferase genes suggests that the conversion of glutathione GSH to R-S-glutathione and further to glutamate is very active in the atg mutants. In addition, shikimate, which is strongly dependent on amino acid metabolism for its biosynthesis, is the entry to the phenylpropanoid pathway that synthesizes many antioxidative compounds. Altogether, the metabolome data suggest that in atg mutants, amino acid interconversion metabolism is focused on the regulation of oxidative stress through the management of methionine, glutamate, glutathione, and shikimate pathways. This would disturb the global nitrogen fluxes in the amino acid metabolic pathways and could explain the changes to individual amino acid concentrations in the atg mutants.
Why Do Autophagy Mutants Accumulate Less Anthocyanin?
The finding that the rosettes of autophagy mutants and RNAi lines do not turn red and contain less anthocyanin and quercetin than the wild type is consistent with Pourcel et al. (2010). However, while their reports concluded that autophagy machinery participates in the formation of AVIs and is important for the trafficking of anthocyanin to the vacuole, our data show that atg mutants do not contain as much anthocyanin and quercetin as the wild type due to the downregulation of the flavonoid biosynthetic pathway. The presence of AVI-like structures in the roots of atg5 pap1-D double mutants shows that anthocyanin trafficking to the vacuole is as efficient in atg5 mutant as in the wild type. If autophagy machinery also plays a role in the trafficking of anthocyanin to the vacuole, our results show that this role is not as essential as Pourcel et al. (2010) suggested. From the atg5 and atg9 transcriptomes, it was clear that the downregulation of the flavonoid pathway was due to lower expression of PAP1, PAP2, and TT8. However, it is not clear why these regulatory genes show lower expression in the atg mutants relative to the wild type. Despite this, the overexpression of the UV-inducible gene MC8 in atg5 and atg9 suggests that the atg mutants are more sensitive to UV, and this could be linked to their lower flavonoid content (He et al., 2008; Nawkar et al. 2013).
Why Do Autophagy Mutants Senesce Earlier?
Flavonoids are antioxidants that can inhibit ROS generation. Their decreased levels in atg mutant/RNAi lines could explain the higher ROS concentrations previously reported in these plants (Yoshimoto et al., 2009; Agati et al., 2012) and the higher ascorbate/dehydroascorbate contents found in the current metabolome data. Interestingly, we found that the atg5 pap1-D double mutants have bigger rosettes than atg5 mutants under all growth conditions. However, this positive effect of anthocyanin production in atg mutants was not sufficient to delay their early leaf senescence symptoms (Figures 3 and 8). Agati et al. (2012) suggested that quercetins are especially important for protecting chloroplasts from the light-induced generation of singlet oxygen. Thus, decreased quercetin could affect chloroplast protection in atg mutants.
All these defects promoting oxidative stress could be triggering factors for SA biosynthesis; SA production is linked with the appearance of leaf yellowing in atg mutants, as shown by Yoshimoto et al. (2009). Our transcriptome and metabolome data confirmed the upregulation of SA biosynthesis and SA accumulation in atg mutants, especially under low nitrate conditions. SA induction could involve signals related to hypersensitivity to stress and ROS management. In addition, as SA is synthesized within the chloroplast during ageing, it is possible that its synthesis is accelerated and higher in the atg mutants compared with the wild type due to some defects in chloroplast quality control and oxidative stress management.
Why Do Autophagy Mutants Overexpress Defense Genes?
Autophagy mutants display stress-related phenotypes, such as the accumulation of raffinose and pipecolate (Nishizawa et al., 2008; Návarová et al., 2012), and the top list of genes upregulated in atg5 exhibited clear similarities with upregulated genes found in cpr5, mkk1/mkk2, and mpk4 transcriptomes. As in these mutants, we observed cell death spots on the leaves of atg mutants (Supplemental Figure 6; Phillips et al., 2008) and defense genes were also highly upregulated in atg5 (Jing et al., 2008; Jing and Dijkwel, 2008; Kong et al., 2012). The common feature of atg mutants and cpr5, mkk1/mkk2, and mpk4 mutants is the increase in SA and the overexpression of plant defense and SA-related genes. This also explains why the atg expression pattern is the opposite of that found in pad4 and npr1 mutants. Oxidative stress is an appropriate background for hyper-reactive responses to stress and SA accumulation. The similarity between the upregulated genes in atg5 and cat2 mutants is also consistent with the hypothesis that atg mutants are suffering from oxidative stress (Mhamdi et al., 2012).
There is not much known about the relationship between autophagy and the proteasome in plants. Interestingly, comparison of the upregulated genes in the atg mutants also showed similarities with genes upregulated in the COP9 (csn3, csn4, and csn5) mutants. Besides the fact that immunity and SA responses are affected in csn3, csn4, and csn5 mutants, the COP9/signalosome (CSN) complex modulates protein degradation in eukaryotes by regulating the ubiquitin-proteasome system. It was shown in mice that the COP9 signalosome regulates autophagosome maturation (Su et al., 2011). The authors observed the accumulation of immature autophagosomes in csn8 mouse cardiomyocytes. In plants, COP9 controls cell cycle progression, plant development, and JA- and SA-mediated defense against biotic stress (Schwechheimer and Isono, 2010; Stratmann and Gusmaroli, 2012). The CSN subunits 4, 5, and 8 interact physically with NPR1, and the loss of CSN8 function (cop9 mutants) results in the stabilization of NPR1 and then in hypersensitivity to SA (Spoel et al., 2009). As a result, PR genes are strongly upregulated in the COP9 Arabidopsis mutants even though the mutants do not significantly accumulate SA. Although there is no evidence so far that COP9 is involved in the maturation of autophagosome in plants, the similarity between csn and atg mutants at the transcriptional level and in their response to SA is interesting.
Are Other Plant Hormone Pathways Affected in Autophagy Mutants?
Overexpression of ATG8 (GFP-ATG8f-HA) resulted in several cytokinin-dependent phenotypes (Slavikova et al., 2008). The GFP-ATG8f-HA plants showed defects in root growth and anthocyanin biosynthesis when zeatin was applied to roots. Upon zeatin treatment, the GFP signal relocalized from autophagosome structures to novel and bigger structures located at the vicinity of the vascular system. Although a link between ATG8 overexpression and autophagy activity has not been established, Slavikova et al. (2008) suggested a link between autophagy and cytokinin signaling. Our transcriptome analysis found that genes upregulated in atg5 are also upregulated in the ahk2 ahk3 ahk4 triple mutant and downregulated in the arr1 arr10 arr12 triple mutant treated with 6-benzyl aminopurine. In addition, the MYB DOMAIN PROTEIN2 (MYB2) regulator, which is significantly upregulated in both atg5 and atg9 under low and high nitrate conditions regardless of the plant age, has been reported to regulate whole-plant senescence by inhibiting cytokinin-mediated branching (Guo and Gan, 2011). Thus, overall, it is likely that autophagy plays a role in cytokinin signal perception.
Conclusion
Changes in primary and secondary metabolites confirmed the increase in amino acid concentrations and the decrease in hexose concentrations previously described for atg mutants by Guiboileau et al. (2013). These metabolite defects are consistent with defects in nitrogen remobilization (Guiboileau et al., 2012) and hypersensitivity to nutrient deficiency. Among the amino acid changes, we found that glutamate, methionine, shikimate, and pipecolate are invariably and significantly increased in atg mutants. The only explanation for glutamate accumulation emerging from this work is related to the increased level of transcripts encoding enzymes involved in the glutathione-to-glutamate interconversion pathway. Altogether, the results suggest a strong link between amino acid metabolism and oxidative stress management in atg mutants, which is consistent with their global phenotype as well as with the increase in stress markers, such as raffinose, galacturonate, phytol, pipecolate, and stigmasterols (Griebel and Zeier, 2010; Návarová et al., 2012; Wang et al., 2012; Watanabe et al., 2013). Our transcriptomic data consistently showed that many genes related to plant defense against biotic and abiotic stresses are overexpressed in atg mutants relative to the wild type.
It was thus surprising to observe that although the atg mutants accumulate several compounds related to stress, they do not actually accumulate anthocyanins that are widely considered as antioxidant molecules involved in the response of plants to biotic and abiotic stress. As we showed that the lower anthocyanin levels in the mutants are due to downregulation of most of the genes involved in the anthocyanin pathway, atg mutants provide a tool for further in-depth investigation of the signals involved in anthocyanin biosynthesis and the modulation of the master genes known to regulate this pathway.
Finally, our transcriptome data also revealed links between autophagy and cytokinin perception or the signalosome that has been previously suggested to exist in plants and animals but remains to be fully explored. Future work will address the role of these underlying mechanisms in determining the various phenotypes of autophagy mutants.
METHODS
Plant Growth
Arabidopsis thaliana Col, atg5-3; [SALK_020601], atg5-1 SAIL line [SAIL_129B07], and atg9 [SALK_130796] were characterized in previous reports (Hanaoka et al., 2002; Inoue et al., 2006; Yoshimoto et al., 2009). The ATG18a RNAi transgenic plants (RNAi18) were kindly provided by Diane C. Bassham (Iowa State University) (Xiong et al., 2005). Plants were cultivated under short days (8 h light) in sand with low nitrogen (2 mM nitrate) or high nitrogen nutrition (10 mM nitrate) as described by Lemaître et al. (2008). Three different bulk samples of rosettes were harvested between 10 and 11 am 30, 60, and 75 DAS. Three independent and consecutive plantings (biological repeats) were performed under the same conditions.
To decipher the role of autophagy in anthocyanin biosynthesis, we crossed the atg5-1 SAIL line [SAIL_129B07] with the pap1-D overexpressing line (Borevitz et al., 2000). We preferred the atg5-1 SAIL line for crossing because previous experiments performed by Yoshimoto et al. (2009) and our own experiments showed that the atg5-3; [SALK_020601] SALK line induces silencing of 35S constructs, which is not the case in the atg5-1 SAIL line [SAIL_129B07]. Double homozygous atg5 pap1-D lines were then selected. Homozygosity for the atg5 mutation was verified using PCR primers (1) 5′-CATAAAGACGCAAGAGAAGATGACGTC-3′ and 5′-GAGGTCCATAGATCCTCTTG-3′ to confirm the absence of the wild -type allele and (2) 5′-CATAAAGACGCAAGAGAAGATGACGTC-3′ and LBb3 (5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′) to test the presence of the T-DNA insertion in ATG5. Homozygosity of the pap1-D allele was verified using primers 5′-TGAGACTTTTCAACAAAGGG-3′ and 5′-AcggAATATCTggACAgCCggA-3′ to confirm the presence of the pap1-D allele and 5′-AcggAATATCTggACAgCCggA-3′ and 5′-TgCAgAgacTACTCCCTCgTgC-3′ to test the absence of the wild-type region at the PAP1 locus.
Metabolite Profiling Using GC-MS
Extraction, derivatization, analysis, and data processing were performed according to Fiehn (2006). Metabolites were analyzed by GC-MS 3 h after derivatization. One microliter of the derivatized samples was injected in splitless mode on an Agilent 7890A gas chromatograph coupled to an Agilent 5975C mass spectrometer. The column was an Rtx-5SilMS from Restek (30 m with 10-m Integra-Guard column). The liner (Restek 20994) was changed before each series of analyses, and 10 cm of column was removed. The oven temperature ramp was 70°C for 7 min then 10°C/min to 325°C for 4 min (run length 36.5 min). The helium constant flow was 1.5231 mL/min. Temperatures were as follows: injector, 250°C; transfer line, 290°C; source: 250°C; and quadripole, 150°C. Samples and blanks were randomized. Amino acid standards were injected at the beginning and end of the analysis to monitor the derivatization stability. An alkane mix (C10, C12, C15, C19, C22, C28, C32, and C36) was injected in the middle of the queue for external calibration. Five scans per second were acquired.
Metabolites were annotated, and their levels on a fresh weight basis were normalized with respect to the ribitol internal standard. The three independent plantings (each planting containing three plant repeats) performed showed similar results as shown by ANOVA.
Since only small sized molecules are determined with GC-MS, we used LC-MS and enzymatic assays to measure flavonoid and glutathione concentrations, respectively.
Glutathione Enzyme Assay
Leaf material (150 mg fresh weight) was extracted in 1 mL of HClO4 (1 M) containing insoluble PVP. After centrifugation (15 min, 14,000g, 2°C), 0.5 mL of supernatant was collected and kept on ice and was added to 0.1 mL of NaH2PO4 (0.12 M, pH 5.6) and buffered to pH 6.0 with 0.09 mL of cold K2CO3 (2.5 M). After centrifugation (15 min, 14,000g, 2°C) to obtain a clear supernatant, glutathione was measured according to Noctor and Foyer (1998) with the exception that volumes were modified and adapted to microtitration plates. To measure GSSG, an earlier treatment of the sample was necessary: 0.2 mL of sample was pretreated by adding 5 μL of vinylpyrimidine and incubating it for 20 min at room temperature. The treated sample was then centrifuged twice (15 min, 14,000g, 2°C), and the supernatant was used to measure total glutathione (GSH + GSSG). Measurements were performed using 5 μL of extracts, 242 μL of the reaction buffer, pH 7.5 [0.12 M NaH2PO4, 0.74 mM EDTA, 5 mM 5,5′-dithiobis(2-nitrobenzoic acid), and 0.4 mM NADPH], and 0.34 units of glutathione reductase in each 96-well microplate. Absorbance at 405 nm was measured during a 20-min kinetic. Standards were prepared as millimolar dilutions of GSH and GSSG in (1 M HClO4/2.5 M K2CO3/0.12 M NaH2PO4, pH 5.6; 1/0.25/0.2, v/v/v).
Flavonoid Analysis Using LC-MS
Samples were ground for 15 s at maximum speed with a FastPrep-24 homogenizer (MP Biomedicals), in 1 mL of methanol/acetone/water/trifluoroacetic acid (30/42/28/0.05, v/v/v/v) solvent mix and sonicated for 15 min at 25 Hz (Elma sonicator). A 4-μg aliquot of apigenin was added as an internal standard. Following centrifugation at 18,000g for 9 min, the pellet was further extracted with 0.8 mL of the same solvent mix for 4 h at 4°C and centrifuged again. The two extracts were pooled and evaporated under vacuum overnight using a Speedvac concentrator (Thermo Scientific Savant SPD121 P). The dry residue was finally dissolved in 300 μL solvent mix, sonicated for 10 min to complete dissolution, and briefly centrifuged for 5 min to remove any insoluble materials. LC-MS analyses of individual flavonoids was performed as previously described (Routaboul et al., 2006, 2012) using a Quattro LC with an ESI Z-Spray interface (MicroMass), an Alliance 2695 RP-HPLC system, and a Waters 2487 UV detector set at 280 nm. Anthocyanins and flavonols were identified based on mass fragmentation in comparison with reported data (Tohge et al., 2005; Nakabayashi et al., 2009). Flavonol contents were expressed relative to quercetin-3-O-rhamnoside, rutin, and epicatechin (Extrasynthese) external standards, or monoglycosylated and diglycosylated flavonols and flavan-3-ols (PAs), respectively. Anthocyanins were expressed relative to cyanidin (Extrasynthese).
Metabolomic Data Processing
Raw Agilent data files were converted in NetCDF format and analyzed with AMDIS (http://chemdata.nist.gov/mass-spc/amdis/). A home retention index/mass spectra library built from the NIST, Golm, and Fiehn databases and standard compounds was used for metabolite identification. Peak areas were then determined using the Quanlynx software (Waters) after conversion of the NetCDF file to masslynx format. TMEV (http://www.tm4.org/mev.html) was used for all statistical analysis. Univariate analysis by permutation (one-way and two-way ANOVA) was first used to select the significant metabolites. Multivariate analysis (hierarchical clustering and principal component analysis) was then performed on this subset. Only metabolites showing repeatable and significant differences (based on t tests) in the mutant/RNAi relative to the wild type in the three replicate plantings are reported.
Microarrays and Statistical Analysis
Total RNA was extracted using the procedure described by Lemaître et al. (2008). Plant material was produced in two independent plantings made at an interval of several months, thus forming two biological replicates R1 and R2. For R1 and R2, total RNA was extracted from three independent plant material bulks representing three biological repeats. For each genotype, RNA samples used for cDNA production and microarray experiment were a mix of the three plants. Then, two independent hybridizations were performed for R1 and R2. Microarray analysis was performed at the RIKEN Plant Science Center using the Affymetrix ATH1 microarrays according to the manufacturer’s instructions. Raw data obtained as CEL files were then processed. The analytical techniques used to normalize data and determine significance levels and fold changes between genotypes was based on the method of Rank products and FDR estimation as described by Breitling et al. (2004). Data sets from R1 and R2 were crossed in order to calculate rank products, FDR%, and fold change ratios of (1) atg5 versus Col wild type (using the comparisons of atg5R1vsColR1, atg5R2vsColR1, atg5R1vsColR2, and atg5R2vsColR2), (2) atg9 versus Col wild type (using the comparisons of atg9R1vsColR1, atg9R2vsColR1, atg9R1vsColR2, and atg9R2vsColR2), and (3) both atg5 and atg9 versus Col wild type (using the comparisons of atg5R1vsColR1, atg5R2vsColR1, atg5R1vsColR2, atg5R2vsCol R2, atg9R1vsColR1, atg9R2vsColR1, atg9R1vsColR2, and atg9R2vsColR2, thus explaining that, in that case, the range of FDR% is much larger). Top lists of significantly up- or downregulated genes in each condition, genotype, and harvest time were computed and cutoffs were based on FDR% ≤10−6 for individual mutant analysis (atg5 versus Col or atg9 versus Col) and on FDR% ≤10−10 for the combination of the two mutants (atg5 and atg9 versus Col).
From these lists, intersections and GOs were analyzed using VirtualPlant1.3 (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/) and AGRIGO (http://bioinfo.cau.edu.cn/agriGO/index.php; Du et al., 2010) bioinformatics tools.
Quantitative RT-PCR
Total RNA (1 μg), extracted as described above on three plant replicates, was used as a template to perform RT reactions using Moloney murine leukemia virus reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Quantitative RT-PCR reactions were performed using 2× Mesa Fast qPCR MasterMix Plus for SYBR assay (Eurogentec) following the manufacturer’s protocols. TRANSLATION ELONGATION FACTOR1A, APT, ACTIN1, and UBIQUITIN1 were used as reference genes. Real-time RT-PCR conditions and primer sets used in this study for TT8, CHS, and DFR analysis are described by Baudry et al. (2004, 2006). QuantiTect Primers were purchased from Qiagen to analyze F3′H (QT00825181) and LDOX (QT00813176) transcript accumulation. PAP1 transcripts were quantified using the forward 5′-TGTAAGAGCTGGGCTAAACC-3′ and reverse 5′-GAAGATCGACTTCATCAGAGC-3′ primers.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: AT5G20250, AT3G57520, AT1G20350, AT1G01340, AT2G22470, AT1G35230, AT1G33960, AT1G79450, AT1G68620, AT1G02220, AT2G17040, AT3G44350, AT5G22380, AT5G20230, AT2G41100, AT3G54420, AT1G02400, AT4G25110, AT3G13100, AT3G52430, AT3G22910, AT1G11190, AT5G56870, AT3G60140, AT3G01830, AT5G47850, AT5G50260, AT2G43570, AT5G42380, AT5G52760, AT3G26830, AT3G49620, AT1G07000, AT1G30700, AT1G26420, AT4G14630, AT4G39670, AT3G50480, AT2G34600, AT1G16420, AT1G62490, AT1G04600, AT3G23250, AT2G47190, AT1G09500, AT2G35710, AT5G64790, AT3G28580, AT5G13320, AT2G26560, AT2G18660, AT2G14610, AT1G48210, AT1G72540, AT5G25440, AT4G37900, AT3G48650, AT4G02380, AT3G51680, AT3G01290, AT3G62950, AT1G57630, AT2G32140, AT4G04300, AT2G13310, AT3G53150, AT2G43820, AT3G11340, AT1G76230, AT1G13340, AT3G13950, AT3G48640, AT4G28460, AT2G04025, AT1G20180, AT3G12510, AT4G32870, AT5G43580, AT1G78410, AT2G22880, AT1G21240, AT4G31800, AT5G22570, AT5G57550, AT1G59590, AT5G04390, AT4G14365, AT1G08050, AT1G56650, and AT5G42800.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Hierarchic Clustering of Metabolite Changes between atg Mutants and Controls Show Salicylic Dependence of Metabolite Classes.
Supplemental Figure 2. Genes Involved in Galacturonate Biosynthesis Are Upregulated in atg5.
Supplemental Figure 3. Phytosterol Pathway Is Upregulated in atg5.
Supplemental Figure 4. PCR Genotyping of atg5.pap1-D Double Mutant.
Supplemental Figure 5. Overexpression of Isochorismate Synthase 1 and PBS3 Genes in atg5 Relative to the Wild Type Explain SA Accumulation in Autophagy Mutants.
Supplemental Figure 6. Cell Death Area on Leaves of Autophagy Mutants.
Supplemental Table 1. Flavonoid Compounds Identified and Quantified Using LC-MS.
Supplemental Table 2. Metabolites Significantly More or Less Concentrated in atg5, atg5 sid2, atg5 NahG Compared with Col, sid2, or NahG Controls Are Listed.
Supplemental Table 3. List of the 86 Genes Significantly Upregulated in atg5 and atg9 Relative to Wild Type under Low Nitrate Conditions for 30 and 60 d.
Supplemental Table 4. Frequency of Gene Ontology Terms (GO) Represented in the 86 Genes Upregulated in Both atg5 and atg9 Independently of Nitrate Conditions or Plant Age.
Supplemental Table 5. Gene Ontology Terms (GO) Represented in the List of Genes Significantly Upregulated in the atg Mutants in Different Conditions.
Supplemental Table 6. Gene Ontology Terms (GO) Represented in the List of Genes Significantly Downregulated in the atg Mutants in Different Conditions.
Supplemental Table 7. Differential Expression in atg5 and atg9 Relative to Wild-Type Genes Involved in the Nitrogen Assimilation Pathway and in Aspartate and Glutamate Metabolism.
Supplemental Table 8. Differential Expression in atg5 Relative to Wild-Type Genes Involved in the Shikimate and Flavonoid Pathways and Reported by Lillo et al. (2008).
Supplemental Table 9. Differential Expression in atg5 and atg9 Relative to the Wild Type of Senescence-Associated Genes (SAG).
Supplemental Table 10. Differential Expression (in atg5 and atg9 Relative to Wild Type) of Genes Responding to Salicylic Acid (SA) and Jasmonic Acid (JA).
Supplemental Table 11. Differential Expression of WRKY Genes in Rosettes of atg5 and atg9 Relative to Wild Type.
Supplemental Table 12. Differential Expression of NAC Genes in atg5 and atg9 Relative to Wild Type.
Supplemental Data Set 1. Area Raw Data for the Metabolome Analysis of the Rosettes of the Col Wild Type and atg5, atg9, and RNAi18 Mutants Grown under Low (2 mM) or High (10 mM) Nitrate and Harvested at 30 DAS, 60 DAS, and 75 DAS.
Supplemental Data Set 2. The Relative Metabolite Concentrations in the Rosettes of atg5, atg9, and RNAi18 Relative to Wild Type Are Presented as Fold Change Values.
Supplemental Data Set 3. Transcriptome of atg5 versus Wild Type, 30 DAS under Low Nitrate Conditions.
Supplemental Data Set 4. Transcriptome of atg9 versus Wild Type, 30 DAS under Low Nitrate Conditions.
Supplemental Data Set 5. Transcriptome of atg5 and/or atg9 versus Wild Type, 30 DAS under Low Nitrate Conditions.
Supplemental Data Set 6. Transcriptome of atg5 versus Wild Type, 60 DAS under Low Nitrate Conditions.
Supplemental Data Set 7. Transcriptome of atg9 versus Wild Type, 60 DAS under Low Nitrate Conditions.
Supplemental Data Set 8. Transcriptome of atg5 and/or atg9 versus Wild Type, 60 DAS under Low Nitrate Conditions.
Supplemental Data Set 9. Transcriptome of atg5 versus Wild Type, 60 DAS under High Nitrate Conditions.
Supplemental Data Set 10. Lists of the Genes Significantly Upregulated in atg5, atg9, and Both atg5 and atg9.
Supplemental Data Set 11. Lists of the Genes Significantly Downregulated in atg5, atg9, and Both atg5 and atg9.
Supplementary Material
Acknowledgments
We thank Joël Talbotec and Philippe Maréchal for taking care of plants, Isabelle Debeaujon (Institut Jean-Pierre Bourgin, INRA, Versailles, France) for helpful discussions, and Patrick Armengaud for teaching FDR and rank product computing from microarray raw data. We also thank Leigh Gebbie for improving English writing and Michèle Reisdorf-Cren for proofreading. This work was supported by a PhD grant from Ministère de l’Education et de la Recherche of France to A.G. and by a Grant-in-Aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (Grants 18770040, 19039033, and 2020061) to K.Y. Collaboration between Institut Jean-Pierre Bourgin and RIKEN was facilitated by the SAKURA program 21124QA for bilateral collaborations supported by the French Ministère des Affaires Etrangères et Européennes and the Japan Society for the Promotion of Science.
AUTHOR CONTRIBUTIONS
P.A., G.C., F.S., J.-M.R., A.G., and C.M.-D. performed the experiments. P.A., G.C., J.-M.R., A.G., and C.M.-D. analyzed and evaluated the data. P.A., G.C., J.-M.R., and C.M.-D. interpreted the data. A.G., K.S., K.Y., and C.M.-D. conceived the study. C.M.-D. coordinated the research and wrote the article. All authors read and approved the final article.
Glossary
- SA
salicylic acid
- Col
Columbia
- DAS
days after sowing
- RNAi
RNA interference
- ROS
reactive oxygen species
- GC-MS
gas chromatography–mass spectrometry
- LC-MS
liquid chromatography–mass spectrometry
- FDR
false discovery rate
- GO
Gene Ontology
- AVI
anthocyanic vacuolar inclusion
- JA
jasmonic acid
- GS
glutamine synthetase
- GOGAT
glutamate oxoglutarate amino transferase
References
- Agati G., Azzarello E., Pollastri S., Tattini M. (2012). Flavonoids as antioxidants in plants: location and functional significance. Plant Sci. 196: 67–76 [DOI] [PubMed] [Google Scholar]
- Araújo W.L., Tohge T., Ishizaki K., Leaver C.J., Fernie A.R. (2011). Protein degradation - an alternative respiratory substrate for stressed plants. Trends Plant Sci. 16: 489–498 [DOI] [PubMed] [Google Scholar]
- Ascencio-Ibáñez J.T., Sozzani R., Lee T.J., Chu T.M., Wolfinger R.D., Cella R., Hanley-Bowdoin L. (2008). Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection. Plant Physiol. 148: 436–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry A., Caboche M., Lepiniec L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46: 768–779 [DOI] [PubMed] [Google Scholar]
- Baudry A., Heim M.A., Dubreucq B., Caboche M., Weisshaar B., Lepiniec L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39: 366–380 [DOI] [PubMed] [Google Scholar]
- Borevitz J.O., Xia Y.J., Blount J., Dixon R.A., Lamb C. (2000). Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeze E., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004). Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engqvist M.K.M., Kuhn A., Wienstroer J., Weber K., Jansen E.E.W., Jakobs C., Weber A.P.M., Maurino V.G. (2011). Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J. Biol. Chem. 286: 11382–11390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. (2006). Metabolite profiling in Arabidopsis. Methods Mol. Biol. 323: 439–447 [DOI] [PubMed] [Google Scholar]
- Floyd B.E., Morriss S.C., Macintosh G.C., Bassham D.C. (2012). What to eat: evidence for selective autophagy in plants. J. Integr. Plant Biol. 54: 907–920 [DOI] [PubMed] [Google Scholar]
- Goyer A., Johnson T.L., Olsen L.J., Collakova E., Shachar-Hill Y., Rhodes D., Hanson A.D. (2004). Characterization and metabolic function of a peroxisomal sarcosine and pipecolate oxidase from Arabidopsis. J. Biol. Chem. 279: 16947–16953 [DOI] [PubMed] [Google Scholar]
- Griebel T., Zeier J. (2010). A role for beta-sitosterol to stigmasterol conversion in plant-pathogen interactions. Plant J. 63: 254–268 [DOI] [PubMed] [Google Scholar]
- Guiboileau A., Avila-Ospina L., Yoshimoto K., Soulay F., Azzopardi M., Marmagne A., Lothier J., Masclaux-Daubresse C. (2013). Physiological and metabolic consequences of autophagy deficiency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 199: 683–694 [DOI] [PubMed] [Google Scholar]
- Guiboileau A., Yoshimoto K., Soulay F., Bataillé M.-P., Avice J.-C., Masclaux-Daubresse C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194: 732–740 [DOI] [PubMed] [Google Scholar]
- Guo Y., Gan S. (2011). AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 156: 1612–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H., Noda T., Shirano Y., Kato T., Hayashi H., Shibata D., Tabata S., Ohsumi Y. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A.P., Dinesh-Kumar S.P. (2011). What can plant autophagy do for an innate immune response? Annu. Rev. Phytopathol. 49: 557–576 [DOI] [PubMed] [Google Scholar]
- Hayward A.P., Tsao J., Dinesh-Kumar S.P. (2009). Autophagy and plant innate immunity: Defense through degradation. Semin. Cell Dev. Biol. 20: 1041–1047 [DOI] [PubMed] [Google Scholar]
- He R., Drury G.E., Rotari V.I., Gordon A., Willer M., Farzaneh T., Woltering E.J., Gallois P. (2008). Metacaspase-8 modulates programmed cell death induced by ultraviolet light and H2O2 in Arabidopsis. J. Biol. Chem. 283: 774–783 [DOI] [PubMed] [Google Scholar]
- Hickman R., et al. (2013). A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 75: 26–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofius D., Schultz-Larsen T., Joensen J., Tsitsigiannis D.I., Petersen N.H.T., Mattsson O., Jørgensen L.B., Jones J.D.G., Mundy J., Petersen M. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137: 773–783 [DOI] [PubMed] [Google Scholar]
- Inoue Y., Suzuki T., Hattori M., Yoshimoto K., Ohsumi Y., Moriyasu Y. (2006). AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol. 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Jing H.-C., Dijkwel P.P. (2008). CPR5: A Jack of all trades in plants. Plant Signal. Behav. 3: 562–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H.C., Hebeler R., Oeljeklaus S., Sitek B., Stühler K., Meyer H.E., Sturre M.J.G., Hille J., Warscheid B., Dijkwel P.P. (2008). Early leaf senescence is associated with an altered cellular redox balance in Arabidopsis cpr5/old1 mutants. Plant Biol. (Stuttg.) 10 (suppl. 1): 85–98 [DOI] [PubMed] [Google Scholar]
- Kong Q., Qu N., Gao M., Zhang Z., Ding X., Yang F., Li Y., Dong O.X., Chen S., Li X., Zhang Y. (2012). The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24: 2225–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Wang F., Zheng Z., Fan B., Chen Z. (2011). A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. 66: 953–968 [DOI] [PubMed] [Google Scholar]
- Lemaître T., Gaufichon L., Boutet-Mercey S., Christ A., Masclaux-Daubresse C. (2008). Enzymatic and metabolic diagnostic of nitrogen deficiency in Arabidopsis thaliana Wassileskija accession. Plant Cell Physiol. 49: 1056–1065 [DOI] [PubMed] [Google Scholar]
- Lenz H.D., et al. (2011). Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. 66: 818–830 [DOI] [PubMed] [Google Scholar]
- Lillo C., Lea U.S., Ruoff P. (2008). Nutrient depletion as a key factor for manipulating gene expression and product formation in different branches of the flavonoid pathway. Plant Cell Environ. 31: 587–601 [DOI] [PubMed] [Google Scholar]
- Liu Y., Bassham D.C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Liu Y., Xiong Y., Bassham D.C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963 [DOI] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Czymmek K., Tallóczy Z., Levine B., Dinesh-Kumar S.P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121: 567–577 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Reisdorf-Cren M., Orsel M. (2008). Leaf nitrogen remobilisation for plant development and grain filling. Plant Biol. (Stuttg.) 10 (suppl. 1): 23–36 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Reisdorf-Cren M., Pageau K., Lelandais M., Grandjean O., Kronenberger J., Valadier M.H., Feraud M., Jouglet T., Suzuki A. (2006). Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhamdi A., Noctor G., Baker A. (2012). Plant catalases: peroxisomal redox guardians. Arch. Biochem. Biophys. 525: 181–194 [DOI] [PubMed] [Google Scholar]
- Nakabayashi R., Kusano M., Kobayashi M., Tohge T., Yonekura-Sakakibara K., Kogure N., Yamazaki M., Kitajima M., Saito K., Takayama H. (2009). Metabolomics-oriented isolation and structure elucidation of 37 compounds including two anthocyanins from Arabidopsis thaliana. Phytochemistry 70: 1017–1029 [DOI] [PubMed] [Google Scholar]
- Návarová H., Bernsdorff F., Döring A.C., Zeier J. (2012). Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24: 5123–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawkar G.M., Maibam P., Park J.H., Sahi V.P., Lee S.Y., Kang C.H. (2013). UV-induced cell death in plants. Int. J. Mol. Sci. 14: 1608–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A., Yabuta Y., Shigeoka S. (2008). Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol. 147: 1251–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G., Foyer C.H. (1998). Simultaneous measurement of foliar glutathione, gamma-glutamylcysteine, and amino acids by high-performance liquid chromatography: comparison with two other assay methods for glutathione. Anal. Biochem. 264: 98–110 [DOI] [PubMed] [Google Scholar]
- Ono Y., Wada S., Izumi M., Makino A., Ishida H. (2013). Evidence for contribution of autophagy to rubisco degradation during leaf senescence in Arabidopsis thaliana. Plant Cell Environ. 36: 1147–1159 [DOI] [PubMed] [Google Scholar]
- Phillips A.R., Suttangkakul A., Vierstra R.D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178: 1339–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L., Irani N.G., Lu Y., Riedl K., Schwartz S., Grotewold E. (2010). The formation of Anthocyanic Vacuolar Inclusions in Arabidopsis thaliana and implications for the sequestration of anthocyanin pigments. Mol. Plant 3: 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poustka F., Irani N.G., Feller A., Lu Y., Pourcel L., Frame K., Grotewold E. (2007). A trafficking pathway for anthocyanins overlaps with the endoplasmic reticulum-to-vacuole protein-sorting route in Arabidopsis and contributes to the formation of vacuolar inclusions. Plant Physiol. 145: 1323–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul J.-M., Dubos C., Beck G., Marquis C., Bidzinski P., Loudet O., Lepiniec L. (2012). Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J. Exp. Bot. 63: 3749–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul J.M., Kerhoas L., Debeaujon I., Pourcel L., Caboche M., Einhorn J., Lepiniec L. (2006). Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224: 96–107 [DOI] [PubMed] [Google Scholar]
- Scheible W.R., Krapp A., Stitt M. (2000). Reciprocal diurnal changes of phosphoenolpyruvate carboxylase expression and cytosolic pyruvate kinase, citrate synthase and NADP-isocitrate dehydrogenase expression regulate organic acid metabolism during nitrate assimilation in tobacco leaves. Plant Cell Environ. 23: 1155–1167 [Google Scholar]
- Schwechheimer C., Isono E. (2010). The COP9 signalosome and its role in plant development. Eur. J. Cell Biol. 89: 157–162 [DOI] [PubMed] [Google Scholar]
- Slavikova S., Ufaz S., Avin-Wittenberg T., Levanony H., Galili G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59: 4029–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Mou Z., Tada Y., Spivey N.W., Genschik P., Dong X. (2009). Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137: 860–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratmann J.W., Gusmaroli G. (2012). Many jobs for one good cop - the COP9 signalosome guards development and defense. Plant Sci. 185-186: 50–64 [DOI] [PubMed] [Google Scholar]
- Su H., Li F., Ranek M.J., Wei N., Wang X. (2011). COP9 signalosome regulates autophagosome maturation. Circulation 124: 2117–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T., et al. (2005). Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Watanabe M., Balazadeh S., Tohge T., Erban A., Giavalisco P., Kopka J., Mueller-Roeber B., Fernie A., Hoefgen R. (2013). Comprehensive dissection of spatiotemporal metabolic shifts in primary, secondary, and lipid metabolism during developmental senescence in Arabidopsis. Plant Physiol. 162: 1290–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Senthil-Kumar M., Ryu C.-M., Kang L., Mysore K.S. (2012). Phytosterols play a key role in plant innate immunity against bacterial pathogens by regulating nutrient efflux into the apoplast. Plant Physiol. 158: 1789–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Contento A.L., Bassham D.C. (2005). AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J. 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Yoshimoto K., Jikumaru Y., Kamiya Y., Kusano M., Consonni C., Panstruga R., Ohsumi Y., Shirasu K. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K., Lukomska J., Moniuszko G., Gwozdecki R., Surowiecki P., Lewandowska M., Liszewska F., Wawrzyńska A., Sirko A. (2011). Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.