This work identifies genomic targets of the far-red light receptor phytochrome A, finding a broad set of light-regulated and other genes, including hormone- and stress-responsive transcription factors, providing insights into how plants rapidly fine-tune their growth strategy upon changes in their light environment.
Abstract
The far-red light (FR) photoreceptor phytochrome A (phyA) contains no DNA binding domain but associates with the CHALCONE SYNTHASE promoter through its chaperone FAR-RED ELONGATED HYPOCOTYL1 and transcription factors. Here, we performed a genome-wide identification of phyA targets using a combination of phyA chromatin immunoprecipitation and RNA sequencing methods in Arabidopsis thaliana. Our results indicate that phyA signaling widely affects gene promoters involved in multiple FR-modulated aspects of plant growth. Furthermore, we observed an enrichment of hormone- and stress-responsive elements in the phyA direct target promoters, indicating that a much broader than expected range of transcription factors is involved in the phyA signaling pathway. To verify our hypothesis that phyA regulates genes other than light-responsive ones through the interaction with corresponding transcription factors, we examined the action of phyA on one of its direct target genes, NAC019, which encodes an abscisic acid–dependent transcription factor. The phyA signaling cascade not only targets two G-boxes on the NAC019 promoter for subsequent transcriptional regulation but also positively coordinates with the abscisic acid signaling response for root elongation inhibition under FR. Our study provides new insight into how plants rapidly fine-tune their growth strategy upon changes in the light environment by escorting photoreceptors to the promoters of hormone- or stress-responsive genes for individualized modulation.
INTRODUCTION
When germinating seedlings emerge from subterranean darkness, the perception of the ambient light environment, under either direct sunlight (a high ratio of red light [R] to far-red light [FR]) or canopy (a low ratio of R to FR), is critical for their survival. A group of plant photoreceptors known as phytochromes detect the changing ratio of R to FR. Among the five Arabidopsis thaliana phytochromes, designated as phyA to phyE, phyA is responsible for sensing FR (Kami et al., 2010) as well as mediating early R responses (Tepperman et al., 2006). Conversely, phyB to phyE play a predominant role upon sustained R exposure. Under FR, phyA continuously translocates from the cytoplasm to the nucleus via its chaperones FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and FHY1-LIKE (FHL) (Hiltbrunner et al., 2005, 2006). Under R, phyA changes from the R-absorbing Pr form to the FR-absorbing Pfr form. The conformational change leads to both FHY1 phosphorylation and FHY1/FHL sequestration, which in turn prevents further phyA nuclear accumulation (Rausenberger et al., 2011; Chen et al., 2012).
Upon light-induced nuclear translocation, phyA controls the stability of light-responsive transcription factors, such as HY5 and PIF3 (Osterlund et al., 2000; Al-Sady et al., 2006). FHY1/FHL also facilitates the association between phyA and the transcription factors HFR1 and LAF1 (Yang et al., 2009). Furthermore, our previous study demonstrated that the phyA-FHY1 complex is recruited to the promoter of CHALCONE SYNTHASE (CHS; a gene whose expression is critical to anthocyanin biosynthesis) in tandem with the transcription factors HY5 or PIF3 to coregulate CHS transcription. Lastly, we have shown that a deficiency in phyA signaling abolishes the association of the phyA complex with the DNA and vice versa (Chen et al., 2012). However, whether the “phyA–promoter association” model is a universal mechanism for gene regulation throughout the entire genome of Arabidopsis, and whether FR influences multiple biological processes beyond anthocyanin accumulation, remain to be determined.
Interestingly, recent reports have identified crosstalk networks involving phyA and numerous internal and external stimuli, such as brassinosteroid, auxin, and various stresses resulting from water loss or wounds (Robson et al., 2010; Liu et al., 2011; Auge et al., 2012; Sandhu et al., 2012). Moreover, abscisic acid (ABA) signaling interferes with phyA-dependent seed germination (Lee et al., 2012). On the other hand, phyA synergistically coordinates with ABA to inhibit root elongation under white light (Jiang et al., 2010). Yet, the mechanism by which phyA influences the expression of ABA-responsive genes and collaborates with ABA responses under FR awaits further investigation.
A glimpse at the magnitude of the gene network regulated by phyA was first revealed a decade ago though transcriptome profiling using microarrays (Tepperman et al., 2001), a methodology recently reported to overlook important changes in transcript levels (Cloix and Jenkins, 2008). A subsequent study identified nine unique phyA-regulated motifs (SORLIP1 to SORLIP5 and SORLREP2 to SORLREP5) distinct from the light-responsive motif G-box by analyzing the promoters of 812 phyA-regulated genes (Hudson and Quail, 2003). In addition, genes directly targeted by the light-responsive transcription factors PIF1, PIF3, PIF4, PIF5, HY5, and FHY3 have recently been characterized (Oh et al., 2009, 2012; Ouyang et al., 2011; Zhang et al., 2011; 2013; Hornitschek et al., 2012). Among them, PIF1 and PIF3 are known to physically interact with phyA (Leivar and Quail, 2011). This study specifically aims to discern a comprehensive transcriptional network that integrates the phyA-regulated motifs and the entire set of transcription factors conceivably involved in phyA signaling. Accordingly, we first performed a genome-wide identification of phyA-associated genes through chromatin immunoprecipitation sequencing (ChIP-seq) and a newly developed data-processing method. We then performed RNA sequencing (RNA-seq) to provide more comprehensive transcriptomic information. The combination of ChIP-seq and RNA-seq data successfully identified genes that are both associated and regulated by phyA.
This work not only provides a technical advance in ChIP-seq by using a target protein that indirectly binds to DNA but also introduces the concept that the photoreceptor phyA acts on individual genes throughout the genome to directly mediate FR-modulated plant responses. To verify this hypothesis, the influence of phyA on the ABA-dependent transcription factor NAC019, one of the phyA direct target genes, is delineated. NAC019 acts as an ABA-dependent transcription factor, and its expression level is ABA inducible (Tran et al., 2004; Puranik et al., 2012). It is one of the 117 members of the NAC (for no apical meristem [NAM], Arabidopsis transcription activation factor [ATAF], cup-shaped cotyledon [CUC]) superfamily in Arabidopsis. This strategy allowed us to determine that an increase in phyA signaling further aggravated the ABA-mediated inhibition of root growth and vice versa. Together with the fact that enriched hormone- and stress-responsive elements were found on the promoters of phyA direct targets, our results clearly suggest an extensive role for phyA signaling in multiple biological processes. The mechanism of photoreceptors directly acting on numerous promoters appears to be a relatively rapid strategy for plants to better adapt to their changing environments without relying on the establishment of a complicated and delicate network between light and its crosstalking pathways.
RESULTS
Determination of phyA Association Sites by ChIP-seq
A chromatin immunoprecipitation (ChIP) PCR time-course experiment demonstrated that phyA did not significantly accumulate at the CHS promoter until FR irradiation reached 3 h or longer (Supplemental Figure 1). As the phyA association with promoters theoretically occurs prior to the observed change in gene transcript abundance, we selected the shortest FR treatment time (3 h) for ChIP-seq for an accurate identification of phyA association sites. PphyA:phyA-GFP phyA201 transgenic seedlings (Kim et al., 2000), which exhibited a similar phenotype to wild-type seedlings in our FR conditions (>10 µmol m−2 s−1) (Supplemental Figure 2), were grown in the dark for 4 d and irradiated with 3 h of FR for ChIP. For ChIP, we used an anti–green fluorescent protein (GFP) antibody, which detects phyA-GFP with high specificity upon FR irradiation (see Methods; Chen et al., 2012). To obtain reliable sequencing results, three biologically distinct phyA-associated DNA samples were subjected to library construction and high-throughput sequencing (Illumina). A high-value Pearson correlation coefficient (>0.97) (Barski et al., 2007) indicated excellent repeatability between biological replicates (Supplemental Figure 3). In addition, an input DNA sample (genomic DNA before antibody immunoprecipitation in ChIP) was sequenced in parallel to exclude false-positive signals caused by preferential PCR on certain genomic regions in the process of library construction (Park, 2009). A total of 205, 211, 204, and 213 million reads (75 bp per read) were obtained from three phyA libraries and the input library, respectively. Following adapter and low-quality reads removal, there were 71, 58, 52, and 90 million reads uniquely mapped to the Arabidopsis genome by the ultrafast, memory-efficient short read aligner Bowtie (http://bowtie.cbcb.umd.edu). The vast amount of reliable reads provided statistical significance to call true phyA-associated genes.
The indirect binding between phyA and DNA led to higher redundancy and weaker signals of phyA peaks when compared with conventional transcription factors. We optimized the parameters “tag number” and “mfold” by setting the well-characterized phyA direct target gene CHS as a positive control (see Methods). All putative phyA peaks with low false discovery rate (<5%) and high enrichment (>3-fold) were screened to separate closely located subpeaks. phyA peaks (also called phyA association sites hereafter) were defined as subpeaks found in all three biological replicates with a 50% minimum overlap.
The reliability of our approach was confirmed following the detection of two phyA peaks at the CHS promoter. These peaks were nearly identical in the three biological replicates, and they precisely covered two G-boxes involved in phyA association (Chen et al., 2012). By contrast, no phyA peak met our criteria in the transcribed region of CHS (Supplemental Figure 4).
Distribution and Gene Assignment of phyA Association Sites
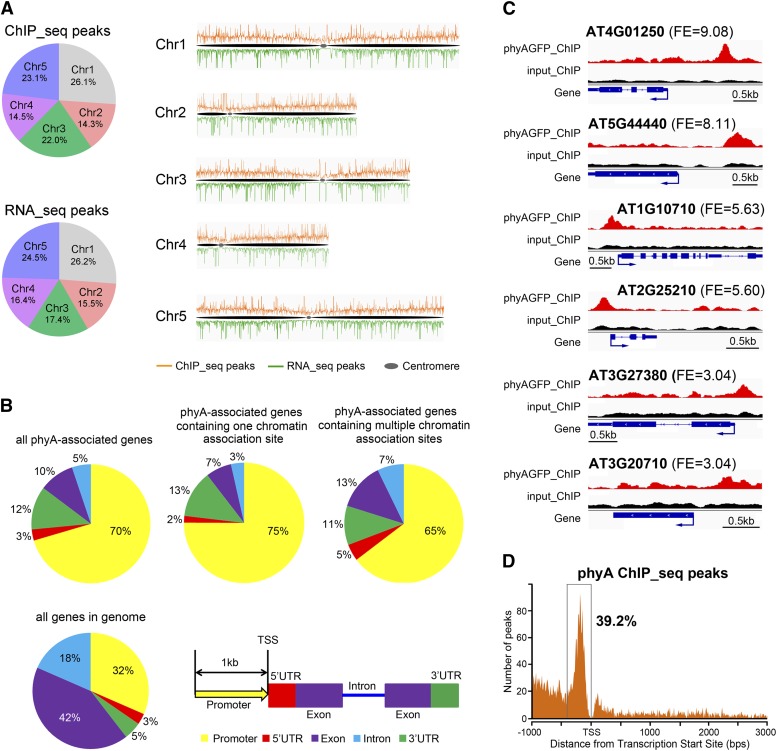
A total of 3798 phyA peaks identified in our study were distributed across the five Arabidopsis chromosomes. The percentages of phyA peaks on each chromosome were positively correlated with the chromosome size (Figure 1A). Centromeric regions showed few or no phyA peaks, indicating that the phyA association with DNA preferentially occurs within gene-rich regions.
Figure 1.
Genome-Wide Distribution of phyA Association Sites and phyA-Regulated Genes.
(A) Distribution of phyA association sites and phyA-regulated genes across the five Arabidopsis chromosomes. ChIP-seq peaks indicate the phyA association sites; RNA-seq peaks indicate the phyA-regulated genes. The numbers in the pie charts indicate the percentage of peaks in each chromosome.
(B) Distribution of phyA association sites within genic regions. A 1-kb region before the transcription start site (TSS) is defined as the promoter region. If a peak covers two adjacent gene regions, it is counted into the region that contains more than 50% of the peak length. If each of two adjacent regions contains exactly 50% of a peak length, the peak is counted following this priority: 5′ UTR > promoter > 3′ UTR > exon > intron. The distribution of individual gene regions in all genomic genes is shown as a control.
(C) Representative phyA association sites on the gene promoter region. The top, middle, and bottom two genes exhibit phyA peaks with high, moderate, and low fold enrichment (FE), respectively. For each gene, phyA peaks in the phyA ChIP sample (red), the input control sample (black), and the gene structure (blue) are shown in the top, middle, and bottom rows, respectively. Bars = 0.5 kb.
(D) phyA association sites are highly enriched in a 400-bp region immediately upstream of the transcription start site.
Among the 2987 phyA peaks assigned to gene regions (from −1000 bp of the transcription site to the 3′ untranslated region [UTR]), 1183 (40%) had a 1:1 ratio with their associated genes. The remaining 60% of phyA peaks either shared their associated genes with several other phyA peaks or were assigned to multiple genes closely located in a defined genomic region (Supplemental Table 1). In the latter case, all of the genes identified by a single phyA peak were designated as phyA-associated genes. Therefore, 2987 phyA peaks were assigned to 3017 phyA-associated genes (Supplemental Data Sets 1 and 2), of which 2241 (74%) had unique phyA association sites and 776 (26%) contained multiple sites. Eleven randomly selected phyA peaks were validated by chromatin immunoprecipitation–quantitative PCR (ChIP-qPCR) assays (Supplemental Figure 5). This validation not only demonstrated the high level of fidelity of our global approach but also suggested that the phyA association with DNA only occurred under FR and not in darkness. These data agreed with previous reports showing that light induces phyA nuclear translocation (Kircher et al., 2002; Genoud et al., 2008).
We next explored the distribution of phyA association sites over genic regions. Interestingly, while the promoter and 5′ UTR areas only account for 35% of all the genes within the genome, a large portion of phyA-associated genes (73%) were associated with phyA in their promoter or 5′ UTR area (Figures 1B and 1C). This proportion reached 77% in phyA-associated genes containing unique phyA peaks (Figure 1B). Furthermore, the locations of 39.2% of the phyA association sites were found to be concentrated within a 400-bp window immediately upstream of the transcription start sites (Figure 1D). This distribution pattern of phyA association sites suggests that the phyA association with DNA is not due to a light-inducible random aggregation on chromatin and is consistent with its potential role as a transcriptional regulator.
phyA and Transcription Factors Bind to Common Target Genes for Multiple Plant Responses
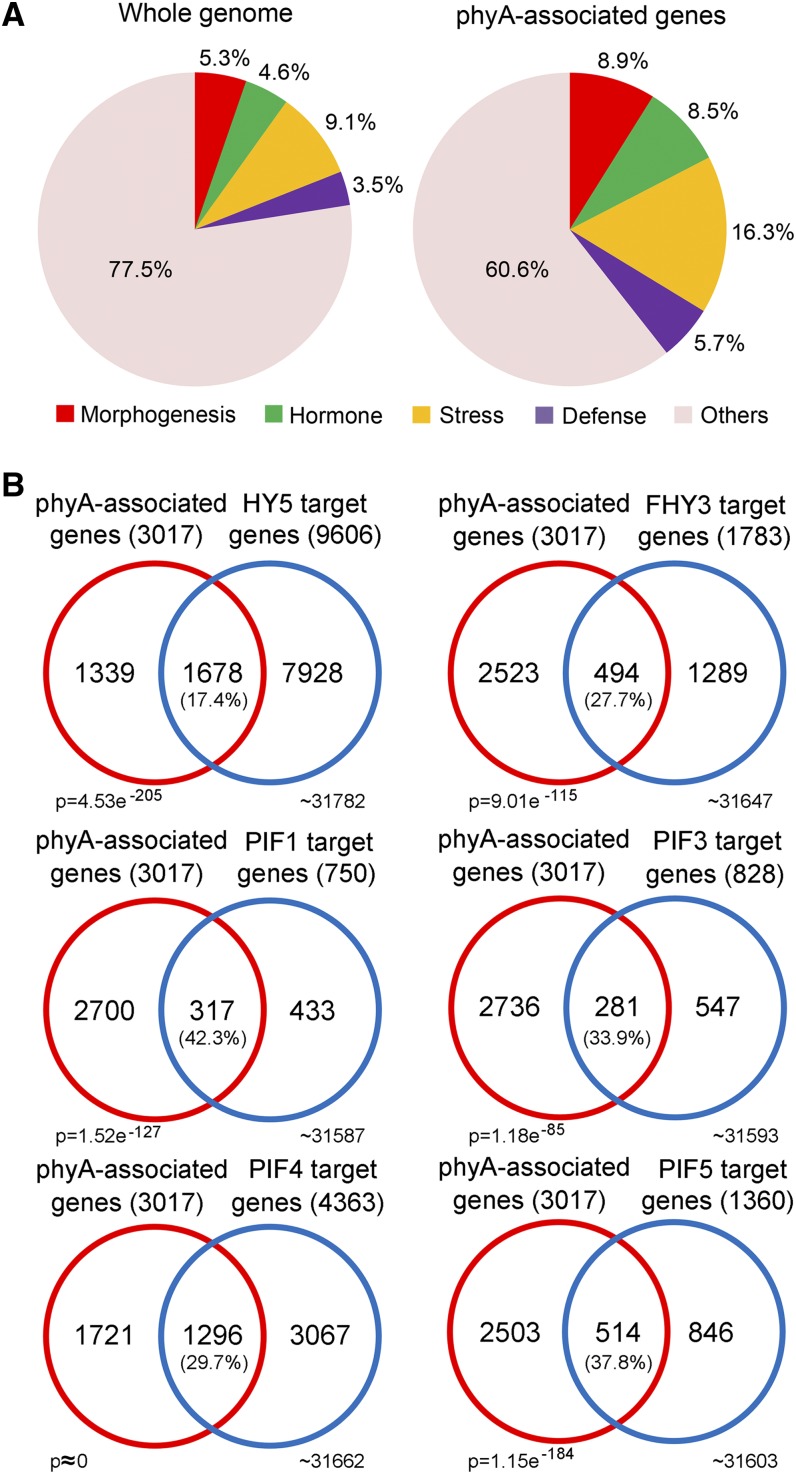
The possibility that phyA not only serves as a photoreceptor to mediate light signaling but also acts as a common intermediate in multiple plant responses was assessed, as phyA-associated genes were found to be enriched in several light crosstalking responses, including morphogenesis, hormone, stress, and defense signaling pathways (Figure 2A). Comparison of phyA-associated genes and reported target genes of HY5, FHY3, PIF1, PIF3, PIF4, and PIF5 revealed that hundreds of common genes were cotargeted by phyA in combination with each of the above transcription factors (Figure 2B). This result demonstrates that, in addition to PIF1 and PIF3 containing an active phyA binding region (Leivar and Quail, 2011), phyA also associates with downstream genes in combination with a broad range of transcription factors. PIF1 was reported to bind phyA with more affinity than PIF3 (Leivar and Quail, 2011). Consistent with this, PIF1 shared more target genes with phyA (43.2%) than with PIF3 (33.9%).
Figure 2.
Functional Analysis of phyA-Associated Genes.
(A) Functional classification of phyA-associated genes enriched compared with the whole genome.
(B) Overlap of phyA-associated genes with known target genes of several light-responsive transcription factors. Percentage values indicate the percentages of genes targeted by transcription factors that overlapped with phyA. P values were calculated using the hypergeometric distribution.
phyA Is Involved in Multiple Biological Processes by Transcriptionally Regulating Its Associated Genes
We next sought to identify genes regulated by phyA on a genome-wide scale by RNA-seq analysis. The wild-type seedlings and the phyA-1 mutant (Whitelam et al., 1993) (both of the Landsberg erecta [Ler] ecotype) were grown in the same conditions used for the phyA ChIP-seq analysis (D4d+FR3h) prior to RNA isolation. Three independent biological replicates were subjected to RNA-seq analysis (each of them generated more than 22 million reads) and showed high-value (>0.994) Pearson correlation coefficients (Supplemental Figure 6).
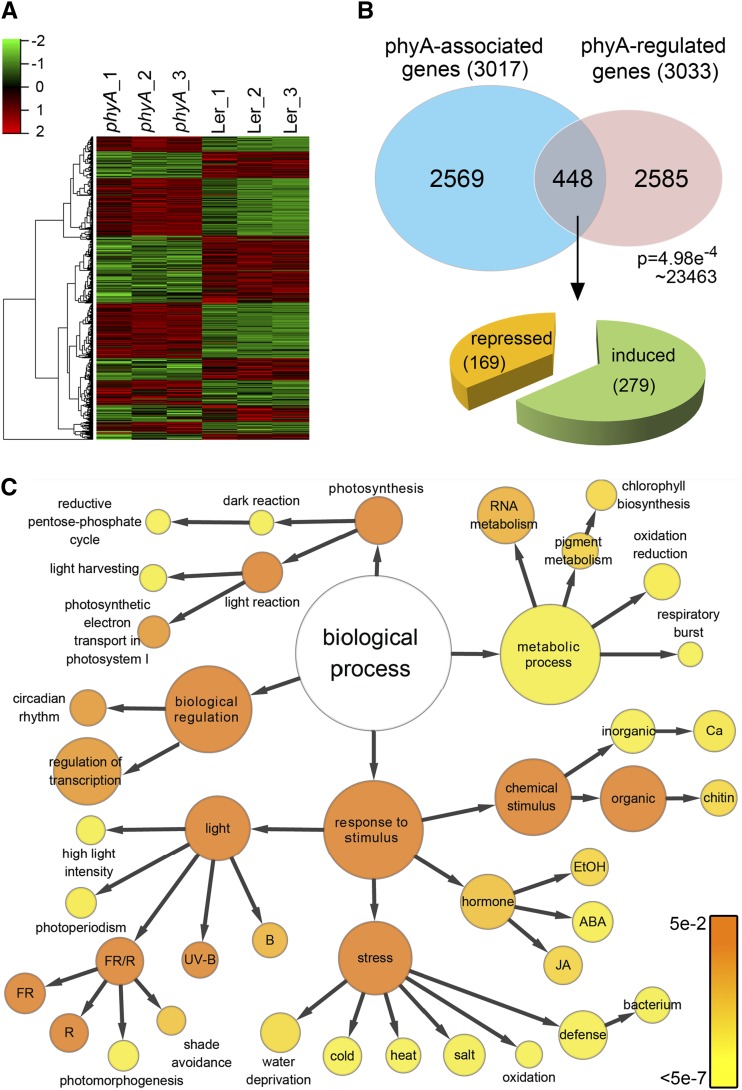
Our RNA-seq data identified 3033 differentially expressed genes that were represented in all three biological replicates (Figure 3A; Supplemental Data Set 3) after strict screening (see Methods). Similar to phyA association sites, phyA-regulated genes were evenly distributed across the five Arabidopsis chromosomes and were proportional to chromosome size (Figure 1A). Of these genes, 1401 (46.2%) were phyA-repressed genes while 1632 (53.8%) were phyA-induced genes (Supplemental Data Set 2). To validate the RNA-seq data, we conducted quantitative RT-PCR (qRT-PCR) analyses and validated 12 randomly selected phyA-regulated genes (Supplemental Figure 7).
Figure 3.
Identification of phyA Direct Target Genes by Merging RNA-seq Data and ChIP-seq Data.
(A) Hierarchical clustering of differentially expressed genes in wild-type (Ler) seedlings and phyA mutant lines. All three biological replicates of RNA-seq are shown.
(B) Venn diagram showing the overlap of phyA-associated genes (ChIP-seq data) and phyA-regulated genes (RNA-seq data). Genes in the overlap (448 genes) are identified as phyA direct target genes. The P value of the Venn diagram was calculated using the hypergeometric distribution.
(C) GO analysis of phyA direct target genes. Each circle represents an enriched category compared with the whole genome after false discovery rate correction. The size of each circle is proportional to the number of genes annotated to the node. The color of each circle indicates the P value from 5 × 10−2 to 5 × 10−7.
To determine which of the phyA-associated genes were transcriptionally regulated by phyA, we superimposed our ChIP-seq and RNA-seq data. As shown in Figure 3B, 448 genes that physically associate with phyA displayed a phyA-regulated expression pattern (hereafter defined as phyA direct target genes). We performed principal component analysis using these genes, and the phyA ChIP-seq data appeared to match the gene expression data (Supplemental Figure 8).
A number of phyA-regulated genes were not associated with phyA, likely due to the phyA-dependent COP1-mediated degradation or stabilization of transcription factors that can indirectly regulate gene expression profiles (Osterlund et al., 2000; Bauer et al., 2004). Conversely, there are many possibilities for why a number of phyA-associated genes were not regulated by phyA. First, phyA association sites were identified in 27% of phyA-associated genes through their exons, introns, or 3′ UTRs (Figure 1B), which may not be beneficial to the phyA function for transcriptional regulation. Second, similar to transcription factors that can activate, suppress, or appear to have no detectable effect on their target genes (Loh et al., 2006), phyA may associate with target genes that do not respond in the absence of critical transcriptional regulators. Of the phyA direct target genes, 37.7% were phyA repressed while the rest were phyA induced (Figure 3B; Supplemental Data Set 2). These results suggest that phyA can act either as a transcriptional activator or repressor to regulate its direct target genes.
We next performed a Gene Ontology (GO) analysis using BiNGO (Maere et al., 2005) to further characterize the newly identified phyA direct target genes. As shown in Figure 3C, genes responsive to light, stresses, hormones, photosynthesis, metabolism, and the biological regulations were significantly enriched in phyA direct target genes when compared with the entire genome (P < 0.05). This result suggests a direct involvement of phyA in these processes.
Numerous Motifs Mediate the phyA Association and Regulation of Target Genes
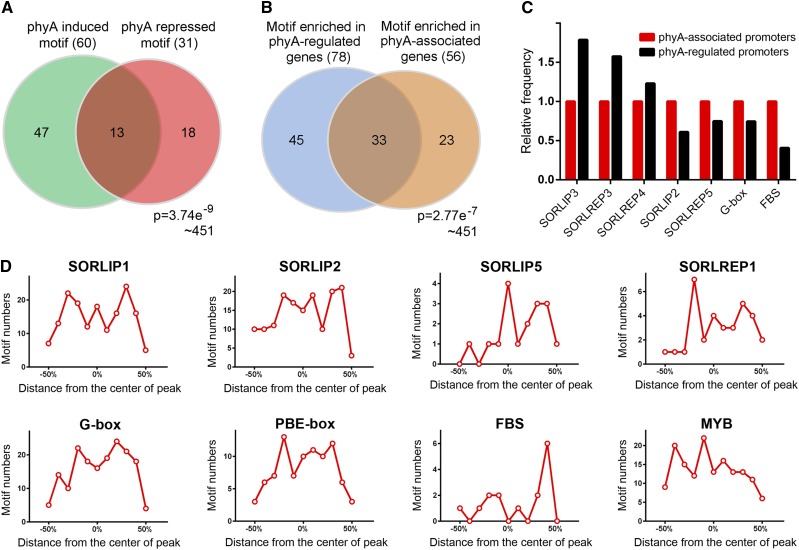
As functional components of the promoter, cis-elements specify protein binding for transcriptional regulation. To further understand the interaction of phyA with chromatin, we examined the enrichment of motifs in phyA-associated and -regulated promoters by using the PLACE (for plant cis-acting regulatory DNA elements) database (Higo et al., 1999). This analysis revealed that 56 motifs appeared at a higher frequency in phyA-associated promoters than in the random genome (Supplemental Table 2). Similarly, 78 motifs were enriched in phyA-regulated promoters, of which 13 motifs were present in the promoters of both phyA-induced and -repressed genes (Figure 4A; Supplemental Table 2). In addition, there are 33 motifs that are overrepresented in both phyA-associated promoters and phyA-regulated promoters (Figure 4B). Consistent with the predicted functions of phyA direct target genes, numerous cis-elements that are responsive to light, ABA, auxin, gibberellic acid (GA), ethylene, and plant defense were observed (Supplemental Table 3). These results further suggested that the phyA association with DNA is a universal mechanism with preferences for particular DNA sequences.
Figure 4.
Motif Analysis of phyA-Regulated and phyA-Associated Promoters.
(A) Venn diagram showing the overlap of motifs enriched in phyA-induced and -repressed genes. The frequency of each motif in the PLACE database was calculated. It is identified as an enriched motif if its frequency on phyA-regulated promoters is 1.5-fold or higher compared with that on a random genome. The P value was calculated using the hypergeometric distribution.
(B) Venn diagram showing the overlap of motifs enriched in phyA-regulated and -associated promoters. The P value was calculated using the hypergeometric distribution.
(C) Frequency of several phyA-responsive motifs in phyA-associated and -regulated promoters.
(D) Distribution of well-characterized highly redundant phyA-responsive motifs within phyA association regions (ChIP-seq peaks).
We next investigated the abundance of well-characterized phyA-responsive motifs and other cis-elements involved in phyA signaling in phyA-associated promoters and phyA-regulated promoters. SORLIP3, SORLREP3, and SORLREP4 (Hudson and Quail, 2003) had greater frequencies in phyA-regulated promoters than in phyA-associated promoters (Figure 4C), indicating that phyA-dependent mechanisms other than a physical association are involved in activating promoters harboring these motifs. Conversely, SORLIP2, SORLREP5, G-box, and FHY3 binding motifs (Lin et al., 2007) were found to be more enriched in phyA-associated promoters than in phyA-regulated promoters, suggesting that the association of phyA with DNA is not sufficient for the transcriptional regulation of all promoters containing one of these four motifs.
Interestingly, we observed a large number of phyA association patterns in abundant motifs. The motif distribution peaked either at the center of the phyA association region or some distance away from it (Figure 4D), revealing that phyA does not associate with the promoter directly in the motif, unlike the transcription factors FHY3 and PIF3 (Ouyang et al., 2011; Zhang et al., 2013). These data are consistent with the concept that phyA indirectly binds promoters as a member of a regulatory complex.
Involvement of phyA in ABA Metabolism and Signaling
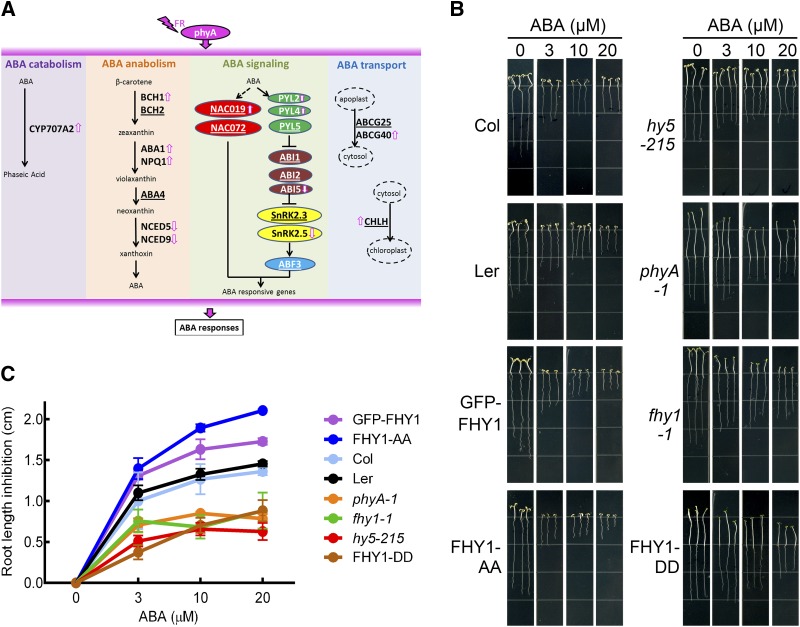
Together, the functional classifications of phyA direct target genes and phyA-associated genes (Figure 3C; Supplemental Figure 9) and the large number of ABA response elements identified in phyA direct target motifs (Supplemental Table 3) laid emphasis on ABA as one of the hormone signals that is directly coordinated by phyA. Therefore, we chose the ABA pathway as a model to investigate how phyA is directly involved in biological processes beyond the light response. Through the ABA-related genes revealed by our ChIP-seq and RNA-seq data, we hypothesized that, in addition to ABA signaling, both the internal levels and subcellular localization of ABA could also be influenced by phyA (Figure 5A), although the precise mechanism responsible for the phyA association or regulation of ABA-related genes awaits further investigation.
Figure 5.
Synergy between phyA Signaling and ABA Response in the Inhibition of Root Elongation under FR.
(A) ABA-related genes associated with or regulated by phyA revealed by genomic analysis. Underlined gene symbols indicate phyA-associated genes. Arrows indicate phyA-induced genes.
(B) Root elongation inhibition in phyA signaling-related mutants upon ABA treatment. Four-day-old FR-grown seedlings were transferred to vertical plates containing the indicated concentrations of ABA for 5 d in FR. Col, wild-type Columbia-0; GFP-FHY1, 35S:GFP-FHY1/fhy1-1; FHY1-AA, 35S:GFP-FHY1S39AT61A/fhy1-1; FHY1-DD, 35S:GFP-FHY1S39DT61D/fhy1-1. Ler was the background for all mutants and transgenic lines used in this experiment except for the hy5-215 null mutant, which was in the Columbia-0 background.
(C) Statistical analysis of the root elongation inhibition shown in (B). Relative root growth on ABA plates was normalized with seedlings that were grown in the absence of ABA for each line. Error bars represent se (n = 20) of three biological replicates.
We then investigated the integral effect of phyA signaling on ABA responses under FR. When compared with wild-type plants, ABA-mediated inhibition of root elongation under FR was observed to be more severe in seedlings with enhanced phyA signaling, such as transgenic lines overexpressing FHY1 or its constitutively active variant FHY1S39AT61A. Conversely, abolishment of phyA signaling, either by mutating phyA or FHY1 or by expressing a constitutively inactive FHY1 (FHY1S39DT61D), decreased the seedling sensitivity to ABA-mediated inhibition of root elongation (Figures 5B and 5C). Notably, the hy5 mutant also exhibited ABA tolerance, suggesting its involvement in the ABA response mediated by phyA. This observation implies that phyA signaling positively regulates the ABA response under FR even if phyA activates several negative regulators of the ABA response, such as CYP707A1/2 and ABI1/2, likely by feedback regulation.
The ABA-Dependent Transcription Factor NAC019 Is a Direct Target of phyA
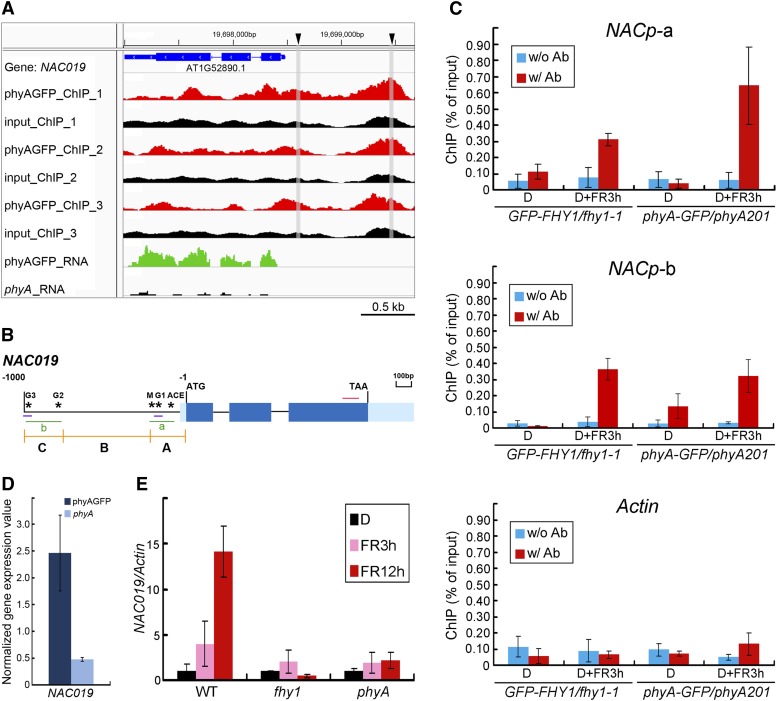
Our ChIP-seq data revealed two phyA peaks on the NAC019 promoter (Figure 6A). As NAC019 encodes an ABA-dependent transcription factor (Tran et al., 2004), we selected it for study in order to understand how phyA directly acts on ABA-responsive genes and synergistically coordinates the ABA response under FR. Using ChIP-qPCR assays, we confirmed that the regions corresponding to the two phyA peaks on the NAC019 promoter (a and b) were associated with phyA in a FR-dependent manner (Figures 6B and 6C). Both RNA-seq (Figures 6A and 6D) and qRT-PCR (Figure 6E) assays demonstrated that the FR-inducible NAC019 expression was phyA dependent, further identifying NAC019 as a phyA direct target gene. It is interesting that FHY1 was also associated with the promoter of NAC019 (Figure 6C) and is critical to NAC019 expression (Figure 6E).
Figure 6.
Validation of phyA and FHY1 Association and Regulation of the ABA-Responsive Transcription Factor NAC019.
(A) phyA ChIP-seq and RNA-seq peaks on NAC019. phyA ChIP-seq peaks (in red) of all three biological replicates are mainly located on the NAC019 promoter region. The input controls are shown in black. Representative RNA-seq peaks cover the gene exon regions. Triangles represent the positions of two G-boxes within the NAC019 promoter, which are experimentally proven association sites of phyA and FHY1. The gray bars indicate the positions of the two G-boxes in the phyA ChIP-seq peaks.
(B) Schematic diagram of the NAC019 gene and promoter. Stars represent five possible motifs responsible for the regulation of NAC genes. NAC019 5′ UTR and promoter regions are divided into three fragments (A, B, and C) for yeast assays in Figures 7B and 8D. Two green lines (a and b) and the red line indicate the locations of amplicons used for ChIP-qPCR in (C) and qRT-PCR in (E), respectively. Two short blue lines indicate the locations of probes used for EMSA in Figure 8A.
(C) ChIP-qPCR analysis showing that both phyA and FHY1 associate with NACp-a and NACp-b regions on the NAC019 promoter in response to FR. Four-day-old etiolated GFP-FHY1/fhy1-1 and phyA-GFP/phyA-1 seedlings were left untreated (D) or irradiated with 3 h of FR (D+FR3h). ChIP using anti-FHY1 antibody (w/ Ab; for GFP-FHY1/fhy1-1 seedlings), anti-GFP antibody (for phyA-GFP/phyA-1 seedlings), or no antibody (w/o Ab) was followed by amplification of NACp-a, NACp-b, and Actin (negative control). Data were normalized with corresponding input samples.
(D) Transcript levels of NAC019 defined by RNA-seq analyses. Means and error values are derived from RNA-seq data.
(E) qRT-PCR showing that the transcription of NAC019 is induced upon FR in a phyA/FHY1-dependent manner. Materials were treated as described in (C) for 3 or 12 h of FR irradiation, as indicated. The values of the NAC019 transcript were normalized to that of Actin in the qRT-PCR.
All error bars represent sd (n = 3) of three biological replicates.
HY5 Regulates NAC019 Expression
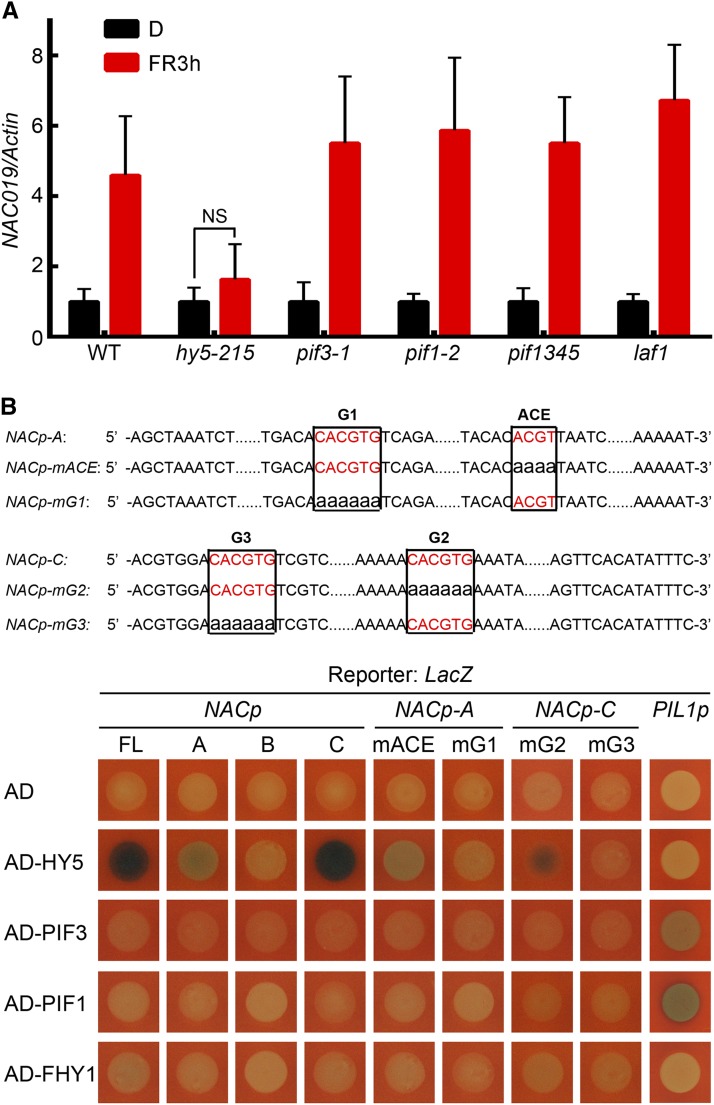
To understand how phyA influences the transcription of NAC019, we sought to identify the transcription factor that regulates NAC019 expression while recruiting phyA/FHY1 to its promoter. We first searched within the promoter region of NAC019 for FR-responsive motifs and other cis-elements involved in NAC gene regulation (Nakashima et al., 2012). As a result of this analysis, a MYB motif, an ACE motif, and three G-boxes were identified (Figure 6B). Given that MYB is a putative binding site for LAF1 and that ACE and G-box can be recognized by HY5 and PIFs, we examined NAC019 transcript accumulation in mutants of these transcription factors. Only the hy5 mutant attenuated the FR-inducible NAC019 expression (Figure 7A), suggesting HY5 as a key transcription factor. This finding was further substantiated by a yeast one-hybrid assay in which HY5, but not PIF1 or PIF3, was able to bind to the 1-kb region upstream of the NAC019 translation start site (Figure 7B).
Figure 7.
HY5 Is Responsible for NAC019 Transcription in FR.
(A) The induction of NAC019 transcription is attenuated only in the hy5 mutant upon FR. Four-day-old etiolated seedlings of the indicated genotypes were left in the dark (D) or exposed to FR for 3 h (FR3h), followed by qRT-PCR. The values of the NAC019 transcript were normalized to that of Actin. NS, not significant. Error bars represent sd (n = 3) of three biological replicates.
(B) Yeast one-hybrid assay showing that HY5, but not other tested transcription factors, binds to the G1 and G3 motifs on the NAC019 promoter. At top are diagrams of the wild-type and mutant NAC019 promoter fragments used to drive the expression of the LacZ reporter gene in yeast. Wild-type ACE and G-boxes are shown in red. Nucleotide substitutions for ACE (mACE) and three G-boxes (mG1, mG2, and mG3) are in lowercase. At bottom, mutation of the G1 and G3 motifs abolished the HY5 binding in the A and C regions, respectively, of the NAC019 promoter. The LacZ reporter driven by the PIL1 promoter was used as a positive control for transcription factors PIF1 and PIF3 in the yeast one-hybrid assay. AD, activation domain; FL, full length.
The phyA Pathway Targets Two G-Boxes on the NAC019 Promoter
We next divided the 1-kb region containing both the 5′ UTR and the promoter of NAC019 into three fragments in order to identify specific regions responsible for HY5 binding (Figure 6B). HY5 bound to regions A and C (Figure 7B), which were also found to associate with phyA and FHY1 (Figure 6C). Subsequently, the four cis-elements recognized by HY5 within the A and C regions were mutated to identify the precise binding site(s). Mutation of G1, but not of the ACE motif, eliminated HY5 binding on the NAC019 promoter A region (Figure 7B). Similarly, HY5 binding to the C region was dependent on the presence of the cis-element G3 but not G2. Coincidentally, G1 and G3 were both located in the phyA association sites revealed by the ChIP-seq data (Figure 6A). In addition, the association of phyA with two G-box–containing regions on the NAC019 promoter was abolished in the absence of HY5 (Supplemental Figure 10). These results suggest that these two G-boxes may guide both HY5 and phyA to the NAC019 promoter.
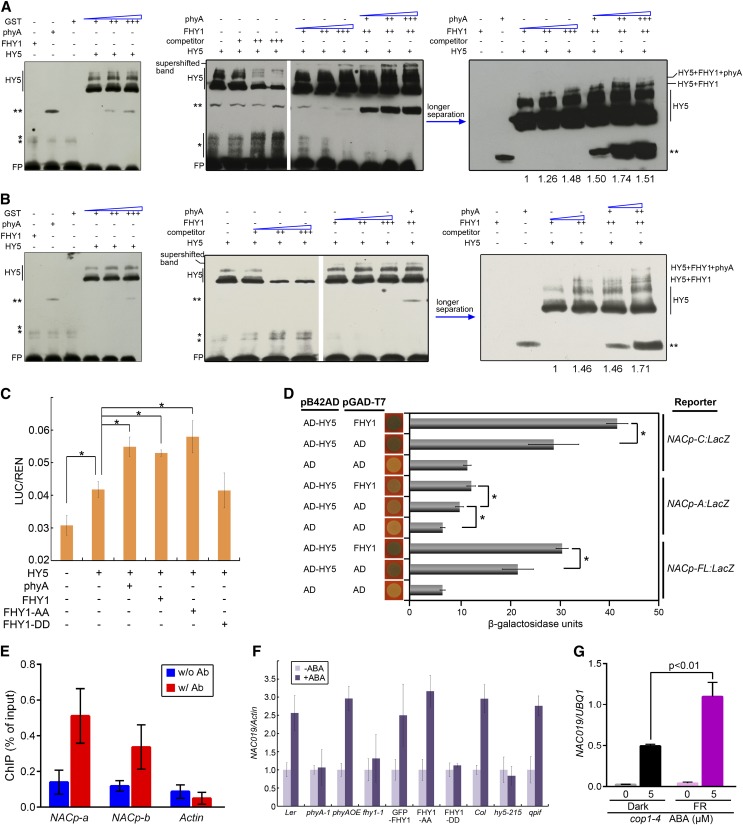
To verify that phyA and FHY1 are recruited through HY5 to the G1 and G3 motifs inside the NAC019 promoter, we performed a supershift electrophoretic mobility shift assay (EMSA). The HY5 and NAC019 promoter G3-containing fragment complex was supershifted in the presence of FHY1 but not glutathione S-transferase (GST) (Figure 8A). Moreover, inclusion of phyA in the reaction further shifted the FHY1-HY5-G3 complex band and increased the total amount of probe bound by the supercomplexes. Likewise, the phyA-FHY1-HY5 complex was also observed with the G1 motif (Figure 8B). A mutant probe without a G-box motif failed to recruit the HY5-FHY1-phyA complex and impeded its ability to compete with wild-type probes for complex formation (Supplemental Figure 11), suggesting that the binding motif of HY5 is critical for phyA-DNA binding.
Figure 8.
phyA and FHY1 Are Recruited on the NAC019 Promoter through HY5 and Activate NAC019 Expression in an ABA-Dependent Manner.
(A) and (B) Supershifted EMSA showing that phyA, FHY1, and HY5 form a complex on the G3 (A) and G1 (B) regions of the NAC019 promoter. Left, GST protein fails to shift the HY5-DNA band; middle and right, the HY5-DNA band is supershifted in the presence of FHY1 and phyA proteins. The two films in the middle are from the same gel. The EMSA reactions contained 1 µg (+), 4 µg (++), or 6 µg (+++) of the indicated proteins and labeled probes (for locations, see Figure 6B) without (−) or with 200-fold (+), 1000-fold (++), or 2000-fold (+++) competitors (unlabeled probes). Single asterisks represent nonspecific bands, and double asterisks represent an unknown band. FP, free probe. Numbers below the gel indicate the quantification of complex-bound probes, which were normalized against the value in the first lane.
(C) phyA and FHY1 enhance the transcriptional activities of HY5. The luciferase transcription reporter driven by the NAC019 promoter was coinfiltrated into tobacco leaves with constructs expressing HY5, phyA, FHY1, FHY1-AA (FHY1S39AT61A), or FHY1-DD (FHY1S39DT61D) as indicated. The reporter activity was measured after incubating the leaves in the dark for 2 d and then exposing them to FR for 1 d. Data were normalized to the internal control REN. *P < 0.05; n = 6.
(D) FHY1 enhances the transcriptional activities of HY5 in quantitative yeast β-galactosidase activity assays. Promoters driven by the LacZ reporter gene are indicated on the right, and the combinations of proteins transformed into the yeast are indicated on the left. *P < 0.05; n = 3.
(E) phyA associates with NACp-a and NACp-b regions on the NAC019 promoter in the absence of FHY1 in response to FR. Four-day-old etiolated seedlings of the cross line PphyA:phyA-GFP fhy1-1 were irradiated with 3 h of FR. ChIP using anti-GFP antibody (w/ Ab) or no antibody (w/o Ab) was followed by amplification of NACp-a, NACp-b, and Actin (negative control). Data were normalized with corresponding input samples. Error bars represent sd of three biological replicates.
(F) Mutation of phyA, FHY1, or HY5 eliminates the ABA-dependent induction of NAC019 in FR. Four-day-old seedlings grown in FR with the indicated genotypes were transferred onto vertical GM plates without (−ABA) or with (+ABA) 5 µM ABA for 5 d in FR, followed by RNA extraction and qRT-PCR. qpif, pif1345. Normalized values of NAC019 transcripts by Actin are shown. Error bars represent sd (n = 3).
(G) phyA coactivates the HY5 activity for ABA-inducible NAC019 transcription in vivo. Four-day-old cop1-4 mutants grown in the dark or FR were transferred onto vertical GM plates without (−ABA) or with (+ABA) 5 µM ABA for 5 d under darkness or FR, respectively. Normalized values of NAC019 transcripts by UBQ1 are shown in the qRT-PCR. Error bars represent sd (n = 3) of three biological replicates. P values are from Student’s t tests.
The phyA Pathway Directly Activates NAC019 Expression
We performed a dual-luciferase reporter assay in Nicotiana benthamiana to examine how the phyA pathway activates the transcription of NAC019. HY5 alone was able to activate NAC019 expression (Figure 8C). Moreover, coexpression of phyA, wild-type FHY1, or constitutive active FHY1 (FHY1S39AT61A) with HY5 potentiated the activity of the NAC019 promoter (Figure 8C). These results indicated that each component of the phyA pathway may contribute to the activation of NAC019 expression and that phyA and FHY1 may potentially increase HY5 transcriptional activity. Furthermore, the coactivation of FHY1 with HY5 was observed on the full-length NAC019 promoter as well as on its A and C regions (Figure 8D). These results suggest that the phyA pathway could activate the expression of NAC019 through either one of these two G-boxes.
Since either phyA or FHY1 alone can enhance the activity of the NAC019 promoter, we next investigated whether the phyA association with the NAC019 promoter could occur independently of FHY1. Through an anti-GFP ChIP-qPCR using the PphyA:phyA-GFP fhy1-1 line, we found that phyA is able to associate with both the NACp-a and NACp-b promoter regions in the absence of FHY1 (Figure 8E). Therefore, FHY1 is not necessary for the phyA association with the NAC019 promoter, although phyA and FHY1 are able to interact with each other on DNA fragments in the in vitro EMSA.
The phyA Pathway Is Necessary for NAC019 Sensitivity to ABA
Our results suggest that transcriptional regulation of the ABA-dependent transcription factor NAC019 via the phyA pathway is one of the mechanisms capable of rapidly transmitting the FR signal to the ABA downstream signaling network. To further demonstrate how the phyA pathway directly influences the plant’s response to ABA, we examined the ABA-inducible NAC019 expression in mutants that were defective in phyA signaling. Seedlings expressing an inactive FHY1 (FHY1S39DT61D) or phyA, fhy1, and hy5 mutants were unable to induce NAC019 expression upon ABA treatment (Figure 8F), indicating that a deficiency in the phyA pathway renders NAC019 insensitive to ABA.
To exclude the possibility that the attenuated NAC019 expression in phyA signaling-defective mutants is due to the lack of HY5 protein instead of the loss of HY5 activity, we tested ABA-inducible NAC019 expression in the cop1 mutant, in which HY5 degradation is blocked (Osterlund et al., 2000). When the cop1 mutant is grown in darkness, HY5 accumulates in the nucleus in the absence of active phyA and mediates ABA-inducible NAC019 expression at a moderate level (Figure 8G). This result is consistent with our previous observations that HY5 alone is able to activate NAC019 expression, as demonstrated in the tobacco and yeast assays (Figures 8C and 8D). In the FR-grown cop1 mutant, however, both HY5 and active phyA accumulate in the nucleus. Consequently, the NAC019 transcription level increased further upon ABA treatment compared with that in the dark-grown cop1 mutant (Figure 8G). This result suggests that phyA not only contributes to the HY5 accumulation upon FR (Osterlund et al., 2000) but also serves as a transcriptional coactivator with HY5. Therefore, we conclude that the phyA pathway ensures the appropriate expression of ABA signaling components through multiple mechanisms and, as a result, optimal ABA response under FR.
DISCUSSION
Phytochromes Directly Associate with Gene Promoters
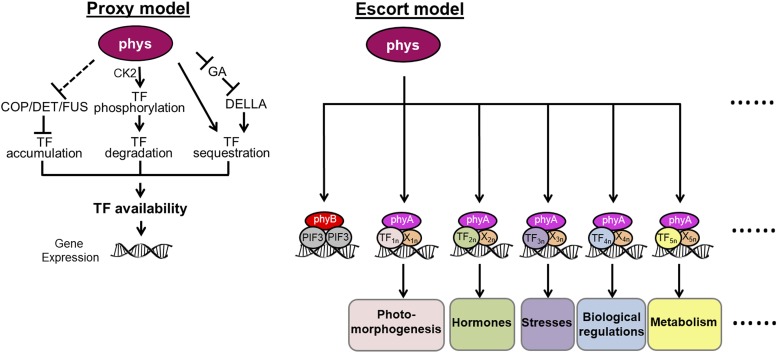
Substantial efforts have been made to examine the nuclear signal transduction pathway of phytochromes and their effects on downstream genes. The PIF transcription factors negatively regulate photomorphogenesis. The interaction of PIF1, PIF3, PIF4, and PIF5 with phytochromes triggers their phosphorylation, in some cases by multiple kinases, and leads to proteosome-mediated degradation (Al-Sady et al., 2006; Lorrain et al., 2008; Bu et al., 2011). The repression of GA signaling by phytochromes facilitates the formation of inactive complexes between DELLA proteins and PIF3 or PIF4, resulting in the inhibition of PIF-DNA binding (de Lucas et al., 2008; Feng et al., 2008). Recent studies demonstrated that phyB could also sequester PIF1 and PIF3 and prevent them from binding to DNA (Park et al., 2012). phyB could also interact with phosphorylated PIF7 to prevent its dephosphorylation and subsequent binding to the promoters of its target genes (Li et al., 2012). phyA promotes the accumulation of the required level of HY5 protein through the inhibition of the COP/DET/FUS degradation machinery under FR (Osterlund et al., 2000). All of the above models revolve around the notion that phytochromes control the availability of transcription factors for their target genes (Figure 9, proxy model).
Figure 9.
Schematic Models Show the Nuclear Behaviors of Phytochromes.
The proxy model summarizes how phytochromes (phys) regulate gene expression by controlling the availability of transcription factors (TF). The escort model suggests that phyA and phyB directly associate with gene promoters by interacting with TF and coregulators (X). Consequently, phyA is directly involved in multiple plant responses that are coordinated with the FR signal. n represents different TFs or Xs involved in the phyA signaling.
An emerging model that phytochromes directly associate with the promoters of their target genes in order to regulate their expression was first unveiled by the report that phyB and PIF3 can form complexes on DNA in vitro (Martínez-García et al., 2000). Our previous study revealed that phyA can be directly recruited to the gene promoter and coactivate transcription in the case of CHS (Chen et al., 2012). Here, by globally identifying and analyzing phyA direct target genes (Figures 1 to 3), we demonstrated that the association of phyA with promoter DNA is one of the general mechanisms through which the FR signal can directly regulate multiple aspects of plant growth and development, including photomorphogenesis, hormone-related pathways, stress response pathways, and metabolic processes (Figures 3 and 9). Since the number of phyA direct target genes only accounts for around 15% of all phyA-regulated genes, we assume that the newly proposed phyA-DNA association mechanism, mechanisms of phyA-controlled stability, the activity and sequestration of transcription factors, and other unknown mechanisms are complementary strategies for phyA to regulate ∼10% of the whole genome in response to FR (Tepperman et al., 2001). This assumption is consistent with the low correlation between all phyA-regulated genes and the phyA-associated genes revealed in our study (Pearson correlation coefficient = 0.101).
The phyA transcriptional complex that targets gene promoters consists of phyA, transcription factors, and coregulators. While different transcription factors might be involved in each plant response, some transcription factors could conceivably participate in multiple responses. For example, both HY5 and PIF3 are involved in phyA signaling for anthocyanin biosynthesis (Chen et al., 2012), whereas HY5 is also required for an optimal ABA response (Lau and Deng, 2010; Figure 8). Although FHY1 might act as the cofactor to bridge the interaction between phyA and transcription factors for the potential coactivation on gene promoters (Yang et al., 2009; Chen et al., 2012), we cannot exclude the possibility that there are other cofactors in addition to FHY1 and its homolog FHL. This prediction is consistent with the observation that the phyA association with the NAC019 promoter is independent of FHY1 and that FHY1/FHL deficiency does not completely abolish phyA signaling (Kami et al., 2012).
The DNA Association Patterns of phyA Differ from That of Transcription Factors
As a protein that indirectly binds DNA, phyA must interact with the chromatin using a different strategy than direct DNA binding proteins. First, phyA specifically associates with gene-rich regions within the genome (Figure 1), while transcription factors (like FHY3) will bind to centromeric or other gene-poor regions as long as the appropriate motifs exist in those regions (Ouyang et al., 2011). Second, phyA associates preferentially with gene promoters or 5′ UTRs (73%) when compared with the transcription factor FHY3 (55%) (Figure 1; Ouyang et al., 2011). These results suggest that phyA’s association with the genome is biased toward the transcriptional regulation of associated genes, while transcription factors may have less or no effect on gene transcription when they bind to gene-poor or transcribed genic regions (Loh et al., 2006). Consequently, more associated genes are transcriptionally regulated by phyA (14.8%) than by the transcription factors FHY3 (8.5%) and PIF3 (2.7%) (Figure 3) (Ouyang et al., 2011; Zhang et al., 2013). This suggests that there is an in vivo mechanism guiding protein complexes involving phyA and other transcription factors preferentially to regulatory regions.
A Broad Range of Transcription Factors Act Downstream of phyA
The PIFs are the most intensively studied subset of transcription factors that directly and physically interact with phytochromes. Among them, PIF1 and PIF3 contain binding motifs not only for phyB but also for phyA (Leivar and Quail, 2011). PIFs are thought to serve as nodes that integrate phytochrome signals and signals from other pathways by recognizing the G-box and PBE-box motifs (Leivar and Quail, 2011; Zhang et al., 2013). However, our study reveals that, in addition to these motifs, there are dozens of cis-elements that are enriched in phyA direct target promoters (Figure 4; Supplemental Table 3). This result suggests that numerous transcription factors other than PIFs may act immediately downstream of phyA for phyA’s association with various genes by specifically recognizing these enriched motifs. Furthermore, beyond the target genes of PIF1 and PIF3, phyA-associated genes overlap extensively with the target genes of HY5, FHY3, PIF4, and PIF5 (Figure 2), transcription factors that might not directly interact with phyA.
Our escort model (Figure 9) proposes that a variety of transcription factors (including PIFs) can act in the phyA signaling pathway by indirect interaction with phyA if there is a coregulator that serves as a platform (Yang et al., 2009), effectively expanding the role of phyA in the cell. In this mechanism, all transcription factors responsive to light, such as LAF1 and HFR1 (both known to indirectly interact with phyA) (Yang et al., 2009), and those transcription factors responsive to other internal or external stimuli (Supplemental Table 3) could also be candidates for guiding phyA to a gene promoter.
Many signaling pathways overlap due to the roles of common transcription factors. For example, the transcription factor PIF1 regulates phyA- and ABA-mediated seed germination; PIF3 and PIF4 integrate light and GA signals in seedling photomorphogenesis; HY5 functions in crosstalk between light and GA, ABA, and auxin; and light-responsive FHY3 and PIF5 participate in chloroplast division and auxin biosynthesis, respectively (Lau and Deng, 2010; Ouyang et al., 2011; Hornitschek et al., 2012). In all of these findings, the transcription factors serve as pivotal points for distinct signaling pathways. Our study brings forth the concept that transcription factors can act downstream of phyA in two separate mechanisms: either by linking phyA signaling with other signaling pathways prior to their targeting on gene promoters or by recruiting phyA to the promoters that directly respond to multiple internal or external stimuli.
The Role of phyA in the ABA Response
The R signal decreases the endogenous ABA level by repressing the expression of ABA biosynthetic genes, such as NCED6 and NCED9 (Seo et al., 2009). Our study at the genome-wide level reveals that the FR signal also directly controls the ABA level by phyA-mediated repression of the ABA anabolic genes NCED5 and NCED9 and by induction of the ABA catabolic gene CYP707A2 (Figure 5). By contrast, three ABA anabolic genes, BCH1, NPQ1, and ABA1, are also induced by phyA, suggesting that the role of phyA in regulating the ABA level is the additive result of two counteractive processes. In addition, ABA localization and signaling in the cytosol are also regulated by FR (Figure 5).
Despite substantial evidence of the antagonism between FR and ABA in the regulation of seed germination, the mechanism through which the FR signal controls root elongation synergistically with ABA remains unknown. Our data demonstrate that FR-induced phyA signaling further inhibits ABA-mediated root elongation (Figure 5). NAC019 is an ABA-responsive intermediate, and a G-box motif on its promoter at position −159 is predicted to be responsible for NAC019 expression (Tran et al., 2004). Furthermore, our genomic analysis and molecular assays uncovered two phyA association sites on the NAC019 promoter at positions −159 and −988 (Figures 6 to 8). Interestingly, the association of phyA with the NAC019 promoter mediates the transcriptional regulation of NAC019 in response to both FR and ABA (Figures 6 and 8). As NAC019 is a transcription factor, it is conceivable that At GSTU17, a GST that participates in ABA-mediated inhibition of root elongation under white light (Jiang et al., 2010), is subsequently regulated by NAC019 under FR. It is likely that more ABA-responsive intermediates in addition to NAC019 are also involved in FR-modulated root elongation, since NAC019 has multiple functionally redundant homologs, such as NAC072 and NAC055 (Tran et al., 2004). The phyA-mediated expression of ABA-responsive factors, including NAC019, is critical for FR to positively coordinate with the ABA response. The evolution of a synergy between FR and ABA for the inhibition of root elongation likely emerged to ensure that plants maintain their root growth under a canopy until more optimal ambient conditions for further development become available.
METHODS
Plant Materials and Growth Conditions
The wild-type Arabidopsis thaliana used in this study was of the Ler ecotype, unless indicated otherwise. The phyA-1 (Whitelam et al., 1993), fhy1-1 (Desnos et al., 2001), hy5-215 (Oyama et al., 1997), and qpif (Leivar et al., 2008) mutants and the phyA-GFP/phyA201 (Kim et al., 2000), phyAOE (Boylan and Quail, 1991), GFP-FHY1 (Shen et al., 2005), and GFP-FHY1S39AT61A and GFP-FHY1S39DT61D (Chen et al., 2012) transgenic plants have been described previously. The growth conditions and light sources were as described by Chen et al. (2012).
ChIP
Four-day-old etiolated seedlings of phyAGFP/phyA201 and phyA-1 were treated by FR for 3 h and cross-linked with 1% formaldehyde in a vacuum for 30 min under dim green light. Subsequently, the chromatin DNA was isolated and sonicated as described previously (Chen et al., 2012). For ChIP-seq, the DNA was immunoprecipitated by anti-GFP (Clontech) antibody (ChIP DNA) or not (input). The GFP antibody allows for the detection of nuclear phyAGFP signals in FR-irradiated phyAGFP/phyA201 seedlings but not in dark-treated phyAGFP transgenic seedlings or FR-treated wild-type seedlings. Moreover, the phyAGFP signals obtained with the GFP antibody were present only on gene promoter regions but not on exons or in the no-antibody control sample (Chen et al., 2012). The construction of libraries and sequencing were conducted by the Yale Center for Genome Analysis. For ChIP-qPCR, anti-FHY1 (Shen et al., 2005) and anti-GFP antibodies were used for DNA precipitation. An equal amount of sample without antibody was used as a mock control. ChIP DNA was analyzed by quantitative PCR with Power SYBR Green PCR Master Mix (Applied Biosystems). Primer information can be found in Supplemental Table 4. Each ChIP value was normalized to its respective input DNA value (defined as 100%) (Guo et al., 2008). ChIP-qPCR experiments were independently performed in triplicate, and representative results are shown.
ChIP-seq Data Analysis
Sequencing adaptors were first removed from raw reads generated by ChIP-seq. Next, nucleotides with sequencing Phred quality scores below 28 were trimmed by FASTX-Tookit (http://hannonlab.cshl.edu/fastx_toolkit/). All sequencing reads were then mapped to the Arabidopsis genome reference (TAIR10 from www.arabidopsis.org). Bowtie software (http://bowtie-bio.sourceforge.net) was used for mapping under default parameters except for discarding multiple loci matching reads that might introduce error signals by repeat counting. phyA peaks were identified afterward by the model-based analysis software MACS (http://liulab.dfci.harvard.edu/MACS/) using input DNA as a control. MACS default parameters were optimized by referring to a previously confirmed phyA-associated gene, CHS (Chen et al., 2012). In detail, -keep-dup was set to 17 to distinguish CHS association peaks from noise as well as to reduce PCR amplification and sequencing bias; -m was set to 0-20 instead of the default 0-30 because phyA association signals were conceivably weaker than transcription factors; fold enrichment against random Poisson distribution was set to equal or larger than 3. PeakSplitter (www.ebi.ac.uk/research/bertone/software) was used to subdivide MACS called peaks into discrete signal subpeaks by default parameters except that each peak was required to have more than 40 reads in its summit region. Finally, phyA association sites were identified if overlapped subpeaks were discovered among all three biological replicates and their overlapped region covered more than 50% of each. The sequence ontology of phyA association sites was analyzed based on the TAIR10 genome annotation. Multiple gene-localized sites were all kept for further verification. The GO information on phyA-associated genes was extracted from the TAIR10 database. GO results were viewed with the WEGO website, and GO enrichment analysis was performed by BiNGO software (www.psb.ugent.be/cbd/papers) with default parameters. P values for the Venn diagram analyses were calculated as described previously (Zhou et al., 2007).
RNA-seq and qRT-PCR
For RNA-seq, wild-type (Ler) and phyA-1 plants were treated as for ChIP-seq (darkness for 4 d + FR for 3 h) and used for total RNA extraction using the RNeasy Plant Mini Kit (Qiagen). Sequencing was conducted by the Yale Center for Genome Analysis using Illumina HiSeq 2000. For qRT-PCR, RNA was reverse transcribed via the SuperScript II Reverse Transcriptase Kit (Invitrogen) followed by quantitative PCR using Power SYBR Green PCR Master Mix (Applied Biosystems). Primer information can be found in Supplemental Table 4. Expression levels were normalized to that of the Actin gene. qRT-PCR experiments were independently performed in triplicate, and representative results are shown.
RNA-seq Data Analysis
RNA-seq data analysis followed a well-developed transcriptome sequencing analysis pipeline, Cufflink (Trapnell et al., 2013). Briefly, RNA-seq raw reads were mapped to the TAIR10 Arabidopsis genome reference by TopHat (http://tophat.cbcb.umd.edu/) after adaptor removal and trimming of low-quality nucleotides. According to the Arabidopsis genome annotation, all mapped reads were then assembled into known transcripts by Cufflink software. Next, the expression of transcripts was calculated in fragments per kilobase of exon model per million mapped fragments (Mortazavi et al., 2008). Afterward, Cuffmerge software merged all transcripts discovered among replicates and controls into one file for expression difference analysis. Finally, Cuffdiff was used to calculate the expression changes of transcripts based on a Poisson fragment distribution. The P value was required to be lower than 0.05. Besides statistical cutoffs, significant expression changes needed to satisfy the criterion that fragments per kilobase of exon model per million mapped fragments fold changes must be equal to or larger than ±1.5, since early FR-responsive genes, like CHS, exhibit a slight ratio change in expression level (Chen et al., 2012). The Cufflink, Cuffmerge, and Cuffdiff software were from the Cufflink package (http://cufflinks.cbcb.umd.edu).
Root Elongation Inhibition
Stratified seeds were irradiated with white light for 12 h to induce germination and then grown vertically under FR for 4 d. Around 20 seedlings for each line were transferred to Murashige and Skoog plates containing indicated concentrations of ABA and grown vertically under FR for another 5 d. Root elongation on the ABA plates was measured using ImageJ (http://rsb.info.nih.gov/ij/). Three independent experiments were performed, and representative results are shown.
Yeast Assays
For the yeast one-hybrid assay, full-length open reading frames of FHY1, HY5, PIF1, and PIF3 were cloned into the pB42AD vector (Clontech) and cotransformed with the p8op-lacZ plasmid, which contains the LacZ reporter driven by full-length, wild-type, or mutated A or C regions of NAC019 promoters in the EGY48 yeast strain. Transformants were grown on SD/-Ura-Trp plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside for blue color development. For transcription activation assays in yeast, full-length FHY1 and HY5 were cloned into both pB42AD and pGADT7 vectors (Clontech). Full-length A or C regions of NAC019 promoters were cloned into pLacZi2µ vector (Lin et al., 2007) to generate the reporter plasmids. The indicated combinations of AD fusion plasmid, empty vector, and reporter plasmid were cotransformed according to the Yeast Protocols Handbook (Clontech). Transformants were selected on SD/-Leu-Trp-Ura plates. The transcription activity was checked by measuring β-galactosidase activity using the Yeast β-Galactosidase Assay Kit (Thermo Scientific). All of the primer information for the constructs generated is shown in Supplemental Table 4.
Transcription Dual-Luciferase Assay
The NAC019 promoter (−1000 to −1) was cloned into pGreenII-800 vector (Hellens et al., 2005) to induce the LUC reporter gene, while the internal control REN reporter gene was driven by the 35S promoter. Primer information for the constructs generated is shown in Supplemental Table 4. The procedure for Nicotiana benthamiana infiltration was performed as described previously (Chen et al., 2012). The LUC/REN activity was measured via the Dual-Luciferase Reporter Assay System (Promega) on a GLOMAX 20/20 luminometer (Promega). Six biological replicates were performed for each combination.
EMSA
EMSA was performed using the Lightshift Chemiluminescent EMSA Kit (Pierce) according to the manufacturer’s instructions. Oligonucleotides corresponding to the HY5 binding regions of the NAC019 promoter (−142 to −184 for G1 and −967 to −1013 for G3) were biotin labeled as probes or unlabeled as corresponding competitors. Indicated proteins were incubated with probes or competitors in 20-μL reaction mixtures containing 10 mM Tris-HCl, pH 7.5, 50 mM KCl, 1 mM DTT, and 50 ng/μL poly(dI-dC) for 20 min at room temperature and separated on a 6% native polyacrylamide gel.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: phyA (AT1G09570), FHY1 (AT2G37678), HY5 (AT5G11260), and NAC019 (AT1G52890). High-throughput sequencing data analyzed in this study are available in the Gene Expression Omnibus database under accession number GSE48770.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Time Course of phyA Association with Its Known Direct Target Gene CHS by ChIP-qPCR.
Supplemental Figure 2. Phenotypes of 5-d-old FR-grown phyA-GFP Line, phyA-1 Null Mutant, and Wild-Type Seedlings (of Ler and Wassilewskija Ecotypes).
Supplemental Figure 3. Scatterplot of the Pearson Correlation of Three phyA ChIP-seq Replicates.
Supplemental Figure 4. phyA ChIP-seq and RNA-seq Peaks on Its Known Direct Target Gene CHS.
Supplemental Figure 5. Validation of Several phyA-Associated Genes by ChIP-qPCR Analysis.
Supplemental Figure 6. Scatterplot of the Pearson Correlation of Three RNA-seq Replicates for Both the Wild Type (Ler) and the phyA Mutant.
Supplemental Figure 7. Validation of Several phyA-Induced Genes (Top) and phyA-Repressed Genes (Bottom) by qRT-PCR Analysis.
Supplemental Figure 8. Principal Component Analysis of 448 phyA Direct Target Genes.
Supplemental Figure 9. WEGO Analysis of phyA-Associated Genes Compared with Random.
Supplemental Figure 10. phyA Fails to Associate with Two G-Box–Containing Regions on the NAC019 Promoter in the Absence of HY5.
Supplemental Figure 11. Supershifted EMSA Shows that the HY5 Binding Site G-Box Is Critical to the Recruitment of the phyA-FHY1-HY5 Complex.
Supplemental Table 1. Correspondence between phyA Peaks and phyA-Associated Genes.
Supplemental Table 2. List of Motifs Enriched in Promoters of phyA-Associated or phyA-Regulated Genes.
Supplemental Table 3. List of Motifs Enriched in Both phyA-Associated and phyA-Regulated Promoters.
Supplemental Table 4. List of Primers Used in This Study.
Supplemental Data Set 1. phyA Peaks by ChIP-seq.
Supplemental Data Set 2. Annotation of phyA-Associated Genes, phyA-Regulated Genes, and phyA Direct Target Genes under FR.
Supplemental Data Set 3. phyA-Regulated Genes after 3 h of Exposure to FR.
Supplementary Material
Acknowledgments
We thank Matthew Davis and Cynthia Nezames for critical reading of the article. This work was supported by the National Institutes of Health (Grant GM47850 to X.W.D.), the Ministry of Science and Technology of China (Grant 2012CB910900), the State Key Laboratory of Protein and Plant Gene Research at Peking University, the Peking-Tsinghua Center for Life Sciences, and the Next-Generation BioGreen 21 Program (Grant PJ00901003), Rural Development Administration, Republic of Korea.
AUTHOR CONTRIBUTIONS
X.W.D. and F.C. designed the project. F.C. performed the research. B.L. and F.C. conducted bioinformatics analysis. F.C., B.L., G.L., M.D., X.S., and X.W.D. analyzed the data. F.C., J.-B.C., B.L., X.S., and X.W.D. wrote the article.
Glossary
- R
red light
- FR
far-red light
- ABA
abscisic acid
- ChIP-seq
chromatin immunoprecipitation sequencing
- RNA-seq
RNA sequencing
- ChIP
chromatin immunoprecipitation
- UTR
untranslated region
- ChIP-qPCR
chromatin immunoprecipitation–quantitative PCR
- Ler
Landsberg erecta
- qRT-PCR
quantitative RT-PCR
- GO
Gene Ontology
- GA
gibberellic acid
- EMSA
electrophoretic mobility shift assay
Footnotes
Online version contains Web-only data.
References
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Auge G.A., Rugnone M.L., Cortés L.E., González C.V., Zarlavsky G., Boccalandro H.E., Sánchez R.A. (2012). Phytochrome A increases tolerance to high evaporative demand. Physiol. Plant. 146: 228–235 [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Bauer D., Viczian A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adam E., Fejes E., Schafer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan M.T., Quail P.H. (1991). Phytochrome A overexpression inhibits hypocotyl elongation in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 88: 10806–10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q., Zhu L., Huq E. (2011). Multiple kinases promote light-induced degradation of PIF1. Plant Signal. Behav. 6: 1119–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Shi X., Chen L., Dai M., Zhou Z., Shen Y., Li J., Li G., Wei N., Deng X.W. (2012). Phosphorylation of FAR-RED ELONGATED HYPOCOTYL1 is a key mechanism defining signaling dynamics of phytochrome A under red and far-red light in Arabidopsis. Plant Cell 24: 1907–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloix C., Jenkins G.I. (2008). Interaction of the Arabidopsis UV-B-specific signaling component UVR8 with chromatin. Mol. Plant 1: 118–128 [DOI] [PubMed] [Google Scholar]
- de Lucas M., Davière J.M., Rodríguez-Falcón M., Pontin M., Iglesias-Pedraz J.M., Lorrain S., Fankhauser C., Blázquez M.A., Titarenko E., Prat S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Desnos T., Puente P., Whitelam G.C., Harberd N.P. (2001). FHY1: A phytochrome A-specific signal transducer. Genes Dev. 15: 2980–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T., Schweizer F., Tscheuschler A., Debrieux D., Casal J.J., Schäfer E., Hiltbrunner A., Fankhauser C. (2008). FHY1 mediates nuclear import of the light-activated phytochrome A photoreceptor. PLoS Genet. 4: e1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Zhou J., Elling A.A., Charron J.B., Deng X.W. (2008). Histone modifications and expression of light-regulated genes in Arabidopsis are cooperatively influenced by changing light conditions. Plant Physiol. 147: 2070–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo K., Ugawa Y., Iwamoto M., Korenaga T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltbrunner A., Tscheuschler A., Viczián A., Kunkel T., Kircher S., Schäfer E. (2006). FHY1 and FHL act together to mediate nuclear accumulation of the phytochrome A photoreceptor. Plant Cell Physiol. 47: 1023–1034 [DOI] [PubMed] [Google Scholar]
- Hiltbrunner A., Viczián A., Bury E., Tscheuschler A., Kircher S., Tóth R., Honsberger A., Nagy F., Fankhauser C., Schäfer E. (2005). Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr. Biol. 15: 2125–2130 [DOI] [PubMed] [Google Scholar]
- Hornitschek P., Kohnen M.V., Lorrain S., Rougemont J., Ljung K., López-Vidriero I., Franco-Zorrilla J.M., Solano R., Trevisan M., Pradervand S., Xenarios I., Fankhauser C. (2012). Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Hudson M.E., Quail P.H. (2003). Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 133: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.W., Liu M.J., Chen I.C., Huang C.H., Chao L.Y., Hsieh H.L. (2010). A glutathione S-transferase regulated by light and hormones participates in the modulation of Arabidopsis seedling development. Plant Physiol. 154: 1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C., Hersch M., Trevisan M., Genoud T., Hiltbrunner A., Bergmann S., Fankhauser C. (2012). Nuclear phytochrome A signaling promotes phototropism in Arabidopsis. Plant Cell 24: 566–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami C., Lorrain S., Hornitschek P., Fankhauser C. (2010). Light-regulated plant growth and development. Curr. Top. Dev. Biol. 91: 29–66 [DOI] [PubMed] [Google Scholar]
- Kim L., Kircher S., Toth R., Adam E., Schäfer E., Nagy F. (2000). Light-induced nuclear import of phytochrome-A:GFP fusion proteins is differentially regulated in transgenic tobacco and Arabidopsis. Plant J. 22: 125–133 [DOI] [PubMed] [Google Scholar]
- Kircher S., Gil P., Kozma-Bognár L., Fejes E., Speth V., Husselstein-Muller T., Bauer D., Adám E., Schäfer E., Nagy F. (2002). Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm. Plant Cell 14: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Deng X.W. (2010). Plant hormone signaling lightens up: Integrators of light and hormones. Curr. Opin. Plant Biol. 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lee K.P., Piskurewicz U., Turečková V., Carat S., Chappuis R., Strnad M., Fankhauser C., Lopez-Molina L. (2012). Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 26: 1984–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Quail P.H. (2011). PIFs: Pivotal components in a cellular signaling hub. Trends Plant Sci. 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P., Monte E., Oka Y., Liu T., Carle C., Castillon A., Huq E., Quail P.H. (2008). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., et al. (2012). Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R., Ding L., Casola C., Ripoll D.R., Feschotte C., Wang H. (2007). Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Cohen J.D., Gardner G. (2011). Low-fluence red light increases the transport and biosynthesis of auxin. Plant Physiol. 157: 891–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y.H., et al. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38: 431–440 [DOI] [PubMed] [Google Scholar]
- Lorrain S., Allen T., Duek P.D., Whitelam G.C., Fankhauser C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. (2005). BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Martínez-García J.F., Huq E., Quail P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288: 859–863 [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Nakashima K., Takasaki H., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819: 97–103 [DOI] [PubMed] [Google Scholar]
- Oh E., Kang H., Yamaguchi S., Park J., Lee D., Kamiya Y., Choi G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Zhu J.Y., Wang Z.Y. (2012). Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Ouyang X., et al. (2011). Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T., Shimura Y., Okada K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E., Park J., Kim J., Nagatani A., Lagarias J.C., Choi G. (2012). Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 72: 537–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park P.J. (2009). ChIP-seq: Advantages and challenges of a maturing technology. Nat. Rev. Genet. 10: 669–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranik S., Sahu P.P., Srivastava P.S., Prasad M. (2012). NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 17: 369–381 [DOI] [PubMed] [Google Scholar]
- Rausenberger J., Tscheuschler A., Nordmeier W., Wüst F., Timmer J., Schäfer E., Fleck C., Hiltbrunner A. (2011). Photoconversion and nuclear trafficking cycles determine phytochrome A’s response profile to far-red light. Cell 146: 813–825 [DOI] [PubMed] [Google Scholar]
- Robson F., Okamoto H., Patrick E., Harris S.R., Wasternack C., Brearley C., Turner J.G. (2010). Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu K.S., Hagely K., Neff M.M. (2012). Genetic interactions between brassinosteroid-inactivating P450s and photomorphogenic photoreceptors in Arabidopsis thaliana. G3 (Bethesda) 2: 1585–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., Nambara E., Choi G., Yamaguchi S. (2009). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69: 463–472 [DOI] [PubMed] [Google Scholar]
- Shen Y., Feng S., Ma L., Lin R., Qu L.J., Chen Z., Wang H., Deng X.W. (2005). Arabidopsis FHY1 protein stability is regulated by light via phytochrome A and 26S proteasome. Plant Physiol. 139: 1234–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman J.M., Hwang Y.S., Quail P.H. (2006). phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 48: 728–742 [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Zhu T., Chang H.S., Wang X., Quail P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L.S., Nakashima K., Sakuma Y., Simpson S.D., Fujita Y., Maruyama K., Fujita M., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. (2013). Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31: 46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam G.C., Johnson E., Peng J., Carol P., Anderson M.L., Cowl J.S., Harberd N.P. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.W., Jang I.C., Henriques R., Chua N.H. (2009). FAR-RED ELONGATED HYPOCOTYL1 and FHY1-LIKE associate with the Arabidopsis transcription factors LAF1 and HFR1 to transmit phytochrome A signals for inhibition of hypocotyl elongation. Plant Cell 21: 1341–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., He H., Wang X., Wang X., Yang X., Li L., Deng X.W. (2011). Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J. 65: 346–358 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Mayba O., Pfeiffer A., Shi H., Tepperman J.M., Speed T.P., Quail P.H. (2013). A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 9: e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Chipperfield H., Melton D.A., Wong W.H. (2007). A gene regulatory network in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 104: 16438–16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.