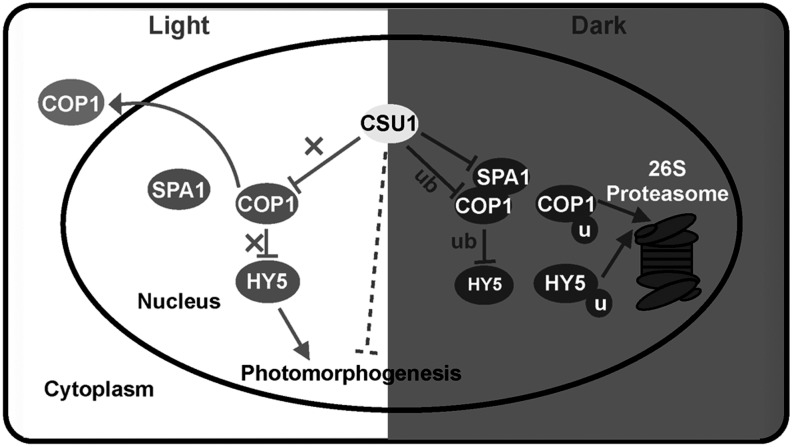
This study identifies the E3 ubiquitin ligase COP1 SUPPRESSOR1 (CSU1) as a negative regulator of COP1. A CSU1 mutant suppresses cop1-6 phenotypes in the dark. CSU1 colocalizes with COP1 and decreases COP1 protein accumulation in darkness, fine-tuning COP1 homeostasis by targeting COP1 for ubiquitination and degradation to ensure tight control of photomorphogenic development in darkness.
Abstract
CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) functions as an E3 ubiquitin ligase in both plants and animals. In dark-grown Arabidopsis thaliana seedlings, COP1 targets photomorphogenesis-promoting factors for degradation to repress photomorphogenesis. Little is known, however, about how COP1 itself is regulated. Here, we identify COP1 SUPPRESSOR1 (CSU1), a RING-finger E3 ubiquitin ligase, as a regulator of COP1. Genetic evidence demonstrates that csu1 mutations suppress cop1-6 phenotypes completely in the dark. Furthermore, CSU1 colocalizes with COP1 in nuclear speckles and negatively regulates COP1 protein accumulation in darkness. CSU1 can ubiquitinate COP1 in vitro and is essential for COP1 ubiquitination in vivo. Therefore, we conclude that CSU1 plays a major role in maintaining COP1 homeostasis by targeting COP1 for ubiquitination and degradation in dark-grown seedlings.
INTRODUCTION
In the dark and light, plants undergo two distinct developmental programs. The dark developmental program, known as etiolation or skotomorphogenesis, allows seedlings to penetrate soil and reach the surface, where photoautotrophic growth can commence. Upon reaching light, seedling development switches to photomorphogenesis, a phase characterized by the inhibition of hypocotyl elongation, expansion of the cotyledons, and acquisition of photosynthetic capacity (Sullivan and Deng, 2003). During deetiolation, light perceived by photoreceptors induces a cascade resulting in massive transcriptional reprogramming (Ma et al., 2001).
Forward genetic screens have identified a group of loss-of-function mutants that display photomorphogenic growth, even in darkness. The recessive nature of these cop/det/fus mutants indicates that COP/DET/FUS proteins function to suppress deetiolation in the dark (Wei and Deng, 1996). For example, the E3 ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1) acts as a master regulator of the transition from etiolated to deetiolated development. COP1 mediates the ubiquitin-dependent degradation of many key players in light-regulated plant development, such as LONG HYPOCOTYL5 (HY5), HY5 HOMOLOG, LONG AFTER FAR-RED LIGHT1 (LAF1), LONG HYPOCOTYL IN FAR-RED1 (HFR1), CONSTANS, SALT TOLERANCE HOMOLOG3 (STH3/BBX22), EARLY FLOWERING3, and GIGANTEA, as well as phytochrome photoreceptors (Osterlund et al., 2000; Holm et al., 2002; Saijo et al., 2003; Seo et al., 2003, 2004; Jang et al., 2005, 2008, 2010; Datta et al., 2008; Yu et al., 2008). However, in the case of phytochrome A (phyA), a recent study showed that COP1 plays a conditional role in phyA degradation, depending on the presence of metabolizable sugar in the growth medium (Debrieux et al., 2013).
Light regulates the nuclear abundance of COP1, as the abundance of COP1 protein in the nucleus increases in darkness and decreases in response to light. Thus, it has been suggested that nucleocytoplasmic partitioning may regulate COP1 function. In the dark, COP1 ubiquitinates a number of regulators for degradation via the 26S-proteasome pathway, thereby suppressing photomorphogenic development and promoting skotomorphogenic development. Upon light illumination, COP1 shuttles from the nucleus to the cytoplasm, which, in turn, releases the suppression of photomorphogenic development (von Arnim and Deng, 1994; von Arnim et al., 1997; Pacín et al., 2013).
The differential gene expression induced by far-red (FR) and red (R) light indicates that phytochromes might be involved in the downregulation of COP1 (Tepperman et al., 2001, 2004). The mechanism by which phytochromes regulate COP1 activity, however, remains largely unknown. FIN219, a positive regulator of phyA-mediated FR light signaling, negatively regulates COP1 protein levels by physically interacting with COP1 under continuous FR light and modulating the subcellular location of COP1 in FR light and dark conditions (Wang et al., 2011). In the absence of a light signal, COP1 forms stable core complexes with SPA family proteins (Zhu et al., 2008) and works in concert with CUL4 and DDB1 to ubiquitinate photomorphogenesis-promoting transcriptional regulators for degradation (Chen et al., 2010). In response to blue (B) light, photoactivated CRY1 and CRY2 both interact with SPA1. In order to suppress COP1 activity, the CRY1–SPA1 interaction, in turn, promotes the dissociation of COP1 from SPA1, while the association of CRY2 and SPA1 enhances the CRY2–COP1 interaction (Lian et al., 2011; Liu et al., 2011; Zuo et al., 2011). In contrast with its functions under R, FR, and B light, COP1 acts as a positive regulator in UV-B light–induced photomorphogenesis (Oravecz et al., 2006). Upon exposure to photomorphogenic UV-B light, the UV-B light receptor UVR8 monomerizes and interacts with COP1 (Favory et al., 2009; Rizzini et al., 2011; Wu et al., 2012). This UVR8–COP1 interaction, in turn, promotes the expression and stability of HY5, which then promotes COP1 expression via a positive feedback loop (Huang et al., 2012, 2013). In addition, FHY3 positively regulates UV-B light–induced photomorphogenesis by directly activating COP1 transcription in response to UV-B light (Huang et al., 2012).
COP1 functions as a central repressor of seedling photomorphogenesis, but how COP1 itself is regulated remains elusive. In this article, we identify a COP1 suppressor, named CSU1, via a genetic screen for mutants that suppress the constitutive photomorphogenic phenotype of cop1-6 in darkness. We show that CSU1 colocalizes with COP1 in the nucleus in plant cells. Like COP1, CSU1 is an E3 ubiquitin ligase and negatively regulates COP1 protein accumulation by ubiquitinating COP1 in the dark. Collectively, our genetic and biochemical data demonstrate that Arabidopsis thaliana CSU1 plays a key role in regulating COP1 homeostasis in darkness.
RESULTS
csu1 Mutations Suppress the Constitutive Photomorphogenic Phenotype of cop1-6 in Darkness
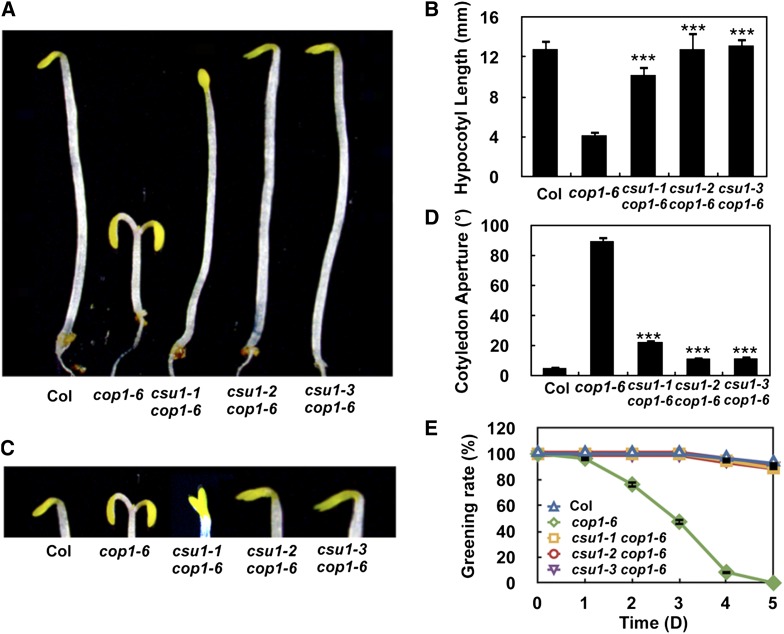
cop1 mutants undergo constitutive photomorphogenesis in the dark, exhibiting short hypocotyls and expanded cotyledons (Deng et al., 1991). To explore the factors regulating COP1 in dark-grown seedlings, we performed a genetic screen for mutants that suppress cop1-6, a weak allele of cop1. This screen identified an extragenic, recessive suppressor of cop1-6, which we named csu1, and genetic complementation tests revealed that we had isolated three independent alleles of csu1 (see Methods). The csu1 alleles showed better suppression of cop1 than most suppressors identified in our genetic screen, and consequently, we analyzed csu1 in detail. The cop1-6 mutation changes the splicing junction AG at the 3′ end of intron 4 to GG and leads to four cryptically spliced products at intron 4 (McNellis et al., 1994a). PCR analysis of the splicing pattern of COP1 showed that the mutations in CSU1 had no effect on the splicing of COP1-6 mRNA and little effect on the abundance of COP1-6 transcripts (Supplemental Figure 1).
In the dark, the csu1 cop1-6 double mutants displayed phenotypes that resemble the wild type (Figure 1). Similarly, for all three independent alleles of csu1, the csu1 cop1-6 mutant lines exhibited long hypocotyls similar to those of wild-type seedlings (Figures 1A and 1B). Although csu1 cop1-6 displayed slightly more expanded cotyledons than did the wild type, they exhibited a significantly smaller cotyledon aperture than did the cop1-6 mutant (Figures 1C and 1D). Furthermore, the three csu1 alleles completely rescued the greening defects observed in cop1 mutants grown in darkness at various time points (Figure 1E). Interestingly, csu1 cop1-6 double mutant seedlings exhibited intermediate hypocotyl lengths compared with wild-type and cop1-6 single mutant seedlings under continuous white, B, FR, and R light conditions (Supplemental Figure 2). Moreover, csu1 partially rescued the dwarf phenotype of cop1 under long-day conditions (16 h of light/8 h of dark) for 15 or 30 d (Supplemental Figure 3). Taken together, these genetic data demonstrate that csu1 completely suppresses cop1 in the dark but only partially suppresses cop1 in the light.
Figure 1.
csu1 Suppresses cop1 in Darkness.
(A) and (B) Hypocotyl lengths of 5-d-old wild-type, cop1-6, and csu1 cop1-6 seedlings grown in the dark. Error bars represent se (n ≥ 20). ***P < 0.001 (Student’s t test) for the differences between csu1 cop1-6 and cop1-6.
(C) and (D) Cotyledon phenotypes and separation angle of 5-d-old wild-type, cop1-6, and csu1 cop1-6 seedlings grown in the dark. Data represent means and se of at least 30 seedlings. ***P ≤ 0.001 (Student’s t test) relative to cop1-6.
(E) Quantification of photobleaching. Seedlings grown in the dark for various time periods were transferred to white light for 3 d, and the number of green seedlings was counted. The degree of photobleaching was expressed as the percentage of green seedlings. A total of 100 seedlings were used for each time point.
Positional Cloning of CSU1
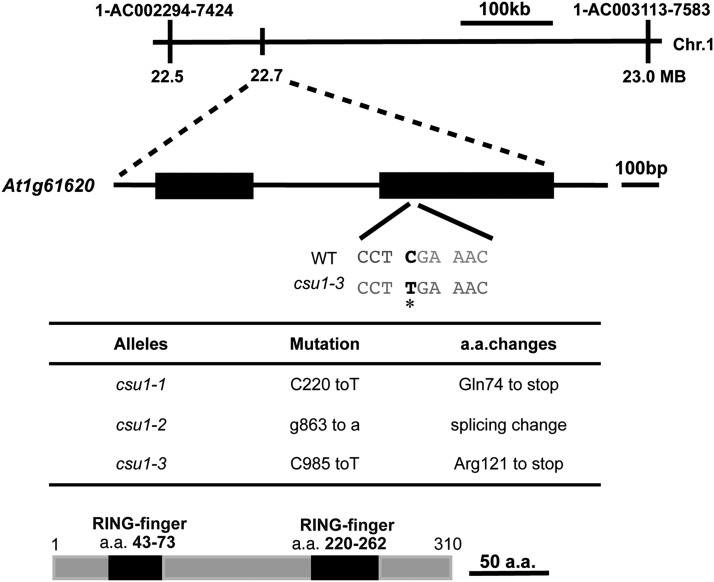
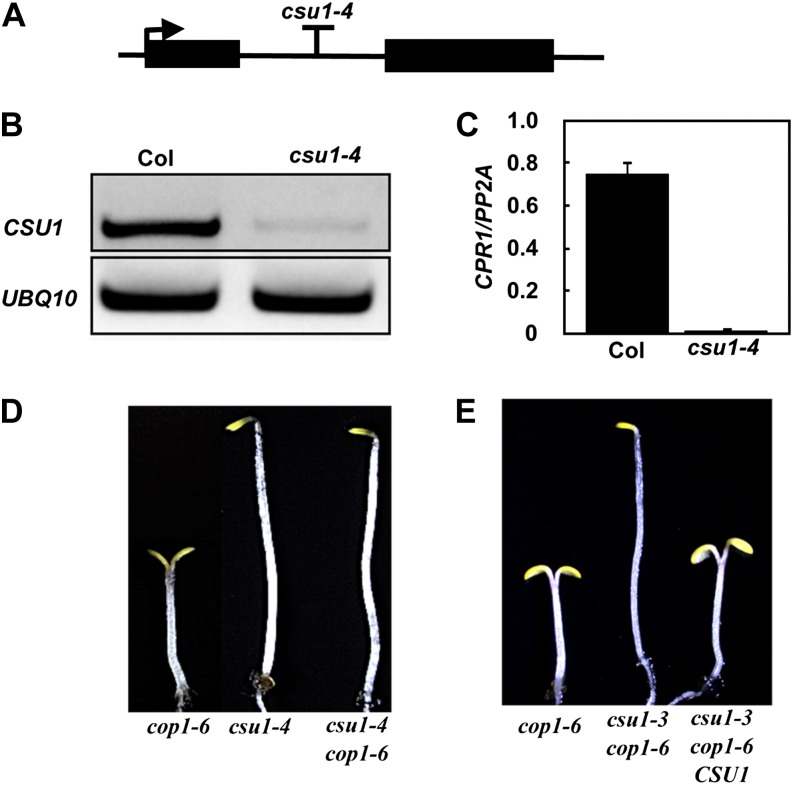
Using a map-based cloning strategy, we mapped CSU1 to an ∼500-kb region between markers 1-AC002294-7424 and 1-AC003113-7583 on the bottom arm of chromosome 1 (Figure 2A). Using SOLiD System Sequencing, we identified a mutation in At1g61620 in csu1-3 (Figure 2A). The mutation in csu1-3 is a C-to-T substitution at position 985 from the start codon, which creates a premature stop codon (Figure 2A). To determine whether other csu1 alleles also contain mutations in the same gene, we used PCR to amplify At1g61620 from csu1-1 and csu1-2 mutants and then sequenced the PCR products. This identified a premature stop codon in csu1-1 and a splice site change in csu1-2 in At1g61620 (Figure 2B). To further confirm that the observed suppression of cop1-6 was indeed caused by the disruption of At1g61620, we obtained a T-DNA insertion mutant (SALK_060493; named csu1-4) from the European Arabidopsis Stock Centre (Figures 3A to 3C) and generated the double mutant, csu1-4 cop1-6. As expected, csu1-4 in the double mutant completely suppressed the cop1-6 mutation in the dark (Figure 3D). Furthermore, csu1-3 cop1-6 transformed with the genomic fragment containing the full-length CSU1 gene (csu1-3 cop1-6 CSU1) displayed a cop1-6 phenotype in the dark (Figure 3E), indicating that a functional CSU1 gene indeed complements the phenotype conferred by csu1 cop1-6 in darkness.
Figure 2.
Map-Based Cloning of CSU1.
(A) Map of the CSU1 locus and CSU1 gene structure. The exon is represented by boxes, and the intron is represented by the line. The interval linked to the mutation was sequenced and the base substitution identified. The mutation in csu1-3 is marked in black with an asterisk.
(B) Mutations identified in the csu1 alleles, and the consequences of mutations to the CSU1 protein.
(C) Protein structure of CSU1. a.a., amino acids.
Figure 3.
Genomic Complementation Test.
(A) Schematic representation of the CSU1 gene (At1g61620). The arrow indicates the position of the start Met, and the T indicates the T-DNA insertion position.
(B) and (C) RT-PCR and quantitative real-time RT-PCR showing CSU1 transcript accumulation in wild-type and csu1-4 seedlings 5 d after germination under long-day conditions.
(D) csu1-4 suppresses cop1-6 in the dark. Seedlings were grown on GM medium plates for 5 d in darkness.
(E) Genomic complementation test. Representative cop1-6, csu1-3 cop1-6, and csu1-3 cop1-6 CSU1 seedlings grown on GM medium plates for 5 d under dark conditions are shown.
[See online article for color version of this figure.]
CSU1 encodes a RING finger protein (Figure 2C) and has no specified function in Arabidopsis. Database searches identified CSU1 as a single-copy gene in Arabidopsis, encoding a protein that shares 29.1% amino acid sequence identity with the nitric oxide synthase–interacting protein (NOSIP)/mammalian receptor-associated ubiquitin ligase (RUL), which interacts with the erythropoietin receptor (EpoR) in mammalian cells and mediates the ubiquitination of EpoR (Friedman et al., 2003). Further alignment analysis of CSU1 with its homologs from Caenorhabditis elegans, Drosophila melanogaster, Mus musculus, Danio rerio, and Oryza sativa revealed that these homologous proteins are highly conserved, exhibiting 25, 26.6, 28.4, 30.7, and 70.8% amino acid sequence identity with these homologs, respectively (Supplemental Figure 4).
csu1 and hy5 Cosuppress cop1
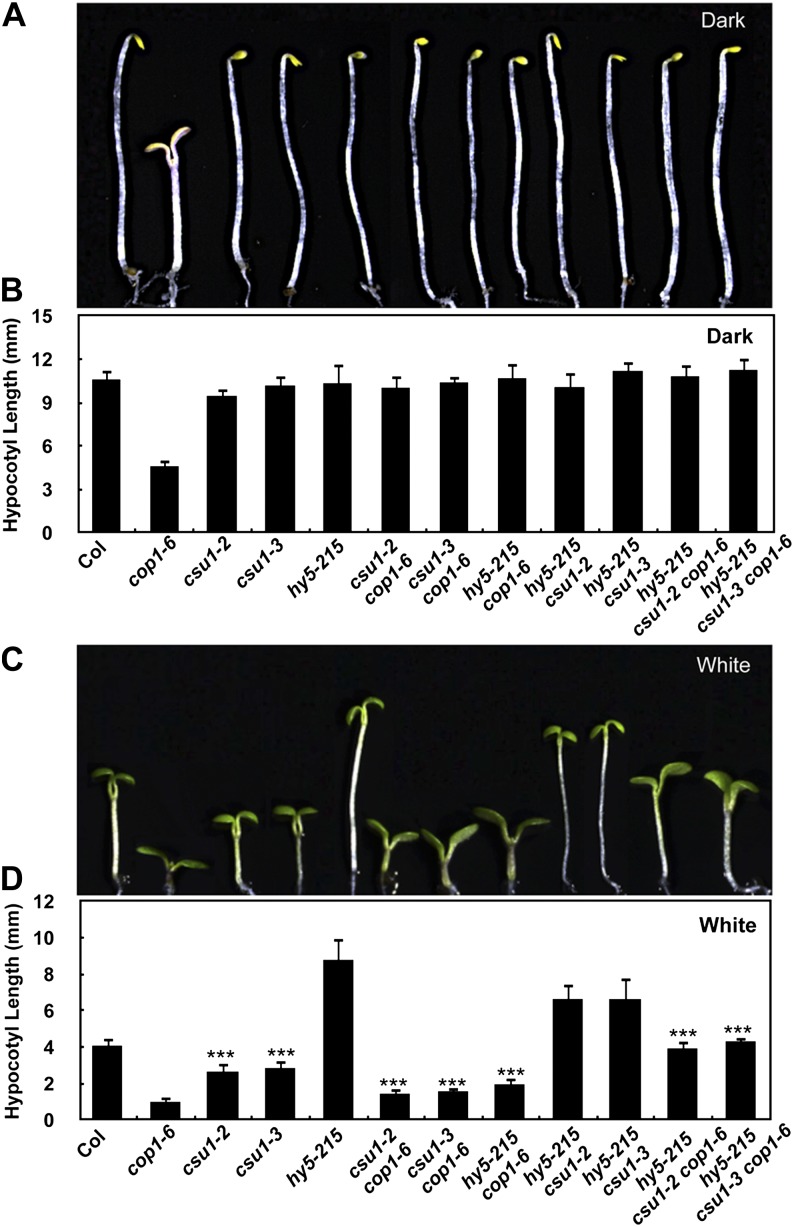
hy5 has been characterized as a recessive cop1 suppressor (Ang and Deng, 1994). HY5 acts downstream of COP1 and is ubiquitinated by COP1 for degradation in darkness (Ang et al., 1998; Osterlund et al., 2000; Saijo et al., 2003). Further genetic characterization has likewise demonstrated that csu1, together with hy5, continued to suppress cop1 in the dark (Figures 4A and 4B). However, under various light conditions (white, B, FR, and R light), either csu1 or hy5 alone only partially suppressed cop1-6, whereas csu1 and hy5 together completely suppressed cop1-6 (Figures 4C and 4D; Supplemental Figure 5). Interestingly, the csu1 single mutants developed shorter hypocotyls than did the wild type under continuous white, B, R, and FR light conditions (Figures 4C and 4D; Supplemental Figure 5), suggesting that CSU1 itself functions as a negative regulator of hypocotyl growth in the light. Thus, CSU1 and HY5 appear to play opposite roles in light-mediated inhibition of hypocotyl elongation. When grown in continuous white, B, FR, and R light conditions, the hypocotyl length of the hy5 csu1 double mutant was similar to that of hy5-215, indicating that hy5 is epistatic to csu1 (Figures 4C and 4D; Supplemental Figure 5). These results suggest that the short-hypocotyl phenotype of csu1 is dependent on a functional HY5 protein in the light.
Figure 4.
csu1 and hy5 Cosuppress cop1.
(A) and (B) Hypocotyl lengths of 5-d-old dark-grown wild-type and various mutant seedlings. Error bars represent se (n ≥ 20).
(C) and (D) Hypocotyl lengths of 5-d-old wild-type and various mutant seedlings grown under white light conditions (15 μmol m−2 s−1). Error bars represent se (n ≥ 20). ***P < 0.001 (Student’s t test) for the differences between the wild type and csu1, csu1 cop1-6 and cop1-6, hy5-215 csu1 and csu1, or hy5-215 csu1 cop1-6 and csu1 cop1-6.
[See online article for color version of this figure.]
CSU1 Is a Nuclear Protein and Colocalizes with COP1 in Nuclear Bodies in Darkness
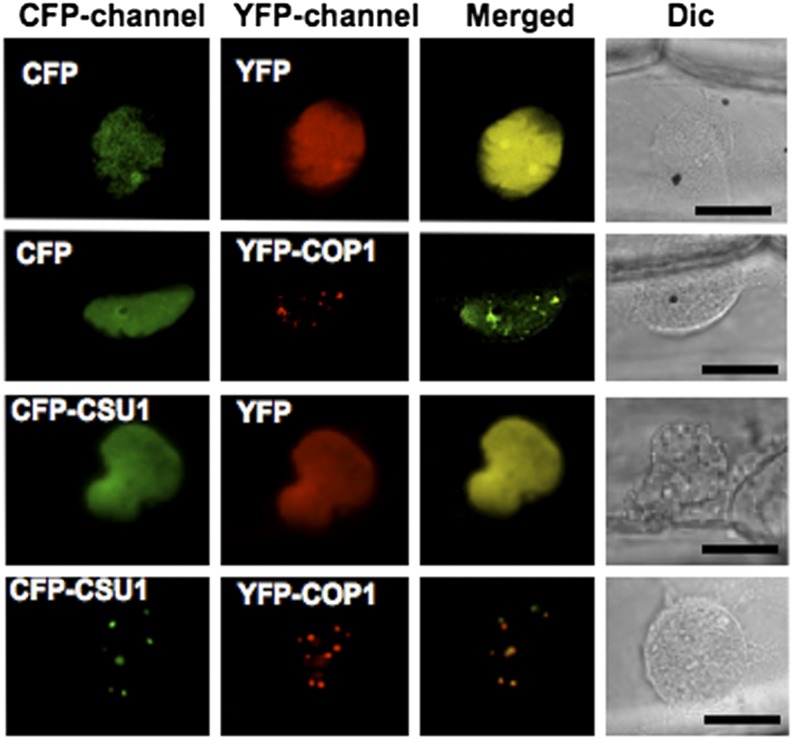
Previous studies showed that COP1 is capable of recruiting several interacting proteins to nuclear speckles (Ang et al., 1998; Holm et al., 2002; Seo et al., 2003; Jang et al., 2005; Datta et al., 2006, 2007, 2008). To determine whether CSU1 colocalizes with COP1 in vivo, we coexpressed yellow fluorescent protein (YFP)-tagged COP1 (YFP-COP1) and cyan fluorescent protein (CFP)-tagged CSU1 (CFP-CSU1) in onion (Allium cepa) epidermal cells, which were then incubated in darkness for 24 h. A strong uniform fluorescence with consistent nuclear speckles was observed when both CFP-CSU1 and YFP-COP1 were present (Figure 5). CFP-CSU1 produced a uniform fluorescence when it was expressed alone, and its localization in the nuclear speckles was detected only when coexpressed with YFP-COP1, suggesting that CSU1 is recruited into the nuclear speckles by COP1 in the dark.
Figure 5.
CSU1 Colocalizes with COP1 in NBs in the Dark.
Onion peels were cobombarded with equal amounts of the DNA constructs, as indicated, and incubated in darkness for 24 h. Epidermal cells were imaged using the CFP and YFP channels of a confocal microscope. CFP-channel, CFP channel image; YFP-channel, YFP channel image; Merged, merged images of CFP and YFP; Dic, differential interference contrast in light microscope mode. Bars = 20 μm.
CSU1 Negatively Regulates COP1 and SPA1 Protein Levels in Darkness
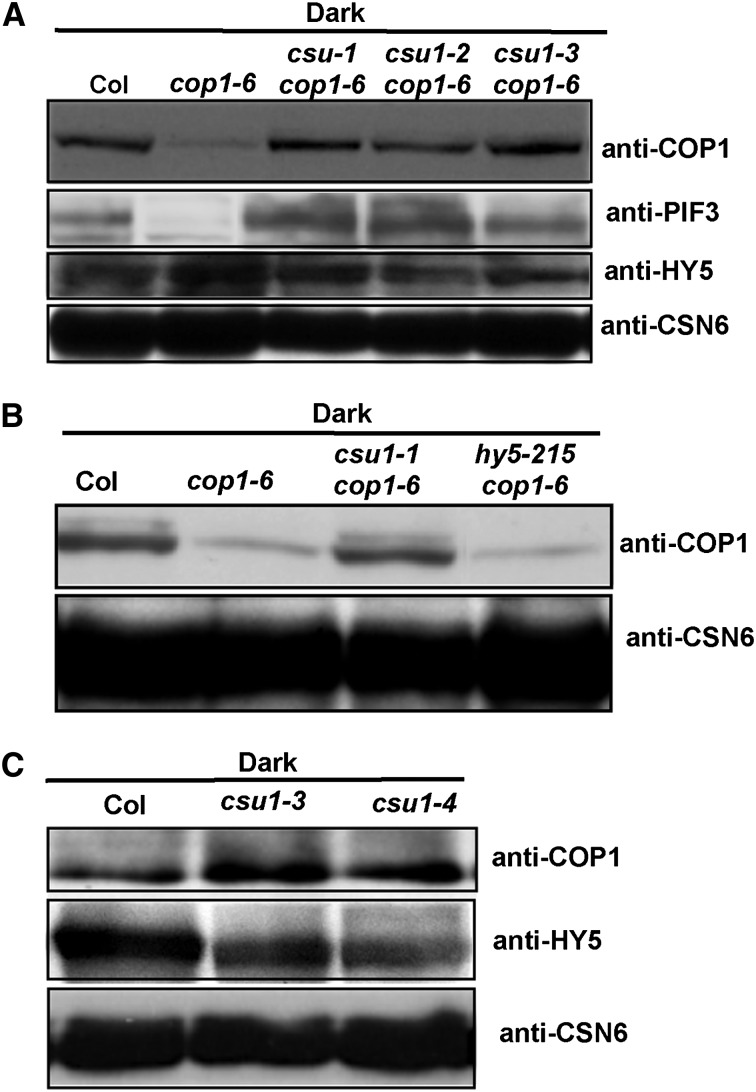
cop1-6 is a weak allele that encodes a mutant COP1 protein with five additional amino acids inserted between the amino acids 301 and 302 (McNellis et al., 1994a), albeit with significantly reduced abundance of COP1-6 protein in vivo (Figure 6A). In order to determine whether CSU1 regulates COP1 protein, we tested the COP1-6 protein level in csu1 cop1-6 double mutants. In 5-d-old dark-grown cop1-6 single mutants, significantly less COP1-6 protein accumulated. By contrast, an increased level of COP1-6 protein, similar to that observed in the wild type, was detected in all three csu1 cop1-6 double mutant lines (Figure 6A). COP1 positively regulates PIF3 protein accumulation (Bauer et al., 2004) and targets HY5 for degradation (Ang et al., 1998; Osterlund et al., 2000; Saijo et al., 2003) in dark-grown seedlings. Consistent with this notion, we detected less PIF3 but more HY5 protein in cop1-6 mutants. As expected, the amount of PIF3 protein increased whereas that of HY5 protein decreased in the csu1 cop1-6 double mutants (Figure 6A). These results suggest that the COP1-6 protein is functional, to a large extent, in the dark in vivo. Specifically, the restoration of COP1-6 protein levels by csu1 is likely the major reason why csu1 suppresses the cop1-6 phenotype in the dark. Notably, given that HY5 is a downstream substrate of COP1 (Ang et al., 1998; Osterlund et al., 2000; Saijo et al., 2003), mutation in HY5 completely suppresses cop1-6 in the dark (Ang and Deng, 1994) but has no effect on the COP1-6 protein level in cop1-6 mutant seedlings (Figure 6B). Therefore, CSU1 appears to behave like an upstream regulator of COP1 in darkness. Consistent with this notion, more COP1 but less HY5 protein accumulated in csu1 single mutants in 5-d-old seedlings grown in the dark (Figure 6C).
Figure 6.
CSU1 Negatively Regulates COP1 in the Dark.
(A) Protein levels of COP1, PIF3, and HY5 in wild-type, cop1-6, and csu1 cop1-6 seedlings as detected by COP1, PIF3, and HY5 antibodies, respectively.
(B) COP1 protein levels in wild-type, cop1-6, hy5-215 cop1-6, and csu1 cop1-6 seedlings.
(C) COP1 protein levels in wild-type and csu1 seedlings.
Plant total proteins were extracted from 5-d-old seedlings grown in the dark. CSN6 levels were used as loading controls.
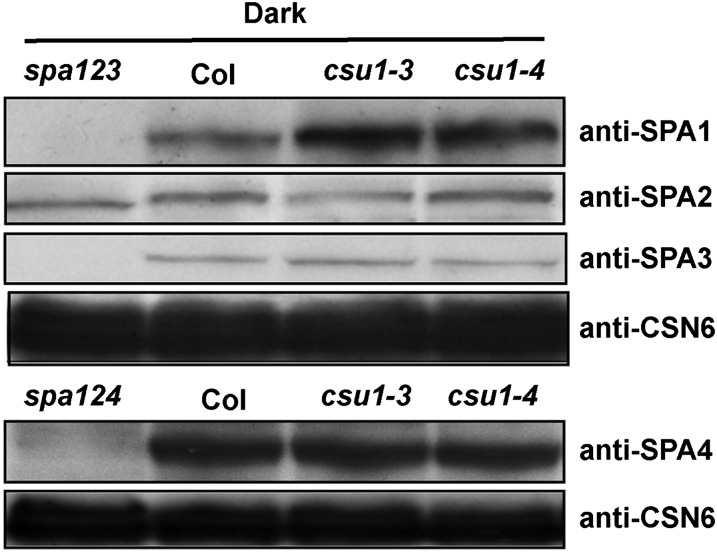
Two COP1 and two SPA proteins (SPA1 to SPA4) form a functional SPA-COP1 complex in darkness (Zhu et al., 2008), and SPA1 promotes the E3 ubiquitin ligase activity of COP1 (Saijo et al., 2003; Seo et al., 2003). Therefore, we determined the protein levels of four SPAs in etiolated wild-type and csu1 mutant seedlings. Interestingly, higher levels of SPA1 accumulated in csu1 mutants, whereas the abundance of SPA2, SPA3, and SPA4 was not obviously altered in csu1 mutants compared with that in the wild type in darkness (Figure 7), indicating that, out of four SPAs, CSU1 specifically negatively regulates SPA1 in the dark.
Figure 7.
CSU1 Negatively Regulates SPA1 in the Dark.
Protein levels of SPA1, SPA2, SPA3, and SPA4 in wild-type and csu1 seedlings were detected by SPA1, SPA2, SPA3, and SPA4 antibodies, respectively. Plant total proteins were extracted from 5-d-old seedlings grown in the dark. spa123 and spa124 were used as negative controls. CSN6 levels were used as loading controls.
To further investigate how COP1 is regulated by CSU1, we examined the COP1 protein levels in light-grown csu1 mutant seedlings. Interestingly, under white light, the protein levels of COP1, HY5, and phyB in csu1 were similar to those in the wild type (Supplemental Figure 6). In addition, no obvious changes were observed for phyA levels in csu1 and the wild type in FR light (Supplemental Figure 7A). Similarly, phyB accumulated at comparable levels in the wild type and csu1 mutants under R light (Supplemental Figure 7B). In response to FR and R light, PIF3 protein was degraded in both wild-type and csu1 mutant seedlings (Supplemental Figure 7). Together, all these results indicate that mutation of CSU1 did not change the protein levels of COP1, HY5, phyA, phyB, or PIF3 in the light.
Ubiquitination of COP1 by CSU1
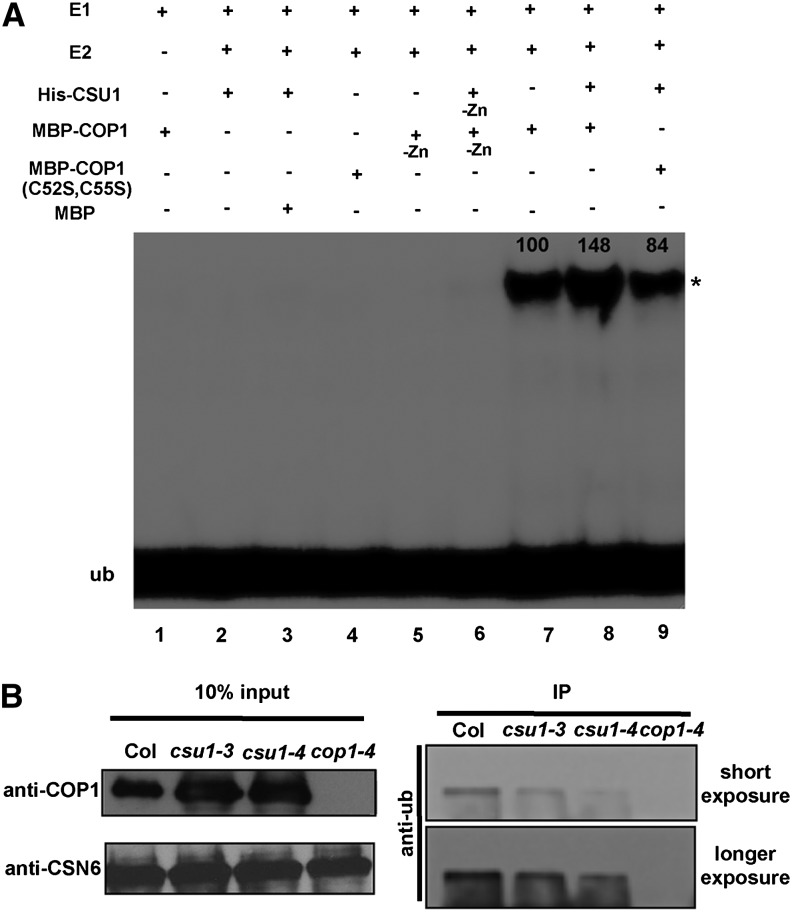
CSU1 contains two RING-finger domains (Figure 2C) and thus likely functions as an E3 ubiquitin ligase (Kraft et al., 2005; Stone et al., 2005). To test this hypothesis, we examined the self-ubiquitination activity of CSU1 in an in vitro ubiquitination assay. Interestingly, we observed that CSU1 was unable to ubiquitinate itself (Figure 8A, lane 2). Given that CSU1 negatively regulates COP1 at the protein level, we next performed a series of in vitro ubiquitination assays to investigate whether CSU1 could ubiquitinate COP1. Consistent with a previous report (Seo et al., 2003), COP1 possessed self-ubiquitination activity dependent on the presence of Zn2+ ions (Figure 8A, lanes 5 and 7). However, when we added CSU1 together with COP1 in the reaction, the amount of ubiquitinated COP1 increased significantly (Figure 8A, lane 8). Similarly, CSU1 was also able to ubiquitinate the COP1 mutant (C52S and C55S) proteins previously shown to lack E3 ubiquitin ligase activity (Seo et al., 2003) (Figure 8A, lanes 4 and 9). Thus, these in vitro data suggest that CSU1 possesses the ability to ubiquitinate COP1. To further test whether CSU1 contributes to COP1 ubiquitination in vivo, we performed an immunoprecipitation assay as illustrated in Figure 8B. As expected, more ubiquitinated COP1 protein was detected in the wild type than in csu1 single mutant seedlings grown in the dark, implying that the loss of CSU1 results in reduced COP1 ubiquitination in dark-grown seedlings. Together, these data support the conclusion that CSU1 is able to ubiquitinate COP1 both in vitro and in vivo.
Figure 8.
CSU1 Ubiquitinates COP1.
(A) Ubiquitination assays were performed using UBE1 (E1), UbcH5b (E2), and HA-tagged ubiquitin (HA-Ub). Recombinant 6× His-CSU1 and MBP-COP1 were preincubated with 20 μM zinc acetate. MBP, nonincubated 6× His-CSU1 (-Zn), and MBP-COP1 (-Zn) were used as negative controls. Ubiquitinated MBP-COP1 was detected using anti-HA antibody. Asterisk indicates ubiquitinated MBP-COP1 (anti-HA). Values were calculated using ImageJ software.
(B) Immunoblots against ubiquitin showing that loss of CSU1 results in the reduction of COP1 ubiquitination in vivo. COP1 protein was immunoprecipitated by COP1 antibody and then blotted with monoclonal ubiquitin antibody (anti-Ub). Wild-type and csu1 seedlings were grown in the dark for 5 d. cop1-4 was used as a negative control. IP, immunoprecipitation.
DISCUSSION
Mammalian COP1 is a known tumor suppressor whose downregulation promotes prostatic epithelial cell proliferation and tumorigenesis. Specifically, it regulates p53 protein level by acting as an E3 ubiquitin ligase of p53 (Dornan et al., 2004; Migliorini et al., 2011; Vitari et al., 2011). Human NOSIP/RUL, a homolog of Arabidopsis CSU1, also acts as an E3 ubiquitin ligase and mediates the ubiquitination of EpoR. NOSIP/RUL mutants lacking the ubiquitin ligase activity are known to inhibit the cytokine-induced expression of immediate-early genes associated with the growth and survival of factor-dependent cells (Friedman et al., 2003).
In Arabidopsis, COP1, an E3 ubiquitin ligase, colocalizes with HY5, HFR1, and LAF1 in nuclear bodies (NBs) and targets these proteins for degradation in darkness (Ang et al., 1998; Osterlund et al., 2000; Saijo et al., 2003; Seo et al., 2003, 2004). COP1 also colocalizes with phyA in NBs (Jang et al., 2005) and contributes to its degradation in the presence of metabolizable sugar (Debrieux et al., 2013). phyB also localizes in NBs in a PIF3-dependent manner (Bauer et al., 2004) and is ubiquitinated by COP1 for degradation by the 26S-proteasome pathway. It has been proposed that the phyB–PIFs–COP1 interaction takes place in NBs (Jang et al., 2010). Similar to the clastosomes in animal cells (Lafarga et al., 2002), these NBs have also been proposed as sites for protein degradation (Seo et al., 2003). Consistent with this hypothesis, we observed that CSU1 and COP1 colocalize in NBs (Figure 5). In this case, COP1 was unable to ubiquitinate CSU1 (Figure 8A, lanes 8 and 9) but was ubiquitinated by CSU1 (Figures 8A and 8B). In mammalian cells, the ataxia telangiectasia mutated protein kinase directly phosphorylates COP1 on Ser-387 and initiates the auto-E3 degradation of COP1 (Dornan et al., 2006). In Arabidopsis, COP1 also can self-ubiquitinate (Seo et al., 2003). Therefore, it is possible that COP1 might be responsible for its own ubiquitination in dark-grown csu1 single mutant seedlings (Figure 8B). The extent to which photomorphogenic development of seedlings is inhibited by COP1 correlates quantitatively with the in vivo COP1 protein levels, while a moderate abundance of COP1 is essential for the appropriate regulation of photomorphogenic development (McNellis et al., 1994b; Stoop-Myer et al., 1999). Both COP1 and its dominant negative version with mutations in four Cys residues (C52S, C55S, C86S, and C89A) of the RING motif were labile and stabilized by the proteasome inhibitor MG132 in vivo, indicating that COP1 is proteasomally regulated (Seo et al., 2003, 2004). Thus, based on our results showing COP1-CSU1 colocalization in NBs in darkness in planta (Figure 5), COP1 ubiquitination by CSU1 in vitro (Figure 8A), and decreased COP1 ubiquitination (Figure 8B) but increased COP1 protein levels (Figure 6C) in dark-grown csu1 single mutant seedlings, we conclude that COP1 is targeted for proteasomal degradation by CSU1, another E3 ubiquitin ligase, in the nucleus in darkness.
Mutations in several B-box proteins that are involved in COP1-regulated processes, such as BBX4/COL3, BBX21/STH2, and BBX22/STH3, partially suppress cop1 in the dark (Datta et al., 2006, 2007, 2008). The recessive hy5 mutation completely suppressed cop1-6 (Ang and Deng, 1994) but did not affect COP1-6 protein levels in cop1-6 (Figure 5B). These data further demonstrate that HY5 is a downstream substrate of COP1. csu1 also completely restored the cop1-6 constitutive photomorphogenic phenotype (Figure 1). However, in contrast to hy5, this suppression was likely caused by the restoration of COP1-6 protein (Figure 6A), indicating that CSU1 possibly acts upstream of COP1. It is not surprising that the loss of HY5 and CSU1 together can completely suppress cop1 in the dark. In addition to darkness, COP1 also plays a critical role in regulating the development of light-grown plants (Deng et al., 1991, 1992; McNellis et al., 1994a); however, its exact molecular roles in the light remain largely unknown. The phenotypes of hy5 cop1, csu1 cop1, and hy5 csu1 cop1 mutants under various light conditions (white, R, FR, and B light) imply that either HY5 or CSU1 alone is capable of partially suppressing COP1 action and that the activities of both proteins completely suppress COP1 function in the light (Figures 4C and 4D; Supplemental Figure 5).
cop1 mutants display dramatically short hypocotyls (Deng et al., 1991, 1992). By contrast, overexpression of COP1 results in an increase in seedling hypocotyl length in the light (McNellis et al., 1994b). As shown in Figure 6C, more COP1 accumulated in the dark-grown csu1 single mutant seedlings. If the loss of CSU1 still results in more COP1 accumulation in the light, csu1 mutant seedlings should develop longer hypocotyls than do wild-type seedlings in the light. csu1 mutants, however, were hypersensitive to various light conditions and displayed shorter hypocotyls than the wild type (Figures 4C and 4D; Supplemental Figure 5). A possible explanation for this discrepancy is that the light-gown wild-type and csu1 mutant seedlings accumulate comparable levels of COP1, HY5, phyA, phyB, and PIF3 proteins (Supplemental Figures 6 and 7), which implies that the shorter hypocotyl phenotype of csu1 mutants is COP1 independent in the light. The fact that CSU1 does not appear to regulate COP1 in the light, at least at the COP1 protein level, might be due to the cytoplasmic localization of COP1 in the light (von Arnim and Deng, 1994). In darkness, COP1 is abundant in the nuclei, where it has the potential to target many photomorphogenesis-promoting factors, including HY5, LAF1, and HFR1, for ubiquitination and degradation and thus is capable of repressing photomorphogenesis. In the light, nuclear depletion of COP1 physically separates COP1 from its nuclear targets (von Arnim and Deng, 1994). This, in turn, would conceivably permit the accumulation of HY5, LAF1, and HFR1 and, consequently, allow the expression of the target genes of these proteins (Osterlund et al., 2000; Saijo et al., 2003; Seo et al., 2003; Jang et al., 2005). CSU1, however, may remain localized in the nucleus (Supplemental Figure 9) in the light. Therefore, it seems likely that the exportation of COP1 from the nucleus abolishes COP1-CSU1 colocalization in NBs and, subsequently, terminates the negative regulation of COP1 by CSU1 via proteasomal degradation in the light.
In conclusion, our findings demonstrate that CSU1 represses COP1 by ubiquitinating COP1 in a manner that fine-tunes COP1 homeostasis in the dark. This regulation, in turn, serves to ensure a tight control of photomorphogenic development in darkness (Figure 9).
Figure 9.
A Proposed Model Showing How CSU1 Regulates COP1.
In the dark, COP1 ubiquitinates downstream substrates, such as HY5, for degradation in order to repress photomorphogenesis. To maintain COP1 homeostasis, CSU1 targets COP1 for ubiquitination and degradation. In addition, CSU1 negatively regulates SPA1. In the light, COP1 is exported from the nucleus to the cytoplasm, whereas CSU1 still localizes in the nucleus. u, ubiquitin; ub, ubiquitination. Solid and dotted lines denote direct and indirect regulation, respectively.
METHODS
Plant Materials and Growth Conditions
The cop1-6 (McNellis et al., 1994a), hy5-215 (Ang and Deng, 1994), csu1-1 cop1-6, csu1-2 cop1-6, csu1-3 cop1-6, csu1-3, and csu1-4 (this study) mutants of Arabidopsis thaliana are in the Columbia (Col) ecotype background. Seeds were surface-sterilized with 30% commercial Clorox bleach and 0.02% Triton X-100 for 15 min. The seeds were then washed three times with sterile water and plated on 1× Murashige and Skoog medium supplemented with 0.4% Bacto-agar (Difco) and 1% Suc. The plates were kept at 4°C for 3 d for stratification and then transferred to light chambers that were maintained at a temperature of 22°C.
Genetics Screen, Identification, and Characterization
Genetic complementation tests showed that three different csu1 (csu1-1 cop1-6, csu1-2 cop1-6, and csu1-3 cop1-6 lines) ethyl methanesulfonate (EMS) mutations were allelic to each other. Homozygous mutant suppressor plants were crossed to wild-type plants (Col), and segregation in the F2 generations was analyzed in the dark to distinguish between intragenic and extragenic suppressors among three csu1 cop1-6 lines. The ratio of wild-type or suppressed phenotype to cop1-6 phenotype in the F2 population was around 13:3, indicating that three csu1 cop1-6 lines are extragenic suppressors. The three extragenic suppressor mutants were backcrossed to cop1-6. F1 generations showing the cop1-6 phenotype and the segregation patterns in the F2 generations (suppressor phenotype:cop1-6 phenotype ≈ 1:3) in the dark confirmed that the suppression phenotype is caused by a monogenic recessive mutation in all three alleles.
Map-Based Cloning of csu1-3
To generate the mapping population, we crossed csu1-3 cop1-6 (Col background) with cop1-6, which was backcrossed to Landsberg, eight times. F2 generation seeds were plated onto plates and grown in the dark for 5 d. The suppressed-phenotype seedlings (long hypocotyl and apical hook) were then picked for mapping. The markers used for mapping were designed based on the Arabidopsis Mapping Platform (http://amp.genomics.org.cn) and the standards described previously (Jander et al., 2002).
SOLiD Sequencing and Mutation Identification
The fragment libraries were created using the SOLiD Fragment library construction procedures according to the manufacturer’s instructions (Life Technologies). The libraries were sequenced on a SOLiD5500 sequencer according to the manufacturer’s instructions (Life Technologies). Mapping of sequencing reads to the Arabidopsis reference genome (TAIR10) and single nucleotide polymorphism (SNP) calling were accomplished using LifeScope version 2.5. SNPs were then sorted into four categories (EMS-induced homozygous, EMS-induced heterozygous, other homozygous, other heterozygous). Candidate homozygous EMS-induced SNPs were identified in windows with reduced heterozygosity in the regions identified by physical mapping using in-house scripts: a C→T transition at position 22,738,637 (TAIR10) within At1g61620.
For genetic complementation tests to determine allelism, three EMS mutant lines (csu1-1 cop1-6, csu1-2 cop1-6, and csu1-3 cop1-6) were crossed to each other. A full-length DNA fragment of At1g61620 was amplified from these two mutants by PCR and then sequenced.
Genomic Complementation Test
For the transgenic complementation test, a 3.1-kb genomic fragment containing the full-length CSU1 gene was inserted into the KpnI and PstI sites of the pFP100 vector (Bensmihen et al., 2004) containing an At2S3:E-GFP:35Ster cassette driving the expression of E-GFP in seeds in order to generate pFP100-CSU1. This construct was used to transform Agrobacterium tumefaciens GV3101 by the freeze-thaw method, which was then introduced into csu1-3 cop1-6 mutant plants via the floral dip method (Clough and Bent, 1998). Transgenic T1 seeds were selected using a Leica MZFL III stereomicroscope equipped with a GFP filter. T3 transgenic seeds were used for phenotypic analyses.
RT-PCR and Quantitative Real-Time RT-PCR
Total RNA was extracted from Arabidopsis seedlings using the RNeasy Plant Mini Kit (Qiagen). Then, cDNAs were synthesized from 2 μg of total RNA using the SuperScript II first-strand cDNA synthesis system (Fermentas) according to the manufacturer’s instructions. Quantitative PCR was performed using the CFX96 real-time PCR detection system (Applied Biosystems) and SYBR Green PCR Master Mix (Takara). PCR was performed in triplicate for each sample, and the expression levels were normalized to that of a PP2A gene. For the cop1-6 splicing test, PCR products were amplified using primers corresponding to the adjacent exons of COP1 and separated on a 4% agarose gel. All primers used for this assay are listed in Supplemental Table 1.
Measurement of Hypocotyl Length
To measure the hypocotyl length of seedlings, seeds were cold treated at 4°C for 3 d and then transferred to continuous white light for 8 h in order to induce uniform germination. The seeds were then transferred to various light conditions (white, B, R, and FR light) and incubated at 22°C for 5 d (Datta et al., 2008). The hypocotyl length of seedlings was measured using ImageJ software.
Colocalization Assay
Colocalization assay experiments were performed according to the standards outlined in previous research (Datta et al., 2008). The pAM-PAT-35SS-YFP-COP1 (Datta et al., 2008) and pAM-PAT-35SS-CFP-CSU1 constructs were introduced into onion (Allium cepa) epidermal cells by particle bombardment and incubated in darkness for 24 h. The cells were then analyzed by confocal microscopy.
Immunoblot Analysis and Immunoprecipitation
For all immunoblots, Arabidopsis wild-type or mutant seedlings were homogenized in a protein extraction buffer containing 50 mM Tris, pH 7.5, 200 mM NaCl, 1 mM EDTA, 10% glycerol, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and 1× complete protease inhibitor cocktail (Roche). For immunoprecipitation, the extracts were preincubated with Protein A-Sepharose (Sigma). The recovered supernatants were then incubated with the COP1 antibodies at 4°C for 3 h, followed by further incubation for 2 h with Protein A-Sepharose. The primary antibodies used in this study were anti-HY5 (Osterlund et al., 2000), anti-PIF3 (Al-Sady et al., 2006), anti-COP1 (Saijo et al., 2003), anti-SPA1, anti-SPA2, anti-SPA3, and anti-SPA4 (Zhu et al., 2008), and anti-CSN6 (Peng et al., 2001) antibodies.
In Vitro Ubiquitination Assays
In vitro ubiquitination assays were performed as described previously (Saijo et al., 2003) with minor modifications. Ubiquitination reaction mixtures (60 μL) contained 30 ng of UBE1 (E1; Boston Biochem), 30 ng of UbcH5b (E2; Boston Biochem), 10 μg of HA-tagged ubiquitin (HA-Ub; Boston Biochem), 200 ng of 6×His-CSU1, and 0.5 μg of MBP-COP1 (previously incubated with 20 μM zinc acetate) in a reaction buffer containing 50 mM Tris, pH 7.5, 10 mM MgCl2, 2 mM ATP, and 0.5 mM DTT. Either 6×His-CSU1 or MBP-COP1 that had not been incubated with zinc acetate and MBP were used as negative controls. After 2 h of incubation at 30°C, the reactions were stopped by adding 5× sample buffer. One-half of each mixture (30 μL) was then separated onto 8% SDS-PAGE gels. Ubiquitinated MBP-COP1 was detected using anti-HA antibody (Roche).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: CSU1 (AT1G61620), COP1 (AT2G32950), HY5 (AT5G11260), and PIF3 (AT1G09530).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. csu1 Has No Effect on the Splicing Pattern of COP1-6 mRNA.
Supplemental Figure 2. csu1 Partially Suppresses cop1 in the Light.
Supplemental Figure 3. The Morphology of Adult Wild-Type and Mutant Plants.
Supplemental Figure 4. Alignment of CSU1 with Its Homologs from Other Species.
Supplemental Figure 5. csu1 and hy5 Cosuppress cop1 in B, R, and FR Light.
Supplemental Figure 6. COP1 Protein Levels in the Light-Grown csu1 Mutant Seedlings.
Supplemental Figure 7. phyA, phyB, and PIF3 Protein Levels in csu1 Mutant Seedlings Grown in FR or R Light.
Supplemental Figure 8. Five Percent Input Used in Vitro Ubiquitination Assays.
Supplemental Figure 9. CSU1 Localizes to the Nucleus in the Light.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Abigail Coplin for critically reading the article. This work was supported by the National Basic Research Program of China (973 Program Grant 2012CB910900), the National Institutes of Health (Grant GM-47850), and the Peking-Tsinghua Center for Life Sciences (to X.W.D.); the Swedish Research Council, the Magnus Bergvalls Foundation, the Royal Physiographic Society in Lund, the Royal Swedish Academy of Agriculture and Forestry, the Adlerbert Research Foundation, and the Olle Engkvist Byggmästare Foundation (to M.H.); the PA Larsson Foundation (to D.X.), the Wenner-Gren Foundation (to Y.J.); the Next-Generation BioGreen 21 Program (Grant PJ00901003), Rural Development Administration, Republic of Korea; the Uppsala Genome Center and UPPMAX for providing assistance in massive parallel sequencing and computational infrastructure; and the National Genomics Infrastructure and Science for Life Laboratory in Uppsala.
AUTHOR CONTRIBUTIONS
D.X., M.H., and X.W.D. designed the project. D.X., F.L., Y.J., X.H., J.J.L., and C.H. performed the research. J.G.L. contributed to revision of the article. C.T.-R. conducted the bioinformatics analysis. D.X. and X.W.D. analyzed the data. D.X. and X.W.D. wrote the article.
Glossary
- FR
far-red
- R
red
- B
blue
- NB
nuclear body
- Col
Columbia
- EMS
ethyl methanesulfonate
- SNP
single nucleotide polymorphism
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Al-Sady B., Ni W., Kircher S., Schäfer E., Quail P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Ang L.H., Deng X.W. (1994). Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6: 613–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L.H., Chattopadhyay S., Wei N., Oyama T., Okada K., Batschauer A., Deng X.W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213–222 [DOI] [PubMed] [Google Scholar]
- Bauer D., Viczián A., Kircher S., Nobis T., Nitschke R., Kunkel T., Panigrahi K.C., Adám E., Fejes E., Schäfer E., Nagy F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S., To A., Lambert G., Kroj T., Giraudat J., Parcy F. (2004). Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Lett. 561: 127–131 [DOI] [PubMed] [Google Scholar]
- Chen H., Huang X., Gusmaroli G., Terzaghi W., Lau O.S., Yanagawa Y., Zhang Y., Li J., Lee J.H., Zhu D., Deng X.W. (2010). Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR OF PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell 22: 108–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi C., Johansson H., Holm M. (2007). SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell 19: 3242–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Hettiarachchi G.H., Deng X.W., Holm M. (2006). Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell 18: 70–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S., Johansson H., Hettiarachchi C., Irigoyen M.L., Desai M., Rubio V., Holm M. (2008). LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell 20: 2324–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrieux D., Trevisan M., Fankhauser C. (2013). Conditional involvement of CONSTITUTIVE PHOTOMORPHOGENIC1 in the degradation of phytochrome A. Plant Physiol. 161: 2136–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.W., Caspar T., Quail P.H. (1991). cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5: 1172–1182 [DOI] [PubMed] [Google Scholar]
- Deng X.W., Matsui M., Wei N., Wagner D., Chu A.M., Feldmann K.A., Quail P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71: 791–801 [DOI] [PubMed] [Google Scholar]
- Dornan D., Shimizu H., Mah A., Dudhela T., Eby M., O’Rourke K., Seshagiri S., Dixit V.M. (2006). ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 313: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Dornan D., Wertz I., Shimizu H., Arnott D., Frantz G.D., Dowd P., O’Rourke K., Koeppen H., Dixit V.M. (2004). The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429: 86–92 [DOI] [PubMed] [Google Scholar]
- Favory J.J., et al. (2009). Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28: 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A.D., Nimbalkar D., Quelle F.W. (2003). Erythropoietin receptors associate with a ubiquitin ligase, p33RUL, and require its activity for erythropoietin-induced proliferation. J. Biol. Chem. 278: 26851–26861 [DOI] [PubMed] [Google Scholar]
- Holm M., Ma L.G., Qu L.J., Deng X.W. (2002). Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O.S., Chen L., Wei N., Deng X.W. (2013). Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc. Natl. Acad. Sci. USA 110: 16669–16674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Ouyang X., Yang P., Lau O.S., Li G., Li J., Chen H., Deng X.W. (2012). Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell 24: 4590–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jander G., Norris S.R., Rounsley S.D., Bush D.F., Levin I.M., Last R.L. (2002). Arabidopsis map-based cloning in the post-genome era. Plant Physiol. 129: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Henriques R., Seo H.S., Nagatani A., Chua N.H. (2010). Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell 22: 2370–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I.C., Yang J.Y., Seo H.S., Chua N.H. (2005). HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 19: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Marchal V., Panigrahi K.C., Wenkel S., Soppe W., Deng X.W., Valverde F., Coupland G. (2008). Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 27: 1277–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft E., Stone S.L., Ma L., Su N., Gao Y., Lau O.S., Deng X.W., Callis J. (2005). Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarga M., Berciano M.T., Pena E., Mayo I., Castaño J.G., Bohmann D., Rodrigues J.P., Tavanez J.P., Carmo-Fonseca M. (2002). Clastosome: A subtype of nuclear body enriched in 19S and 20S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell 13: 2771–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian H.L., He S.B., Zhang Y.C., Zhu D.M., Zhang J.Y., Jia K.P., Sun S.X., Li L., Yang H.Q. (2011). Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 25: 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Zuo Z., Liu H., Liu X., Lin C. (2011). Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 25: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Li J., Qu L., Hager J., Chen Z., Zhao H., Deng X.W. (2001). Light control of Arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 13: 2589–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Araki T., Komeda Y., Miséra S., Deng X.W. (1994a). Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis T.W., von Arnim A.G., Deng X.W. (1994b). Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 6: 1391–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini D., Bogaerts S., Defever D., Vyas R., Denecker G., Radaelli E., Zwolinska A., Depaepe V., Hochepied T., Skarnes W.C., Marine J.C. (2011). Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J. Clin. Invest. 121: 1329–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oravecz A., Baumann A., Máté Z., Brzezinska A., Molinier J., Oakeley E.J., Adám E., Schäfer E., Nagy F., Ulm R. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell 18: 1975–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M.T., Hardtke C.S., Wei N., Deng X.W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Pacín M., Legris M., Casal J.J. (2013). COP1 re-accumulates in the nucleus under shade. Plant J. 75: 631–641 [DOI] [PubMed] [Google Scholar]
- Peng Z., Serino G., Deng X.W. (2001). Molecular characterization of subunit 6 of the COP9 signalosome and its role in multifaceted developmental processes in Arabidopsis. Plant Cell 13: 2393–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzini L., Favory J.J., Cloix C., Faggionato D., O’Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G.I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332: 103–106 [DOI] [PubMed] [Google Scholar]
- Saijo Y., Sullivan J.A., Wang H., Yang J., Shen Y., Rubio V., Ma L., Hoecker U., Deng X.W. (2003). The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes Dev. 17: 2642–2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Watanabe E., Tokutomi S., Nagatani A., Chua N.H. (2004). Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev. 18: 617–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang J.Y., Ishikawa M., Bolle C., Ballesteros M.L., Chua N.H. (2003). LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995–999 [DOI] [PubMed] [Google Scholar]
- Stone S.L., Hauksdóttir H., Troy A., Herschleb J., Kraft E., Callis J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoop-Myer C., Torii K.U., McNellis T.W., Coleman J.E., Deng X.W. (1999). The N-terminal fragment of Arabidopsis photomorphogenic repressor COP1 maintains partial function and acts in a concentration-dependent manner. Plant J. 20: 713–717 [DOI] [PubMed] [Google Scholar]
- Sullivan J.A., Deng X.W. (2003). From seed to seed: The role of photoreceptors in Arabidopsis development. Dev. Biol. 260: 289–297 [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Hudson M.E., Khanna R., Zhu T., Chang S.H., Wang X., Quail P.H. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38: 725–739 [DOI] [PubMed] [Google Scholar]
- Tepperman J.M., Zhu T., Chang H.S., Wang X., Quail P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98: 9437–9442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitari A.C., et al. (2011). COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature 474: 403–406 [DOI] [PubMed] [Google Scholar]
- von Arnim A.G., Deng X.W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035–1045 [DOI] [PubMed] [Google Scholar]
- von Arnim A.G., Osterlund M.T., Kwok S.F., Deng X.W. (1997). Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 114: 779–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.G., Chen C.H., Chien C.T., Hsieh H.L. (2011). FAR-RED INSENSITIVE219 modulates CONSTITUTIVE PHOTOMORPHOGENIC1 activity via physical interaction to regulate hypocotyl elongation in Arabidopsis. Plant Physiol. 156: 631–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N., Deng X.W. (1996). The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112: 871–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Hu Q., Yan Z., Chen W., Yan C., Huang X., Zhang J., Yang P., Deng H., Wang J., Deng X.W., Shi Y. (2012). Structural basis of ultraviolet-B perception by UVR8. Nature 484: 214–219 [DOI] [PubMed] [Google Scholar]
- Yu J.W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D., Maier A., Lee J.H., Laubinger S., Saijo Y., Wang H., Qu L.J., Hoecker U., Deng X.W. (2008). Biochemical characterization of Arabidopsis complexes containing CONSTITUTIVELY PHOTOMORPHOGENIC1 and SUPPRESSOR OF PHYA proteins in light control of plant development. Plant Cell 20: 2307–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo Z., Liu H., Liu B., Liu X., Lin C. (2011). Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis. Curr. Biol. 21: 841–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.