Two homeodomain transcription factors that are implicated in cell-type differentiation of the epidermis were found to have redundant functions in driving trichome morphogenesis in Arabidopsis. A detailed molecular genetic analysis led to the discovery of a regulatory link to a MYB-related transcription factor required for maintenance of trichome cell fate.
Abstract
The class IV homeodomain leucine zipper transcription factor GLABRA2 (GL2) acts in a complex regulatory circuit that regulates the differentiation of trichomes in Arabidopsis thaliana. We describe a genetic interaction with HOMEODOMAIN GLABROUS11 (HDG11), previously identified as a negative regulator of trichome branching. gl2 hdg11 double mutants display enhanced trichome cell-type differentiation defects. Transgenic expression of HDG11 using the GL2 promoter partially suppresses gl2 trichome phenotypes. Vice versa, expression of GL2 under the control of its native promoter partially complements hdg11 ectopic branching. Since gl2 hdg11 and gl2 myb23 double mutants and the triple mutant display similar trichome differentiation defects, we investigated a connection to the R2R3-MYB transcription factor MYB23. We show that MYB23 transcript levels are significantly reduced in shoots from gl2 mutants and that GL2 can drive the expression of a MYB23-promoter fusion to green fluorescent protein. Yeast one-hybrid, chromatin immunoprecipitation, and in planta reporter gene experiments indicate that an L1-box in the MYB23 promoter acts as a GL2 binding site. Taken together, our findings reveal a functional redundancy between GL2 and HDG11, two homeodomain leucine zipper transcription factors previously thought to mediate opposing functions in trichome morphogenesis. A model is proposed in which GL2 transcript levels are maintained through a positive feedback loop involving GL2 activation of MYB23.
INTRODUCTION
In plants, the epidermis of leaves is derived from the outer L1 layer of the shoot apical meristem. As leaf primordia develop, the epidermal cell layer differentiates to form an organized pattern of distinct cell types, including pavement cells, stomata, and trichomes. In Arabidopsis thaliana, mature leaf trichomes are characteristically large (300 to 500 μm) branched hair cells whose nuclei have undergone multiple rounds of endoreduplication and are present on the leaf surface in a nonrandom regular distribution (Hülskamp et al., 1994; Balkunde et al., 2010). Among other plant species, leaf trichomes are highly variable and may be branched or unbranched, single celled or multicellular, and may function as glandular organs. Proposed functions of trichomes include protection from herbivores and shielding from abiotic environmental factors such as frost, evaporation, and UV radiation (Balkunde et al., 2010; Yang and Ye, 2013).
Transcription factors of the class IV homeodomain leucine-zipper (HD-Zip) family play central roles in the differentiation of the epidermis during both embryonic and postembryonic development. This protein family is characterized by a homeodomain (HD) N terminal to a plant-specific Leu zipper having an internal loop (zipper-loop-zipper [ZLZ]) (also called truncated leucine zipper [Yang et al., 2002]), followed by a steroidogenic acute regulatory protein–related lipid transfer (START) domain (Schrick et al., 2004). The Arabidopsis genome encodes a total of 16 class IV HD-Zip family members, several of which are known to be involved in cell fate determination within layer-specific contexts (Nakamura et al., 2006).
The class IV HD-Zip transcription factor GLABRA2 (GL2) plays a pivotal role in regulating the differentiation of the epidermis in numerous tissues, including the root (Di Cristina et al., 1996) and seed coat (Western et al., 2001). Its role in trichomes is the major focus of this article. gl2 mutants display aborted trichomes with thin cell walls that lack papillae, and some trichomes aberrantly expand in the plane of the pavement cells (Koornneef et al., 1982; Rerie et al., 1994). GL2 is highly expressed in mature trichomes (Marks et al., 2009), and late activity of GL2 seems to be required for proper trichome development. While GL2 is required for the differentiation and maintenance of trichome cell fate, it is dispensable for trichome initiation. In a current model, GL2 acts as a downstream transcription factor in a regulatory circuit together with the mitosis inhibitor SIM that is required for endoreduplication (Grebe, 2012). The initiation of trichome cell fate and transcriptional activation of GL2 and SIM involves an activator complex composed of a WD40 protein encoded by TRANSPARENT TESTA GLABRA1 (TTG1), an R2R3-MYB transcription factor encoded by GL1 and the partially redundant gene MYB23, a basic helix-loop-helix (bHLH) protein encoded by two genes, GL3 and ENHANCER OF GL3 (EGL3), and the possible addition of another component encoded by MYC1 (Grebe, 2012). This complex is counteracted by inhibitory R3 MYB transcription factors encoded by a group of at least six genes, including CAPRICE and TRIPTYCHON, which diffuse between cells, competing with the R2R3-MYB proteins for binding to the bHLH protein.
Based on mutant phenotypes and expression profiling, three other class IV HD-Zip transcription factors in addition to GL2 are implicated in trichome cell-type differentiation. HOMODOMAIN GLABROUS2 (HDG2) is strongly expressed in mature trichomes, similar to the level observed for GL2 (Marks et al., 2009). While hdg2 mutants display no obvious defects in overall trichome cell morphology, their trichome surfaces appear more transparent and their cell wall composition is abnormal (Marks et al., 2009). Based on mutant analysis, HDG11 and HDG12 play negative regulatory roles in trichome branching. hdg11 mutants result in excess branching of trichomes, a phenotype that is enhanced in combination with hdg12 (Nakamura et al., 2006). While promoter–β‑glucuronidase (GUS) reporter lines for both exhibit strong expression in trichomes (Nakamura et al., 2006), transcript levels of HDG11 and HDG12 are low and not detectable, respectively, in mature trichomes (Marks et al., 2009), consistent with expression being mostly restricted to the branching phase of trichome development.
In this article, we uncover a genetic interaction and functional redundancy between the class IV HD-Zip transcription factors GL2 and HDG11 in trichomes. The phenotype of gl2 hdg11 double mutants led us to investigate a connection to the R2R3-MYB transcription factor MYB23. Members of the expansive R2R3-MYB gene family play key roles in various plant-specific processes, including the regulation of cell fate in development (Stracke et al., 2001; Dubos et al., 2010). MYB23 had previously been found to act redundantly with GL1 in trichome cell-type specification (Kirik et al., 2005). In the root, MYB23 had been shown to function in cell fate determination and to bind its own promoter in a MYB23-mediated positive feedback loop (Kang et al., 2009). Here, we provide evidence for an additional positive feedback mechanism involving GL2 transcriptional activation of MYB23 in the maintenance of cell fate during trichome morphogenesis.
RESULTS
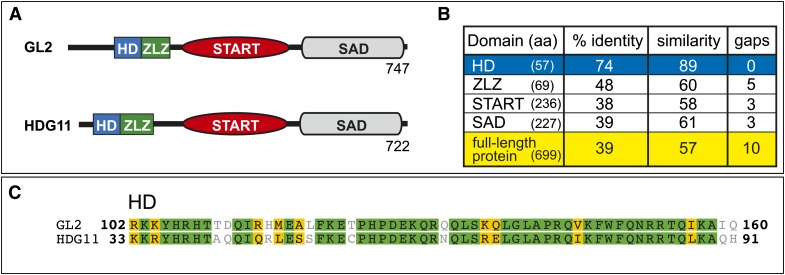
Sequence Analysis of GL2 and HDG11 Reveals Highly Related HDs
GL2 and HDG11 are linked loci on Arabidopsis chromosome 1. Phylogenetic analysis of the 16 members of the class IV HD-Zip family does not reveal a high degree of relatedness between GL2 and HDG11 in comparison with other family members (Nakamura et al., 2006). To further probe sequence similarity between GL2 and HDG11, we divided their protein sequences into four distinct conserved domains: (1) HD, (2) ZLZ, (3) START, and (4) START adjacent domain (SAD) (Figures 1A and 1B). The N-terminal HD implicated in DNA binding is followed by the ZLZ plant-specific Leu zipper dimerization domain conserved among members of the class IV family (Schrick et al., 2004). The START domain implicated in ligand binding (Ponting and Aravind, 1999; Schrick et al., 2004) lies in the middle, followed by the SAD of unknown function at the C terminus. We determined the percentage amino acid identities and similarities between the four domains of the two proteins in pairwise alignments from protein–protein BLAST (Figure 1B) and found the HD domain to be most conserved, exhibiting 74 and 89% identity and similarity, respectively (Figure 1C). Pairwise alignments were used to compare the HD domain of GL2 with each of the 15 other members of the class IV HD-Zip family (Supplemental Table 1). Ranking by amino acid identity to GL2, HDG11 placed first, followed by HDG5 and PROTODERMAL FACTOR2 (PDF2). Among the other class IV members whose mRNA transcripts are expressed in leaf trichomes (HDG2, HDG11, and HDG12), HDG11 exhibits the highest degree of amino acid identity and similarity to GL2.
Figure 1.
Sequence Relationships between Class IV HD-Zip Transcription Factors GL2 and HDG11.
(A) Domain arrangements of the GL2 and HDG11 proteins composed of 747 and 722 amino acids, respectively.
(B) Percentage amino acid (aa) identities and similarities between GL2 and HDG11 for the four conserved domains and the respective full-length proteins. The HD domains exhibit the highest degree of amino acid identity and similarity.
(C) Amino acid alignment of the HD domains from the GL2 and HDG11 proteins, generated with Geneious 5.4.6 software (Biomatters). Green and yellow shading indicate identical and similar residues, respectively.
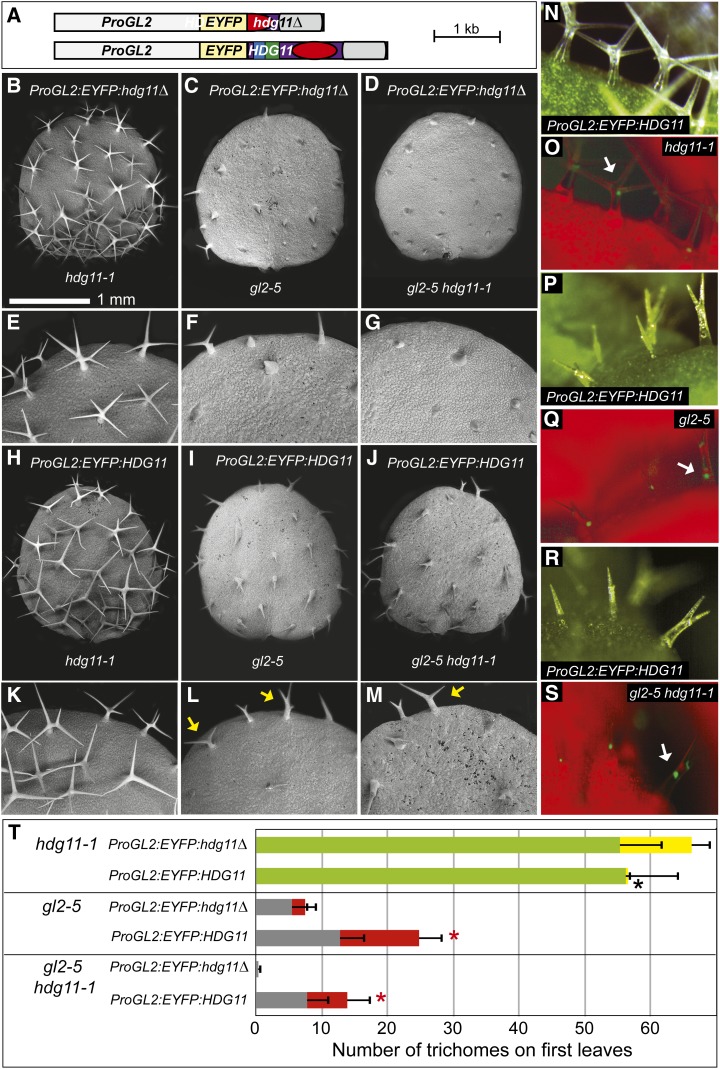
Double Mutants for gl2 and hdg11 Display Enhanced Trichome Differentiation Defects
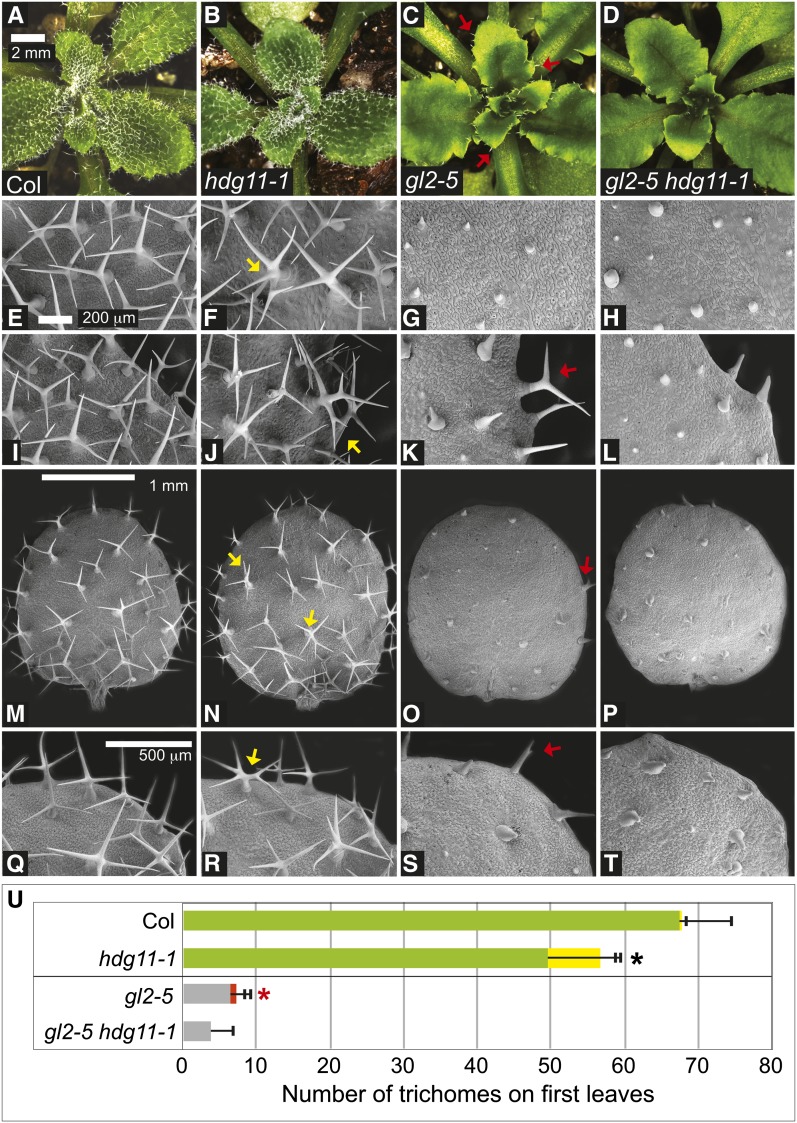
Crosses between gl2-5 and hdg11-1 plants of the Columbia (Col) ecotype were performed to construct homozygous double mutants. In comparison with gl2-5 single mutants, the double mutants exhibited enhanced trichome differentiation defects (Figure 2). Trichome phenotypes were visualized on rosette leaves by stereomicroscopy (Figures 2A to 2D). Leaf trichome morphologies were observed under higher magnification using scanning electron microscopy (Figures 2E to 2T). While trichome initiation appears not to be altered in gl2-5 single and gl2-5 hdg11-1 double mutants, outgrowth and differentiation of the trichomes are affected. Trichomes are generally aborted on leaves from gl2-5 mutants, whereas trichomes at the leaf margins are more differentiated and typically branched with two (or rarely three) branches. Quantification of trichome outgrowth revealed a significant difference in the number of branched trichomes at the leaf margins in gl2-5 mutants versus gl2-5 hdg11-1 double mutants (Figure 2U; Supplemental Table 2). We observed ectopic branching in hdg11-1 mutants, as reported previously (Nakamura et al., 2006). On first leaves, trichomes with five branches or more were very rare in wild-type Col (∼0.4%), while hdg11-1 mutants exhibited ∼13% of the total trichomes with five or more branches. In contrast, in gl2-5 hdg11-1 double mutants, there was a complete absence of branched trichomes, while gl2-5 mutants displayed branched trichomes among ∼11% of the total trichomes on first leaves. Trichomes also form on the epidermis of sepals from Arabidopsis flowers. Analogous deviations in trichome morphogenesis were detected on sepals of hdg11-1 and gl2-5 mutants (Supplemental Figure 1). Wild-type sepals exhibited unbranched trichomes, while hdg11-1 sepals showed both unbranched and branched trichomes. gl2-5 mutants exhibited fewer and smaller trichomes, while gl2-5 hdg11-1 mutants typically lacked trichomes on sepals.
Figure 2.
gl2-5 hdg11-1 Double Mutants Display an Enhanced Trichome Differentiation Defect.
(A) to (D) Rosettes from 3-week-old Arabidopsis plants.
(A) and (B) Col wild-type (A) and hdg11-1 (B) plants display trichomes covering leaf surfaces.
(C) and (D) gl2-5 plants appear mostly glabrous except for trichomes at the leaf margins (arrows) (C), while in comparison, gl2-5 hdg11-1 double mutants display an enhanced glabrous phenotype (D).
(E) to (L) Scanning electron micrographs of leaves ([E] to [H]) and leaf margins ([I] to [L]) reveal detailed trichome morphologies.
(E) and (I) Col wild-type trichomes exhibit three to four branches.
(F) and (J) hdg11-1 leaves show five or more branches for some trichomes (arrows).
(G) and (K) gl2-5 trichomes are small and undifferentiated except at leaf margins, where branching (arrow) is observed.
(H) and (L) gl2-5 hdg11-1 double mutants exhibit smaller undifferentiated trichomes at leaf margins.
(M) to (T) First leaves ([M] to [P]) and detailed edges of first leaves ([Q] to [T]).
(M) and (Q) Col wild-type trichomes have three to four branches.
(N) and (R) hdg11-1 mutants exhibit ectopic trichome branching (arrows).
(O) and (S) gl2-5 mutants display undifferentiated trichomes except at leaf edges, where some trichomes display branching (arrows).
(P) and (T) gl2-5 hdg11-1 double mutants have undifferentiated trichomes throughout the leaf.
(U) Quantification of trichomes and trichome branching on first leaves. For the Col wild type and hdg11-1, green bars show the number of trichomes with three to four branches and yellow bars indicate the number of trichomes with five or more branches. For gl2 and gl2 hdg11, gray bars show the number of unbranched single-spiked trichomes while the red bar indicates branched trichomes. Positive error bars indicate sd for n ≥ 20 plants. Asterisks indicate significant differences in trichome branching (two-tailed t test, P < 0.0001).
Using the Landsberg erecta (Ler) ecotype as the genetic background, we crossed gl2-1 and hdg11-3 mutants to produce the corresponding double mutant plants (Supplemental Figure 2). The quantitative difference between trichome branching on first leaves for gl2-1 single mutants versus gl2-1 hdg11-3 double mutants was more striking in the Ler genetic background as compared with the Col ecotype (Supplemental Figure 2M and Supplemental Table 2). We also constructed gl2-5 hdg11-3 double mutants and found their trichome cell-type differentiation defects enhanced in comparison with gl2-5 siblings (Supplemental Figure 3).
gl2 hdg11 Trichome Defects Resemble Those of gl2 myb23 Mutants
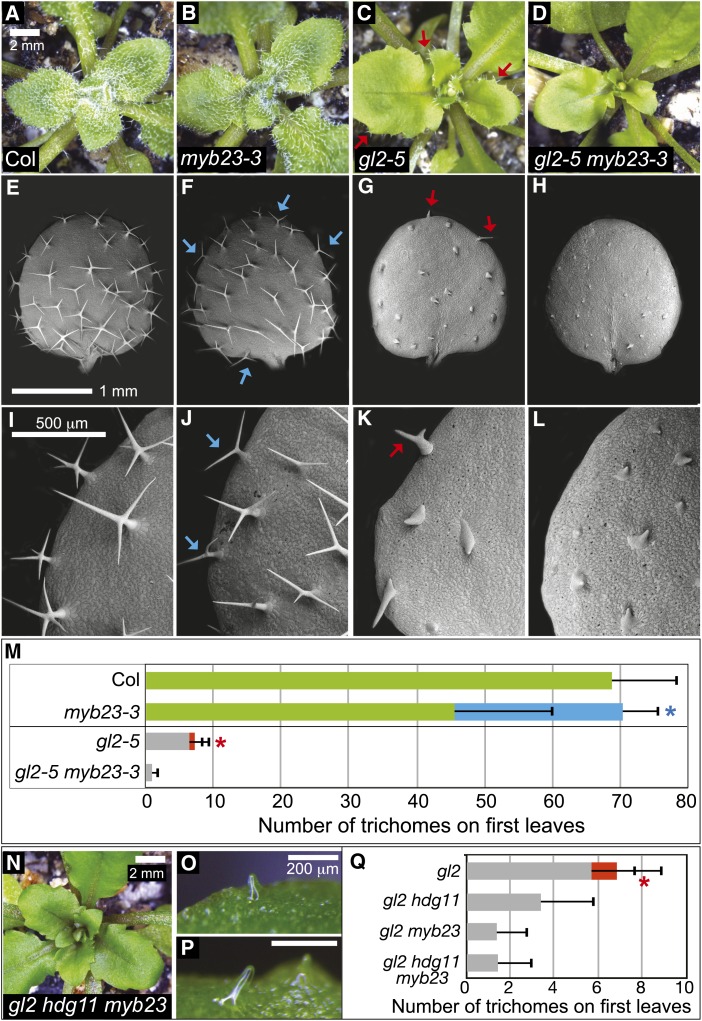
Previously it was shown that the trichome differentiation defect of gl2 mutants is enhanced in a double mutant for the Col alleles gl2-4AA and myb23-2 (Kirik et al., 2005). It was reported that myb23 mutants exhibit reduced trichome branching but no obvious defect in trichome patterning (Kirik et al., 2005) and display smaller trichomes (Li et al., 2009). We confirmed the previously reported trichome defects using the myb23-3 mutant allele (Figure 3). The gl2-5 myb23-3 double mutant was constructed, and enhanced trichome defects were confirmed for this new allelic combination in the Col ecotype. Similar to gl2 hdg11 mutants, gl2-5 myb23-3 mutants exhibited trichomes that were distinctly smaller and less expanded than gl2-5 trichomes, and this was especially apparent at the leaf margins (Figures 3A to 3D). Using scanning electron microscopy to view trichome morphologies in detail, we observed trichome differentiation defects in gl2-5 myb23-3 mutants that were analogous to those seen in gl2 hdg11 mutants (Figures 2 and 3E to 3L). Quantification of the trichome defects on first leaves indicated that while ∼11% of the total trichomes in gl2-5 mutants were branched, none of the trichomes in the gl2-5 myb23-3 mutants were branched (Figure 3M; Supplemental Table 2).
Figure 3.
Enhanced Trichome Differentiation Defects in the gl2-5 myb23-3 Double Mutant and the gl2 hdg11 myb23 Triple Mutant.
(A) to (D) Rosettes from 3-week-old plants.
(A) and (B) Col wild-type (A) and myb23-3 (B) plants display trichomes covering leaf surfaces.
(C) and (D) gl2-5 plants display trichomes at leaf margins (arrows) (C), while gl2-5 myb23-3 double mutants exhibit glabrous leaves (D).
(E) to (L) Scanning electron micrographs of first leaves reveal trichome morphology details.
(E) and (I) Col wild-type trichomes display three to four branches.
(F) and (J) myb23-3 trichomes exhibit two to three branches. Arrows mark abnormal trichomes with two branches.
(G) and (K) gl2-5 trichomes display differentiation defects, although some trichomes at leaf edges show branching (arrows).
(H) and (L) gl2-5 myb23-3 double mutants exhibit small undifferentiated trichomes that lack branching.
(M) Quantification of trichomes on first leaves. For Col and myb23-3, green bars indicate the number of trichomes with three to four branches and the blue bar indicates the number of abnormal trichomes with fewer than three branches. For gl2-5 and gl2-5 myb23-3, gray bars show the number of unbranched trichomes while the red bar indicates branched trichomes. Positive error bars indicate sd for n ≥ 20 plants, and asterisks indicate significant deviations in trichome branching (two-tailed t test, P < 0.0001).
(N) Rosette from a gl2-5 hdg11-1 myb23-3 triple mutant.
(O) and (P) Details of leaf trichomes from the triple mutant.
(Q) Quantification of trichomes on first leaves as in (M) from gl2-5, gl2-5 hdg11-1, gl2-5 myb23-3, and gl2-5 hdg11-1 myb23-3.
Construction of the gl2 hdg11 myb23 Triple Mutant Reveals Genetic Interactions
Given the similarity between the leaf trichome phenotypes of gl2 hdg11 and gl2 myb23 double mutants, we investigated genetic interactions between hdg11 and myb23 in the gl2 background. If the hdg11 and myb23 mutations affect the same biological pathway in the absence of GL2, then the triple mutant gl2 hdg11 myb23 would be expected to display similar trichome differentiation defects to either double mutant. Indeed, we found that the gl2-5 hdg11-1 myb23-3 triple mutant displays a trichome phenotype resembling the double mutants (Figures 3N to 3P). Quantification of trichomes on first leaves indicated that the triple mutant has a phenotype that is indistinguishable from gl2-5 myb23-3 (Figure 3Q), and examination of first leaves by scanning electron microscopy shows striking similarity between the double and triple mutants (Supplemental Figure 4).
During construction of the triple mutant, we observed trichome defects in the F1 progeny from reciprocal crosses between gl2 hdg11 and gl2 myb23, whose genotype was homozygous for gl2-5 and heterozygous for both hdg11-1 and myb23-3 (Supplemental Figure 5). Unlike gl2 mutants that exhibit trichomes on leaf margins, the gl2/gl2 hdg11/+ myb23/+ plants exhibited enhanced trichome defects similar to those of gl2 hdg11 and gl2 myb23 double mutants (Supplemental Figures 5A to 5F). Unexpectedly, we also uncovered fertility defects that were associated with inviable pollen from plants heterozygous for hdg11-1 or myb23-3 or both. However, these phenotypes were found to be independent of GL2 (Supplemental Figures 5G to 5L). Double mutants for hdg11 and myb23 were found to exhibit an intermediate trichome phenotype, lacking the ectopic branching of hdg11 mutants, while showing significantly fewer two-branched trichomes than myb23 mutants (Supplemental Figure 6 and Supplemental Table 2). This suggests that in the presence of GL2, HDG11 and MYB23 act in parallel to orchestrate trichome cell differentiation, and MYB23 is not required for this inhibitory function of HDG11.
Interactions between GL2, HDG11, and MYB23 Are Tissue Specific
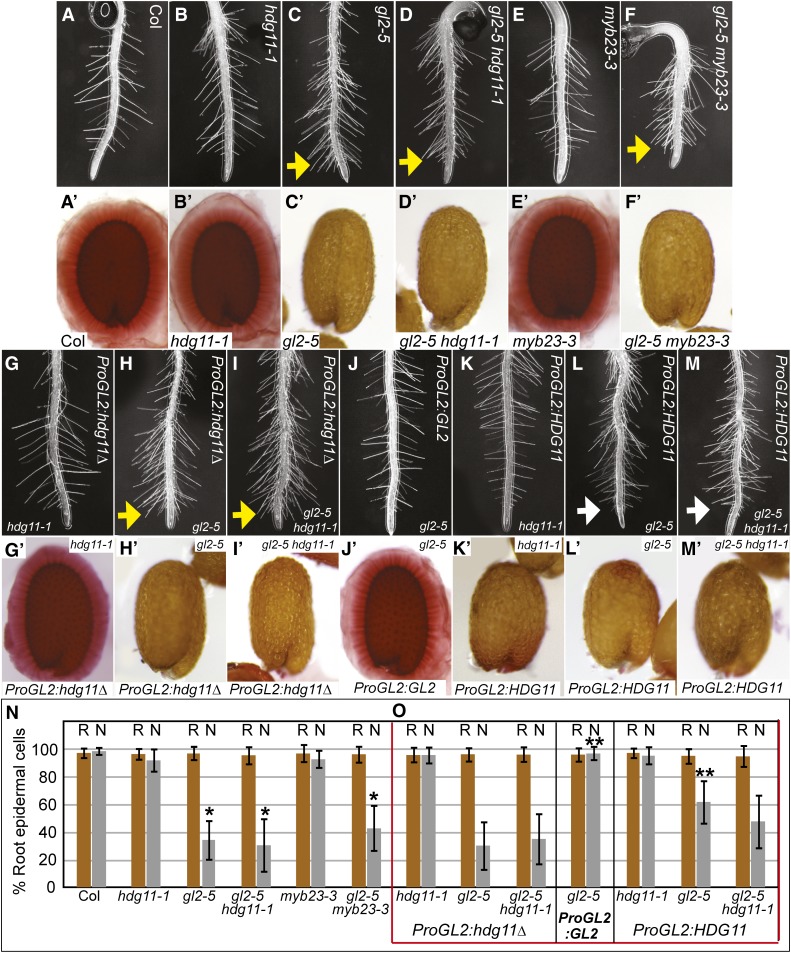
After establishing the enhanced trichome cell differentiation defects in gl2 hdg11 double mutants, we investigated phenotypes in roots and seeds, other tissues that are affected in gl2 single mutants. In the root epidermis, gl2 mutants exhibit an increase in the proportion of cells that are specified for root hair fate (Di Cristina et al., 1996). We observed excess root hair formation on primary roots of gl2 hdg11 double mutants from both the Col and Ler backgrounds but did not detect an enhanced defect as compared with gl2 single mutants (Figures 4A to 4D and 4N; Supplemental Figure 7 and Supplemental Table 3). gl2 mutants also exhibit a defect in the production of seed coat mucilage upon imbibition (Western et al., 2000). Using ruthenium red staining for pectin in the seed coat mucilage, we observed that gl2 hdg11 double mutants lack the mucilage layer in a manner that is indistinguishable from gl2 single mutants (Figures 4A′ to 4D′; Supplemental Figure 7). In addition to defects in the epidermis, gl2 mutants accumulate higher levels of oil in the seed (Shen et al., 2006), and this phenotype is associated with the lack of seed coat mucilage (Shi et al., 2012). We also found that gl2 hdg11 double mutants display a similar increase in seed oil levels to gl2 single mutants (Supplemental Figure 8). Similarly, examination of gl2 myb23 mutants for root hair and seed mucilage phenotypes revealed no obvious enhanced or novel phenotypes relative to the gl2 single mutants, consistent with the idea that the genetic interaction with MYB23 is also specific to trichomes (Figure 4; Supplemental Table 3).
Figure 4.
Root Hair Density and Seed Coat Mucilage Phenotypes Reveal Tissue-Specific Interactions.
(A) to (M) Primary roots from 3- to 4-d-old seedlings.
(A) to (F) Col wild-type (A), hdg11-1 (B), and myb23-3 (E) root hair density appears normal in comparison with excess root hair formation in gl2-5 (C), gl2-5 hdg11-1 (D), and gl2-5 myb23-3 (F).
(G) to (I) Transgenic lines expressing ProGL2:EYFP:hdg11Δ do not appear to alter root hair density in hdg11-1 (G), gl2-5 (H), or gl2-5 hdg11-1 (I).
(J) Expression of ProGL2:EYFP:GL2 rescues the root hair phenotype of gl2-5.
(K) to (M) Expression of ProGL2:EYFP:HDG11 has no effect on root hair density in hdg11-1 seedlings (K). Partial complementation is observed for gl2-5 (L) and possibly gl2-5 hdg11-1 (M) roots (white arrows).
Yellow arrows point to abnormal root hair formation near the tips of the roots.
(A′) to (M′) Seeds were stained with ruthenium red to detect mucilage production upon imbibition.
(A′) to (F′) Col wild-type (A′), hdg11-1 (B′), and myb23-3 (E′) seeds exhibit a normal outer mucilage, while gl2-5 (C′), gl2-5 hdg11-1 (D′), and gl2-5 myb23-3 (F′) lack mucilage.
(G′) to (I′) ProGL2:EYFP:hdg11Δ expression does not affect seed mucilage production in hdg11-1 (G′), gl2-5 (H′), or gl2-5 hdg11-1 (I′).
(J′) Expression of ProGL2:EYFP:GL2 rescues the mucilage phenotype of gl2-5.
(K′) ProGL2:EYFP:HDG11 expression negatively affects mucilage production in hdg11-1 seeds.
(L′) and (M′) gl2-5 (L′) and gl2-5 hdg11-1 (M′) seeds expressing ProGL2:EYFP:HDG11 lack the outer mucilage.
(N) and (O) Quantification of root epidermal cells in genotypes lacking transgenes (N) and expressing EYFP:HDG11 or EYFP:GL2 from transgenes (boxed in red) (O). Percentages of trichoblast cells in root hair cell files (R, brown bars) and atrichoblast cells in non-root-hair cell files (N, gray bars) are indicated (n ≥ 20). Error bars indicate sd. Single asterisks in (N) mark significant differences from the Col wild type, while double asterisks in (O) mark significant differences between mutants with ProGL2:EYFP:hdg11Δ control and ProGL2:EYFP:GL2 or ProGL2:EYFP:HDG11 transgenes expressing wild-type GL2 or HDG11, respectively (two-tiered t test, P < 0.0001).
Expression of HDG11 under the Control of the GL2 Promoter Rescues Ectopic Branching of hdg11 Mutants and Partially Suppresses Trichome Differentiation Defects of gl2 Mutants
We used the 2.1-kb promoter upstream of GL2 to express HDG11 in trichomes from hdg11, gl2, and gl2 hdg11 mutants (Figure 5). An N-terminal enhanced yellow fluorescence protein (EYFP) tag was used to monitor the expression of HDG11 in transgenic lines. As a control, we utilized an hdg11 mutant construct that is missing the first 305 residues of the predicted protein and therefore lacking the HD and ZLZ domains as well as part of the START domain (Figure 5A). The trichome phenotypes of representative hdg11, gl2, and gl2 hdg11 lines, as visualized by scanning electron microscopy, were unchanged, indicating that this construct is null for HDG11 activity (Figures 5B to 5G). By contrast, scanning electron microscopy analysis showed that the wild-type ProGL2:EYFP:HDG11 construct fully complements the trichome-branching defect of hdg11-1 mutants (Figures 5H and 5K). Its expression was also sufficient to induce increased branching of trichomes at leaf margins in the gl2-5 hdg11-1 double mutant, consistent with complementation of the hdg11 mutant phenotype (Figures 5L and 5M; Supplemental Figures 9B, 9D, and 9F). However, the ProGL2:EYFP:HDG11 construct only partially rescued the trichome differentiation defect of gl2-5 mutants on first leaves (Figures 5I and 5J) and leaves from rosettes (Supplemental Figures 9A, 9C, and 9E). Unexpectedly, novel transient dwarf phenotypes were observed at the rosette stage for both gl2-5 and gl2-5 hdg11-1 mutants transformed with ProGL2:EYFP:HDG11 (Supplemental Figures 9G to 9L).
Figure 5.
ProGL2:EYFP:HDG11 Rescues hdg11 Ectopic Branching and Partially Rescues the gl2 Trichome Differentiation Defects.
(A) Transgene constructs ProGL2:EYFP:hdg11Δ and ProGL2:EYFP:HDG11.
(B) to (M) Scanning electron micrographs of first leaves ([B] to [D] and [H] to [J]) and magnification of leaf margins ([E] to [G] and [K] to [M]) are shown for mutant construct ProGL2:EYFP:hdg11Δ ([B] to [G]) and wild-type construct ProGL2:EYFP:HDG11 ([H] to [M]) in hdg11-1 ([B], [E], [H], and [K]), gl2-5 ([C], [F], [I], and [L]), and gl2-5 hdg11-1 ([D], [G], [J], and [M]) mutant backgrounds. ProGL2:EYFP:HDG11 complements hdg11-1 ectopic branching ([H] and [K]) while conferring partial rescue of the gl2-5 trichome phenotype ([I], [J], [L], and [M]). In (L), arrows mark an internal branched trichome and a three-branched trichome, indicative of partial rescue of the gl2 phenotype. In (M), the arrow shows branched trichomes on the leaf margin.
(N) to (S) Nuclear localization of EYFP:HDG11 in trichomes. Matching white light and fluorescence images are from hdg11-1 ([N] and [O]), gl2-5 ([P] and [Q]), and gl2-5 hdg11 ([R] and [S]) leaves expressing EYFP:HDG11. Arrows indicate the brightest nuclear signal in each image.
(T) Quantification of trichomes and trichome branching on first leaves. For hdg11-1, green bars show the number of trichomes with three to four branches and yellow bars indicate the number of trichomes with five or more branches. For gl2 and gl2 hdg11, gray bars show the number of unbranched trichomes while red bars indicate branched trichomes. Positive error bars indicate sd for n ≥ 20 plants. Asterisks indicate significant differences in branching phenotypes for mutant (hdg11Δ) versus wild-type HDG11 transgene (two-tiered t test, P < 0.00001).
In all three genetic backgrounds, expression of the ProGL2:EYFP:HDG11 fusion protein was monitored in trichomes and found to be localized in nuclei of trichome cells (Figures 5N to 5S). Quantification of trichomes on first leaves revealed a significant difference in the branching and/or number of trichomes for mutant lines expressing ProGL2:EYFP:HDG11 (Figure 5T; Supplemental Table 2). For hdg11 mutant plants expressing the wild-type HDG11 transgene, ∼0.6% of trichomes had five or more branches in comparison with the control, in which ∼17% of the trichomes exhibited five or more branches. Thus, expression of HDG11 under the control of the GL2 promoter is sufficient for rescue of the hdg11 branching phenotype. In gl2 mutant plants expressing the wild-type EYFP:HDG11 transgene, an ∼3.3-fold increase in average trichome numbers and more branching (∼48 versus ∼27% in the control) on first leaves were observed, but overall trichome formation was partial in comparison with the wild type. For the gl2-5 hdg11-1 double mutant, branching and trichome cell-type differentiation were restored to approximately the levels in gl2-5 mutants, as indicated by an ∼35-fold increase in visible trichomes and more branching (∼44 versus 0% in the control).
We also expressed HDG11 in gl2-5 mutants using the epidermis-specific ∼3.4-kb ATML1 promoter from class IV HD-Zip transcription factor ATML1 (Supplemental Figure 10). ProATML1:HDG11 transgenic lines exhibited increased trichome cell-type differentiation in comparison with untransformed gl2-5 control plants. However, the trichome cell-type differentiation was partial, resulting in an increase in mainly one- to two-branched and rarely three-branched trichomes (Supplemental Figures 10D to 10G). Additional phenotypes, such as leaf curling and novel pointy leaf shapes, were also observed in conjunction with trichome formation, suggesting that the ectopic expression of HDG11 either induces or interferes with other cell-type differentiation pathways in the epidermis.
Expression of HDG11 under the Control of the GL2 Promoter Partially Rescues the Root Hair Phenotype of gl2 Mutants but Negatively Affects Seed Mucilage Production
In roots, we detected a partial rescue of the excess root hair phenotype of gl2 mutants in transgenic lines expressing ProGL2:EYFP:HDG11 in comparison with the ProGL2:EYFP:hdg11Δ negative control (Figures 4H, 4L, and 4O; Supplemental Table 3). A complete rescue of the gl2 root hair phenotype was conferred by the ProGL2:EYFP:GL2 transgene (Figures 4J and 4O). This construct, in which EYFP-tagged GL2 is expressed under the control of its native promoter (Figure 6A), was shown previously to completely rescue the trichome defects of gl2-5 mutants (K. Schrick, B.P. Srinivas, and M. Hülskamp, unpublished data). The expression of mutant or wild-type ProGL2:EYFP:HDG11 did not alter the root hair phenotype of hdg11 mutants, which appears indistinguishable from that of the wild type (Figures 4G, 4K, and 4O). However, in seeds, the expression of ProGL2:EYFP:HDG11 in hdg11 had a negative effect on mucilage formation in comparison with the control, resulting in a “gl2” phenotype (Figures 4G′ and 4K′). Although ProGL2:EYFP:GL2 was able to complement the seed mucilage phenotype of gl2 (Figure 4J′), there was no visible rescue of the seed coat mucilage phenotype in gl2 mutants by the ProGL2:EYFP:HDG11 transgene (Figures 4H′, 4I′, 4L′, and 4M′). Taken together, these data indicate that the ability of HDG11 to partially rescue the gl2 phenotype and/or interfere with GL2 activity is tissue specific.
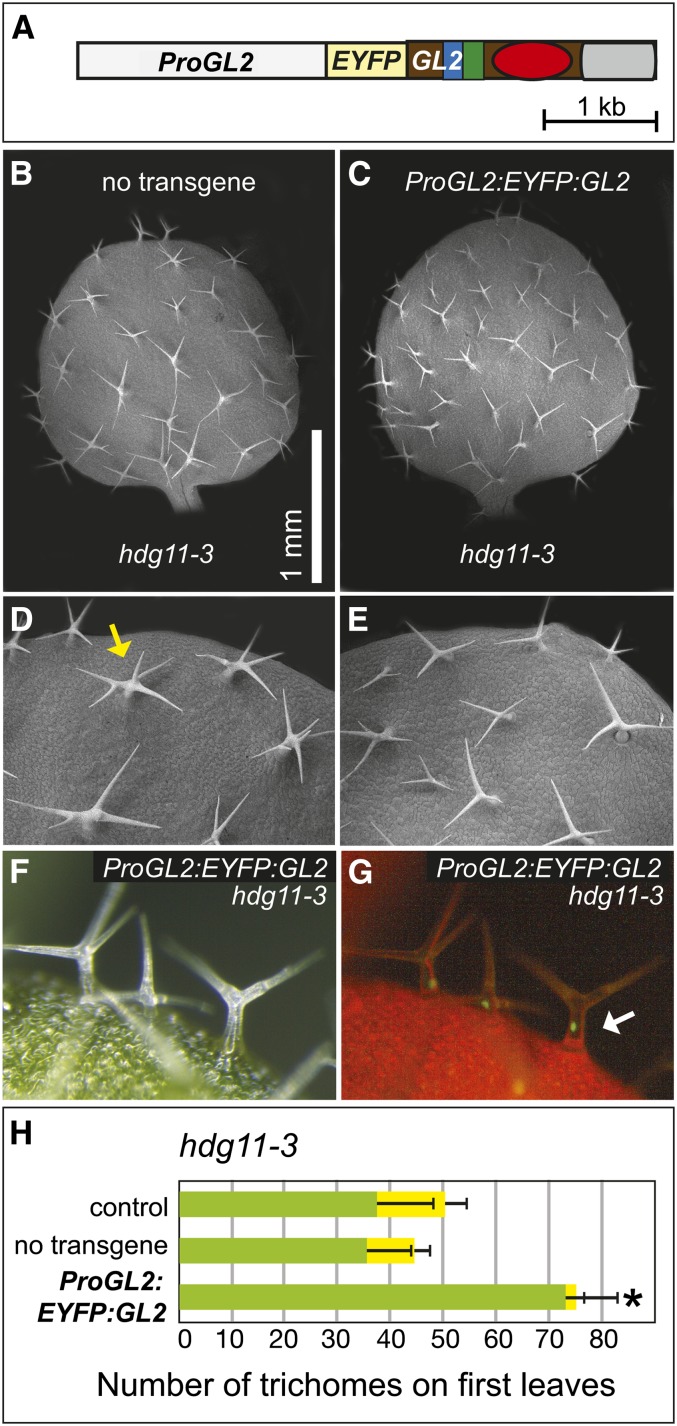
Figure 6.
ProGL2:EYFP:GL2 Rescues Ectopic Branching of hdg11.
(A) Transgene construct ProGL2:EYFP:GL2, in which the cDNA of GL2 is fused to an EYFP tag.
(B) and (C) Scanning electron micrographs of first leaves.
(D) and (E) Magnification of leaf margins are shown for an hdg11-3 mutant lacking the transgene (D) and an hdg11-3 mutant expressing ProGL2:EYFP:GL2 (E). In (D), the yellow arrow marks a trichome with ectopic branching.
(F) and (G) Matching white light and fluorescence images for the hdg11-3 mutant expressing ProGL2:EYFP:GFP. The white arrow indicates a bright nuclear signal.
(H) Quantification of trichomes and trichome branching on first leaves of hdg11-3 plants. The control is the hdg11-3 parent from a cross to a line containing the transgene. Progeny of F2 offspring lacking the transgene were designated as “no transgene,” while pure lines expressing the transgene were designated as ProGL2:EYFP:GL2. Green bars show the number of trichomes with three to four branches and yellow bars indicate the number of trichomes with five or more branches. Positive error bars indicate sd for n ≥ 20 plants. The asterisk indicates a significant reduction in branched trichomes for hdg11-3 plants expressing ProGL2:EYFP:GFP (two-tiered t test, P < 0.0001).
GL2 Overexpression Partially Rescues Ectopic Trichome Branching of hdg11 Mutants
Since HDG11 overexpression in trichomes can partially rescue the trichome and root hair phenotypes of gl2, we asked whether GL2 overexpression is able to complement the ectopic branching phenotype of hdg11 mutants. We expressed ProGL2:EYFP:GL2 in hdg11 mutants and observed partial complementation of the ectopic branching phenotype (Figure 6). Scanning electron microscopy analysis showed that in comparison with the control lacking the transgene (Figures 6B and 6D), hdg11 mutants expressing ProGL2:EYFP:GL2 exhibited fewer trichomes having five or more branches. Expression of the transgene was associated with nuclear localization of the EYFP:GL2 protein in trichomes (Figures 6E and 6F). Quantification of trichomes on first leaves revealed a significant difference in hdg11 mutants expressing the transgene (∼20% ectopically branched trichomes) versus the controls lacking the transgene (∼3% ectopically branched trichomes) (Figure 6H; Supplemental Table 2). Expression of the GL2 transgene also led to an increase in the average total number of trichomes on first leaves of hdg11 mutants (∼75 versus ∼45 in the control), suggesting that GL2 expression can drive trichome cell-type specification in the absence of HDG11.
GL2 Activates MYB23 Transcription in Trichomes by Binding an L1-Box in the MYB23 Promoter
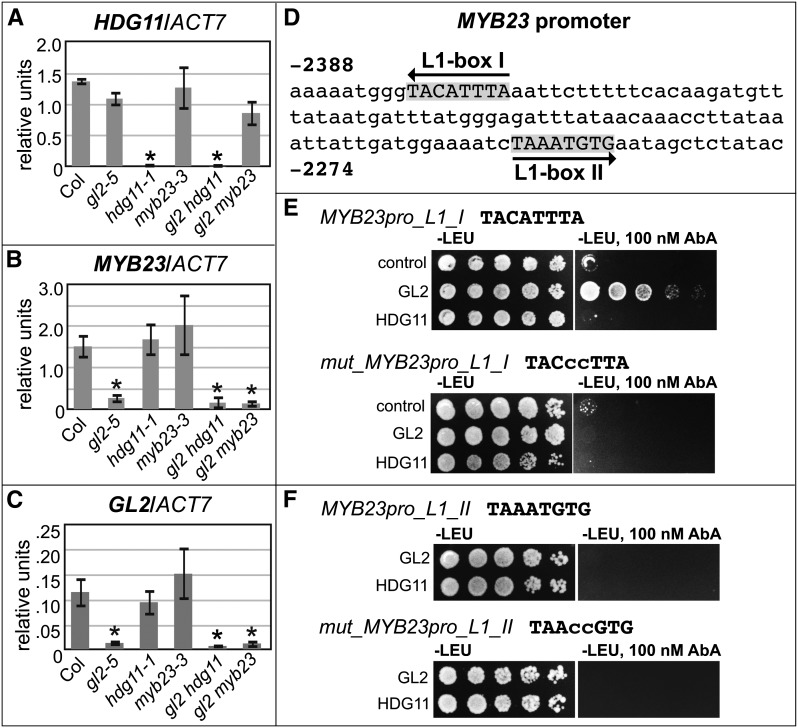
To further dissect the genetic interactions between GL2, HDG11, and MYB23, we monitored their respective mRNA transcripts in seedlings from the Col wild type, gl2-5, hyd11-1, and myb23-3 as well as the gl2 hdg11 and gl2 myb23 double mutants. mRNA was isolated from 14-d-old seedling shoots to enrich for transcript levels in trichomes. Quantitative real-time PCR confirmed that the gl2-5 and hdg11-1 alleles are RNA knockdown alleles, while myb23-3 mutants do not exhibit a reduced level of mRNA expression in shoots (Figures 7A to 7C). It was shown previously that for root tips from myb23-3, transcript levels in myb23-3 are reduced to ∼58% of wild-type levels (Kang et al., 2009). The myb23-3 root hair phenotype was reported to be indistinguishable from that of the transcript-null allele myb23-1, indicating that although a transcript is expressed in myb23-3, it is not functional (Kang et al., 2009). HDG11 mRNA levels were decreased only in hdg11-1 and gl2-5 hdg11-1 (Figure 7A). By contrast, our analysis revealed that MYB23 mRNA levels were significantly reduced in gl2-5 single mutants (∼5-fold) as well as in gl2-5 hdg11-1 and gl2-5 myb23-3 double mutants (Figure 7B).
Figure 7.
GL2 Is Required for MYB23 Transcription, and GL2 Binds MYB23 L1-Box I in Yeast.
(A) to (C) Quantitative real-time PCR was performed with cDNA from 14-d-old seedling shoots with ACT7 as the reference gene. Data represent averages of three biological replicates, and normalized units are plotted on the y axis. Error bars indicate sd. Significant differences (two-tiered t test, P < 0.05) from the wild type (Col) are indicated by asterisks.
(A) HDG11 transcript is downregulated in hdg11-1 mutants.
(B) MYB23 transcript is downregulated in gl2-5 mutants.
(C) GL2 transcript is downregulated in gl2-5 mutants.
(D) The 3.1-kb MYB23 promoter region upstream of the ATG start codon contains two predicted L1-box sequences.
(E) and (F) Yeast one-hybrid analysis was performed with MYB23 promoter–derived L1-boxes I and II. Nucleotide base substitutions are indicated in lowercase for the corresponding mutant L1-boxes.
(E) GL2 but not HDG11 binds to the wild-type L1-box I but not its mutant version.
(F) Neither GL2 nor HDG11 binds to the wild-type or mutant L1-box II.
The real-time PCR data are consistent with the possibility that MYB23 is a direct transcriptional target of GL2. Previously, it was shown that a ProMYB23:MYB23 construct containing the MYB23 promoter (3.1 kb N terminal to the start codon and 1.0 kb C terminal to the stop codon) was sufficient to rescue the trichome defect in myb23-1 mutants (Kirik et al., 2005). The 3.1-kb promoter of MYB23 contains two putative L1-boxes (Figure 7D) that are heptamer/octamer elements shown to be required for DNA binding of GL2 (Ohashi et al., 2003; Tominaga-Wada et al., 2009). The first, L1-box I, located ∼2.4 kb upstream of the start codon of MYB23, is a perfect match, in reverse orientation, to the L1-box (TAAATGTA) described for the GL2-regulated CELLULOSE SYNTHASE5 (CESA5) and XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE17 (XTH17) promoters (Tominaga-Wada et al., 2009). The second, L1-box II (TAAATGTG), located 76 bp downstream from L1-box I, has a one-base mismatch (G) adjacent to the conserved heptamer sequence.
We used a yeast one-hybrid assay to test whether GL2 is able to bind to the L1-box motifs found in the MYB23 promoter (Figures 7E and 7F). The results indicate that GL2 binds to the wild-type L1-box I sequence but not a mutated version thereof. In contrast, GL2 was not able to bind to the imperfect sequence represented by L1-box II. We also examined whether HDG11 is able to bind to the motifs and failed to detect binding activity to either L1-box. The experiments indicate that GL2 binds one of the L1-boxes in the MYB23 promoter, whereas HDG11 is unable to bind that same sequence in the yeast system.
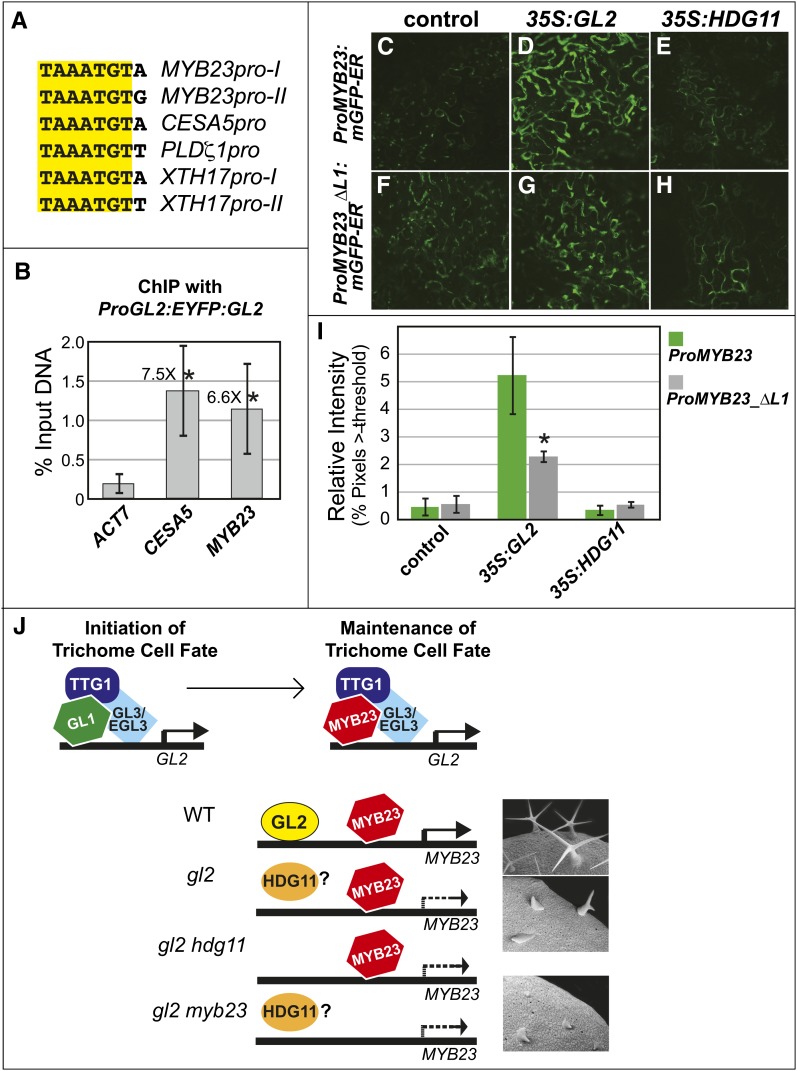
The L1-box motifs in the MYB23 promoter were compared with motifs identified for other known direct targets of GL2 (CESA5, PHOSPHOLIPASE D ζ1, and XTH17) (Figure 8A) (Ohashi et al., 2003; Tominaga-Wada et al., 2009). To further test the idea that GL2 binds to an L1-box in the MYB23 promoter, we performed chromatin immunoprecipitation (ChIP) on seedlings from a ProGL2:EYFP:GL2 transgenic line (Figure 8B). Using an anti–green fluorescent protein (GFP) antibody to purify protein-DNA complexes containing EYFP:GL2, we isolated ChIP DNA and performed quantitative real-time PCR. The results indicate that DNA containing the L1-box region of the MYB23 promoter is enriched to a similar level as a positive control, DNA corresponding to an L1-box–containing region from the CESA5 promoter. The percentage input values are significantly increased relative to the ACT7 negative control, indicating that GL2 binds to both the MYB23 and CESA5 promoters in seedlings.
Figure 8.
MYB23 Is Transcriptionally Activated by GL2, and Full Activation Requires Binding of GL2 to an L1-Box in the MYB23 Promoter.
(A) Alignment of L1-box sequences for GL2-regulated genes.
(B) ChIP with seedlings expressing ProGL2:EYFP:GL2, followed by quantitative real-time PCR of ChIP DNA. Primers that amplify L1-box regions in the CESA5 and MYB23 promoters, and ACT7 control primers, were used. The average percentage input DNA and corresponding sd were calculated for four biological replicates. Fold differences are indicated, and asterisks mark significant differences from the ACT7 control (two-tiered t test, P < 0.05).
(C) to (H) Representative images of N. benthamiana leaf epidermal cells expressing either a wild-type MYB23 promoter fusion to GFP, ProMYB23:mGFP-ER ([C] to [E]), or a mutant reporter, ProMYB23_ΔL1:mGFP-ER, in which the L1-box I is deleted ([F] to [H]). Controls ([C] and [F]) had the indicated reporter constructs but lacked effector genes. Reporter constructs in combination with Pro35S:GL2 ([D] and [G]) or Pro35S:HDG11 ([E] and [H]) effector genes are indicated.
(I) Quantification of the results shown in (C) to (H), using images from three biological replicates. Error bars indicate sd. The asterisk marks a significant decrease in activation of the mutant reporter in comparison with the wild-type reporter by GL2 (two-tiered t test, P < 0.005).
(J) Model for the relationship between the GL2, HDG11, and MYB23 transcription factors in trichome development. An activator complex composed of TTG1, GL1, and GL3/EGL3 positively regulates GL2 transcription for initiation of the trichome cell fate. GL2 transcription is maintained as trichomes differentiate via the action of MYB23, which is functionally redundant with GL1. Both MYB23 and GL2 are required for full transcriptional activation of MYB23. In the absence of GL2, HDG11 can partially activate MYB23 at leaf margins, either through direct binding to cis-regulatory elements normally occupied by GL2 or via an alternative mechanism. Trichome phenotypes are shown for the Col wild type (top), gl2-5 (middle), and gl2-5 myb23-3 (bottom).
GL2 Drives a MYB23-Promoter Fusion, and L1-Box I Is Required for Maximal Activation
To test whether L1-box I is required for transcriptional activation of MYB23 by GL2, we transiently expressed a reporter composed of the 3.1-kb promoter upstream of MYB23 fused to GFP in Nicotiana benthamiana. Low levels of GFP fluorescence were observed in controls that expressed the reporter alone (Figures 8C and 8I). A marked increase in GFP fluorescence was observed in epidermal cells that coexpressed GL2 (Figures 8D and 8I). However, no activation above the basal level was detected for cells that coexpressed HDG11 (Figures 8E, 8H, and 8I). An ∼2-fold decrease in activation by GL2 was observed for a mutant reporter that lacked L1-box I (Figures 8G and 8I), indicating that the L1-box is required for maximal activation.
A naturally occurring mutation is found within L1-box I of the Arabidopsis accession Guckingen-0 (Gu-0) (Supplemental Figure 11A). Hierarchical clustering based on pairwise sequence differences among 96 accessions indicated that Col is the most closely related accession to Gu-0 (Nordborg et al., 2005). Strikingly, we observed significantly fewer four-branched trichomes on first leaves from Gu-0 in comparison with Col (Supplemental Figures 11B to 11F), consistent with compromised MYB23 function. Real-time PCR experiments indicated that Gu-0 shoots express lower levels of MYB23 transcripts (Supplemental Figures 11G to 11I), consistent with a function for L1-box I in the full activation of MYB23 by GL2.
DISCUSSION
Class IV HD-Zip Transcription Factors Drive Trichome Cell-Type Differentiation in Plants
In this work, we uncovered a functional redundancy between GL2 and HDG11 in trichome cell-type differentiation. Both genes are members of the class IV HD-Zip transcription factor family from Arabidopsis, and although the full-length sequences are not closely related, we found that their 59-amino acid HD domains display a high degree of amino acid identity (Figure 1; Supplemental Table 1). Class IV HD-Zip transcription factors are highly conserved among the land plants (Floyd et al., 2006; Prigge and Clark, 2006). The role of these transcription factors in the differentiation of epidermis in relation to trichome formation seems to be maintained in both dicot and monocot angiosperms. Variation in the form and function of leaf trichomes may be attributed to evolutionary adaptation and likely occurred by functional diversification of ancestral members of the class IV HD-Zip family. HOMEOBOX1, a family member from Gossypium arboreum, appears to be a functional homolog of GL2 in trichome development (Guan et al., 2008). In tomato (Solanum lycopersicum), a dominant allele of a PDF2-related gene, Woolly, confers a hairy trichome phenotype (Yang et al., 2011). In an opposing fashion, mutations in maize (Zea mays) OUTER CELL LAYER4 and corresponding RNA interference plants indicate that its function is necessary for the inhibition of trichome formation on leaf blades (Vernoud et al., 2009).
Overexpression of the Arabidopsis class IV HD-Zip member ATML1 is sufficient to induce epidermal identity, including the formation of trichomes and stomata in internal cell layers (Takada et al., 2013), providing evidence for its role as a master regulator of shoot epidermis cell fate. Intriguingly, overexpression of the related family member HDG2 is also able to trigger the development of stomata in nonepidermal tissues, and ATML1 and HDG2 bind the same DNA elements to activate transcription in yeast (Peterson et al., 2013). Here, we show that overexpression of HDG11 in the epidermis by either the GL2 or ATML1 promoters induces trichome cell fate in gl2 mutants in a similar manner to GL2 (Figure 5; Supplemental Figure 10). Thus, our study provides additional evidence for combinatorial, overlapping, albeit distinct, functions for members of the class IV HD-Zip transcription factor family.
GL2-Independent Trichome Cell-Type Differentiation Mediated by HDG11 and the R2R3-MYB Transcription Factor MYB23
GL2 was previously identified to play a pivotal role in trichome cell-type differentiation (Rerie et al., 1994). However, gl2 mutants are not completely devoid of trichomes, and they express trichomes that are partially differentiated and branched at leaf margins, suggesting GL2-independent mechanisms for trichome development. Here, we show that one GL2-independent pathway requires HDG11, which encodes a class IV HD-Zip transcription factor first identified as a negative regulator of trichome branching (Nakamura et al., 2006). Since GL2 appears to act upstream of HDG11 in trichome cell-type differentiation, one expectation would be that gl2 is epistatic to hdg11 in the double mutant. In this view, it is surprising that gl2 hdg11 double mutants exhibit enhanced trichome differentiation defects (Figure 2; Supplemental Figures 2 and 3).
The ability of the ProGL2:EYFP:HDG11 construct to rescue trichome defects in the gl2 hdg11 double mutant and partially rescue the gl2 single mutants demonstrates that HDG11 can function as a positive regulator of trichome differentiation when GL2 is absent. HDG11’s role as a negative regulator appears to be directly or indirectly dependent on wild-type GL2 activity. The opposite functions of HDG11 may be explained in a model in which cis-regulatory elements specific for GL2 may be bound by HDG11 when GL2 is absent (Figure 8J). However, the affinity of HDG11 for GL2 binding sites is likely to be significantly lower, since HDG11 did not bind the MYB23 L1-box I sequence in the yeast one-hybrid assay nor did it activate the MYB23 promoter–GFP fusion in N. benthamiana. It is intriguing that overexpression of GL2 under the control of its native promoter can rescue the hdg11 branching phenotype (Figure 6). An analogous explanation is that in hdg11 mutants, GL2 has affinity for HDG11 binding sites. Functional redundancy at the level of DNA binding is consistent with the high degree of similarity in the HD domains from GL2 and HDG11 (Figure 1).
MYB23 Is a Direct Target of GL2 as Part of a Positive Feedback Loop to Maintain Trichome Cell Fate
In gl2 shoots, transcript levels of MYB23 are significantly decreased (Figure 7B), indicating that GL2 is required for full transcription of MYB23. Indeed, GL2 drives the expression of a MYB23 promoter fusion to GFP in the N. benthamiana leaf epidermis (Figure 8D). The additional data presented here, including ChIP and yeast one-hybrid experiments, strongly suggest that GL2 binds to an L1-box in the MYB23 promoter (Figures 7 and 8). A natural mutation in this L1-box that is associated with reduced trichome branching and lower levels of MYB23 transcripts in shoots (Supplemental Figure 11) further corroborates this conclusion. The GFP reporter experiments indicate that although this L1-box is required for full transcriptional activation of MYB23, other factors are involved. One possibility is that GL2 binds other cis-regulatory elements in the 3.1-kb MYB23 promoter. Alternatively, GL2 may activate transcription via physical interaction with another DNA binding protein. Taken together, our findings are consistent with a previous study in which it was shown that overexpression of MYB23 in trichomes using the GL2 promoter partially rescues gl2 trichome defects (Kirik et al., 2005). Contrary to the conventional model, in which GL2 is thought to act downstream of the transcriptional coregulators GL1 and MYB23, these data demonstrate that MYB23 also acts downstream of GL2.
A model for the maintenance of trichome cell fate through the action of GL2 and MYB23 is proposed in Figure 8J. The activator complex is composed of the R2R3-MYB transcription factor GL1, the bHLH proteins GL3/EGL3, and the WD40 protein TTG. Transcriptional activation of GL2 occurs through concurrent binding of GL1 and GL3 (or the redundant protein EGL3) to the GL2 promoter (Wang and Chen, 2008). In turn, GL2 activates MYB23 transcription by binding to the MYB23 promoter. Consistent with GL2 regulation of the MYB23 transcript, the activity of the MYB23 promoter–GUS fusion persists in mature trichomes and is similar to the expression pattern seen for GL2 (Kirik et al., 2001). Promoter and protein-coding region swap experiments corroborate that MYB23 plays the dominant role in trichome branching, although GL1 and MYB23 appear functionally redundant with respect to trichome initiation (Kirik et al., 2005). In the root epidermis, it was shown that MYB23 binds to its own promoter (Kang et al., 2009). In the model presented here (Figure 8J), MYB23 participates in the activator complex and binds to the GL2 promoter as well as to the promoter of its own encoding gene as part of a positive feedback loop to maintain trichome cell fate. In gl2 mutants in which GL2 is absent, DNA binding sites specific for GL2 may be bound by HDG11. Alternatively, HDG11 may promote MYB23 activity directly or indirectly by another mechanism. In this model, the phenotypes for gl2 hdg11 and gl2 myb23 double mutants are similar because in both types of mutants, MYB23 is not fully transcriptionally activated.
Tissue-Specific Relationships between GL2, HDG11, and MYB23
Our results point to a tissue-specific regulation of MYB23 transcript by GL2 in trichomes. The enhanced phenotypes of the gl2 hdg11 and gl2 myb23 double mutants relative to the gl2 single mutants were observed in trichomes but were not apparent in seeds or roots (Figure 4; Supplemental Figure 4). gl2 mutants lack mucilage formation altogether, so it is difficult to envision an enhanced phenotype in the seed. Whereas MYB23 together with MYB5 are implicated in mucilage synthesis and seed coat development (Li et al., 2009), HDG11 expression in seeds is very low. We analyzed the expression pattern of HDG11 versus GL2 using the publicly available transcriptome database Bio-Analytic Resource Arabidopsis eFP browser. Differences between GL2 and HDG11 expression are striking in developing seeds. While GL2 expression is strong, especially in mucilage secretory cells that constitute the seed coat, HDG11 expression is relatively low or absent in developing seeds (Supplemental Figure 12), consistent with our observation that hdg11 mutants are not defective in mucilage production. Intriguingly, expression of the ProGL2:EYFP:HDG11 transgene in hdg11 mutants resulted in a dominant negative inhibition of mucilage formation (Figure 4), analogous to its inhibitory role in limiting trichome branching in the shoot epidermis.
In roots, we observed that expression of the ProGL2:EYFP:HDG11 transgene partially rescued the excess root hair phenotype of gl2 (Figure 4). The differential behavior of HDG11 in seeds versus roots is likely due to tissue-specific interactions with coexpressed genes. The analysis of myb23 mutants has shown that MYB23 is necessary for precise establishment of the root epidermal pattern (Kang et al., 2009). It is noteworthy that transcript levels of MYB23 appear not to be regulated by GL2 in the root. It was previously shown that in roots from gl2 mutants, MYB23 mRNA levels are unchanged relative to those of the wild type (Kang et al., 2009). A genome-wide transcriptome profiling of the Arabidopsis root epidermis also failed to show that MYB23 transcript levels are changed in gl2 root tissues (Bruex et al., 2012).
Several observations suggest crosstalk between class IV HD-Zip transcription factors in regulatory and developmental pathways. In transgenic lines that expressed HDG11 under the control of the GL2 promoter, we observed a novel transient dwarf phenotype (Supplemental Figure 8). Moreover, when HDG11 was expressed under the control of the epidermis-specific ATML1 promoter in gl2 mutants, several of the transgenic lines displayed leaf curling and/or pointy leaves. In relation to these observations, the WW domain protein CURLY FLAG LEAF1, corresponding to a curly leaf mutant phenotype in rice (Oryza sativa), has been shown to physically interact with a class IV HD-Zip family member, HDG1 (Wu et al., 2011). Other striking observations included fertility defects related to pollen viability that were seen in plants heterozygous for either hdg11 or myb23 or both (Supplemental Figure 5), indicating that genetic interactions between these transcription factors are highly sensitive to dosage effects. Relevant to these findings, it was shown recently that a special mutant allele of PDF2 (pdf2-1) in combination with loss-of-function mutations in other family members (HDG1, HDG2, HDG5, or HDG12) result in developmental defects in floral organs (Kamata et al., 2013). Future studies will unravel the molecular basis for these and other interactions involving transcription factors of the class IV HD-Zip family in relation to regulatory circuits underlying the differentiation of specified cell types in plants.
METHODS
Plant Material and Growth Conditions
The wild-type Arabidopsis thaliana ecotypes were Col and Ler. The hdg11-1 (Col) (SAIL_865_G09) (Nakamura et al., 2006) and hdg11-3 (Ler) alleles were described previously (Roeder et al., 2012). The myb23-3 (Col) (SALK_018613) allele contains a T-DNA insertion in the 5′ promoter (Kang et al., 2009). gl2-1 (Ler) is a fast-neutron–induced allele (Koornneef et al., 1982; Di Cristina et al., 1996). gl2-5 (Col) is a En-1 insertion allele of GL2 (Ohashi et al., 2003). The genomic region was sequenced using En205 and En8130 primers (Baumann et al., 1998) and GL2-specific primers (Wang et al., 2007), revealing an insertion following the Ser-encoded triplet AGC (bases 121 to 123), after the first 37 amino acids of the predicted protein. A TGA stop codon is located 29 amino acids downstream, within the En-1 transposon, indicating that the mutant allele results in a truncated protein of 63 amino acids, of which the first 37 amino acids are derived from the N terminus of GL2. En-1 lies at the beginning (33 bp) of the second exon of the gene, upstream of the HD (amino acids 106 to 157). Supplemental Table 4 lists oligonucleotides used for genotyping. Plants were stratified at 4°C for 3 to 5 d and grown on soil containing Metro-Mix 380, vermiculite, and perlite (Hummert International) in a 7:3:2 ratio at 23°C under continuous light. Agrobacterium tumefaciens (GV3101 pMP90)–mediated transformation of plants was performed using the floral dip method as described previously (Clough and Bent, 1998).
DNA Constructs
Construction of ProGL2:EYFP:GL2 (SR54, a derivative of the binary vector pCAMBIA1300) will be described elsewhere (A. Khosla, B.K.P. Venkata, P.N. Cox, D.F. Stucky, P.M. Stephens, S.A. Marlatt, B.P. Srinivas, M. Hülskamp, and K. Schrick, unpublished data). An HDG11 cDNA clone was obtained from the RIKEN Arabidopsis full-length clone collection. Internal SalI and KpnI restriction sites interfering with the cloning scheme were removed prior to amplification by site-directed mutagenesis using a one-step PCR-based method as described previously (Scott et al., 2002). To construct ProGL2:EYFP:HDG11, the cDNA sequences for wild-type HDG11 and hdg11Δ305 were PCR amplified with Phusion High-Fidelity DNA Polymerase (Thermo Scientific) using primers engineered to contain SalI and KpnI restriction sites at the 5′ and 3′ ends, respectively. hdg11Δ305 (referred to as hdg11Δ in Figures 4 and 5) is a deletion mutant missing the N-terminal 305 residues of the predicted protein, resulting in an in-frame 417 amino acid protein that lacks the first 78 residues of the START domain. The amplified PCR fragment was restriction digested with SalI/KpnI and subcloned into the corresponding restriction sites in binary vector SR54 to make ProGL2:EYFP:HDG11 and ProGL2:EYFP:hdg11Δ305. The constructs were transformed into the gl2-5 and hdg11-1 single mutant and gl2-5 hdg11-1 double mutant plants. For ProGL2:EYFP:HDG11, Mendelian segregation of EYFP fluorescence was seen for the T2 progeny from 6 of 19 hdg11-1, 29 of 43 gl2-5, and 28 of 34 gl2-5 hdg11-1 plants. Strong EYFP expression was observed in trichomes from 3 hdg11-1, 16 gl2-5, and 9 gl2-5 hdg11-1 lines carrying ProGL2:EYFP:HDG11. For the ProGL2:EYFP:hdg11Δ construct, none of the transgenic lines from a total of 75 (20 in hdg11, 15 in gl2, and 40 in gl2 hdg11) independent transformants exhibited EYFP expression. The ProATML1:HDG11 construct was made using pAR176 (Roeder et al., 2010). The HDG11 cDNA was cloned into pENTR-D-TOPO (Life Technologies) and moved into pAR176 by Gateway LR Clonase II (Life Technologies). For ProATML1:HDG11, ∼150 T1 plants survived BASTA selection, 24 independent trichome-bearing transformants were characterized, and 8 of 24 T1 progeny segregated in a Mendelian fashion. Oligonucleotides used for plasmid construction, site-directed mutagenesis, and sequence verification are listed in Supplemental Table 4.
RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
Arabidopsis seeds were surface-sterilized and sown onto 0.8% (plant tissue culture grade) agar containing 1× Murashige and Skoog medium (Murashige and Skoog, 1962), 1% Suc, and 0.5% MES buffer at pH 5.8. Seeds were stratified for 3 to 5 d at 4°C and then transferred to 23°C and continuous light for 14 d. After removal of roots and hypocotyl, shoots were flash frozen in liquid nitrogen and stored at −80°C. Approximately 50 mg of each sample was used for RNA extraction using the RNeasy Plant Mini Kit and on-column RNase-Free DNase Set (Qiagen). Furthermore, 0.5 to 2 μg of RNA was used as a template for cDNA synthesis. cDNA synthesis was performed using SuperScript II Reverse Transcriptase (Life Technologies) or GoScript Reverse Transcriptase (Promega) according to the manufacturer’s protocol. Real-time PCR was performed using IQ SYBR Green Supermix with the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Each reaction contained 10 μL of 2× SYBR Green Supermix, 1 μL of forward and reverse 10 mM gene-specific primers, and 5 μL of cDNA (diluted 5-fold) in 20 µL. Standard curves were generated from 10-fold dilutions of amplicons for each primer pair. ACT7 served as the reference gene. The data represent three biological samples of three to four seedling shoots per sample with three technical replicates of each biological replicate. Primers used for PCR are listed in the Supplemental Table 4.
Phenotypic Analysis and Microscopy
Arabidopsis lines were confirmed by PCR and assayed for leaf trichomes, seed coat mucilage, and root hairs. For root hair density analysis, seedlings were grown vertically on 0.8% agar/water medium under continuous light. Primary roots were viewed 3 to 4 d after germination. For quantification of root epidermal cells, seedlings were stained in a 0.05% aqueous solution of toluidine blue, destained in water, mounted in 10% glycerol, and examined for the numbers of trichoblasts and atrichoblasts in the corresponding respective cell files. For seed mucilage analysis, seeds were stained with a 0.02% aqueous solution of ruthenium red (Sigma) for 2 h followed by replacement with deionized water prior to observation. Seeds, rosettes, and roots were imaged with a Leica M125 fluorescence stereomicroscope, and images were captured using a Leica DFC295 digital camera and Leica Application Suite 4.1. For the quantification of trichomes on first leaves, the leaves were placed on 1% agar plates and counting of trichomes on first leaves was performed manually upon viewing with the stereomicroscope. Scanning electron microscopy imaging was performed on fresh leaves and sepals using cold stage (−30°C) with an S-3500N instrument from Hitachi Science Systems equipped with an S-6542 Absorbed Electron Detector, a C1005 SEM Cold/Hot Stage (Oxford Instruments Microanalysis), and backscattered Robinson detector (ETP-USA/Electron Detectors). Images were processed as TIF files using Adobe Photoshop CS4.
Yeast One-Hybrid Analysis
Three tandem copies of L1-box sequences and mutated versions thereof were cloned into the pAbAi vector (Clontech) by annealing of oligonucleotides having KpnI and SalI restriction sites at their ends. Reporter bait constructs were integrated into the yeast genome at the ura3-52 locus using URA3+ selection in the yeast strain SUB62 (MATa lys2-801 leu2-3, 2-112 ura3-52 his3-Δ200 trp1-1). Reporter strains were transformed by standard LiAc transformation with pGAD-T7 (Clontech) containing HD and ZLZ domains from GL2 (amino acids 1 to 231) or HDG11 (amino acids 1 to 169) that been inserted using the EcoRI and BamHI restriction sites. A construct containing the C-terminal START and SAD domains of GL2 (amino acids 253 to 747), also inserted by these restriction sites, was used as a control. Oligonucleotide sequences for this work are listed in Supplemental Table 4.
ChIP–Quantitative PCR
Transgenic seeds expressing ProGL2:EYFP:GL2 were germinated on 0.8% (plant tissue culture grade) agar containing 1× Murashige and Skoog medium (Murashige and Skoog, 1962), and seedlings were harvested at 10 d. Cross-linking of seedlings followed by extraction of chromatin were performed as described (Zhu et al., 2012) with minor modifications. Chromatin was sonicated with the S220 Focused-Ultrasonicator (Covaris) to <500-bp DNA fragments. A 10-fold dilution of total chromatin was prepared to serve as the input control. Immunoprecipitations were performed overnight at 4°C with 2 μL of anti-GFP antibody (ab290; Abcam) bound to Dynabeads Protein A (Life Technologies). After washing sequentially with low-salt wash buffer (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 20 mM Tris-HCl, pH 8), high-salt wash buffer (500 mM NaCl and otherwise identical to the first buffer), LiCl buffer (0.25 mM LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8), and TE buffer (10 mM Tris-HCl, pH 8, and 1 mM EDTA), the immunocomplexes were eluted. Cross-linking was reversed by overnight incubation at 65°C, followed by proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation. ChIP DNA was quantified using iTaq Universal SYBR Green Supermix with the CFX96 Touch Real-Time PCR Detection System (Bio-Rad). PCR was performed in triplicate with the gene-specific primers listed in Supplemental Table 4. ACT7 was used for normalization.
Transient Expression in Nicotiana benthamiana
The ProMYB23:mGFP5-ER reporter contains the 3.1-kb MYB23 promoter driving the expression of an endoplasmic reticulum–targeted modified GFP (mGFP-ER) in pCB302 (Kang et al., 2009). To construct ProMYB23_∆L1:mGFP5-ER, L1-box I was deleted using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs) using the oligonucleotides listed in Supplemental Table 4. To construct the Pro35S:GL2 and Pro35S:HDG11 effector genes, cDNA sequences for GL2 and HDG11 were PCR amplified using Phusion High-Fidelity DNA Polymerase (Thermo Scientific), cloned into pENTR-D-TOPO (Life Technologies), and moved to the pK7WG2 expression vector by Gateway LR Clonase II (Life Technologies). N. benthamiana plants (2 to 4 weeks old) were used to express the various construct combinations by Agrobacterium (GV3101 pMP90)–mediated transient transformation of lower epidermal leaf cells as described previously (Sparkes et al., 2006). Fluorescence images were captured using a Zeiss LSM 5 PASCAL system at 3 d postinfiltration. GFP was excited at 488 nm and detected at 493 to 530 nm. Images of 512 × 512 pixels over an area of 307.1 × 307.1 μm were analyzed from three biological replicates for each effector and reporter combination. Quantification of the image data was performed by calculating the percentage of the measurement area for which the relative GFP fluorescence exceeded a threshold of 20% of the maximum overall signal intensity.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: At1g79840 (GL2), At1g73360 (HDG11), and At5g40330 (MYB23).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Trichomes on Sepals from Arabidopsis Flowers.
Supplemental Figure 2. gl2-1 hdg11-3 Double Mutants from the Ler Ecotype Exhibit Enhanced Trichome Differentiation Defects.
Supplemental Figure 3. Trichome Phenotype of gl2-5 hdg11-3 Mutants.
Supplemental Figure 4. Trichomes on First Leaves of the gl2-5 hdg11-1 myb23-3 Triple Mutant.
Supplemental Figure 5. Developmental Defects of Plants Heterozygous for hdg11-1 and/or myb23-3.
Supplemental Figure 6. Intermediate Trichome Phenotype of hdg11-1 myb23-3 Double Mutants.
Supplemental Figure 7. Root Hair Density and Seed Mucilage Phenotypes in the Ler Ecotype.
Supplemental Figure 8. Crude Oil Levels in gl2 Single and gl2 hdg11 Double Mutant Seeds Are Similar.
Supplemental Figure 9. Leaf and Rosette Phenotypes of gl2 and gl2 hdg11 Mutant Lines Expressing ProGL2:EYFP:HDG11.
Supplemental Figure 10. Expression of HDG11 under the Control of the ATML1 Epidermis-Specific Promoter Induces Trichome Differentiation in gl2 Mutants.
Supplemental Figure 11. A Natural Polymorphism in the MYB23 L1-Box I from the Gu-0 Accession Is Correlated with Reduced Trichome Branching.
Supplemental Figure 12. Comparison of GL2 and HDG11 Expression in the Developing Seed.
Supplemental Table 1. Pairwise Alignments between the Homeodomains from GL2/At1g79840 and Each of the 15 Other Class IV HD-Zip Family Members.
Supplemental Table 2. Trichome Quantification on the First Two Leaves of Arabidopsis Plants.
Supplemental Table 3. Quantification of Trichoblast and Atrichoblast Hair Cell Files on Roots.
Supplemental Table 4. Oligonucleotides Used in This Study.
Supplementary Material
Acknowledgments
We thank Kent Hampton and Lloyd Willard for technical assistance with scanning electron microscopy, Paige Cox for the construction of ProATML1:HDG11, Dave Trumble for seed oil analysis, Michael Veeman for assistance with the microscopy of anthers, Martin Hülskamp and Bhylahalli Srinivas for providing SR54, Adrienne Roeder for pAR176 and hdg11-3 seeds, Myeong Min Lee for ProMYB23:mGFP5-ER, Jeroen Roelofs for the SUB62 yeast strain, and Chris Toomajian for Gu-0 seeds. A.K. was partially supported by the Johnson Cancer Research Center. J.M.P. was supported by the National Science Foundation (Grant MCB-1122016). This project was supported by the National Research Initiative of the USDA National Institute of Food and Agriculture (Grant 2007-35304-18453). This is contribution 14-161-J from the Kansas Agricultural Experiment Station.
AUTHOR CONTRIBUTIONS
K.S. conceived the project and performed crosses, phenotypic analyses, and microscopy. K.S., A.K., and T.R.N. analyzed data and wrote the article. A.K. made DNA constructs and transgenic plants, performed microscopy, and conducted the yeast one-hybrid and ChIP experiments. J.M.P. and A.K. performed real-time PCR experiments. A.P.B. and A.M.B. conducted segregation analysis, genotyping, microscopy, and phenotypic studies relating to the mutants and transgenic lines.
Glossary
- bHLH
basic helix-loop-helix
- Col
Columbia
- Ler
Landsberg erecta
- ChIP
chromatin immunoprecipitation
- Gu-0
Guckingen-0
Footnotes
Online version contains Web-only data.
References
- Balkunde R., Pesch M., Hülskamp M. (2010). Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr. Top. Dev. Biol. 91: 299–321 [DOI] [PubMed] [Google Scholar]
- Baumann E., Lewald J., Saedler H., Schulz B., Wisman E. (1998). Successful PCR-based reverse genetic screens using an En-1-mutagenised Arabidopsis thaliana population generated via single-seed descent. Theor. Appl. Genet. 97: 729–734 [Google Scholar]
- Bruex A., Kainkaryam R.M., Wieckowski Y., Kang Y.H., Bernhardt C., Xia Y., Zheng X., Wang J.Y., Lee M.M., Benfey P., Woolf P.J., Schiefelbein J. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Di Cristina M., Sessa G., Dolan L., Linstead P., Baima S., Ruberti I., Morelli G. (1996). The Arabidopsis Athb-10 (GLABRA2) is an HD-Zip protein required for regulation of root hair development. Plant J. 10: 393–402 [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Floyd S.K., Zalewski C.S., Bowman J.L. (2006). Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173: 373–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe M. (2012). The patterning of epidermal hairs in Arabidopsis—Updated. Curr. Opin. Plant Biol. 15: 31–37 [DOI] [PubMed] [Google Scholar]
- Guan X.Y., Li Q.J., Shan C.M., Wang S., Mao Y.B., Wang L.J., Chen X.Y. (2008). The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol. Plant. 134: 174–182 [DOI] [PubMed] [Google Scholar]
- Hülskamp M., Misŕa S., Jürgens G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76: 555–566 [DOI] [PubMed] [Google Scholar]
- Kamata N., Okada H., Komeda Y., Takahashi T. (2013). Mutations in epidermis-specific HD-ZIP IV genes affect floral organ identity in Arabidopsis thaliana. Plant J. 75: 430–440 [DOI] [PubMed] [Google Scholar]
- Kang Y.H., Kirik V., Hulskamp M., Nam K.H., Hagely K., Lee M.M., Schiefelbein J. (2009). The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V., Lee M.M., Wester K., Herrmann U., Zheng Z., Oppenheimer D., Schiefelbein J., Hulskamp M. (2005). Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development 132: 1477–1485 [DOI] [PubMed] [Google Scholar]
- Kirik V., Schnittger A., Radchuk V., Adler K., Hülskamp M., Bäumlein H. (2001). Ectopic expression of the Arabidopsis AtMYB23 gene induces differentiation of trichome cells. Dev. Biol. 235: 366–377 [DOI] [PubMed] [Google Scholar]
- Koornneef M., Dellaert L.W., van der Veen J.H. (1982). EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat. Res. 93: 109–123 [DOI] [PubMed] [Google Scholar]
- Li S.F., Milliken O.N., Pham H., Seyit R., Napoli R., Preston J., Koltunow A.M., Parish R.W. (2009). The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21: 72–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks M.D., Wenger J.P., Gilding E., Jilk R., Dixon R.A. (2009). Transcriptome analysis of Arabidopsis wild-type and gl3-sst sim trichomes identifies four additional genes required for trichome development. Mol. Plant 2: 803–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T., Skoog F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15: 473–497 [Google Scholar]
- Nakamura M., Katsumata H., Abe M., Yabe N., Komeda Y., Yamamoto K.T., Takahashi T. (2006). Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol. 141: 1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Oka A., Rodrigues-Pousada R., Possenti M., Ruberti I., Morelli G., Aoyama T. (2003). Modulation of phospholipid signaling by GLABRA2 in root-hair pattern formation. Science 300: 1427–1430 [DOI] [PubMed] [Google Scholar]
- Peterson K.M., Shyu C., Burr C.A., Horst R.J., Kanaoka M.M., Omae M., Sato Y., Torii K.U. (2013). Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development 140: 1924–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C.P., Aravind L. (1999). START: A lipid-binding domain in StAR, HD-ZIP and signalling proteins. Trends Biochem. Sci. 24: 130–132 [DOI] [PubMed] [Google Scholar]
- Prigge M.J., Clark S.E. (2006). Evolution of the class III HD-Zip gene family in land plants. Evol. Dev. 8: 350–361 [DOI] [PubMed] [Google Scholar]
- Rerie W.G., Feldmann K.A., Marks M.D. (1994). The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev. 8: 1388–1399 [DOI] [PubMed] [Google Scholar]
- Roeder A.H., Chickarmane V., Cunha A., Obara B., Manjunath B.S., Meyerowitz E.M. (2010). Variability in the control of cell division underlies sepal epidermal patterning in Arabidopsis thaliana. PLoS Biol. 8: e1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder A.H., Cunha A., Ohno C.K., Meyerowitz E.M. (2012). Cell cycle regulates cell type in the Arabidopsis sepal. Development 139: 4416–4427 [DOI] [PubMed] [Google Scholar]
- Schrick K., Nguyen D., Karlowski W.M., Mayer K.F. (2004). START lipid/sterol-binding domains are amplified in plants and are predominantly associated with homeodomain transcription factors. Genome Biol. 5: R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.P., Teh A., Peng C., Lavin M.F. (2002). One-step site-directed mutagenesis of ATM cDNA in large (20kb) plasmid constructs. Hum. Mutat. 20: 323. [DOI] [PubMed] [Google Scholar]
- Shen B., Sinkevicius K.W., Selinger D.A., Tarczynski M.C. (2006). The homeobox gene GLABRA2 affects seed oil content in Arabidopsis. Plant Mol. Biol. 60: 377–387 [DOI] [PubMed] [Google Scholar]
- Shi L., Katavic V., Yu Y., Kunst L., Haughn G. (2012). Arabidopsis glabra2 mutant seeds deficient in mucilage biosynthesis produce more oil. Plant J. 69: 37–46 [DOI] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4: 447–456 [DOI] [PubMed] [Google Scholar]
- Takada S., Takada N., Yoshida A. (2013). ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140: 1919–1923 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R., Iwata M., Sugiyama J., Kotake T., Ishida T., Yokoyama R., Nishitani K., Okada K., Wada T. (2009). The GLABRA2 homeodomain protein directly regulates CESA5 and XTH17 gene expression in Arabidopsis roots. Plant J. 60: 564–574 [DOI] [PubMed] [Google Scholar]
- Vernoud V., Laigle G., Rozier F., Meeley R.B., Perez P., Rogowsky P.M. (2009). The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 59: 883–894 [DOI] [PubMed] [Google Scholar]
- Wang S., Chen J.G. (2008). Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 49: 1792–1804 [DOI] [PubMed] [Google Scholar]
- Wang S., Kwak S.H., Zeng Q., Ellis B.E., Chen X.Y., Schiefelbein J., Chen J.G. (2007). TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882 [DOI] [PubMed] [Google Scholar]
- Western T.L., Burn J., Tan W.L., Skinner D.J., Martin-McCaffrey L., Moffatt B.A., Haughn G.W. (2001). Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 127: 998–1011 [PMC free article] [PubMed] [Google Scholar]
- Western T.L., Skinner D.J., Haughn G.W. (2000). Differentiation of mucilage secretory cells of the Arabidopsis seed coat. Plant Physiol. 122: 345–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R., Li S., He S., Wassmann F., Yu C., Qin G., Schreiber L., Qu L.J., Gu H. (2011). CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23: 3392–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Li H., Zhang J., Wang T., Ye Z. (2011). Fine-mapping of the woolly gene controlling multicellular trichome formation and embryonic development in tomato. Theor. Appl. Genet. 123: 625–633 [DOI] [PubMed] [Google Scholar]
- Yang C.X., Ye Z.B. (2013). Trichomes as models for studying plant cell differentiation. Cell. Mol. Life Sci. 70: 1937–1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.Y., Chung M.C., Tu C.Y., Leu W.M. (2002). OSTF1: a HD-GL2 family homeobox gene is developmentally regulated during early embryogenesis in rice. Plant Cell Physiol. 43: 628–638 [DOI] [PubMed] [Google Scholar]
- Zhu, J.-Y., Sun, Y., and Wang, Z.-Y. (2012). Genome-wide identification of transcription factor-binding sites in plants using chromatin immunoprecipitation followed by microarray (ChIP-chip) or sequencing (ChIP-seq). Methods Mol. Biol. 876: 173–188 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.