This work identifies a physiological substrate and a physiological function of the Arabidopsis oligopeptide transporter, OPT3, in iron (Fe) homeostasis, provides a mechanistic explanation of the role of OPT3 in systemic Fe signaling, and uncovers an aspect of crosstalk between Fe homeostasis and cadmium partitioning.
Abstract
Iron is essential for both plant growth and human health and nutrition. Knowledge of the signaling mechanisms that communicate iron demand from shoots to roots to regulate iron uptake as well as the transport systems mediating iron partitioning into edible plant tissues is critical for the development of crop biofortification strategies. Here, we report that OPT3, previously classified as an oligopeptide transporter, is a plasma membrane transporter capable of transporting transition ions in vitro. Studies in Arabidopsis thaliana show that OPT3 loads iron into the phloem, facilitates iron recirculation from the xylem to the phloem, and regulates both shoot-to-root iron signaling and iron redistribution from mature to developing tissues. We also uncovered an aspect of crosstalk between iron homeostasis and cadmium partitioning that is mediated by OPT3. Together, these discoveries provide promising avenues for targeted strategies directed at increasing iron while decreasing cadmium density in the edible portions of crops and improving agricultural productivity in iron deficient soils.
INTRODUCTION
Iron (Fe) is essential for plant growth and development and is an important component of the human diet. Cadmium (Cd), on the other hand, is a nonessential and highly toxic element that competes with Fe for uptake and partitioning in plant tissues, thus posing a threat to crop productivity and human health. The ability of Fe to change its oxidation state (Fe3+ ↔ Fe2+) highlights its importance in biological processes that involve electron transfer reactions (e.g., respiration and photosynthesis). However, the same property imposes toxicity when Fe is accumulated in cells in excess due to its ability to promote the formation of reactive oxygen species (Valko et al., 2005). Bioavailability of Fe in aerobic soils with neutral to basic pH is below the limits required to sustain plant growth and development because insoluble Fe(III) chelates prevail under these conditions. Consequently, alkaline soils, occupying ∼30% of the world’s arable lands, are considered Fe-limiting for plant growth (Marschner, 1995). To increase Fe bioavailability, Arabidopsis thaliana and other dicotyledonous and nongraminaceous monocotyledonous plants use a reduction strategy (Hindt and Guerinot, 2012; Kobayashi and Nishizawa, 2012). In brief, this mechanism includes the acidification of the rhizosphere by the H+-ATPases of the Arabidopsis H+-ATPase family (Santi and Schmidt, 2009), the reduction of Fe(III) chelates to soluble Fe(II) by the root surface-localized ferric chelate reductase FRO2 (Robinson et al., 1999), and the subsequent uptake of Fe(II) into root epidermal cells by the high-affinity Fe(II) transporter IRT1 (Eide et al., 1996). The FRO2/IRT1 system constitutes the major pathway for Fe entry into root epidermal cells. Given the essential and toxic nature of Fe, expression of FRO2 and IRT1 is under tight local and long-distance regulation (Hindt and Guerinot, 2012; Kobayashi and Nishizawa, 2012). Interestingly, Fe itself plays a signaling role and is regarded as a positive regulator of local signaling (Vert et al., 2003). In contrast, it is suggested to act as a negative regulator of shoot-to-root signaling via the phloem (Hindt and Guerinot, 2012; Kobayashi and Nishizawa, 2012). However, the latter suggestion has not been substantiated experimentally.
After initial uptake from the soil into root epidermal cells, Fe moves symplastically toward the vasculature and is effluxed into the xylem vessels where it is chelated with citrate to form a tri-Fe(III) tricitrate complex that undergoes long-distance transport to the shoot (Durrett et al., 2007). Another strong Fe ligand that is responsible for the translocation of Fe in the phloem is a nonproteinogenic amino acid, nicotianamine (NA) (Curie et al., 2009). The molecular machinery contributing to root-to-shoot partitioning of Fe, its redistribution between source and sink tissues, and events involved in shoot-to-root communication of shoot Fe status are much less understood. The key identified players in Arabidopsis are the multidrug and toxin efflux family member FRD3, the ferroportin (FPN)-like protein FPN1 (alias IREG1 [IRON REGULATED1]), and members of two distinct clades of the Oligopeptide Transporter family, the Yellow Stripe-like (YSL) proteins and the Oligopetide Transporters (OPTs), for which the family was named (Rogers and Guerinot, 2002; Green and Rogers, 2004; Morrissey et al., 2009; Lubkowitz, 2011). FRD3 mediates citrate release into the apoplastic space and is essential for xylem-based Fe movement and Fe nutrition in tissues lacking symplastic connections (Rogers and Guerinot, 2002; Green and Rogers, 2004; Roschzttardtz et al., 2011), FPN1 is proposed to load Fe into the xylem (Morrissey et al., 2009), while YSL2 is suggested to be involved in lateral distribution of Fe-NA complexes from the xylem into neighboring cells (DiDonato et al., 2004; Schaaf et al., 2005).
The phloem-based long-distance transport to sink tissues, such as young leaves, developing seeds, and roots, involves apoplastic loading of Fe into the companion cells/sieve elements complex (CC/SE) as well as unloading into corresponding sink tissues. Knowledge of transporters contributing to phloem loading/unloading is scarce. The main contributors in this process are At-YSL1, At-YSL3, and Os-YSL2, which mediate Fe-NA transport and facilitate Fe loading into seeds of Arabidopsis and rice (Oryza sativa) (Waters et al., 2006; Chu et al., 2010; Ishimaru et al., 2010). In this regard, a member of the OPT family in Arabidopsis, OPT3, has been under scrutiny for more than a decade since the discovery of its role in Fe homeostasis (Stacey et al., 2002, 2006, 2008). Studies in heterologous systems have implicated Arabidopsis and rice OPT family members, and the closest OPT3 homolog from Brassica juncea, Bj-GT1, in transport of synthetic peptides, as well as of the ubiquitous tripeptide GSH (Bogs et al., 2003; Cagnac et al., 2004; Osawa et al., 2006). However, the physiological substrate(s) of OPTs, including OPT3, has not yet been identified. With respect to Fe homeostasis, it was shown that (1) OPT3 mRNA is expressed in the vasculature where it is transcriptionally upregulated by Fe deficiency; (2) OPT3 is involved in Fe accumulation in seeds, and loss of this function is suggested to cause embryo lethality in the opt3-1 null mutant; (3) the opt3-2 knockdown mutant harboring a T-DNA insertion in the promoter region is viable but accumulates high levels of Fe in both shoots and roots while exhibiting constitutive Fe starvation phenotypes (e.g., upregulated expression of IRT1 and FRO2) even when the mutant is grown under Fe-sufficient conditions (Stacey et al., 2002, 2008). However, the mechanistic basis of the misregulated Fe signaling in the opt3-2 knockdown mutant and the physiological substrate of OPT3 are unknown.
It is noteworthy that disrupted Fe signaling observed in the opt3-2 mutant has been also found in several other mutants, such as the frd3 (man1) mutant and the quadruple nicotianamine synthase mutant (nas4x-1) of Arabidopsis, the dgl and brz iron homeostasis mutants of pea (Pisum sativum), and the chloronerva (chln) mutant of tomato (Solanum lycopersicum), all showing constitutive activation of Fe acquisition genes even when grown under Fe-sufficient conditions (Scholz et al., 1985; Grusak et al., 1990; Kneen et al., 1990; Grusak and Pezeshgi, 1996; Schuler et al., 2012). Importantly, foliar application of Fe blocked the expression of Fe-acquisition genes in the wild-type cultivars and in frd3-3, brz, and chln mutants, but not in opt3-2 and dgl mutants, reinforcing the previous suggestion of the existence of an Fe-related repressive signal moving from leaves to roots and pointing to distinct molecular mechanisms of shoot-to-root communication of Fe status in different mutants (García et al., 2013, Maas et al., 1988). Therefore, the identification of the physiological substrate of OPT3 is among the key questions to be addressed with respect to its role in Fe homeostasis and shoot-to-root Fe signaling.
Iron homeostasis is tightly linked with homeostasis of essential elements such as zinc (Zn), manganese (Mn), cobalt (Co), and the nonessential and potentially toxic element cadmium (Cd) due to the low substrate specificity of IRT1 (Eide et al., 1996; Baxter et al., 2008). The crosstalk between essential elements and Cd has been under scrutiny over past decades because Cd is increasingly released into the environment as industrial and consumer waste and poses a threat to crop productivity and human health (Järup, 2003). Cadmium causes stunting and chlorosis in plants and affects major biochemical processes, including redox balance, photosynthesis, and water status (Hasan et al., 2009). Cadmium is detoxified in the cytosol by forming a bidentate Cd-GS2 complex with GSH (Li et al., 1997), which then facilitates synthesis of strong Cd ligands, phytochelatins (PC) (Rea 2012). Cd-PC complexes, as well as free Cd ions, are either sequestrated into the vacuole in the root or bypass the vacuole and instead load into xylem vessels of the root to travel into shoot with the transpiration stream and are eventually loaded into vacuoles in the shoot (Salt et al., 1995; Wong and Cobbett, 2009; Park et al., 2012). With respect to Fe nutrition, it has been shown that Cd competes with Fe(II) for the uptake into root epidermal cells via IRT1 and that IRT1-overexpressing Arabidopsis plants accumulate more Cd (Eide et al., 1996; Connolly et al., 2002). A recent finding that co-overexpression of a master regulator of Fe homeostasis, FIT, with its binding partners, the transcription factor bHLH38 or bHLH39, enhances Cd tolerance in Arabidopsis by increasing Cd sequestration in roots and improving Fe homeostasis of shoots (Wu et al., 2012) further emphasizes important relationship between Cd resistance and Fe homeostasis.
Here, we report that a member of the oligopeptide transporter family in Arabidopsis, OPT3, is a phloem-specific transporter that mediates Fe loading into the phloem, and, unlike other members of this family, transports transition metal ions rather than small peptides as the name oligopeptide transporter implies. Our findings also suggest that by loading of Fe into the phloem in leaves, OPT3 regulates both signaling of Fe demand from shoots to roots and Fe transport to developing tissues. We also present data showing an aspect of crosstalk between Fe homeostasis and Cd partitioning that is mediated by OPT3.
RESULTS
OPT3 Localizes to the Plasma Membrane in Arabidopsis Protoplasts
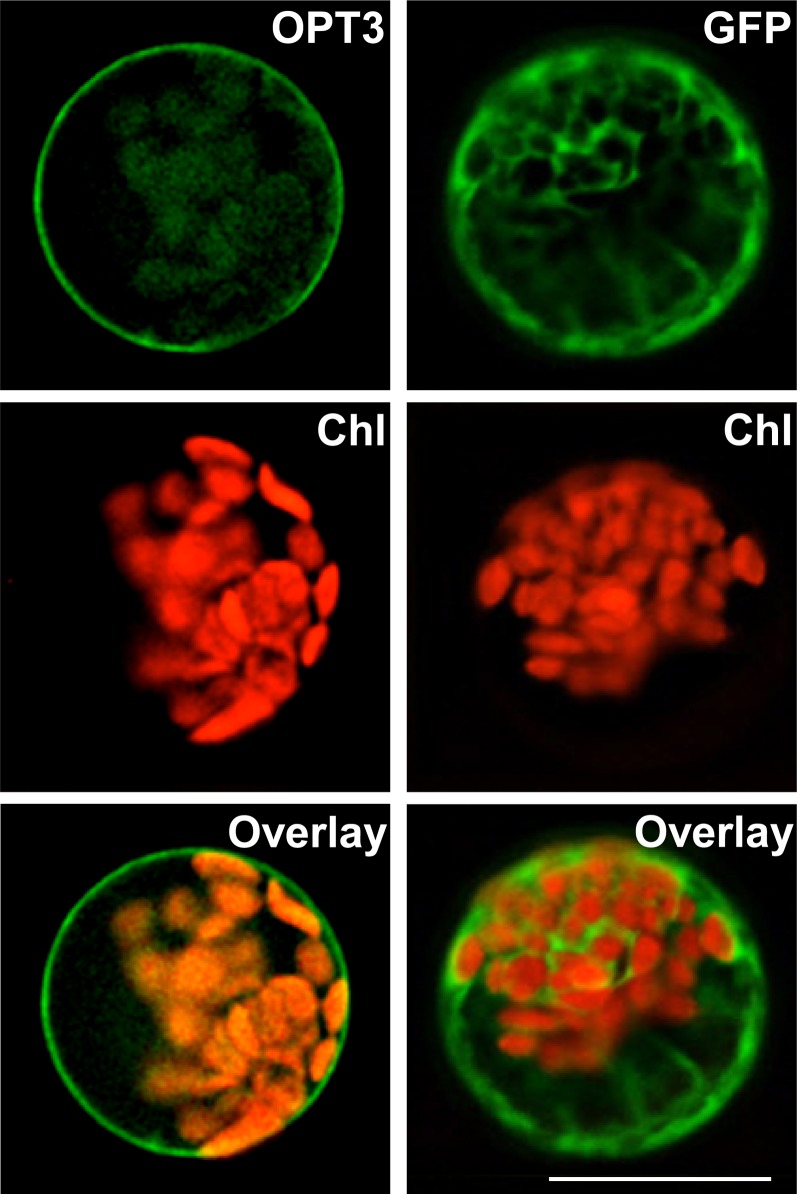
To begin the investigation of the role of OPT3 in the regulation of Fe deficiency responses, its physiological substrate and function in Arabidopsis, we first determined its subcellular localization. We fused the OPT3 cDNA with a coding sequence of green fluorescent protein (GFP) and transiently expressed the OPT3-GFP construct under the control of the cauliflower mosaic virus 35S promoter in Arabidopsis protoplasts. GFP-mediated fluorescence was present in the cytosol and did not overlap with chlorophyll autofluorescence in protoplasts transfected with GFP only (Figure 1). In contrast, OPT3-GFP–mediated fluorescence was present at the periphery of transfected protoplasts, did not overlap with chlorophyll autofluorescence, and was absent in the tonoplast (Figure 1). The plasma membrane localization of OPT3 was further confirmed by transient expression of the OPT3-GFP construct in onion (Allium cepa) epidermal cells (Supplemental Figure 1). Therefore, we concluded that OPT3 localizes to the plasma membrane in Arabidopsis and thus is involved in movement of substrate(s) into or out of the cell rather than in subcellular (e.g., vacuolar) sequestration.
Figure 1.
OPT3 Localizes to the Plasma Membrane in Arabidopsis Protoplasts.
GFP-mediated fluorescence, derived from the OPT3-GFP (OPT3) constructor or the empty GFP vector (GFP), and chlorophyll autofluorescence (Chl) were visualized using FITC or rhodamine filter sets. Superimposed images of chlorophyll autofluorescence and GFP-mediated fluorescence (Overlay) were created to demonstrate that green fluorescence was derived from GFP. Bar = 20 μm.
OPT3 Does Not Complement the GSH Uptake Deficiency of the S. cerevisiae GSH Uptake Mutant, opt1, and Does Not Mediate GSH Uptake in Yeast
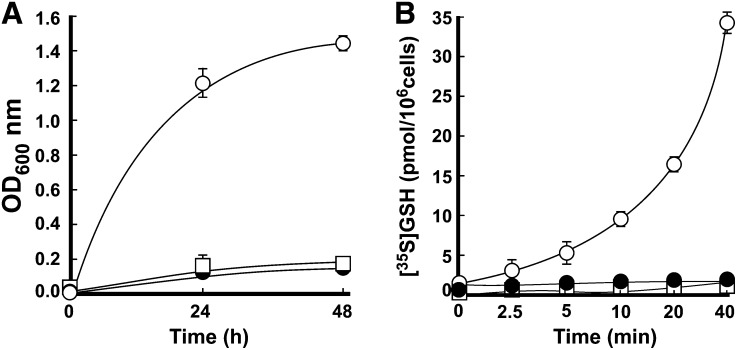
The closest OPT3 homolog from B. juncea, Bj-GT1, transports GSH (Bogs et al., 2003). To test whether OPT3 could transport GSH as well, we used functional complementation and in vitro transport assays in the S. cerevisiae mutant lacking a plasma membrane–localized GSH transporter, Opt1p (alias Hgt1p) (Bourbouloux et al., 2000). As opt1 mutant cells do not grow on medium with GSH as the sole source of sulfur, we first tested whether heterologously expressed At-OPT3 could complement the GSH uptake deficiency of the opt1 mutant. We transformed opt1 mutant cells with the vector expressing Sc-OPT1 or At-OPT3 (opt1/OPT1 and opt1/OPT3, respectively) or with the empty vector (opt1/EV) and analyzed the ability of transformants to grow in liquid minimal medium with GSH as the only sulfur source. As a control, we replaced GSH with ammonium sulfate. We found that while the growth of opt1/OPT3 and opt1/EV yeast lines was undistinguishable from opt1/OPT1 in control medium (Supplemental Figure 2), only cells transformed with Sc-OPT1 were able to grow in medium with GSH as the only source of sulfur (Figure 2A). These data indicated that heterologously expressed At-OPT3 could not complement the GSH uptake defect of the opt1 mutant. We also performed in vitro uptake assays and found that while opt1/OPT1 cells absorb [35S]GSH, opt1/OPT3 and opt1/EV yeast lines were not able to accumulate [35S]GSH (Figure 2B). These data suggested that, in this heterologous system, At-OPT3 does not mediate GSH transport.
Figure 2.
OPT3 Does Not Mediate GSH Transport in S. cerevisiae.
(A) S. cerevisiae opt1 mutant cells expressing Sc-OPT1cDNA (open circles), the empty vector (closed circles), or vector with the At-OPT3 cDNA insert (open squares) were grown in SC-S media supplemented without or with 200 μM GSH. OD values were measured after 24 and 48 h of culturing at 30°C.
(B) Time course of in vitro [35S]GSH uptake by opt1 mutant cells expressing Sc-OPT1cDNA (open circles), the empty vector (closed circles), or vector with the At-OPT3 cDNA insert (open squares). Error bars represent se (n = 3).
Heterologously Expressed OPT3 Partially Rescues the Fe Uptake Defect of the S. cerevisiae fet3 fet4 Mutant
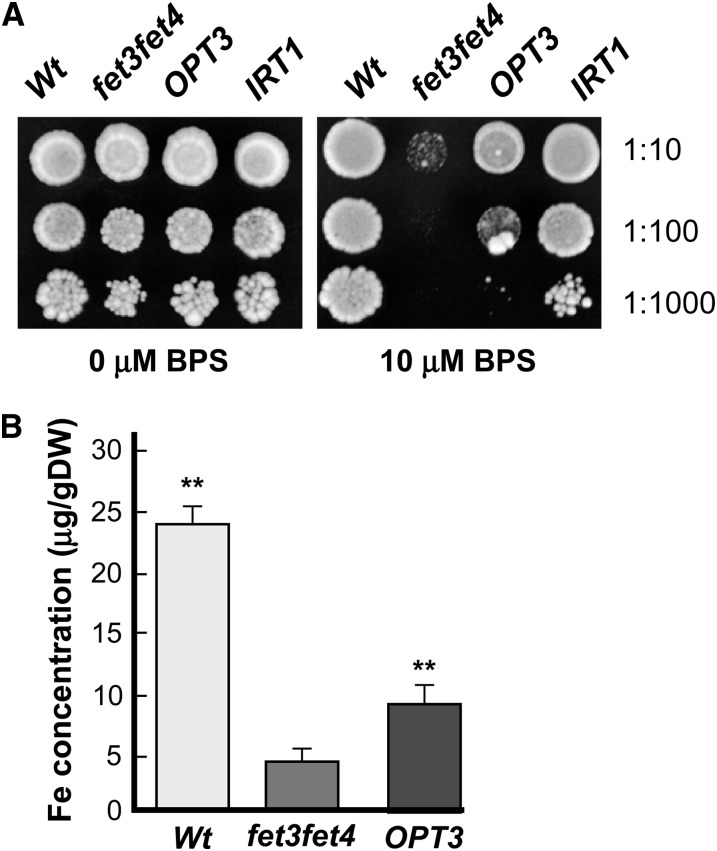
Because OPT3 has been linked to Fe homeostasis in Arabidopsis (Stacey et al., 2002, 2008; Wintz et al., 2003), we next tested whether it would be able to rescue Fe accumulation defects of the S. cerevisiae fet3 fet4 mutant, which lacks both high- and low-affinity Fe uptake systems and cannot grow in Fe-limited conditions (Dix et al., 1994). DY1457 wild-type and fet3 fet4 mutant yeast cells were transformed with an empty vector, and the fet3 fet4 mutant was transformed with a vector containing At-OPT3 cDNA or At-IRT1 cDNA (used here as a positive control). Cell lines were grown on solid medium with or without the Fe chelator bathophenanthroline disulfonate (BPS). All yeast strains grew well on medium without BPS (Figure 3A). In contrast to wild-type cells expressing the empty vector, the fet3 fet4 mutant expressing the empty vector was not able to grow on medium supplemented with BPS. Consistent with the function of At-IRT1 in high-affinity Fe(II) transport (Eide et al., 1996), its heterologous expression rescued the growth defect of the fet3 fet4 mutant on Fe-limited medium. Heterologous expression of At-OPT3 partially rescued the growth defect of fet3 fet4 on Fe-limited medium, suggesting that it might be involved in Fe transport.
Figure 3.
OPT3 Partially Rescues Fe Deficiency of the fet3 fet4 S. cerevisiae Mutant.
(A) The wild type and the fet3 fet4 mutant, transformed with the empty vector (Wt and fet3fet4, respectively), and the fet3 fet4 mutant transformed with At-OPT3 or At-IRT1 cDNAs (OPT3 and IRT1) were serially 10-fold diluted and spotted onto solid medium supplemented with the indicated concentrations of the Fe chelator BPS. Colonies were visualized after incubating plates for 6 d at 30°C. Dilution series are indicated on the left.
(B) Iron concentration in different S. cerevisiae lines, designated as in (A). Shown are mean values ± se (n = 5 to 8); asterisks indicate statistically significant differences (P ≤ 0.001) from the empty vector–expressing opt2 or fet3 fet4 mutant cells.
We then compared the Fe concentration of the wild-type cells expressing the empty vector with that of the fet3 fet4 mutant expressing either the empty vector or the vector with At-OPT3 cDNA. We found that the Fe concentration was 2-fold higher in At-OPT3–expressing cells in comparison with fet3 fet4 cells expressing the empty vector (Figure 3B). We note that in these experiments, cells were grown in medium lacking Fe ligands such as nicotianamine. Therefore, the increased Fe concentration of the At-OPT3–expressing cells (Figure 3B) could be due to the ability of At-OPT3 to mediate uptake of Fe ions. This suggestion challenges the prevailing view that At-OPT3 is involved in the transport of peptides, as was shown for other OPT family members (Lubkowitz, 2011).
OPT3 Mediates Transition Ions Transport in Xenopus laevis Oocytes
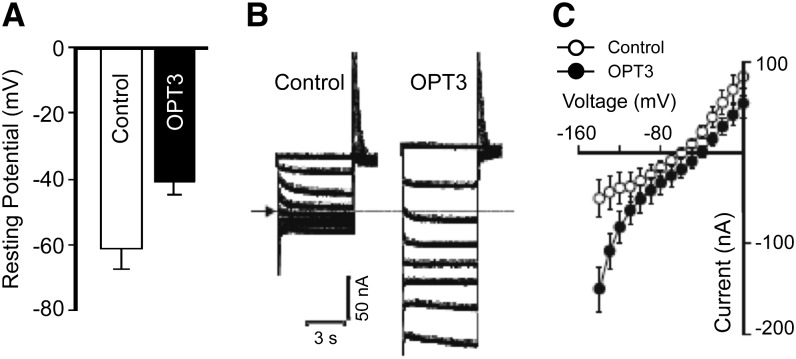
Transport properties of OPT3 were examined in X. laevis oocytes. On the onset of uptake studies, we examined the localization of OPT3-GFP in oocytes, characterized electrophysiological properties of OPT3-expressing oocytes, and determined suitable uptake conditions. We found that OPT3 resided at the plasma membrane in oocytes, as shown by the localization of the OPT3-GFP–mediated fluorescence at the cell periphery (Supplemental Figure 3). When bathed in ND96 recording solution, oocytes injected with OPT3 cRNA had significantly less negative resting membrane potentials than control cells in the same ND96 recording solution (Figure 4A), suggesting that the expression of OPT3 resulted in an OPT3-mediated electrogenic transport. Further investigations were performed by measuring ion transport under voltage clamp conditions, using the conventional two-electrode voltage-clamp method (Figure 4B). In ND96 recording solution, OPT3-expressing cells mediated larger inward (i.e., negative) currents relative to those recorded in control cells. Additionally, the OPT3-mediated currents reversed at less negative membrane potentials than those recorded in control cells (about −40 mV in contrast to −60 mV observed for control cells; Figure 4C). By convention, the recorded inward (e.g., negative) currents are the product of net positive charge influx (or net negative charge efflux). Although we propose that OPT3 mediates cation influx, the exact nature of the ion(s) carrying the OPT3-mediated current in this ionic environment remains unknown. However, we speculate that under these conditions, the inward currents and decreased resting membrane potential observed in OPT3-expressing oocyte cells are the product of a broad substrate specificity of OPT3, which results in OPT3-mediated uptake of the high level of Ca2+ present in the ND96-recording solution. Unfortunately, efforts to reduce this large Ca2+ background (e.g., 1.8 mM CaCl2) present in the ND96 recording solution below 0.9 mM resulted in an increased activity of endogenous inward currents in control cells (Supplemental Figure 4A). Furthermore, total removal of CaCl2 from the bath solution resulted in the activation of a large endogenous inward conductance (>1 μA at −140 mV) similar to the reported large Ca2+-inactivated Cl− conductance (CaIC) (Amasheh and Weber, 1999). Therefore, based on these preliminary electrophysiological findings, we modified the ionic conditions of the uptake solution to maximize our chances to characterize metal uptake in OPT3-expressing oocytes. We maintained 0.9 mM CaCl2 in the medium used for the cation uptake experiments to minimize the activity of endogenous transporters. Likewise, as moderate acidification of the basal uptake media (from 7.5 to 6.0) did not alter the basal transport characteristics of control cells (Supplemental Figure 4B), the pH of the uptake media was adjusted to 6.0. We note that in contrast to control cells, extracellular acidification resulted in a stimulation of the OPT3-mediated inward currents, particularly at holding potentials more negative than −100 mV (Supplemental Figure 4C). This is consistent with the proton-coupled nature of OPT-mediated transport (Osawa et al., 2006). Taken together, our data show that OPT3 is functional at the plasma membrane of oocytes, making this heterologous system suitable for cation uptake experiments.
Figure 4.
OPT3 Is Functional in X. laevis Oocytes.
(A) Resting membrane potentials of OPT3-expressing (OPT3) and water-injected (Control) cells measured in standard ND96 recording solution.
(B) Example of OPT3-mediated currents (right panel) elicited in response to holding potentials ranging from 0 to −140 mV (shown only in 20-mV increments for clarity) in standard ND96 recording solution. Endogenous currents recorded in control cells are shown for reference on the left panel. The arrow and dotted line on the left margin indicates the zero current level.
(C) Mean current-voltage (I/V) curves constructed from the steady state currents recordings such as those shown in (B) for holding potentials ranging from 0 to −140 mV in 10-mV steps. Error bars represent se (n = 5).
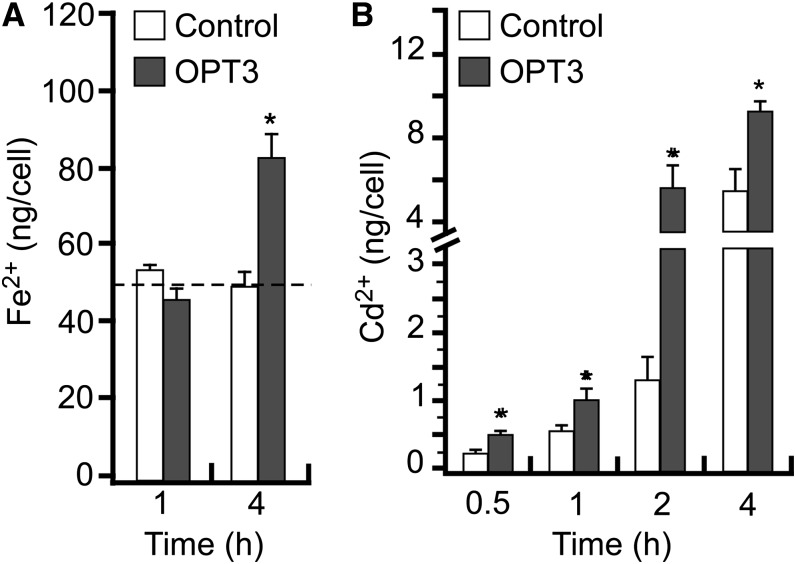
Having established the uptake conditions that would not lead to changes in the endogenous transport in oocytes, we analyzed Fe2+ uptake in OPT3-expressing cells. Significant uptake of Fe2+ was observed in oocytes expressing OPT3 cRNA only after 4 h of incubation possibly due to the high endogenous levels of Fe2+ found in oocytes (∼55 ng/cell) (Figure 5A). Since our electrophysiological studies (Figure 4) suggest that OPT3 might have broad substrate specificity and because Fe transporters such as IRT1 can also transport other divalent metals among which is the toxic metal Cd (Eide et al., 1996), we examined the ability of OPT3 to transport Cd2+ ions as well. Differences in Cd2+ uptake between control and OPT3-expressing cells were detected after 30 min of incubation of oocytes in the Cd2+-containing uptake solution (Figure 5B), with the magnitude of OPT3-mediated Cd2+ uptake increasing in a time-dependent manner. It is worth noting that although these results indicate that OPT3 is capable of mediating the transport of Fe2+ and Cd2+, they do not provide an estimate of the relative affinity of the transporter for these ions (e.g., selectivity), which would ultimately determine their role in planta. Nonetheless, these results provide functional proof that OPT3 is a multispecific transporter of transition metal ions unlike other OPT family members that transport peptides.
Figure 5.
OPT3 Mediates Cation Uptake in X. laevis Oocytes.
Uptake of Fe2+ (A) and Cd2+ (B) in OPT3-expressing oocytes (OPT3) or water-injected cells (Control) at different time points. The basal uptake solution supplemented with 0.4 mM FeSO4 + 1 mM l-ascorbic acid (A) or 0.5 mM CdCl2 (B), yielded free extracellular ionic activities of 35 and 150 μM of {Cd2+}out and {Fe2+}out, respectively, as determined by GEOCHEM-EZ (Shaff et al., 2010). The dashed line in (A) indicates the basal concentration of endogenous Fe2+. Error bars represent se (n = 5). Asterisks indicate statistically significant differences (P ≤ 0.05).
OPT3 Is Expressed in the Phloem and the Majority of Its Expression Is Associated with the Minor Veins of Leaves and Nodes of Stems
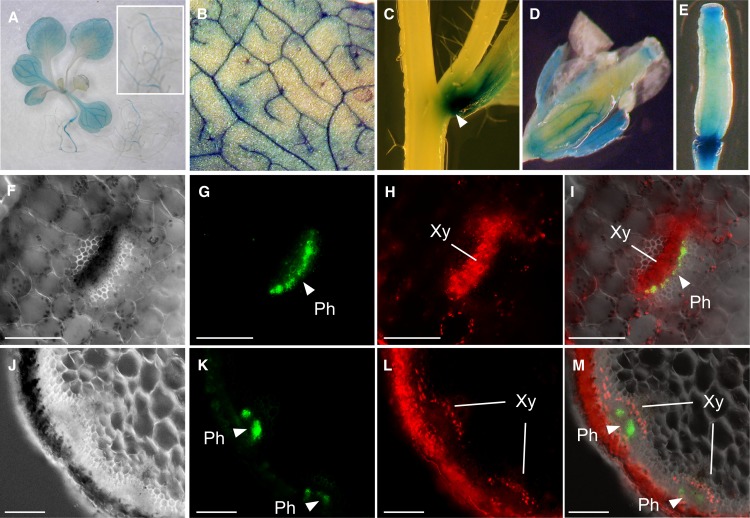
We next examined the cell-type specificity of OPT3 expression since this knowledge is critical for building the predictive models of the physiological function of OPT3 in planta. Analysis of the expression pattern of OPT3 in transgenic plants expressing an OPT3pro-GUS construct revealed β-glucuronidase (GUS) activity predominantly in the vascular tissues of leaves and reproductive organs in Arabidopsis (Figures 6A to 6E) and not in roots (Figure 6A), which is consistent with previous findings (Stacey et al., 2006). We noticed, however, that the majority of GUS activity in leaves was localized to minor veins of the vasculature (Figure 6B). We also found the bulk of OPT3 expression at the nodes of the stem (Figure 6C). Microscopy analysis of the cell-type specificity of OPT3 expression was conducted in transgenic plants expressing the OPT3pro-GFP construct. Transverse cross sections through petioles (Figures 6F to 6I) and a nodal section of a stem (Figures 6J to 6M) showed that the majority of OPT3pro-GFP–mediated fluorescence is associated with the phloem. Consistent with our data, profiling of the translatome of discrete cell populations identified OPT3 in CCs in leaves (Mustroph et al., 2009). The tissue- and cell-type specificity of OPT3 expression, along with results from subcellular localization and uptake studies (Figures 1, 3, and 5) suggest that OPT3 may function in apoplastic loading of transition ions into the phloem companion cells/sieve element complex (CC/SE) for subsequent long-distance transport. In addition, because xylem-to-phloem transfer is suggested to occur at the nodal regions in the stem as well as in minor veins in leaves (Bouche-Pillon et al., 1994; Andriunas et al., 2013), which are the sites of the preferential expression of OPT3 (Figures 6B and 6C), we hypothesized that OPT3 participates in xylem-to-phloem transfer of transition elements.
Figure 6.
Tissue- and Cell-Type Specificity of OPT3 Expression in Arabidopsis.
(A) to (E) Histochemical analysis of the OPT3 promoter activity in transgenic plants expressing the OPT3pro-GUS construct.
(A) Representative expression pattern for OPT3 in a whole seedling. Note the bulk of OPT3 expression in shoots (main figure) but not in roots (inset).
(B) Close-up of the leaf area to demonstrate OPT3 expression in minor veins.
(C) Expression of OPT3 at the node (arrow).
(D) and (E) Pattern of OPT3 expression in reproductive organs.
(F) to (M) Hand-cut cross sections through the petiole ([F] to [I]) or inflorescence stem ([J] to [M]) of 21-d-old transgenic plants expressing the OPT3pro-GFP construct.
(F) and (J) Differential interference contrast images of sections through the petiole and stem at the nodal region, respectively. Overlay images ([I] and [M]) were created to show that GFP-mediated fluorescence ([G] and [K]) does not overlap with phenolics-mediated autofluorescence in xylem vessels and chlorophyll-mediated autofluorescence in parenchyma cells ([H] and [L]). Xy, xylem; Ph, phloem. Bar = 100 μm.
Characterization of a Nonlethal Mutant Allele of OPT3
To further examine the physiological function of OPT3 in Fe and Cd transport in Arabidopsis, we obtained two T-DNA insertion alleles, SALK_021168C (alias opt3-2), possessing a T-DNA insertion 36 bp upstream of the OPT3 start codon (Stacey et al., 2008) and SALK_058794C, designated opt3-3, bearing a T-DNA insertion 41 bp upstream of the OPT3 start codon (Supplemental Figure 5A). Quantitative RT-PCR (qRT-PCR) studies revealed more than 90% reduction of the OPT3 transcript in plants carrying either allele in comparison with the wild type (Supplemental Figure 5B). We then evaluated the effect of the T-DNA insertion using opt3-3 mutant plants. Consistent with previous findings with the opt3-2 allele (Stacey et al., 2008), opt3-3 mutant plants were smaller than wild-type plants (Supplemental Figure 6A), developed necrotic lesions in cotyledons and older leaves (Supplemental Figures 6B and 6C), had significantly higher concentrations of Fe compared with the wild type (Supplemental Table 1), and exhibited a constitutive Fe deficiency response that was manifested by increased expression of IRT1 and FRO2 (Supplemental Figure 6D). Since opt3-2 and opt3-3 mutants exhibited similar phenotypes and fold decreases in OPT3 transcript abundance, we used the opt3-3 mutant for subsequent studies.
OPT3 Mediates Partitioning of Fe, but Not Cd, from Source to Sink Tissues
OPT3 is expressed in the phloem, which delivers nutrients, including mineral elements, from source to sink tissues. Therefore, to determine the contribution of OPT3 to Fe, and, possibly, Cd partitioning, we examined concentrations of Fe and Cd in old leaves (sources) and young leaves and seeds (sinks) of the opt3-3 mutant and wild-type plants. We reasoned that if Fe and Cd are physiological substrates of OPT3, then young leaves and seeds of the opt3-3 mutant would have a lower concentration of Fe and Cd than young leaves and seeds of wild-type plants.
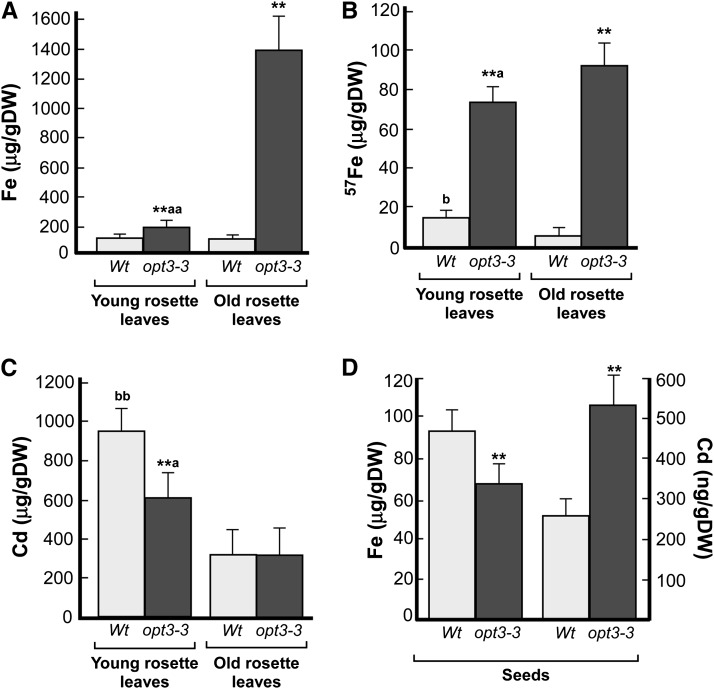
For analyses of ion concentrations in young and old leaves, we used plants at the late vegetative stage and performed long-term (for Fe; Figure 7A) and short-term (for Fe and Cd; Figures 7B and 7C, respectively) uptake and transport assays. The long-term Fe uptake and accumulation experiment revealed that while there were no significant differences in the concentration of Fe in young and old leaves of wild-type plants, the concentration of Fe was 8-fold higher in old versus young rosette leaves of opt3-3 mutant plants and 14-fold higher in old leaves when compared with the corresponding leaves in wild-type plants (Figure 7A). Consistent with these data, we were able to detect Fe using the Perls’ staining only in old leaves of the opt3-3 mutant (Supplemental Figure 7). It is noticeable that young leaves of the opt3-3 mutant accumulated 1.6-fold more Fe than corresponding leaves of the wild type (Figure 7A). This result was not entirely surprising because the Fe concentration in leaves of the opt3-3 mutant is significantly higher than in the wild type (Supplemental Table 1), which is likely due to increased expression of IRT1/FRO2 (Supplemental Figure 6D; Stacey et al., 2008). Consistent with the role of OPT3 in Fe partitioning from source to sink leaves and the fact that young leaves are more susceptible to Fe deficiency (Marschner, 1995), young leaves of the opt3-2 mutant were more sensitive to Fe deficiency than young leaves of the wild type (Supplemental Figure 8).
Figure 7.
OPT3 Mediates Fe Transport from Source to Sink Tissues.
ICP-MS analysis of Fe and Cd concentrations in old and young leaves ([A] to [C]) and seeds (D) of wild-type and opt3-3 mutant plants. Asterisks indicate statistically significant differences from the corresponding leaves in the wild type (P ≤ 0.001). Letters (a and aa) indicate statistically significant differences between old and young rosette leaves of the opt3-3 mutant (aP ≤ 0.05 or aaP ≤ 0.01). Statistically significant differences between old and young rosette leaves in the wild type are indicated as (bbP ≤ 0.001). DW, dry weight.
(A) to (C) Young and old rosette leaves were harvested at the late vegetative stage from hydroponically grown plants. In (A), plants were grown in 10 μM 56FeHBED (Fe) until tissues were collected for the ICP-MS analysis. In (B), plants were grown in 56Fe until the late vegetative stage and then transferred for 24 h to a fresh hydroponic medium containing 25 μM 57FeHBED (57Fe), while in (C), plants were grown for additional 24 h with 25 μM CdCl2 before sink and source leaves were harvested and subjected to the ICP-MS analysis.
(D) Iron and Cd concentrations in plants grown in soil with 7.5 nM Cd or 10 μM Fe. Error bars represent se (n = 3).
In short-term Fe and Cd transport and loading experiments, plants were grown hydroponically until the late vegetative stage and then transferred into a fresh hydroponic medium in which the most common isotope of 56Fe (alias Fe) was replaced with the stable isotope, 57Fe (provided as 57FeHBED [25 μM]), or to hydroponic medium, supplemented with 25 μM CdCl2. After 24 h, roots and shoots were harvested and analyzed for 57Fe and Cd by inductively coupled plasma–mass spectrometry (ICP-MS). In these experiments, young leaves of wild-type accumulated significantly more 57Fe than old leaves (Figure 7B). In contrast, the concentration of 57Fe in young leaves of the opt3-3 mutant was still significantly lower than in old leaves of the mutant, further supporting the role of OPT3 in Fe partitioning between source and sink tissues. The concentration of 57Fe in young leaves of the opt3-3 mutant was significantly higher than in young leaves of the wild type as well (Figure 7B). In contrast to 57Fe accumulation pattern, the concentration of Cd was similar in young and old leaves of wild-type and opt3-3 mutant plants (Figure 7C), suggesting that opt3-3 might not be involved in the phloem-based remobilization of Cd from source to sink tissues.
We then compared Fe and Cd concentrations in seeds of the wild type and the opt3-3 mutant, grown in soil with 7.4 nM of Cd or 10 μM Fe. Consistent with previous findings (Stacey et al., 2008), the concentration of Fe in seeds of the opt3-3 mutant was 1.4-fold lower in comparison with the seeds of wild-type plants (Figure 7D). In contrast, the concentration of Cd in seeds of the opt3-3 mutant was 2.1-fold higher in comparison with seeds of wild-type plants (Figure 7D). Collectively, these results suggest that OPT3 functions in Fe but not Cd remobilization from source to sink tissues via the phloem in Arabidopsis; thus, Fe but not Cd is a physiological substrate of OPT3.
The Phloem Sap of the opt3 Mutant Contains Less Fe, While the Xylem Sap of the opt3-3 Mutant Contains More Fe and Less Cd Than the Wild Type
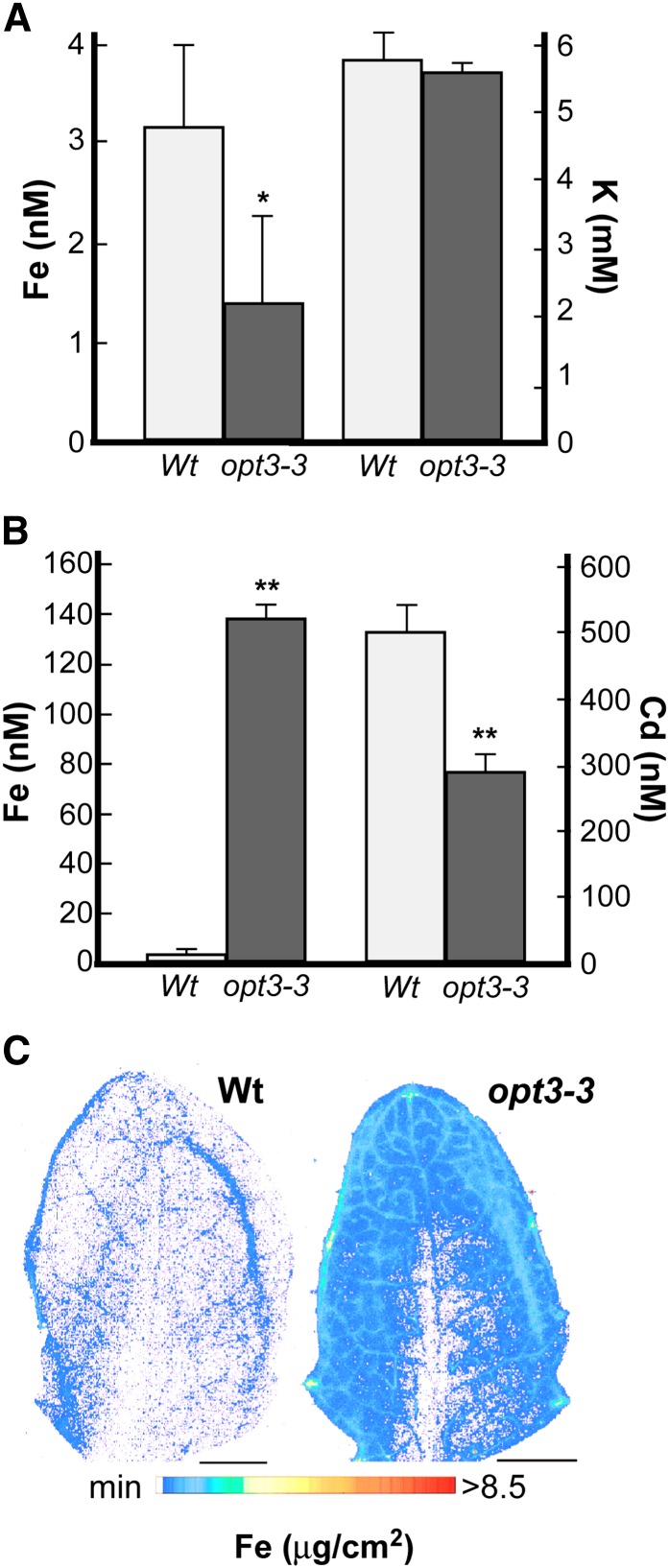
Localization of OPT3 expression in minor veins of leaves, at the nodal section of stems (Figures 6A and 6B), and in the phloem throughout the plant (Figures 6F to 6M), and the inability of the opt3-3 mutant to remobilize Fe from the sources to sinks (Figure 7) suggested a function of OPT3 in apoplastic Fe phloem loading and, possibly, xylem-to-phloem Fe transfer along the transport pathway. If this hypothesis is correct, then the concentration of Fe in the phloem would be lower in plants lacking functional OPT3 in comparison with the wild type and, accordingly, the Fe concentration in xylem sap would be higher in opt3 mutant versus the wild type.
Analysis of phloem sap revealed that the Fe concentration in the phloem was 2.3-fold lower in the mutant than in the wild type (Figure 8A). To ensure that the lower concentration of Fe in the phloem of the opt3-3 mutant was due to its inability to load Fe into the CC/SE complex rather than lower amounts of phloem exudates, we also compared the concentration of one of the major osmolytes in the phloem, potassium (K), which plays an important role in maintaining hydraulic pressure and pressure flow–based translocation of solutes from source leaves to sink tissues, such as developing leaves, seeds, and roots (Marschner, 1995). Since there was no statistically significant difference between the concentration of K in the phloem of the opt3-3 mutant and wild-type plants (Figure 8A), we concluded that the decreased Fe concentration in the phloem of the opt3-3 mutant is due to its inability to load Fe into the CC/SE complex. Due to technical difficulties with phloem sap collection from Cd-treated plants, we were not able to determine the Cd concentration in the phloem.
Figure 8.
OPT3 Mediates Xylem-to-Phloem Fe Transfer.
(A) and (B) The concentration of Fe and K in phloem sap (A) or Fe and Cd in xylem sap (B), both collected from wild-type and opt3-3 mutant plants grown hydroponically under Fe-sufficient conditions. Error bars represent se (n = 3). Asterisks indicate statistically significant differences (*P ≤ 0.05 and **P ≤ 0.001).
(C) Synchrotron x-ray fluorescence map of Fe distribution in leaves of the wild type and the opt3-3 mutant in Arabidopsis. Note that the line of Fe in the leaf of the wild type is an artifact of the leaf folding. Bar = 1 mm.
Analysis of xylem sap revealed that the concentration of Fe in the sap of the opt3-3 mutant was 42-fold higher in comparison with the wild type (Figure 8B). This result could be explained using the following not mutually exclusive scenarios: It is possible that lack of an Fe sufficiency signal in the phloem causes upregulation of FRO2 and IRT1, driving enhanced Fe uptake and loading into the xylem. Alternatively, the opt3-3 mutant may lack the ability to remove Fe from the xylem and load it into the phloem. Consistent with the latter suggestion, analysis of the spatial distribution of Fe using synchrotron x-ray fluorescence microscopy (SXRF) revealed that Fe was associated primarily with the minor veins of the opt3-3 mutant and was located at the hydathodes and toward the leaf blade periphery, while leaves of the wild type contained significantly less Fe (Figure 8C). Iron was also associated with minor veins of leaves of the opt3-3 mutant stained with Perls’ stain (Supplemental Figure 7). Because minor veins and the leaf periphery are the sites of the preferential expression of OPT3 (Figure 6B) and are the sites of the intensive xylem-to-phloem transfer (Bouche-Pillon et al., 1994; Turgeon and Webb 1973; Andriunas et al., 2013), we propose that OPT3 loads Fe into the phloem by mediating xylem-to-phloem cycling.
In contrast to Fe, the concentration of Cd was 1.7-fold lower in xylem sap of the mutant in comparison with wild-type plants (Figure 8B). This finding further supports the notion that although OPT3 is capable of transporting Cd ions in vitro, Cd is not the physiological substrate of OPT3 in planta.
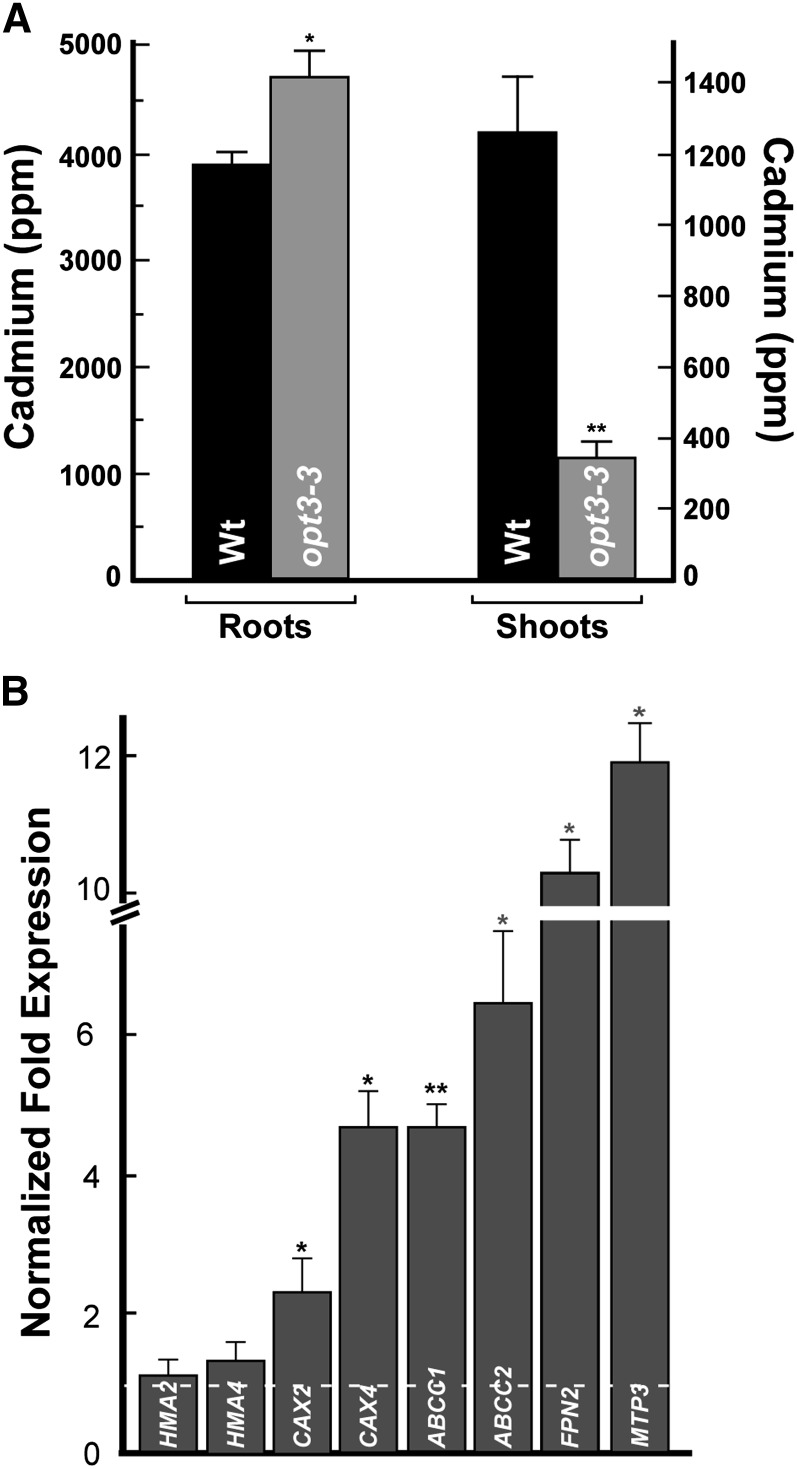
Shoots of Cd-Treated the opt3-3 Mutant Accumulate Less Cd and Are Less Sensitive to Cd Than Shoots of Wild-Type Plants
The decreased concentration of Cd in xylem sap of the opt3-3 mutant suggests that Cd delivery from roots to shoots might be affected in the mutant, which would result in altered Cd sensitivity. To test this prediction, we grew plants hydroponically to a late vegetative stage and exposed them to Cd for 4 d. We found that roots of the Cd-grown opt3-3 mutant accumulated significantly more Cd, while leaves accumulated significantly less Cd compared with corresponding organs in wild-type plants (Figure 9A). As a result of the decreased Cd concentration, leaves of the opt3-3 mutant were less sensitive to Cd than leaves of the wild type, both grown under the same conditions (Supplemental Figure 9).
Figure 9.
Leaves of opt3-3 Plants Accumulate Less Cd Than Leaves of Wild-Type Plants.
(A) The concentration of Cd in roots and shoots of wild-type and opt3-3 mutant plants grown for 4 d with 25 μM CdCl2. Shown are mean values ± se; n = 3. Asterisks indicate statistically significant differences (*P ≤ 0.05 and **P ≤ 0.001).
(B) Transcript abundance of the indicated genes was analyzed in roots of wild-type and opt3-3 plants, both grown hydroponically to the vegetative stage. Results are presented relative to expression of these genes in roots of wild-type plants designated as 1 (dashed line). Error bars indicate se (n = 6). Asterisks indicate statistically significant differences (*P ≤ 0.05 and **P ≤ 0.001, respectively).
Expression of Vacuolar Heavy Metal Transporters Is Upregulated in Roots of the opt3-3 Mutant
Given that the opt3-3 mutant has altered root-to-shoot signaling of Fe deficiency that is manifested by the constitutive upregulation of the expression of IRT1 and FRO2 in roots (Supplemental Figure 6D; Stacey et al., 2008), we hypothesized that expression of genes encoding transporters implicated in vacuolar sequestration or xylem loading of Cd might be altered in roots of the opt3-3 mutant as well. To test this hypothesis, we examined the steady state mRNA level for genes encoding HMA2 and HMA4, which are responsible for Cd loading into the xylem in roots (Wong and Cobbett, 2009), FPN2 (alias IREG2), MTP3, CAX2, and CAX4, which are suggested to play a role in vacuolar heavy metal sequestration (Arrivault et al., 2006; Schaaf et al., 2006; Korenkov et al., 2007), or ABCC1 and ABCC2, which are involved in the vacuolar sequestration of Cd-PC complexes (Park et al., 2012). The qRT-PCR analyses did not reveal statistically significant differences in transcript abundance of HMA2 and HMA4 in roots of opt3-3 versus wild-type plants (Figure 9B). In contrast, transcript abundance of CAX2, CAX4, ABCC1, ABCC2, FPN2, and MTP3 were significantly higher in roots of the mutant compared with the wild type. These data suggest that increased abundance of these and, possibly, other vacuolar heavy metal transporters is among the reasons for Cd retention in roots, decreased Cd loading into the xylem, and accumulation in leaves of the opt3-3 mutant versus the wild type (Figures 8B and 9A), leading to Cd resistance phenotypes in shoots of the opt3-3 mutant (Supplemental Figure 9).
DISCUSSION
OPT3 Transports Transition Ions
OPT3 belongs to the oligopeptide transporter family, whose members transport synthetic tetra- and pentapeptides when expressed in heterologous systems; however, the physiological substrates and the physiological role of OPTs, including OPT3, in Arabidopsis are unknown (Lubkowitz, 2011). Therefore, after finding that OPT3 localizes to the plasma membrane (Figure 1), we examined if OPT3, similar to is closest homolog from B. juncea, Bj-GT1, as well as other OPTs from Arabidopsis, would transport GSH. We found that heterologously expressed OPT3 did not complement the GSH uptake defect of the Saccharomyces cerevisiae GSH uptake mutant opt1 nor did it transport GSH (Figure 2), suggesting that GSH might not be the physiological substrate of OPT3. In contrast, heterologously expressed OPT3 partially rescued the growth of the fet3 fet4 mutant in Fe-limited conditions (Figure 3A; Wintz et al., 2003). Importantly, we showed that the ability of OPT3 to rescue Fe deficiency of the fet3 fet4 mutant is associated with the OPT3-dependent increase in accumulation of Fe in cells of the mutant, even though yeast were grown in medium that lacked the Fe ligand NA (Figure 3B). These data suggested that OPT3 might mediate transport of ions rather than peptides or ion-ligand complexes. Consistent with this suggestion, chelation of Fe2+ with the addition of NA to the growth medium abolished the ability of OPT3 to rescue growth of the fet3 fet4 mutant (Wintz et al., 2003).
The nature of OPT3-mediated transport was examined in X. laevis oocytes. After establishing that OPT3 resides at the plasma membrane in oocytes as it does in Arabidopsis, we employed uptake conditions that would minimize activation of endogenous oocyte transporters and found that OPT3 likely mediates proton-coupled inward currents as was documented for other OPT family members (Osawa et al., 2006) (Figure 4; Supplemental Figure 4). Subsequently, we showed that OPT3 indeed mediates uptake of Fe2+ into X. laevis oocytes in a time-dependent manner in medium lacking metal ligands (Figure 5A). Because several characterized Fe transporters have broad substrate specificity, and IRT1 transports Cd in addition to Fe, we also examined whether OPT3 would be able to transport Cd2+ ions as well. We found that in addition to Fe, OPT3-mediated Cd2+ uptake in X. laevis oocytes in medium lacking metal ligands (Figure 5B).
OPT3 Functions in the Translocation of Fe but Not Cd to Sink Organs
To begin the investigation of the role of OPT3 in transition ion transport in planta, we first determined the cell-type specificity of its expression. Using transgenic plants expressing OPT3pro-GUS or OPT3pro-GFP fusions, we found that OPT3 is expressed in minor veins of the vasculature in leaves and nodes of stems and that the bulk of its expression is associated with the phloem (Figures 6B, 6C, and 6F to 6M). Furthermore, OPT3 was found in CC of the phloem (Mustroph et al., 2009). Because OPT3 mediated Fe and Cd influx in X. laevis oocytes as well as Fe accumulation in yeast cells (Figures 3 and 5), we hypothesized that OPT3 is involved in loading of Fe and, possibly, Cd into the CC/SE complex. Because the phloem plays a key role in redistribution of mineral elements from source to sink tissues, we compared Fe and Cd accumulation in older and younger rosette leaves and seeds of the opt3-3 mutant versus the wild type. We found that the Fe concentration was 8-fold higher in old versus young rosette leaves of opt3-3 mutant plants, and 14-fold higher in old leaves when compared with the corresponding leaves in wild-type plants (Figure 7A). This result suggested that OPT3 plays a significant role in the delivery of Fe to sink tissues. Because symptoms of Fe deficiency are more prominent in younger tissues (Marschner, 1995), we expected that if OPT3 functions in the phloem-based Fe partitioning then young leaves of the opt3-3 mutant would be more susceptible to Fe deficiency than young leaves of the wild type. Consistent with this hypothesis, we observed that young leaves of the opt3-3 mutant were significantly more chlorotic compared with the wild type grown in the same conditions (Supplemental Figure 8). Finally, OPT3 was important for the delivery of Fe to developing seeds (Figure 7D; Stacey et al., 2008). Based on these results, we concluded that OPT3 functions in the phloem-based delivery of Fe to sink tissues and that Fe is a physiological substrate of OPT3.
In contrast to Fe, Cd content was similar in young and old leaves of wild-type and opt3-3 mutant plants (Figure 7C), suggesting that opt3-3 might not be involved in the phloem-based remobilization of Cd from source to sink tissues. This suggestion was substantiated by distinctively different kinetics of 57Fe and Cd accumulation in young and old leaves of plants (Figures 7B and 7C) and finding that OPT3 is not involved in Cd loading into seeds (Figure 7D). Together, these results support the notion that although OPT3 can mediate transport of Cd in vitro, it is not involved in Cd partitioning in planta under physiological conditions. It is possible that increased accumulation of Cd in seeds of the mutant versus the wild type is due to the effect of the OPT3 knockdown on the function/abundance of transporters, such as YSL1 or YSL3, or other transporters that are responsible for metal loading into seeds (Waters et al., 2006; Chu et al., 2010).
The Role of OPT3 in Xylem-to-Phloem Transfer in Arabidopsis
It is recognized that young leaves, seeds, and other sink organs receive the majority of their mineral nutrients via the phloem (Marschner, 1995). In addition to symplastic and apoplastic loading of the CC/CE complex, efficient xylem-to-phloem transfer may occur along the long-distance transport pathway with hubs at the nodal regions in the stem and minor veins in the leaf blade (Bouche-Pillon et al., 1994; Andriunas et al., 2013). The companion cells in these places differentiate into transfer cells to enhance membrane transport capacity and phloem loading and redistribution of resources (Andriunas et al., 2013). We found that (1) OPT3 is expressed in the phloem of minor veins in leaves and at the nodal sections of the stem (Figure 6), where it is associated with companion cells (Mustroph et al., 2009). (2) The opt3-3 mutant accumulates Fe in the vicinity of minor veins at the leaf blade periphery (Figure 8C; Supplemental Figure 7), which is suggested to have the strongest source strength (Turgeon and Webb, 1973). (3) OPT3 mediates uptake of Fe ions in yeast and X. laevis oocytes (Figures 3 and 5). (4) The xylem sap of the opt3-3 mutant hyperaccumulates Fe, while phloem sap contains less Fe compared with xylem and phloem saps of the wild type (Figures 8A and 8B). Collectively, these data suggest that OPT3 might contribute to xylem-to-phloem transfer of Fe for subsequent partitioning from source-to-sink tissues.
OPT3 Contributes to Shoot-to-Root Signaling of Fe Status
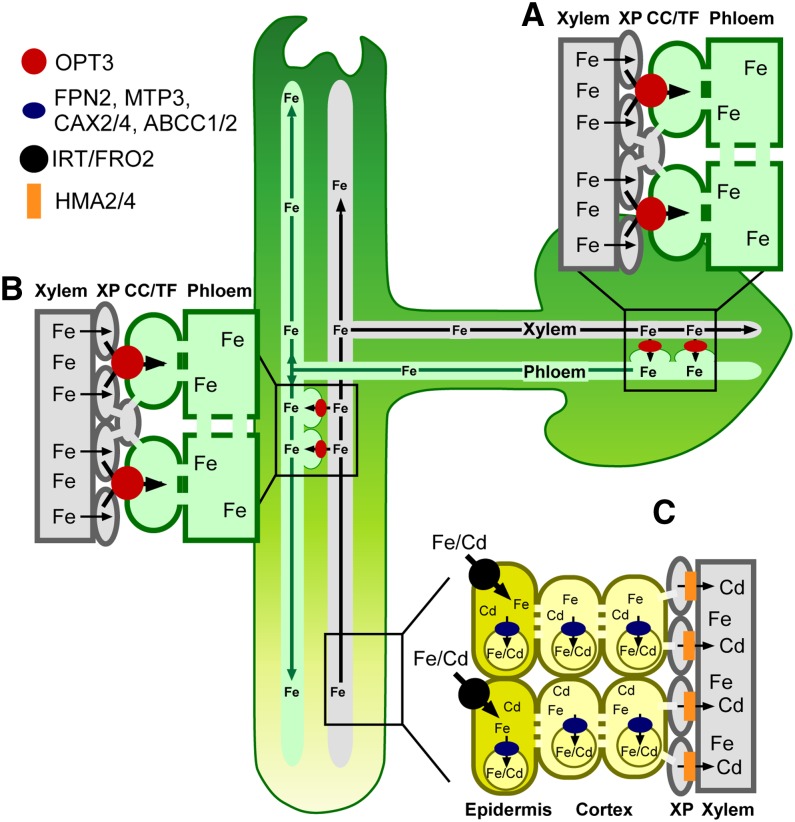
Revealing the physiological substrate and function of OPT3 has provided fundamental insights that have implications for the nature and mechanism of systemic Fe signaling in plants (summarized in Figure 10). First, our finding that the concentration of Fe is significantly lower in the phloem sap of the opt3-3 mutant (Figure 8A), which overexpresses IRT1 and FRO2 in the root even under Fe-sufficient conditions, provides experimental evidence that is consistent with the hypothesis that Fe availability in the phloem is essential for the regulation of Fe deficiency responses in the root. Second, it has been suggested that OPT3 plays a role in communicating Fe status from shoots to roots (Stacey et al., 2008), and we provide a mechanistic basis for this phenomenon where OPT3 mediates Fe recirculation from the xylem to the phloem (Figure 8). Loss of this ability results in Fe hyperaccumulation in leaves of opt3 knockdown mutants (Supplemental Table 1; Stacey et al., 2008). The mechanistic explanation of foliar Fe hyperaccumulation phenotypes of the opt3 knockdown mutant provided by our data also rationalize findings by García et al. (2013) that foliar application of Fe rescues constitutive Fe acquisition responses manifested in roots of the Arabidopsis frd3, pea brz, and tomato chln mutants, but not in the root of the opt3-2 mutant. We reason that knockdown of OPT3 results in plants incapable of loading Fe into the phloem for the long-distance transport into the root; thus, foliar application of Fe does not rescue its Fe deficiency responses. In contrast, we found that grafting wild-type shoots onto opt3-3 mutant roots downregulated FRO2 and IRT1 expression relative to their expression in control grafts (wild-type shoots onto wild-type roots) (Supplemental Figure 10A). This finding emphasizes that OPT3-mediated Fe transport function in the shoot is sufficient to regulate the transcriptional Fe deficiency responses in the root. Consistent with the suggestion that OPT3 function is required in shoots, its transcript abundance in shoots was 10-fold higher than in roots of wild-type plants (Supplemental Figure 10B). Collectively, these data provide molecular evidence that Fe availability in the phloem regulates expression of Fe acquisition genes in the root and identify OPT3 as an important contributor to the systemic signaling of Fe status.
Figure 10.
Model of OPT3 Function in Arabidopsis.
This figure summarizes the proposed dual roles of OPT3 in the redistribution of Fe from source to sink tissues, in shoot-to-root signaling of shoot Fe status and its contribution to Cd partitioning in Arabidopsis.
(A) OPT3 is expressed in CCs of the phloem that might differentiate into the transfer cells (TF) in minor veins of leaves and in nodes of stems. In these sites, OPT3 facilitates Fe loading into the phloem, possibly by direct xylem-to-phloem Fe transport.
(B) At the whole-plant level, OPT3 is involved in redistribution of Fe from sources to sinks and Fe recirculation into roots. Iron recirculation into roots via OPT3 plays a signaling role by conveying Fe status of the shoot.
(C) Although OPT3 transports Cd in vitro, it is likely that it mediates root-to-shoot partitioning of Cd indirectly by orchestrating shoot-to-root Fe signaling that, in turn, alters expression of genes encoding multispecific transition ion transporters (e.g., FPN2, MTP3, CAX2/4, ABCC1/2, and possibly others). These transporters facilitate vacuolar sequestration of Cd and its retention in the root, which in turn, affects root-to-shoot Cd partitioning and Cd resistance. XP, xylem parenchyma cells.
Our data also suggest that the decreased Cd concentration in xylem sap and shoots of the opt3-3 mutant and increased Cd tolerance of leaves of the mutants versus the wild type (Figures 8B and 9A; Supplemental Figure 9) is a consequence of OPT3 contribution to phloem-based systemic Fe signaling (Figure 10). Indeed, we found that expression of genes encoding transporters that are implicated in vacuolar sequestration of Cd ions (FPN2, CAX2, CAX4, and MTP3) and vacuolar sequestration of Cd-PC complexes (ABCC1 and ABCC2) is significantly upregulated in roots of the opt3-3 mutant versus the wild type (Figure 9B). We hypothesize that these and possibly other transporters contribute to the retention of Cd in roots of the opt3-3 mutant, subsequently affecting Cd loading into the xylem, root-to-shoot Cd partitioning, and increasing Cd tolerance of the shoot.
In conclusion, data presented in this article uncovered a central role of OPT3 in Fe loading into the phloem, provided mechanistic explanation of OPT3-mediated shoot-to-root communication of Fe status in Arabidopsis, and highlighted the role of Fe in the phloem-based systemic signaling, a long-debated area in plant Fe homeostasis research. Our results also indicate that OPT3 contributes to Fe recycling from the xylem and acts as a functional link between the xylem and the phloem. We show that OPT3 is a multispecific transition element transporter. This functional activity distinguishes OPT3 from other peptide-transporting OPT family members. Given the acute Fe deficiency responses of OPT3 knockdown and knockout mutants, and the indirect effect of OPT3 on Cd partitioning in Arabidopsis, we propose that Fe, but not Cd, is a physiological substrate of OPT3, although it is capable of transporting both Cd2 and Fe2+ ions when expressed in X. laevis oocytes. Another important implication from this work is finding that loss of OPT3 function significantly decreases Fe while increasing Cd concentration in seeds, suggesting that manipulation of the expression of this transporter can provide promising avenues for targeted biofortification strategies directed at increasing Fe density, while omitting Cd, in the edible portions of crops.
METHODS
Plant Material and Growth Conditions
All plant lines used in the study were in the Arabidopsis thaliana Columbia background. Seeds of opt3-2 (SALK_021168C) and opt3-3 (SALK_058794C) T-DNA insertion alleles were obtained from the ABRC (Alonso et al., 2003). The opt3-2 allele is also described (Stacey et al., 2002, 2008). opt3-2 and opt3-3 homozygous lines were identified by PCR using the left border T-DNA–specific and the gene-specific primers (Supplemental Table 2).
For growing plants in hydroponics, 8-d-old seedlings were transferred from half-strength Murashige and Skoog (MS) agar plates to Arabidopsis hydroponic solution described by Arteca and Arteca (2000). The hydroponic solution was changed every 5 d. For Fe deficiency assays, plants were grown hydroponically for 3 weeks and transferred to medium without FeHBED for the indicated time. In all experiments, the growth conditions were as follows: 12-h-light/12-h-dark photoperiod (at a photosynthetic photon flux density of 120 μmol photons m–2 s–1) at 23°/19°C light/dark temperature regime and 75% relative humidity.
Subcellular Localization and Fluorescent Microscopy
The full-length OPT3 cDNA was isolated by RT-PCR using total RNA from wild-type Arabidopsis leaves and primers sets that would introduce attB sites on the resulting PCR product (Supplemental Table 2). The PCR product was introduced into the DONR222 entry vector (Invitrogen) before recombination cloning into the GWB406 destination vector (Nakagawa et al., 2007) to fuse GFP in frame with the N termini of OPT3 under the control of the cauliflower mosaic virus 35S promoter. The resulting 35Spro-OPT3-GFP construct or GWB406, lacking the cDNA insert, was transfected into Arabidopsis protoplasts (Zhai et al., 2009). GFP-mediated fluorescence and chlorophyll autofluorescence were visualized using fluorescein isothiocyanate (FITC) or rhodamine filter sets, respectively, of an Axio Imager M2 microscope equipped with the motorized Z-drive (Zeiss). Z-stack (1.3 μm thick) images were collected with the high-resolution AxioCam MR camera and then 3D deconvoluted using an inverse filter algorithm of the Zeiss AxioVision 4.8 software.
Expression of OPT3-GFP in Onion Epidermal Cells
For visualizing OPT3 localization in onion (Allium cepa) cells, the 35Spro-OPT3-GFP construct in the GWB406 vector and the plasma membrane marker PIP2A fused to mCherry in the BIN vector (Nelson et al., 2007) were introduced into onion epidermal cells by biolistic transformation as described (Jung et al., 2012). Briefly, 2 μg of each construct in 10 μL of distilled water was mixed with 10 μL of solution containing 50 mg/mL of 1.0-μm gold particles, 10 μL of 2.5 mM CaCl2, and 4 μL of 0.1 M spermidine. The mixture was incubated for 30 min at room temperature. Gold particles coated with plasmid DNAs were rinsed with cold ethanol and then gently suspended in 20 μL of ethanol. Onion pieces were placed onto agar plates containing 1× MS medium and bombarded using a double-barreled extension of the Bio-Rad He/1000 particle delivery system (PDS-1000/He) with 1100 p.s.i. rupture discs under a vacuum of 0.04 bar. Onion pieces were left to recover after bombardment in the dark at 25°C for 16 h. Onion skin epidermal layers were peeled from the onion pieces and placed on glass slides for analyses. When indicated, onion epidermal cells were plasmolyzed by incubating in 20% (w/v) sucrose solution for 10 min.
Tissue- and Cell-Type Specificity of OPT3 Expression in Arabidopsis
A 3.5-kb genomic fragment including sequence upstream of OPT3 start codon was amplified by PCR using indicated primer pairs (Supplemental Table 2). The resulting amplicon was fused to the bacterial uidA gene encoding GUS of the GUS1-Gate vector (Jung et al., 2012) or to GFP of the YXT2 destination vector (Xiao et al., 2010). The resulting OPT3pro-uidA (OPT3pro-GUS) or OPT3pro-GFP constructs were transformed into wild-type Arabidopsis (Clough and Bent, 1998). GUS staining was performed for 16 h with 2 mM X-Gluc (5-bromo-4-chloro-3-indolyl-β-d-glucuronide) as described (Jefferson et al., 1987). Staining patterns were analyzed using Leica S6E stereomicroscope. Hand-cut sections were prepared from stems and petioles of transgenic Arabidopsis plants expressing OPT3pro-GFP using a feather double-edge razor blade. GFP and autofluorescence were visualized using FITC (for GFP) or rhodamine (for autofluorescence) filter sets of the Axio Imager M2 microscope equipped with the motorized Z-drive (Zeiss). Images were processed using the Adobe Photoshop software package, version 12.0.
Functional Complementation Assays in Saccharomyces cerevisiae
S. cerevisiae strains used in this study were BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0); opt1(alias ABC822) (MATa; ura3-52; leu2∆1; lys2-801; his3∆200; trp1∆63; ade2-101; hgt1∆::LEU2); DY1457 (MATa ade6 can1 his3 leu2 trp1 ura3); and DEY1453 (MATa/MATα ade2/+ canl/canl his3/his3 leu2/leu2 trpl/trpl ura3/ura3 fet3-2::HIS3/fet3-2::HIS3fet4-1::LEU2/fet4-1::LEU2). For details of functional complementation assays, see the Supplemental Methods. Briefly, the full-length OPT3 or IRT1 cDNAs were cloned by recombination into YES3-Gate vector, and resulting vectors, YES3-Gate-OPT3, YES3-Gate-IRT1, and YES3-Gate, lacking the cDNA insert, were transformed into different yeast strains using the Frozen-EZ yeast Transformation II Kit (Zymo Research).
To test if OPT3 would rescue GSH uptake deficiency of the opt1 mutant in medium with GSH as the only sulfur (S) source, the opt1 mutant expressing either OPT3 or the empty vector were grown in YNB medium with supplements to an OD600 = 1.2. Cells were then washed in ice-cold water three times and inoculated (1:200) into SC-S media supplemented with 200 μM of GSH. SC-S was prepared according to the YNB recipe (Bacto Yeast Nitrogen Base without amino acids and ammonium sulfate; DIFCO Laboratories), with the modification that all sulfur containing reagents in macroelements and microelements were substituted with equal amounts of the corresponding chloride salts (Zhang et al., 2004). The medium was supplemented with His, Trp, Ade, and Lys.
For functional complementation assays in the fet3 fet4 Fe uptake-deficient mutant, YES3-Gate-OPT3, YES3-Gate-IRT1, and YES3-Gate lacking cDNA inserts were transformed into the fet3 fet4 mutant or the empty vector was transformed into the wild-type strain DY1457. The transformants were selected on YNB medium (pH 4.0) lacking uracil but supplemented with adenine sulfate (20 mg/L), tryptophan (20 mg/L), and ferric chloride (10 μM). Yeast colonies were inoculated into the same liquid medium and grown overnight to an OD600 of 1.0. Yeast cells were washed in sterile water to desorb excess FeCl3 from cell walls and aliquots of cell suspension were serially diluted and spotted onto solid YNB medium (pH 4.0) lacking uracil but supplemented with adenine sulfate (20 mg/L), tryptophan (20 mg/L), and 10 μM BPS. Colonies were visualized after incubating plates for 6 d at 30°C.
Elemental Analysis
Elemental composition was analyzed in Arabidopsis and S. cerevisiae. For studies in Arabidopsis, plants were grown hydroponically as described above. In long-term experiments of Fe accumulation in old and young leaves, plants were grown until the late vegetative stage. For short-term Fe and Cd uptake and transport studies, plants were grown in 10 μM of Fe-HBED (in 56Fe as common isotope of Fe, natural abundance 91.7%) until the late vegetative stage and then transferred for an additional 24 h to a fresh hydroponic medium in which 56Fe was replaced with 25 μM 57Fe (Isoflex USA; 95% enrichment) or to a fresh hydroponic medium supplemented with 25 μM CdCl2 for 24 h before sink and source leaves were harvested and subjected to the ICP-MS analysis. Roots and leaves were harvested and roots were desorbed of Cd and Fe as well as other elements by washing with 10 mM EDTA followed by washing in a solution with 0.3 mM bathophenanthroline disulphonate and 5.7 mM sodium dithionite and then rinsed with deionized water. Shoots were rinsed with deionized water. In analyses of Fe and Cd accumulation in young and old leaves, the two bottommost rosette leaves (old) and uppermost rosette leaves >3 mm (young) were collected from three to five plants. For analyses of Fe and Cd concentration in seeds, plants were grown in soil with 7.4 nM Cd or 10 μM Fe. Elemental analysis was performed using ICP-MS as described (Lahner et al., 2003). The ICP-MS–based analysis of the Fe concentration in yeast cells was performed as described (Jung et al., 2012; Gayomba et al., 2013) except that liquid YNB media used for growing yeast cells was at pH 4.0, was lacking uracil, and was supplemented with adenine sulfate (20 mg/L), tryptophan (20 mg/L), and 10 μM FeCl3.
Expression of OPT3 in Xenopus laevis Oocytes and Electrophysiological Recordings
The coding region of OPT3 was cloned into the T7TS plasmid (Cleaver et al., 1996), at the EcoRV unique site flanked by the 3′ and 5′ untranslated regions of a X. laevis β-globin gene. The construct was fully sequenced and checked for sequence accuracy. cRNA was synthesized from 1 μg SmaI-linearized plasmid DNA template using a mMessage in vitro transcription kit (Ambion) according to the manufacturer’s recommendations and stored at −80°C. Stage V to VI X. laevis oocytes were harvested, defolliculated, and cultured in ND96 solution containing 96 mM NaCl, 2 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 2.5 mM Na-pyruvate, 50 μg/mL gentamycin, 0.4 mg/mL BSA, and 5 mM HEPES/NaOH to adjust pH to 7.5, as described previously (Piñeros et al., 2008). X. laevis oocytes were injected with 50 nL of water (control) or 50 nL of water containing 50 ng of OPT3 cRNA and incubated in ND96 solution at 18°C for 2 to 4 d prior to the uptake and electrophysiological measurements.
Electrophysiological recordings were done under constant bath perfusion with both GeneClamp 500 and Axoclamp 900A amplifiers (Axon Instruments) using the two-electrode voltage-clamp technique. Recording electrodes were filled with 0.5 M K2SO4 and 30 mM KCl and had resistances between 0.5 and 1.5 MΩ. Cells were bathed under constant perfusion in a ND96-recording solution consisting of (in mM) 96 NaCl, 1 KCl, and 1.8 CaCl2 with the pH adjusted to 7.5 with 5 mM HEPES/NaOH. Given the presence of variable endogenous inward current at hyperpolarizing holding potentials (Amasheh and Weber, 1999; Kuruma et al., 2000) currents under voltage clamp conditions were elicited by a 6-s voltage pulses (in 10-mV step increments) restricted between 0 and −140 mV, with a 2-s rest at 0 mV between each voltage pulses. The output signal was digitized and analyzed using Digidatas 1320A and 1440A PClamp 10 data acquisition systems (Axon Instruments). The steady state current-voltage (I/V) relationships were constructed by measuring the current amplitude at the end of the test pulse. All data points represent the mean of at least eight different cells from three to four donor frogs. Error bars denote se and are not shown when they are smaller than the symbol.
Metal Uptake in Oocytes
The basal uptake solution consisted of a modified ND96 solution containing 96 mM NaCl, 1 mM KCl, and 0.9 mM CaCl2, buffered with 5 mM MES/NaOH to pH 6.0. The uptake solutions was supplemented with 0.5 mM CdCl2 or 0.4 mM FeSO4 + 1 mM l-ascorbic acid (freshly prepared) to prevent Fe oxidation. Given the strong interaction of Cd with Cl (e.g., 80% of the Cd present in the uptake solution is complexed with Cl), the free Cd2+ activity in the uptake solution was estimated to be 35 μM Cd2+, while Fe2+ free activity was 150 μM as determined by GEOCHEM-EZ (Shaff et al., 2010). Each sample contained six oocytes, with five replicates per time point. At a given time point, the uptake was terminated by washing oocytes through six consecutive ice-cold basal uptake solution. Samples were digested in 100 μL of 70% HClO4, resuspended in 10 mL of 0.1 M nitric acid, and analyzed using ICP-MS (Perkin-Elmer Sciex). Uptake data are expressed “per oocyte” and are representative of at least five independent experiments.
Collection and Analysis of Xylem Sap
Xylem sap was collected as previously described (Sunarpi et al., 2005). Briefly, wild-type and opt3-3 mutant plants were cultured on hydroponic media until the late vegetative stage. For analyses of Cd concentration, plants were subjected to 25 μM CdCl2 for 48 h. After removal of rosette leaves, the inflorescence stem was cut with a fresh razor blade. Xylem sap exudation was facilitated through a high humidity environment by covering the plants with a plastic dome. The first droplets were excluded to avoid contamination and then xylem sap was collected quantitatively with a micropipette. Cadmium and Fe concentrations in the sap were analyzed by ICP-MS and normalized per volume of the collected sample.
Collection and Analysis of Phloem Sap
Phloem sap was collected from hydroponically grown wild-type and opt3-3 mutant plants at the late vegetative stage. To prevent entrance of air bubbles into the vasculature, whole rosettes were removed from the root using a razor blade and immersed in deionized water before individual leaves were detached at the petiole. Three leaves (leaf numbers 9 and 10) collected from one plant were pooled together and flushed of xylem sap by placing the petioles in a tube filled with 300 μL of deionized water and incubated in an illuminated growth chamber for 15 min before further incubation in darkness for 1 h. The petioles were then recut under 5 mM Na2-EDTA (pH 7.5) under low light before placing the petioles in 250 μL of 5 mM Na2-EDTA (pH 7.5). The leaves where then incubated in darkness for 1 h in a high-humidity chamber lined with wet paper towels and sealed with Vaseline. Samples were diluted with 5 mL of 5% HNO3 for subsequent detection of K or Fe by ICP-MS.
SXRF
Wild-type and opt3-3 mutant plants were grown on solid half-strength MS medium for 23 d. Fully developed leaves (second from the bottom) were detached immediately prior to analysis using Teflon-coated forceps and placed adaxial side uppermost, on 35-mm slide mounts across which Kapton metal-free tape was stretched. The distribution of Fe in hydrated leaf tissue was imaged via SXRF with hard x-ray microprobe X26A of the National Synchrotron Light Source as detailed in the Supplemental Methods.
Perls’ Staining
Ferric iron was visualized in leaves of Arabidopsis using Perls’ staining as described (Green and Rogers, 2004). Briefly, wild-type and opt3-3 mutant plants were grown hydroponically in Fe-replete conditions until the late vegetative stage. Shoots from 5-week-old hydroponically grown wild-type and opt3-3 plants were vacuum infiltrated with Perls’ stain solution (4% HCl [v/v] and 4% [w/v] potassium ferrocyanide) and allowed to incubate under vacuum for 1 h. Samples were then removed from the vacuum chamber and incubated for 1.5 h. The reaction was stopped by rinsing samples three times with distilled water.
Reciprocal Grafting
Five- or seven-day-old seedlings were used for grafting experiments, performed as described (Rus et al., 2006). After recovery for 5 d, robust grafts lacking adventitious roots on the scion were transferred to hydroponics and cultured for 2.5 weeks before CdCl2 (25 μM) was added to the hydroponic medium. Grafts were photographed and tissues were collected at the indicated time points. RNA for qRT-PCR analysis was isolated from roots of grafted plants grown to the late vegetative stage.
RNA Isolation and qRT-PCR
Arabidopsis wild-type and opt3-3 mutant plants were grown hydroponically until late vegetative stage. Total RNA was isolated from roots and shoots of hydroponically grown plants using the Trizol reagent (Invitrogen) according to the manufacturer’s recommendations. DNase I (Roche) digestion of genomic DNA prior to first-strand cDNA synthesis and qRT-PCR thermocycling procedures were as described (Jung et al., 2012; Gayomba et al., 2013). Data were normalized to the expression of F14F8.90 (AT5G15710) and ACTIN2 (AT3G18780). The fold difference (2−ΔΔCt) or relative quantities were calculated using the CFX Manager Software, version 1.5 (Bio-Rad).
Statistical Analysis
Statistical analyses of the majority of experimental data were performed using the ANOVA single-factor analysis. Statistical analysis of qRT-PCR data was performed using the Relative Expression Software Tool (Qiagen; Pfaffl et al., 2002).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers (accession numbers in parenthesis): At-OPT3 (AT4G16370), At-FIT1 (AT2G28160), At-IRT1 (AT4G19690), At-FRO2 (AT1G01580), At-HMA2 (AT4G30110), At-HMA4 (AT2G19110), At-FPN2 (AT5G03570), At-ACT2 (AT3G18780), At-F14F8.90 (AT5G15710), At-CAX2 (AT3G13320), At-CAX4 (AT5G01490), At-ABCC1 (AT1G30400), At-ABCC2 (AT2G34660), At-MTP3 (AT3G58810), and Sc-OPT1 (NM_001181645)
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. OPT3-GFP Localizes to the Plasma Membrane in Epidermal Onion Cells.
Supplemental Figure 2. OPT3 Does Not Alter Growth of the opt1 Mutant of S. cerevisiae under Control Conditions.
Supplemental Figure 3. OPT3 Localizes to the Plasma Membrane in X. laevis Oocytes.
Supplemental Figure 4. Electrophysiological Properties of X. laevis Oocytes Expressing OPT3.
Supplemental Figure 5. T-DNA Insertions Upstream of the OPT3 Start Codon Reduce the OPT3 Transcript.
Supplemental Figure 6. The opt3-3 Knockdown Allele (opt3-3) Exhibits Constitutive Fe Deficiency Symptoms.
Supplemental Figure 7. Iron Localizes at Minor Veins of Old Rosette Leaves in the opt3-3 Mutant (opt3-3).
Supplemental Figure 8. Young Leaves of the opt3-3 Mutant Are More Sensitive to Fe Deficiency Than Corresponding Leaves of Wild-Type Plants.
Supplemental Figure 9. Leaves of opt3-3 Plants Accumulate Less Cd and Are Less Sensitive to Cd Than Leaves of the Wild-Type.
Supplemental Figure 10. OPT3 Functions Primarily in Shoots in Arabidopsis.
Supplemental Table 1. Leaf Element Profile of the Wild-Type and the opt3-3 (opt3-3) Allele.
Supplemental Table 2. List of Oligos.
Supplementary Material
Acknowledgments
We thank Walter Gassmann (University of Missouri) and Serge Delrot (University of Bordeaux, France) for S. cerevisiae mutant strains; Christopher D. Town (J. Craig Venter Institute) for the YXT2 vector; Julian Schroeder (University of California, San Diego) and David Mendoza-Cozatl (University of Missouri) for discussing the cell-type specificity of OPT3 expression; the Scanlon Lab (Cornell University) for help with Arabidopsis tissue sectioning; and the Turgeon Lab (Cornell University) for discussing phloem exudate collection. SXRF studies were performed at Beamline X26A, National Synchrotron Light Source (NSLS), Brookhaven National Laboratory. X26A is supported by the Department of Energy-Geosciences (DE-FG02-92ER14244 to the University of Chicago-CARS). Use of the NSLS was supported by the Department of Energy under Contract DE-AC02-98CH10886. Work in the Vatamaniuk lab was supported by the National Science Foundation (MCB-0923731) and Cornell University Agricultural Experiment Station CUAES Hatch (NYC-125433). Work in the Guerinot lab was funded by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the U.S. Department of Energy through Grant DE- FG02-06ER15809, and work in the Salt lab was funded by the U.S. National Science Foundation through Grant IOS 0701119.
AUTHOR CONTRIBUTIONS
Z.Z., H.J., S.R.G., N.K.V., M.P., and O.K.V. designed experiments. D.E.S., M.L.G., and L.V.K. contributed to discussion on the article and provided analytical tools for the article. Z.Z., H.J., S.R.G., M.P., N.K.V., M.R., T.P., E.C., B.L., and J.D. performed experiments. The article was written by O.K.V., S.R.G., and M.P. All authors analyzed data and contributed constructive comments on the article.
Glossary
- NA
nicotianamine
- CC
companion cell
- SE
sieve element
- BPS
bathophenanthroline disulfonate
- ICP-MS
inductively coupled plasma–mass spectrometry
- SXRF
synchrotron x-ray fluorescence microscopy
- qRT-PCR
quantitative RT-PCR
- MS
Murashige and Skoog
- FITC
fluorescein isothiocyanate
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amasheh S., Weber W. (1999). Further characteristics of the Ca(2+)-inactivated Cl(-) channel in Xenopus laevis oocytes. J. Membr. Biol. 172: 169–179 [DOI] [PubMed] [Google Scholar]
- Andriunas F.A., Zhang H.-M., Xia X., Patrick J.W., Offler C.E. (2013). Intersection of transfer cells with phloem biology-broad evolutionary trends, function, and induction. Front Plant Sci 4: 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrivault S., Senger T., Krämer U. (2006). The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46: 861–879 [DOI] [PubMed] [Google Scholar]
- Arteca R.N., Arteca J.M. (2000). A novel method for growing Arabidopsis thaliana plants hydroponically. Physiol. Plant. 108: 188–193 [Google Scholar]
- Baxter I.R., Vitek O., Lahner B., Muthukumar B., Borghi M., Morrissey J., Guerinot M.L., Salt D.E. (2008). The leaf ionome as a multivariable system to detect a plant’s physiological status. Proc. Natl. Acad. Sci. USA 105: 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogs J., Bourbouloux A., Cagnac O., Wachter A., Rausch T., Delrot S. (2003). Functional characterization and expression analysis of a glutathione transporter, BjGT1, from Brassica juncea: evidence for regulation by heavy metal exposure. Plant Cell Environ. 26: 1703–1711 [Google Scholar]
- Bouche-Pillon S., Fleurat-Lessard P., Fromont J.C., Serrano R., Bonnemain J.L. (1994). Immunolocalization of the plasma Mmembrane H+-ATPase in minor veins of Vicia faba in relation to phloem loading. Plant Physiol. 105: 691–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbouloux A., Shahi P., Chakladar A., Delrot S., Bachhawat A.K. (2000). Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 275: 13259–13265 [DOI] [PubMed] [Google Scholar]
- Cagnac O., Bourbouloux A., Chakrabarty D., Zhang M.-Y., Delrot S. (2004). AtOPT6 transports glutathione derivatives and is induced by primisulfuron. Plant Physiol. 135: 1378–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H.H., Chiecko J., Punshon T., Lanzirotti A., Lahner B., Salt D.E., Walker E.L. (2010). Successful reproduction requires the function of Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol. 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver O.B., Patterson K.D., Krieg P.A. (1996). Overexpression of the tinman-related genes XNkx-2.5 and XNkx-2.3 in Xenopus embryos results in myocardial hyperplasia. Development 122: 3549–3556 [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Connolly E.L., Fett J.P., Guerinot M.L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Cassin G., Couch D., Divol F., Higuchi K., Le Jean M., Misson J., Schikora A., Czernic P., Mari S. (2009). Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. (Lond.) 103: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato R.J., Jr, Roberts L.A., Sanderson T., Eisley R.B., Walker E.L. (2004). Arabidopsis Yellow Stripe-Like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J. 39: 403–414 [DOI] [PubMed] [Google Scholar]
- Dix D.R., Bridgham J.T., Broderius M.A., Byersdorfer C.A., Eide D.J. (1994). The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J. Biol. Chem. 269: 26092–26099 [PubMed] [Google Scholar]
- Durrett T.P., Gassmann W., Rogers E.E. (2007). The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 144: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D., Broderius M., Fett J., Guerinot M.L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M.J., Romera F.J., Stacey M.G., Stacey G., Villar E., Alcántara E., Pérez-Vicente R. (2013). Shoot to root communication is necessary to control the expression of iron-acquisition genes in Strategy I plants. Planta 237: 65–75 [DOI] [PubMed] [Google Scholar]
- Gayomba S.R., Jung H.I., Yan J., Danku J., Rutzke M.A., Bernal M., Krämer U., Kochian L.V., Salt D.E., Vatamaniuk O.K. (2013). The CTR/COPT-dependent copper uptake and SPL7-dependent copper deficiency responses are required for basal cadmium tolerance in A. thaliana. Metallomics 5: 1262–1275 [DOI] [PubMed] [Google Scholar]
- Green L.S., Rogers E.E. (2004). FRD3 controls iron localization in Arabidopsis. Plant Physiol. 136: 2523–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak M.A., Pezeshgi S. (1996). Shoot-to-root signal transmission regulates root Fe(III) reductase activity in the dgl mutant of pea. Plant Physiol. 110: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusak M.A., Welch R.M., Kochian L.V. (1990). Physiological characterization of a single-gene mutant of Pisum sativum exhibiting excess iron accumulation: I. Root iron reduction and iron uptake. Plant Physiol. 93: 976–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan S.A., Fariduddin Q., Ali B., Hayat S., Ahmad A. (2009). Cadmium: toxicity and tolerance in plants. J. Environ. Biol. 30: 165–174 [PubMed] [Google Scholar]
- Hindt M.N., Guerinot M.L. (2012). Getting a sense for signals: Regulation of the plant iron deficiency response. Biochim. Biophys. Acta 1823: 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y., Masuda H., Bashir K., Inoue H., Tsukamoto T., Takahashi M., Nakanishi H., Aoki N., Hirose T., Ohsugi R., Nishizawa N.K. (2010). Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 62: 379–390 [DOI] [PubMed] [Google Scholar]
- Järup L. (2003). Hazards of heavy metal contamination. Br. Med. Bull. 68: 167–182 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H.I., Gayomba S.R., Rutzke M.A., Craft E., Kochian L.V., Vatamaniuk O.K. (2012). COPT6 is a plasma membrane transporter that functions in copper homeostasis in Arabidopsis and is a novel target of SQUAMOSA promoter-binding protein-like 7. J. Biol. Chem. 287: 33252–33267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneen B.E., Larue T.A., Welch R.M., Weeden N.F. (1990). Pleiotropic effects of brz: a mutation in Pisum sativum (L.) cv; Sparkle’ conditioning decreased nodulation and increased iron uptake and leaf necrosis. Plant Physiol. 93: 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., Nishizawa N.K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63: 131–152 [DOI] [PubMed] [Google Scholar]
- Korenkov V., Hirschi K., Crutchfield J.D., Wagner G.J. (2007). Enhancing tonoplast Cd/H antiport activity increases Cd, Zn, and Mn tolerance, and impacts root/shoot Cd partitioning in Nicotiana tabacum L. Planta 226: 1379–1387 [DOI] [PubMed] [Google Scholar]
- Kuruma A., Hirayama Y., Hartzell H.C. (2000). A hyperpolarization- and acid-activated nonselective cation current in Xenopus oocytes. Am. J. Physiol. Cell Physiol. 279: 1401–1413 [DOI] [PubMed] [Google Scholar]
- Lahner B., Gong J., Mahmoudian M., Smith E.L., Abid K.B., Rogers E.E., Guerinot M.L., Harper J.F., Ward J.M., McIntyre L., Schroeder J.I., Salt D.E. (2003). Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana. Nat. Biotechnol. 21: 1215–1221 [DOI] [PubMed] [Google Scholar]
- Li Z.S., Lu Y.P., Zhen R.G., Szczypka M., Thiele D.J., Rea P.A. (1997). A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl. Acad. Sci. USA 94: 42–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubkowitz M. (2011). The oligopeptide transporters: a small gene family with a diverse group of substrates and functions? Mol. Plant 4: 407–415 [DOI] [PubMed] [Google Scholar]
- Maas F.M., van de Wetering D.A., van Beusichem M.L., Bienfait H.F. (1988). Characterization of Phloem iron and its possible role in the regulation of fe-efficiency reactions. Plant Physiol. 87: 167–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner, H. (1995). Mineral Nutrition of Higher Plants. (London; San Diego: Academic Press). [Google Scholar]
- Morrissey J., Baxter I.R., Lee J., Li L., Lahner B., Grotz N., Kaplan J., Salt D.E., Guerinot M.L. (2009). The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell 21: 3326–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustroph A., Zanetti M.E., Jang C.J., Holtan H.E., Repetti P.P., Galbraith D.W., Girke T., Bailey-Serres J. (2009). Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc. Natl. Acad. Sci. USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., et al. (2007). Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nelson B.K., Cai X., Nebenführ A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Osawa H., Stacey G., Gassmann W. (2006). ScOPT1 and AtOPT4 function as proton-coupled oligopeptide transporters with broad but distinct substrate specificities. Biochem. J. 393: 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Song W.-Y., Ko D., Eom Y., Hansen T.H., Schiller M., Lee T.G., Martinoia E., Lee Y. (2012). The phytochelatin transporters AtABCC1 and AtABCC2 mediate tolerance to cadmium and mercury. Plant J. 69: 278–288 [DOI] [PubMed] [Google Scholar]
- Pfaffl M.W., Horgan G.W., Dempfle L. (2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeros M.A., Cançado G.M., Kochian L.V. (2008). Novel properties of the wheat aluminum tolerance organic acid transporter (TaALMT1) revealed by electrophysiological characterization in Xenopus oocytes: functional and structural implications. Plant Physiol. 147: 2131–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea P.A. (2012). Phytochelatin synthase: of a protease a peptide polymerase made. Physiol. Plant. 145: 154–164 [DOI] [PubMed] [Google Scholar]
- Robinson N.J., Procter C.M., Connolly E.L., Guerinot M.L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rogers E.E., Guerinot M.L. (2002). FRD3, a member of the multidrug and toxin efflux family, controls iron deficiency responses in Arabidopsis. Plant Cell 14: 1787–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H., Séguéla-Arnaud M., Briat J.-F., Vert G., Curie C. (2011). The FRD3 citrate effluxer promotes iron nutrition between symplastically disconnected tissues throughout Arabidopsis development. Plant Cell 23: 2725–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rus A., Baxter I., Muthukumar B., Gustin J., Lahner B., Yakubova E., Salt D.E. (2006). Natural variants of AtHKT1 enhance Na+ accumulation in two wild populations of Arabidopsis. PLoS Genet. 2: 1964–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt D.E., Prince R.C., Pickering I.J., Raskin I. (1995). Mechanisms of cadmium mobility and accumulation in indian mustard. Plant Physiol. 109: 1427–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S., Schmidt W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Schaaf G., Honsbein A., Meda A.R., Kirchner S., Wipf D., von Wirén N. (2006). AtIREG2 encodes a tonoplast transport protein involved in iron-dependent nickel detoxification in Arabidopsis thaliana roots. J. Biol. Chem. 281: 25532–25540 [DOI] [PubMed] [Google Scholar]
- Schaaf G., Schikora A., Häberle J., Vert G., Ludewig U., Briat J.F., Curie C., von Wirén N. (2005). A putative function for the Arabidopsis Fe-Phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant Cell Physiol. 46: 762–774 [DOI] [PubMed] [Google Scholar]
- Scholz G., Schlesier G., Seifert K. (1985). Effect of nicotianamine on iron uptake by the tomato mutant chloronerva. Physiol. Plant. 63: 99–104 [Google Scholar]
- Schuler M., Rellán-Álvarez R., Fink-Straube C., Abadía J., Bauer P. (2012). Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24: 2380–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaff J.E., Schultz B.A., Craft E.J., Clark R.T., Kochian L.V. (2010). GEOCHEM-EZ: a chemical speciation program with greater power and flexibility. Plant Soil 330: 207–214 [Google Scholar]
- Stacey M.G., Koh S., Becker J., Stacey G. (2002). AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. Plant Cell 14: 2799–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey M.G., Osawa H., Patel A., Gassmann W., Stacey G. (2006). Expression analyses of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta 223: 291–305 [DOI] [PubMed] [Google Scholar]
- Stacey M.G., Patel A., McClain W.E., Mathieu M., Remley M., Rogers E.E., Gassmann W., Blevins D.G., Stacey G. (2008). The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol. 146: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunarpi Y., et al. (2005). Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 44: 928–938 [DOI] [PubMed] [Google Scholar]
- Turgeon R., Webb J.A. (1973). Leaf development and phloem transport in Cucurbita pepo: Transition from import to export. Planta 113: 179–191 [DOI] [PubMed] [Google Scholar]
- Valko M., Morris H., Cronin M.T. (2005). Metals, toxicity and oxidative stress. Curr. Med. Chem. 12: 1161–1208 [DOI] [PubMed] [Google Scholar]
- Vert G.A., Briat J.-F., Curie C. (2003). Dual regulation of the Arabidopsis high-affinity root iron uptake system by local and long-distance signals. Plant Physiol. 132: 796–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters B.M., Chu H.-H., Didonato R.J., Roberts L.A., Eisley R.B., Lahner B., Salt D.E., Walker E.L. (2006). Mutations in Arabidopsis yellow stripe-like1 and yellow stripe-like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 141: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintz H., Fox T., Wu Y.Y., Feng V., Chen W., Chang H.S., Zhu T., Vulpe C. (2003). Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 278: 47644–47653 [DOI] [PubMed] [Google Scholar]
- Wong C.K., Cobbett C.S. (2009). HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytol. 181: 71–78 [DOI] [PubMed] [Google Scholar]
- Wu H., Chen C., Du J., Liu H., Cui Y., Zhang Y., He Y., Wang Y., Chu C., Feng Z., Li J., Ling H.Q. (2012). Co-overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 158: 790–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y.L., Redman J.C., Monaghan E.L., Zhuang J., Underwood B.A., Moskal W.A., Wang W., Wu H.C., Town C.D. (2010). High throughput generation of promoter reporter (GFP) transgenic lines of low expressing genes in Arabidopsis and analysis of their expression patterns. Plant Methods 6: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z., Sooksa-nguan T., Vatamaniuk O.K. (2009). Establishing RNA interference as a reverse-genetic approach for gene functional analysis in protoplasts. Plant Physiol. 149: 642–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.-Y., Bourbouloux A., Cagnac O., Srikanth C.V., Rentsch D., Bachhawat A.K., Delrot S. (2004). A novel family of transporters mediating the transport of glutathione derivatives in plants. Plant Physiol. 134: 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.