Abstract
Background
DNA barcode differences within animal species are usually much less than differences among species, making it generally straightforward to match unknowns to a reference library. Here we aim to better understand the evolutionary mechanisms underlying this usual “barcode gap” pattern. We employ avian barcode libraries to test a central prediction of neutral theory, namely, intraspecific variation equals 2 Nµ, where N is population size and µ is mutations per site per generation. Birds are uniquely suited for this task: they have the best-known species limits, are well represented in barcode libraries, and, most critically, are the only large group with documented census population sizes. In addition, we ask if mitochondrial molecular clock measurements conform to neutral theory prediction of clock rate equals µ.
Results
Intraspecific COI barcode variation was uniformly low regardless of census population size (n = 142 species in 15 families). Apparent outliers reflected lumping of reproductively isolated populations or hybrid lineages. Re-analysis of a published survey of cytochrome b variation in diverse birds (n = 93 species in 39 families) further confirmed uniformly low intraspecific variation. Hybridization/gene flow among species/populations was the main limitation to DNA barcode identification.
Conclusions/Significance
To our knowledge, this is the first large study of animal mitochondrial diversity using actual census population sizes and the first to test outliers for population structure. Our finding of universally low intraspecific variation contradicts a central prediction of neutral theory and is not readily accounted for by commonly proposed ad hoc modifications. We argue that the weight of evidence–low intraspecific variation and the molecular clock–indicates neutral evolution plays a minor role in mitochondrial sequence evolution. As an alternate paradigm consistent with empirical data, we propose extreme purifying selection, including at synonymous sites, limits variation within species and continuous adaptive selection drives the molecular clock.
Introduction
DNA barcoding is a practical method for distinguishing species using a short DNA sequence from a standardized location on the genome. In animals, the agreed-upon standard is a 648 base pair fragment of mitochondrial cytochrome oxidase c subunit I (COI) [1], [2]. COI barcode differences within animal species are usually much less than differences among, a pattern often referred to as a “barcode gap,” making it generally straightforward to match unknowns to reference sequences [3]–[5]. A decade of DNA barcoding has generated libraries representing hundreds of thousands of vertebrate and invertebrate species, offering an unprecedented window into genetic variation [6].
To better understand the limits to DNA barcoding and the evolutionary mechanisms that underlie the usual barcode gap pattern, here we use birds to test whether differences within and among species conform to neutral theory, the reigning null hypothesis for mitochondrial sequence evolution [7]–[9]. We analyze apparent barcode gap exceptions in detail–those with unusually large intraspecific differences and those lacking interspecific differences. From a practical point of view exceptions may help define limits to COI barcodes as a marker of speciation. In the context of evolutionary theory, exceptions may give valuable insight into the mechanisms controlling variance within and among species. Birds are uniquely suited this task: they are well represented in barcode libraries, have the best-known species limits of any large animal group, and, most critically, are the only large group with known census population sizes, a key parameter in neutral theory [10]–[13].
Neutral theory posits most sequence differences within and among species are selectively neutral and so invisible to both adaptive and purifying selection. Several lines of evidence support neutrality. First, nearly all mitochondrial protein-coding gene (including COI) differences within and among closely related animal species are synonymous, i.e., do not change the amino acid sequence, indicating the observed changes are at least relatively neutral compared to non-synonymous substitutions [14], [15]. Second, viable hybrids and introgression imply most substitutions are functionally silent rather than species-specific adaptations [16]. Finally, variation in mitochondrial protein-coding region synonymous sites is the same magnitude as in the non-coding control region (e.g., [17]), which is thought to be under highly relaxed selection.
Neutral theory predicts intraspecific variation equals 2 Nµ, where N is population size and µ is mutation rate per generation [7]–[9]. Although textbooks and scientific reports recognize a multitude of exceptions to this predicted relationship, deviations are subsumed under the rubric of “effective population size” and accounted for by ad hoc modifications to the theory, which is assumed operative (e.g., [18]). Here we harness the unique resources of avian barcode libraries and census population data to look at the question the other way around, namely, do the empirical data show any signature of variance proportional to population size? If not, does the observed range of variation fit with commonly proposed modifications to neutral theory? In addition, we examine whether molecular clock measurements conform to neutral theory prediction that clock rate equals µ [19]–[21].
Taxonomic classifications are potential stumbling blocks in analyzing genetic variation. While a DNA sequence is an objective feature of an individual organism, species names reflect expert judgment. Even in a well-studied group like birds, ornithologists describe new species every year by “splitting” taxa previously considered single species [11], [22]. A compilation of DNA sequences may include as yet unsplit species, with the result that variation appears anomalously large. More generally, many species have genetically distinct, geographically isolated subpopulations [23]. Regardless of whether these merit species status, they fit the theoretical model of species as reproductively isolated groups [24], [25]. Another difficulty is hybridization between species or gene flow between regional populations, which may blur both genetic and taxonomic boundaries. Additional sources of inflated variation within species include mislabeled sequence records, sequencing error, and unrecognized pseudogenes [26]. The converse difficulty occurs when species previously considered distinct are “lumped” as single taxa based on new information. In these cases, apparent differences within and among may incorrectly appear low. Here we address potential taxonomic confounders by examining outliers in detail.
In the following we demonstrate uniformly low intraspecific mitochondrial DNA variation in birds regardless of population size. Nearly all apparent exceptions reflect lumping of reproductively isolated populations (many of which represent distinct species) or hybrid lineages. To our knowledge, this is the first large test of neutral theory applied to mitochondrial diversity using actual census population measurements rather than crude proxies of population size such as phylogeny or body weight [27]–[30], and the first to test outliers for population structure. In contrast to prior analyses, we find uniformly low intraspecific variation regardless of census population size. We conclude that this finding together with the molecular clock phenomenon are strong evidence that neutral processes play a minor role in animal mitochondrial evolution. We argue a radically different view of evolution–extreme purifying selection and continuous adaptive evolution–is needed to account for the widespread pattern of limited variation within species and larger differences among that underlies the general effectiveness of DNA barcoding.
Results
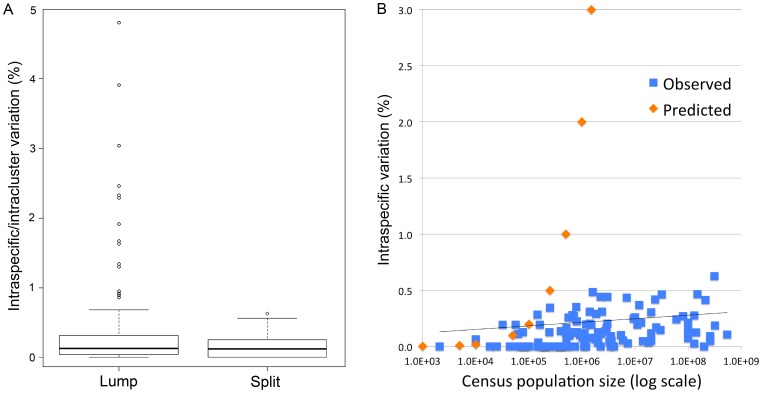
We examined COI barcode records from two avian families representing the two major divisions of birds, one non-passerine (Scolopacidae, sandpipers; 61 species analyzed) and one passerine (Parulidae, New World warblers; 63 species analyzed). These families were chosen as being among the best characterized in terms of barcode records (number of species and number of individuals per species [26]) and census data [12], [13]. We reasoned that analyzing multiple species within a family would give the clearest evidence for a population size effect since other factors postulated to influence intraspecific variation, such as proportion of breeding adults, number of offspring, generation time, and mating system [7]–[9] are generally similar within avian families [10]. In addition, in case a population size effect was evident only at the extreme of abundance, we analyzed birds with 100 million or more individuals for which 10 or more barcode records were available (18 additional species in 13 additional families). Intraspecific variation measured as average pairwise Kimura-2-parameter (K2P) difference was generally very low (Fig. 1, Table S1) and was unrelated to sample size (Fig. S1). Nearly all cases with high variation, defined as greater than 0.5% average pairwise difference, had geographic or hybrid lineages (Fig. 1, Table S1, Figs. S2, S3, S4, and S5). Geographic lineages were defined as monophyletic branches in NJ trees that mapped to well-established avian biogeographic regions, such as eastern and western North America [25], [31]. In most cases, inferences of reproductive isolation between geographic populations were supported by published studies including analyses using other mitochondrial and nuclear loci (Table S2). In neutral theory a model species is a reproductively isolated population. Thus in terms of variation a reproductively isolated population is expected to behave as an independent entity equivalent to a species. When geographic and hybrid lineages were considered as independent entities, intraspecific diversity appeared tightly constrained with an average about 0.1% and maximum about 0.5% (Fig. 1, Tables S1, S2). The observed average and maximum levels were what would be expected at equilibrium under a neutral model for a species with about 50,000 and 250,000 individuals, respectively [32]. However, our results demonstrated uniformly low variation in birds with census sizes ranging from a few thousand up to several hundred million (Fig. 1).
Figure 1. Low ceiling on intraspecific mtDNA variation in birds regardless of population size.
A) Intraspecific variation (average pairwise percent K2P difference) in 142 avian species listed in Table S1. Lower and upper bounds of boxes are first and third quartiles respectively, center bar is median, and lower and upper whiskers represent 1.5 times the interquartile range or the minimum or maximum value respectively. Residual values are shown as dots. Right, same dataset except that outliers were separated into geographic or hybrid clusters where present (Table S2). B) Intraspecific variation and population size among 111 species with census estimates; species with geographic or hybrid clusters were excluded. Note y-axis scale differs between A and B. Inclusion of geographic and hybrid clusters as representing equal fractions of parent population gave a similar result. Orange markers indicate predicted variation (2 Nµ) for a model species with census population size N and mutation rate µ of 10−8 per nucleotide site per generation (equivalent to 1% per lineage per My).
To test whether these findings were representative of mitochondrial coding region variation in a more diverse set of birds, we re-analyzed a published series reporting up to 5.0% intraspecific variation in avian cytochrome b [30]. Of 92 avian species in 39 families with greater than 0.5% average pairwise difference, review of original reports demonstrated nearly all (92%) reflected geographic clusters (Table S3, Fig. S6). Furthermore, about half (55%) with geographic clusters are currently split or recommended to be split into distinct species. Hybridization and overlooked pseudogenes were present in single cases. The few remaining cases had relatively modest variation (0.5%–1.1%) and small sample sizes (4–6 individuals).
To assess exceptions lacking interspecific differences, we examined all species highlighted as problematic in four recent avian barcode surveys [33]–[36] covering over 1000 species in total. Of 33 pairs or sets of species flagged as difficult to distinguish, four in fact have diagnostic sequence differences, three are proposed to represent single species, and eight lack sufficient data for analysis (single paraphyletic sequences) (Table S4). Of 18 evaluable cases with shared barcode clusters, 14 (78%) are associated with hybridization or introgression.
Discussion
Our analysis shows uniformly low intraspecific mtDNA variation in birds ranging in population size from 103 to 108 individuals. Apparent outliers demonstrate low variation when reproductively isolated populations and hybridization are taken into account (Fig. 1). This is the first large study of animal mitochondrial diversity that tests for geographic and hybrid lineages and employs actual census population measurements rather than inferring population size from a crude proxy such as phylogeny or body weight [27]–[30]. Our results contrast prior studies in finding variation is uniformly low, rather than differing according to mutation rate [29], [30] or recency of selective sweeps [27]. Re-analysis of a published series [30] further confirmed that outliers reported to have high mitochondrial variation in fact reflect lumping of reproductively isolated populations or overlooked species (Table S3).
Implications for neutral theory
The universally low intraspecific variation observed in this study does not support a central prediction of neutral theory, namely variance equals 2 Nµ, where N is population size and µ is mutation rate per generation [7]. Taken at face value the results imply µ is inversely proportional to population size across a 100,000-fold range. In contrast, direct measurements of mitochondrial mutation rate per generation are roughly similar in animals analyzed so far including organisms with very different population sizes and generation times such as fruit flies and humans [37].
The ad hoc modifications to neutral theory commonly proposed to account for low variation in individual cases, namely, recurrent bottlenecks or selective sweeps, struggle as general mechanisms. If bottlenecks limit variation, then a universal low ceiling implies recent population crashes for all species. This appears unlikely–almost a Noah’s Ark hypothesis–although perhaps long-term climate cycles might cause widespread periodic bottlenecks [38]. If selective sweeps limit variation, then a universal low ceiling implies a dynamic view of evolution, with all species adapting all the time [39], in contrast to the equilibrium model at the core of neutral theory.
However, even if bottlenecks or selective sweeps do occur regularly in all species, neutral theory still predicts a wide range of variation–with higher levels in organisms that reproduce more rapidly, as the rate of diversity recovery after a selective sweep or bottleneck is proportional to generation time (time to equilibrium variation is N generations) [7], [40]. In contrast to this prediction, the widespread effectiveness of DNA barcoding reflects similarly low levels of intraspecific variation across across the diversity of animal life, including insects and vertebrates that differ 100-fold in generation time (e.g., [41], [42]).
Finally, regardless of selective sweep or bottleneck frequency, there remains a paradox related to the molecular clock. In neutral theory, the molecular clock is proposed to be powered by drift and clock rate equals µ (mutation rate per generation) [19]–[21], [43]. As noted above, µ appears to be approximately the same in different animals. Neutral theory therefore predicts a chronologic clock rate proportional to generation time. However, published studies show the mitochondrial molecular clock ticks at roughly the same rate (∼1–2%/site/My) in diverse animals with very different generation times (e.g., [44], [45]).
Thus two very well documented phenomena, namely, universal limited intraspecific variation and a universal molecular clock, contradict key predictions of neutral theory. We suggest a new paradigm is needed [46]. As a step in that direction, we critically examine two assumptions underlying the neutral model as it is usually applied: most sequence differences are neutral and species are relatively stable over long periods of time, i.e., adaptive evolution is infrequent.
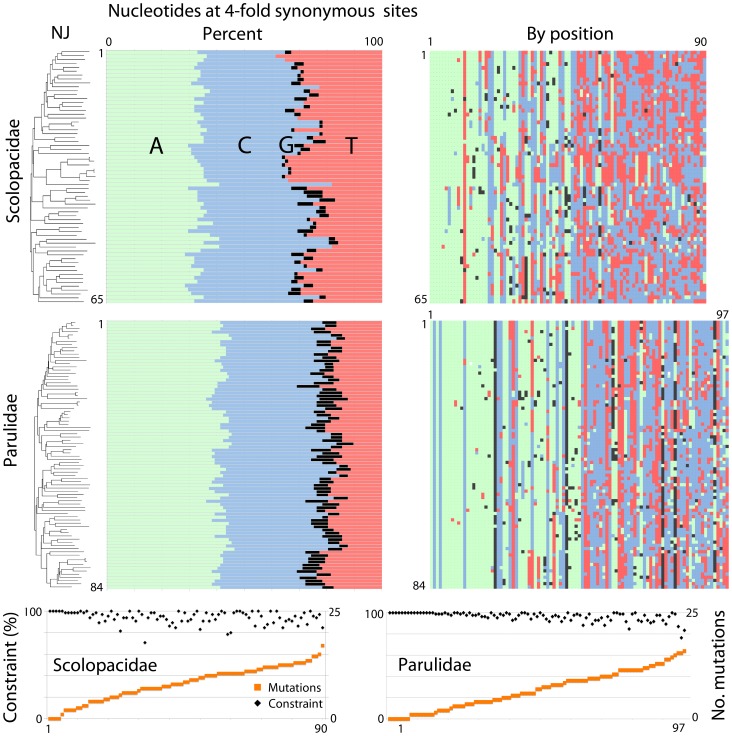
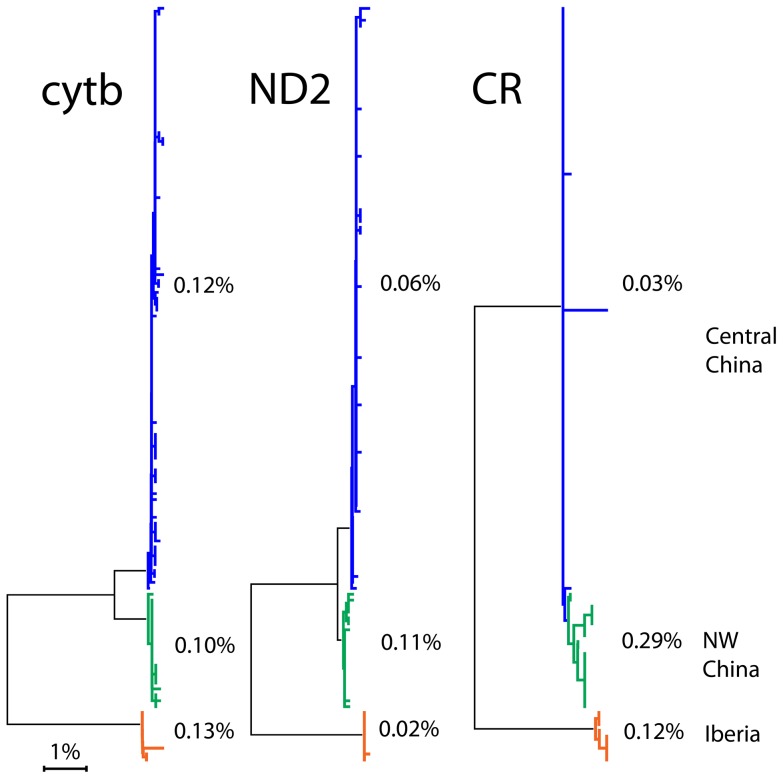
Synonymous substitutions are not necessarily selectively silent [47], [48]. As a test we analyzed patterns of interspecific substitution at four-fold synonymous sites, which theoretically can accommodate all four bases. Nucleotide diversity was significantly constrained (Fig. 2). This pattern might indicate translational efficiency limits synonymous substitutions. However, variation appears roughly uniform across mtDNA coding genes and the non-coding control region (e.g., Fig. 3), suggesting a mechanism unrelated to translation limits diversity, although there is much less data on control region than for COI barcodes. Similarly skewed nucleotide composition at four-fold synonymous sites in human and other animal mtDNAs is attributed to strand- and site-specific DNA mutation and repair biases [50]. Regardless of mechanism, an emerging view is exceptions to neutrality in mitochondrial evolution may be the rule [51]–[53].
Figure 2. Constrained synonymous variation.
From left, COI NJ tree (n = 65 Scolopacidae, 84 Parulidae species), nucleotide composition at four-fold synonymous sites by species (A, adenine, green; C, cytosine, blue; G, guanine, black; T, thymine, red), and by position are shown (n = 90 Scolopacidae, 97 Parulidae sites in 519 nt COI segment). For the latter, positions are sorted by number of apparent mutation events according to NJ tree. At bottom, number of apparent mutation events and percent constraint (limited to one or two nucleotides) for each position are shown. Evidence for restricted variation includes unequal nucleotide composition; unequal distribution of mutation events according to predominant nucleotide (positions with predominantly A have fewer mutation events than those with T or C); and nucleotide composition largely constrained to one or two nucleotides at all sites regardless of number of mutation events.
Figure 3. Mitochondrial coding genes and non-coding control region show similar patterns of variation and divergence (adapted from [49]).
Azure-winged magpie (Cyanopica cyanus) (n = 128 individuals) K2P NJ trees for cytochrome b (cytb), ND2, and control region (CR) generated in MEGA are shown at the same scale. Termini representing geographic populations are colorized and average pairwise percent K2P difference within each geographic population are shown.
We hypothesize an absolute limit to intraspecific variation, including at synonymous sites, due to extreme purifying selection. In this scenario, mutations (and reversions) occur at a very small number of positions, resulting in low variation even in large populations over long times. In support, somatic mtDNA mutations in normal human cells usually match variants found in unrelated individuals and rarely involve protein coding regions, suggesting a very limited repertoire of allowable substitutions [54]. In contrast, cancer cells show diverse somatic mutations in coding and non-coding regions that are not represented in mtDNA databases. Further evidence of limited allowable variation is that homoplasy at synonymous sites is common within species (including humans) and between closely-related species [55], [56]. Moreover, much of what appears to be neutral intraspecific variation may instead reflect distinct lineages resulting from local adaptations in heterogeneous environments [48].
Second, we hypothesize species are adapting more or less continuously, with mitochondrial genes evolving together with nuclear genome through genetic draft and epistasis [39], [57], [58]. In this scenario, selective sweeps power the molecular clock [59]. Assuming, as posited above, that allowable variation is tightly restricted, then all species reach the same maximum but low level of variation after a small number of generations, and chronologically-timed selective sweeps produce a chronologic clock.
A universal selection-driven mtDNA clock implies all organisms are evolving at about the same rate [39], [44], [45], [58]. What could cause similar rates of change for diverse organisms in diverse environments? We speculate all life is adapting together at more or less the same rate, tied together by a multitude of food web, predator-prey, and parasite-host interactions, whether on land, lakes, or sea, with long-term planetary climate cycles as the ultimate driver of evolution. This leads us to ask whether there are any environments unchanged over the past hundreds of thousands of years. For example, there might be deep-sea environments with little physical change (although the biological environment might still be dynamic). If so, it would be of interest to examine intraspecific variation and the mitochondrial clock in those settings.
Limitations of this study include most species examined are Nearctic or Palearctic birds. Low intraspecific diversity in these regions may be due to population expansions associated with glacial cycles [60]. However, recent studies of tropical birds show similarly low variation when geographic lineages are taken into account [61], [62]. Another limitation is sample sizes are relatively small. For species with high variation and geographic clustering, the inferred divisions are supported by published work using other mtDNA loci in most cases and correspond to known biogeographic boundaries (Tables S2, S3, Figs. S2, S3, S4, and S5). For species with low variation, further study may reveal greater genetic diversity but this is likely to be in the form of geographic or hybrid lineages.
On the other hand, our results may overestimate variation. Geographic differences were analyzed only in species with greater than 0.5% average or 1.5% maximum variation, and some showed distinct regional clusters below those thresholds. Existing records are insufficient to detect small-scale geographic clustering, particularly as collection locations include migration ranges. Variation in some species may be elevated due to gene flow, i.e., mixing of historically distinct populations.
The findings presented here could in principle be particular to birds. Sexual selection is common in birds and females are the heterogametic sex, with both factors hypothesized to restrict mitochondrial DNA variation [57]. However, barcode surveys abundantly document low intraspecific variance in thousands of species from diverse animal phyla, with exceptions usually carrying biological differences that indicate reproductively isolated populations or overlooked species (e.g., [41], [42]). It may be of interest to conduct similar comprehensive analyses of mtDNA variation in other taxonomically well-studied groups with large DNA barcode databases, such as fish (Actinopterygii) or butterflies and moths (Lepidoptera).
A critical question is whether these findings reflect special features of mitochondrial biology [63]–[65]. A largely unexplored potential mechanism is male contribution to mitochondrial inheritance via gene conversion, which might play a role in limiting variation within species and promoting divergence at speciation. Recent studies call attention to an old riddle of limited intraspecific variation in nuclear genes [66], [67], suggesting the relevant processes are common to both genetic compartments. On the other hand Bazin and colleagues report evidence for neutral evolution in nuclear but not mitochondrial genes [27].
Implications for DNA barcoding
Based on our analysis, the main limitation to DNA barcode taxonomy is hybridization/gene flow between species/populations that were previously isolated (Table S4). When species or populations with divergent mtDNA genomes interbreed, complex patterns may occur, ranging from occasional hybrid individuals to stable hybrid zones [68] to complete replacement, i.e., introgression [16]. In such cases, assigning individuals to a species with a single mitochondrial locus and even with more extensive mitochondrial and nuclear data may not be possible [69]. From a practical point of view, this scenario appears relatively uncommon, involving a few percent of individuals and species. In fact, the general rarity of large differences within species suggests such reproductive mixing is short-lived from an evolutionary point of view. Such scenarios may similarly challenge taxonomists deciding what constitutes a species vs. a set of populations. Young species per se and retention of ancestral polymorphisms per se do not appear important limitations to DNA barcoding unless there is hybridization or gene flow, since shared polymorphisms are otherwise uncommon. In birds at least, most species with greater than 0.5% variation are composites of reproductively isolated populations, many of which are candidate species. As noted above, deciding whether such populations merit species status necessarily involves expert judgment and additional biological information. In any case, it is evident that the commonly applied 2% threshold is insufficient to capture all animal species [3]. Even single base pair differences may signal reproductively isolated lineages worthy of further study.
Conclusion
COI barcode variation within avian species is uniformly low regardless of census population size. This finding directly contradicts a central prediction of neutral theory and is not readily accounted for by commonly proposed ad hoc modifications. As an alternative model consistent with empirical data including the molecular clock, we propose extreme purifying selection, including at synonymous sites, limits variation within species and continuous adaptive evolution drives the molecular clock.
Materials and Methods
COI sequences were downloaded from GenBank using search terms “family or species name”(organism) AND (COI[gene name] OR COX1[gene name]), aligned in MEGA [70] with MUSCLE, and trimmed to 648 bp barcode region corresponding to mouse mitochondrial genome positions 52–699 [2]. Sequences not covering at least 80% of barcode region were omitted. Average and maximum pairwise intraspecific distances were calculated in MEGA after excluding 10% at either end of barcode region (final segment 519 nucleotides, barcode positions 64–582) to minimize contribution of sequencing error [26]. Box and dot plots were generated in R and Excel, respectively. Global population estimates [12], [13] were averaged if given as ranges. To examine nucleotide composition at synonymous sites, representative barcode sequences of Scolopacidae and Parulidae species were aligned and re-ordered using TreeParser following NJ K2P template trees [71]. The re-ordered FASTA files were opened in MEGA and nucleotides at four-fold synonymous sites were exported to Excel for nucleotide composition analysis. To compare coding and control region variation, sequences reported by Zhang and colleagues [49] were downloaded from GenBank and analyzed in MEGA. The analyzed dataset includes cytochrome b, ND2 and control region sequences from 128 Azure-winged Jay (Cyanopica cyanus) individuals.
Supporting Information
Intraspecific COI barcode variation is unrelated to sample size.
(PDF)
Collection locations for Scolopacidae species with high intraspecific variation, geographic clusters.
(PDF)
Collection locations for Parulidae species with high intraspecific variation, geographic clusters.
(PDF)
Collection locations for highly abundant species with high intraspecific variation, geographic clusters.
(PDF)
NJ trees with GenBank accession nos. for birds with high intraspecific variation, geographic clusters.
(PDF)
Intraspecific cytochrome b variation is unrelated to sample size (adapted from [30] ).
(PDF)
Intraspecific mitochondrial DNA variation in birds is generally low.
(PDF)
Variation within avian geographic and hybrid clusters is low.
(PDF)
Re-analysis of intraspecific variation in Nabholz 2009 [30] avian cytochrome b dataset.
(PDF)
Avian species with shared DNA barcode clusters.
(PDF)
(DOCX)
Acknowledgments
We thank Benoit Nabholz for generously sharing datasets, Jesse Ausubel, Fiona Doetsch, Michael Hickerson, and David Lohman for helpful comments, and Alan Curry for assistance with statistical analysis.
Funding Statement
This work was supported by The Alfred P. Sloan Foundation (www.sloan.org) (MYS, DST). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hebert PDN, Cywinkska A, Ball SL, de WaardJR (2003) Biological identifications through barcodes. Proc Biol Sci 270: 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Consortium for the Barcode of Life Database Working Group (2009) Data standards for BARCODE records in INSDC (BRIs). Available: http://www.barcoding.si.edu/pdf/dwg_data_standards-final.pdf.
- 3. Hebert PDN, Stoeckle MY, Zemlak TS, Francis CM (2004) Identification of birds through DNA barcodes. PLOS Biol 2: e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Frézal L, Leblois R (2008) Four years of DNA barcoding: current advances and prospects. Infect Genet Evol 8: 727–736. [DOI] [PubMed] [Google Scholar]
- 5. Jingo U, Kato T, Ito M (2011) Current progress in DNA barcoding and future implications for entomology. Entomol Sci 14: 107–124. [Google Scholar]
- 6. Ratnasingham S, Hebert PDN (2007) BOLD: The Barcode of Life Data System (www.barcodinglife.org). Mol Ecol Notes. 7: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hartl DL, Clark AG (2007) Principles of population genetics, fourth edition. Sunderland: Sinauer Associates, Inc. 565 p. [Google Scholar]
- 8.Crow JF, Kimura M (2009) An introduction to population genetics theory. Devon: Blackburn Press. 608 p. [Google Scholar]
- 9.Hedrick PW (2011) Genetics of populations, fourth edition. Sudbury: Jones and Bartlett Publishers. 675 p. [Google Scholar]
- 10.del Hoyo J, Elliott A, Sargatal J, Christie (eds.) (2013) Handbook of the birds of the world, special volume. Barcelona: Lynx Edicions. 812 p. [Google Scholar]
- 11.International Ornithologists’ Union (2014) IOC World Bird List (v 4.1) Gill F, Donsker D (eds). Available: http://www.worldbirdnames.org. Accessed 2013 Nov 15.
- 12.Partners in Flight Science Committee (2013) Population estimates database, version 2013. Available: http://rmbo.org/pifpopestimates. Accessed 2013 Nov 15.
- 13.Wetlands International (2002) Waterbird population estimates, 3rd edition. Wageningen: Wetlands International Global Series, no. 12. Wageningen, The Netherlands.
- 14. Kocher TD, Thomas WK, Meyer A, Edwards SV, Paabo S, et al. (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci U S A 86: 6196–6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kerr KCR (2011) Searching for evidence of selection in avian DNA barcodes. Mol Ecol Resources 11: 1045–1055. [DOI] [PubMed] [Google Scholar]
- 16. Rheindt FE, Edwards SV (2011) Genetic introgression: an integral but neglected component of speciation in birds. Auk 128: 620–632. [Google Scholar]
- 17. Barker FK, Benesh MK, Vandergon AJ, Lanyon SM (2012) Contrasting evolutionary dynamics and information content of the avian mitochondrial control region and ND2 gene. PLOS ONE 7: e46403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bortolotto E, Bucklin A, Mezzvilla M, Zane L, Patarnello T (2011) Gone with the currents: lack of genetic differentiation in the Antarctic krill Euphausia superba . BMC Genet 12: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimura M (1968) Evolutionary rate at the molecular level. Nature 217: 624–626. [DOI] [PubMed] [Google Scholar]
- 20. Kumar S (2005) Molecular clocks: four decades of evolution. Nature Rev Genet 6: 654–662. [DOI] [PubMed] [Google Scholar]
- 21. Ho S (2008) The molecular clock and estimating species divergence. Nature Education 1: 168. [Google Scholar]
- 22. Chesser RT, Banks RC, Barker FK, Cicero C, Dunn FL, et al. (2013) Fifty-fourth supplement to the American Ornithologists’ Union Check-List of North American birds. Auk 130: 558–571. [Google Scholar]
- 23. Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, et al. (1987) Intraspecific phylogeography: the mitochondrial bridge between population genetics and systematics. Ann Rev Ecol Syst 18: 489–522. [Google Scholar]
- 24.Coyne JA, Orr HA (2004) Speciation. Sunderland: Sinauer Associates, Inc. 545 p. [Google Scholar]
- 25.Newton I (2003) The speciation and biogeography of birds. London: Elsevier Science Ltd. 668 p. [Google Scholar]
- 26. Stoeckle MY, Kerr KCR (2012) Frequency matrix approach demonstrates high sequence quality in avian BARCODEs and highlights cryptic pseudogenes. PLOS ONE 7: e43992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bazin E, Glémin S, Galtier N (2006) Population size does not influence mitochondrial genetic diversity in animals. Science 312: 570–572. [DOI] [PubMed] [Google Scholar]
- 28. Nabholz B, Mauffrey J-F, Bazin E, Galtier N, Glémin S (2008) Determination of mitochondrial genetic diversity in mammals. Genetics 178: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Piganeau G, Eyre-Walker A (2009) Evidence for variation in the effective population size of animal mitochondrial DNA. PLOS ONE 4: e4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nabholz B, Glémin S, Galtier N (2009) The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across mammals and birds. BMC Evol Biol 9: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pyle P (1997) Identification guide to North American birds. Part I: Columbidae to Ploceidae. Bolinas: Slate Creek Press. 732 p. [Google Scholar]
- 32. Duret L (2008) Neutral theory: the null hypothesis of molecular evolution. Nature Education 1: 218. [Google Scholar]
- 33. Kerr KCR, Lijtmaer DA, Barreira AS, Hebert PDN, Tubaro PL (2009) Probing evolutionary patterns in Neotropical birds through DNA barcodes. PLOS ONE 4: e4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnsen A, Rindal E, Ericson PGP, Zuccon D, Kerr KCR, et al. (2010) DNA barcoding of Scandinavian birds reveals divergent lineages in trans-Atlantic species. J Ornithol 151: 565–578. [Google Scholar]
- 35. Kerr KCR, Stoeckle MY, Dove CJ, Weigt LA, Francis CM, et al. (2007) Comprehensive DNA barcode coverage of North American birds. Mol Ecol Notes 7: 535–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tavares ES, Goncalves P, Miyaki CY, Baker AJ (2011) DNA barcode detects high genetic structure within neotropical bird species. PLOS ONE 6: e28543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kondrashov FA, Kondrashov AS (2010) Measurements of spontaneous rates of mutations in the recent past and the near future. Philos Trans R Soc London B Biol Sci 365: 1169–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shackleton NJ (2000) The 100,000-year Ice-Age cycle identified and found to lag temperature, carbon dioxide, and orbital eccentricity. Science 289: 1897–1902. [DOI] [PubMed] [Google Scholar]
- 39.Thompson JN (2013) Relentless evolution. Chicago: University of Chicago Press. 499 p. [Google Scholar]
- 40. Bedford T, Cobey S, Pascual M (2011) Strength and tempo of selection revealed in viral gene genealogies. BMC Evol Biol 11: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN (2006) DNA barcodes distinguish species of tropical Lepidoptera. Proc Natl Acad Sci U S A 103: 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN (2005) DNA barcoding Australia’s fish species. Philos Trans R Soc Lond B Biol Sci 360: 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lanfear R, Kokko H, Eyre-Walker A (2014) Population size and the rate of evolution. Trends Ecol Evol 29: 33–41. [DOI] [PubMed] [Google Scholar]
- 44. Weir JT, Schluter D (2008) Calibrating the avian molecular clock. Mol Ecol 17: 2321–2328. [DOI] [PubMed] [Google Scholar]
- 45. Papadopoulou A, Anastasiou I, Vogler AP (2010) Revisiting the insect mitochondrial molecular clock: the Mid-Aegean Trench calculation. Mol Biol Evol 27: 1659–1672. [DOI] [PubMed] [Google Scholar]
- 46.Kuhn TS (1996) The structure of scientific revolutions, third edition. Chicago: University of Chicago Press. 226 p. [Google Scholar]
- 47. Stergachis AB, Haugen E, Shafer A, Fu W, Vernot B, et al. (2013) Exonic transcription factor binding directs codon choice and affects protein evolution. Science 342: 1367–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kazancioğlu E, Arnqvist G (2014) The maintenance of mitochondrial genetic variation by negative frequency-dependent selection. Ecol Letters 17: 22–27. [DOI] [PubMed] [Google Scholar]
- 49. Zhang R, Song G, Qu Y, Alström P, Ramos R, et al. (2012) Comparative phylogeography of two widespread magpies: importance of habitat preference and breeding behavior on genetic structure in China. Mol Phylogenet Evol 65: 562–572. [DOI] [PubMed] [Google Scholar]
- 50. Perna NT, Kocher TD (1995) Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol 41: 353–358. [DOI] [PubMed] [Google Scholar]
- 51. Meiklejohn CD, Montooth KL, Rand DM (2007) Positive and negative selection on the mitochondrial genome. Trends Genetics 23: 259–263. [DOI] [PubMed] [Google Scholar]
- 52. Galtier N, Nabholz B, Glémin S, Hurst GDD (2009) Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol 18: 4541–4550. [DOI] [PubMed] [Google Scholar]
- 53. Wares JP (2010) Natural distributions of mitochondrial sequence diversity support new null hypotheses. Evolution 64: 1136–1142. [DOI] [PubMed] [Google Scholar]
- 54. He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, et al. (2010) Heteroplasmic mitochondrial DNA mutations in normal and tumor cells. Nature 464: 610–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Galtier N, Enard D, Radondy Y, Bazin E, Belkhir K (2006) Mutation hot spots in mammalian mitochondrial DNA. Genome Res 16: 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Levin L, Zhidkov I, Gurman Y, Hawlena H, Mishmar D (2013) Functional recurrent mutations in human mitochondrial phylogeny–dual roles in evolution and disease. Genome Biol Evol 5: 876–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hill GE, Johnson JD (2013) The mitonuclear compatibility hypothesis of sexual selection. Proc Biol Sci 280: 20131314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Neher RA (2013) Genetic draft, selective interference, and populations genetics of rapid adaptation. Ann Rev Ecol Evol Systematics 44: 195–215. [Google Scholar]
- 59. Grenfell BT, Pybus OG, Gog JR, Wood JLN, Dally JM, et al. (2004) Unifying the epidemiological and evolutionary dynamics of pathogens. Science 303: 327–332. [DOI] [PubMed] [Google Scholar]
- 60. Moritz C, Cicero C (2004) DNA barcoding: promise and pitfalls. PLOS Biol 2: 1529–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lohman DJ, Ingram KK, Prawiradilaga DM, Winker K, Sheldon FJ, et al. (2010) Cryptic genetic diversity in “widespread” Southeast Asian bird species suggest that Philippine avian endemism is gravely underestimated. Biological Conservation 143: 1885–1890. [Google Scholar]
- 62. Milá B, Tavares ES, Saldaña AM, Karubian J, Smith TB, et al. (2012) A trans-Amazonian screening of mtDNA reveals deep intraspecific divergence in forest birds and suggests a vast underestimation of species diversity. PLOS ONE 7: e40541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Parmakelis A, Kotsakiozi P, Rand D (2013) Animal mitochondria, positive selection and cyto-nuclear co-evolution: insights from pulmonates. PLOS ONE 8: e61970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang F, Broughton RE (2013) Mitochondrial-nuclear interactions: compensatory evolution or variable functional constraint among vertebrate oxidative phosphorolation genes? Genome Biol Evol 5: 1781–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boore JL (1999) Animal mitochondrial genomes. Nucl Acids Res 27: 1767–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewontin RC (1974) The genetic basis of evolutionary change. Columbia University Press, London, 346 pp.
- 67. Leffler EM, Bullaughey K, Matute DR, Meyer WK, Ségurel L, et al. (2012) Revisiting an old riddle: what determines genetic diversity levels within species? PLOS Biol 10: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dabrowski A, Fraxer R, Confer JL, Lovette IJ (2005) Geographic variability in mitochondrial introgression among hybridizing populations of Golden-winged (Vermivora chrysoptera) and Blue-winged (V. pinus) Warblers. Conservation Genet 6: 843–853. [Google Scholar]
- 69. Sonsthagen SA, Chesser RT, Bell DA, Dove CJ (2012) Hybridization among arctic white-headed gulls (Larus ssp.) obscures the genetic legacy of the Pleistocene. Ecol Evol 2: 1278–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5: 150–163. [DOI] [PubMed] [Google Scholar]
- 71. Stoeckle MY, Coffran C (2013) TreeParser aided Klee diagrams display taxonomic clusters in DNA barcode and nuclear gene datasets. Scientific Reports 3: 2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intraspecific COI barcode variation is unrelated to sample size.
(PDF)
Collection locations for Scolopacidae species with high intraspecific variation, geographic clusters.
(PDF)
Collection locations for Parulidae species with high intraspecific variation, geographic clusters.
(PDF)
Collection locations for highly abundant species with high intraspecific variation, geographic clusters.
(PDF)
NJ trees with GenBank accession nos. for birds with high intraspecific variation, geographic clusters.
(PDF)
Intraspecific cytochrome b variation is unrelated to sample size (adapted from [30] ).
(PDF)
Intraspecific mitochondrial DNA variation in birds is generally low.
(PDF)
Variation within avian geographic and hybrid clusters is low.
(PDF)
Re-analysis of intraspecific variation in Nabholz 2009 [30] avian cytochrome b dataset.
(PDF)
Avian species with shared DNA barcode clusters.
(PDF)
(DOCX)