Abstract
Background
Both an excess of toxic lead (Pb) and an essential iron disorder have been implicated in many diseases and public health problems. Iron metabolism genes, such as the hemochromatosis (HFE) gene, have been reported to be modifiers for lead absorption and storage. However, the HFE gene studies among the Asian population with occupationally high lead exposure are lacking.
Objectives
To explore the modifying effects of the HFE genotype (wild-type, H63D variant and C282Y variant) on the Pb load and iron metabolism among Asian Pb-workers with high occupational exposure.
Methods
Seven hundred and seventy-one employees from a lead smelter manufacturing company were tested to determine their Pb intoxication parameters, iron metabolic indexes and identify the HFE genotype. Descriptive and multivariate analyses were conducted.
Results
Forty-five H63D variant carriers and no C282Y variant carrier were found among the 771 subjects. Compared with subjects with the wild-type genotype, H63D variant carriers had higher blood lead levels, even after controlling for factors such as age, sex, marriage, education, smoking and lead exposure levels. Multivariate analyses also showed that the H63D genotype modifies the associations between the blood lead levels and the body iron burden/transferrin.
Conclusions
No C282Y variant was found in this Asian population. The H63D genotype modified the association between the lead and iron metabolism such that increased blood lead is associated with a higher body iron content or a lower transferrin in the H63D variant. It is indicated that H63D variant carriers may be a potentially highly vulnerable sub-population if they are exposed to high lead levels occupationally.
Introduction
There is considerable variability in the toxic response to lead (Pb) exposure among both the general population and occupational workers. For exposed persons, the outcome may be affected by factors such as the degree of exposure, age, and genetically determined differences in uptake, elimination, or sensitivity to the toxic effects of the substance. Genetic variants have occurred in enzymes known to influence or regulate lead metabolism. A potential candidate gene for susceptibility to lead exposure is the hemochromatosis (HFE) gene encoding the HFE protein. The presence of mutations in the HFE gene (major in C282Y variant and less common in H63D variant) leads to hemochromatosis. Hemochromatosis is an autosomal recessive hereditary disease in which an abnormality in iron metabolism, consisting in increased iron absorption by the gut and increased export of iron by the reticulum-endothelial system, leads to iron accumulation and deposits in multiple internal organs. The result is the progressive tissue damage as fibrosis/cirrhosis, diabetes, arthropathy, cardiomiopathy, hipopituitarism among others. The HFE gene is localized in chromosome 6p21.3 and two predominant variants have been described, the C282Y variant and the H63D variant and the C282Y variant is associated in homozygous in more than 80% with Hereditary Hemochromatosis [1].
Pb is non-essential to humans. Therefore, it is unlikely that a specific transporter exists for it. Pb shares the same pathway as Fe because they are both divalent cations, and our previous study has shown that Pb and Fe may share the same transporters, such as the divalent metal transporter 1 (DMT1) and ferroportin 1 (FP1) [2]. Iron status may over- or under-regulate intestinal divalent cation transporters and has been suggested specifically with the HFE genotype [3], [4]. Because of the associations between iron and lead transport, it is possible that polymorphisms in the HFE gene may also influence lead absorption and storage. Studies have shown mixed results.
Considering the clinical association between Fe deficiency and Pb toxicity, several studies have shown that the HFE mutations (H63D variant and C282Y variant) were significantly associated with increased blood lead (BPb) levels in young children [5]. However, in a cohort study of elderly men, HFE mutation predicted lower patella and BPb levels [4]. Additionally, a study of 2,926 Australian adult male and female twins showed that the HFE genotype did not contribute to variations in the BPb levels in adults [6]. Hopkins et al. [5] proposed that the HFE gene affects the BPb levels, which vary by age because of the differences in iron needs and storage that correlate with age and sex. Furthermore, they proposed that in younger populations, which have higher iron needs, HFE mutation may increase Pb absorption among the variant's carriers. However, the results did not demonstrate evidence of the BPb levels' modification by serum ferritin or hemoglobin in children or adults. Conversely, in an older population of elderly males, the effect of the HFE mutation may contribute to the down-regulation of Fe and Pb absorption. However, this study lacked data on the analysis concerning iron storage. Iron status is plausibly related to BPb levels and HFE genotype and should theoretically be a modifier of the association of HFE with BPb rather than a confounder. The conclusions of three previous studies differ from each other, suggesting that gene's environmental interactions may vary by life stage and Pb exposure.
The existing research about the association between HFE genotype and Pb exposure was performed on the general population and cannot directly illuminate the causes of variation in more highly exposed groups of Pb workers. The Meta analyses of the published data show that individuals with ALAD polymorphism have higher BPb levels at high Pb exposures, whereas there is no difference at low Pb exposures [7], [8], [9]. Moreover, owing to the wide variability in the prevalence rates of HFE mutation, such as C282Y, in particular the rare reports of the C282Y variant in China, it will be important to replicate the study of these genetic associations in different populations.
In the present study, we measured the biomarkers associated with the exposure, distribution, and excretion of Pb [BPb and urine lead (UPb)] and the body iron metabolic indexes in Chinese Pb-workers. In this study, we used unexposed factory workers as controls. In particular, we examined the distribution of the HFE genotype and the potential genetic modifications of the biomarkers of toxicokinetics and their associations with toxic effects. The analysis includes a consideration of covariates such as sex, age, smoking, alcohol intake, and education level to better understand the effects of the HFE genotype on the association between Pb exposure and Fe metabolism.
Materials and Methods
Studied Populations
Eight hundred and thirty-one individuals were identified from a lead smelter. Those who were exposed to lead smoke/lead dust directly, such as through lead extractive metallurgy or electrolytic lead, were recruited into the exposed group. Other individuals who worked in the offices distant from the workplaces associated with lead exposure were recruited into the control group, and the controls were workers of the same lead smelter but unexposed. Subjects who did not complete a questionnaire, failed to provide biological samples, or were diagnosed with a disorder of the peripheral nervous system, diabetes mellitus, limb injuries and/or drug intoxication were excluded. Seven hundred and seventy-one individuals were finally recruited for the study, of which 705 were lead-exposed individuals (576 males and 129 females, with a median employment of 10.63 years) and 66 controls (43 males and 23 females). The survey was approved by The Clinical Ethics Committee of First Affiliated Hospital of Nanchang University. Written informed consent was obtained from each individual prior to the study.
A standardized questionnaire was used to elicit their ethnic backgrounds, alcohol and smoking habits (yes/no), protective measures, self-reported symptoms, medical history and occupational history, such as work unit (department), type of work, and exposure duration.
Sample Collection
All of the samples were obtained from the participants during a fasting state at the same time of the morning. Blood samples were obtained by venipuncture after overnight (>12 h) fasting: 2 ml of blood was collected in a vacutainer with K2EDTA anticoagulant for extracting the DNA of the lymphocytes, and 1 ml was collected in a plastic “metal-free” evacuated tube containing sodium heparin anticoagulant for determining the BPb and zinc protoporphyrin (ZPP) immediately. At the same time, 2 ml of blood was collected in a tube with a non-anticoagulant for extracting serum samples. Spot urine samples were collected in acid-washed polyethylene bottles. Each urine sample was acidified with concentrated nitric acid and was used for Pb analysis. All of the samples except for the blood samples used to determine BPb and ZPP were frozen at −20°C until the survey was finished (2–3 days) and then stored at −80°C until the beginning of the analysis.
Evaluation of Lead Intoxication Parameters
BPb and UPb were measured by a stripping out analyzer (MP-2, Shandong, China), using the differential potentiometer stripping method (WS/T 21-1996) and the oscillo-polarographic method (WS/T 91-1996), respectively. A quality control sample was inserted in each run of the 10 samples. The limits of detection for Pb were 0.9 µg/L in blood and 5 µg/L in urine, and the precision was 6.1% and 5.4%, respectively. The accuracy was assured with certified blood and urine samples from the National Center for Disease Control and Prevention of China; 303±5.8 µg/L in blood and 260±6.5 µg/L in urine (recommended 299±11 µg/L and 252±30 µg/L).
The ZPP concentration was assayed directly by a ZPP blood fluorescence determinator (model 2002, Xian, China), using the hematofluorometer method (WS/T 92-1996), which measured the ratio of the fluorescence of ZPP to the absorption of the light by sample (by hemoglobin (Hb)) and is presented as the µg ZPP/g of the Hb (µg/g Hb). The blood concentration of Hb was assayed by the cyanomethemoglobin colorimetric assay method.
Measurement of Serum Iron Indices
Serum samples were obtained in the fasting state at the same time of day, stored at −20°C, and then sent to the Clinical Laboratory Department of Tongji Hospital (Wuhan, China) together. The spectrophotometric determination of serum iron (sFe, reference interval, 10.6–28.3 µmol/L for men and 6.6–26.0 µmol/L for women) and unsaturated iron-binding capacity (UIBC, reference interval, 7.5–65.3 µmol/L) were measured using the Roche (Modular P) automatic biochemical analyzer (Roche Diagnostics, Mannheim, German). The turbidimetric immunoassay of the serum ferritin (sFn, reference interval, 20–290 µg/L for men and 4.5–170 µg/L for premenopausal women, 24–260 µg/L for postmenopausal women), transferrin (Tf, reference interval, 2.0–3.6 g/L) and soluble transferrin receptor (sTfR, reference interval, 0.76–1.76 mg/L) were determined using GmbH (Marburg, Germany). The total iron binding capacity (TIBC) was calculated from these results as follows: TIBC = Fe+UIBC. The serum transferrin saturation (TfS) was calculated based on the formula TfS (%) = Fe/TIBC×100. The body iron content was calculated based on the formula (BIC, mg/kg) = −[log(sTfR/sFn ratio)-2.8229]/0.1207.
The CVs for inter-day precision near the upper bounds of the reference intervals for ferritin, Tf and sTfR were 3.1%, 2.6%, and 1.2%, respectively. The inter-day CV was 1.2% for sFe and 1.6% for UIBC, with means of 9.31 µmol/L and 32.9 µmol/L, respectively.
Identification of the H63D and C282Y Variant in the Hemochromatosis Gene
The genomic DNA was extracted from the subjects' peripheral white blood cells using the improved NaI method [10]. Briefly, this method involves the treatment of whole blood with an equal volume of NaI [3 M final concentration, analytical grade, purchased from Bodi Chemical Reagent Co., Ltd (Tianjing, China)] followed by a chloroform-isoamyl alcohol extraction to clear the Hb and cell debris. The clear aqueous layer is then mixed with isopropanol to obtain the DNA. Finally, the DNA was dissolved in a TE buffer (containing Tris, a common pH buffer, and EDTA, a chelating agent). After the DNA quantification, the samples were adjusted to the TE buffer, partitioned into aliquots, and stored at −20°C.
The C282Y (rs1800562) and H63D (rs1799945) variants (Reference Sequence NM 139011, GenBank) were genotyped by a PCR and a restriction fragment length polymorphism (RFLP) analysis as previously described [11]. Briefly, two portions of the HFE gene surrounding both mutations were amplified separately using PCR with HFE-specific primers. These primers was designed using Primer Express v5.0 software (Applied Biosystem) based on the published sequence of the HFE gene and synthesized from Sangon Biotech Co., Ltd (Shanghai, China). The PCR reactions consisted of 20 µM of each primer, forward PCR primer, 5′- TGT GGA GCC TCA ACA TCCT-3′; reverse PCR primer, 5′- TGA AAA GCT CTG ACA ACC TCA-3′ for the H63D variant (rs1799945), and forward PCR primer, 5′ -TCC AGT CTT CCT GGC AA-3′; reverse PCR primer, 5′-TTC TAG CTC CTG GCT CTCA-3′ for the C282Y variant (rs1800562), 2.5 µl 10×Reaction Buffer (containing Mg2+), 2 µl dNTP (2.5 mM), 0.125 µl Ex Taq DNA polymerase(5 U/µl), and 4 µl genomic DNA; then sterile water was added to 25 µl of Ex Taq DNA polymerase, 10×Reaction Buffer, dNTP (100 mM) and 6×Loading Buffer, which were purchased from Takara (Dalian, China). The PCR conditions were 3 min at 94°C, followed by 35 cycles of 30 sec at 94°C, 30 sec at 56°C and 1 min at 72°C, and finally, 5 min at 72°C, performed using the autorisierter thermocycler (eppendorf Mastercycler personal, Germany).
After the PCR amplification, restriction digests were performed directly with 10 µl of the PCR mixtures by the addition of 5 U SnaBI for C282Y (BioLabs, New England, MA, USA) or BclI (BioLabs) for H63D and the corresponding buffers (2 µl 10×NEB4 Buffer added to 2 µl 10×BSA, 5 µl sterile water and 2 µl 10×NEB3 Buffer and adding sterile water to reach 20 µl) and incubated for 2 h at 37°C for the SnaB I restriction or overnight at 50°C for the Bcl I restriction. The products were resolved on 2% agarose gel and stained by ethidium bromide (EB). A random sample of 10% of the subjects was run in duplicate as a quality control measure. The genotypes were also determined in control blood known to be from subjects homozygous for the wild-type genotype and heterozygous and homozygous for each of the variant genotypes.
Statistical Methods
We conducted a cross-sectional analysis of the association between the HFE mutation and the BPb/UPb concentrations and the iron metabolic indexes among Pb-exposed and unexposed workers in a Pb smelter. We first compared the characteristics of subjects who had all of the data of interest, including the genotype, BPb and UPb levels, and the iron metabolic index with the subjects who were not included because of missing data. The allele and genotype frequencies were determined and the Hardy-Weinberg test for equilibrium was performed. The univariate distributions of the continuous variables were examined to determine departures from normality. The distributions of the demographic and lifestyle characteristics and the BPb/UPb/ZPP levels and iron metabolic index by genotype (wild-type vs. C282Y variant or H63D variant) were examined, and the differences were tested by the Chi-square test, Student's t-test or the nonparametric test, as appropriate. The distribution of ZPP evidenced departures from normality and was thus transformed to a natural logarithm (ln) in the hypothesis tests between the groups of exposed and non-exposed workers. The adequacy of the ln-transformation of these measures was confirmed by verification that the distributions of the residuals after the linear regression modeling were normal.
Multiple linear regressions were used to model the effect of the genotype on the Pb burden and the iron metabolic index in cross-sectional analyses, while controlling for the same covariates used in the final models. Cross-product terms for the genotype and Pb burden variables or for the genotype and the iron metabolic indexes were used to evaluate the effect of the modification by genotype on the association between Pb exposure and body iron status. In consideration of the correlation among the iron metabolic indexes (sFe, UIBC, sFn, Tf, sTfR, TIBC, TfS, BIC, Hb), these variables were used separately in the final regression models to predict the Pb burden (BPb/UPb/ZPP), and each of these regressions was repeated, adding an indicator variable for the genotype. Covariates for the final regression model were identified by stepwise forward modeling (based on the P value) of the age, gender (male vs. female), education (lower than high school vs. higher than high school), tobacco use (yes vs. no), alcohol consumption (yes vs. no), occupational Pb exposure (unexposed, dissolved Pb operations or electrolytic Pb operations) and exposed/unexposed work duration.
The statistical analysis was performed using SPSS 19.0 software. Statistical significance was considered to be present at a P<0.05.
Results
The lead biomarkers BPb, UPb, ZPP of the exposed workers were 364.90±150.83 µg/L, 36.00 (17.00, 75.00) µg/L and 6.10 (3.40, 10.85) µg/g Hb, respectively, and were significantly higher than those of the unexposed workers (142.99±58.62 µg/L, 2.50 (1.00, 7.00) µg/L and 0.90 (0.00, 1.95) µg/g Hb), respectively. The means and the distributions of the iron metabolism indexes (sFe, TfS, sFn, BIC) and education, drinking, and the years that the males were employed were different between the exposed and unexposed workers. However, the distributions of smoking and the HFE genotypes, as well as the means of UIBC, Tf, TIBC, sTfR, and Hb, among all of the subjects were similar (Table 1, 2). The overall prevalence values for the H63D genotypes were wild-type, 94.16% (726/771); heterozygous, 5.71% (44/771), with 5.39% (38/705) for exposed group and 9.09% (6/66) for unexposed group; and homozygous, 0.13% (1/771), with 0.14% (1/705) for exposed group. Allelic frequency in global was 2.98% (46/1542) in this population, with 2.84% (40/1410) for exposed workers and 4.55% (6/132) for unexposed workers. The prevalence values of the C282Y genotypes were wild-type, 100%; heterozygous and homozygous, 0%. The distribution of the H63D genotype conformed to the Hardy-Weinberg expected frequencies (H63D: Chi-square = 0.011, P = 0.995).
Table 1. Demographic Characteristics of Subjects by Exposure [M(P25, P75)or N(%)].
| Grouping | Exposed | Unexposed | ||
| Males | Females | Males | Females | |
| Number | 576(81.7)** | 129(18.3)** | 43(65.2) | 23(34.8) |
| Age (years) | 32.0(23.0, 39.0) | 38.0(34.0, 41.0)** | 26.5(24.0, 41.5) | 26.0(23.0, 39.0) |
| Exposed/unexposed work duration (years) exposure | 2.6(1.6, 18.2) | 18.0(12.0, 21.0)* | 2.0(1.6, 2.8) | 2.6(2.4, 21.0) |
| Education (≥high school) | 79(14.3)** | 11(8.8)** | 28(68.3) | 15(65.2) |
| Married | 377(67.2) | 109(87.2)** | 23(54.8) | 14(60.9) |
| Smoker | 351(62.9) | 0(0.0) | 24(58.5) | 0(0.0) |
| Drinker | 320(57.7)* | 1(0.8) | 31(75.6) | 0(0.0) |
*(**) p<0.05 (0.01) (Chi-square test or nonparametric test) compared with the unexposed group of the same sex.
Results were expressed as the mean ± SD when the continuous variables followed a normal distribution.
Results were expressed as M (P25, P75) when the continuous variables did not follow a normal distribution.
Table 2. Lead Biomarkers, Serum Iron Indices and Genotypes by Exposure [Mean ± SD or N (%) or M (P25, P75)].
| Grouping | Exposed | Unexposed | ||
| Males | Females | Males | Females | |
| BPb(µg/L) | 384.54±151.98** | 276.16±107.93** | 144.72±60.22 | 139.91±56.84 |
| UPb(µg/L) | 39.00(18.00,80.00)** | 28.50(12.00,51.50)** | 3.00(1.00,7.00) | 1.00(1.00,7.50) |
| ZPP(µg/gHb) | 9.00±8.92** | 9.03±8.24** | 2.49±1.24 | 4.98±5.79 |
| sFe (µmol/L) | 20.97±7.90* | 17.08±7.13 | 24.00±7.93 | 14.74±7.00 |
| UIBC(µmol/L) | 37.4±12.4 | 44.2±13.9* | 35.4±12.7 | 51.1±14.8 |
| TIBC(µmol/L) | 56.90(51.79, 62.73) | 61.3±10.8 | 57.83(52.65, 65.29) | 65.9±12.1 |
| Tf (g/L) | 2.73±0.49 | 2.92±0.54 | 2.87±0.42 | 2.96±0.58 |
| TfS (%) | 34.80(26.10, 44.96)* | 28.92±12.84 | 41.37(30.31, 49.68) | 23.39±11.65 |
| sFn (µg/L) | 160.00(92.30, 230.00)** | 38.90(20.95, 59.00) | 238.00(148.00, 343.00) | 42.20(20.70, 64.50) |
| sTfR (mg/L) | 1.30(1.13, 1.55) | 1.38(1.20, 1.56)* | 1.36(1.12, 1.55) | 1.26(0.99, 1.44) |
| BIC (mg/kg) | 40.30±2.63** | 35.22±3.28 | 41.74±2.67 | 35.46±2.69 |
| Hb(g/L) | 139.8±15.0 | 120.5±12.2 | 138.3±13.3 | 115.9±14.2 |
| H63D | ||||
| HH | 546(94.8) | 120(93.0) | 39(90.7) | 21(91.3) |
| HD | 29(5.0) | 9(7.0) | 4(9.3) | 2(8.7) |
| DD | 1(0.2) | 0 | 0 | 0 |
| C282Y | ||||
| CC | 576(100) | 129(100) | 43(100) | 23(100) |
| CY | 0 | 0 | 0 | 0 |
| YY | 0 | 0 | 0 | 0 |
Abbreviations: BPb, blood lead; UPb, urine lead; ZPP, zinc protoporphyrin; sFe, serum iron; UIBC, unsaturated iron-binding capacity; TIBC, total iron binding capacity; Tf, transferrin; TfS, serum transferrin saturation; sFn, serum ferritin; sTfR, soluble transferrin receptor; BIC, body iron content; Hb, haemoglobin. For H63D genotype, HH, wild-type; HD, H63D heterozygous variant; DD, H63D homozygous variant. For C282Y genotype, CC, wild-type; CY, C282Y heterozygous variant; YY, C282Y homozygous variant.
*(**) p<0.05 (0.01) (Chi-square test, Student's t-test or nonparametric test) compared with the unexposed group of the same sex.
Results were expressed as the mean ± SD when continuous variables followed a normal distribution.
Results were expressed as M (P25, P75) when continuous variables did not follow a normal distribution.
Table 3 shows the distribution of the BPb, UPb and covariates stratified by genotype. The overall trend of the lead in the body levels was for the blood levels in the carriers of the H63D variant to be slightly higher, but there was no statistically significant difference compared with the wild-type subjects. The UPb/ZPP levels for the carriers of the H63D variant did not differ from those of the wild-type subjects. The covariates, except for UIBC, TIBC, and Tf, were similar between the subjects with one copy of the allele and the wild-type.
Table 3. Lead Biomarkers and Serum Iron Indices by HFE Genotypes [Mean ±SD or N or M (P25, P75)].
| All | HH | HD or DD | |||||
| Na | Mean ±SD or M (P25, P75) | N | Mean ±SD or M (P25, P75) | N | Mean ±SD or M (P25, P75) | P | |
| BPb (µg/L) | 748 | 345.90±157.95 | 705 | 344.75±158.70 | 43 | 364.87±145.52 | 0.418 |
| UPb (µg/L) | 727 | 32.00(12.300,71.00) | 686 | 31.00(12.00,71.00) | 41 | 40.00(18.00,77.00) | 0.536 |
| ZPP(µg/gHb) | 757 | 5.60(3.00,10.20) | 714 | 5.70(3.00,10.33) | 43 | 4.90(2.80,9.40) | 0.406 |
| sFe (µmol/L) | 769 | 20.30±7.98 | 724 | 20.27±7.96 | 45 | 20.72±8.31 | 0.728 |
| UIBC(µmol/L) | 768 | 37.45(30.60,46.38) | 723 | 37.80(30.90,46.80) | 45 | 33.40(26.15, 42.25) | 0.012* |
| TIBC (µmol/L) | 768 | 57.43(52.40,63.87) | 723 | 57.90(52.55,64.18) | 45 | 54.85(49.11, 60.05) | 0.008** |
| Tf (g/L) | 768 | 2.72(2.44,3.04) | 723 | 2.74(2.46,3.05) | 45 | 2.54(2.23, 2.85) | 0.005** |
| TfS (%) | 768 | 33.41(24.47,44.09) | 723 | 33.13(24.41,43.57) | 45 | 37.19(25.47, 47.17) | 0.195 |
| sFn (µg/L) | 768 | 129.00(66.08,214.00) | 723 | 129.00(65.80,214.00) | 45 | 130.00(66.95, 227.00) | 0.983 |
| sTfR(mg/L) | 755 | 1.31(1.14,1.55) | 710 | 1.31(1.14,1.55) | 45 | 1.34(1.15, 1.54) | 0.844 |
| BIC(mg/kg) | 753 | 39.91(37.53,41.71) | 708 | 39.90(37.49,41.75) | 45 | 40.03(37.77, 41.64) | 0.853 |
| Hb (g/L) | 762 | 135.8±16.3 | 717 | 135.7±16.3 | 45 | 137.0±15.8 | 0.606 |
Abbreviations: BPb, blood lead; UPb, urine lead; ZPP, zinc protoporphyrin; sFe, serum iron; UIBC, unsaturated iron-binding capacity; TIBC, total iron binding capacity; Tf, transferrin; TfS, serum transferrin saturation; sFn, serum ferritin; sTfR, soluble transferrin receptor; BIC, body iron content; Hb, haemoglobin; HH, wild-type; HD, H63D heterozygous variant; DD, H63D homozygous variant.
*(**) p<0.05 (0.01) compared with HH (student's t-test or nonparametric test).
Because of missing data, the numbers do not equal 771.
Results were expressed as the mean ± SD when continuous variables followed a normal distribution.
Results were expressed as M (P25, P75) when continuous variables did not follow a normal distribution.
However, the above analysis does not take into account several potential confounders. Some factors in distribution were unbalanced between the group of Pb-exposed and of unexposed, such as gender, educational levels and iron metabolism indexes et al. From Table 2, Pb-exposed women had lower BPb and UPb values than men, as well as lower sFn and Hb values. Thus, those factors were considered in the multivariate analyses.
Multiple regression models for evaluating the effect of the H63D genotype on the association between Pb exposure and iron metabolism are presented in Table 4. In the multiple regression models, no evidence was found that the H63D variants directly influenced either the Pb exposure or the iron metabolism. However, the H63D genotype did modify associations between the BPb levels, and BIC/Tf was observed. Per unit joint change in BIC with wild-type, there was an −1.149 unit impact to BPb levels (P = −0.008), while per unit joint change in BPb with wild-type, there was an −0.002 unit impact to BIC levels (P<0.001) (the first and second analysis in Table 4).
Table 4. Linear regression models evaluating effect of HFE genotypes on the association between lead exposure and iron metabolism.
| β | SE | Bcoff | t value | P-value1 | R 2 | F value | P-value2 | |
| Variable | BPb(µg/L) | |||||||
| Content | 340.588 | 65.512 | 5.199 | 0.000 | ||||
| BIC | −5.507 | 1.953 | −0.119 | −2.280 | 0.005 | |||
| BIC×HH or HD DD | −1.149 | 0.432 | −0.099 | −2.661 | 0.008 | 0.594 | 135.187 | 0.000 |
| BIC(mg/kg) | ||||||||
| Content | 40.661 | 0.532 | 76.374 | 0.000 | ||||
| BPb×HH or HD DD | −0.002 | 0.000 | −0.284 | −9.254 | 0.000 | 0.444 | 135.408 | 0.000 |
| Tf(g/L) | ||||||||
| Content | 3.649 | 0.109 | 33.474 | 0.000 | ||||
| BPb | −0.002 | 0.001 | −0.538 | −2.877 | 0.004 | |||
| BPb×HH or HD DD | 0.000 | 0.000 | 0.414 | 2.237 | 0.026 | 0.141 | 26.626 | 0.000 |
Abbreviations: BPb, blood lead; Tf, transferrin; BIC, body iron content; HH, wild-type; HD, H63D heterozygous variant; DD, H63D homozygous variant.
Linear models were adjusted stepwise for age (year), gender (male vs. female), education (lower than high school vs. higher than high school), marriage (yes vs. no), tobacco use (yes vs. no), alcohol consumption (yes vs. no), occupational lead exposure (unexposed, dissolved lead operations or electrolytic lead operations) and work years. H63D genotype (HH vs. HD or DD), iron metabolic index/BPb and the cross-product with the genotype and each iron metabolic index/BPb. While BPb was independent factor, each iron metabolic index and the cross-product with the genotype were put separately into the model.
P value for each statistic;
P value for each regression model.
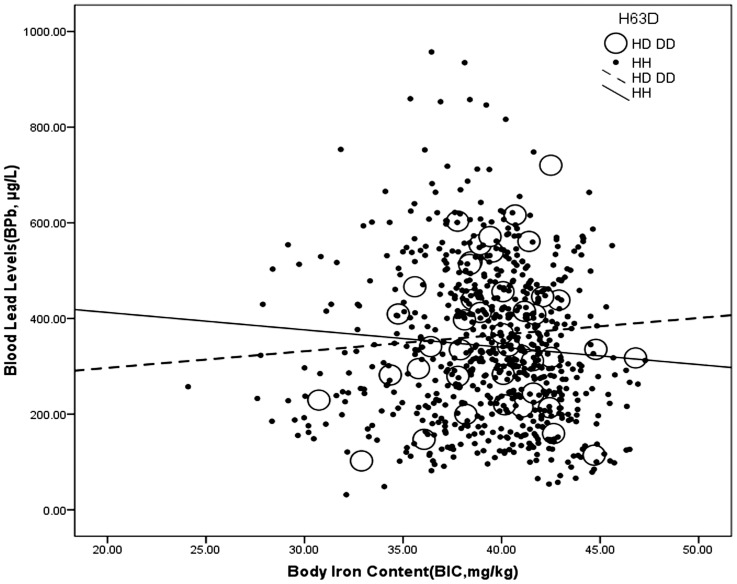
The visual inspection of polymorphism was performed on the plots depicting associations between the BPb and BIC or the BPb and Tf stratified by genotype for evidence of a dominant or recessive effect of an allele. The plot depictions also revealed that there was an interactive effect between the H63D genotype and the BIC on the BPb levels. Open dots and dashed line regression line (BPb = 227.201+3.478*BIC) denote H63D variant carriers, which means a higher BIC was predicted to mean higher BPb levels, whereas close dots and full line regression line (BPb = 485.247–3.634*BIC) denote workers with wild-type, a higher BIC was predicted to lower the BPb levels (Fig. 1).
Figure 1. Interactive effect between the H63D genotype and body iron content on blood lead levels.
H63D genotype: HD or DD: open dots, dashed line; HH: close dots, full line. The data are analyzed using multivariate analysis.
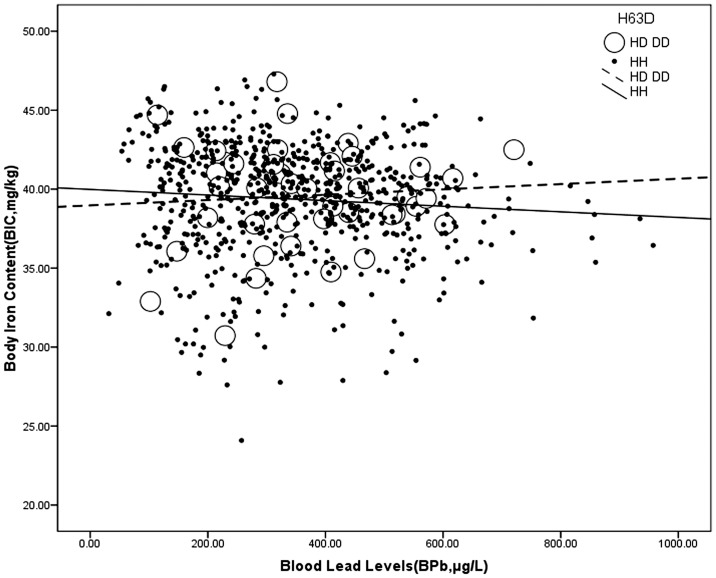
Interestingly, the evidence showed that there was an interactive effect between the H63D genotype and environmental exposure on the BIC, which revealed that higher BPb levels were associated with a higher BIC in the carriers of the H63D variant (open dots and dashed line regression line, BIC = 38.973+0.002*BPb), whereas higher BPb levels were associated with a lower BIC in those without the H63D variant (close dots and full line regression line regression line, BIC = 39.984–0.002*BPb) (Fig. 2).
Figure 2. Interactive effect between the H63D genotype and blood lead levels on body iron content.
H63D genotype: HD or DD: open dots, dashed line; HH: close dots, full line. The data are analyzed using multivariate analysis.
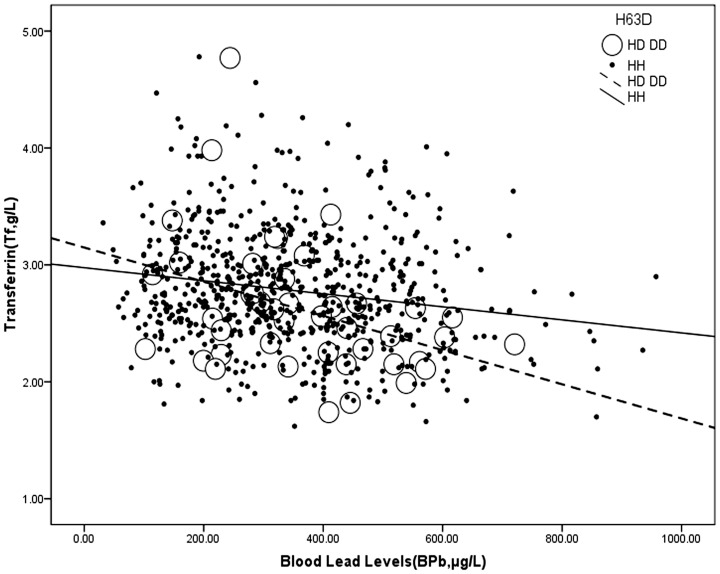
Additionally, our results revealed that higher BPb levels were associated with a lower Tf, and a steeper slope was observed in workers with the H63D variant (Table 4 and Fig. 3).
Figure 3. Interactive effect between the H63D genotype and blood lead levels on transferrin.
H63D genotype: HD or DD: open dots, dashed line; HH: close dots, full line. The data are analyzed using multivariate analysis.
The data also revealed that BIC is an independent predictor of significantly lower BPb levels (βcoff = −0.199, P = 0.005) in the final regression models that were adjusted for age, sex, smoking, drinking, education, marriage and lead exposure (Table 4).
We also compared the results of the effect modification by genotype on the associations between the iron metabolic index and the same outcomes (UPb and ZPP). No evidence of effect modification by genotype on associations between the iron metabolic index and UPb or ZPP was found.
Discussion
In our current study of Chinese Pb-exposed workers, we determined the effect of the HFE genotype on the relationship between Pb exposure and iron metabolism. First, the effect modification by genotype on association between BPb and BIC was observed. A higher BIC predicted higher BPb levels in the workers with the H63D variant, whereas a higher BIC predicted lower BPb levels in the workers without the H63D variant. The results also showed the converse to be true, that higher BPb levels were associated with a higher BIC in the workers with the H63D variant, while higher BPb levels were associated with a lower BIC in the subjects with the wild-type. Second, in contrast with BIC, higher BPb levels were associated with a lower Tf in the workers with the H63D variant. Our results suggest that the H63D genotype modifies the association between iron and lead in Chinese lead workers with high occupational exposure, independent of the presence of the C282Y allele, although the H63D variant has been regarded as nonfunctional for iron overload in Europe. Moreover, it is more likely for workers with the H63D variant to have excess iron stores, and the Pb absorption may promote lead poisoning and increase the AD risk.
To the best of our knowledge, this is the first study to examine the association between iron metabolism and lead exposure modified by the HFE gene in Asian workers who are occupationally exposed to Pb. The wide age range of 18 to 55 years has allowed us to observe the associations between Pb exposure and iron metabolism at almost every stratified age in adults. Pb shares the same pathway as Fe, as they are both divalent cations, and the iron status may up- or down-regulate the intestinal divalent cation transporters [12], [13]. Numerous studies have shown that the regulatory mechanisms that cause an increase in iron absorption accompany an increase in the amount of ingested lead that is absorbed during iron deficiency [14], [15], [16], [17]. Body iron status is inversely associated with the Pb burden because elevations in the BIC prevent the body from controlling iron's absorption to prevent its systemic excess in humans, and the Pb absorption is subsequently lower [18], [19]. Polymorphisms of the HFE gene are of interest in the studies of human Pb exposure because the iron metabolism impacts the Pb absorption from the gastrointestinal tract and the Pb transfer and/or storage. One research group found, based on this hypothesis, that the HFE mutation modifies lead metabolism in children and elderly men, revealing a higher blood lead in children and lower blood/bone lead in elderly men carrying the HFE mutation. They explained that the HFE gene's effect on the blood lead may vary by life stage because of the differences in body iron needs and storages that correlate with age and sex [4], [5]. The conclusions of these previous studies differ from each other and also from our results.
In contrast with other studies, the C282Y variant was not detected, whereas 5.8% of the Chinese lead workers in our study carried the H63D variant. Although there is no clear data establishing whether the H63D variant leads to the phenotypic manifestations of hemochromatosis or significant iron overload, some studies have suggested that the H63D variant is associated with a milder form of iron overload or hereditary hemochromatosis [20], [21]. In addition, evidence has been reported in a previous functional study that the H63D variant alters the normal HFE product's affinity for its ligand [22], which leads to relatively small increments of cellular iron content [23]. An increased hepatic iron deposition was observed in the H63D heterozygous variant carriers (with 3 compound heterozygous C282Y/H63D in 58 H63D heterozygous variant) [24]. Iron absorption often accompanies an increase in lead absorption [25]. A case-control study of hereditary hemochromatosis indicated an up-regulation of iron absorption with a subsequent increase in lead absorption [3]. Our multivariate analysis results revealed that BIC was positively associated with BPb levels in individuals with the H63D variant, and higher lead values were found in the individuals with a higher body iron content, whereas the reverse occurred in the workers without the H63D variant.
Our study differs from previous cohort-based studies assessing lead exposure among the general population when the major HFE mutation was C282Y variant because we assess high-level occupational lead exposure and focus on Chinese subjects that are H63D heterozygous variant for the HFE mutation. All of these factors may play a role in explaining the differing results. In a study by Wright et al. [4], iron status was related to blood lead and the H63D genotype, but unfortunately, the study lacked data on the iron status, and there were no effect modifications by either serum ferritin or hemoglobin in the Hopkins et al. [5] study. In a general population-based cohort study, Whitfild et al. [6] found no significant associations between the H63D genotype and blood lead in adults, and blood lead levels were inversely associated with ferritin. Using serum ferritin or hemoglobin concentrations to estimate body iron has certain limitations, the most important of which is the influence of inflammation or liver disease on the serum ferritin level independent of the body iron stores; however, the sTfR can still be used to detect concurrent iron deficiencies in persons with inflammation. At the same time, quantitative estimates of the body iron content calculated from the R/F ratio greatly enhance the evaluation of the iron status [26], [27]. We examined various iron metabolism indicators in the workers' blood, and our results showed that BIC was an independent predictor of significantly lower blood lead levels in the multivariate analyses that were adjusted for age, sex, smoking, drinking, education, and lead exposure. The mean iron metabolism indexes were in the normal range for both lead exposed and unexposed workers. Evidence was found of an interaction between the H63D genotype and the body iron status.
Our current study also found that the H63D genotype has an interactive effect with BPb on Tf: higher BPb levels were associated with a lower Tf, and with an elevation in BPb, the Tf dropped more in workers with the H63D variant than with the subjects with the wild-type, although the R square for the multiple linear analysis that was adjusted for the confounders only to 0.141. It is known that Tf is a powerful chelator, capable of binding iron tightly but reversibly; a molecule of Tf can bind two atoms of ferric iron with high affinity, and a significant serum Tf decrease indicates that there may be a clinical iron overload in the subject's body [28]. Additionally, a study of twins in Jahanshad et al. [29] revealed that the H63D variant was associated with decreased transferrin levels and that AD patients who carry only the H63D heterozygous variant showed higher levels of iron and lower levels of transferrin than the wild-type AD patients [30]. A higher blood lead was also associated with the HFE mutation in young children (with 16.7% H63D heterozygous variant and 3.3% C282Y variant), with an elevation in the need for body iron [5]. Furthermore, Pb has been shown to bind directly to transferrin, down-regulating Tf gene expression [31]. Our results suggest that the H63D variant affects both lead metabolism and iron metabolism, and our investigation further enriches the knowledge of the interaction and relationship between Pb exposure and iron metabolism.
We tested for the effects of a number of covariants that have been reported to affect blood lead levels in other general population-based studies. There were no significant effects of smoking and alcohol use, which is different from previously reports, except for sex [6], [32]. The mean values of the blood lead were significantly higher than those reported elsewhere because subjects in our study were workers who were occupationally exposed to lead. The measures of environmental exposure (exposure duration and the type of work) show independent effects on lead concentrations.
The conclusions of this study are subject to several limitations. First, our sample was restricted to a homogeneous sample of Chinese workers who were occupationally exposed to lead, who displayed a lower prevalence of the allelic frequency of the H63D variant (2.98%) than people from Europe (12∼15%), the America (14∼15%), North Africa (13∼17%) and South Korea (3.8%) but displayed a higher prevalence than people from Japan (0.99%) [33], [34]. Furthermore, our sample contained no C282Y variant carriers. Although studies outside Asia have found that the C282Y variant is the most common variant genotype and is highly prevalent (50–100%) in C282Y homozygous variant [35], the frequency of the H63D variant is higher than the C282Y variant in Asia. The roles of the H63D and C282Y variants in producing hemochromatosis were believed to different [36]; with regard to the H63D variant, its effect on the body iron stores was much milder than the C282Y variant, particularly the heterozygous and iron overload was scarcely observed [37]. This finding is in line with our study within the normal range of iron metabolism indexes in our subjects. Therefore, it will be important to replicate this study in different occupationally lead-exposed populations. Second, we measured Pb only in the blood and urine. Although the BPb and UPb levels are the most common method for establishing the degree of exposure in humans, the factors described may affect either the lead absorption and body burden or only its distribution within the body. In the latter case, some people could have high blood lead levels but an acceptable total body lead content, or vice versa. Distinguishing between these possibilities would require the measurement of lead in the bones. Third, the control population was not engaged in lead operations in the same lead smelter, and their blood lead levels were higher than in the general population, even though there was no likelihood of a greater risk to lead exposure within the control population. It would be preferable to choose controls from the general population or use double controls, using subjects from both the lead smelter and the general population, to determine the association between lead exposure and iron metabolism.
Conclusions
In summary, in a cross-sectional study of occupationally lead-exposed workers, the H63D genotype modified the association between lead exposure and iron metabolism with a positive association between the body iron content and blood lead in subjects carrying the H63D variant.
Funding Statement
This work was supported by The National Nature Science Foundation of China (Nos.30960324, 81273120, 21267017), Jiangxi Provincial Natural Science Foundation (No. 20132BAB205069). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, et al. (1996) A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet 13: 399–408. [DOI] [PubMed] [Google Scholar]
- 2. Zhu G, Fan G, Feng C, Li Y, Chen Y, et al. (2013) The effect of lead exposure on brain iron homeostasis and the expression of DMT1/FP1 in the brain in developing and aged rats. Toxicol Lett 216: 108–123. [DOI] [PubMed] [Google Scholar]
- 3. Barton JC, Patton MA, Edwards CQ, Griffen LM, Kushner JP, et al. (1994) Blood lead concentrations in hereditary hemochromatosis. J Lab Clin Med 124: 193–198. [PubMed] [Google Scholar]
- 4. Wright RO, Silverman EK, Schwartz J, Tsaih SW, Senter J, et al. (2004) Association between hemochromatosis genotype and lead exposure among elderly men: The normative aging study. Environ Health Perspect 112: 746–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hopkins MR, Ettinger AS, Hernandez-Avila M, Schwartz J, Tellez-Rojo MM, et al. (2008) Variants in iron metabolism genes predict higher blood lead levels in young children. Environ Health Perspect 116: 1261–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Whitfield JB, Dy V, McQuilty R, Zhu G, Montgomery GW, et al. (2007) Evidence of genetic effects on blood lead concentration. Environ Health Perspect 115: 1224–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao A, Lu XT, Li QY, Tian L (2010) Effect of the delta-aminolevulinic acid dehydratase gene polymorphism on renal and neurobehavioral function in workers exposed to lead in China. Sci Total Environ 408: 4052–4055. [DOI] [PubMed] [Google Scholar]
- 8. Scinicariello F, Murray HE, Moffett DB, Abadin HG, Sexton MJ, et al. (2007) Lead and delta-aminolevulinic acid dehydratase polymorphism: Where does it lead? A meta-analysis. Environ Health Perspect 115: 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao Y, Wang L, Shen H-B, Wang Z-X, Wei Q-Y, et al. (2007) Association between δ-aminolevulinic acid dehydratase (ALAD) polymorphism and blood lead levels: A meta-regression analysis. J Toxicol Environ Health A, Part A 70: 1986–1994. [DOI] [PubMed] [Google Scholar]
- 10. Loparev V, Cartas M, Monken C, Velpandi A, Srinivasan A (1991) An efficient and simple method of DNA extraction from whole blood and cell lines to identify infectious agents. J Virol Methods 34: 105–112. [DOI] [PubMed] [Google Scholar]
- 11. Cardoso EMP, Stal P, Hagen K, Cabeda JM, Esin S, et al. (1998) HFE mutations in patients with hereditary haemochromatosis in Sweden. J Intern Med 243: 203–208. [DOI] [PubMed] [Google Scholar]
- 12. Dupic F, Fruchon S, Bensaid M, Loreal O, Brissot P, et al. (2002) Duodenal mRNA expression of iron related genes in response to iron loading and iron deficiency in four strains of mice. Gut 51: 648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leong W-I, Bowlus CL, Tallkvist J, Lönnerdal B (2003) Iron supplementation during infancy—effects on expression of iron transporters, iron absorption, and iron utilization in rat pups. Am J Clin Nutr 78: 1203–1211. [DOI] [PubMed] [Google Scholar]
- 14. Barton JC, Conrad M, Nuby S, Harrison L (1978) Effects of iron on the absorption and retention of lead. J Lab Clin Med 92: 536–5. [PubMed] [Google Scholar]
- 15. Bradman A, Eskenazi B, Sutton P, Athanasoulis M, Goldman LR (2001) Iron deficiency associated with higher blood lead in children living in contaminated environments. Environ Health Perspect 109: 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammad TA, Sexton M, Langenberg P (1996) Relationship between blood lead and dietary iron intake in preschool children - A cross sectional study. Ann Epidemiol. 30–33. [DOI] [PubMed]
- 17. Six KM, Goyer R (1972) The influence of iron deficiency on tissue content and toxicity of ingested lead in the rat. J Lab Clin Med 79: 128–136. [PubMed] [Google Scholar]
- 18. Marcus AH, Schwartz J (1987) Dose—Response curves for erythrocyte protoporphyrin vs blood lead: Effects of iron status. Environ Res 44: 221–227. [DOI] [PubMed] [Google Scholar]
- 19. Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, et al. (2001) A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 276: 7811–7819. [DOI] [PubMed] [Google Scholar]
- 20. Beutler E (1997) The significance of the 187G (H63D) mutation in hemochromatosis. Am J Hum Genet 61: 762–764. [PMC free article] [PubMed] [Google Scholar]
- 21. Bacon BR, Olynyk JK, Brunt EM, Britton RS, Wolff RK (1999) HFE genotype in patients with hemochromatosis and other liver diseases. Ann Intern Med 130: 953–962. [DOI] [PubMed] [Google Scholar]
- 22. Feder JN, Penny DM, Irrinki A, Lee VK, Lebron JA, et al. (1998) The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci USA 95: 1472–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Njajou OT, Houwing-Duistermaat JJ, Osborne RH, Vaessen N, Vergeer J, et al. (2003) A population-based study of the effect of the HFE C282Y and H63D mutations on iron metabolism. Eur J Hum Genet 11: 225–231. [DOI] [PubMed] [Google Scholar]
- 24. Geier A, Reugels M, Weiskirchen R, Wasmuth HE, Dietrich CG, et al. (2004) Common heterozygous hemochromatosis gene mutations are risk factors for inflammation and fibrosis in chronic hepatitis C. Liver Int 24: 285–294. [DOI] [PubMed] [Google Scholar]
- 25. Wright RO, Tsaih SW, Schwartz J, Wright RJ, Hu H (2003) Association between iron deficiency and blood lead level in a longitudinal analysis of children followed in an urban primary care clinic. J Pediatr 142: 9–14. [DOI] [PubMed] [Google Scholar]
- 26. Cook JD, Flowers CH, Skikne BS (2003) The quantitative assessment of body iron. Blood 101: 3359–3363. [DOI] [PubMed] [Google Scholar]
- 27. Punnonen K, Irjala K, Rajamäki A (1997) Serum transferrin receptor and its ratio to serum ferritin in the diagnosis of iron deficiency. Blood 89: 1052–1057. [PubMed] [Google Scholar]
- 28. Barnum-Huckins K, Adrian GS (2000) Iron regulation of transferrin synthesis in the human hepatoma cell line HepG2. Cell Biol Int 24: 71–77. [DOI] [PubMed] [Google Scholar]
- 29. Jahanshad N, Kohannim O, Hibar DP, Stein JL, McMahon KL, et al. (2012) Brain structure in healthy adults is related to serum transferrin and the H63D polymorphism in the HFE gene. Proc Natl Acad Sci USA 109: E851–E859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Giambattistelli F, Bucossi S, Salustri C, Panetta V, Mariani S, et al. (2012) Effects of hemochromatosis and transferrin gene mutations on iron dyshomeostasis, liver dysfunction and on the risk of Alzheimer's disease. Neurobiol Aging 33: 1633–1641. [DOI] [PubMed] [Google Scholar]
- 31. Adrian GS, Rivera EV, Adrian EK, Lu Y, Buchanan J, et al. (1993) Lead Suppresses Chimeric Human Transferrin Gene-Expression in Transgenic Mouse-Liver. Neurotoxicology 14: 273–282. [PubMed] [Google Scholar]
- 32. Weyermann M, Brenner H (1997) Alcohol consumption and smoking habits as determinants of blood lead levels in a national population sample from Germany. Arch Environ Health 52: 233–239. [DOI] [PubMed] [Google Scholar]
- 33. Sassi R, Hmida S, Kaabi H, Hajjej A, Abid A, et al. (2004) Prevalence of C282Y and H63D mutations in the haemochromatosis (HFE) gene in Tunisian population. Ann Genet 47: 325–330. [DOI] [PubMed] [Google Scholar]
- 34. Lin A, Yan WH, Xu HH, Zhu M, Zhou MY (2007) Analysis of the HFE gene (C282Y, H63D and S65C) mutations in a general Chinese Han population. Tissue Antigens 70: 252–255. [DOI] [PubMed] [Google Scholar]
- 35. Burke W, Imperatore G, McDonnell SM, Baron RC, Khoury MJ (2000) Contribution of different HFE genotypes to iron overload disease: a pooled analysis. Genet Med 2: 271–277. [DOI] [PubMed] [Google Scholar]
- 36. Pietrangelo A, Montosi G, Totaro A, Garuti C, Conte D, et al. (1999) Hereditary hemochromatosis in adults without pathogenic mutations in the hemochromatosis gene. N Engl J Med 341: 725–732. [DOI] [PubMed] [Google Scholar]
- 37. Gochee PA, Powell LW, Cullen DJ, Du Sart D, Rossi E, et al. (2002) A population-based study of the biochemical and clinical expression of the H63D hemochromatosis mutation. Gastroenterology 122: 646–651. [DOI] [PubMed] [Google Scholar]