Abstract
An extensive sequence comparison of the chloroplast ndhF gene from all major clades of the largest flowering plant family (Asteraceae) shows that this gene provides approximately 3 times more phylogenetic information than rbcL. This is because it is substantially longer and evolves twice as fast. The 5' region (1380 bp) of ndhF is very different from the 3' region (855 bp) and is similar to rbcL in both the rate and the pattern of sequence change. The 3' region is more A+T-rich, has higher levels of nonsynonymous base substitution, and shows greater transversion bias at all codon positions. These differences probably reflect different functional constraints on the 5' and 3' regions of ndhF. The two patterns of base substitutions of ndhF are particularly advantageous for phylogenetic reconstruction because the conserved and variable segments can be used for older and recent groups, respectively. Phylogenetic analyses of 94 ndhF sequences provided much better resolution of relationships than previous molecular and morphological phylogenies of the Asteraceae. The ndhF tree identified five major clades: (i) the Calyceraceae is the sister family of Asteraceae; (ii) the Barnadesioideae is monophyletic and is the sister group to the rest of the family; (iii) the Cichorioideae and its two basal tribes Mutisieae and Cardueae are paraphyletic; (iv) four tribes of Cichorioideae (Lactuceae, Arctoteae, Liabeae, and Vernonieae) form a monophyletic group, and these are the sister clade of the Asteroideae; and (v) the Asteroideae is monophyletic and includes three major clades.
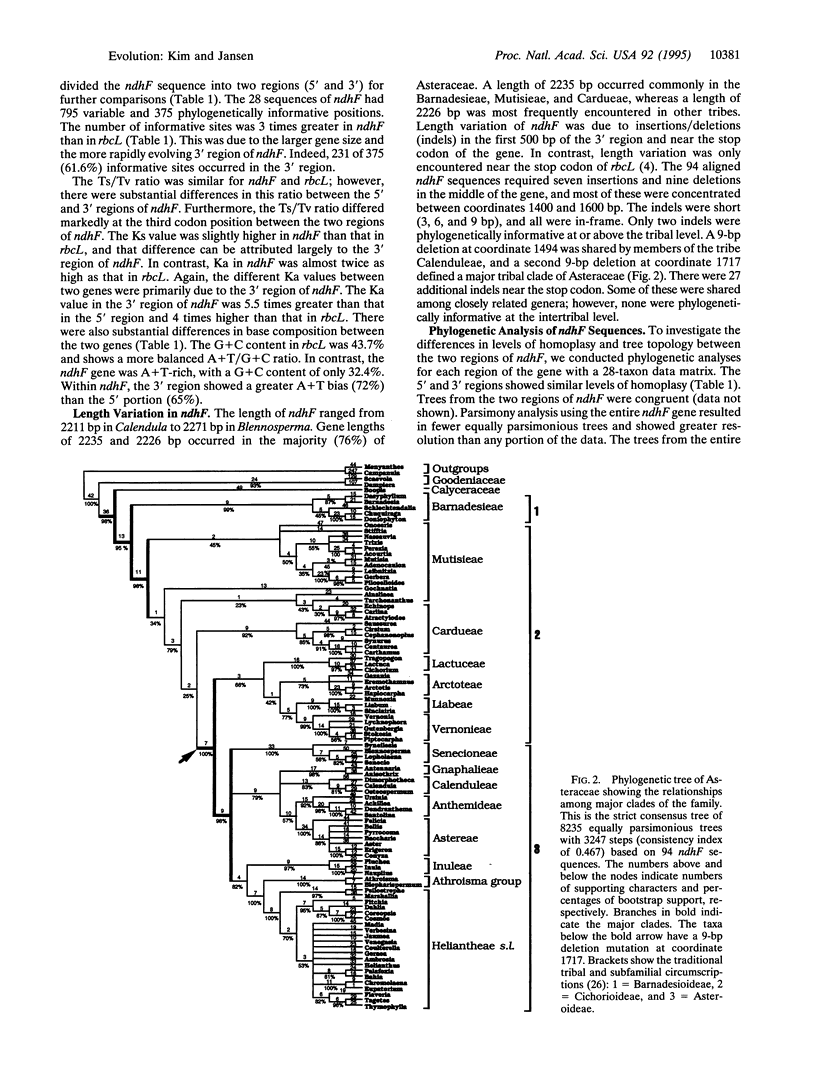
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clegg M. T. Chloroplast gene sequences and the study of plant evolution. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):363–367. doi: 10.1073/pnas.90.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka J., Shimada H., Whittier R., Ishibashi T., Sakamoto M., Mori M., Kondo C., Honji Y., Sun C. R., Meng B. Y. The complete sequence of the rice (Oryza sativa) chloroplast genome: intermolecular recombination between distinct tRNA genes accounts for a major plastid DNA inversion during the evolution of the cereals. Mol Gen Genet. 1989 Jun;217(2-3):185–194. doi: 10.1007/BF02464880. [DOI] [PubMed] [Google Scholar]
- Jansen R. K., Palmer J. D. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc Natl Acad Sci U S A. 1987 Aug;84(16):5818–5822. doi: 10.1073/pnas.84.16.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno A., Hirai A. A transcription map of the chloroplast genome from rice (Oryza sativa). Curr Genet. 1993 Feb;23(2):166–174. doi: 10.1007/BF00352017. [DOI] [PubMed] [Google Scholar]
- Levinson G., Gutman G. A. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987 May;4(3):203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Li W. H., Wu C. I., Luo C. C. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 1985 Mar;2(2):150–174. doi: 10.1093/oxfordjournals.molbev.a040343. [DOI] [PubMed] [Google Scholar]
- Matsubayashi T., Wakasugi T., Shinozaki K., Yamaguchi-Shinozaki K., Zaita N., Hidaka T., Meng B. Y., Ohto C., Tanaka M., Kato A. Six chloroplast genes (ndhA-F) homologous to human mitochondrial genes encoding components of the respiratory chain NADH dehydrogenase are actively expressed: determination of the splice sites in ndhA and ndhB pre-mRNAs. Mol Gen Genet. 1987 Dec;210(3):385–393. doi: 10.1007/BF00327187. [DOI] [PubMed] [Google Scholar]
- Morton B. R. Codon use and the rate of divergence of land plant chloroplast genes. Mol Biol Evol. 1994 Mar;11(2):231–238. doi: 10.1093/oxfordjournals.molbev.a040105. [DOI] [PubMed] [Google Scholar]
- Olmstead R. G., Sweere J. A., Wolfe K. H. Ninety extra nucleotide in ndhF gene of tobacco chloroplast DNA: a summary of revisions to the 1986 genome sequence. Plant Mol Biol. 1993 Sep;22(6):1191–1193. doi: 10.1007/BF00028992. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Ohme M., Tanaka M., Wakasugi T., Hayashida N., Matsubayashi T., Zaita N., Chunwongse J., Obokata J., Yamaguchi-Shinozaki K. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986 Sep;5(9):2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M. The chloroplast chromosomes in land plants. Annu Rev Cell Biol. 1989;5:51–70. doi: 10.1146/annurev.cb.05.110189.000411. [DOI] [PubMed] [Google Scholar]


