Abstract
The CBM signalosome plays a pivotal role in mediating antigen-receptor induced NF-κB signaling to regulate lymphocyte functions. The CBM complex forms filamentous structure and recruits downstream signaling components to activate NF-κB. MALT1, the protease component in the CBM complex, cleaves key proteins in the feedback loop of the NF-κB signaling pathway and enhances NF-κB activation. The aberrant activity of the CBM complex has been linked to aggressive lymphoma. Recent years have witnessed dramatic progresses in understanding the assembly mechanism of the CBM complex, and advances in the development of targeted therapy for aggressive lymphoma. Here, we will highlight these progresses and give an outlook on the potential translation of this knowledge from bench to bedside for aggressive lymphoma patients.
Keywords: CBM complex, Lymphoma, NF-κB, Targeted therapy
1. Introduction
NF-κB family of transcription factors plays a critical role in regulating lymphocytes activation, proliferation, survival and effector functions in innate and adaptive immune responses [1,2]. Defects in NF-κB signaling have been linked to immunodeficiency, and aberrant constitutive activation of NF-κB results in autoimmune diseases or neoplastic disorders [3]. Upon antigen stimulation, the T-cell receptor (TCR) engages MHC-bound antigen peptides and the B-cell receptor (BCR) interacts with antigens, which initiate downstream signaling cascades including activation of a series of kinases, adaptor proteins that culminate in NF-κB activation. Protein kinase C θ (PKCθ) of T-cells and protein kinase β (PKCβ) of B-cells are activated as a part of the antigen-receptor signaling cascade, which further activate their downstream signaling components [1].
The trimolecular protein complex composed of CARMA1 (CARD- and membrane-associated guanylate kinase-like domain-containing protein 1, also called CARD11), Bcl10 (B-cell lymphoma/leukemia 10) and MALT1 (mucosa-associated lymphoid tissue lymphoma translocation protein 1), referred to as the CBM complex, was identified to function downstream of PKCθ/PKCβ and plays a critical role in mediating NF-κB activation in B and T cells upon antigen-receptor stimulation (reviewed in [1,4]). As a direct target of PKCβ/PKCθ, the CBM complex is activated by PKCβ/PKCθ and recruited to the lipid raft immunological synapse, which subsequently recruits other downstream signaling components such as TRAF2/TRAF6, TAK1 (transforming growth factor-β-activated kinase 1) and TAB (TAK1 binding protein) to activate the inhibitor of nuclear factor-κB (NF-κB) kinase (IKK) complex [1]. In the canonical NF-κB signaling pathway, IKK complex in turn phosphorylates inhibitor of NF-κB (IκB), enables its proteosomal degradation and the release of NF-κB from its sequestration in the cytosol. NF-κB’s nuclear localization signal (NLS) is thus exposed, resulting in NF-κB translocation into the nucleus and activation of its target genes [4]. The assembly of the CBM complex also activates the proteolytic activity of paracaspse MALT1, which cleaves and inactivates the negative regulators of NF-κB and further enhances NF-κB activity [5–7].
Enormous studies have indicated that the genes encoding CARMA1, Bcl10 and MALT1 are bona fide oncogenes. Frequent missense mutations and translocations of these genes have been found in patients with non-Hodgkin’s lymphoma [8]. Given the important role that the CBM complex plays in the antigen-receptor signaling pathway as well as other pathways that lead to NF-κB activation, intensive studies have been initiated to delineate the activation mechanisms of CBM in NF-κB signaling. In this review, we will focus on the advances in the structural architecture and assembly mechanism of the CBM complex in the context of antigen-receptor induced NF-κB signaling and progresses in drug discovery research targeting CBM complex as a potential therapeutic target for aggressive lymphomas.
2. The structural and functional features of CARMA1, Bcl10 and MALT1
2.1 CARMA1
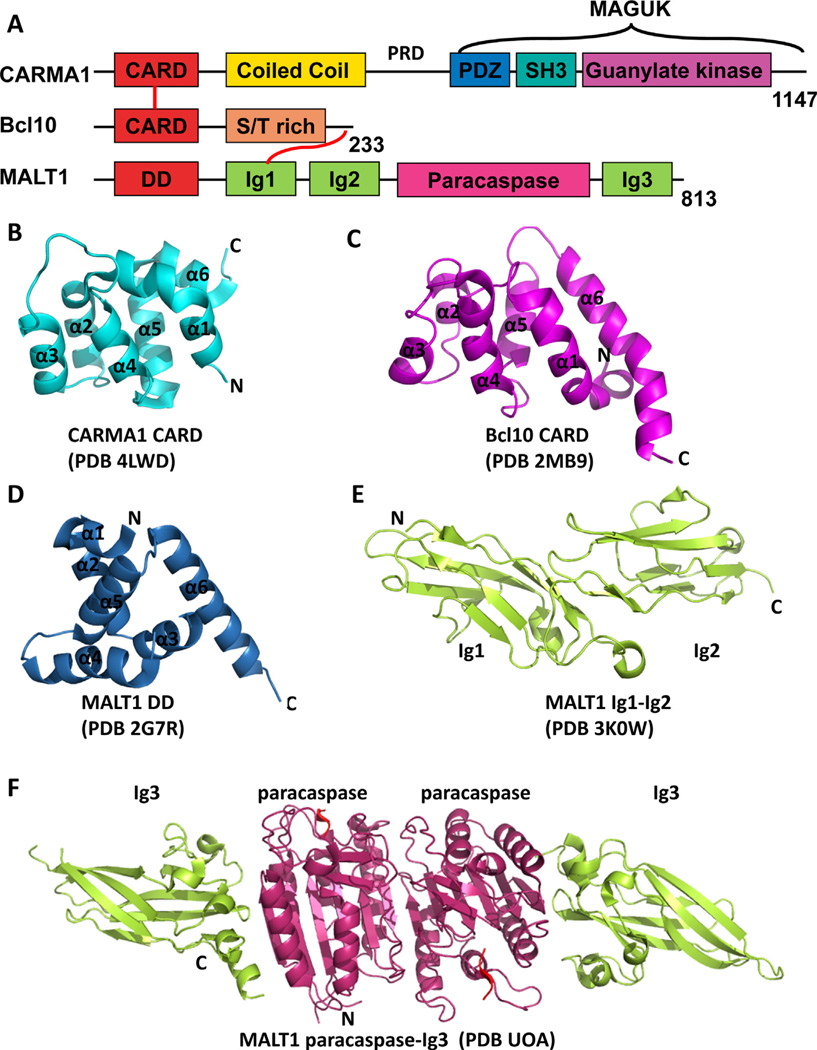
CARMA1 (also known as CARD11 and Bimp3) contains an N-terminal CARD (caspase recruitment domain) domain followed by a long coiled-coil (CC) domain, linker region and a C-terminal MAGUK (membrane-associated guanylate kinase) domain (Fig. 1A). CARD is a subfamily member of death domain (DD) superfamily which usually mediates protein–protein interactions via DD–DD or CARD–CARD interaction between two or more proteins [9,10]. The CARD domain is the most-studied domain of CARMA1. CARMA1 CARD presents as a monomer in solution. The structure of CARMA1 CARD has recently been solved by X-ray crystallography [11,12] (Fig. 1B). Like other CARD structures, CARMA1 CARD exhibits a conserved compact six-helical bundle fold. However, CARMA1 CARD has a kink in its first helix and an extra-extended loop between helix 3 and 4 [11,12]. This long loop is involved in the interaction with its binding partner Bcl10 CARD [12].
Fig. 1.
Structures of CARMA1, Bcl10 and MALT1. (A) Domain organization of CARMA1, Bcl10 and MALT1. CARD, caspase recruitment domain; PDZ, PSD95, DLG and ZO1 homology domain; SH3, Src homology 3 domain; MAGUK, membrane-associated guanylate kinase domain; S/T rich, Ser/Thr rich region; DD, death domain; Ig, immunoglobulin-like domain. The number of residues in each protein is labeled at the end. (B) Crystal structure of CARMA1 CARD domain (PDB 4LWD). (C) NMR structure of Bcl10 mutant E53D (PDB 2MB9). (D) Crystal structure of MALT1 death domain (PDB 2G7R). (E) Crystal structure of MALT1 tandem Ig1-Ig2 domains (PDB 3K0W). (F) Crystal structure of MALT1 paracaspase-Ig3 domains in complex with irreversible peptide inhibitor Z-VRPR-FMK shown in red (PDB UOA).
Coiled-coil (CC) domain usually functions as an oligomerization motif for a wide variety of proteins. CARMA1’s long CC domain has four predicted coiled-coil segments and is responsible for self-oligomerization. Our recent study showed that CARMA1 CARD alone exists as a monomer in solution, while in the presence of additional CC segments, CARMA1 presents from soluble trimer to insoluble large aggregations suggesting high-order oligomerization and aggregation [12].
The linker region between CC domain and MAGUK domain is a flexible region that has been identified as the target for PKCu/PKCβ. Signal-dependent phosphorylation of serine residues in this region (e.g. Ser552 and Ser645) triggers the conformational change of CARMA1 that activates it; therefore this linker region has also been termed as PKC-regulated domain (PRD) [4]. In resting cells, CARMA1 adopts an auto-inhibitory conformation in which PRD interacts with CARD and blocks CARD-dependent protein–protein interactions; signal induced phosphorylation of CARMA1 in the PRD results in conformational changes that enable the accessibility of CARD for the interaction with its downstream binding partner Bcl10 and MALT1 [3,13].
The MAGUK domain of CARMA1 shares the typical features of MAGUK family proteins as scaffolding proteins. It contains three subdomains named PDZ (named after the domain-containing PSD-95, Dlg and ZO-1 proteins), SH3 (src homology 3) and GUK (guanylate kinase) (Fig. 1A). The function of MAGUK domain is not currently clear; however, it is suggested that MAGUK domain is responsible for tethering CARMA1 to the cytosolic side of its membrane-bound binding partners [3,13]. The MAGUK domain also plays an essential role in polyubiquitination and proteasomedependent degradation of CARMA1 [14]. Elimination of the MUGUK ubiquitination sites in CARMA1 leads to an increased NF-κB and JNK activity as a result of a defect in K48 ubiquitination-dependent CARMA1 degradation. Therefore, ubiquitination of the MAGUK domain may serve as an important negative feed-back regulation mechanism to balance CARMA1-dependent activation of NF-κB signaling pathway [14].
2.2. Bcl10
Bcl10 is a relatively small protein with 233 amino acids. It also has an N-terminal CARD domain as CARMA1 and a likely unstructured C-terminal serine/threonine rich motif (Fig. 1A). Bcl10 CARD is responsible for the interaction with CARMA1 CARD. Unlike CARMA1 CARD, Bcl10 CARD does not behave well in solution and has a tendency to aggregate. Our recent study showed that Bcl10 CARD forms filaments spontaneously in vitro and in cells and this process is greatly enhanced by the presence of CARMA1 [12]. This filamentous structure is very stable and confers resistance to limited protease digestion and tolerates high salt condition [12]. Many attempts to solve the crystal structure of Bcl10 CARD had failed because of its intrinsic property. Therefore, we determined the structure of a mutant form of Bcl10 (E53R) that exists as monomer in solution by NMR [12] (Fig. 1C). Unexpectedly, the structure of Bcl10 CARD is quite different from that of other known CARD family proteins. Although Bcl10 CARD also contains six α-helices, its α1 and α6 are unusually long resulting in an overall relatively relaxed pear shape. In addition, 1H-15N-HSQC experiments indicate that about one-third of Bcl10 CARD residues are in dynamic chemical exchange, suggesting that Bcl10 may be in a highly dynamic state in the monomeric form [12]. Interaction with other binding partners may be needed for Bcl10 to stabilize its structure.
The Bcl10 C-terminal S/T rich region is responsible for the interaction with MALT1. In addition, it has been well established that this region is the posttranslational regulation region in response to antigen-receptor activation. Zeng et al. showed that phosphorylation of Bcl10 at Ser138 targets Bcl10 for ubiquitination and degradation. When Ser138 was mutated to alanine to prevent TCR-induced phosporylation on this residue and its associated subsequent ubiquitination and degradation of Bcl10, it led to a prolonged NF-κB activation and IL-2 production, suggesting that phosphorylation of Ser138 mostly plays a negative regulatory role in TCR-mediated NF-κB signaling [15].
2.3. MALT1
MALT1 consists of an N-terminal death domain (DD), two Ig domains followed by a caspase-like domain (also referred to as paracaspase domain) and a third Ig domain at the C-terminus (Fig. 1A). The N-terminal part (DD-Ig2) of MALT1 is responsible for interaction with Bcl10, while the C-terminal part (paracaspase-Ig3) possesses proteolytic activity. The structures of MALT1’s individual domains have been solved by X-ray crystallography (see Table 1) (Fig. 1D–F); however, the overall structure still remains to be determined.
Table 1.
Summary of CAMRMA1, Bcl10 and MALT1 structure information.
| Protein | PDB ID | Note |
|---|---|---|
| CARMA1 | ||
| CARD | 4LWD (Qiao, 2013) | Human |
| 4I16 (Li, 2012) | Mouse | |
| Bcl10 | ||
| CARD | 2MB9 (Qiao, 2013) | Human |
| MALT1 | ||
| DD | 2G7R | Human |
| Ig1–Ig2 | 3K0W (Qiu, 2011) | Human |
| Paracaspase-Ig3 | 3V55 (Wiesmann, 2012) |
Human, apo structure |
| 3V4O (paracaspase) (Wiesmann, 2012) |
Human, in complex | |
| with z-VRPR-FMK | ||
| 3V4L (Wiesmann, 2012) | Mouse, in complex with z-VRPR-FMK |
|
| 3UOA (Yu, 2011) | Human, in complex with z-VRPR-FMK |
|
| 4I1R (Schlauderer, 2013) | Human, in complex with thioridazine |
MALT1 is a distant relative of caspases and it was identified through weak sequence homology to caspases [16]. Like initiator caspases, MALT1 paracaspase presents as inactive monomer in solution and requires dimerization for its proteolytic activity [17]. Substrate binding promotes dimerization and activation of MALT1. The structure of the paracaspase-Ig3 domain alone or in complex with its inhibitor has been solved [18,19]. As expected, the overall structure of MALT1 paracaspase domain is very similar to typical caspase structures. Each monomer of the paracaspase-Ig3 domain contains a six-stranded β-sheet which is sandwiched by three α-helices at one side and two α-helices at the other side. The b6 strand mediates the anti-parallel homodimer formation by forming a contiguous twelve-stranded β-sheet extended from one MALT1 protomer to the other (Fig.1F). Upon substrate binding, paracaspase domain together with Ig3 domain undergoes substantial conformational changes [19]. Despite the similarity between MALT1 and caspases, MALT1 is distinguished from canonical caspases in three major characteristics that define its catalytic specificity: First, MALT1 is an arginine-specific protease which cleaves after arginine at the P1 site, unlike canonical caspases that cleave after aspartic acid [17]. Second, the intersubunit linker that contains loop L2 and L2′ remains intact, while in canonical caspases it is usually auto-processed upon maturation to facilitate the active site formation. Therefore, loops L2 and L2′ sit at the center of the homodimer interface and cannot assemble the loop-bundle interaction that is associated with the high catalytic activity seen in other caspases. Third, in addition to the β6 strand, α5 helix also contributes significantly to the dimerization interface in MALT1, which presumably adds dimerization specificity of MALT1 [18,19].
Besides its proteolytic function, MALT1 also has a major role as a scaffolding protein as a member of the CBM signalosome. After signal stimulation and the assembly of the CBM complex, MALT1 recruits TRAF6, the E3 ubiquitin ligase, into the CBM signalosome. Subsequently, TRAF6 polyubiquitinates itself as well as MALT1 and Bcl10, which provides a platform for the recruitment of IKK complex leading to NF-κB activation [20–22].
3. The high order assembly of the CBM signalosome
Upon antigen-receptor ligation and signal cascade transduction, CARMA1 CARD is released from the auto-inhibition conformation and becomes accessible for the association with its downstream partners Bcl10 and MALT1 to assemble into the CBM complex, leading to NF-κB activation in T and Bcells. CARMA1 interacts with Bcl10 through CARD–CARD interaction and the C-terminus of Bcl10 is responsible for the interaction with the N-terminal Ig domains of MALT1 (Fig. 1A).
Previously, several studies reported that Bcl10, diffusely distributed in the cytosol of rest cells, formed filament-like structures when it is overexpressed [23,24], or punctuate structures termed POLKADOTS when it is activated upon TCR signaling [25,26]. Sun et al. reported that a small fraction of reconstituted Bcl10 and MALT1 complex forms oligomers and only the oligomer form of the Bcl10-MALT1 complex can activate IKK complex via oligomerizing and activating TRAF6-the E3 ligase, suggesting that the high-molecular weight complex plays a crucial role in signal transduction [20]. Therefore, an oligomerization cascade-based activation model was proposed that culminates in NF-κB activation [4,20]. However, the exact molecular mechanism of how these supramolecular structures are assembled together remained unclear until recently we unraveled the structural architecture of the CBM complex [12].
Using a combination of Electron Microscopy (EM), X-ray crystallography and Nuclear Magnetic Resonance (NMR) techniques, we have unveiled the assembly mechanism and structural architecture of the CBM complex [12]. The CBM complex is a helical filamentous assembly where CARMA1 presents as a substoichiometric component in the complex and functions as the nucleator for the filament formation. Bcl10 CARD forms the core of the filament and MALT1 is brought to the periphery of the filament by interacting with the C-terminus of Bcl10 and thus becomes oligomerized and activated [12] (Fig. 2). The interaction between CARMA1 CARD and Bcl10 CARD is driven by charge–charge interaction, as suggested by Li et al. based on docking studies using Bcl10 homology models [11]. Bcl10 CARD forms filaments in a highly cooperative manner, and this process is greatly promoted by oligomerized CARMA1, supporting the role of CARMA1 as a nucleator. We have demonstrated that CARMA1 forms oligomers through its long CC domain and the CC domain is required for its function as a nucleator. CARMA1 CARD alone failed to promote Bcl10 filament formation. This filamentous assembly of the CBM complex exists both in vitro and in cells. Only the high-order filamentous CBM complex can activate MALT1 and NF-κB in vitro and in cells. Bcl10 mutants that failed to form filaments exert dominant negative effects on wild type Bcl10, and could not activate MALT1 or NF-κB in cells. All together, these data supported a paradigm of nucleation-induced higher-order protein complex machinery assembly that promotes signal transduction in response to cell signaling [12].
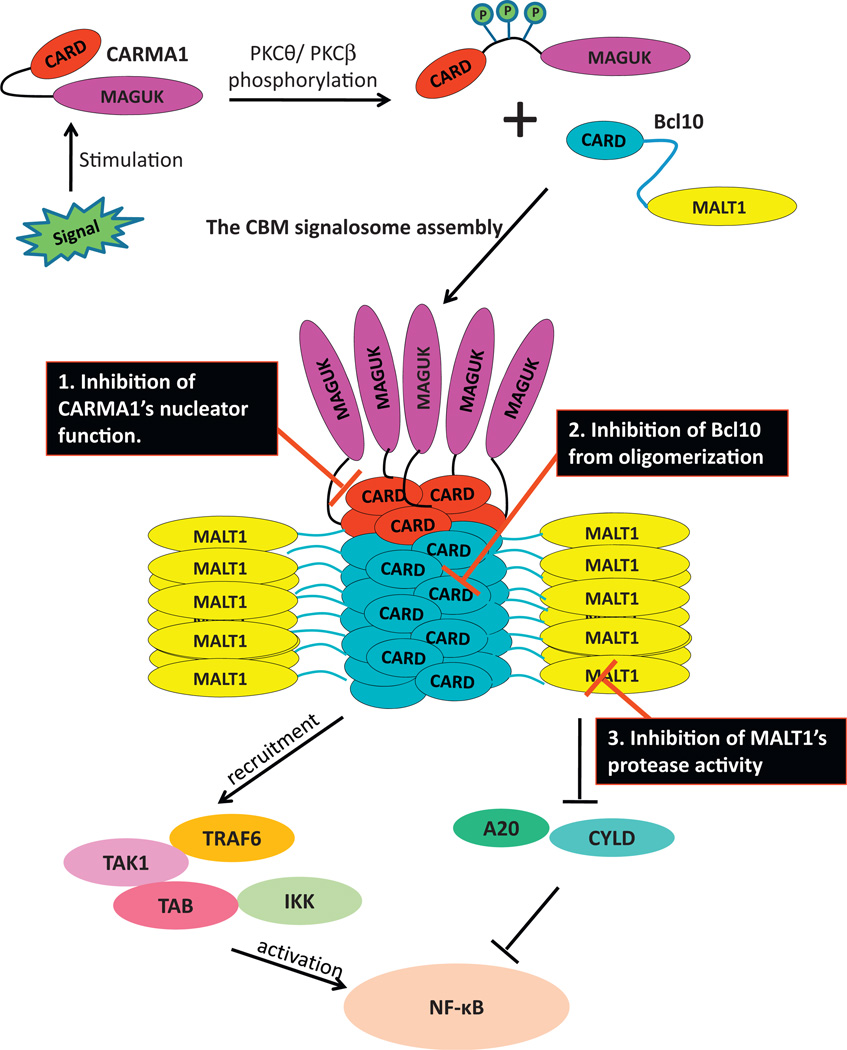
Fig. 2.
Model for the CBM signalosome assembly and potential inactivation approaches. Upon signaling stimulation, PKCθ/PKCβ phosphorylate CARMA1 at the PRD region and enable its transformation from an auto-inhibitory confirmation to an active confirmation. CARMA1then functions as nucleators and promotes Bcl10 filament formation. Bcl10 recruits MALT1 to the periphery of the filament and therefore oligomerizes and activates MALT1. The assembled CBM complex further recruits downstream signaling components such as TRAF6, TAK1, TAB and the IKK complex which ultimately activates NF-κB. MALT1 also inactivates NF-κB inhibitors like A20 and CYLD and enhances NF-kB activity. The black boxes suggest three approaches to inactivate the CBM complex and tune down NF-κB activity.
This high-order filamentous signaling machinery assembly for cellular signal transduction is not unique to the CBM complex mediated NF-κB activation pathway; it has also been observed in other pathways [27]. For example, PIDDosome composed of PIDD/RAIDD that functions in caspase-2 activation [28], and Myddosome made of MyD88/IRAK4/IRAK2 that mediates TLR/IL-1R signaling [29], are both helical filamentous assemblies. These high-order structure complex assemblies bring the enzymatic components into proximity for activation and signal amplification. This mechanism may represent a new paradigm of signal transduction in which signaling components are assembled into one high-order structure signaling machine that resembles mechanical machines where all parts are integrated into one to increase efficiency, and thus provide a temporal and spatial control of signal transduction in cells [27].
4. Functions of the CBM signalosome in mediating NF-κB activation
The CARMA1-Bcl10-MALT1 complex was initially found to function as part of the immune system by mediating the B-cell receptor (BCR) and T-cell receptor (TCR) induced NF-κB activation. CARMA1 is primarily expressed in hematopoietic tissues, while both Bcl10 and MALT1 are expressed in all tissues [13]. In addition to the CARMA1-Bcl10-MALT1 complex that functions in hematopoietic cells, other CARMA family members that have different tissue distribution profiles can also form complexes with Bcl10 and MALT1 to transduce NF-κB signaling downstream of multiple receptor types [13,30,31]. For example, CARMA2 (also known as CARD14 or Bimp2) [32], expressed in placental and mucosal tissues and CARMA3 (also called CARD10 or Bimp1), widely expressed in non-hematipoietic systems, can also form complexes with Bcl10 and MALT1. While the role of CARMA2-Bcl10-MALT1 complex is not specified yet, CARMA3-Bcl10-MALT1 was shown to play crucial roles in GPCR and RTK-induced NF-κB signaling and IL-8 production [13,31]. CARD9, which lacks the C-terminal MAGUK domain, was shown to complex with Bcl10 and MALT1 to mediate NF-κB signaling in innate immune response following bacteria, fungus and virus infection [13,31]. Therefore, the CBM complex has become a molecular hub for NF-κB activation by multiple signaling pathways [31].
As a filamentous supramolecular complex, the CBM complex functions as a major scaffold that serves as the central platform for the recruitment of effector proteins downstream of the CBM complex to mediate NF-κB signaling. Various studies have reported that CARMA1, Bcl10 and MALT1 can all interact with and recruit IKKγ (also named as NEMO), the regulatory component of the IKK complex. It has been reported that CARMA1 can directly associate with NEMO [33], and is essential for mediating NEMO ubiquitination upon TCR engagement [34]. Bcl10, as the core of the filamentous structure, can also be K63-linked polyubiquitinated at residue Lys31 and Lys63 that might be recognized by NEMO following simulation of different receptors [22,35]. In response to T-cell activation, MALT1 can recruit TRAF6, an E3 ubiquitin ligase, at its C-terminus [36] and promote TRAF6 oligomerization and activation [20], which in turn facilitate K63-linked polyubiquitylation of multiple proteins including the NEMO, TRAF6 itself, Bcl10, and MALT1. Polyubiquitination of these proteins provides a docking surface for the recruitment of other downstream signaling components such as the transforming growth factor b-activated kinase 1 (TAK1), TAK1 binding protein (TAB) and IKK complex that leads toIKK/NF-κB activation [21]. The detailed mechanism of how CARMA1, Bcl10 or MALT1 recruit the IKK complex and the contribution of each individual protein in the CBM complex needs to be further elucidated. It could be possible that they have a redundant function in recruiting the IKK complex, or they may recruit the IKK complex as a complex platform.
MALT1, the only enzymatic component in the CBM complex that possesses proteolytic activity, enchants the CBM complex with another mechanism in activating NF-κB signaling by regulating the negative feedback pathways. Currently, five substrates of MALT1 have been identified: A20, CYLD, Bcl10, RelB and NIK (reviewed in [37]). A20 and RelB are negative regulators of NF-κB [6,7]. Cleavage of A20 and RelB by MALT1 promotes their proteasomal degradation of them and enhances NF-κB signaling [6,7]. Cleavage of CYLD, a negative regulator of JNK, by MALT1 promotes TCR-induced JNK activation [38]. NIK (NF-κB inducing kinase) is an essential mediator of IKKα dependent noncanonical NF-κB signaling stimulated by a subset of TNF receptor family members. Cleavage of NIK by MALT1 generates a C-terminal kinase domain of NIK that is resistant to proteasomal degradation which leads to constitutive noncanonical NF-κB activation [37,39]. As the first identified MALT1 substrate, Bcl10 cleavage by MALT1 does not activate NF-κB; instead, it is required for TCR-induced integrin-mediated T-cell adhesion to fibronectin [5].
5. Regulation of the CBM signalosome activity
CARMA1, Bcl10 and MALT1 are subject to posttranscriptional regulation to limit the antigen-receptor induced NF-κB signaling and restore homeostasis. As an example, Moreno-Garcia et al. found that phosphorylations of Ser564, Ser657 and Ser649 of CARMA1 have distinct peak profiles as well as different outcomes. Phospho-Ser657 peaked rapidly and declined rapidly after antigen-receptor stimulation, leading to CARMA1-mediated NF-κB activation; however, phosphorylation of Ser649 occurred later but sustained longer most likely resulting in down-regulation of CARMA1 activity [40]. Similarly, several studies also reported that TCR-induced NF-κB signaling initially activates Bcl10 but later promotes Bcl10 to ubiquitination-dependent degradation, thereby negatively regulating NF-κB activity [15,41–43]. Pelzer et al. reported that the protease activity of MALT1 is regulated by monoubiquitination [44]. Monoubiquitination on Lys644 activated the cleavage activity of MALT1, while an ubiquitination-deficient mutant of MALT1 significantly reduced the proteolytic activity of MALT1, which led to impaired NF-κB activity and IL-2 induction.
In addition, both Bcl10 and MALT1 could be the targets for nondegradative K63-linked polyubiquitination that positively activates NF-κB [21,22]. Upon T-cell activation, Bcl10 undergoes K63-linked polyubiquitination at K31 and K63, which subsequently contributes to the interaction with NEMO [22]. TRAF6 associates with MALT1 and mediates K63-linked auto-ubiquitination and polyubiquitination of MALT1 at the C-terminus. Polyubiquitin chain provides a docking site for the recruitment of IKK complex components such as NEMO and the TAK1-TAB2 complex [21].
6. CBM signalosome and lymphomas
The CBM signalosome serves as the supramolecular hub where it integrates different receptor-induced signaling pathways that lead to NF-κB activation. Its aberrant activation has been associated with many NF-κB signaling dependent lymphocytic neoplasms. For example, CARMA1 overexpression has been found in different lymphocyte malignancies including diffuse large B-cell lymphoma (DLBCL), primary gastric B-cell lymphoma as well as in adult T-cell leukemia [2]. Gain of function mutations of CARMA1 at the coiled-coil region that result in constitutive NF-κB activation have also been found in DLBCL, the most common type of non-Hodgkin’s lymphoma [45]. Chromosomal translocations of Bcl10 and MALT1 have been frequently found in MALT lymphoma [46–49]. These translocations, t(14;18) of MALT1 and t(1;14)(p22:q32) of Bcl10, position MALT1 and BCL10 gene next to the Ig heavy chain loci, leading to MALT1 and Bcl10 overexpression and NF-κB activation [49]. Chromosomal translocation t(11;18)(q21:q21) is another common translocation found in MALT lymphoma [50–52]. This translocation creates a fusion protein that contains three BIR domains of cellular apoptosis inhibitor-2 (cIAP2) at the N-terminal and paracaspase domain of MALT1 at the C-terminal, which constitutively activates NF-κB [48]. Collectively, these studies established the critical roles of the CBM complex in neoplastic disorders of the lymphocytes and suggest that the CBM complex would be an attractive therapeutic target for lymphocytic disorders.
7. Targeting CBM signalosome for lymphoma
DLBCL is a heterogeneous group of diseases. According to its gene expression profiles, it can be subclassified into three major subtypes: germinal center B cell-like (GCB), activated B cell-like (ABC) and primary mediastinal B cell lymphoma (PMBL) DLBCL. Patients with GCB or PMBL subtype respond relatively well to standard immunochemotherapy and have a better survival rate than patients with ABC subtype. Therefore, ABC-DLBCL subtype is the most clinically challenging subtype and relies on constitutive activation of NF-κB signaling [8]. Using RNA interference screening, Ngo et al. identified CARMA1, Bcl10, MALT1 and IKKα as essential components for the survival of ABC-DLBCL [53], suggesting that inhibition of the CBM complex activity would be an appealing therapeutic method for ABC-DLBCL. According to the structure and function of the CBM complex, there are two major ways to target the complex: interfering with the assembly of the complex to inhibit the scaffold function or directly inhibiting the proteolytic activity of MALT1 to enhance the negative regulation of NF-κB activity.
7.1. Targeting the scaffold aspect of CBM complex
The molecular assembly mechanism and the structural information of the CBM complex are now unraveled [12], which paves the way for the development of novel targeted therapy for malignancy diseases that rely on CBM-mediated NF-κB signaling. As previously mentioned, the CBM complex is a high-order filamentous assembly in which CARMA1 serves as the nucleator and Bcl10 is the core of the filament. MALT1 is brought to the periphery of the filament via interaction with Bcl10 and as a result becomes oligomerized and activated [12]. The intactness of this high-order structure is essential for the function of the CBM complex in mediating NF-κB activation. Therefore, inhibition of the high-order structure assembly may provide a novel avenue for CBM-dependent constitutive activation of NF-κB in lymphomas. Theoretically, we can target the CBM complex as a scaffold platform in three ways (Fig. 2): First, we could develop inhibitors that block the interaction between CARMA1 and Bcl10 to inhibit CARMA1’s function as a nucleator, thus prevent CARMA1 from promoting Bcl10 filamentous structure formation. Alternately, inspired by the auto-inhibitory conformation of CARMA1, antagonist that mimics the structure of PRD motif (also termed as peptidomimetics) that competitively interacts with CARMA1 and prevents it from interacting with Bcl10 could also be a potential therapeutic method that inhibits the nucleator function of CARMA1. Successful examples of the peptidomimetics are the SMAC-mimicking IAP antagonists that are currently in preclinical and clinical trials [54]. Second, we can directly block the Bcl10 filamentous formation by developing inhibitors that disrupt Bcl10–Bcl10 interaction. Third, the interaction between Bcl10 and MALT1 is important for the oligomerization and activation of MALT1 as well as the recruitment and activation of TRAF6. Disruption of this interaction could have dual functions: inhibition of both the enzymatic activity and the scaffold function of MALT1.
However, targeting protein–protein interactions may be very challenging largely because of technological hurdles. For example, protein–protein interaction interface may be very large and diffuse, there is no well-defined binding pockets for small molecule inhibitors to target; therefore, it may be difficult to target one interface and achieve complete interruption effects. Nevertheless, there has been significant progress in this endeavor in recent years [55–57].
7.2. Targeting MALT1 – the enzymatic component of the CBM complex
MALT1, as the only enzyme in the CBM complex, is a unique paracaspase in humans. Although it has similar caspase fold as other caspases, it cleaves after arginine residue and distinguishes itself from conventional caspases [17]. MALT1 cleavage activity is linked to the pathogenesis of ABC-DLBCL, the most chemotherapy-resistant and clinically challenging form of DLBCL [58,59]. Targeting MALT1 proteolytic activity for ABC-DLBCL has become a hot research topic recently [60,61]. Notably, MALT1 knockout mice are generally healthy, except that they are defective in T-cell and B-cell activation in response to antigen stimulation [62]. Therefore, targeting MALT1 may be a safe and effective approach without causing significant side effects for patients.
Currently, several MALT1 inhibitors have been developed. z-VRPR-FMK is a peptide inhibitor that specifically and irreversibly inhibits the proteolytic activity of MALT1 (Fig. 1F) [58,59]. It has been shown that inhibition of MALT1 proteolytic activity by z-VRPR-FMK significantly reduced NF-κB activity and selectively inhibited the growth and survival of ABC-DLBCL, thus demonstrating the key role of the proteolytic activity of MALT1 in ABC-DLBCL and supporting the potential of MALT1 as a promising therapeutic target for aggressive lymphoma [58,59]. However, despite the efficacy that z-VRPR-FMK has demonstrated toward ABC-DLBCL, it may not be suitable to be used as a clinical drug due to its low cell permeability and low potency.
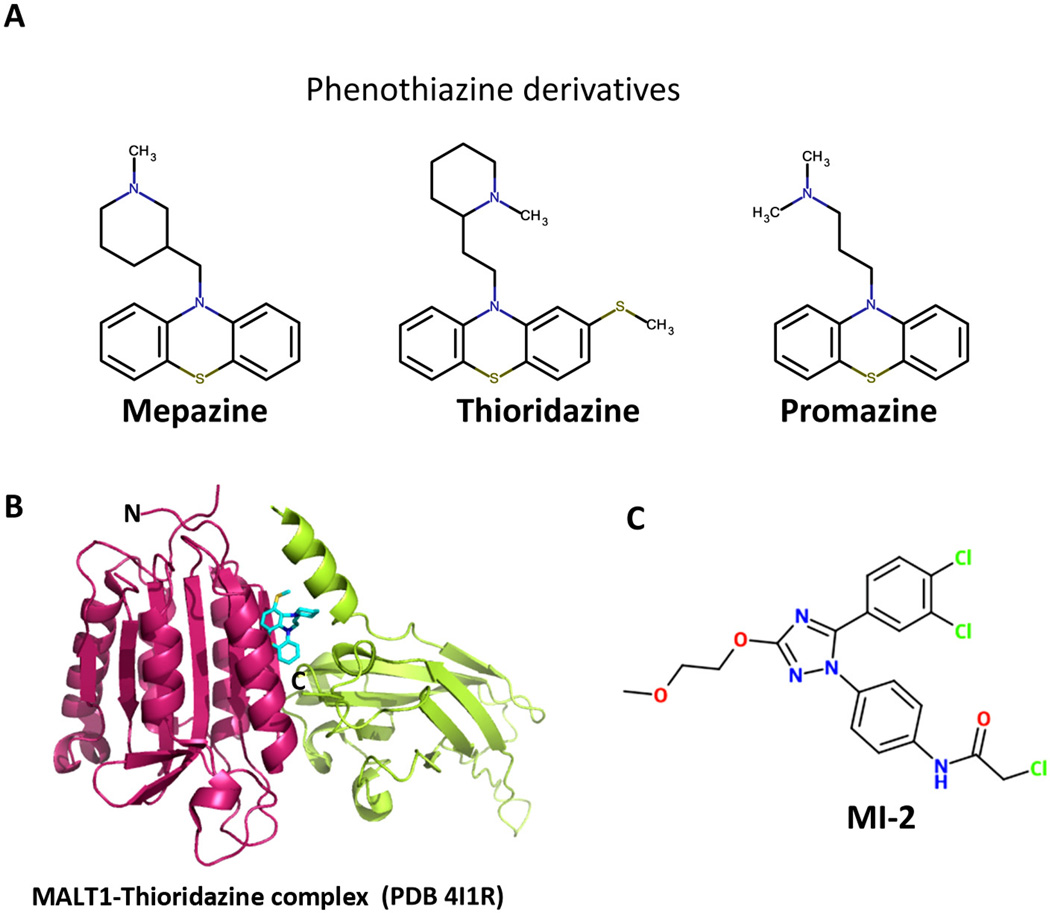
In order to search for more druggable inhibitors of MALT1, recently two groups have applied high-throughput screening using engineered dimeric form of MALT1 and have identified several small molecule compounds that could be potentially used as therapeutic agents for the treatment of ABC-DLBCL (Fig. 3) [60,61]. Using a library of approved drugs, Nagel et al. identified three phenothiazine derivatives named mepazine, thioridazine, and promazine as allosteric inhibitors of MALT1 (Fig. 3A) [60]. These compounds are noncompetitive and reversible MALT1 inhibitors. They bind MALT1 allosterically at the interface between paracaspase domain and the Ig3 domain far away from the catalytic center and function by interfering with the conformational change of MALT1 into an active enzyme (Fig. 3B) [63]. These phenothiazine derivatives showed selective inhibitory activity in MALT1-dependent ABC-DLBCL cells and xenograft mice models [60]. While these phenothiazine derivatives may offer favorable moves in clinical trials for the treatment of ABC-DLBCL since they are already in clinical use as anti-psychotic drugs by acting as a dopamine receptor antagonist, they may be limited by the known side effects such as tardive dyskinesia as well as off-target effects.
Fig. 3.
The structure of current inhibitors of MALT1. (A) Phenothiazine derivatives (Mepazine, Thioridazine and Promazine) that inhibit MALT1 activity. (B) Crystal structure of MALT1 in complex with Thioridazine shown in cyan stick. Thioridazine binds at the interface between paracaspase and Ig3 domains and allosterically inhibits the activity of MALT1. (C) MI-2 is a novel compound that specifically inhibits MALT1 protease activity.
Another group, Fontan et al. identified a novel compound named ‘‘MI-2’’ that binds MALT1 at the active site and irreversibly inhibits its enzymatic activity (Fig. 3C) [61]. MI-2 selectively inhibited the growth of ABC-DLBCL cells in vitro as well as in xenotransplanted mouse models by induction of apoptosis. It is important to point out that it did not cause any detectable physiological or histological signs of toxicity to MI-2 treated mice. Furthermore, MI-2, as an irreversible inhibitor, may offer pharmacokinetic advantage by providing prolonged inhibition effect since the activity of MALT1 will return only when new MALT1 is synthesized. Therefore, MI-2 may represent a novel class of therapeutic agents for the treatment of ABC-DLBCL. Notably, currently MI-2 is a lead compound and will need further optimization to improve its pharmacological properties before it can be used in clinic settings.
8. Conclusions and perspectives
The CBM complex is a key player in NF-κB signaling pathways and it activates NF-κB with two different mechanisms: it provides a central scaffold platform where downstream signaling proteins are recruited to the scaffold and become activated; its enzymatic component MALT1 inactivates the negative feedback loop of NF-κB signaling which further enhances NF-κB activity (Fig. 2). The CBM complex is a nucleation-induced helical filamentous assembly, where CARMA1 acts as the nucleator to promote Bcl10 filament formation and MALT1 polymerization and activation. It is well accepted that TRAF6 is recruited to the CBM complex and connects the CBM complex to the IKK complex to activate NF-κB in response to antigen-receptor signaling; however, the individual roles of CARMA1, Bcl10 and MALT1 in recruiting TRAF6 are not entirely clear yet. Further studies are needed to delineate in detail the entire signaling pathway, which is important for the development of targeted therapies and rational combinational therapies to treat lymphoma patients with aberrant NF-κB activity.
The intactness of the CBM supramolecular complex assembly is critical in transducing signals to NF-κB. Mutations in the CBM complex that failed to assemble filamentous structure dominantnegatively reduce NF-κB activity, indicating that interfering with high-order molecular structure assembly could be a promising therapeutic approach for aggressive lymphoma that rely on NF-κB signaling through the CBM complex. In addition, agents that inhibit the proteolytic activity of MALT1 could also be appealing for the treatment of aggressive lymphoma. Recently, two types of small molecule inhibitors of MALT1 have been identified to selectively inhibit the growth of ABC-DLBCL in vitro and in xenograft mouse models as well as in primary patient tumors [60,61], which validated the hypothesis that the CBM complex would be a promising therapeutic target for aggressive lymphomas that depend on signaling transduction through the CBM complex. Since single agent therapy for cancer is not curative and usually generates resistance fast, combinational therapy has become the trend for cancer these days. Therefore, rational combination of agents targeting the CBM complex with other targeted therapies may be beneficial for aggressive lymphoma patients.
Acknowledgements
We acknowledge the support from Cancer Research Institute to Q.Q. and L.D. and National Institutes of Health (R01AI089882) to H.W.
Biographies

Chenghua Yang completed her Ph.D. at Colorado State University in 2009 investigating the structure and function of the MeCP2-nucleosome complex. Between 2009 and 2012, she worked as a postdoctoral associate at Weill Cornell Medical College investigating the assembly mechanism of the CBM complex in NF-κB signaling and developing targeted therapy against the CBM complex for aggressive lymphoma. Then she worked as a Research Scholar at the Memorial Sloan-Kettering Cancer Center investigating the action mechanism of heat shock protein inhibitors on cancer cells. Now she is transiting to a Professor/Investigator position at the Joint Center for Translational Research of Chronic Diseases in the Institute of Nutritional Science, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences and Changhai Hospital, The Second Military Medical University in Shanghai. Her research interest focuses on the molecular mechanisms of cancers and the development of targeted therapies against cancers.

Liron David is a Postdoctoral Research Fellow at Boston Children’s Hospital and Harvard Medical School, USA. She completed her Ph.D. at the Technion, Israel Institute of Technology, Israel in 2012 investigating the structure of the Phycobilisome complex, a gigantic light harvesting complex. Since 2012 she has studied the molecular mechanism of the CBM complex in NF-κB signaling.

Qi Qiao received his B.S. degree of Biotechnology from Peking University in 2006 and Ph.D. degree from Institute of Biophysics, Chinese Academy of Sciences, in 2011. During his Ph.D. training, he worked in the laboratory of Professor Rui-Ming Xu, mainly focused on structural and functional elucidations of histone methyltransferases. In 2012, Dr. Qiao joined Dr. Hao Wu’s lab in Weill Cornell Medical Center as a postdoctoral fellow, and later moved to Boston Children’s Hospital with the lab. In his postdoc training, he began his studies on the molecular mechanism of signal transduction pathways in both innate and adaptive immune responses, CBM signalosome and activation-induced cytidinedeaminase (AID)related studies respectively. In 2013, Dr. Qiao was awarded the Cancer Research Institute postdoctoral fellowship to support his mechanistic study of AID in immunity and cancer.

Ermelinda Damko is a Lab Supervisor in the laboratory of Professor Frederick Maxfield at Weill Cornell Medical College since 2012. She received her MS degree from Columbia University in 2008. Between 2008 and 2012 she worked as a Research Specialist at the Professor Hao Wu’s lab at Weill Cornell Medical College.

Hao Wu is Asa and Patricia Springer Professor of Pediatrics and of Biological Chemistry and Molecular Pharmacology at Harvard Medical School, and Senior Investigator in the Program in Cellular and Molecular Medicine at Boston Children’s Hospital. She completed her Ph.D. with Dr. Michael Rossmann at Purdue University in 1992 and worked as a Postdoctoral Fellow in the laboratory of Professor Wayne Hendrickson at Columbia University between 1992 and 1997. She became an Assistant Professor at Weill Cornell Medical College in 1997 and was promoted to Professor in 2003. In 2012, she moved to Harvard Medical School. Her lab focuses on elucidating the molecular mechanism of signal by immune receptors, especially innate immune receptors.
References
- 1.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 2.Blonska M, Lin X. CARMA1-mediated NF-κB and JNK activation in lymphocytes. Immunol Rev. 2009;228:199–211. doi: 10.1111/j.1600-065X.2008.00749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-κB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2:a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rawlings DJ, Sommer K, Moreno-Garcia ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006;6:799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 5.Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 6.Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-κB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 7.Hailfinger S, Nogai H, Pelzer C, Jaworski M, Cabalzar K, Charton JE, et al. Malt1-dependent RelB cleavage promotes canonical NF-κB activation in lymphocytes and lymphoma cell lines. Proc Natl Acad Sci U S A. 2011;108:14596–14601. doi: 10.1073/pnas.1105020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med. 2010;362:1417–1429. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber CH, Vincenz C. The death domain superfamily: a tale of two interfaces. Trends Biochem Sci. 2001;26:475–481. doi: 10.1016/s0968-0004(01)01905-3. [DOI] [PubMed] [Google Scholar]
- 10.Ferrao R, Li J, Bergamin E, Wu H. Structural insights into the assembly of large oligomeric signalosomes in the Toll-like receptor-interleukin-1 receptor superfamily. Sci Signal. 2012;5:re3. doi: 10.1126/scisignal.2003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li S, Yang X, Shao J, Shen Y. Structural insights into the assembly of CARMA1 and BCL10. PLoS ONE. 2012;7:e42775. doi: 10.1371/journal.pone.0042775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qiao Q, Yang C, Zheng C, Fontan L, David L, Yu X, et al. Structural architecture of the CARMA1/Bcl10/MALT1 signalosome: nucleation-induced filamentous assembly. Mol Cell. 2013;51:766–779. doi: 10.1016/j.molcel.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blonska M, Lin X. NF-κB signaling pathways regulated by CARMA family of scaffold proteins. Cell Res. 2011;21:55–70. doi: 10.1038/cr.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno-Garcia ME, Sommer K, Shinohara H, Bandaranayake AD, Kurosaki T, Rawlings DJ. MAGUK-controlled ubiquitination of CARMA1 modulates lymphocyte NF-κB activity. Mol Cell Biol. 2010;30:922–934. doi: 10.1128/MCB.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng H, Di L, Fu G, Chen Y, Gao X, Xu L, et al. Phosphorylation of Bcl10 negatively regulates T-cell receptor-mediated NF-κB activation. Mol Cell Biol. 2007;27:5235–5245. doi: 10.1128/MCB.01645-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uren AG, O’Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, et al. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 17.Hachmann J, Snipas SJ, van Raam BJ, Cancino EM, Houlihan EJ, Poreba M, et al. Mechanism and specificity of the human paracaspase MALT1. Biochem J. 2012;443:287–295. doi: 10.1042/BJ20120035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu JW, Jeffrey PD, Ha JY, Yang X, Shi Y. Crystal structure of the mucosa-associated lymphoid tissue lymphoma translocation 1 (MALT1) paracaspase region. Proc Natl Acad Sci U S A. 2011;108:21004–21009. doi: 10.1073/pnas.1111708108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wiesmann C, Leder L, Blank J, Bernardi A, Melkko S, Decock A, et al. Structural determinants of MALT1 protease activity. J Mol Biol. 2012;419:4–21. doi: 10.1016/j.jmb.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 21.Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, et al. Malt1 ubiquitination triggers NF-κB signaling upon T-cell activation. EMBO J. 2007;26:4634–4645. doi: 10.1038/sj.emboj.7601897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu CJ, Ashwell JD. NEMO recognition of ubiquitinated Bcl10 is required for T cell receptor-mediated NF-κB activation. Proc Natl Acad Sci U S A. 2008;105:3023–3028. doi: 10.1073/pnas.0712313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan M, Lee J, Schilbach S, Goddard A, Dixit V. mE10, a novel caspase recruitment domain-containing proapoptotic molecule. J Biol Chem. 1999;274:10287–10292. doi: 10.1074/jbc.274.15.10287. [DOI] [PubMed] [Google Scholar]
- 24.Guiet C, Vito P. Caspase recruitment domain (CARD)-dependent cytoplasmic filaments mediate bcl10-induced NF-κB activation. JCell Biol. 2000;148:1131–1140. doi: 10.1083/jcb.148.6.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaefer BC, Kappler JW, Kupfer A, Marrack P. Complex and dynamic redistribution of NF-κB signaling intermediates in response to T cell receptor stimulation. Proc Natl Acad Sci U S A. 2004;101:1004–1009. doi: 10.1073/pnas.0307858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossman JS, Stoicheva NG, Langel FD, Patterson GH, Lippincott-Schwartz J, Schaefer BC. POLKADOTS are foci of functional interactions in T-Cell receptor-mediated signaling to NF-κB. Mol Biol Cell. 2006;17:2166–2176. doi: 10.1091/mbc.E05-10-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu H. Higher-order assemblies in a new paradigm of signal transduction. Cell. 2013;153:287–292. doi: 10.1016/j.cell.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, et al. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDo-some core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wegener E, Krappmann D. CARD-Bcl10-Malt1 signalosomes: missing link to NF-κB. Sci STKE. 2007;2007:e21. doi: 10.1126/stke.3842007pe21. [DOI] [PubMed] [Google Scholar]
- 31.Jiang C, Lin X. Regulation of NF-κB by the CARD proteins. Immunol Rev. 2012;246:141–153. doi: 10.1111/j.1600-065X.2012.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertin J, Wang L, Guo Y, Jacobson MD, Poyet JL, Srinivasula SM, et al. CARD11 and CARD14 are novel caspase recruitment domain (CARD)/membrane-associated guanylate kinase (MAGUK) family members that interact with BCL10 and activate NF-kappa B. J Biol Chem. 2001;276:11877–11882. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 33.Stilo R, Liguoro D, Di Jeso B, Formisano S, Consiglio E, Leonardi A, et al. Physical and functional interaction of CARMA1 and CARMA3 with Ikappa kinase gamma-NFkappaB essential modulator. J Biol Chem. 2004;279:34323–34331. doi: 10.1074/jbc.M402244200. [DOI] [PubMed] [Google Scholar]
- 34.Shambharkar PB, Blonska M, Pappu BP, Li H, You Y, Sakurai H, et al. Phosphorylation and ubiquitination of the IkappaB kinase complex by two distinct signaling pathways. EMBO J. 2007;26:1794–1805. doi: 10.1038/sj.emboj.7601622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou H, Wertz I, O’Rourke K, Ultsch M, Seshagiri S, Eby M, et al. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 36.Noels H, van Loo G, Hagens S, Broeckx V, Beyaert R, Marynen P, et al. A novel TRAF6 binding site in MALT1 defines distinct mechanisms of NF-κB activation by API2middle dotMALT1 fusions. J Biol Chem. 2007;282:10180–10189. doi: 10.1074/jbc.M611038200. [DOI] [PubMed] [Google Scholar]
- 37.McAllister-Lucas LM, Baens M, Lucas PC. MALT1 protease: a new therapeutic target in B lymphoma and beyond. Clin Cancer Res. 2011;17:6623–6631. doi: 10.1158/1078-0432.CCR-11-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staal J, Driege Y, Bekaert T, Demeyer A, Muyllaert D, Van Damme P, et al. T-cell receptor-induced JNK activation requires proteolytic inactivation of CYLD by MALT1. EMBO J. 2011;30:1742–1752. doi: 10.1038/emboj.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosebeck S, Madden L, Jin X, Gu S, Apel IJ, Appert A, et al. Cleavage of NIK by the API2-MALT1 fusion oncoprotein leads to noncanonical NF-κB activation. Science. 2011;331:468–472. doi: 10.1126/science.1198946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moreno-Garcia ME, Sommer K, Haftmann C, Sontheimer C, Andrews SF, Rawlings DJ. Serine 649 phosphorylation within the protein kinase C-regulated domain down-regulates CARMA1 activity in lymphocytes. J Immunol. 2009;183:7362–7370. doi: 10.4049/jimmunol.0902438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-kappa B signaling. Mol Cell Biol. 2004;24:3860–3873. doi: 10.1128/MCB.24.9.3860-3873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lobry C, Lopez T, Israel A, Weil R. Negative feedback loop in T cell activation through IkappaB kinase-induced phosphorylation and degradation of Bcl10. Proc Natl Acad Sci U S A. 2007;104:908–913. doi: 10.1073/pnas.0606982104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-κB. Immunity. 2012;36:947–958. doi: 10.1016/j.immuni.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pelzer C, Cabalzar K, Wolf A, Gonzalez M, Lenz G, Thome M. The protease activity of the paracaspase MALT1 is controlled by monoubiquitination. Nat Immunol. 2013;14:337–345. doi: 10.1038/ni.2540. [DOI] [PubMed] [Google Scholar]
- 45.Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science. 2008;319:1676–1679. doi: 10.1126/science.1153629. [DOI] [PubMed] [Google Scholar]
- 46.Willis TG, Jadayel DM, Du H, Peng MQ, Perry AR, Abdul-Rauf M, et al. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell. 1999;96:35–45. doi: 10.1016/s0092-8674(00)80957-5. [DOI] [PubMed] [Google Scholar]
- 47.Streubel B, Lamprecht A, Dierlamm J, Cerroni L, Stolte M, Ott G, et al. T(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood. 2003;101:2335–2339. doi: 10.1182/blood-2002-09-2963. [DOI] [PubMed] [Google Scholar]
- 48.Zhou H, Du MQ. Dixit VM. Constitutive NF-κB activation by the t(11;18)(q21;q21) product in MALT lymphoma is linked to deregulated ubiquitin ligase activity. Cancer Cell. 2005;7:425–431. doi: 10.1016/j.ccr.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 49.Ye H, Gong L, Liu H, Hamoudi RA, Shirali S, Ho L, et al. MALT lymphoma with t(14;18)(q32;q21)/IGH-MALT1 is characterized by strong cytoplasmic MALT1 and BCL10 expression. J Pathol. 2005;205:293–301. doi: 10.1002/path.1715. [DOI] [PubMed] [Google Scholar]
- 50.Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, et al. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- 51.Akagi T, Motegi M, Tamura A, Suzuki R, Hosokawa Y, Suzuki H, et al. A novelgene, MALT1 at 18q21, is involved in t(11;18) (q21;q21) found in low-grade B-cell lymphoma of mucosa-associated lymphoid tissue. Oncogene. 1999;18:5785–5794. doi: 10.1038/sj.onc.1203018. [DOI] [PubMed] [Google Scholar]
- 52.Morgan JA, Yin Y, Borowsky AD, Kuo F, Nourmand N, Koontz JI, et al. Breakpoints of the t(11;18)(q21;q21) in mucosa-associated lymphoid tissue (MALT) lymphoma lie within or near the previously undescribed gene MALT1 in chromosome 18. Cancer Res. 1999;59:6205–6213. [PubMed] [Google Scholar]
- 53.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 54.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 55.Arkin MR, Wells JA. Small-molecule inhibitors of protein–pprotein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 56.Mullard A. Protein–protein interaction inhibitors get into the groove. Nat Rev Drug Discov. 2012;11:173–175. doi: 10.1038/nrd3680. [DOI] [PubMed] [Google Scholar]
- 57.Cesa LC, Patury S, Komiyama T, Ahmad A, Zuiderweg ER, Gestwicki JE. Inhibitors of difficult protein–protein interactions identified by high-throughput screening of multiprotein complexes. ACS Chem Biol. 2013;8:1988–1997. doi: 10.1021/cb400356m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferch U, Kloo B, Gewies A, Pfander V, Duwel M, Peschel C, et al. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagel D, Spranger S, Vincendeau M, Grau M, Raffegerst S, Kloo B, et al. Pharmacologic inhibition of MALT1 protease by phenothiazines as a therapeutic approach for the treatment of aggressive ABC-DLBCL. Cancer Cell. 2012;22:825–837. doi: 10.1016/j.ccr.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 61.Fontan L, Yang C, Kabaleeswaran V, Volpon L, Osborne MJ, Beltran E, et al. MALT1 small molecule inhibitors specifically suppress ABC-DLBCL in vitro and in vivo. Cancer Cell. 2012;22:812–824. doi: 10.1016/j.ccr.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-κB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- 63.Schlauderer F, Lammens K, Nagel D, Vincendeau M, Eitelhuber AC, Verhelst SH, et al. Structural analysis of phenothiazine derivatives as allosteric inhibitors of the MALT1 paracaspase. Angew Chem Int Ed Engl. 2013;52:10384–10387. doi: 10.1002/anie.201304290. [DOI] [PubMed] [Google Scholar]