Abstract
Background: Previously, we studied the genetic basis for variability in total thyroxine (TT4) as part of investigating induced Graves' hyperthyroidism in panels of genetically diverse recombinant inbred (RI) mice. Because Graves' disease occurs predominantly in women, we used female mice. A limitation of this approach is that thyrotropin (TSH) is undetectable by some assays in most female mice.
Method: Variation in levels of serum TSH, TT4, and free thyroxine (FT4) was measured in males from three related RI families (CXB, BXH, and AXBXA) followed by quantitative genetic analysis and mapping of these traits.
Results: In general, TSH levels were higher in males than females. FT4 levels were also higher in males than in females, but TT4 sex differences were absent or inconsistent. Chromosomal linkage was only observed for TSH in BXH males and for FT4 in AXBXA males. Different chromosomes were linked to TT4 in males of the three RI sets. The most striking finding came from genetic linkages in males versus our previous data for females. TT4 was linked to the same chromosomal loci in CXB males and females. In contrast, TT4, FT4, and TSH were linked to different “sex-specific chromosomes” in AXBXA and BXH families.
Conclusions: In three RI mouse families, TSH and FT4 were significantly higher in males than females. Linkage analysis revealed chromosomal overlap for TT4 in males and females for one RI set but striking sex differences for TT4, FT4, and TSH linkage in two RI sets. Our findings provide a cautionary note: genetic linkage analysis of thyroid hormones traits in mice should be studied separately in males and females.
Introduction
Thyroid function is evaluated by measuring serum levels of thyrotropin (TSH) and the thyroid hormones thyroxine (T4) and triiodothyronine (T3). Because T4 and T3 are bound to plasma proteins, assay data can include both total T4 (TT4) and T3 (TT3) and free T4 (FT4) and free T3. Within the disease-free population, differences in some of these parameters are evident among races/ethnicities (1,2). In humans, differences in thyroid hormone and TSH levels between males and females are more difficult to assess because of the influence of pregnancy and estrogen ingestion, and because of the greater prevalence of autoimmune thyroiditis in women than in men (1,3). In addition, TSH levels increase with age (1,4). Nevertheless, after adjusting for these confounding factors, small but statistically significant differences in thyroid function parameters have been described between the sexes. To illustrate the magnitude of these differences, in a group of 2169 individuals, serum levels of TSH were significantly higher in women than in men (1.78 vs. 1.66 mU/L respectively) and TT4 levels were also higher in women than in men (7.53 vs. 7.43 μg/dL respectively) (5). In a very large cohort of 153,127 individuals, median TSH levels in thyroid disease-free adults were slightly but significantly higher in males than in females: 1.72 versus 1.70 mU/L (6). Because this sex difference in thyroid function parameters is comparatively modest, sex effects do not appear to have been analyzed or reported in previous genetic studies (7–9). However, earlier this year, a meta-analysis highlighted several sex-specific genetic loci associated with serum levels of TSH and (FT4) (10).
Recombinant inbred (RI) strains of mice, for which extensive phenotype data and high-resolution genetic maps are openly available, complement genetic studies in humans and enable more tightly controlled experimental perturbations across cohorts of diverse lines. These RI families are generated from the progeny of two or more inbred strains followed by repeated sib matings for at least 20 generations to establish multiple, stable homozygous lines (11). Previously, we used three families of RI strains to investigate the genetic contribution to induced Graves' hyperthyroidism (12–14). These studies, which required determining baseline concentrations of TT4 in serum, confirmed the known variability in TT4 values among mice (15,16). For these studies, we used female mice because Graves' disease in humans, like many autoimmune diseases, occurs predominantly in this sex. A surprising observation was that there appeared to be a genetic basis for variability in the efficacy of thyroid stimulation by an agonist (TSH receptor autoantibodies) (12–14). We explored this phenomenon further by quantifying bovine TSH-induced T4 release in endogenous TSH-suppressed (T3 injection) females of three RI families (17). This investigation revealed a limitation of focusing on females: presuppression TSH levels were undetectable in most of the strains (17), as previously observed using the same radio immunoassay for female mice of other strains (16). In contrast, with the same assay, TSH levels were considerably higher and readily detectable in males (16).
Because of the major difference in TSH levels between male and female mice of other strains, as well as three RI families, we have now complemented previous studies in females by (i) measuring baseline TSH, FT4, and TT4 levels in sera from males of the same three families, and (ii) by performing matched linkage analysis. These families share C57/BL/6 as one parental strain, and BALB/c, C3He, or the A strains as the other parent. We observed the anticipated male–female differences in serum TSH levels, and found other male–female differences in thyroid hormones. Unexpectedly, for some RI families, we observed differences in patterns of linkage for TT4 in males and females.
Materials and Methods
Mouse strains
Males of the following strains (7–9 weeks old; two mice per strain) were obtained from Jackson Laboratory (Bar Harbor, ME): (i) BXH (derived from C3H/HeJ and C57/BL6J): BXH2TyJ, 4, 6 to 11, 14, 19 and BXH20 KccJ and BXH22KccJ (abbreviated BXH2,4 to BXH22); (ii) CXB (derived from BALB/cByJ and C57/BL6ByJ): CXB1/ByJ to CXB7/ByJ, CXB8/HiAJ to CXB13/HiAJ (abbreviated CXB1 to 13); (iii) AXB/BXA (derived from A/J and C57/BL6J): AXB/PgnJ 1, 2, 4, 5, 6, 8, 10, 12, 13, 15, 19a, 23, 24; BXA/PgnJ 1, 2, 4, 7, 8, 11, 12, 13, 14, 16, 24, 25, 26 (abbreviated A1 to A24 for AXB strains and B1 to B26 for BXA strains; as a group, they are referred to as “AXBXA”).
Female mice of the same strains and similar age, also purchased from the Jackson Laboratory, have been studied previously (12–14,17). Female and male mice were obtained between January 2005 and July 2012 (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/thy). Thyroid function was investigated in two cohorts of females: (i) sera from the first female cohort were tested for TT4 (12–14) and later tested for FT4 (18); (ii) the second cohort of female mice was investigated for TT4 and TSH (17). TT4 levels were significantly correlated in the two female studies (Supplementary Table S1). CXB11 females in the second cohort had higher baseline TT4 levels than mice of the same strain in the first female cohort. Nevertheless, these CXB11 mice had normal L-T3 suppression and responded to bovine TSH (17). TSH, not measured in the first female cohort, was elevated in the BXH9 strain compared with all other females, and these mice were resistant to T3 suppression (17). In other respects, female and male mice appeared to have normal thyroid function. Although the males used in the present study were not from the same cohorts as the previously studied females, in all studies, males and females were of comparable age (6–9 weeks), and sera were analyzed with the same assays and same internal standards. Because of the relatively large volumes of serum required for these assays (see below), it was not possible to retest sera from females in parallel with sera from male mice.
Mice at the Jackson Laboratory were fed LabDiet 5K52/5K67 or LabDiet 5K54 (containing 2.1 and 2.2 ppm iodine respectively). At Cedars–Sinai Medical Center, animals were maintained (1–3 weeks) on LabDiet Mouse Chow 5015 (1.01 ppm iodine). All mice were housed in the same facility under standardized conditions of temperature, light, and unlimited access to food and water. When mice were 7–9 weeks old, they were euthanized, and blood was drawn (mid-morning to mid-afternoon). These studies were approved by the Cedars–Sinai Institutional Animal Care and Use Committee.
TT4 and FT4 and TSH
All assays used for males were the same as those used previously for female mice, including the same standards. TT4 and FT4 in individual mice were measured by radioimmunoassay kits (“Coat-a-Count total T4” and “Coat-a-Count free T4”; Siemens Healthcare Diagnostics, Inc., Los Angeles, CA). The TT4 assay required 25 μL, and the FT4 assay required 50 μL undiluted serum. Standards for TT4 ranged from 1 to 24 μg/dL (sensitivity 0.25 μg/dL) and for FT4 from 0 to 14 ng/dL (sensitivity 0.01 ng/dL). TT4 and FT4 values were computed from kit standards and reported as μg/dL TT4 and ng/dL FT4. Two male mice per strain were studied; the average coefficient of variation within each RI set was less than 20%.
TSH levels were determined by radioimmunoassay (16) by Dr. Roy Weiss (Thyroid Unit, University of Chicago, Chicago, IL). For this purpose, pooled samples from two cases per strain were analyzed in duplicate aliquots of undiluted serum (50 μL). TSH in female mice was also studied in a serum pool from each strain assayed in duplicate (17). All male sera were tested in a single assay, and all female sera were tested in one assay. Sera with values <10 mU/L were assigned an arbitrary value of 5.0 for linkage analysis (see below).
Statistical analysis and linkage analysis
Within each RI set (CXB, BXH, or AXBXA), the statistical significance of differences between males and females for TT4, FT4, and TSH levels was evaluated using paired t-tests (when normally distributed) or by a signed-rank test. Comparisons of TT4, FT4, and TSH levels in males of the three RI sets were tested by analysis of variance (ANOVA; SigmaStat; Systat software, San Jose, CA).
Quantitative trait loci (QTL) for TT4, FT4, and TSH were mapped using genotype files for AXBXA, BXH, and CXB sets generated by Williams et al. (19,20) embedded in GeneNetwork (www.genenetwork.org). The probability of linkage between TT4 and FT4 and previously mapped genotypes was estimated at ∼1 cM intervals along the entire genome, except for the Y chromosome. To establish criteria for suggestive and significant linkage, a permutation test was performed (24,000 permutations) (21). This test compares the peak likelihood ratio statistics (LRS; LRS=LOD×4.6, where LOD is the logarithm of the odds) obtained for correctly ordered data set with peak LRS scores obtained for permutations of the same data set.
The phenotypes have been entered into GeneNetwork (GN) under the following trait accession identifiers: AXB/BXA GN 10275, 10276, 10277; CXB GN 10743, 10744, 10746; BXH GN 10170, 10171, and 10172. Data can be found by searching the CXB, BXH, or AXB/BXA databases for the name “McLachlan.”
Linkage values were combined, when appropriate, to provide data sets for two or three RI sets. For example, as previously described (13,18,22), we calculated the probability associated with a chi-square value equal to: −2 (ln pBXH+ln pCXB) with four degrees of freedom where ln pBXH and ln pCXB are the natural logarithms of the probabilities derived independently for the two families in the same chromosomal interval. When combining data for three families, the same approach involved using six degrees of freedom.
Results
TT4, FT4, and TSH in male versus female BXH, CXB, and AXBXA RI mice
TSH, TT4, and FT4 in the sera of untreated young adult male RI CXB, BXH, and AXBXA lines were compared with our previous data for these parameters in females. Data for male mice will be considered first because all animals were obtained over the same time period and all sera were tested in the same assays. TSH was significantly higher in BXH males than in males of either the CXB or BXH RI families (Fig. 1A; ANOVA, p<0.05). Moreover, FT4 was significantly higher in BXH than in CXB males (Fig. 1B; t-test, p=0.045). However, TT4 levels did not differ significantly in male BXH, CXB, or AXBXA strains (Fig. 1C). Of interest, FT4 and TSH were positively correlated in BXH male mice (Spearman's rank, ρs=0.67, p=0.015).
FIG. 1.
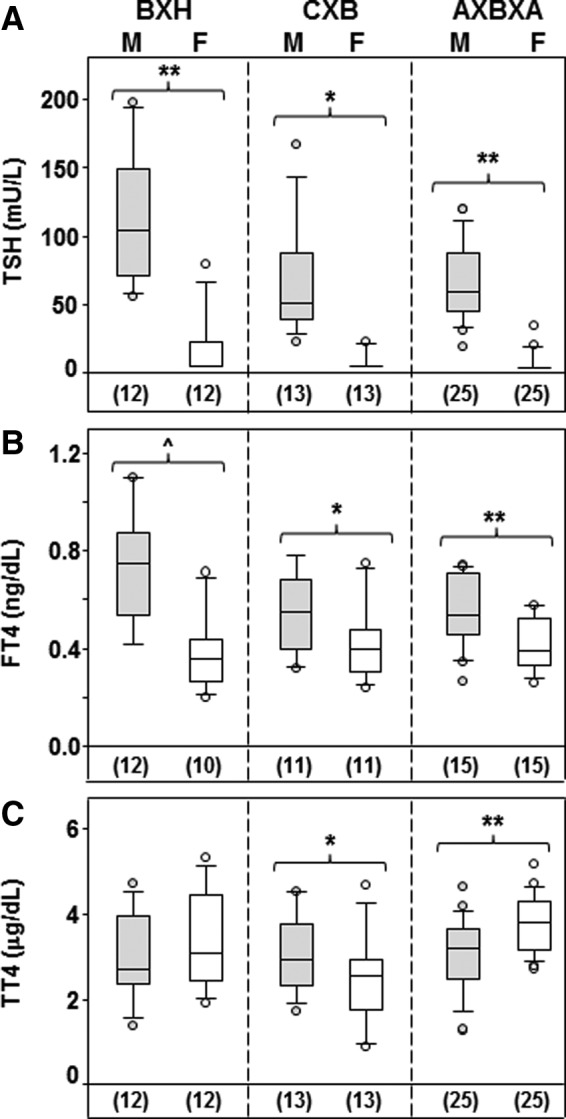
Thyrotropin (TSH), free thyroxine (FT4), and total thyroxine (TT4) in male versus female recombinant inbred mice (strains BXH, CXB, and AXBXA). Box plots indicate the median, 25th, and 75th percentiles and, as error bars, the 10th and 90th percentiles; “outliers” are shown as open circles. The numbers of strains investigated is given in parentheses. FT4 and TT4 data for female mice from references (12–14) and for TSH from (17). Values significantly different between males and females: TSH, **p<0.001 (paired t-test); *p<0.001 (signed-rank test); FT4, ^p=0.018 (t-test); *p=0.009 and **p<0.001 (paired t-tests); TT4: **p=0.003 (paired t-test); *p<0.001 (signed-rank test).
Turning to the comparison between male and female mice, marked sex differences were observed for TSH: males of all three RI families had significantly higher serum TSH than the low or undetectable levels in female mice of the corresponding RI sets (Fig. 1A). Although not tested in the same cohort of males and females, sera for TSH assays were obtained from the two sexes over a similar time period (2010 to 2011 for females, and the first half of 2012 for males).
Sex differences in thyroid hormone levels were less consistent than for TSH. FT4 and TT4 were both measured in the first female cohort, obtained up to seven years before males of the corresponding strains. With this proviso, FT4 levels were significantly higher in males than in females of all three RI families, particularly in the BXH set (Fig. 1B). TT4 levels were not significantly different in BXH males and females but were significantly lower in CXB females than males and significantly higher in AXBXA females than males (Fig. 1C).
Whole genome interval mapping for TSH, FT4, and TT4 in male RI mice
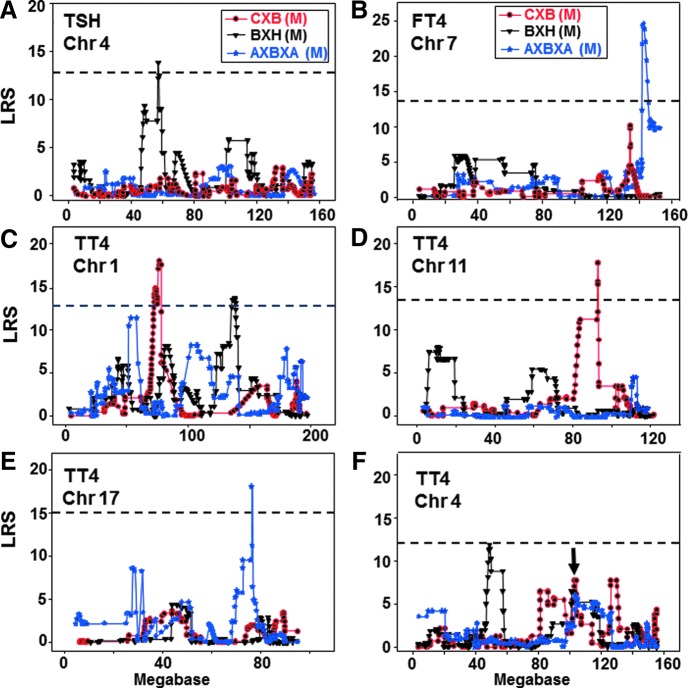
TSH was suggestively linked in BXH males to a chromosome (Chr) 4 locus (Fig. 2A). No linkage was observed for TSH in the CXB and AXBXA sets. However, combining the data from different RI families offers a more powerful statistical analysis and revealed linkage of TSH to Chr 14 in the CXB and AXBXA sets (Table 1). FT4 was significantly linked to a Chr 7 locus in AXBXA males (Fig. 2B, Table 1). Although no FT4 linkage was observed for the CXB or BXH sets, combined analysis for these sets provided evidence of linkage to Chr 1 (Table 1).
FIG. 2.
Interval mapping on chromosomes (Chr) 4 for TSH, Chr 7 for FT4, and Chr 1, 11, 17, and 4 for TT4 in male BXH, CXB, and AXBXA RI strains. LRS values are on the vertical axis; Mb on the horizontal axis. The broken horizontal lines indicate the mean LRS value above which there is suggestive linkage for the three RI sets. Number of strains studied: CXB, n=13; BXH, n=11; and AXBXA, n=26. GeneNetwork (GN) identification numbers: BXH GN 10172; CXB GN 10746; AXBXA GN 10277.
Table 1.
Chromosomal Linkage for Thyrotropin and Free Thyroxine in Male BXH, CXB, and AXBXA Mice
| Strain | Chr | LRSInd | LRSComb | Mb | Closest genes | Hu Chr |
|---|---|---|---|---|---|---|
| TSH | ||||||
| BXH | 4 | 13.8 | 57.44278 | Ptpn3 | 9 | |
| (n=12) | 13.8 | 57.62658 | Palm2-akap2 | 9 | ||
| Combined linkage analysis χ2=16.66271; p=0.002247 | ||||||
| CXB | 14 | (7.7) | 12.2 | 101.26 | Klf12 | 13 |
| AXBXA | (8.9) | 104.13 | Scel Klf12 | 13 | ||
| FT4 | ||||||
| AXBXA | 7 | 24.3* | 141.8777 | Adam 12 | 10 | |
| (n=26) | ||||||
| Combined linkage Chr 1: χ2=26.13734; p=2.9E-05 | ||||||
| CXB | 1 | (12.9) | 16.1 | 76.380 | Slc4a3 | 2 |
| thru | Pax 3 | 2 | ||||
| BXH | 1 | (8.2) | 80.948 | Cul3 | 2 | |
Suggestive or significant (bold) linkage ratio statistic (LRS) values are given together with locations of loci (megabases, Mb), the closest genes (regardless of function) and the corresponding human Chr. No linkage was observed individually (LRSInd) for thyrotropin (TSH) in CXB and AXBXA sets or for free thyroxine (FT4) in CXB and BXH sets. However, combined analysis provides evidence for linkage (LRSComb; the highest LRSInd values are given in parentheses).
GeneNetwork numbers for TSH: BXH 10172, AXBXA 10277, CXB 10746; for FT4: BXH 10275, AXBXA 10276, CXB 10744.
Determined using SD.
Chr 9: Ptpn3, protein tyrosine phosphatase (57.20371 Mb); Palm2-akap2, Palm-akap2 protein 2 (57.72268 Mb). Chr 13: Klf12, Kruppel-like factor 12 (100.26986 Mb); Scel, sciellin (103.9126 Mb). Chr 10: Adam 12, a disintegrin and metalloprotein (141.074882 Mb). Chr 2: Slc4a3, solute carrier 4A3 (75.54284 Mb); Pax3, paired box gene 3 (78.09784 Mb); Cul3, cullin 3 (80.26339 Mb).
TT4 was linked to different chromosomes or different loci on the same Chr in the three RI families (Table 2): Chr 1 in the CXB set and a more distal Chr 1 locus in the BXH set (Fig. 2C); Chr 11 in the CXB set (Fig. 2D); and Chr 17 in the AXBXA set (Fig. 2E). The only overlap in LRS values among the three RI sets was on Chr 4 (Fig. 2F). Combined analysis demonstrated linkage in CXB, BXH, and AXBXA families between TT4 and a Chr 4 region downstream of the deiodinase type 1 (Dio1) gene (Table 2). There was no overlap in the chromosomes or loci linked to variation in TT4 or FT4 analyzed for a single RI set or after combined analysis.
Table 2.
Chromosomal Linkage for Total Thyroxine in Male CXB, BXH, and AXBXA Mice
| Strain | Chr | LRSInd | LRSComb | Mb | Closest gene | Hu Chr |
|---|---|---|---|---|---|---|
| CXB | 1 | 17.6 | 76.525 | Epha4 | 2q34.1 | |
| (n=13) | 78.164 | Pax 3 | 2q35–q37 | |||
| 11 | 17.6 | 92.910 | Kif2b | 17q22 | ||
| 92.925 | Mbtd1 | 17 | ||||
| AXBXA | 17 | 18.1* | 76.11434 | Rasgrp3 | 2p25.1-p24.1 | |
| (n=26) | 76.27829 | |||||
| BXH | 1 | 13.5 | 136.1073 | Chi3l1 | 1q32.1 | |
| (n=12) | to | Adipor1 | 1q32.1 | |||
| 137.7065 | Kif14 | 1q32.1 | ||||
| Combined linkage χ2=20.36086; p=0.002388 | ||||||
| CXB | 4 | (7.8) | 12.0 | 101.103 | ||
| BXH | (6.5) | to | Insl5 | |||
| AXBXA | (6.0) | 103.26 | Slc35d1 | 1p31.1 | ||
Suggestive or significant (bold) LRS values for individual sets (LRSInd) are shown together with the number of strains studied, the locations of loci (megabases, Mb), the closest gene (regardless of function) and the corresponding human Chr. Combined linkage analysis (LRSComb) demonstrated sharing of Chr 4 loci between the three strains (brackets indicate the highest LRS values for individual strains).
GeneNetwork numbers for TT4: CXB 10743; BXH 10170; AXBXA 10275.
Determined using SD.
Chr, chromosomal; TT4, total thyroxine; Epha4, Eph receptor A4 (77.363759 Mb); Pax 3, paired box gene 3 (78.09784 Mb); Chi3l1, chitinase 3 like (136.07906 Mb); Adipor1, adiponectin receptor 1 (136.312043 Mb); Kif14, kinesin family member 14 (138.364534 Mb); Insl5, Insulin-like 5 (102.690477 Mb); Slc35d1, solute carrier family 35 (102.844322 Mb). Mbtd1, mbt domain containing 1 (93.74753 Mb); Rasgrp3, Ras, guanyl releasing protein (75.86495 Mb); Kif2b, kinesin family member 2B (91.4366 Mb); Note: Dio1; deiodinase type 1 (106.964069 Mb) outside the inverval for combined linkage.
It should be emphasized that the genes included in Tables 1 and 2 are genes closest to the LRS peaks and are unrelated to thyroid function. No candidate genes have yet been identified.
Linkages for TSH, FT4, and TT4 in males versus females
A comparison of TSH linkage between sexes, only possible in BXH strains, showed no overlap between loci. TSH was linked to a Chr 4 locus in males versus a Chr 18 locus in females (Fig. 3). In AXBXA strains, FT4 was linked to a Chr 7 locus in males but to a Chr 2 locus in females (Supplementary Table S2). As mentioned, there was no significant linkage for FT4 in male CXB or BXH strains, and there was no overlap between the highest LRS values for males and the Chr linkage previously observed in CXB or BXH females (Supplementary Table S2).
FIG. 3.
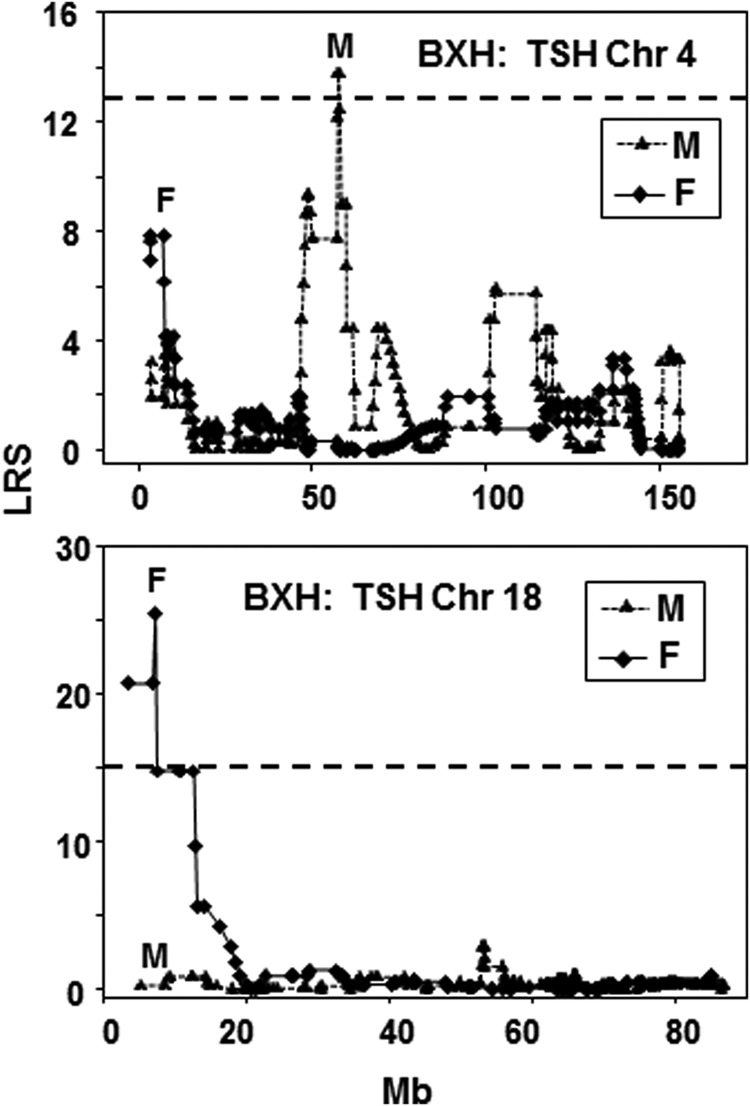
Difference in interval mapping for TSH on chromosomes 4 and 18 for TSH in male and female BXH RI families. LRS values are on the vertical axis; Mb on the horizontal axis. The broken horizontal lines indicate the mean LRS value above which there is suggestive linkage for the male and female mice. GN identifications: BXH males GN10172; data for female BXH mice from reference (17). Number of strains studied: males n=12; females n=12.
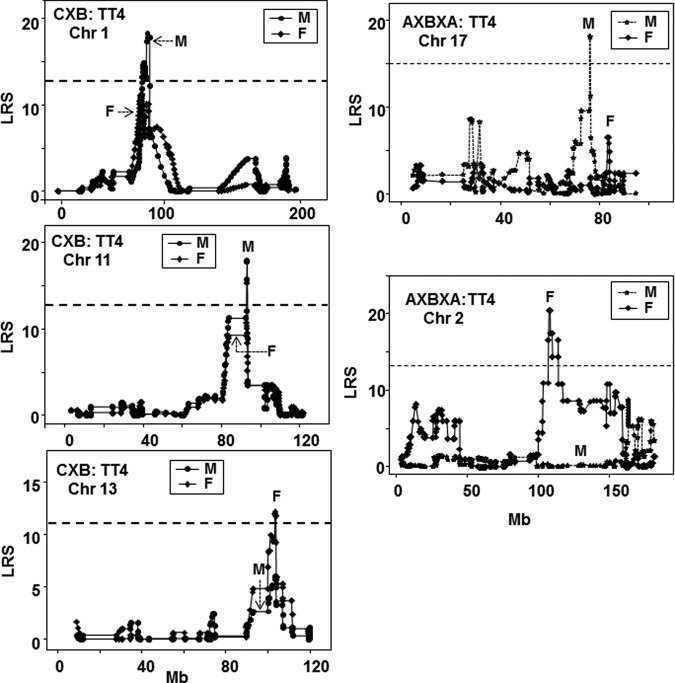
We noted some similarities for TT4 linkage in males and females in the CXB family: TT4 was linked to the same Chr 1 and 11 loci (Fig. 4, upper and middle left panels). A Chr 13 locus linked to TT4 in CXB females overlapped with a minor peak in males (Fig. 4, lower left panel). In contrast, in AXBXA strains, TT4 was linked to different chromosomes in males and females: Chr 17 in males and Chr 2 in females (Fig. 4, right panels). For the BXH set, TT4 was linked to Chr 1 in males and Chr 13 in females (Supplementary Table S3).
FIG. 4.
Comparison of interval mapping for TT4 in male and female mice. Left panels: CXB mice, chromosomes 1, 11, 13; right panels: AXBXA mice, chromosomes 2 and 17. LRS values are on the vertical axis; Mb on the horizontal axis. The broken horizontal lines indicate the mean LRS value above which there is suggestive linkage for male and female mice. Number of strains studied: CXB, males n=13; females n=13 (12,17); AXBXA, males n=25; females n=25 (14). GN identification: CXB males GN 10743, AXBXA Male GN 10277. Number of strains studied: CXB, males n=13; females n=13; AXBXA, males n=25; females n=25.
It should be emphasized that TT4 linkage was similar for three chromosomal loci in male and female CXB strains, despite the seven-year interval between obtaining CXB females versus CXB males (2005 vs. 2012; Supplementary Table S1). However, there was no overlap in TT4 linkage between male and female AXBXA strains that were obtained about three years apart (Supplementary Table S1). Moreover, the major TT4 linkages (to Chr 2) were similar in both the first AXBXA female cohort (Fig. 4, right panels) and the second female AXBXA cohort (17) obtained at almost the same time as the males. For the AXBXA set, therefore, disparate TT4 sex linkages could not be ascribed to cohort differences.
Discussion
We investigated serum levels and chromosomal linkages for TSH, FT4, and TT4 in young adult male mice of three recombinant inbred (RI) families, namely CXB, BXH, and AXBXA. These observations, compared with our previous findings for female mice of the same RI families (12–14,17), provided confirmatory and novel data.
The driving force for the present study was the inability to obtain measurements of serum TSH in most female mice in the three RI sets of interest to us (17). Our observation of markedly higher serum TSH in males than females of these RI sets confirms and extends previous data for C57BL/6 and CD-1 mouse strains (16). In addition, the higher TSH levels we observed in BXH than CXB or AXBXA males are consistent with TSH differences in male mice of five other strains (16). Sex differences for serum TT4 were variable. TT4 levels were higher in male than female AXBXA mice, not significantly different in the BXH set, and lower in male than female CXB mice. Variable sex differences were also reported for TT4, higher in C57BL/6 males than females, and not significantly different in CD-1 mice (16).
As in a previous study (16), we found no relationship between TT4 and TSH in male mice. However, FT4 and TSH were correlated in BXH strain mice. Although the significance of this correlation is unclear, it may be relevant that the C3H/He strain is one of the parental strains used to generate the BXH set. Unlike C57BL/6, the other parental strain, C3H/He mice have a defect in the Dio1 gene (23,24). TSH and T3 levels are similar in male C3H/He and C57BL/6 mice, although TT4 is elevated in C3H/He males (23). From studies of Dio1-knockout mice, which resemble C3H/He mice in terms of TSH, T3, and T4 levels, it seems likely that Dio1 is not essential to the “well-being” of euthyroid mice unless challenged with iodine deficiency or induced to develop hyperthyroidism by injecting L-T3 (25).
Turning to the genetic basis for serum levels of TT4, FT4, and TSH, there was no similarity in the major chromosomal linkages for TT4 in male CXB, BXH, or AXBXA strains. Linkage in males for TSH was only observed in the BXH set and for FT4 in the AXBXA set. However, combined linkage analysis revealed linkage for TSH (CXB and AXBXA sets) and FT4 (CXB and BXH sets). Importantly, TT4 and FT4 levels were linked to different chromosomes. Circulating T4 is bound to transthyretin and albumin in rodents (reviewed in Palha (26)). Consequently, genetic control of these T4 binding proteins likely contributes to variability in serum TT4 levels. It should be emphasized that no candidate genes were identified. However, these findings in male RI sets recapitulate our observations of “distinct genetic signatures” in females of the same RI sets (18), as well as the strain-specific differences in T4 released in response to TSH in T3-suppressed mice (17).
Because sex differences in RI mice were small for FT4 and absent for TT4, genetic linkage differences between males and females might not have been expected. However, as noted in the Introduction, in humans, despite slight differences in TSH and FT4 in men and women (only evident on statistical analysis), sex-specific genetic loci were associated with serum TSH and FT4 (10). Our most striking current findings relate to genetic linkage comparisons between male and female mice of the same RI sets. In CXB strains, TT4 linkage was shared by males and females for three Chr loci. However, TSH in the BXH set, and TT4 in the AXBXA and BXH sets, were linked to different chromosomes in males and females. Male–female differences in the genetic control of TT4 could be related to sex-dependence of the levels of T4 binding globulins (e.g., Vranckx et al. (27)).
The importance of sex-specific candidate genes in recombinant inbred mice was previously demonstrated by a study of severe murine influenza in the AXBXA family (28). In contrast, in studying TT4 levels in UM-HET3 mice, generated by crossing the F1 offspring of (BALBcJ X B6J)×(C3H/HeJ X DBA/2J), mouse age was taken into consideration but linkage data for males and females were pooled (29). Similarly, as mentioned in the Introduction, genetic studies of thyroid function in humans were performed in populations of men and women apparently without considering addressing sex effects (7–9). After completion of our study in mice, sex-specific linkages for thyroid-related traits were reported in a meta-analysis for humans (10).
To summarize, we observed sex-specific differences for some thyroid function traits in male versus female RI mice. These differences included markedly higher TSH levels in males than females, as well as moderately increased FT4 levels in males than females. However, our major findings relate to genetic linkage. Unlike the strong overlap between the chromosomal loci linked to TT4 in males and females in one RI set (CXB), we observed striking sex differences for TT4 and FT4 linkage in two other RI sets (BXH and AXBXA). Our findings are consistent with very recent observations of some sex-specific thyroid hormone linkages in humans. More importantly, they provide a cautionary note, which we are currently implementing in ongoing genetic studies, even for traits that are apparently not sex specific. In conclusion, focusing on either males (or females) is critical for genetic linkage analysis of traits involving thyroid hormones, and possibly other traits, in mice.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health Grants DK54684 (S.M.M.), U01AA13499, U24AA13513, P20-DA-21131, U01CA105417, and U24RR021760 (R.W.W.).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Hollowell JG, Staehling NW, Flanders WD, Hannon WH, Gunter EW, Spencer CA, Braverman LE.2002Serum TSH, T(4), and thyroid antibodies in the United States population (1988 to 1994): National Health and Nutrition Examination Survey (NHANES III). J Clin Endocrinol Metab 87:489–499 [DOI] [PubMed] [Google Scholar]
- 2.Surks MI, Boucai L.2010Age- and race-based serum thyrotropin reference limits. J Clin Endocrinol Metab 95:496–502 [DOI] [PubMed] [Google Scholar]
- 3.Vanderpump MPJ, Tunbridge WMG, French JM, Appleton D, Bates D, Clark F, Grimley Evans J, Hasan DM, Rodgers H, Tunbridge F, Young ET.1995The incidence of thyroid disorders in the community: a twenty-year follow-up of the Whickham survey. Clin Endocrinol 43:55–68 [DOI] [PubMed] [Google Scholar]
- 4.Surks MI, Hollowell JG.2007Age-specific distribution of serum thyrotropin and antithyroid antibodies in the US population: implications for the prevalence of subclinical hypothyroidism. J Clin Endocrinol Metab 92:4575–4582 [DOI] [PubMed] [Google Scholar]
- 5.Beydoun MA, Beydoun HA, Kitner-Triolo MH, Kaufman JS, Evans MK, Zonderman AB.2013Thyroid hormones are associated with cognitive function: moderation by sex, race, and depressive symptoms. J Clin Endocrinol Metab 98:3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vadiveloo T, Donnan PT, Murphy MJ, Leese GP.2013Age- and gender-specific TSH reference intervals in people with no obvious thyroid disease in Tayside, Scotland: the Thyroid Epidemiology, Audit, and Research Study (TEARS). J Clin Endocrinol Metab 98:1147–1153 [DOI] [PubMed] [Google Scholar]
- 7.Arnaud-Lopez L, Usala G, Ceresini G, Mitchell BD, Pilia MG, Piras MG, Sestu N, Maschio A, Busonero F, Albai G, Dei M, Lai S, Mulas A, Crisponi L, Tanaka T, Bandinelli S, Guralnik JM, Loi A, Balaci L, Sole G, Prinzis A, Mariotti S, Shuldiner AR, Cao A, Schlessinger D, Uda M, Abecasis GR, Nagaraja R, Sanna S, Naitza S.2008Phosphodiesterase 8B gene variants are associated with serum TSH levels and thyroid function. Am J Hum Genet 82:1270–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teumer A, Rawal R, Homuth G, Ernst F, Heier M, Evert M, Dombrowski F, Volker U, Nauck M, Radke D, Ittermann T, Biffar R, Doring A, Gieger C, Klopp N, Wichmann HE, Wallaschofski H, Meisinger C, Volzke H.2011Genome-wide association study identifies four genetic loci associated with thyroid volume and goiter risk. Am J Hum Genet 88:664–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panicker V, Wilson SG, Walsh JP, Richards JB, Brown SJ, Beilby JP, Bremner AP, Surdulescu GL, Qweitin E, Gillham-Nasenya I, Soranzo N, Lim EM, Fletcher SJ, Spector TD.2010A locus on chromosome 1p36 is associated with thyrotropin and thyroid function as identified by genome-wide association study. Am J Hum Genet 87:430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Porcu E, Medici M, Pistis G, Volpato CB, Wilson SG, Cappola AR, Bos SD, Deelen J, den Heijer M, Freathy RM, Lahti J, Liu C, Lopez LM, Nolte IM, O'Connell JR, Tanaka T, Trompet S, Arnold A, Bandinelli S, Beekman M, Bohringer S, Brown SJ, Buckley BM, Camaschella C, de Craen AJ, Davies G, de Visser MC, Ford I, Forsen T, Frayling TM, Fugazzola L, Gogele M, Hattersley AT, Hermus AR, Hofman A, Houwing-Duistermaat JJ, Jensen RA, Kajantie E, Kloppenburg M, Lim EM, Masciullo C, Mariotti S, Minelli C, Mitchell BD, Nagaraja R, Netea-Maier RT, Palotie A, Persani L, Piras MG, Psaty BM, Raikkonen K, Richards JB, Rivadeneira F, Sala C, Sabra MM, Sattar N, Shields BM, Soranzo N, Starr JM, Stott DJ, Sweep FC, Usala G, van der Klauw MM, van Heemst D, van Mullem A, Vermeulen SH, Visser WE, Walsh JP, Westendorp RG, Widen E, Zhai G, Cucca F, Deary IJ, Eriksson JG, Ferrucci L, Fox CS, Jukema JW, Kiemeney LA, Pramstaller PP, Schlessinger D, Shuldiner AR, Slagboom EP, Uitterlinden AG, Vaidya B, Visser TJ, Wolffenbuttel BH, Meulenbelt I, Rotter JI, Spector TD, Hicks AA, Toniolo D, Sanna S, Peeters RP, Naitza S.2013A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet 9:e1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey DW.1971Recombinant-inbred strains. An aid to finding identity, linkage, and function of histocompatibility and other genes. Transplantation 11:325–327 [DOI] [PubMed] [Google Scholar]
- 12.Aliesky HA, Pichurin PN, Chen CR, Williams RW, Rapoport B, McLachlan SM.2006Probing the genetic basis for thyrotropin receptor antibodies and hyperthyroidism in immunized CXB recombinant inbred mice. Endocrinology 147:2789–2800 [DOI] [PubMed] [Google Scholar]
- 13.McLachlan SM, Aliesky HA, Pichurin PN, Chen C-R, Williams RW, Rapoport B.2008Shared and unique susceptibility genes in a mouse model of Graves' disease determined in BXH and CXB recombinant inbred mice. Endocrinol 149:2001–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLachlan SM, Aliesky H, Chen C-R, Williams RW, Rapoport B.2011Exceptional hyperthyroidism and a role for both major histocompatibility class I and class II genes in a murine model of Graves' disease. PLoS One 6:e21378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Heyniningen HE.1961Differences in thyroid function of several strains of mice. Proc Soc Exp Biol Med 106:37–40 [DOI] [PubMed] [Google Scholar]
- 16.Pohlenz J, Maqueem A, Cua K, Weiss RE, Van Sande J, Refetoff S.1999Improved radioimmunoassay for measurement of mouse thyrotropin in serum: strain differences in thyrotropin concentration and thyrotroph sensitivity to thyroid hormone. Thyroid 9:1265–1271 [DOI] [PubMed] [Google Scholar]
- 17.Hamidi S, Aliesky H, Williams RW, Rapoport B, McLachlan SM.2012. Genetic linkages for thyroxine released in response to TSH stimulation in three sets of recombinant inbred mice provide evidence for shared and novel genes controlling thyroid function. Thyroid 23:360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLachlan SM, Lu L, Aliesky HA, Williams RW, Rapoport B.2011Distinct genetic signatures for variability in total and free serum thyroxine levels in four sets of recombinant inbred mice. Endocrinology 152:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams RW, Gu L, Qi S, Lu L.2001The genetic structure of recombinant inbred mice: high-resolution consensus maps for complex trait analysis. Genome Biol 2:Research0046.1–0046.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shifman S, Bell JT, Copley RR, Taylor MS, Williams RW, Mott R, Flint J.2006A high-resolution single nucleotide polymorphism genetic map of the mouse genome. PLoS Biol 4:e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Churchill GA, Doerge RW.1994. Empirical threshold values for quantitative trait mapping. Genetics 138:963–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams RW, Strom RC, Goldowitz D.1998. Natural variation in neuron number in mice is linked to a major quantitative trait locus on Chr 11. J Neurosci 18:138–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry MJ, Grieco D, Taylor BA, Maia AL, Kieffer JD, Beamer W, Glover E, Poland A, Larsen PR.1993Physiological and genetic analyses of inbred mouse strains with a type I iodothyronine 5′ deiodinase deficiency. J Clin Invest 92:1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schoenmakers CH, Pigmans IG, Poland A, Visser TJ.1993Impairment of the selenoenzyme type I iodothyronine deiodinase in C3H/He mice. Endocrinology 132:357–361 [DOI] [PubMed] [Google Scholar]
- 25.Schneider MJ, Fiering SN, Thai B, Wu SY, St Germain E, Parlow AF, St Germain DL, Galton VA.2006Targeted disruption of the type 1 selenodeiodinase gene (Dio1) results in marked changes in thyroid hormone economy in mice. Endocrinology 147:580–589 [DOI] [PubMed] [Google Scholar]
- 26.Palha JA.2002Transthyretin as a thyroid hormone carrier: function revisited. Clin Chem Lab Med 40:1292–1300 [DOI] [PubMed] [Google Scholar]
- 27.Vranckx R, Savu L, Maya M, Rouaze-Romet M, Nunez EA.1990Immunological quantitation of rat and mouse thyroxine-binding globulins. Ontogenesis and sex-dependence of the circulating levels of the thyroxine-binding globulins. Acta Endocrinol (Copenh) 123:649–656 [DOI] [PubMed] [Google Scholar]
- 28.Boivin GA, Pothlichet J, Skamene E, Brown EG, Loredo-Osti JC, Sladek R, Vidal SM.2012Mapping of clinical and expression quantitative trait loci in a sex-dependent effect of host susceptibility to mouse-adapted influenza H3N2/HK/1/68. J Immunol 188:3949–3960 [DOI] [PubMed] [Google Scholar]
- 29.Harper JM, Galecki AT, Burke DT, Pinkosky SL, Miller RA.2003Quantitative trait loci for insulin-like growth factor I, leptin, thyroxine, and corticosterone in genetically heterogeneous mice. Physiol Genomics 15:44–51 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.