Rapid changes in protein phosphorylation across abiotic stress, hormone, and pathogen treatments reveal proteins undergoing phosphoregulation in response to dehydration.
Abstract
Elucidating how plants sense and respond to water loss is important for identifying genetic and chemical interventions that may help sustain crop yields in water-limiting environments. Currently, the molecular mechanisms involved in the initial perception and response to dehydration are not well understood. Modern mass spectrometric methods for quantifying changes in the phosphoproteome provide an opportunity to identify key phosphorylation events involved in this process. Here, we have used both untargeted and targeted isotope-assisted mass spectrometric methods of phosphopeptide quantitation to characterize proteins in Arabidopsis (Arabidopsis thaliana) whose degree of phosphorylation is rapidly altered by hyperosmotic treatment. Thus, protein phosphorylation events responsive to 5 min of 0.3 m mannitol treatment were first identified using 15N metabolic labeling and untargeted mass spectrometry with a high-resolution ion-trap instrument. The results from these discovery experiments were then validated using targeted Selected Reaction Monitoring mass spectrometry with a triple quadrupole. Targeted Selected Reaction Monitoring experiments were conducted with plants treated under nine different environmental perturbations to determine whether the phosphorylation changes were specific for osmosignaling or involved cross talk with other signaling pathways. The results indicate that regulatory proteins such as members of the mitogen-activated protein kinase family are specifically phosphorylated in response to osmotic stress. Proteins involved in 5′ messenger RNA decapping and phosphatidylinositol 3,5-bisphosphate synthesis were also identified as targets of dehydration-induced phosphoregulation. The results of these experiments demonstrate the utility of targeted phosphoproteomic analysis in understanding protein regulation networks and provide new insight into cellular processes involved in the osmotic stress response.
Plants experiencing drought in natural environments must respond to the osmotic and ionic stresses that result from dehydration and exposure to high-salt concentrations. The response to osmotic stress is currently best understood at the level of intermediate and long-term adaptation, where changes in gene expression reorient growth and metabolism for survival. Less is known, however, about the mechanisms of initial stress perception and adaptation that occur within the first few minutes of dehydration.
During the initial phases of osmotic stress, plant cells experience a transient loss of water and turgor pressure that is recovered within minutes through increased inorganic ion fluxes into the cytoplasm (Shabala and Lew, 2002; Zonia and Munnik, 2007). Along with turgor recovery, adjustments of vital cellular processes occur, including arrest of cell growth and expansion, restructuring of the cytoskeleton, and shifts in active protein translation, that suggest heightened levels of posttranscriptional regulation (Kawaguchi et al., 2004; Fricke et al., 2006; Urano et al., 2010). Second messengers such as Ca2+, reactive oxygen species (ROS), phospholipids, and protein kinases are thought to play important signaling roles during this initial response phase (Dodd et al., 2010; Munnik and Vermeer, 2010; Steinhorst and Kudla, 2013).
In contrast to plants, the molecular nature of an osmotic stress signaling pathway is fairly well understood in yeast. The osmotic stress response is initiated by two yeast transmembrane osmosensors that act upstream of a mitogen-activated protein kinase (MAPK) signaling cascade to control gene expression in the Hog1 (for high-osmolality glycerol response1) signaling pathway (Hohmann, 2002). There is evidence to suggest that a similar pathway may be present in plants (Urao et al., 1999; Tran et al., 2007; Wohlbach et al., 2008; Kumar et al., 2013), and MAPK phosphorylation likely plays an important role in the osmotic stress response, as MAPK cascades are known to regulate several abiotic and biotic stress response pathways (Rodriguez et al., 2010).
Abscisic acid (ABA) is a hormone that controls the cellular response to water loss through the activation of subfamily 2 sucrose nonfermenting1-related kinases (SnRK2), although changes in ABA synthesis and transport generally occur much later than the initial dehydration perception and turgor response. In the absence of ABA, clade A protein phosphatase 2Cs suppress SnRK2 autophosphorylation, keeping SnRK2s in an inactive state (Park et al., 2009; Umezawa et al., 2009). In the presence of ABA, protein phosphatase 2C suppression is relieved and SnRK2s become active, phosphorylating known downstream targets, including transcription factors that bind to ABA-responsive elements (AREBs) and ion channels (Geiger et al., 2009; Sato et al., 2009; Yoshida et al., 2010). The 10-member SnRK2 family includes kinases that act directly in this ABA pathway (SnRK2.2, SnRK2.3, and SnRK2.6) as well as others that have not been implicated in ABA signaling but are activated during osmotic stress (Fujii and Zhu, 2009; Fujii et al., 2011; McLoughlin et al., 2012).
The significance of MAPK signaling in the yeast paradigm and the role of SnRK2 proteins in ABA- and non-ABA-mediated dehydration responses point to the importance of protein phosphorylation in osmotic stress signaling. In addition to MAPKs and SnRK2s, kinase families that are responsive to changes in cytosolic Ca2+ concentration likely play active roles during the early stages of a plant’s osmotic stress response (Cheong et al., 2010; Franz et al., 2011; Boudsocq and Sheen, 2013). The number and individual complexity of these gene families, along with the various second messengers associated with osmotic perturbation, suggest a multifaceted network of proteins undergoing phosphoregulation during the stress response, even in the earliest stage of perception.
Untangling these important phosphorylation networks has been a major challenge for scientists in recent years. Advances in quantitative proteomic techniques have improved our ability to carefully analyze signaling networks via quantitative in vivo measurements of phosphorylation changes on a large proteome-wide scale (Ross et al., 2004; Kline et al., 2010). Identifying proteins differentially phosphorylated in response to specific perturbations has led to key discoveries in signaling pathways, such as receptor-ligand interactions and mechanisms of transcription factor activation (Umezawa et al., 2013; Haruta et al., 2014).
These studies have used the most frequently employed proteomic method of untargeted data acquisition, also referred to as shotgun or discovery proteomics, to analyze phosphopeptides. In this method, peptides are selected for mass spectrometry (MS) sequence analysis based on real-time determination of peptide ion abundance. Any peptide ion meeting an abundance threshold at a given point in time is measured and sequenced through tandem mass spectrometry (MS/MS) fragmentation using an ion-trap mass spectrometer. Because this sampling feature depends on a peptide’s abundance rather than its identity, acquiring information does not require any prior knowledge of a peptide’s mass or charge state. This makes untargeted proteomics appropriate for survey studies where one does not know the exact peptides being sought but an assessment of the global proteome is being queried.
Despite its appeal for analyzing a very large number of peptides in a short period of time, a common limitation of untargeted proteomic methods is the inability to routinely measure lower abundance peptides. This is because the ion intensity thresholds used to select peptides for MS/MS sequencing can cause low-abundance species to be excluded from analysis. For protein phosphorylation measurements, where phosphorylated species exist at low stoichiometry in the cell, routine peptide detection can be extremely problematic. This impedes the ability to determine the statistical significance of observed changes and to make comparative measurements of phosphorylation changes across various experimental conditions.
To address these limitations, we employ a targeted MS method known as Selected Reaction Monitoring (SRM) to specifically measure select protein phosphorylation events in a highly sensitive, routine fashion. SRM uses knowledge of a peptide’s mass, charge, and fragmentation behavior to enhance peptide detection via filtered selection using a triple quadrupole mass spectrometer. In contrast to untargeted MS, where all possible peptides are analyzed, targeted MS examines a predetermined list of peptides with improved sensitivity and throughput. This makes it an amenable platform for comparative analysis of protein phosphorylation across different samples and treatment conditions. This method has been recently used in the field to successfully measure the abundance of a protein across genotypes (Su et al., 2013; Taylor et al., 2014) and the phosphorylation state of a specific target protein (Dubiella et al., 2013).
In this study, we investigate proteins involved in the early response to hyperosmotic stress in Arabidopsis (Arabidopsis thaliana) by measuring stress-induced phosphorylation using untargeted and targeted proteomics. Phosphorylation responses were first quantified using untargeted MS and then validated and characterized with respect to specificity and onset using SRM. Our data implicate new proteins and points of regulation in the osmotic stress response and provide a new method of comparing phosphorylation networks in Arabidopsis that is also applicable with any plant whose proteome sequence is known.
RESULTS
Untargeted Phosphoproteomic Measurements Identify Early Phosphorylation Changes after Osmotic Stress Treatment
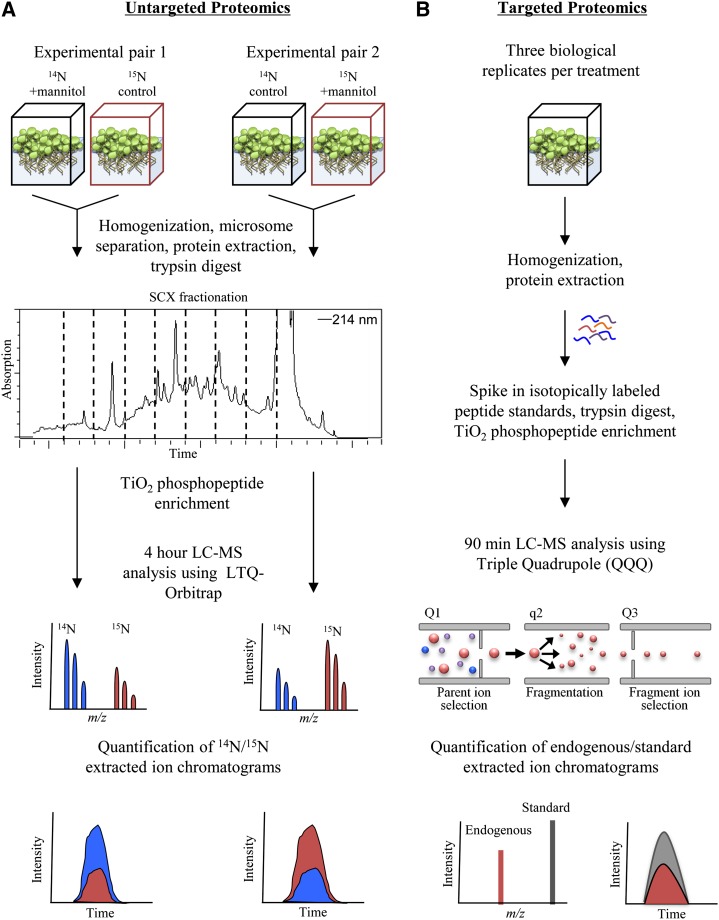
To identify proteins involved in the initial stages of the osmotic stress response, we performed an untargeted proteomics experiment measuring protein phosphorylation following 5 min of perturbation with 0.3 m mannitol. To quantify phosphorylation changes, we used full metabolic labeling of Arabidopsis seedlings in the reciprocal experimental strategy outlined in Figure 1A. Plants were grown in liquid culture in experimental pairs consisting of one sample grown in natural abundance 14N medium and the other grown in isotopically heavy 15N medium. After 10 to 11 d of growth, the sample medium was replaced with either treatment or control medium for 5 min. Treated and control sample pairs were combined and processed together for tandem liquid chromatography (LC)-MS analysis using an LTQ-Orbitrap mass spectrometer. A reciprocal experimental pair was processed in parallel to serve as a biological replicate and correct for any isotopic effects.
Figure 1.
Overview of quantitative proteomic methods used in this study. A, Untargeted proteomics experimental design using 15N full metabolic labeling. Phosphorylation changes were determined from 14N to 15N area ratios of phosphopeptide extracted ion chromatograms. B, Targeted proteomics experimental design using SRM and stable isotope-labeled phosphopeptide standards. Parent ion-to-fragment ion transitions are measured for endogenous and standard synthetic peptides. Phosphorylation changes are determined from endogenous to standard area ratios of fragment ion extracted ion chromatograms.
In total, two sample pairs were analyzed, constituting one set of reciprocally labeled experiments. To reduce sample complexity prior to MS analysis, these samples were prefractionated by first separating the cytoplasmic soluble proteins from microsomal membranes. Following tryptic digestion, the soluble fraction was further fractionated using HPLC and a strong cation exchange (SCX) column. Microsomal pellets and SCX fractions from each sample were enriched for phosphopeptides and analyzed on the mass spectrometer. From the acquired data, only peptides meeting a 1% false discovery rate cutoff and observed in each of the two samples were included in our analysis.
Overall, 1,299 phosphorylated peptides, representing 833 unique proteins, were identified in both 14N/15N experimental pairs. From this data set, we identified 29 proteins showing reciprocal changes in phosphorylation greater than 2-fold, of which 12 proteins possessed greater than 3-fold change in phosphorylation. The annotated functions of these proteins, displayed in Table I, indicate that dehydration could be influencing diverse cellular functions, including vesicle trafficking, RNA degradation, ubiquitination, and membrane transport. A complete list of changing phosphopeptides (1.5-fold or greater change) identified in this study can be found in Supplemental Table S1. Since the chosen time points (5 min) are short and most likely precede changes in de novo protein synthesis, it is reasonable to suggest that the observed changes in phosphopeptide abundance represent phosphorylation increases rather than changes in protein abundance. However, this assumption can be experimentally tested by the identification of additional, nonchanging peptides derived from the same protein. For approximately one-third of the osmotic stress-responsive phosphoproteins listed in Supplemental Table S1, we identified an additional peptide that indicates protein levels were not changing between treated and control samples. These additional peptides are listed in Supplemental Table S2.
Table I. Phosphorylation changes in response to 5 min of osmotic stress.
Phosphopeptides showing 2-fold or greater change in phosphorylation in response to 5 min of 0.3 m mannitol treatment and additional phosphopeptides selected for targeted proteomic analysis are shown. Phosphorylation sites that could not be localized to one residue are indicated by multiple residues inside brackets. MS/MS spectra for the listed phosphopeptides are documented in Supplemental Figure S1. n/a, Not applicable.
| ATG Accession No. | Protein | Phosphopeptide | Fold Change | se | Targeted MS Validation |
|---|---|---|---|---|---|
| Kinases/signaling related | |||||
| AT2G01690 | Vac14 | AT[pS]GVPFSQYK | 12.55 | 1.01 | Yes |
| AT1G10940a | SnRK2.1/4/5/10 | S[pT]VGTPAYIAPEVLSR | 6.18 | 2.06 | Yes |
| AT1G53165 | MAP4Kα1 | SS[pS]ASEDSISNLAEAK | 4.87 | 0.72 | Yes |
| AT1G16270 | MAP3K Raf18 | TV[pS]GGGIETEAR | 2.50 | 1.24 | Yes |
| Proteasome/ubiquitin related | |||||
| AT2G26590 | RPN13 | AGNLVVPNLSSEV[pS]DVTSSSGPVK | 3.84 | 0.23 | n/a |
| AT2G47970 | NPL4 family protein | SIPGAPPVTPAG[pS]FGR | 2.29 | 0.18 | n/a |
| AT3G63000 | NPL41 (NPL4-LIKE) | GGPAVTPAG[pS]FGR | 2.06 | 0.09 | n/a |
| 5′ mRNA decapping | |||||
| AT5G13570 | DCP2 | [pTS]VGGNGTATVESQNR | 4.48 | 0.04 | n/a |
| AT3G13300 | VCS (varicose) | TP[pS]ADYSVDR | 5.69 | 0.95 | Yes |
| ESITSAS[pS]VAQALSR | 5.50 | 2.01 | n/a | ||
| NLDVS[pS]VEEISR | 2.18 | 0.002 | n/a | ||
| TSGLPSQTSGAGSAYATLPQLPL[pS]PR | 0.49 | 0.06 | n/a | ||
| AT3G13290 | VCR (varicose related) | TS[pS]ADYFYVR | 3.98 | 0.53 | n/a |
| Cytoskeleton | |||||
| AT1G04820a | TUA2/4/6 | TIQFVDWCP[pT]GFK | 2.80 | 0.30 | Yes |
| AT5G19770a | TUA1/3/5 | TVQFVDWCP[pT]GFK | 1.98 | 0.06 | Yes |
| AT2G21380 | Kinase motor related | [pSSS]TPTSTVYNSGGVTGSR | 2.53 | 0.93 | n/a |
| Channels/pumps | |||||
| AT3G58730 | Vacuolar ATPase D subunit | GI[pS]INAAR | 2.91 | 0.66 | Yes |
| AT5G60660 | Aquaporin PIP2F | ALGSFGSFG[pS]FR | 2.48 | 0.11 | Yes |
| ALGSFG[pS]FGSFR | 2.09 | 0.13 | No | ||
| AT2G36380 | ATP-binding cassette transporter PDR6 | LP[pT]YDRLR | 2.49 | 0.78 | n/a |
| Vesicle trafficking related | |||||
| AT1G08800 | MyoB1 | ALLTQI[pS]ASR | 5.65 | 0.07 | Yes |
| AT4G19490 | VPS54 | SI[pS]DASSQSLSSILNNPHGGK | 2.68 | 0.14 | n/a |
| AT4G11740 | SAY1 | AASG[pS]LAPPNADR | 1.92 | 0.09 | Yes |
| AT1G79830 | GC5 | QN[pS]AFENGSLPR | 2.78 | 0.004 | Yes |
| Metabolism related | |||||
| AT1G77120 | ADH1 | IIGVDFN[pS]K | 0.22 | 0.10 | No |
| AT5G05600 | Oxidoreductase | VQ[pS]LAESNLSSLPDR | 3.20 | 0.49 | No |
| AT5G40390 | SIP1 | SD[pS]GINGVDFTEK | 1.51 | 0.06 | Yes |
| AT3G58460 | RBL15 | [pTLS]TARDPTAPAGETDPNLHAR | 2.04 | 0.13 | n/a |
| Other/unknown | |||||
| AT2G21230 | bZIP30 | SI[pS]GEDTSDWSNLVK | 2.70 | 0.19 | Yes |
| AT1G72410 | COP1-interacting related | LSLGGG[pS]ADFSK | 4.73 | 0.26 | Yes |
| AT3G03050 | Cellulose synthase-like CSLD3 | [pS]NLSTNSDAAEAER | 2.10 | 0.01 | n/a |
| AT2G01190 | PB1 domain containing | GF[pS]DSDTNVNR | 2.29 | 0.25 | Yes |
| AT5G64430 | PB1 domain containing | LFLFPASSGFG[pS]QSSTQSDRDR | 2.00 | 0.42 | n/a |
| AT2G32240 | PICC defense responsive | DIDLSFS[pS]PTK | 1.49 | 0.001 | Yes |
| AT5G56980 | A70 defense responsive | AP[pS]IIDR | 1.55 | 0.02 | Yes |
| AT5G35430 | TPR domain containing | [pTSS]LLSSSVASDTLR | 2.61 | 0.12 | n/a |
| AT1G22060 | Unknown | SVV[pS]GDLSGLAQSPQK | 2.37 | 0.54 | n/a |
| AT5G58510 | Unknown | TN[pS]VNQSPTDAIR | 2.10 | 0.01 | n/a |
| AT5G42950 | GFY domain containing | [pSPSS]DLLSILQGVTDR | 2.00 | 0.23 | n/a |
Peptide matches multiple family members.
Coordinate Regulation of Protein Phosphorylation among Gene Family Members
Several proteins undergoing dynamic changes in phosphorylation in response to osmotic stress are closely related members of large gene families. Observing a similar pattern of regulation in related proteins indicates that particular subfamilies may be playing important roles in the stress response. An example of a coordinated phosphorylation response is seen in the MAPKKK (MAP3K) and basic leucine zipper (bZIP) transcription factor proteins identified in this study.
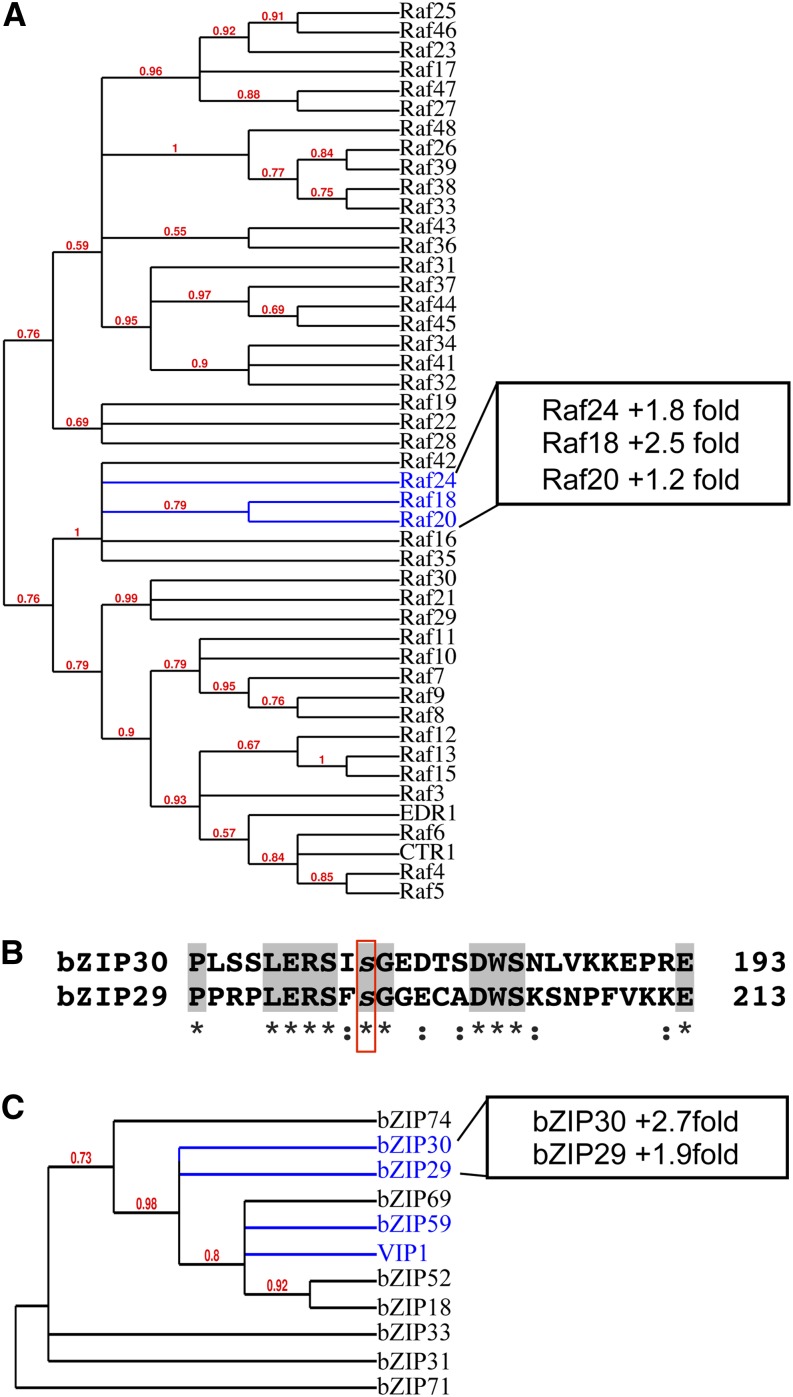
The Arabidopsis MAP3K family is very large, consisting of 60 genes that are broken up into two main divisions: the MAPK/ERK kinase kinase and Raf kinase subfamilies. The MAP3K proteins showing mannitol-responsive phosphorylation in this study (Raf18, Raf20, and Raf24) all belong to the RAF B4 subgroup, which is one of 11 subdivisions in the Raf family (Ichimura et al., 2002). Phylogenic analysis of 46 Raf members shows close grouping of all three RAF proteins (Fig. 2A). Phosphorylation increases of Raf18 and Raf24 are around 2-fold and occur at nearly identical centralized regions in each protein and not in any conserved domains. Although the Raf20 phosphorylation increase is small, its close relationship to Raf18 and Raf24 indicates that these proteins may be coordinately regulated at the posttranslational level during osmotic stress. Currently, no known function has been ascribed to these kinases or to any of the RAF B4 subgroup members.
Figure 2.
Coordinate regulation of related gene family members. A, MAPKKK RAF family phylogenetic tree. RAF proteins showing osmotic stress-induced phosphorylation increases are highlighted in blue. B, Alignment of bZIP29 and bZIP30 protein sequences using Clustal Omega. The conserved phosphorylation site is highlighted in the red box. Gray highlights and asterisks indicate identical residues, and colons indicate highly similar residues. C, Phylogenetic tree of bZIP group I family members. Proteins showing osmotic stress-induced phosphorylation and proteins previously implicated in salt and osmotic stress are highlighted in blue. For A and C, trees were made from protein sequence alignment using Phylogeny.fr (Dereeper et al., 2008).
As with the MAP3Ks, the Arabidopsis bZIP transcription factor family is quite large, consisting of approximately 70 genes. Two closely related proteins of the bZIP class I subfamily (Jakoby et al., 2002), bZIP29 and bZIP30, show increased phosphorylation at a conserved Ser residue (Fig. 2B). bZIP30 phosphorylation has been shown to be responsive to exogenous ABA treatment to a lesser extent (Kline et al., 2010; Xue et al., 2013) and to 30 min of mannitol treatment (Xue et al., 2013), but an earlier osmotic challenge has not been examined previously. Within the bZIP class I subfamily, two proteins closely related to bZIP29 and bZIP30 have been reported to play a role in the abiotic stress response (Fig. 2C). VirE2-interacting protein1 (VIP1), which has been associated with the pathogen response in the past, has recently been shown to be involved in the osmotic stress response (Tsugama et al., 2012), and bZIP59 is reported to function downstream of the salt overly sensitive signaling pathway (Van Oosten et al., 2013). VIP1 and bZIP59 were not detected in our data set, but taken together, our observations suggest that bZIP class I proteins may be important components of the dehydration stress-related phosphosignaling pathway.
Phosphorylation of the 5′ mRNA Decapping Protein Complex
Several proteins involved in cytosolic mRNA regulation through the 5′ to 3′ exonuclease degradation pathway experienced large changes in phosphorylation following a 5-min hyperosmotic challenge. These proteins include members of the 5′ decapping complex: VARICOSE (VCS), VCS-related (VCR), and decapping enzyme DCP2 (Table I). Removing the 5′ cap of mRNA is an important step in posttranscriptional regulation, as it represents movement away from active translation and functions as the rate-limiting prerequisite for exoribonuclease degradation (Jonas and Izaurralde, 2013). VCS is a scaffold protein that holds together DCP1 and DCP2 in a protein complex that is conserved in plants and animals (Franks and Lykke-Andersen, 2008). Our results demonstrate that VCS is phosphorylated at multiple Ser residues, with some sites increasing and some decreasing their levels of phosphorylation in response to mannitol (Fig. 3, A–C), reflecting a complex mechanism of phosphoregulation. Nearly all of the dynamic phosphorylation changes occur in the flexible Ser-rich linker region residing between protein-binding domains (Fig. 3D). VCS has an essential function in the posttranscriptional regulation of mRNA through both 5′ decapping and microRNA degradation (Brodersen et al., 2008). VCS associates with processing bodies, which are protein-RNA complexes that function as sites of transcript repression and degradation and are known to accumulate in response to abiotic stress (Weber et al., 2008). Currently, the molecular mechanisms controlling VCS association with 5′ decapping complex proteins and with processing bodies are unknown.
Figure 3.
VCS is phosphorylated at multiple Ser residues in response to osmotic stress. A, Reciprocal change in phosphorylation at Ser-629. Graphs show extracted ion chromatograms for reciprocal experiments generated by Census software. Blue lines represent 14N peptide, red lines represent 15N peptide, and boldface, italic labels indicate the treated sample in each reciprocal pair. Yellow shading indicates the area used to calculate the peptide abundance ratio in Census. Vertical green lines indicate the scan at which MS/MS data were acquired. B, No change was seen in the VCS nonphosphorylated peptide, indicating that phosphorylation changes are not due to differences in protein abundance. C, Log2 graph of VCS phosphorylation changes. Gray bars indicate no change, blue bars indicate decreasing phosphorylation, and red bars indicate increasing phosphorylation. Red striped bars indicate that phosphopeptide was only identified in one experimental pair. Error bars represent se between experiment 1 and 2 averages. Error could not be calculated for phosphopeptides identified in only one experiment. D, Diagram of the VCS protein with phosphorylation site locations. Most phosphorylation changes occur in the Ser-rich linker region. The DCP2-binding site is located in the conserved protein domains α-helix domain 1 (D1) and D2. D2 contains conserved residues required for processing body localization (Jinek et al., 2008). Phosphorylation sites are indicated by colored diamonds. Color coding is the same as in B.
Limitations of Untargeted Proteomic Analysis
Untargeted data acquisition is a valuable tool for the initial identification of proteins involved in the cellular stress response but offers limited utility in subsequent analysis of low-abundance species. This is due to the indiscriminate nature of MS sampling, in which any peptide meeting abundance thresholds in a certain time interval is selected for analysis, a process called data-dependent acquisition. Acquiring intensity and sequence information for all possible peptides as they enter the mass spectrometer results in frequent sampling of high-abundance species at the expense of others, because instrument space is finite and sampling occurs in a serial fashion. In our study, nearly half of all identified phosphopeptides had to be excluded from analysis because they were found in only one experimental sample (Fig. 4A). Even repeated injections of the same sample yield poor overlap in peptide detection, a common feature inherent in the technique when complex biological samples are analyzed (Fig. 4B). The consequence of this inadequate sample overlap is a reduction in usable data and weak statistical support for observed phosphorylation changes.
Figure 4.
Reproducibility in untargeted phosphoproteomic data. A, Number of proteins and phosphopeptides identified in both experimental samples or only in one sample. B, Phosphopeptide identification overlap between two injection replicates of the same sample.
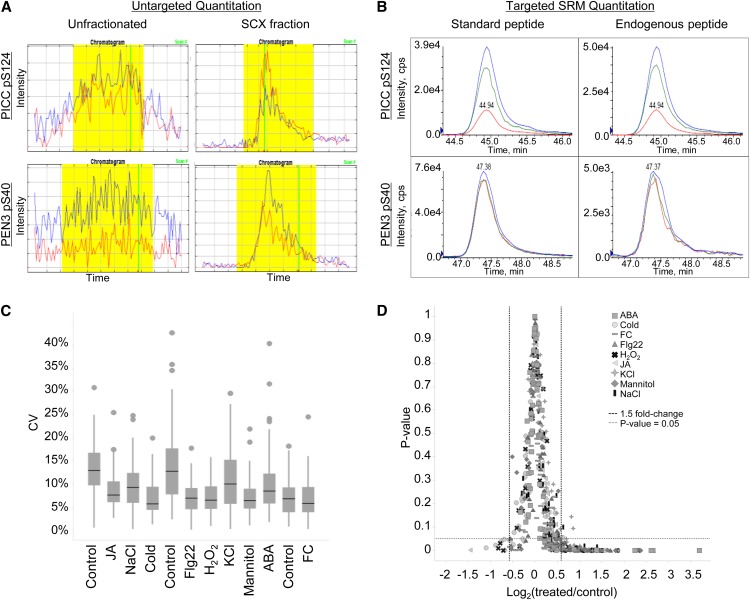
One approach to improving peptide detection is to reduce sample complexity prior to MS analysis. In this study, samples were prefractionated by SCX chromatography and separated over a 4-h reverse-phase HPLC gradient in tandem with MS analysis (Fig. 1A). The addition of SCX chromatography significantly improved sample detection and quantification (Fig. 5A), so much so that nearly all phosphopeptides identified in Table I were not found in our unfractionated data set (K.E. Stecker and G.A. Barrett-Wilt, unpublished data). Improvements in detection, however, come at the cost of significantly increased labor and instrument time.
Figure 5.
Improved phosphopeptide detection using targeted SRM analysis. A and B, Quantification of the PAMP-Induced Coiled-Coil (PICC) phosphopeptide DIDLSFS[pS]PTK (top) and the PEN3 phosphopeptide NIEDIFSSG[pS]R (bottom) across different MS platforms. A, Extracted ion chromatograms from phosphopeptide enrichment of unfractionated, total protein extract (left) compared with phosphopeptide enrichment of SCX-fractionated samples (right) using untargeted proteomic methods. The PEN3 peptide was only identified in one experimental sample in SCX-fractionated data. Extracted ion chromatograms were generated by Census software. Blue lines indicate 14N peptide, and red lines indicate 15N peptide. Yellow shading indicates the area used to calculate the peptide abundance ratio in Census. Vertical green lines indicate the scan at which MS/MS data were acquired. B, Extracted ion chromatograms from phosphopeptide enrichment of unfractionated, total protein extract using the targeted SRM method. Peptide standards (left) and endogenous peptides (right) coelute with identical fragment ion patterns. Three fragment ions were measured for each peptide. C and D, Statistical analysis of targeted proteomic data. C, Box plot of the coefficient of variance (CV) for each phosphopeptide quantified across three biological replicates in 12 different treatments. A total of 58 phosphopeptides are plotted in each treatment. The control treatment is presented three times because samples were grown and processed in three separate batches. Black bars indicate the median value for each treatment. D, Phosphorylation changes of 1.5-fold or greater are statistically significant (P ≤ 0.05, Student’s t test) in almost all targeted experiments. FC, Fusicoccin; JA, methyl jasmonate.
Targeted Proteomics Provides Sensitive and Reproducible Detection of Protein Phosphorylation Changes
To improve the detection of phosphopeptides across multiple experimental samples and to independently evaluate the phosphorylation changes identified in our initial study, SRM methods were developed to specifically target peptides of interest. SRM analysis is performed on a triple quadrupole tandem mass spectrometer and employs a two-step mass filtering strategy in which only peptides and selected peptide fragments with desired mass-to-charge ratios are allowed to pass through the instrument. Target peptides are selected in the first filtering step in quadrupole 1. Peptides are subsequently fragmented in quadrupole 2, and only fragment ions with desired mass-to-charge ratios are selected in the second filtering step in quadrupole 3 (Fig. 1B). The detection of targets using this dual mass filtering significantly reduces background ions, thus enhancing peptide quantitation through improved signal-to-noise ratios. A comparison of peptide detection across experimental platforms is seen in Figure 5, A and B.
To quantify target peptides in SRM analysis, we synthesized stable isotope-labeled peptide for use as a spike in standard. Standards were made for 70 phosphopeptides selected from the mannitol discovery data and several of our untargeted proteomic experiments (Kline et al., 2010; Haruta et al., 2014). From the starting 70 phosphopeptides, 12 phosphopeptides were not included in our final assay for the following reasons: (1) five phosphopeptide standards failed to be detected; (2) four endogenous phosphopeptides were below the levels of detection; and (3) three phosphopeptides possessed very high sample-to-sample variability and could not be reproducibly quantified between biological replicates. Our final assay consisted of 58 phosphopeptides (Supplemental Table S3), of which 21 were derived from the untargeted mannitol data. The selection of mannitol-responsive phosphopeptides for targeted analysis was based on biological interest, the magnitude of phosphorylation response observed in the discovery data, the feasibility of peptide synthesis (20 amino acids or fewer in length), and overlap with other studies in the laboratory. These 58 phosphopeptide standards and corresponding endogenous peptides were measured in a single 90-min LC-MS run. No SCX prefractionation was required, as endogenous peptides could be routinely detected from the phosphopeptide enrichment of crude, total protein extracts (Fig. 5, A and B). This multiplexed targeted assay was used as a secondary experiment to evaluate the discovery data and characterize phosphorylation changes in plants treated with different environmental perturbations.
To better understand if mannitol-responsive phosphorylation events were specific to osmotic stress or were generically responding to perturbations and changes in common second messengers used by several signaling pathways, we measured phosphorylation under nine different treatment conditions (Table II). In each case, the treatment was applied for 5 min. Hormone, biotic, and abiotic treatments that were chosen are predicted to share both overlapping and unique physiology with the osmotic stress response.
Table II. Targeted MS treatment conditions.
Treatment conditions were used for targeted proteomic analysis of 58 phosphopeptides.
| Treatment | Concentration | Rationale |
|---|---|---|
| Mannitol | 300 mm | Osmotic stress, validation of discovery results |
| NaCl | 150 mm | Ionic stress, osmotic stress |
| KCl | 150 mm | Plasma membrane depolarization, osmotic stress |
| S-ABA | 50 μm | Drought stress hormone |
| Cold | 0°C–4°C | Freezing tolerance overlap with dehydration stress; overlapping second messengers with osmotic stress |
| Flg22 | 1 μm | Pathogen elicitor; overlapping second messengers with osmotic stress |
| H2O2 | 5 mm | ROS, second messenger |
| Fusicoccin | 5 μm | Plasma membrane hyperpolarization |
| Methyl jasmonate | 50 μm | Defense response hormone; minimal overlap expected |
In contrast to untargeted MS detection, phosphopeptides were reliably identified in our targeted MS analysis with good statistical support. We observed minimal technical variability and low biological variability, with median coefficient of variance values around 10% for all experiments (Fig. 5C). Due to shorter LC methods (90 versus 240 min) and no HPLC prefractionation, the throughput of targeted peptide analysis is significantly higher. Analysis of nine treatment conditions with three biological replicates each was performed using less instrument time than required for analysis of the two reciprocal experimental samples in our untargeted work.
Hierarchical Clustering of Targeted Phosphoproteomic Data
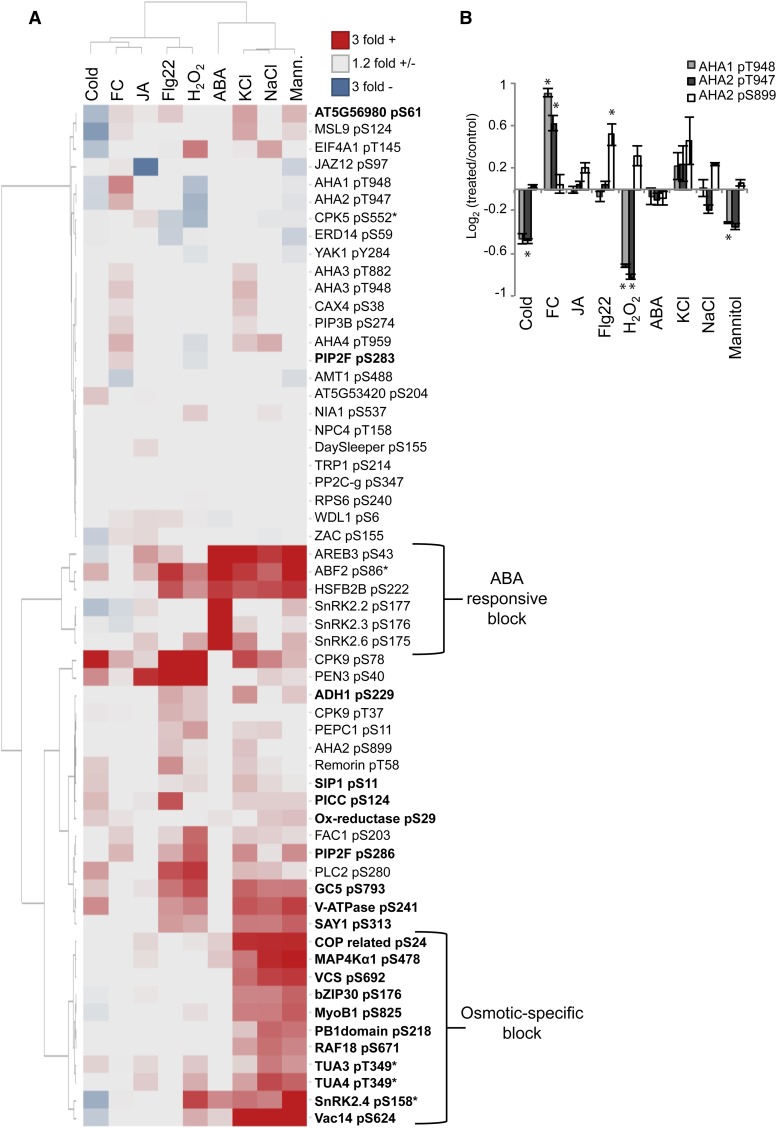
To extract patterns from protein phosphorylation changes induced by various treatments, we performed hierarchical clustering of targeted phosphoproteomic data. Nearly all phosphorylation changes greater than 1.5-fold were statistically significant with P ≤ 0.05 (Fig. 5D). Clustering of treatment conditions revealed some expected trends. The three conditions inducing osmotic stress, mannitol, NaCl, and KCl, cluster together with minimal differences observed between treatments (Fig. 6A). This shared response between the neutral osmolyte, mannitol, and the charged salts, sodium and potassium chloride, indicates that the ionic component of salt stress does not have a unique influence on the phosphorylation of many of the selected proteins. In contrast to the overlap observed among the mannitol and salt phosphorylation responses, ABA treatment appears to have a unique response and little overlap with the other treatments at 5 min. Known components of the ABA receptor signaling pathway show large changes in phosphorylation, while a minimal response is seen with all other protein targets (Fig. 6A). These components include SnRK2 kinases involved in ABA signaling (SnRK2.2, SnRK2.3, and SnRK2.6), AREB transcription factors, and class B heat shock factor HSFB2B. Interestingly, AREBs and HSFB2B experience large changes in phosphorylation under osmotic and defense-related conditions, while ABA-responsive SnRK2s do not. This indicates that these transcription factors are likely regulated by alternate phosphorylation pathways and may represent a mechanism of cross talk between various abiotic and biotic signaling networks.
Figure 6.
Hierarchical clustering of targeted phosphoproteomic data. A, Heat map of 58 phosphopeptides measured using SRM. Values represent log2 area ratios of treatment versus control averages. Averages represent three biological replicates per treatment with two or more injection replicates per sample. Data for the heat map are found in Supplemental Table S4. Asterisks indicate nonunique peptides that match multiple gene family members. Phosphopeptides in boldface were selected from mannitol-untargeted proteomic data (Table I). Clustering was performed using the method by Ward (1963). B, AHA1 and AHA2 phosphorylation across all treatments. Error bars represent se for three biological replicates. Asterisks indicate phosphorylation changes with P < 0.05 (Student’s t test). FC, Fusicoccin; JA, methyl jasmonate.
Clustering shows that the bacterial flagellin flg22 peptide, which acts as a pathogen elicitor, most closely groups with the response to hydrogen peroxide (H2O2). The pathogen-associated molecular pattern (PAMP) response is known to stimulate ROS waves that quickly propagate throughout the plant (Torres et al., 2006; Segonzac and Zipfel, 2011). Our data show that the phosphorylation of PENETRATION3 (PEN3), an ATP-binding cassette transporter involved in PAMP resistance, is dramatically induced by Flg22, H2O2, and methyl jasmonate treatment. Another protein known to be involved in defense signaling, PHOSPHOLIPASE C2 (PLC2), is shown to be responsive to Flg22 and H2O2. PLC2 is a calcium-activated phosphoinositide lipase involved in the production of diacyglycerol and soluble inositol phosphates (Munnik, 2014). Both PEN3 and PLC2 PAMP-responsive phosphorylation have been reported previously (Nühse et al., 2007) and represent positive controls for the treatment conditions and quantitation methods used in this study.
We observed some overlapping phosphorylation patterns between ROS and osmotic perturbations with SnRK2.4, α-tubulin (TUA), and Vac14 (for vacuole morphology and inheritance mutant14). This overlap is consistent with previous reports that osmotic stress induces increased ROS production (Xiong and Zhu, 2002). Notably, these proteins show increased phosphorylation in mannitol, salt, and H2O2 but not in Flg22, demonstrating that they are not responsive to all ROS-inducing conditions. This suggests a responsiveness to ROS that is unique to osmotic stress.
Regulation of the Plasma Membrane Proton Pump (H+-ATPase) by Phosphorylation Changes at Two Distinct Sites
Similar to the dynamic regulation of VCS at multiple phosphorylation sites (Fig. 3C), the plasma membrane H+-ATPase pump is controlled by phosphorylation at several residues in a C-terminal water-soluble domain of approximately 100 amino acids (Nühse et al., 2007). Increased phosphorylation of the most C-terminal Thr, referred to as the penultimate Thr, activates the pump through increased binding with a 14-3-3 protein (Olsson et al., 1998). In contrast, phosphorylation of Ser-899, also located in this C-terminal regulatory domain, is proposed to inhibit pump activity, although the mechanism by which this acts is less well understood (Haruta et al., 2014). In our targeted analysis, the two most highly expressed H+-ATPase (AHA) family members in Arabidopsis, AHA1 and AHA2, display coordinated regulation at the penultimate position (AHA1 pT948 and AHA2 pT947) and are responsive to several treatments (Fig. 6B). We observed phosphorylation increases in response to fusicoccin, a fungal pathogen known to cause pump hyperactivation, and phosphorylation decreases in response to cold, H2O2, and mannitol. Pump inhibition via pS899 phosphorylation is seen in response to Flg22 and to a lesser extent in H2O2 and salt treatments, although these findings have less statistical support. Interestingly, these results indicate that AHA2 down-regulation occurs by two distinct mechanisms (dephosphorylation of Thr-947/948 versus phosphorylation of Ser-899) that vary based on treatment conditions.
It has been speculated that AHA proton pumps are hyperactivated during the osmotic stress response and may play an active role in the initial turgor recovery by providing the proton motive force necessary to drive ion influx into cells (Shabala and Lew, 2002; Shabala and Shabala, 2011). Our phosphorylation data show that AHAs are not being activated through penultimate Thr phosphorylation in the first 5 min of the mannitol stress response. In contrast, AHA activity appears to be down-regulated through decreased penultimate Thr phosphorylation in AHA1 and AHA2. This pattern of phosphoregulation is distinct from the AHA response to KCl treatment, suggesting that these different forms of osmotic stress have unique effects on AHA activity. This observation is supported by genetic data showing that aha2 null mutants are hypersensitive to KCl but not sorbitol or NaCl treatment (Haruta and Sussman, 2012).
Patterns of Osmotic-Specific Phosphorylation Changes Define a Rapid Dehydration-Associated Molecular Phenotype
Our primary interest in this investigation is the quantification of protein phosphorylation events that are selectively responsive to osmotic stress, as they provide insight to cellular processes involved in early signaling and adaptation. From the 21 mannitol-responsive phosphopeptides selected from untargeted data, 19 showed reproducible mannitol-induced phosphorylation (Table I) and 11 possessed a relatively osmotic-specific response and cluster together in our phosphorylation heat map (Fig. 6A). This phosphorylation pattern is characterized as a rapid dehydration-associated molecular phenotype and includes the phosphorylation of VCS, bZIP30, and RAF18 proteins, which were described previously in our untargeted data. In addition to RAF18, another protein in the MAPK superfamily appears highly responsive to osmotic perturbation: MAP4Kα1. MAP4Kα1 is one of three mannitol-responsive MAP4Ks identified in this study (Supplemental Table S1) that belong to the poorly characterized MAP4K 10-member gene family in Arabidopsis (Champion et al., 2004). In yeast (Saccharomyces cerevisiae), the MAP4K Ste20 functions downstream of the osmosensor Sho1 to activate the Hog1 MAPK cascade responsible for osmotic adaptation (Bahn, 2008). It is unknown if a similar osmotic stress MAP4K signaling hierarchy is active in plants.
In addition to MAPK family members, proteins involved in cytoskeleton restructuring and phospholipid production cluster in the osmotic-responsive phosphorylation group. These proteins are the microtubule subunit TUA and a protein involved in phosphatidylinositol bisphosphate production, Vac14.
Tubulin Phosphorylation
We observed approximately 2-fold increases in phosphorylation of the TUA proteins TUA3 and TUA4 at Thr-349 in response to NaCl and mannitol treatment. Recently, TUA phosphorylation at this residue was shown to influence overall microtubule stability (Fujita et al., 2013). This previous work showed that TUA phosphorylation occurred under osmotic stress conditions and resulted in a polymerization-incompetent protein isoform that contributed to microtubule destabilization and depolymerization (Ban et al., 2013; Fujita et al., 2013). This finding was significant because microtubule depolymerization and restructuring are known to occur in response to several abiotic stresses (Nick, 2008; Wang et al., 2011) and are required for seedling survival under salt stress (Wang et al., 2007), but the mechanistic details underlying stress-induced depolymerization were previously unknown. Our data support these recent findings and provide further description of rapid TUA phosphorylation (Fig. 6A).
Vac14 Phosphorylation
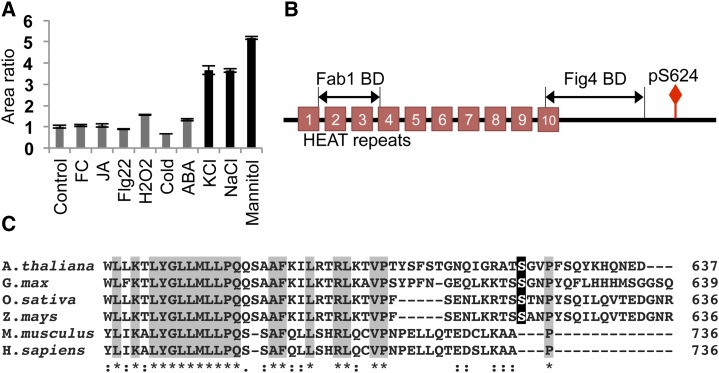
The largest phosphorylation increase observed in our discovery data (approximately 12.5-fold) was of Vac14, a protein involved in the synthesis of the phospholipid phosphatidylinositol 3,5-bisphosphate [PI(3,5)P2]. Vac14 is a scaffold protein that holds together a phosphatidylinositol 3-phosphate 5-kinase Fab1/PIKfyve, and a PI(3,5)P2 phosphatase, Fig4/Sac3, in a conserved complex responsible for PI(3,5)P2 production that is found in all eukaryotes (Dove and Johnson, 2007). The coordination of lipid kinase and phosphatase within the same complex speaks to the highly controlled nature of PI(3,5)P2 metabolism. Although Vac14 function has yet to be described in plants, our data indicate that it is highly responsive to osmotic perturbations (Fig. 7A). PI(3,5)P2 levels are known to rapidly increase in response to osmotic stress in plants, green algae, and yeast (Meijer et al., 1999; Bonangelino et al., 2002; Zonia and Munnik, 2004), with as much as 20-fold induction within 5 min of treatment in Saccharomyces cerevisiae (Duex et al., 2006). Interestingly, Vac14 is required for stress-induced PI(3,5)P2 production in yeast (Bonangelino et al., 2002), although the details regarding its regulation are unknown. The protein sequence of Vac14 is highly conserved from yeast to human, but phosphorylation occurs at a variable C-terminal region of the protein (Fig. 7B). This phosphorylation site is not present in yeast and mammals and is conserved in land plants (Fig. 7C). These data indicate the presence of a Vac14 phosphoregulation mechanism involved in the hyperosmotic response that is unique to plants.
Figure 7.
Vac14 phosphorylation. A, Vac14 phosphorylation is highly responsive to osmotic stress. Black bars represent osmotic stress-inducing conditions. Error bars represent se for three biological replicates. FC, Fusicoccin; JA, methyl jasmonate. B, Vac14 protein domain structure. Phosphorylation occurs at a variable C-terminal region. BD, Binding domain. C, Alignment of the C-terminal Vac14 protein sequences using Clustal Omega. The plant-specific phosphorylation site is highlighted in black. Gray highlights and asterisks indicate identical residues, and colons indicate highly similar residues.
Time-Course Analysis of Osmotic-Responsive Proteins
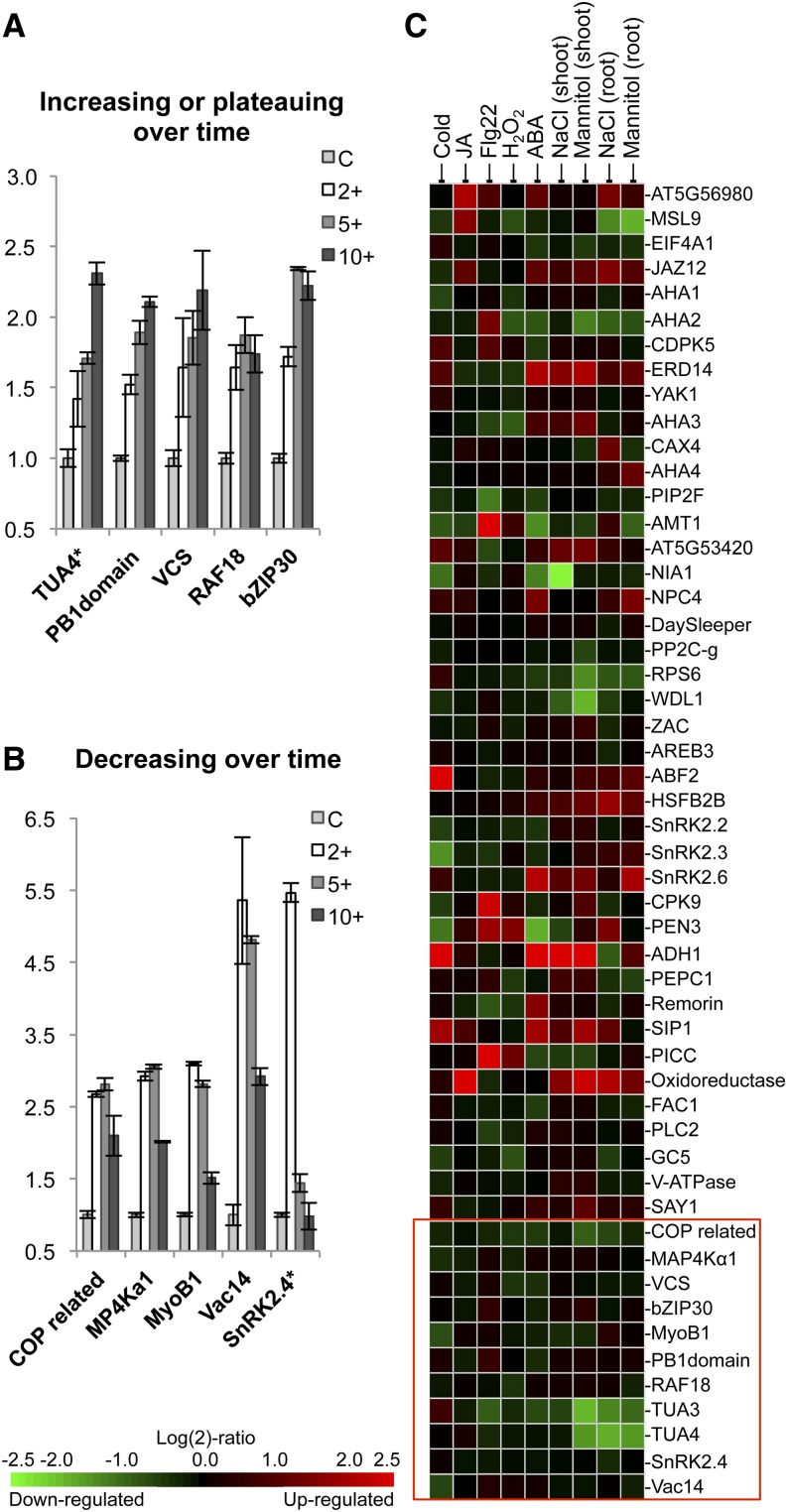
To better understand the temporal nature of osmotic-induced phosphorylation, we preformed a time-course experiment measuring the response to 2, 5, and 10 min of mannitol treatment using SRM. Half of the osmotic-block proteins displayed increasing or plateauing phosphorylation over the entire time period (Fig. 8A), while the other half showed a peak at earlier time points followed by a dropoff at 10 min (Fig. 8B). Vac14, SnRK2.4, and MyoB1, a plant-specific myosin adaptor protein recently shown to be involved in cytoplasmic streaming (Peremyslov et al., 2013), displayed maximal phosphorylation at 2 min. The rapid phosphorylation of SnRK2.4 is consistent with previous in vitro kinase activity results that showed rapid and transient SnRK2.4 activation following salt and mannitol treatment of Arabidopsis roots (McLoughlin et al., 2012). Interestingly, the phosphorylation profile of Vac14 over time closely mirrors stress-induced PI(3,5)P2 production in yeast and Chlamydomonas reinhardtii, which peaks at 5 min and is reduced by 15 min following treatment (Meijer et al., 1999; Duex et al., 2006).
Figure 8.
Phosphorylation time course and gene expression analysis of phosphoproteins used in our targeted proteomics experiment. A and B, SRM measurement of phosphorylation induced over a 0.3 m mannitol treatment time course. Phosphopeptides analyzed belong to the osmotic-specific group in Figure 6A. Error bars represent se for three biological replicates. Asterisks indicate nonunique peptides that match multiple gene family members. C, mRNA expression analysis of genes used in the targeted proteomics study. Gene expression data were analyzed using Genevestigator. mRNA was collected from whole seedlings unless indicated otherwise. Microarray experimental details are found in Supplemental Table S5. The red box contains proteins found in the osmotic-specific phosphorylation block in Figure 6A.
Transcription Data Do Not Identify Phosphoproteins Responsive to Osmotic Stress
To determine if gene expression data can be used to predict proteins undergoing stress-responsive phosphorylation, we analyzed transcriptomics data from available Affymetrix arrays using Genevestigator (Zimmermann et al., 2004). Experiments using whole-seedling and earlier time points (3 h or less) were selected when possible. When minimal to no response was observed at early time points, later sampling values were selected (24 h or less). A complete list of selected microarray experiments is found in Supplemental Table S5. Heat-map arrangement of expression data in the same orientation as phosphopeptide clustering reveals minimal overlap between protein phosphorylation and mRNA measurements (Fig. 8C). This is particularly true for osmotic-block genes, which show no salt- or mannitol-induced changes in gene expression, with the exception of TUA and the unknown protein Constitutive photomorphogenesis protein1-interacting related. Expanding this analysis to all available experiments related to salt, osmotic, and drought stress reveals a similar trend of minimal to no correlation of the phosphorylation and transcriptional responses (Supplemental Fig. S2). However, it is possible that with some genes, the chip-based measurements are misrepresenting the in vivo transcriptional response. For example, increases in MAP4Kα1 gene expression during salt and osmotic stress have been observed previously using real-time quantitative PCR (Charrier et al., 2002), although no response is seen in array data. Nevertheless, genes implicated in stress responses are significantly different in phosphorylation studies compared with expression analyses, indicating that the rapid protein phosphorylation response to dehydration is distinct from the slower molecular phenotypes associated with mRNA changes.
DISCUSSION
The work described in this study provides new insights into cellular processes involved in the early osmotic stress response. Using targeted MS analysis of Arabidopsis seedlings, we compared protein phosphorylation across nine different perturbations to elucidate a response network specific to osmotic stress. This network includes MAP4K and MAP3K family proteins, bZIP transcription factors, 5′ mRNA decapping proteins, and Vac14. Phosphorylation of these targets suggests new points of regulation for stress-induced posttranscriptional reprograming and phosphatidylinositol synthesis. Proteins implicated in the osmotic response through our analysis did not show stress-induced changes in gene expression, demonstrating that phosphoproteomic signaling is an important orthogonal approach to understanding the stress response.
In our untargeted analysis, we identified 79 proteins responsive to osmotic stress with phosphorylation changes of 1.5-fold or greater following 5 min of treatment (Supplemental Table S1). From this list, 21 phosphopeptides were selected for validation and characterization using targeted SRM analysis. The reproducibility of the discovery data was high, with all phosphopeptides showing similar responses except ADH1 pS229, PIP2F pS283, and oxidoreductase pS29 (Table I). These phosphopeptides likely represent false positives in our discovery data sets, as quantification was based on only two biological replicates in which over 1,200 phosphopeptides were quantified. It is possible that the original phosphorylation changes identified for ADH1 and oxidoreductase were the result of differential protein abundance rather than phosphorylation change, as no additional peptides were identified for either of these proteins; thus, no conclusion can be made regarding protein levels in each sample. The inability to comprehensively address protein abundance is an inherent limitation of phosphopeptide enrichment. This fact underscores the importance of rapid phosphorylation analysis to observe phosphorylation responses before the onset of significant changes in protein synthesis/breakdown. Our data set was collected following 5 min of mannitol treatment, but SRM time-course data reveal that some phosphopeptides show the highest response to mannitol after only 2 min of treatment. This suggests a need to sample at more immediate time points, as important phosphorylation responses may have been missed at 5 min.
Hierarchical clustering was applied to SRM data to compare phosphorylation responses across different perturbations. The high sensitivity and throughput of SRM made routine measurements of phosphopeptides possible. This is an analysis that would be difficult to perform using standard untargeted methods because of the low reproducibility of phosphopeptide detection. From our analysis, we identified 11 proteins displaying relatively osmotic-specific phosphorylation responses. These proteins give insight into several cellular processes known to be involved in the early stress response as well as highlight new processes that may play important roles.
The poor correlation of transcript levels with protein abundance following salt treatment (Jiang et al., 2007) and the major readjustment of transcripts away from active polysomes during dehydration stress (Kawaguchi et al., 2004) indicate that mechanisms of posttranscriptional regulation are activated during early stages of the dehydration stress response (Deyholos, 2010; Sunkar et al., 2012). Our data identifying VCS, VCR, and DCP2 phosphorylation increases following mannitol treatment suggest that 5′ mRNA decapping is altered during osmotic stress. Targeted analysis of VCS phosphorylation indicates that this response is specific to hyperosmotic conditions and increases with treatment duration. VCS functions as a scaffold protein for the decapping proteins DCP1 and DCP2. These proteins localize to processing bodies, which are aggregations of mRNA and proteins associated with transcript repression and degradation that increase following stress treatment (Franks and Lykke-Andersen, 2008). It is unclear what regulates 5′ decapping complex assembly and processing body formation, although some evidence exists suggesting that phosphorylation plays an important role. In yeast, phosphorylation of DCP2 by MAP4K Ste20 increases the DCP2 association with processing bodies (Yoon et al., 2010). In plants, DCP1 is phosphorylated by MAPK6 in response to dehydration stress, and work with phosphomimetic mutants suggests that phosphorylation enhances 5′ mRNA decapping (Xu and Chua, 2012). Similar phosphoregulation of VCS has not yet been described, although it has recently been shown to be phosphorylated in response to 30 min of mannitol treatment in Arabidopsis (Xu and Chua, 2012). It is possible that stress-induced VCS phosphorylation modulates VCS activity in a way that promotes decapping complex assembly and association with processing bodies to control posttranscriptional reprogramming during dehydration stress.
Our data highlight another phenomenon associated with the osmotic stress response: rapid accumulation of the phospholipid species PI(3,5)P2. In yeast, PI(3,5)P2 is predominantly localized to the vacuole, where it is required for proper vacuole function and morphology. Osmotic stress causes increased PI(3,5)P2 production, and these increases are essential for vacuolar volume adjustment (Dove et al., 1997; Bonangelino et al., 2002; Ho et al., 2012). Vac14 is a scaffold protein for PI(3,5)P2 synthesis and is required for this stress-induced PI(3,5)P2 production (Bonangelino et al., 2002). Our finding of Vac14 phosphorylation following osmotic perturbation suggests a point of regulation in the PI(3,5)P2 accumulation process. It is possible that phosphorylation mediates complex assembly with PI3P-5-kinase Fab1 during the stress response. Recent work disrupting Fab1 function in plants reveals that PI(3,5)P2 production is required for proper stomata closure in response to ABA (Bak et al., 2013). PI(3,5)P2 is also required for polarized cell growth and influences actin filament rearrangement in moss (van Gisbergen et al., 2012). Thus, PI(3,5)P2 levels have been linked directly to growth- and drought-related processes. Our identification of Vac14 phosphorylation changes is a starting point toward understanding how this protein involved in PI(3,5)P2 production is regulated in plants.
In addition to fortifying our understanding of processes associated with the stress response, some of the untargeted and targeted proteomic results indicate new pathways that may be important for osmotic adaptation. Several family members of a recently characterized group of myosin-binding proteins (MyoB; Peremyslov et al., 2013) were shown to undergo increased phosphorylation in response to mannitol in our untargeted analysis (Supplemental Table S1). These proteins act as plant-specific myosin adaptors that anchor vesicles involved in cytoplasmic streaming to high-speed myosin XI motors (Peremyslov et al., 2013). Our data indicate that MyoB1 is phosphorylated specifically in response to osmotic stress and shows the highest degree of phosphorylation at 2 min. Interestingly, rates of cytoplasmic streaming have recently been identified as a major determinate of plant growth (Tominaga et al., 2013). Little is known about cytoplasmic streaming in the context of the stress response, and it is possible that MyoB1 phosphorylation represents a connection between this important growth-controlling process and osmotic stress.
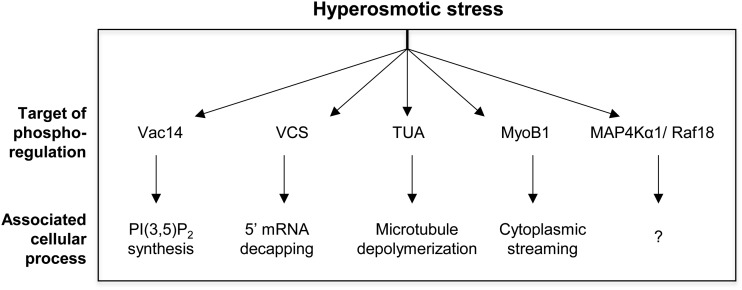
In conclusion, this study demonstrates the use of targeted proteomic analysis as a reliable method for following up on untargeted proteomic experiments and examining rapid protein phosphorylation across multiple treatment conditions to elucidate stress-specific signaling networks. Our results highlight points of both cross talk and specificity in dehydration-induced phosphorylation. We identified several proteins experiencing osmotic stress-induced phosphoregulation (Figure 9) that provide insight into the cellular mechanisms occurring during the initial stress response.
Figure 9.
Summary of phosphorylation targets during the initial osmotic stress response.
MATERIALS AND METHODS
Plant Growth and Sample Preparation
Wild-type Arabidopsis (Arabidopsis thaliana) Columbia-0 seeds were grown in liquid culture under constant light for 10 to 11 d before experimental treatment and protein extraction. For untargeted proteomic experiments using full metabolic labeling, samples were processed as described previously (Minkoff et al., 2014). Briefly, plants were grown in modified Murashige and Skoog medium containing 1% (w/v) Suc, 0.05% (w/v) MES salt, and either ammonium and potassium nitrate or 15N-enriched ammonium nitrate and potassium nitrate (more than 98% 15N; Cambridge Isotope Laboratories) as the sole nitrogen source. After 10 d, mannitol treatment was applied by decanting the existing medium and replacing with control medium or medium containing 0.3 m mannitol. After 5 min of treatment, plant samples were flash frozen and homogenized. Samples were then combined in an experimental pair consisting of one treated sample grown in 14N medium and one control sample grown in 15N medium. For the second reciprocal experimental pair, the samples were combined in the inverse fashion (15N treated and 14N control). Ground frozen tissue was combined at a 1:1 weight ratio prior to further homogenization. For targeted proteomic experiments, sample medium consisted of one-half-strength Murashige and Skoog salts, 1% (w/v) Suc, and 0.05% (w/v) MES salt. After 11 d of growth, sample medium was replaced with fresh medium and allowed to equilibrate for 4 to 5 h prior to experimental treatment. Treatment was applied in the same manner as described for the untargeted experiments. Three biological replicates were processed for each treatment condition. For all experiments, samples were further homogenized in grinding buffer (Huttlin et al., 2007) supplemented with phosphatase inhibitors using a sonicator (1-cm probe, 5 × 10 s, and 50% duty cycle) while kept on ice. The resulting supernatant was filtered through two layers of Miracloth (Calbiochem) and underwent a soft spin (1,500g, 20 min, and 4°C) to remove unbroken cells.
Sample Prefractionation for Untargeted MS Analysis
Sample supernatants from experimental pairs 1 and 2 were further centrifuged at 100,000g for 90 min at 4°C (Beckman Ultracentrifuge) to isolate the microsome pellet. The microsome pellet was resuspended in 50 mm Tris-HCl, pH 8, and proteins were extracted from both the microsome resuspension and soluble fraction using a previously described methanol/chloroform/water method (Wessel and Flügge, 1984; Minkoff et al., 2014). Precipitated proteins were solubilized in 8 m urea-containing phosphatase inhibitor mixture (1× PhosStop; Roche). Samples were diluted to 2 m urea, and protein concentration was measured using a bicinchoninic acid assay kit (BCA, Thermo Scientific Pierce). Five milligrams of protein from microsome and soluble fractions was reduced with 5 mm dithiothreitol (45 min at 50°C) and alkylated using 15 mm iodoacetamide (45 min at room temperature). Samples were then diluted to 1.2 m urea using 50 mm NH4HCO3 and digested with trypsin (Promega) at a 1:100 enzyme:protein ratio overnight at 37°C. Samples were acidified using 0.5% (v/v) formic acid to stop enzymatic digestion and desalted using C-18 solid-phase extraction columns (Waters). Soluble fractions were then subject to further fractionation via SCX chromatographic separation using a polysulfoethyl aspartamide column (4.6 × 200 mm; PolyLC) on a Waters Alliance 2795 HPLC device. Separation was achieved using a 0% to 25% (v/v) buffer B (5 mm KH2PO4, 350 mm KCl, and 30% [v/v] acetonitrile [ACN], pH 2.65) gradient over 33 min at a flow rate of 3 mL min−1. The gradient was followed by a 100% buffer B wash and reequilibration with buffer A (5 mm KH2PO4 and 30% [v/v] ACN, pH 2.65). Independent fractions were collected every 4 min, and blanks were run between samples to ensure no carryover. Sample fractions were flash frozen, lyophilized, and desalted using solid-phase extraction. SCX fractions 3 to 8 and microsome samples were enriched for phosphopeptides using titanium dioxide (TiO2) particles (5 μm; GL Sciences) as described previously (Sugiyama et al., 2007; Minkoff et al., 2014).
Untargeted MS and Data Analysis
Phosphopeptide-enriched samples were analyzed on an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific) using the same method as described by Kline et al. (2010). Briefly, samples were separated by HPLC using an 11-cm C-18 packed analytical column and a 240-min ACN gradient in line with MS analysis. Two to three injection replicates were performed for each sample. Acquired data files containing MS/MS spectra were searched against The Arabidopsis Information Resource 9 protein database using MASCOT software (Matrix Science). Search parameters were set at two allowed missed cleavages, precursor and fragment ion mass tolerances of 15 ppm and 0.6 D, respectively, fixed modifications of Cys carbamidomethylation, and variable modifications of Met oxidation, Ser/Thr/Tyr phosphorylation, and Asn/Gln deamidation. Searches were performed using settings for both 14N and 15N protein masses. MASCOT search results were filtered to maintain a 1% false discovery rate at the peptide level using a reverse-protein sequence database and in-house software. The software determines the minimum MASCOT score in which 1% of MS/MS peptide identifications match the decoy reverse-protein sequence database. All peptide identifications scoring lower than this calculated MASCOT score were excluded from analysis. Quantitative ratio measurements from MS1 peak areas were performed using Census software (Park et al., 2008) as described by Kline et al. (2010). To correct for errors in mixing ratios (i.e. initial combination of 14N and 15N ground, frozen tissue), all data sets for each experimental sample were normalized to the median 14N-15N area ratio value. Only phosphopeptides quantified in both experiments 1 and 2 were considered for analysis. Phosphopeptides showing reciprocal changes of 1.5-fold or greater were manually validated by visual inspection of Census chromatograms. MS/MS spectra were manually validated for all phosphopeptides in Supplemental Tables S1 and S3 and can be found in Supplemental Figure S1.
Targeted SRM Analysis
Proteins were precipitated from sample supernatant with 80% (v/v) acetone overnight at −20°C and then resuspended in 2% (w/v) SDS-Tris-HCl buffer prior to methanol/chloroform/water protein extraction. This initial precipitation step was added to reduce sample volume. Extracted protein was resuspended and quantified by a bicinchoninic acid assay kit as described for the untargeted sample preparation above. For each sample, 3 mg of protein was spiked with isotopically labeled phosphopeptide standards, synthesized by the Sigma-Aldrich PEPscreen platform, Sigma-Aldrich AQUA, or the University of Wisconsin-Madison Biotechnology Center’s peptide synthesis core facility. Phosphopeptide standards were pooled into a master mix, divided into aliquots, and frozen. Each batch of samples received a spike from the same phosphopeptide standard master mix aliquot to avoid the variability that may arise from peptide loss due to freeze-thaw cycles or retention in plastic tubes. The concentration of each phosphopeptide standard in the master mix was individually tailored to generate optimum signal levels during SRM analysis. Calculations were based on empirically determined chromatographic and mass spectrometric behavior of the phosphopeptide standards. Samples were reduced with 5 mm dithiothreitol (45 min at 50°C) and alkylated using 15 mm iodoacetamide (45 min at room temperature). Samples were then diluted to 1.2 m urea using 50 mm NH4HCO3 and digested with trypsin (Promega) at a 1:100 enzyme:protein ratio overnight at 37°C. Samples were acidified using 0.5% (v/v) formic acid to stop enzymatic digestion and desalted using C-18 solid-phase extraction columns (Waters). Phosphopeptide enrichment was performed using homemade TiO2 columns containing 2.4 mg of TiO2 particles (5 μm; GL Sciences), as described by Minkoff et al. (2014).
Phosphopeptide quantitation was performed using the Eksigent NanoLC-Ultra 2D system with the cHiPLC nanoflex microfluidic C18 column (75 μm, 120 Å) coupled to the AB SCIEX 5500 QTRAP mass spectrometer. SRM method development was performed using peptide standards and MRMPilot software (AB SCIEX). Multiple fragment ions were tested for each peptide using both in silico predictions and acquired MS/MS spectra. The three to five most abundant parent ion-to-fragment ion transitions were selected, and collision energies were optimized to maximize fragment ion intensity. The developed method was pilot tested, and the number of transitions for each peptide was reduced to three through the removal of the lowest quality transitions that performed poorly in complex sample backgrounds. In the final method, three transitions per peptide were monitored using scheduled data acquisition with a target scan time of 3.1 s and a detection window of 170 s. A list of SRM transitions used for each endogenous peptide and peptide standard can be found in Supplemental Table S3. Analytical separation was performed using a linear gradient of 2% to 35% (v/v) buffer B (ACN and 0.1% [v/v] formic acid; Honeywell Burdick and Jackson) over 70 min at a flow rate of 300 nL min−1. The gradient was followed by a 90% (v/v) buffer B wash and reequilibration with buffer A (0.1% [v/v] formic acid; Honeywell Burdick and Jackson). Peak areas were integrated using the automatic MQ4 function in MultiQuant software (AB SCIEX). Peak integration parameters were set as follows: 40% noise, a baseline subtraction window of 0.5 min, and peak splitting of 0 to 2 points with a Gaussian smooth width of 0.8 to 1 point. In all cases, identical integration parameters were used for standard and endogenous peptide area quantification. To determine the levels of endogenous peptide in each sample, the peak areas for endogenous peptides were normalized to the peak areas for the corresponding peptide standard for each transition within each run. Transitions that had low signal intensity or contained isobaric interference were not used for quantification and are indicated in Supplemental Table S3. For each treatment condition, peptides were quantified from two to four injection replicates for three independent biological replicates. To determine phosphorylation changes, standard-normalized endogenous phosphopeptide abundances were first averaged across biological replicates and then compared across treated and control conditions as a ratio.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaino et. al, 2010) via the Proteomics Identification Database partner repository with the dataset identifier PXD001057.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. MS/MS spectra for phosphorylation site validation of all phosphopeptides listed in Table I and Supplemental Tables S1 and S2.
Supplemental Figure S2. Extended gene expression analysis of osmotic-responsive phosphoproteins.
Supplemental Table S1. Mannitol-responsive phosphopeptides showing 1.5-fold or greater change in phosphorylation following 5 min of treatment.
Supplemental Table S2. Proteins with secondary peptide identifications for quantifying protein abundance.
Supplemental Table S3. SRM methods for the targeted analysis of 58 phosphopeptides.
Supplemental Table S4. SRM phosphorylation data for the nine experimental conditions.
Supplemental Table S5. Experimental details for gene expression studies used in Genevestigator mRNA analysis.
Supplementary Material
Glossary
- ROS
reactive oxygen species
- MAPK
mitogen-activated protein kinase
- ABA
abscisic acid
- AREB
ABA-responsive element
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
- LC
liquid chromatography
- SCX
strong cation exchange
- SRM
selective reaction monitoring
- PAMP
pathogen-associated molecular pattern
- H2O2
hydrogen peroxide
- PI(3,5)P2
phosphatidylinositol 3,5-bisphosphate
- ACN
acetonitrile
- TiO2
titanium dioxide
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–0929395 and DBI-0701486 to M.R.S. and grant no. DGE–1256259 to K.E.S.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Bahn YS. (2008) Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot Cell 7: 2017–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak G, Lee EJ, Lee Y, Kato M, Segami S, Sze H, Maeshima M, Hwang JU, Lee Y. (2013) Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell 25: 2202–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban Y, Kobayashi Y, Hara T, Hamada T, Hashimoto T, Takeda S, Hattori T. (2013) α-Tubulin is rapidly phosphorylated in response to hyperosmotic stress in rice and Arabidopsis. Plant Cell Physiol 54: 848–858 [DOI] [PubMed] [Google Scholar]
- Bonangelino CJ, Nau JJ, Duex JE, Brinkman M, Wurmser AE, Gary JD, Emr SD, Weisman LS. (2002) Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J Cell Biol 156: 1015–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Sheen J. (2013) CDPKs in immune and stress signaling. Trends Plant Sci 18: 30–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320: 1185–1190 [DOI] [PubMed] [Google Scholar]
- Champion A, Picaud A, Henry Y. (2004) Reassessing the MAP3K and MAP4K relationships. Trends Plant Sci 9: 123–129 [DOI] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M. (2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130: 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Sung SJ, Kim BG, Pandey GK, Cho JS, Kim KN, Luan S. (2010) Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol Cells 29: 159–165 [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos MK. (2010) Making the most of drought and salinity transcriptomics. Plant Cell Environ 33: 648–654 [DOI] [PubMed] [Google Scholar]
- Dodd AN, Kudla J, Sanders D. (2010) The language of calcium signaling. Annu Rev Plant Biol 61: 593–620 [DOI] [PubMed] [Google Scholar]
- Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. (1997) Osmotic stress activates phosphatidylinositol-3,5-bisphosphate synthesis. Nature 390: 187–192 [DOI] [PubMed] [Google Scholar]
- Dove SK, Johnson ZE. (2007) Our FABulous VACation: a decade of phosphatidylinositol 3,5-bisphosphate. Biochem Soc Symp 129–139 [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T. (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duex JE, Nau JJ, Kauffman EJ, Weisman LS. (2006) Phosphoinositide 5-phosphatase Fig 4p is required for both acute rise and subsequent fall in stress-induced phosphatidylinositol 3,5-bisphosphate levels. Eukaryot Cell 5: 723–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J. (2008) The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz S, Ehlert B, Liese A, Kurth J, Cazalé AC, Romeis T. (2011) Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana. Mol Plant 4: 83–96 [DOI] [PubMed] [Google Scholar]
- Fricke W, Akhiyarova G, Wei W, Alexandersson E, Miller A, Kjellbom PO, Richardson A, Wojciechowski T, Schreiber L, Veselov D, et al. (2006) The short-term growth response to salt of the developing barley leaf. J Exp Bot 57: 1079–1095 [DOI] [PubMed] [Google Scholar]
- Fujii H, Verslues PE, Zhu JK. (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc Natl Acad Sci USA 108: 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhu JK. (2009) Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA 106: 8380–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S, Pytela J, Hotta T, Kato T, Hamada T, Akamatsu R, Ishida Y, Kutsuna N, Hasezawa S, Nomura Y, et al. (2013) An atypical tubulin kinase mediates stress-induced microtubule depolymerization in Arabidopsis. Curr Biol 23: 1969–1978 [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sabat G, Stecker K, Minkoff BB, Sussman MR. (2014) A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M, Sussman MR. (2012) The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiol 158: 1158–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Alghamdi TA, Botelho RJ. (2012) Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic 13: 1–8 [DOI] [PubMed] [Google Scholar]
- Hohmann S. (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66: 300–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Hegeman AD, Harms AC, Sussman MR. (2007) Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Mol Cell Proteomics 6: 860–881 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Tena G, Henry Y, Zhang Z, Hirt H, Wilscon C, Morris P, Mundy J, Innes R, Ecker JR. (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7: 301–308 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7: 106–111 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yang B, Harris NS, Deyholos MK. (2007) Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J Exp Bot 58: 3591–3607 [DOI] [PubMed] [Google Scholar]
- Jinek M, Eulalio A, Lingel A, Helms S, Conti E, Izaurralde E. (2008) The C-terminal region of Ge-1 presents conserved structural features required for P-body localization. RNA 14: 1991–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E. (2013) The role of disordered protein regions in the assembly of decapping complexes and RNP granules. Genes Dev 27: 2628–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Girke T, Bray EA, Bailey-Serres J. (2004) Differential mRNA translation contributes to gene regulation under non-stress and dehydration stress conditions in Arabidopsis thaliana. Plant J 38: 823–839 [DOI] [PubMed] [Google Scholar]
- Kline KG, Barrett-Wilt GA, Sussman MR. (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci USA 107: 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MN, Jane WN, Verslues PE. (2013) Role of the putative osmosensor Arabidopsis histidine kinase1 in dehydration avoidance and low-water-potential response. Plant Physiol 161: 942–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin F, Galvan-Ampudia CS, Julkowska MM, Caarls L, van der Does D, Laurière C, Munnik T, Haring MA, Testerink C. (2012) The Snf1-related protein kinases SnRK2.4 and SnRK2.10 are involved in maintenance of root system architecture during salt stress. Plant J 72: 436–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HJ, Divecha N, van den Ende H, Musgrave A, Munnik T. (1999) Hyperosmotic stress induces rapid synthesis of phosphatidyl-D-inositol 3,5-bisphosphate in plant cells. Planta 208: 294–298 [Google Scholar]
- Minkoff BB, Burch HL, Sussman MR. (2014) A pipeline for 15N metabolic labeling and phosphoproteome analysis in Arabidopsis thaliana. Methods Mol Biol 1062: 353–379 [DOI] [PubMed] [Google Scholar]
- Munnik T (2014) PI-PLC: phosphoinositide-phospholipase C in plant signaling. In X Wang, ed, Phospholipases in Plant Signaling. Signaling and Communication in Plants, Vol. 20. Springer-Verlag, Berlin, pp 27–54 [Google Scholar]
- Munnik T, Vermeer JE. (2010) Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant Cell Environ 33: 655–669 [DOI] [PubMed] [Google Scholar]
- Nick P (2008) Microtubules as sensors for abiotic stimuli. In P Nick, ed, Plant Microtubules, Vol 11. Springer, Berlin, pp 175–203 [Google Scholar]
- Nühse TS, Bottrill AR, Jones AM, Peck SC. (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J 51: 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson A, Svennelid F, Ek B, Sommarin M, Larsson C. (1998) A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol 118: 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Venable JD, Xu T, Yates JR., III (2008) A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods 5: 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peremyslov VV, Morgun EA, Kurth EG, Makarova KS, Koonin EV, Dolja VV. (2013) Identification of myosin XI receptors in Arabidopsis defines a distinct class of transport vesicles. Plant Cell 25: 3022–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MC, Petersen M, Mundy J. (2010) Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol 61: 621–649 [DOI] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 3: 1154–1169 [DOI] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, et al. (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J 424: 439–448 [DOI] [PubMed] [Google Scholar]
- Segonzac C, Zipfel C. (2011) Activation of plant pattern-recognition receptors by bacteria. Curr Opin Microbiol 14: 54–61 [DOI] [PubMed] [Google Scholar]
- Shabala S, Shabala L. (2011) Ion transport and osmotic adjustment in plants and bacteria. BioMolecular Concepts 2: 407–419 [DOI] [PubMed] [Google Scholar]
- Shabala SN, Lew RR. (2002) Turgor regulation in osmotically stressed Arabidopsis epidermal root cells: direct support for the role of inorganic ion uptake as revealed by concurrent flux and cell turgor measurements. Plant Physiol 129: 290–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhorst L, Kudla J. (2013) Calcium and reactive oxygen species rule the waves of signaling. Plant Physiol 163: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su SH, Bush SM, Zaman N, Stecker K, Sussman MR, Krysan P. (2013) Deletion of a tandem gene family in Arabidopsis: increased MEKK2 abundance triggers autoimmunity when the MEKK1-MKK1/2-MPK4 signaling cascade is disrupted. Plant Cell 25: 1895–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. (2007) Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics 6: 1103–1109 [DOI] [PubMed] [Google Scholar]
- Sunkar R, Li YF, Jagadeeswaran G. (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17: 196–203 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Fenske R, Castleden I, Tomaz T, Nelson CJ, Millar AH. (2014) Selected reaction monitoring to determine protein abundance in Arabidopsis using the Arabidopsis proteotypic predictor. Plant Physiol 164: 525–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tominaga M, Kimura A, Yokota E, Haraguchi T, Shimmen T, Yamamoto K, Nakano A, Ito K. (2013) Cytoplasmic streaming velocity as a plant size determinant. Dev Cell 27: 345–352 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JD, Dangl JL. (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104: 20623–20628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugama D, Liu S, Takano T. (2012) A bZIP protein, VIP1, is a regulator of osmosensory signaling in Arabidopsis. Plant Physiol 159: 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Takahashi F, Anderson JC, Ishihama Y, Peck SC, Shinozaki K. (2013) Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci Signal 6: rs8. [DOI] [PubMed] [Google Scholar]
- Urano K, Kurihara Y, Seki M, Shinozaki K. (2010) ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr Opin Plant Biol 13: 132–138 [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K. (1999) A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell 11: 1743–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gisbergen PA, Li M, Wu SZ, Bezanilla M. (2012) Class II formin targeting to the cell cortex by binding PI(3,5)P(2) is essential for polarized growth. J Cell Biol 198: 235–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosten MJ, Sharkhuu A, Batelli G, Bressan RA, Maggio A. (2013) The Arabidopsis thaliana mutant air1 implicates SOS3 in the regulation of anthocyanins under salt stress. Plant Mol Biol 83: 405–415 [DOI] [PubMed] [Google Scholar]
- Vizcaino JA, Cote R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L. (2010) The Proteomics Identifications database: 2010 update. Nucleic Acids Res 38: D736–D742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Li J, Yuan M. (2007) Salt tolerance requires cortical microtubule reorganization in Arabidopsis. Plant Cell Physiol 48: 1534–1547 [DOI] [PubMed] [Google Scholar]
- Wang C, Zhang L, Chen W. (2011) Plant cortical microtubules are putative sensors under abiotic stresses. Biochemistry (Mosc) 76: 320–326 [DOI] [PubMed] [Google Scholar]
- Ward JH Jr. (1963) Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58: 236–244 [Google Scholar]
- Weber C, Nover L, Fauth M. (2008) Plant stress granules and mRNA processing bodies are distinct from heat stress granules. Plant J 56: 517–530 [DOI] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Wohlbach DJ, Quirino BF, Sussman MR. (2008) Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell 20: 1101–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. (2002) Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Environ 25: 131–139 [DOI] [PubMed] [Google Scholar]
- Xu J, Chua NH. (2012) Dehydration stress activates Arabidopsis MPK6 to signal DCP1 phosphorylation. EMBO J 31: 1975–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue L, Wang P, Wang L, Renzi E, Radivojac P, Tang H, Arnold R, Zhu JK, Tao WA. (2013) Quantitative measurement of phosphoproteome response to osmotic stress in Arabidopsis based on Library-Assisted eXtracted Ion Chromatogram (LAXIC). Mol Cell Proteomics 12: 2354–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Choi EJ, Parker R. (2010) Dcp2 phosphorylation by Ste20 modulates stress granule assembly and mRNA decay in Saccharomyces cerevisiae. J Cell Biol 189: 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61: 672–685 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L, Munnik T. (2004) Osmotically induced cell swelling versus cell shrinking elicits specific changes in phospholipid signals in tobacco pollen tubes. Plant Physiol 134: 813–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonia L, Munnik T. (2007) Life under pressure: hydrostatic pressure in cell growth and function. Trends Plant Sci 12: 90–97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.