A wax locus negatively regulates ABA response within a small time window during seed germination and also affects ABA levels in seeds.
Abstract
The ECERIFERUM9 (CER9) gene encodes a putative E3 ubiquitin ligase that functions in cuticle biosynthesis and the maintenance of plant water status. Here, we found that CER9 is also involved in abscisic acid (ABA) signaling in seeds and young seedlings of Arabidopsis (Arabidopsis thaliana). The germinated embryos of the mutants exhibited enhanced sensitivity to ABA during the transition from reversible dormancy to determinate seedling growth. Expression of the CER9 gene is closely related to ABA levels and displays a similar pattern to that of ABSCISIC ACID-INSENSITIVE5 (ABI5), which encodes a positive regulator of ABA responses in seeds. cer9 mutant seeds exhibited delayed germination that is independent of seed coat permeability. Quantitative proteomic analyses showed that cer9 seeds had a protein profile similar to that of the wild type treated with ABA. Transcriptomics analyses revealed that genes involved in ABA biosynthesis or signaling pathways were differentially regulated in cer9 seeds. Consistent with this, high levels of ABA were detected in dry seeds of cer9. Blocking ABA biosynthesis by fluridone treatment or by combining an ABA-deficient mutation with cer9 attenuated the phenotypes of cer9. Whereas introduction of the abi1-1, abi3-1, or abi4-103 mutation could completely eliminate the ABA hypersensitivity of cer9, introduction of abi5 resulted only in partial suppression. These results indicate that CER9 is a novel negative regulator of ABA biosynthesis and the ABA signaling pathway during seed germination.
Seed germination is a critical stage in the life cycle of higher plants, during which the imbibed mature seed must shift rapidly from a state of quiescence to one of active metabolism. The transition from embryo dormancy to germination requires the activation of a series of developmental programs with the simultaneous mobilization of seed storage reserves and the loosening of the seed coat; these processes cumulate in radicle emergence, seedling establishment, and subsequent photoautotrophic growth (Bewley, 1997; Finkelstein et al., 2008; Holdsworth et al., 2008; Rajjou et al., 2012). The decision for a seed to germinate depends on certain intrinsic factors and many environmental factors, including water availability, temperature, light, and mineral availability. Among the internal factors, endogenous abscisic acid (ABA) and GA are two important regulators that also mediate interactions with these environmental factors in modulating seed germination. In fact, seed germination or dormancy largely depends on the ratio of these two hormones, which play antagonistic roles in seed germination. ABA inhibits seed germination, and mutants defective in ABA biosynthesis or signaling have enhanced germination efficiency and more easily break dormancy under osmotic stress. Conversely, GA promotes seed germination, and GA-deficient mutants show delayed seed germination (Finkelstein et al., 2008; Holdsworth et al., 2008; Rajjou et al., 2012).
Protein degradation through the ubiquitination pathway is an important mechanism in regulating hormone biosynthesis and signaling. To date, there are over 1,000 predicted E3 ubiquitin ligases in Arabidopsis (Arabidopsis thaliana). Based on the protein structure, these have been classified as, for example, Homologous to E6-AP carboxyl terminus, Really interesting new gene (RING), and U-box (Chen and Hellmann, 2013). Several E3 ligases were found to be involved in ABA signaling pathways and in the regulation of seed germination under various conditions. The RING E3 ligase ABI3-INTERACTING PROTEIN2 (AIP2) was reported to target ABSCISIC ACID-INSENSITIVE3 (ABI3) for degradation. In germination experiments, AIP2 loss of function and overexpression increased and decreased the sensitivity to ABA, respectively (Zhang et al., 2005). SALT- AND DROUGHT-INDUCED RING FINGER1 acts upstream of ABA RESPONSIVE ELEMENT-BINDING FACTOR3 (ABF3), ABF4, and ABI5 in ABA signaling and is a positive regulator of the ABA signaling pathway (Zhang et al., 2007). The RING E3 ligase RING-H2 protein RHA2a regulates ABA-mediated seed germination and early seedling development independently of ABI3, ABI4, and ABI5, these being the main regulators in the ABA signaling pathway in seeds (Bu et al., 2009). KEEP ON GOING (KEG), a multidomain ubiquitin E3 ligase, has been shown to regulate the levels of ABI5 and to act as a negative regulator of ABA signaling. The keg mutation was found to suppress the ABA-insensitive phenotype of ABI5 in a seed germination assay (Stone et al., 2006). In addition, Arabidopsis PROTEOLYSIS6, a ubiquitin recognin box-containing E3 ligase associated with the N-end rule pathway of protein degradation, controls seed ABA sensitivity and seedling establishment (Holman et al., 2009).
The Arabidopsis ECERIFERUM9 (CER9) gene encodes a putative RING domain-containing E3 ubiquitin ligase, which is similar to Doa10 in Saccharomyces cerevisiae, and is expected to target its protein substrates for degradation through the ubiquitination pathway (Lü et al., 2012; Doblas et al., 2013). Recent studies revealed a unique association of CER9 with both cuticular wax and cutin synthesis (Lü et al., 2012). Also identified as SUPPRESSOR OF DRY2 DEFECTS1, CER9 was suggested to be involved in the regulation of sterol biosynthesis in plants (Doblas et al., 2013). We report here that CER9 deficiency has a dramatic effect on plant ABA sensitivity during seed germination and early seedling growth, indicating that CER9 functions in the ABA response pathways. Genome-wide gene expression and proteomics analyses show that CER9 deficiency affects the abundance of gene transcripts and proteins related to ABA responses. We also show that the hypersensitivity of cer9 seed germination to ABA is due partially to the increased ABA levels found in the cer9 mutant and that the inhibition of seed germination imposed by the cer9 mutation requires an intact and functional ABA signaling pathway. These studies indicate that CER9 is a new negative regulator of the ABA response in seed germination and early seedling growth.
RESULTS
CER9 Deficiency Increases ABA Sensitivity during Seed Germination and Early Seedling Establishment
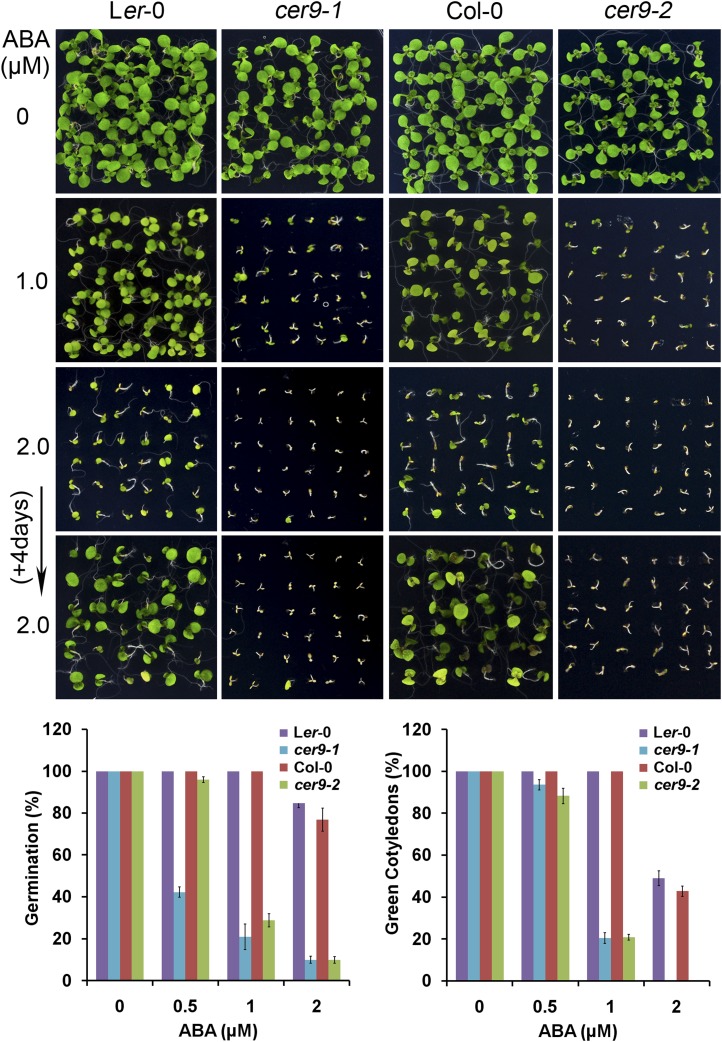
Previous studies showed that the cer9 mutant alters cuticle lipid deposition and whole-plant transpiration (Lü et al., 2012). Since ABA is an important regulator of plant water relations, we investigated whether other processes regulated by ABA are affected by cer9 mutations, including the response of cer9 to ABA during seed germination. As shown in Figure 1, both cer9-1 and cer9-2 were hypersensitive to ABA during seed germination and early seedling establishment. In an ABA dose-response assay, seeds of cer9-1 and cer9-2 were planted on ABA-free or ABA-containing medium and the percentage of seed germination at 3 d after the end of stratification was determined. In the absence of ABA, the seed germination percentages of different genotypes were found to be similar. However, in the presence of ABA, the germination rate of cer9 seeds was reduced dramatically compared with that of wild-type seeds (Fig. 1). Furthermore, cotyledon greenness, as scored at 7 d after the end of stratification, was also notably suppressed in cer9 (Fig. 1).
Figure 1.
The cer9 mutations increase the ABA inhibition of seed germination and postgermination growth. Seeds of cer9-1 (Ler-0 background) and cer9-2 (Col-0 background) along with their wild types were sown onto 1/2 MS agar medium supplemented with 0, 1, or 2 µm ABA. Representative images show the morphology of seedlings 7 d after imbibition. Germination (radicle emergence) percentages were determined 3 d and green cotyledons were scored 7 d after the imbibition. Data are means and sd from four biological replicates each with at least 30 seeds.
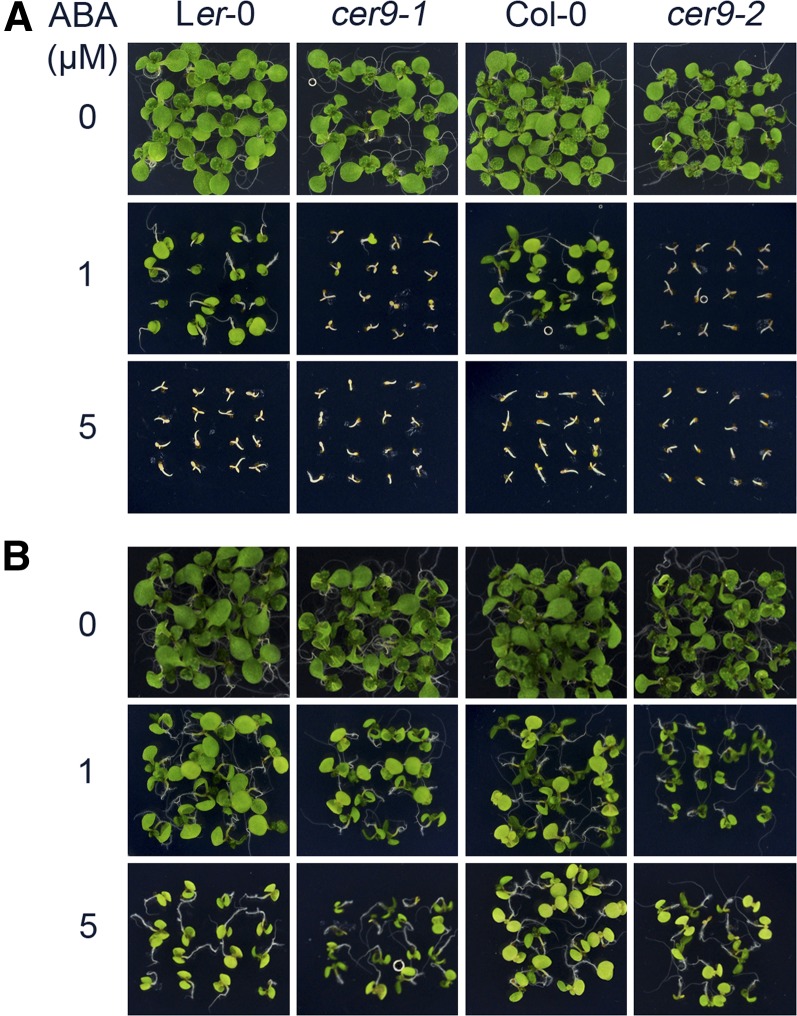
After seed germination (radicle emergence), there is a small time window during which it is determined whether a germinated embryo will remain dormant or make the transition to seedling growth. It is known that ABA also controls this transition. To determine whether CER9 is involved in this germination check-point control, we tested the response of germinating seeds to ABA. As shown in Figure 2A, both the cer9 mutant and the corresponding wild-type seeds were arrested by a higher concentration of ABA (5 µm) at 1 d after stratification. Whereas 1 µm ABA did not arrest the growth of the wild type, it was sufficient to arrest germination in cer9. This concentration of ABA no longer inhibited early seedling growth either for the wild type or cer9 at 2 d after stratification (Fig. 2B). These results indicate that CER9 is involved in the ABA response during the developmental transition from the embryo stage to the seedling stage.
Figure 2.
Seeds of the cer9 mutants are hypersensitive to ABA over a short development window. A, Seeds on 1/2 MS medium were stratified for 1 d and then transferred to 1/2 MS medium supplemented with 0, 1, or 5 μm ABA. Photographs were taken 6 d after the transfer. B, Seeds on 1/2 MS medium stratified for 2 d were transferred to 1/2 MS medium supplemented with 0, 1, or 5 μm ABA. Photographs were taken 5 d after the transfer.
CER9 Expression Is Regulated by ABA
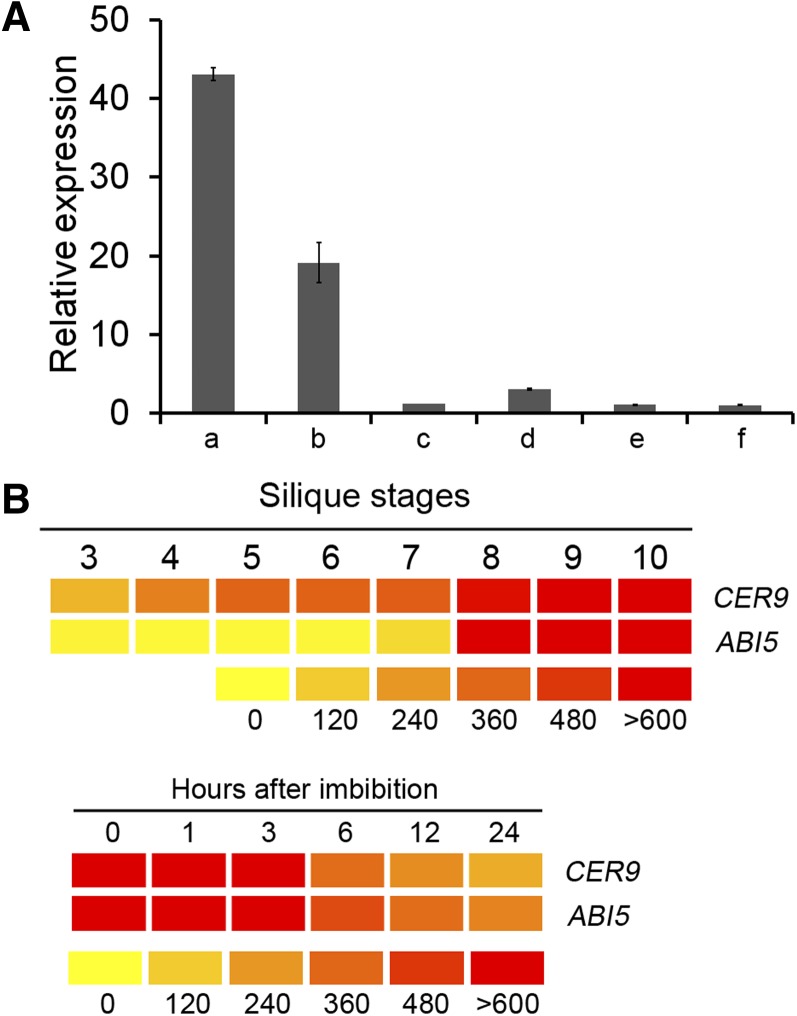
The involvement of CER9 in conditioning ABA sensitivity during seed germination and early seedling establishment prompted us to check whether CER9 is regulated by ABA. RNA was extracted from dry seeds, seeds imbibed for 72 h, and 1- and 3-d light-grown seedlings. The expression level of CER9 was examined using quantitative reverse transcription (qRT)-PCR (Fig. 3A). High levels of CER9 transcripts accumulated in dry seeds, but the level fell rapidly after 3 d of stratification and following germination. This pattern of expression coincides with the changes in the level of ABA, which is also high in dry seeds but decreases rapidly after stratification (Ali-Rachedi et al., 2004). To further demonstrate that CER9 expression was indeed induced by ABA, we also compared expression levels in ABA-treated and untreated 1-d light-grown seeds. The results revealed that CER9 expression was higher in ABA-treated seeds than in the untreated ones. We also compared the expression of CER9 with that of ABI5 as reported in public microarray data. ABI5 is a key regulator of the ABA response in seeds, and its expression is closely related to the relative abundance of ABA (Lopez-Molina et al., 2002). BAR HeatMapper analysis (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper.cgi) indicated that CER9 expression increases during seed maturation and decreases after imbibition, which is similar to the pattern for ABI5 (Fig. 3B). These results revealed a close relationship between CER9 expression and ABA levels in seeds and young seedlings. It was also shown that negative or positive factors coexist to fine-tune the ABA response in planta.
Figure 3.
Transcript abundance of CER9 and ABI5 during various developmental time periods. A, Real-time PCR analysis of CER9 in dry seeds (a) and seeds at the end of 3 d of stratification (b), seeds incubated on 1/2 MS medium for 1 d (c) or 3 d (e), or seeds incubated on 1/2 MS medium containing 1 µm ABA for 1 d (d) or 3 d (f) at 22°C after stratification. B, Expression of CER9 and ABI5 during seed maturation and imbibition. Public microarray data were processed using BAR HeatMapper (http://bar.utoronto.ca/ntools/cgi-bin/ntools_heatmapper.cgi) to visualize gene expression patterns. Higher expression is indicated by more reddish color. Numbers indicate the relative expression levels of the mRNAs.
cer9 Loss of Function Promotes Seed Dormancy
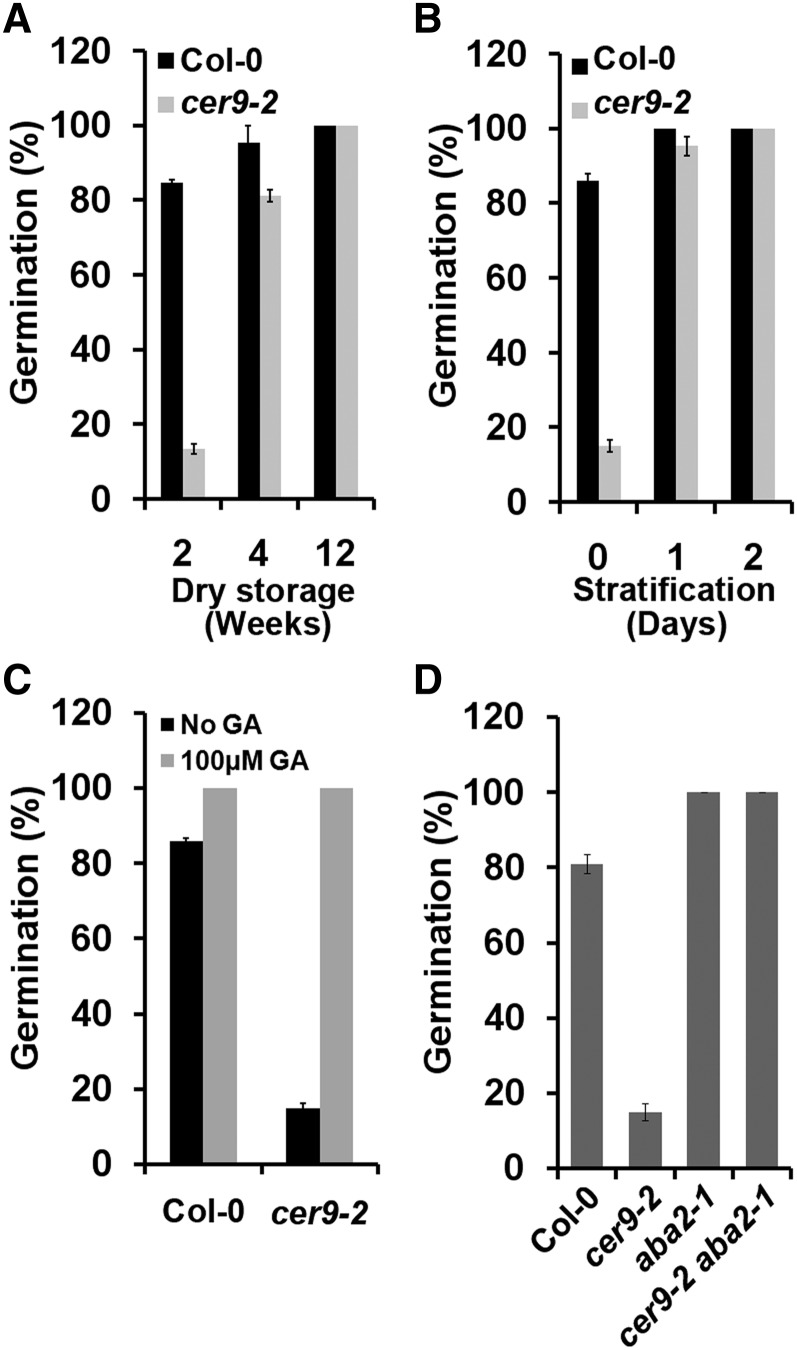
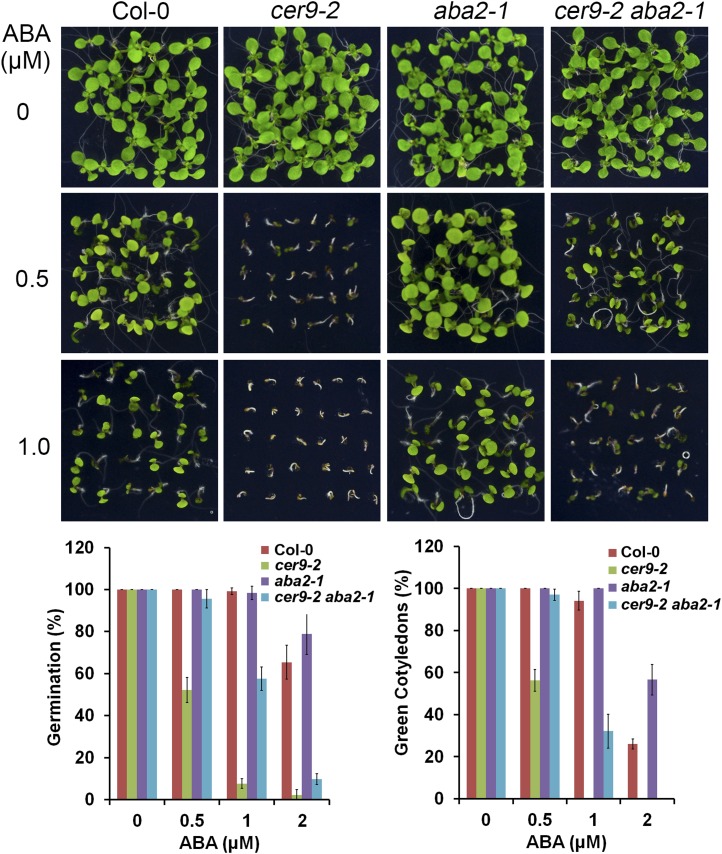
Mature seeds require a period of after-ripening to overcome dormancy (Carrera et al., 2008). Two-week-old freshly harvested cer9-2 seeds exhibited a higher rate (80%) of dormancy than the wild-type Columbia-0 (Col-0; 16%; Fig. 4A). In cer9, increasing the length of dry storage time to 4 weeks decreased the seed dormancy rate to 20%, and 12 weeks of storage led to complete loss of dormancy (Fig. 4A). The high degree of dormancy of cer9-2 seeds can be fully overcome by a 2-d chilling treatment (stratification) or by applying 100 µm GA (Fig. 4, B and C). Furthermore, the enhanced dormancy of cer9-2 seeds could also be fully overcome by the introgression of a second mutation, aba2-1, which is an ABA-deficient locus (Fig. 4D).
Figure 4.
The cer9 mutation increases seed dormancy. A, Germination rates of Col-0 and cer9-2 seeds with different after-ripening times assayed on agar medium 4 d after imbibition. B, Germination potential of 2-week-old seeds imbibed for 4 d on water-agar medium after different stratification times. C, Germination potential of seeds imbibed for 4 d on agar medium in the absence or presence of 100 µm GA3. D, Germination potential of 2-week-old seeds of Col-0, cer9-2, aba2-1, and cer9-2 aba2-1 imbibed for 4 d on water-agar medium. Data are means ± se of three replicates each with at least 30 seeds.
cer9 Seeds Accumulated High Levels of ABA
Since the cer9 mutant has increased seed dormancy that can be reversed by introducing the aba2-1 mutation, it is likely that cer9 seeds have increased ABA levels. We measured ABA levels in dry seeds of the wild type and the mutant using the Phytodetek ABA Test Kit (Agdia Incorporated). It was found that ABA amounts were notably higher in the cer9 mutant compared with the wild type (Fig. 5A). To determine whether higher levels of ABA had caused the delay of seed germination in cer9, we used both pharmacological and genetic methods to test the possibility. Fluridone [1-methyl-3-phenyl-5-(3-trifluoromethylphenyl)-4-(1H)-pyridinone] is an inhibitor of phytoene desaturase, which catalyzes the desaturation step of phytoene to phytofluene in the carotenoid biosynthesis pathway (Bartels and Watson, 1978). Since carotenoids are the main precursors of ABA in plants (Vranová et al., 2013), fluridone can effectively inhibit ABA biosynthesis. To check whether fluridone could improve the seed germination efficiency of cer9, seeds of Col-0 and cer9-2 were imbibed in water or 100 µm fluridone at 4°C for 48 h before being placed at room temperature under light for germination. As shown in Figure 5B, the fluridone treatment slightly but significantly enhanced the germination of cer9-2 seeds compared with the untreated seeds in the presence of 1 or 2 µm ABA. That the fluridone treatment did increase the germination of cer9 seeds but did not fully reverse the ABA suppression of germination suggested that the delayed germination and the hypersensitivity of cer9 seed germination to ABA are at least partially due to the enhancement of ABA biosynthesis in cer9 seeds.
Figure 5.
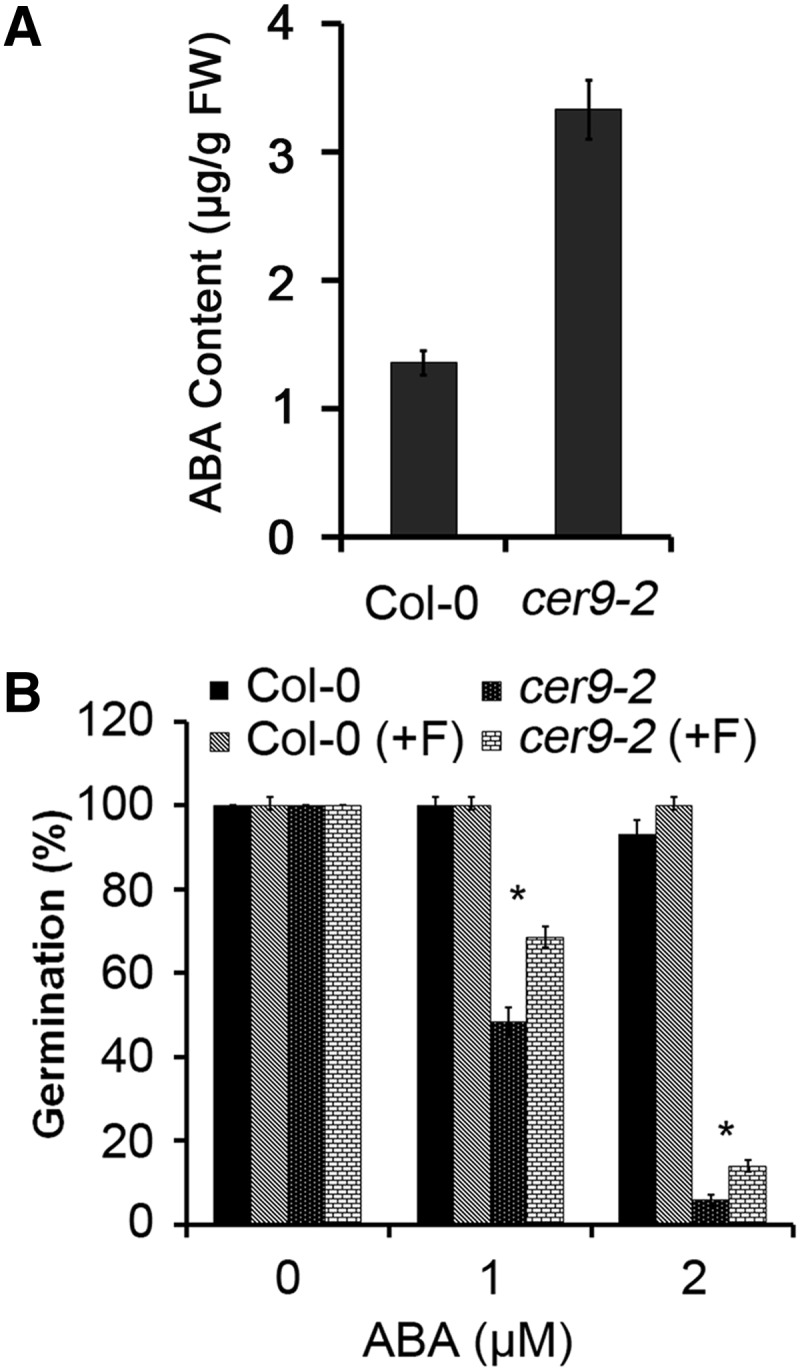
ABA levels in dry seeds and seed germination rates after the fluridone treatment. A, ABA levels in dry seeds of Col-0 and cer9-2 measured using the Phytodetek-ABA kit. FW, Fresh weight. B, Seeds of Col-0 and cer9-2 were immersed in water or 100 µm fluridone at 4°C for 48 h. The treated seeds were washed three times with water and planted on 1/2 MS agar plates supplemented with 0, 1, or 2 µm ABA. Seed germination was scored after 2 d. Seeds were considered germinated when the radicles emerged. *P < 0.05. Col-0 (+F) and cer9-2 (+F) represent Col-0 and cer9-2 with fluridone treatment, respectively.
We also investigated whether the introduced aba2 mutation could decrease the sensitivity of cer9 to ABA. ABA2 encodes a short-chain alcohol dehydrogenase, which catalyzes the conversion of xanthoxin to abscisic aldehyde in the ABA biosynthesis pathway (González-Guzmán et al., 2002). Overexpression of ABA2 promotes seed dormancy (Lin et al., 2007), whereas its loss of function reduces seed ABA levels and seed dormancy (Nambara et al., 1998). Seeds of the wild type, cer9-2, aba2-1, and the cer9-2 aba2-1 double mutant were planted on culture medium containing increasing concentrations of ABA (Fig. 6). In the presence of 0.5 µm ABA, green cotyledon expansion in the wild type was moderately inhibited. Whereas the germination of aba2 seeds was not affected by the ABA treatment, green cotyledon expansion in cer9 was severely inhibited. For the cer9-2 aba2-1 double mutant, the size of the green cotyledons was larger than that of the cer9 mutant but still smaller than those of the wild type and aba2-1. This suggests that the aba2-1 mutation partially rescued the cer9-2 mutant phenotypes, likely by decreasing ABA content and/or ABA sensitivity in the double mutant.
Figure 6.
Sensitivity of the cer9-2 aba2-1 double mutant to ABA during germination. Seeds of Col-0 (wild type), cer9-2, aba2-1, and cer9-2 aba2-1 were sown onto 1/2 MS medium supplemented with the indicated concentrations of ABA. Representative images show the morphology of seedlings 7 d after being sown on the medium plates. Germination (radicle emergence) percentages were determined 3 d and green cotyledons 7 d after imbibition. Data are means and sd from four biological replicates each with more than 30 seeds.
The Sensitivity of cer9 Seed Germination to ABA Treatment Is Not Associated with Seed Coat Permeability
The seed coat (testa) structure plays important roles in the breaking of dormancy. The seed coat can serve as a barrier to prevent water uptake, gas exchange, and inhibitor leakage from the embryo and can also supply seed germination inhibitors, restrain radicle protrusion, and filter light. A number of seed coat or testa mutants have maternally inherited reduced seed dormancy (Debeaujon et al., 2000). Removing the seed coat of GA-deficient mutants could release seeds from dormancy (Debeaujon and Koornneef, 2000). Since CER9 is associated with cuticle biosynthesis and its loss of function causes higher levels of cuticle coating on the epidermal cells of stems and leaves (Lü et al., 2012), we investigated whether the cuticle structure of cer9 mutations was altered and whether the alteration might affect seed coat permeability. As shown in Figure 7A, the cer9-2 seed coat cuticle structure is slightly thicker than that of the wild type. To determine whether the altered seed cuticle is responsible for the dormancy observed in cer9 seeds, the seed coats were perforated with a micro knife using a dissection microscope, and the treated seeds were placed on plates with various concentrations of ABA for germination assays. Compared with seeds without perforation, the perforated seeds showed enhanced sensitivity to ABA treatment (Fig. 7B). Nonetheless, the perforated cer9 seeds still showed higher sensitivity to ABA compared with the wild type (Fig. 7B). These data suggest that the permeability of the seed coat or seed coat structure may not be associated with the cer9 germination phenotype.
Figure 7.
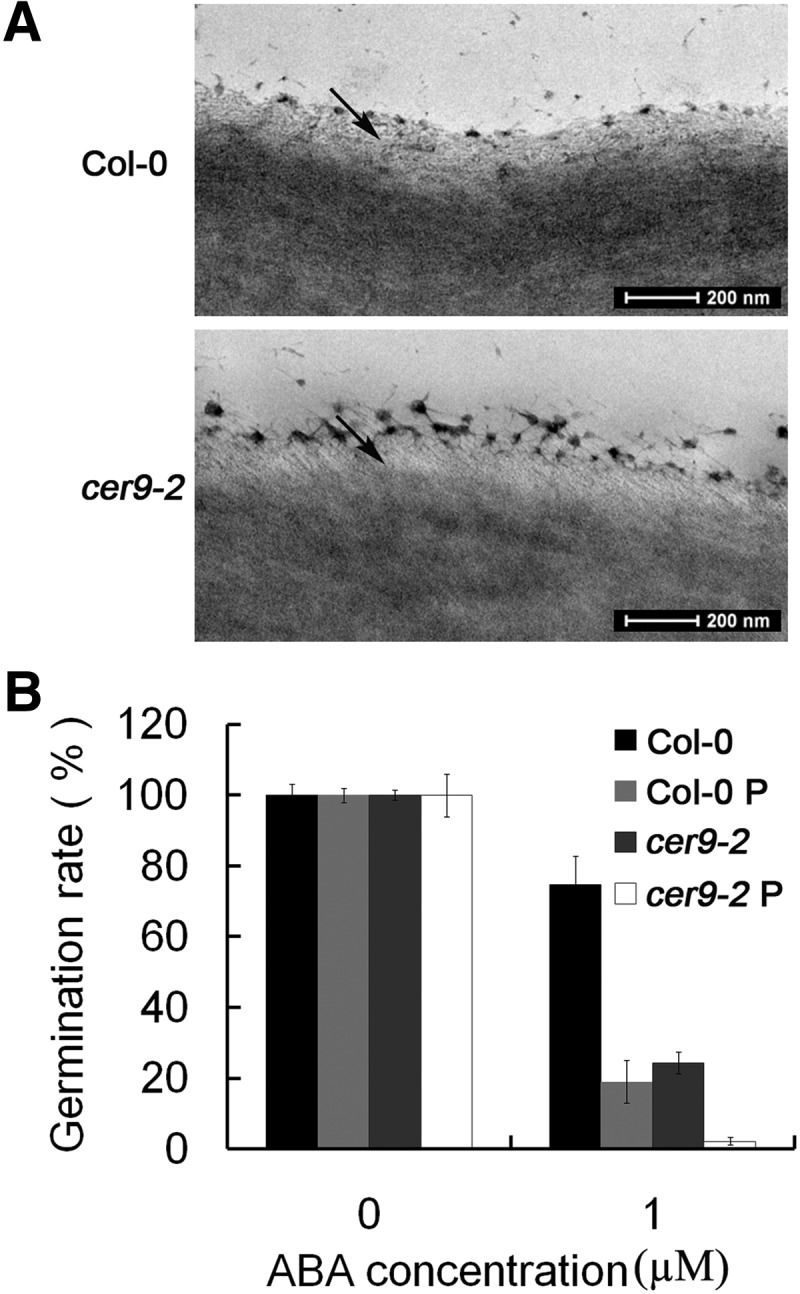
The cuticle of seed coat and the sensitivity of perforated seeds to ABA during germination. A, Transmission electron micrographs of the cuticle layer of Col-0 (top) and cer9-2 (bottom) seed coats. The cuticular layer is indicated by the arrows. Bars = 0.2 μm. B, Germination rates of perforated seeds under ABA treatment. Seeds of Col-0 and cer9-2 were perforated with a surgery stab knife, sown on 1/2 MS medium supplemented with 0 or 1 μm ABA, and incubated at 4°C for 3 d before being moved to 22°C. Seed germination was scored 2 d later. Seeds were considered germinated when the radicles emerged. Col-0 P and cer9-2 P represent perforated Col-0 and cer9-2 seeds, respectively. Data are means ± se of three replicates each with at least 30 seeds.
An Isobaric Tags for Relative and Absolute Quantitation Experiment Reveals That CER9 Is a Negative Regulator of ABA Biosynthesis and Signaling Pathways during Seed Germination
Since CER9 encodes a putative E3 ligase and its mutant seeds showed hypersensitivity to ABA, we speculated that CER9 might regulate the degradation of proteins associated with ABA biosynthesis or signaling pathways during seed germination. Consequently, proteomics analyses were conducted to examine the abundance of proteins. Seeds of Col-0 and cer9-2 were stratified at 4°C for 3 d before being treated with 1 μm ABA at 22°C for 1 d. The germinating seeds were then harvested for isobaric tags for relative and absolute quantitation (iTRAQ) analysis. As shown in Supplemental Table S1, a total of 1,019 proteins were identified by iTRAQ. Using the Col-0 without ABA treatment as a reference, a total of 114 proteins were significantly regulated based on a signal ratio change of 1.5 or greater upon ABA treatment of the Col-0 seeds. Among these significantly regulated proteins, 83 proteins were up-regulated and 31 proteins were down-regulated. Using the same criterion and relative to those of Col-0, 168 and 284 proteins were found to be up-regulated in cer9-2 seeds without ABA and with ABA treatment, respectively. Similarly relative to those of Col-0, 75 and 81 proteins were down-regulated in cer9-2 seeds without and with ABA treatment, respectively.
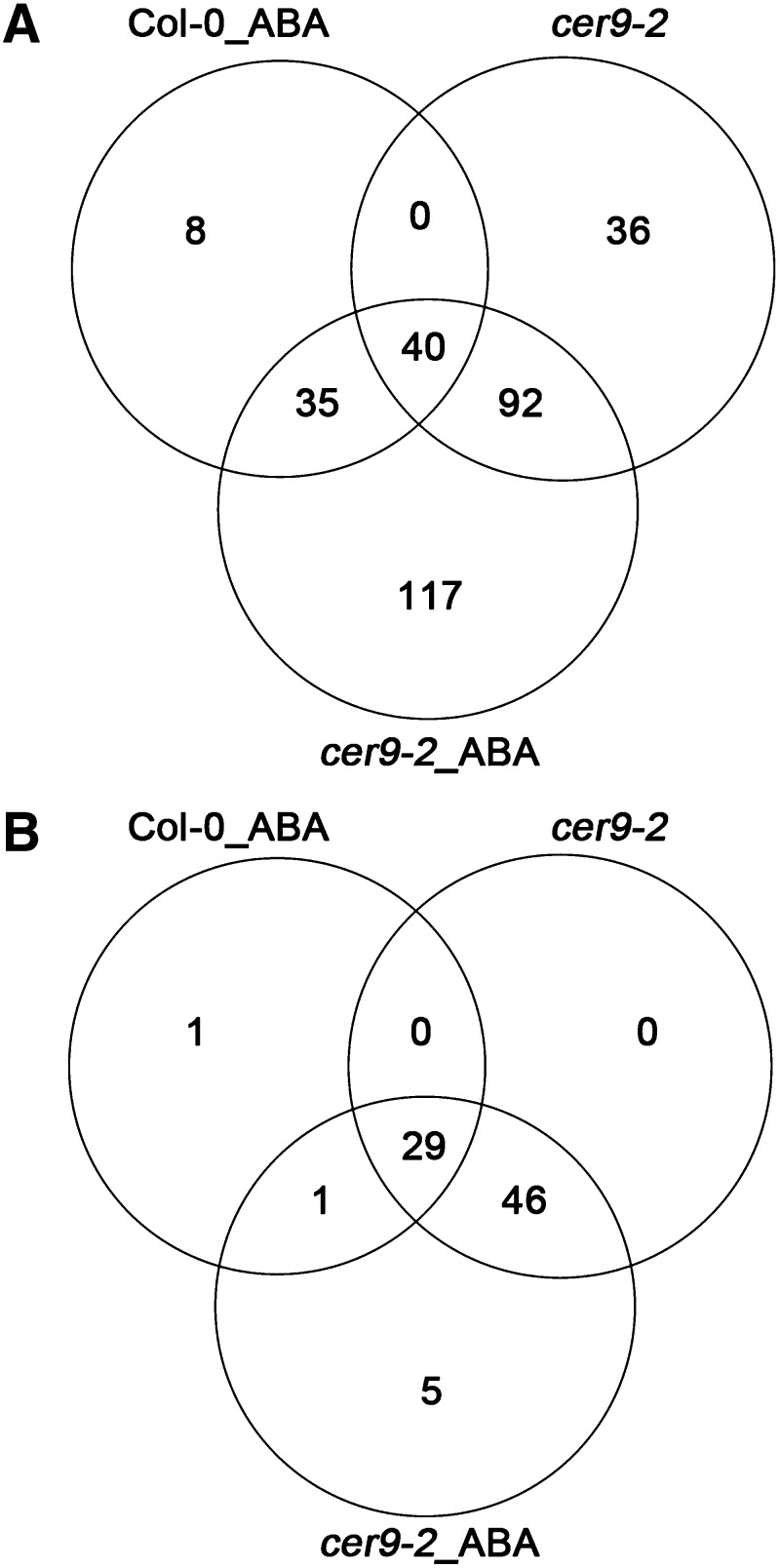
We compared three sets of protein data using the Draw Venn Diagram software (http://bioinformatics.psb.ugent.be/webtools/Venn/). Interestingly, 48% of the up-regulated proteins in the ABA-treated wild type were also up-regulated in cer9-2 without ABA treatment. Furthermore, 201 more proteins were up-regulated in ABA-treated cer9-2 than in ABA-treated Col-0. Notably, 93.5% of the down-regulated proteins in ABA-treated Col-0 were suppressed in cer9-2 (Fig. 8). In fact, most of these down-regulated proteins were suppressed to even greater extents in cer9-2 without ABA treatment (Supplemental Table S1). The high overlapping expression patterns in cer9-2 and the ABA-treated wild type further demonstrated that the impact of the cer9 mutation on protein expression resembles that of ABA treatment during seed germination; this suggests that the regulatory role of CER9 in seed germination is closely related to the ABA response. These results indicate that CER9 is a negative regulator of ABA biosynthesis or the ABA signaling pathway during seed germination.
Figure 8.
Classification of up- and down-regulated proteins in Col-0_ABA (ABA treated), cer9-2 (no ABA), and cer9-2_ABA (ABA treated) seeds. The total numbers of up-regulated proteins (A) and down-regulated proteins (B) were identified by iTRAQ analyses of Col-0 and cer9-2 germinating seeds in the presence or absence of 1 μm ABA. Relative signal intensities of the proteins were calculated by comparing with the control experiment (no-ABA-treated wild type).
Among the up- and down-regulated proteins in cer9-2, certain proteins related to the ABA response had similar regulation in cer9-2. These proteins include putative ABI3 targets (Mönke et al., 2012) and those related to ABA biosynthesis and signaling (Tables I and II). We also noted changes in the levels for proteins involved in lipid metabolism. Lipid is an important storage reserve, providing energy for radicle protrusion and seedling growth, and lipid mobilization is affected by ABA treatment during seed germination (Penfield et al., 2006). Previous reports indicate that oleosin proteins are involved in seed germination, enlargement of intracellular oil bodies, and total lipid and protein accumulation (Siloto et al., 2006; Shimada et al., 2008). We found that several seed storage proteins, including SEED STORAGE ALBUMIN1 (SESA1), SESA12, and SESA3 and two oleosin proteins, were down regulated in the cer9-2 mutant (Table II). These proteins are involved in oil body formation and are also ABI3 putative targets (Mönke et al., 2012; Table II).
Table I. ABA-related proteins up-regulated in cer9-2 germinating seeds.
Up-regulated proteins in cer9-2 that are responsive to ABA based on Gene Ontology analysis or the literature are listed. The values are means of two technical replicates and are shown as relative fold change. Relative fold change ≥ 1.5 indicates significant induction. LEA, Late embryogenesis-abundant.
| Proteins | Function | cer9-2 Versus Col-0 |
|---|---|---|
| Putative ABI3 targets | ||
| AT3G22490 | LEA domain-containing protein | 1.65 |
| AT1G72100 | LEA domain-containing protein | 1.75 |
| AT3G05260 | Oxidoreductase | 1.55 |
| AT5G01670 | Oxidoreductase | 1.50 |
| AT1G54860 | Unknown | 1.55 |
| AT5G22470 | Ribosyltransferase | 1.60 |
| AT2G28490 | Cupins superfamily protein | 1.50 |
| AT3G21370 | BGLU19, β-glucosidase19 | 1.50 |
| AT5G45690 | Unknown | 1.50 |
| AT4G26740 | Caleosin1 | 1.70 |
| AT1G05510 | Unknown | 1.55 |
| AT3G15670 | LEA family protein | 1.65 |
| ABA signaling pathway | ||
| AT4G35790 | Phospholipase Dδ | 1.50 |
| AT1G74100 | ATSOT16, sulfotransferase16 | 1.55 |
| Response to ABA stimulus | ||
| AT1G35720 | ATOXY5, annexin1 | 1.60 |
| AT5G02240 | Oxidoreductase | 1.60 |
| AT5G42950 | GYF domain-containing protein | 1.65 |
| AT5G03740 | HD2C, histone deacetylase2C | 1.50 |
| AT3G60240 | EIF4G | 1.70 |
| AT2G05710 | ACO3, aconitase3 | 1.50 |
Table II. ABA-related proteins down-regulated in cer9-2 germinating seeds that are putative ABI3 targets.
Down-regulated proteins in cer9-2 that are responsive to ABA based on Gene Ontology analysis or the literature are listed. The values are means of two technical replicates and are shown as relative fold change. Relative fold change < 0.6 indicates significant repression.
| Proteins | Function | cer9-2 Versus Col-0 |
|---|---|---|
| AT5G07190 | ATS3, seed gene3 | 0.45 |
| AT4G27160 | SESA3, seed storage albumin3 | 0.45 |
| AT4G28520 | CRU3, CRC, cruciferin3 | 0.4 |
| AT3G01570 | Oleosin family protein | 0.4 |
| AT3G27660 | OLEO4, OLE3, oleosin4 | 0.35 |
| AT5G01300 | Phosphatidylethanolamine-binding protein | 0.3 |
| AT4G25140 | OLEO1, OLE1, oleosin1 | 0.3 |
| AT5G54740 | SESA5, seed storage albumin5 | 0.3 |
| AT4G16160 | Outer envelope protein16-2 | 0.25 |
| AT4G27140 | SESA1, seed storage albumin1 | 0.2 |
Genes Involved in ABA Biosynthesis or Signaling Are Differentially Regulated by CER9
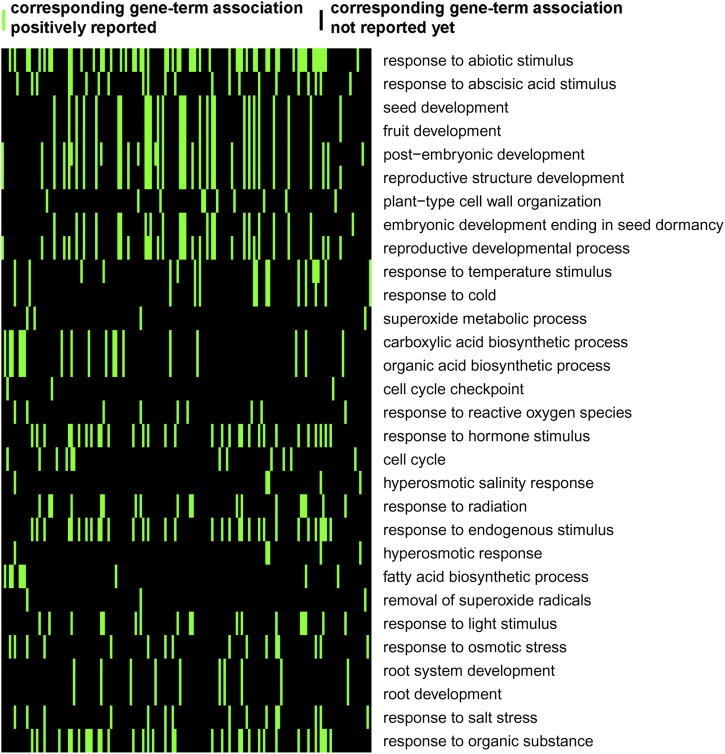
To further understand CER9 regulation of global gene expression during seed germination, we carried out comprehensive transcriptome analyses using the RNA sequencing (RNA-seq) technology. The two seed samples used for iTRAQ analysis, Col-0 and cer9-2, were further employed for RNA-seq analysis. We found that 428 genes were differentially regulated in the cer9 mutant compared with the wild type. Among them, 143 genes were significantly up-regulated and 285 were down-regulated in the mutant. An analysis of functional categories using the software DAVID (Huang et al., 2009) revealed that these genes were significantly enriched in several biological processes, including responses to abiotic and ABA stimuli, seed development, and plant-type cell wall organization (Fig. 9; Supplemental Table S2).
Figure 9.
Functional categories of differentially expressed genes in Col-0 and cer9-2 seeds. RNA-seq analysis was performed in 1-d light-grown seeds of Col-0 and cer9-2. Functional annotations were generated by the DAVID software (Huang et al., 2009). The top 30 functional annotations ordered by the enrichment scores were selected for the two-dimensional view, which indicates that genes with differential expression were strikingly enriched (colored green) in certain categories.
Given that the germination of cer9 mutants exhibited an enhanced sensitivity to ABA inhibition, we looked into the expression of genes involved in ABA biosynthesis, catabolism, and signaling. As shown in Table III and Figure 10, 9-CIS-EPOXYCAROTENOID DIOXYGENASE6 (NCED6), which is involved in ABA biosynthesis, was up-regulated in the mutants. ABI3, ABI4, and ABI5 are transcription factors belonging to the B3, APETALA2, and basic Leu zipper domain families, respectively. They are major positive regulators of the ABA response that largely determine seed germination or the maintenance of dormancy. Our results showed that ABI5 was significantly up-regulated (Supplemental Table S2). The expression of the ABI5-targeted genes Early methionine-labeled1 (EM1) and EM6 was also increased. RNA-seq revealed that ABI3 and ABI4 were slightly up-regulated (Supplemental Table S2), whereas real-time PCR results showed that the expression levels of ABI3 and ABI4 were significantly increased (Fig. 10). In contrast, the expression of genes encoding negative regulators of the ABA signaling pathway, such as ABI2, G-PROTEIN-COUPLED RECEPTOR1 (GCR1), and ABA-hypersensitive germination2 (AHG2), was markedly down-regulated (Fig. 10; Supplemental Table S2). These findings further demonstrate that CER9 significantly affects ABA biosynthesis and/or its signaling pathway.
Table III. Genes exhibiting differential expression in the wild type (Col-0) and cer9-2 mutants in response to ABA treatment.
Data are log2 fold change listed from largest to smallest. ND*, Transcripts not detected in Col-0; ND**, transcripts not detected in cer9-2.
| Gene Identifier | Gene Name | Description | Change (cer9-2/Col-0) |
|---|---|---|---|
| AT3G24220.1 | AtNCED6 | 9-Cis-epoxycarotenoid dioxygenase | ND* |
| AT2G31320.1 | PARP2 | A poly(ADP-Rib) polymerase | 3.23 |
| AT5G52300.1 | RD29B | Responsive to dehydration stress | 2.71 |
| AT5G62490.1 | AtHVA22B | HVA22 homolog B | 2.63 |
| AT3G51810.1 | AtEM1 | ABA-inducible protein | 2.31 |
| AT2G40170.1 | EM6 | Group 1 LEA gene | 1.83 |
| AT3G11050.1 | AtFER2 | Ferritin2 | 1.77 |
| AT1G72770.3 | HAB1 | Protein phosphatase2C | 1.62 |
| AT2G36270.1 | ABI5 | Basic Leu zipper transcription factor | 1.16 |
| AT1G75750.1 | GASA1 | GA-stimulated transcript1 protein homolog | 1.01 |
| AT5G44120.3 | CRU1 | 12S seed storage protein | −1.13 |
| AT4G38620.1 | ATMYB4 | R2R3 MYB protein | −1.73 |
| AT4G28520.1 | CRU3 | 12S seed storage protein | −1.78 |
| AT1G12420.1 | ACR8 | ACT domain repeat8 | −1.91 |
| AT1G48270.1 | GCR1 | G-protein-coupled receptor | −2.03 |
| AT1G48130.1 | AtPER1 | A protein similar to the 1-Cys peroxiredoxin family of antioxidants | −2.20 |
| AT1G55870.1 | AHG2 | Poly(A)-specific RNase | −2.25 |
| AT2G26980.2 | CIPK3 | Ser-Thr protein kinase | −2.93 |
| AT2G47770.1 | AtTSPO | Membrane-bound protein | −3.88 |
| AT5G57050.1 | ABI2 | Protein phosphatase2C | ND** |
Figure 10.
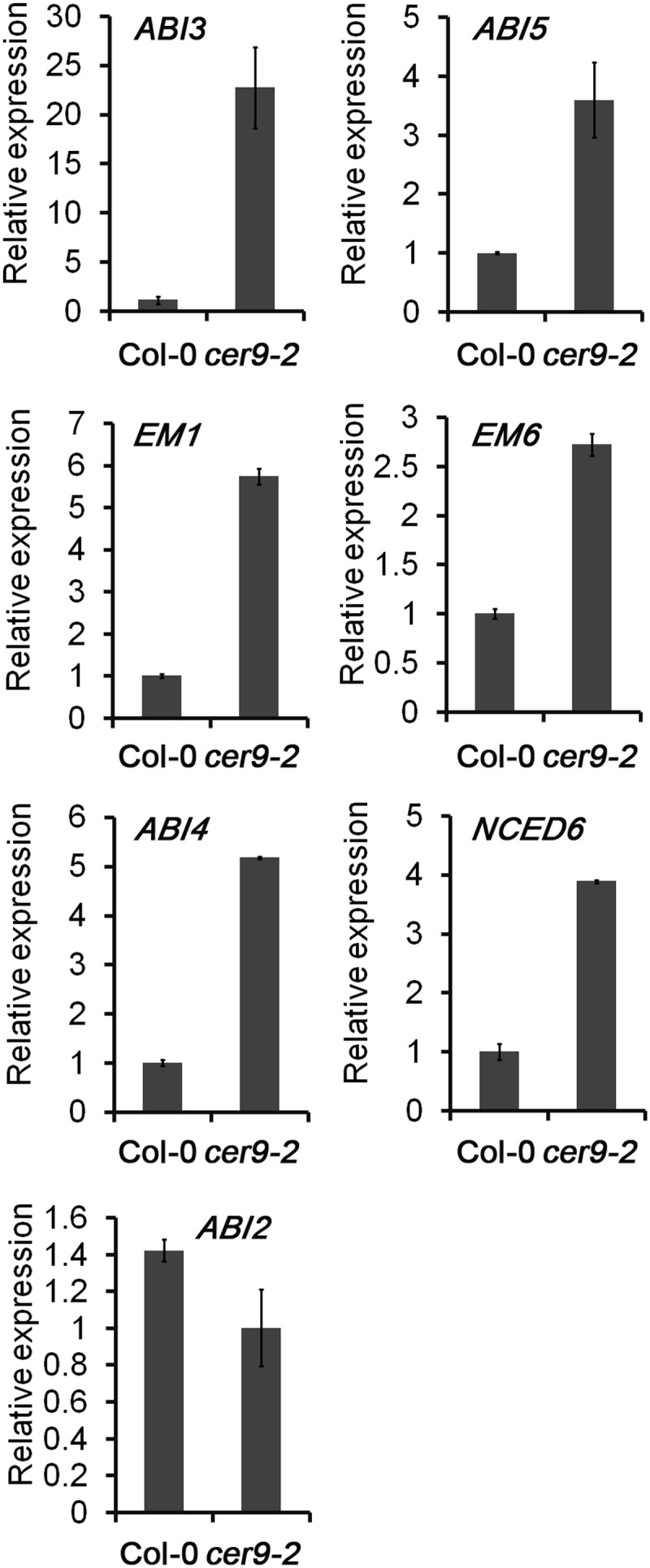
Relative expression levels of selected ABA response and biosynthesis genes in the cer9-2 mutant. RNA was extracted from seedlings grown in 1/2 MS liquid medium for 1 d after the end of stratification. Relative RNA levels of the genes shown were analyzed using gene-specific primers by real-time PCR. Data are means and sd of three biological replicates.
Genetic Analysis of the Regulatory Roles of CER9 in the ABA Signaling Pathway
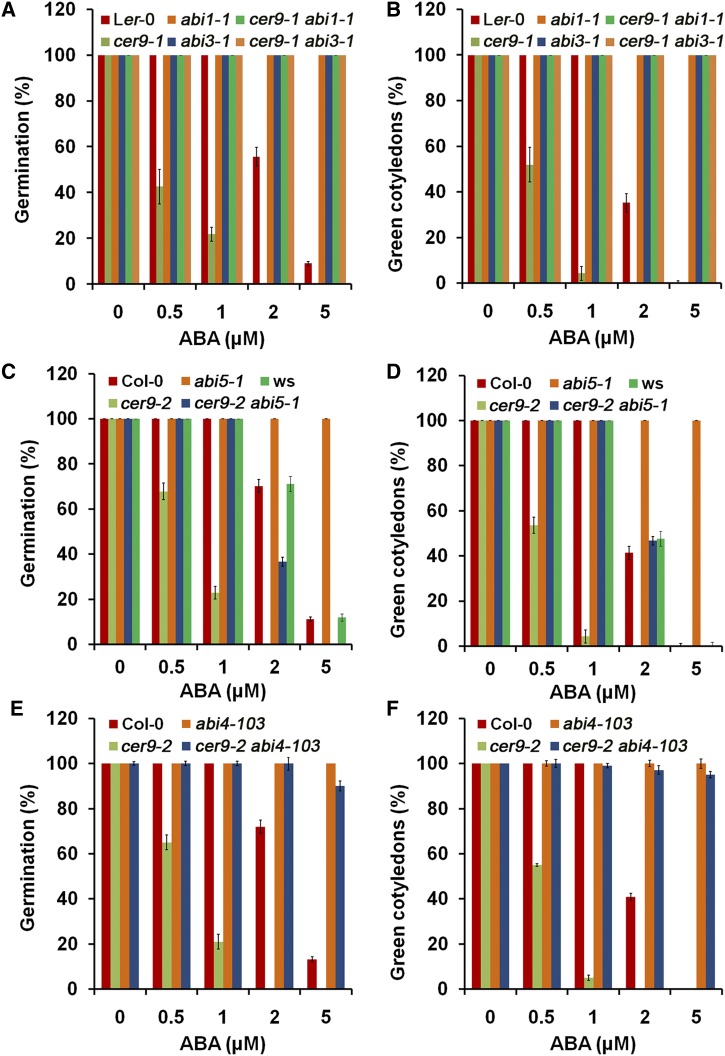
To establish whether the ABA signaling pathway plays any role in CER9-mediated ABA responses, we crossed cer9-1 (Landsberg erecta-0 [Ler-0] background) with abi1-1 and abi3-1 and also crossed cer9-2 (Col-0 background) with abi5-1 and abi4-103 mutants. ABI1 encodes a protein phosphatase type 2C (PP2C) and is a negative regulator of ABA signaling by inhibiting Sucrose nonfermenting1-related protein kinase2 kinases (Weiner et al., 2010). abi1-1 mutants have largely lost responsiveness to ABA. When this mutation was introduced into the cer9 mutant background, the double mutants resembled the abi1-1 single mutant in terms of seed germination response to ABA (Fig. 11, A and B; Supplemental Fig. S1). This result indicates that the dominant mutation of abi1-1 could completely block ABA signaling in the cer9 mutant. The ABI3, ABI4, and ABI5 transcription factors positively regulate the expression of certain ABA-responsive genes in seeds (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2002; Penfield et al., 2006). ABI3 acts upstream of ABI5, and both are located downstream of ABI1 (Lopez-Molina et al., 2002). When abi3-1 was introduced into cer9-1, both the germination rate and the green cotyledon percentage of abi3-1 cer9-1 double mutant seeds were similar to those of abi3-1 under different concentrations of ABA (Fig. 11, A and B). However, after 7 d of germination, the abi3-1 cer9-1 double mutant grew more slowly than abi3-1 but much faster than the wild type (Supplemental Fig. S2). Germination of the abi5-1 cer9-2 double mutant seeds differed in response to different concentrations of ABA. The germination rate and green cotyledon percentage of the abi5-1 cer9-2 double mutant were higher than those of cer9-2 but were similar to those of the wild-type Col-0 and Wassilewskija (Ws) as well as abi5-1 in the presence of 0.5 and 1 µm ABA (Fig. 11, C and D). However, the germination rate of the abi5-1 cer9-2 double mutant was lower than that of the wild type when the seeds were exposed to a higher concentration of ABA (2 and 5 µm; Fig. 11C). After 7 d of germination, the abi5-1 cer9-2 double mutant grew better than cer9-2 but much worse than abi5-1 in the presence of 1 µm ABA and had an intermediate phenotype between abi5-1 and cer9-2 (Supplemental Fig. S3). In the presence of 5 µm ABA, the abi5-1 cer9-2 double mutant showed increased sensitivity to ABA (Supplemental Fig. S3). When abi4-103 was introduced into cer9-2, both the germination rate and the green cotyledon percentage of the cer9-2 abi4-103 double mutant seeds were similar to those of abi4-103 under different concentrations of ABA (Fig. 11, E and F). However, after 7 d of germination, the cer9-2 abi4-103 double mutant grew slightly slower than abi4-103 but much faster than the wild type (Supplemental Fig. S4).
Figure 11.
Genetic analysis between cer9-1 or cer9-2 and abi1-1, abi3-1, abi5-1, or abi4-103 mutants. Seeds of the wild types (Col-0, Ler-0, and Ws), single mutants (cer9-1, cer9-2, abi1-1, abi3-1, abi5-1, and abi4-103), and double mutants (cer9-1 abi1-1, cer9-1 abi3-1, cer9-2 abi5-1, and cer9-2 abi4-103) were sown onto 1/2 MS medium supplemented with 0, 0.5, 1, 2, or 5 µm ABA. Germination (radicle emergence) percentages were determined 3 d and green cotyledons 7 d after imbibition. Data are means and sd from four biological replicates each with at least 30 seeds.
DISCUSSION
The CER9 locus plays important roles in cuticle and wax synthesis as well as leaf transpiration in Arabidopsis (Lü et al., 2012). In this study, we found that CER9 also regulates ABA responses during seed germination and early seedling development. Seeds of the cer9 mutant showed increased dormancy and hypersensitivity to ABA inhibition of germination and seedling growth. One reason for this is that the cer9 mutant accumulated a higher level of ABA. Application of fluridone or introduction of the aba2-1 mutation into the cer9 background suppressed the delayed seed germination of the cer9 mutant, indicating that CER9 is likely to be a negative regulator of ABA biosynthesis. ABA levels are determined by de novo biosynthesis, catabolism, or hydrolysis of conjugated ABA. The enzyme NCED catalyzes a rate-limiting step in the ABA biosynthesis pathway (Nambara and Marion-Poll, 2005). Interestingly, one of the NCED genes, NCED6, was expressed at a higher level in cer9-2 than in the wild type (Fig. 10; Supplemental Table S2). Previous studies indicated that NCED6 is highly expressed in the testa and endosperm, and its overexpression results in increased ABA levels in seeds and enhanced seed dormancy (Lefebvre et al., 2006; Martínez-Andújar et al., 2011). The up-regulation of NCED6 expression in cer9 mutant seeds thus may contribute to the higher ABA level and increased seed dormancy of the mutants. ABA catabolism also plays an important role in regulating seed germination. ABA 8′-hydroxylases, including cytochrome P450 proteins CYP707A1 to CYP707A4, are involved in ABA catabolism. Among them, CYP707A1 and CYP707A2 are required for the proper control of seed dormancy and germination in Arabidopsis. Their double mutants accumulate high levels of ABA and display hyperdormancy in seeds (Okamoto et al., 2006). The transcripts for CYP707A1, CYP707A2, and CYP707A3 were detected in our RNA-seq analyses, yet there was little difference in transcript levels between cer9-2 and the wild type (Supplemental Table S2).
The increased levels of ABA observed in the cer9 mutant could also result from the hydrolysis of glucose-conjugated abscisic acid (ABA-GE). This increased amount of ABA could promote ABA biosynthesis through the autoregulation of ABA biosynthesis genes (Xiong and Zhu, 2003). The enzyme β-GLUCOSIDASE HOMOLOG1 (BG1) can mediate the hydrolysis of ABA-GE and contribute to the increased ABA level (Lee et al., 2006; Xu et al., 2012). Although our iTRAQ analysis failed to detect BG1 and BG2, two β-glucosidases involved in ABA-GE hydrolysis and four other homologous proteins were detected in the proteome profiling. These include BGLU19, BGLU23, BGLU38, and BGLU44. Interestingly, all of these putative β-glucosidases had higher protein levels in the cer9 mutant than in the wild type (Supplemental Table S1). Among these homologs, BGLU19 shares 71% identity with BG1, although it is unclear whether BGLU19 is also involved in ABA-GE hydrolysis. Further investigation into the role of these β-glucosidases in ABA-GE hydrolysis, and whether they are the substrates of CER9, is required.
It should be noted that the increased ABA levels in dry seeds could not fully explain the ABA hypersensitivity phenotypes in seed germination and postgermination growth of the cer9 mutant. Neither fluridone treatment nor the aba2-1 mutation could fully suppress the cer9-2 ABA-hypersensitive phenotype (Figs. 5 and 6). Thus, the cer9 mutant is not simply an ABA overproduction mutant. Rather, CER9 may represent a negative regulator of the ABA response. While it remains unclear which of the ABA signaling components are likely to be direct targets of CER9, the expression of a number of ABA signaling regulators was significantly affected in the cer9 mutant. The transcript levels for several negative regulators of ABA signaling, such as ABI2, GCR1, and AHG2, were markedly down-regulated (Fig. 10; Supplemental Table S2), whereas those encoding positive regulators of the ABA response were increased, as discussed below.
The transcription factors ABI3, ABI4, and ABI5 are well known positive regulators of the ABA responses during seed germination (Finkelstein and Lynch, 2000; Lopez-Molina et al., 2002; Penfield et al., 2006). Our study showed that the expression levels of ABI3, ABI4, and ABI5 were significantly increased in the cer9 mutant. Double mutant analysis showed that the abi3-1, abi4-103, and abi5-1 mutations suppressed the ABA-hypersensitive phenotypes of cer9 seeds to different degrees, suggesting that these regulators may act downstream of CER9 in the ABA response pathways. It has been reported that ABI3 interacts with ABI5 and that ABI5 acts downstream of ABI3 (Lopez-Molina et al., 2002). Our results showed that abi3-1 strongly suppressed the sensitivity of cer9 seeds to ABA, whereas abi5-1 only moderately suppressed it. It is likely that ABI5 homologs may play redundant roles in the ABA signaling pathways and that CER9-mediated ABA signaling may be transduced through both ABI5-dependent and ABI5-independent pathways. This mode of action is similar to that of the AHG3 mutant defective in PP2C, a negative regulator of the ABA response during seed germination. The loss-of-function ahg3 mutant exhibits ABA hypersensitivity during seed germination, and this could nearly be reversed by the abi3 mutation but only partially suppressed by abi5-1 (Yoshida et al., 2006). Interestingly, ahg3 shared some other phenotypes similar to cer9. For example, both mutant seeds contain high levels of ABA. In both cases, the mutation specifically affected seed dormancy without affecting mature plant growth. We checked the expression of AHG3 in RNA-seq and iTRAQ data but could not find this gene (Supplemental Tables S1 and S2), and this may be due to the low abundance of this gene under our experimental conditions. The direct involvement of CER9 in ABA responses in seeds was also demonstrated by the observation that the dominant negative mutation abi1-1 was found to completely repress the ABA hypersensitivity of the cer9 mutant. In fact, the increased ABA level in cer9 may have resulted from its enhanced ABA sensitivity, since most of the ABA biosynthesis genes are up-regulated by ABA through the same or similar ABA signaling pathways (Xiong and Zhu, 2003).
One interesting observation is that there appears to be little correlation between the seed transcriptome and proteome either for cer9 or the wild type. For example, RNA-seq data showed that cer9 deficiency down-regulated the expression of genes associated with the response to ABA, plant cell wall structure, and the response to light. However, these genes were not represented in down-regulated protein groups. This was similarly the case among the genes/proteins in the up-regulated groups. The low concordance between the proteome and transcriptome has also been reported in previous studies (Wegener et al., 2010; Huang et al., 2013). One explanation may be associated with the different sensitivities of the detection technologies. In our experiments, 18,184 transcripts were detected in the RNA-seq experiments, whereas only 1,019 proteins were detected in the proteomics assays. Clearly, RNA-seq is more robust and sensitive in identifying transcript units even at low levels of expression. In contrast, iTRAQ detected only proteins of the highest abundance. Essentially, proteins of low abundance, such as NCED6, ABI3, ABI4, and ABI5, could barely be detected directly by iTRAQ. Second, transcript accumulation and translation may not be coordinated in seeds and during seed germination. It has been well documented in several plant species, including Arabidopsis, that a large number of transcripts are stored in dry seeds as well as in imbibed seeds (Rajjou et al., 2012). For example, 12,470 transcripts were detected in Arabidopsis dry seeds, this being similar to the number of transcripts detected in 24-h imbibed seeds (Nakabayashi et al., 2005). These transcripts may be selectively translated during seed imbibition and germination (Rajjou et al., 2012). In addition, there seems to be a time gap between transcription and translation, in that major translation may occur relatively later during germination while stored proteins could function at the early stage of imbibition (Kimura and Nambara, 2010).
In summary, our genetic analyses combined with transcriptome and proteomics data show that CER9 is an important determinant of seed germination and that this regulation is closely associated with the negative regulatory roles of CER9 in ABA biosynthesis and signaling. Future work will need to focus on the identification of CER9 substrates in order to reveal the precise way in which CER9 regulates ABA accumulation and signaling in seeds and seedlings.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were obtained from the Arabidopsis Biological Resource Center, the European Arabidopsis Stock Centre, or individual researchers. Unless stated otherwise, seeds were stratified for 3 d at 4°C and then moved to 22°C with a 16-h-light/8-h-dark cycle and a light intensity of 110 to 120 mol m−2 s−1 for germination and growth.
Germination Assay
Plants of different genotypes were grown in the same conditions, and seeds were collected at the same time and stored similarly before conducting germination experiments. For germination assays, approximately 30 to 50 seeds were sown on one-half-strength Murashige and Skoog (1/2 MS) medium supplemented with various concentrations of (±)-cis,trans-ABA (Sigma-Aldrich). Germination (emergence of radicles) and postgermination growth (green cotyledon appearance) were scored daily for 3 and 7 d, respectively.
Extraction and Quantification of ABA
Dry seeds were ground in liquid nitrogen, and ABA was extracted as described (Pēna-Cortés et al., 1989). ABA was quantified with the Phytodetek-ABA kit (Idetek) that uses a monoclonal antibody specific to (+)-ABA.
Protein Extraction, iTRAQ Labeling, and Liquid Chromatography-Mass Spectrometry Analysis
Seeds of Col-0 and cer9-2 were first incubated in 1/2 MS liquid medium for 3 d at 4°C and then incubated in the same medium supplemented with or without 1 μm ABA by shaking at 22°C under a 16-h-light/8-h-dark cycle with a light intensity of 110 to 120 mol m−2 s−1 for 1 d. The germinating seeds were ground in liquid nitrogen in a prechilled mortar with a pestle. Proteins were extracted with the extraction buffer (50 mm Tris, pH 8, 2 m urea, and 0.5% [w/v] SDS) supplemented with protease inhibitor (Roche Diagnostics). Protein content was measured using 2D-Quant (GE Healthcare). Proteins were precipitated in acetone (sample:acetone 1:5, v/v) and then resuspended in SDS-PAGE sample buffer. Protein separation by SDS-PAGE and in-gel digestion were performed as described (Meng et al., 2008). The extracted peptides were labeled with 4plexiTRAQ reagents (ABSciex) as follows: the wild type treated with mock, the wild type treated with ABA, the mutant treated with mock, and the mutant treated with ABA were labeled with the iTRAQ reporters 114, 115, 116, and 117, respectively. The combined peptides were fractionated using strong cation-exchange chromatography. Liquid chromatography-mass spectrometry analysis of the resulting 16 fractions as well as mass spectrometry data processing were carried out following our published procedure (Zhang et al., 2010). Briefly, each fraction was analyzed twice using LTQ-OrbitrapVelos. The mass spectrometry spectra were recorded in the Orbitrap, whereas tandem mass spectrometry spectra were recorded in the c-TRAP for higher-energy collisional dissociation (HCD) fragmentation and in the LTQ for the collision-induced dissociation fragmentation. Both HCD and collision-induced dissociation spectra were extracted individually using Proteome Discoverer software and processed by an in-house script before Mascot search against the Arabidopsis protein database The Arabidopsis Information Resource 10 (total 3,5624 sequences). The Mascot search results were further processed by Scaffold software for validation of protein identification and quantitative assessment. For protein identification, we reported only proteins with a minimal 99% possibility and containing at least one peptide with a possibility greater than 95%. As a result, the scaffold local false discovery rate is 0.1% for protein identification and 0.7% for peptide identification for our data set. Protein quantitation was processed using Scaffold Q+ based on i-Tracker (Shadforth et al., 2005). The iTRAQ quantitation using HCD is highly accurate, and a greater than 1.45-fold change could be considered as a significantly differential expression and, therefore, was adopted in this study.
RNA-seq Analysis
Total RNA was extracted from 1-d light-grown seeds using the RNeasy mini kit (Invitrogen), and DNA was cleaned by DNAse I (New England Biolabs). About 2 to 4 μg of cleaned total RNA was used for RNA-seq analysis as described by Chen et al. (2013).
qRT-PCR
Dry seeds, seeds incubated in 1/2 MS medium after a 3-d stratification at 4°C, and germinated seeds of the wild type and cer9-2 mutants in 1/2 MS medium with or without 1 μm ABA for 1 d were harvested and frozen immediately in liquid nitrogen, then stored at −80°C before RNA extraction. For qRT-PCR analysis, total RNA was extracted from the above samples using the RNeasy plant mini kit (Qiagen) following the manufacturer’s instructions. qRT-PCR was performed in 96-well blocks with an Applied Biosystems 7500 Real-Time PCR system using the SYBR Green I master mix in a volume of 20 µL. The reactions were performed in biological triplicates using RNA samples extracted from three independent plant materials and gene-specific primers (Supplemental Table S2). Data were analyzed using the SDS software (Applied Biosystems version 1.0) as described by Lü et al. (2012).
Genetic Analysis of Double Mutants
The cer9-1 mutant (Ler-0) was crossed with abi1-1 (Ler-0). The cer9-2 mutant (Col-0) was crossed with aba2-1 (Col-0) and abi5-1 (Ws). Double mutants were obtained from the F2 progeny by genotyping using the mutation-specific cleaved-amplified polymorphic sequence markers (Supplemental Table S3).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sensitivity of the cer9-1 abi1-1 double mutant to ABA.
Supplemental Figure S2. Sensitivity of the cer9-1 abi3-1 double mutant to ABA.
Supplemental Figure S3. Sensitivity of the cer9-2 abi5-1 double mutant to ABA.
Supplemental Figure S4. Sensitivity of the cer9-2 abi4-103 double mutant to ABA.
Supplemental Table S1. Results of iTRAQ analysis.
Supplemental Table S2. Results of RNA-seq analysis.
Supplemental Table S3. Primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Luis Lopez-Molina (University of Geneva) for providing abi3-1 seeds.
Glossary
- ABA
abscisic acid
- qRT
quantitative reverse transcription
- Col-0
Columbia-0
- iTRAQ
isobaric tags for relative and absolute quantitation
- RNA-seq
RNA sequencing
- Ler-0
Landsberg erecta-0
- Ws
Wassilewskija
- ABA-GE
glucose-conjugated abscisic acid
- 1/2 MS
one-half-strength Murashige and Skoog
Footnotes
This work was supported by King Abdullah University of Science and Technology (to L.X.) and by the Natural Science Foundation of China (grant no. 113137033816 to S.L.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ali-Rachedi S, Bouinot D, Wagner MH, Bonnet M, Sotta B, Grappin P, Jullien M. (2004) Changes in endogenous abscisic acid levels during dormancy release and maintenance of mature seeds: studies with the Cape Verde Islands ecotype, the dormant model of Arabidopsis thaliana. Planta 219: 479–488 [DOI] [PubMed] [Google Scholar]
- Bartels PG, Watson CW. (1978) Inhibition of carotenoid synthesis by fluridone and norflurazon. Weed Sci 26: 198–203 [Google Scholar]
- Bewley JD. (1997) Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Li H, Zhao Q, Jiang H, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Wang D, et al. (2009) The Arabidopsis RING finger E3 ligase RHA2a is a novel positive regulator of abscisic acid signaling during seed germination and early seedling development. Plant Physiol 150: 463–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E, Holman T, Medhurst A, Dietrich D, Footitt S, Theodoulou FL, Holdsworth MJ. (2008) Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J 53: 214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Hellmann H. (2013) Plant E3 ligases: flexible enzymes in a sessile world. Mol Plant 6: 1388–1404 [DOI] [PubMed] [Google Scholar]
- Chen T, Cui P, Chen H, Ali S, Zhang S, Xiong L. (2013) A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet 9: e1003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Koornneef M. (2000) Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol 122: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Léon-Kloosterziel KM, Koornneef M. (2000) Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol 122: 403–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas VG, Amorim-Silva V, Posé D, Rosado A, Esteban A, Arró M, Azevedo H, Bombarely A, Borsani O, Valpuesta V, et al. (2013) The SUD1 gene encodes a putative E3 ubiquitin ligase and is a positive regulator of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity in Arabidopsis. Plant Cell 25: 728–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. (2008) Molecular aspects of seed dormancy. Annu Rev Plant Biol 59: 387–415 [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12: 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guzmán M, Apostolova N, Bellés JM, Barrero JM, Piqueras P, Ponce MR, Micol JL, Serrano R, Rodríguez PL. (2002) The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 14: 1833–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ. (2008) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Holman TJ, Jones PD, Russell L, Medhurst A, Ubeda Tomás S, Talloji P, Marquez J, Schmuths H, Tung SA, Taylor I, et al. (2009) The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proc Natl Acad Sci USA 106: 4549–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Chen L, Te R, Qiao J, Wang J, Zhang W. (2013) Complementary iTRAQ proteomics and RNA-seq transcriptomics reveal multiple levels of regulation in response to nitrogen starvation in Synechocystis sp. PCC 6803. Mol Biosyst 9: 2565–2574 [DOI] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Nambara E. (2010) Stored and neosynthesized mRNA in Arabidopsis seeds: effects of cycloheximide and controlled deterioration treatment on the resumption of transcription during imbibition. Plant Mol Biol 73: 119–129 [DOI] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I. (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Lefebvre V, North H, Frey A, Sotta B, Seo M, Okamoto M, Nambara E, Marion-Poll A. (2006) Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J 45: 309–319 [DOI] [PubMed] [Google Scholar]
- Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH. (2007) Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol 143: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32: 317–328 [DOI] [PubMed] [Google Scholar]
- Lü S, Zhao H, Des Marais DL, Parsons EP, Wen X, Xu X, Bangarusamy DK, Wang G, Rowland O, Juenger T, et al. (2012) Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol 159: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Andújar C, Ordiz MI, Huang Z, Nonogaki M, Beachy RN, Nonogaki H. (2011) Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc Natl Acad Sci USA 108: 17225–17229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W, Zhang H, Guo T, Pandey C, Zhu Y, Kon OL, Sze SK. (2008) One-step procedure for peptide extraction from in-gel digestion sample for mass spectrometric analysis. Anal Chem 80: 9797–9805 [DOI] [PubMed] [Google Scholar]
- Mönke G, Seifert M, Keilwagen J, Mohr M, Grosse I, Hähnel U, Junker A, Weisshaar B, Conrad U, Bäumlein H, et al. (2012) Toward the identification and regulation of the Arabidopsis thaliana ABI3 regulon. Nucleic Acids Res 40: 8240–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Nambara E, Kawaide H, Kamiya Y, Naito S. (1998) Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol 39: 853–858 [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E. (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol 141: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pēna-Cortés H, Sánchez-Serrano JJ, Mertens R, Willmitzer L, Prat S. (1989) Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato. Proc Natl Acad Sci USA 86: 9851–9855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D. (2012) Seed germination and vigor. Annu Rev Plant Biol 63: 507–533 [DOI] [PubMed] [Google Scholar]
- Shadforth IP, Dunkley TP, Lilley KS, Bessant C. (2005) i-Tracker: for quantitative proteomics using iTRAQ. BMC Genomics 6: 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Shimada T, Takahashi H, Fukao Y, Hara-Nishimura I. (2008) A novel role for oleosins in freezing tolerance of oilseeds in Arabidopsis thaliana. Plant J 55: 798–809 [DOI] [PubMed] [Google Scholar]
- Siloto RMP, Findlay K, Lopez-Villalobos A, Yeung EC, Nykiforuk CL, Moloney MM. (2006) The accumulation of oleosins determines the size of seed oilbodies in Arabidopsis. Plant Cell 18: 1961–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18: 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranová E, Coman D, Gruissem W. (2013) Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu Rev Plant Biol 64: 665–700 [DOI] [PubMed] [Google Scholar]
- Wegener KM, Singh AK, Jacobs JM, Elvitigala T, Welsh EA, Keren N, Gritsenko MA, Ghosh BK, Camp DG, II, Smith RD, et al. (2010) Global proteomics reveal an atypical strategy for carbon/nitrogen assimilation by a cyanobacterium under diverse environmental perturbations. Mol Cell Proteomics 9: 2678–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner JJ, Peterson FC, Volkman BF, Cutler SR. (2010) Structural and functional insights into core ABA signaling. Curr Opin Plant Biol 13: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Zhu JK. (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Lee KH, Dong T, Jeong JC, Jin JB, Kanno Y, Kim DH, Kim SY, Seo M, Bressan RA, et al. (2012) A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24: 2184–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Nishimura N, Kitahata N, Kuromori T, Ito T, Asami T, Shinozaki K, Hirayama T. (2006) ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol 140: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhao C, Li X, Zhu Y, Gan CS, Wang Y, Ravasi T, Qian PY, Wong SC, Sze SK. (2010) Study of monocyte membrane proteome perturbation during lipopolysaccharide-induced tolerance using iTRAQ-based quantitative proteomic approach. Proteomics 10: 2780–2789 [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua NH. (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev 19: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang C, Li Y, Zheng N, Chen H, Zhao Q, Gao T, Guo H, Xie Q. (2007) SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19: 1912–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.