Lipoate-protein ligase and octanoyltransferase provide the basal indispensable protein lipoylation network in plant mitochondria.
Abstract
Prosthetic lipoyl groups are required for the function of several essential multienzyme complexes, such as pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGDH), and the glycine cleavage system (glycine decarboxylase [GDC]). How these proteins are lipoylated has been extensively studied in prokaryotes and yeast (Saccharomyces cerevisiae), but little is known for plants. We earlier reported that mitochondrial fatty acid synthesis by ketoacyl-acyl carrier protein synthase is not vital for protein lipoylation in Arabidopsis (Arabidopsis thaliana) and does not play a significant role in roots. Here, we identify Arabidopsis lipoate-protein ligase (AtLPLA) as an essential mitochondrial enzyme that uses octanoyl-nucleoside monophosphate and possibly other donor substrates for the octanoylation of mitochondrial PDH-E2 and GDC H-protein; it shows no reactivity with bacterial and possibly plant KGDH-E2. The octanoate-activating enzyme is unknown, but we assume that it uses octanoyl moieties provided by mitochondrial β-oxidation. AtLPLA is essential for the octanoylation of PDH-E2, whereas GDC H-protein can optionally also be octanoylated by octanoyltransferase (LIP2) using octanoyl chains provided by mitochondrial ketoacyl-acyl carrier protein synthase to meet the high lipoate requirement of leaf mesophyll mitochondria. Similar to protein lipoylation in yeast, LIP2 likely also transfers octanoyl groups attached to the H-protein to KGDH-E2 but not to PDH-E2, which is exclusively octanoylated by LPLA. We suggest that LPLA and LIP2 together provide a basal protein lipoylation network to plants that is similar to that in other eukaryotes.
Lipoic acid (LA; 6,8-dithiooctanoic acid) prosthetic groups are essential for the catalytic activity of four important multienzyme complexes in plants and other organisms: pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGDH), branched-chain α-ketoacid dehydrogenase (BCDH), and the Gly cleavage system (glycine decarboxylase [GDC]; Perham, 2000; Douce et al., 2001; Mooney et al., 2002). In all these multienzyme complexes, LA is covalently attached to the ε-amino group of a particular lysyl residue of the respective protein subunit. Lipoylated E2 subunits of PDH, KGDH, and BCDH are dihydrolipoyl acyltransferases that interact with E1 and E3 subunits to pass acyl intermediates to CoA (Mooney et al., 2002). By contrast, the lipoylated H-protein of GDC acts as a cosubstrate of three other GDC proteins and has no enzymatic activity itself (Douce et al., 2001). In the course of their respective reaction cycles, LA becomes reduced to dihydrolipoic acid. Most of these enzymes are confined to the mitochondrion. As the only exception, PDH is also present in plastids, where it provides acetyl-CoA for fatty acid biosynthesis (Ohlrogge et al., 1979; Lernmark and Gardeström, 1994; Lin et al., 2003).
Mitochondria and plastids each have their own route of de novo LA synthesis, both of which start with the synthesis of protein-bound octanoyl chains (Shimakata and Stumpf, 1982; Ohlrogge and Browse, 1995; Wada et al., 1997; Gueguen et al., 2000; Yasuno et al., 2004). These octanoyl moieties are passed on by organelle-specific octanoyltransferases (Wada et al., 2001a, 2001b) to the respective target apoproteins where lipoyl synthase (LIP1) inserts two sulfur atoms to finally produce functional lipoyl groups (Yasuno and Wada, 1998, 2002; Zhao et al., 2003). A similar pathway has been identified in mammalian mitochondria (Morikawa et al., 2001; Witkowski et al., 2007). In quantitative terms, leaf mesophyll mitochondria have an extraordinarily high requirement for lipoate, because they contain very large amounts of GDC to catalyze the photorespiratory Gly-to-Ser conversion (Bauwe et al., 2010). For this reason, leaf mesophyll mitochondria are the major site of LA synthesis in plants (Wada et al., 1997).
It was thought that the octanoyl chains provided by mitochondrial β-ketoacyl-acyl carrier protein synthase (mtKAS) represent the solitary source for protein lipoylation in plant mitochondria (Yasuno et al., 2004). As we reported earlier, however, leaves of mtKAS-deficient knockout mutants show considerable lipoylation of mitochondrial PDH-E2 and KGDH-E2 subunits and some residual lipoylation of GDC H-protein; roots are not at all impaired. Accordingly, the phenotype of such mutants can be fully cured in the low-photorespiratory condition of elevated CO2 (Ewald et al., 2007). These observations indicated that plant mitochondria, in addition to the mtKAS-LIP2-LIP1 route of protein lipoylation, can resort to an alternative pathway. This would not be uncommon. In Escherichia coli, for example, a salvage pathway utilizes free octanoate or LA in an ATP-dependent two-step reaction catalyzed by the bifunctional enzyme lipoate-protein ligase A (LPLA; Morris et al., 1995). Archaea (Christensen and Cronan, 2009; Posner et al., 2009) and vertebrates (Tsunoda and Yasunobu, 1967) require two separate enzymes to first activate octanoate or LA to lipoyl-nucleoside monophosphate (NMP) and then, in a second step, to convey the activated lipoyl group to the respective target proteins. The lipoate-activating enzyme (LAE) of mammals was identified as a refunctioned medium-chain acyl-CoA synthetase that utilizes GTP to produce lipoyl-GMP (Fujiwara et al., 2001). LIP3 from yeast (Saccharomyces cerevisiae) can use octanoyl-CoA to octanoylate apoE2 proteins (Hermes and Cronan, 2013), whereas octanoyl groups from fatty acid biosynthesis are first attached to H-protein and then passed on to apoE2 proteins (Schonauer et al., 2009).
The physiological significance of lipoyl-protein ligases in plants is not exactly known. Such enzymes do not operate in plastids (Ewald et al., 2014) but could be present in mitochondria. A single-gene-encoded LPLA with predicted mitochondrial localization has been identified in rice (Oryza sativa; Kang et al., 2007). Complementation studies with the lipoylation-deficient E. coli mutant TM137 (Morris et al., 1995) suggested that OsLPLA belongs to the bifunctional type of LPLAs. We report the identification of the homologous enzyme in Arabidopsis (Arabidopsis thaliana), provide evidence for its mitochondrial location, and show that Arabidopsis LPLA requires a separate enzyme for octanoate/lipoate activation. We also examine the interplay between LPLA, LIP2, and the mtKAS route of protein lipoylation and suggest a model for protein lipoylation in plant mitochondria.
RESULTS
AtLPLA Is Uniformly Expressed, AtLIP2 Particularly in Photosynthesizing Organs
We used the only reported plant LPLA amino acid sequence (Kang et al., 2007) to search public databases for the Arabidopsis homolog. Protein At3g29010 (henceforth AtLPLA), which is annotated as a member of the biotin/lipoate A/B protein ligase family, showed 74% sequence identity with OsLPLA and a similar size (for alignment, see Supplemental Fig. S1). The protein harbors known conserved domains for the binding of lipoyl-AMP and target proteins (Fujiwara et al., 2005; Kim et al., 2005) but lacks the C-terminal extension present in bifunctional LPLAs.
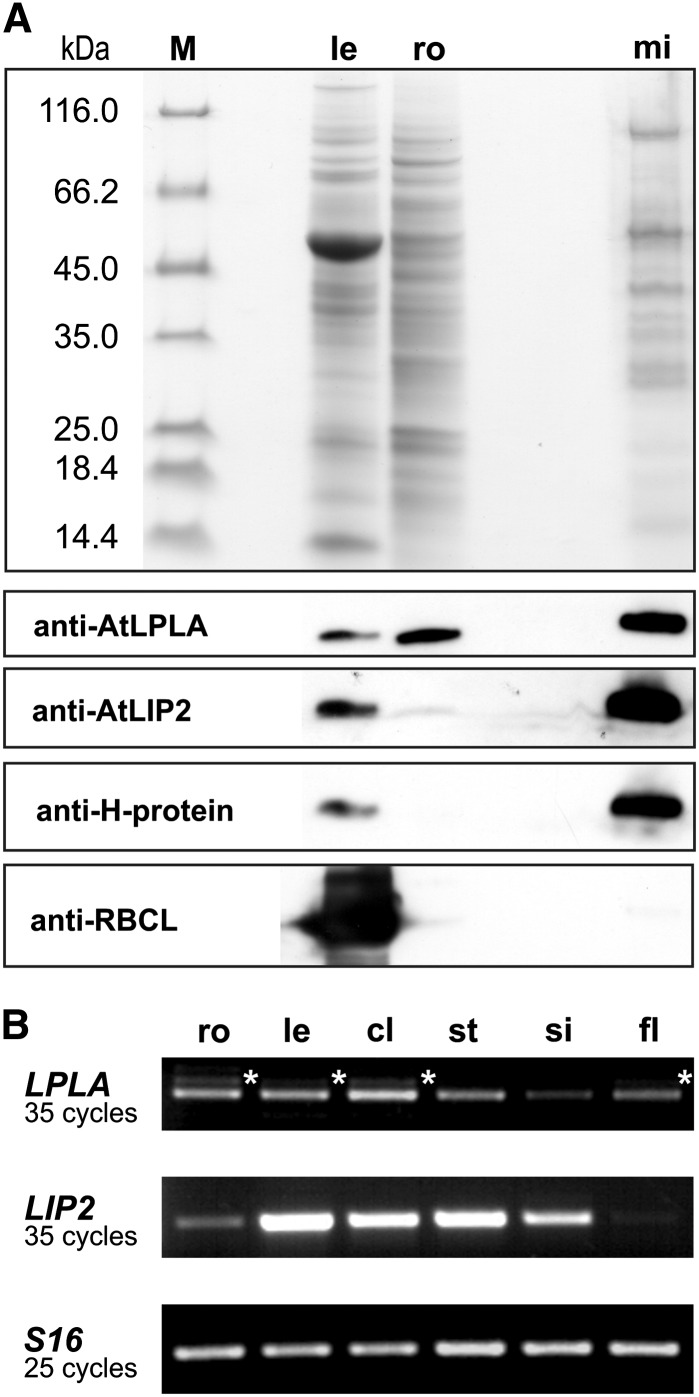
Examination of leaf, root, and mitochondrial matrix proteins (Fig. 1A) by immunoblotting using an immunopurified antibody raised against recombinant AtLPLA and antibodies reacting with control proteins revealed the mitochondrial location of AtLPLA. The purified mitochondria did not show signals with an antibody directed against the Rubisco large subunit, which excludes that the signals obtained by the LPLA antibody could be caused by plastidial proteins. Moreover, the expression pattern of an N-terminal fusion of AtLPLA with GFP also suggested a mitochondrial location of LPLA (Supplemental Fig. S2). Hence, both experiments consistently confirmed the in silico prediction for the location of OsLPLA in rice (Kang et al., 2007). Relative to total soluble protein, the protein is present to about similar levels in leaves (somewhat less) and roots (somewhat more) of Arabidopsis, corresponding to the approximately uniform expression of the AtLPLA gene at the mRNA level in different organs of mature plants (Fig. 1B). The expression profile of AtLPLA in Arabidopsis differs slightly from that in rice, where expression was higher in leaves and seeds and lower in flowers and roots (Kang et al., 2007). Also, a noticeable fraction of LPLA transcripts was incompletely spliced in Arabidopsis and still included the last or even the last two intron sequences present in the pre-mRNA (data not shown). Electronic northern data indicate that AtLPLA is strongly expressed during seed germination and embryo development (Supplemental Fig. S3). By contrast, the AtLIP2 gene is much more strongly expressed in leaves than in heterotrophic organs (Fig. 1) and coregulated with photorespiratory genes (Supplemental Fig. S3), as shown by similar expression patterns in combination with high r values (Toufighi et al., 2005) to At5g36790 (AtPGLP1 = 0.814) and At2g35370 (AtGLDH1 = 0.822).
Figure 1.
AtLPLA and AtLIP2 are mitochondrial enzymes expressed in leaves and roots. A, Protein-stained SDS gel (top) with 10 µg of protein per lane of leaf (le), root (ro), and mitochondrial matrix proteins (mi), and immunoblots (bottom) treated with antibodies raised against recombinant AtLPLA, AtLIP2, H-protein (mitochondrial control), and the large subunit of Rubisco (as a plastidial control). Lane M shows size-marker proteins. B, RT-PCR using total RNA from roots (ro), rosette leaves (le), cauline leaves (cl), stems (st), siliques (si), and flowers (fl). Transcripts of the constitutively expressed 40S ribosomal protein S16 were used for calibration. Oligonucleotide primers are listed in Supplemental Table S1. Stars indicate alternative splicing.
Arabidopsis LPLA Requires a Separate, Unknown Enzyme for Substrate Activation
Putative LPLAs from higher plants and green algae form a sister group to one of the two mitochondrial lipoate ligases of the erythrocytic parasite Plasmodium falciparum, PfLipL2 (Supplemental Fig. S4). Interestingly, this particular enzyme was able to complement the lipoylation-deficient strain E. coli TM136 only in the presence of lipoate, while complementation with the paralogous PfLipL1 was possible without external lipoate (Allary et al., 2007). Using a similar strategy, we tested whether the expression of AtLPLA would cure the lipoylation-deficient E. coli mutant TM137 (Morris et al., 1995). This particular strain, similar to TM136, lacks lipoate-protein ligase and octanoyltransferase and cannot grow on minimal medium with Glc as the only carbon source, even in the presence of external LA. However, TM137 can grow if nonlipoylated PDH and KGDH are bypassed by supplementation with acetate and succinate. This strain was transformed with a pBAD-HisA-based overexpression construct harboring AtLPLA, and the experiment was designed to match the conditions described by Kang et al. (2007) as closely as possible; however, the observed complementation effect was, at most, marginal (Supplemental Fig. S5A).
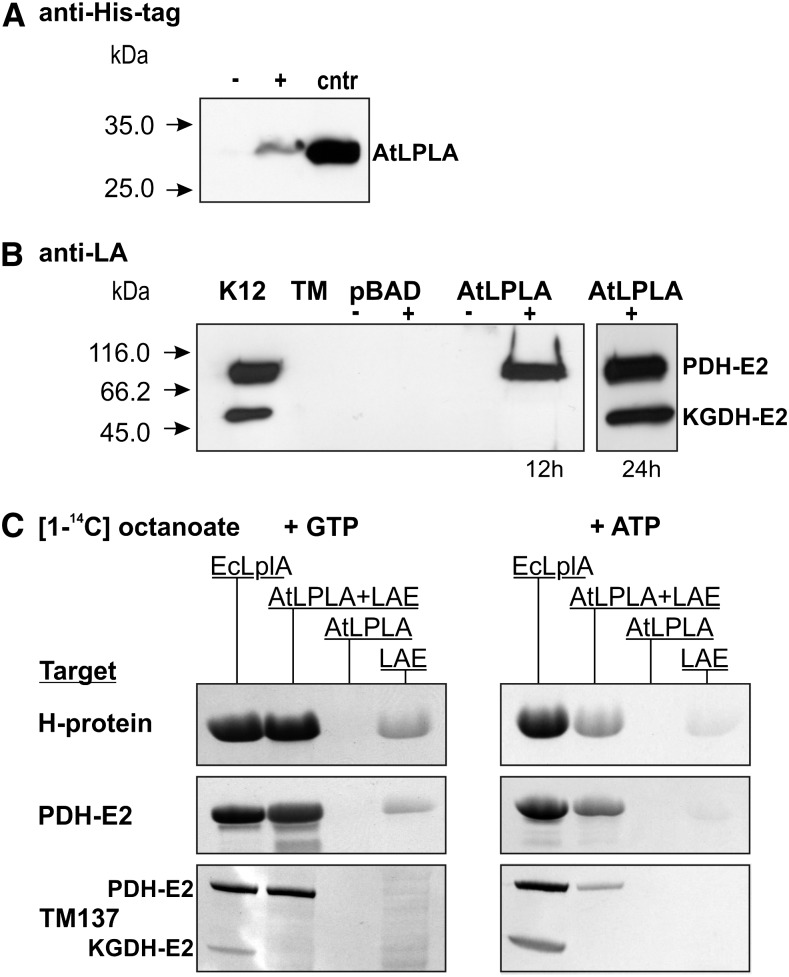
To find out why AtLPLA was unable to restore the growth of TM137, we first confirmed that the transgene is expressed by testing for the presence of the fused His tag with an anti-His tag antibody (Fig. 2A). Usage of an antibody directed against lipoylated epitopes then demonstrated that the lipoylation of apoPDH-E2, but not apoKGDH, was restored in comparison with the E. coli K12 wild-type control and the lipoylation-deficient TM137 control after 12 h of growth in liquid minimal medium (Fig. 2B). Lipoylation of apoKGDH-E2 was observed only in the 24-h sample and is likely due to an uncharacterized enzyme activity in E. coli, as discussed for a related experiment with Thermoplasma acidophilum LPLA (Christensen and Cronan, 2009). Alternatively, AtLPLA could very slowly transfer octanoyl chains bound to octanoyl-acyl carrier protein (ACP), similar to a reported side reactivity of EcLplA (Jordan and Cronan, 2003). This explanation is supported by our observation that the addition of succinate led to higher growth stimulation than the addition of acetate, which also suggests that it is KGDH deficiency that becomes growth limiting after the expression of AtLPLA (Supplemental Fig. S5B).
Figure 2.
Activity of AtLPLA in vivo and in vitro. A, AtLPLA is expressed in TM137, as shown with a His tag-specific antibody. Ten micrograms of total soluble protein was loaded per lane from uninduced (−) and induced (+) TM137 pBAD-HisA-LPLA cells. Five micrograms of affinity-purified His-tagged AtLPLA was loaded as a control (cntr). B, AtLPLA expression in TM137 results in rapid lipoylation of E. coli PDH-E2 and very slow lipoylation of KGDH-E2. K12, E. coli K12 control; TM, LplA− LipB− strain TM137; pBAD, empty pBAD-HisA vector transformed into TM137; AtLPLA, pBAD-HisA-LPLA transformed into TM137 after 12 and 24 h of growth without (−) and with (+) induction of expression with 0.2% Ara and appropriate antibiotics. C, AtLPLA requires substrate activation by a separate enzyme. The autoradiograms show [14C]octanoyl signals after SDS-PAGE of in vitro assay mixtures with 0.19 mm [1-14C]octanoic acid and 5 mm GTP (left) or ATP (right) and the three different target proteins Arabidopsis H-protein (5 µg), Arabidopsis PDH-E2 (2.5 µg), and TM137 soluble proteins (50 µg). Octanoate is not attached without NTP. Assay variants were 2.5 µg of E. coli LPLA as a positive control, 2.5 µg of AtLPLA plus 0.5 µg of bovine LAE, 2.5 µg of AtLPLA without LAE, and 0.5 µg of bovine LAE.
To confirm the absence of reactivity with KGDH-E2, we tested the target protein specificity of AtLPLA in vitro, which excludes ill-characterized side effects that can occur in vivo. This was done by using [14C]octanoic acid in combination with recombinant plant H-protein and PDH-E2 protein and soluble TM137 proteins (Fig. 2C). E. coli EcLplA, which was used as a positive control for a bifunctional LPLA, efficiently octanoylated the two plant proteins and also PDH-E2 and KGDH-E2 present in the TM137 protein mix. AtLPLA alone did not show any activity in this assay. However, AtLPLA octanoylated the two plant proteins and the bacterial apoPDH-E2, but not bacterial apoKGDH-E2, in the presence of recombinant Bos taurus lipoate-activating enzyme and GTP or ATP. The signal observed with ATP was distinctly weaker in comparison with GTP but not as much (1,000-fold) as reported for bovine LAE (Fujiwara et al., 2001). LAE produced a very small degree of unspecific background, which cannot be due to contamination by bacterial LplA or LipB. This possibility can be excluded because, except for EcLplA, all proteins used in this experiment, including LAE, were produced in TM137. Hence, these results clearly show that AtLPLA is an octanoate-protein ligase that, unlike EcLplA and other bifunctional LPLAs, requires octanoate/lipoate activation by a separate enzyme. In comparison with EcLPLA, AtLPLA is also more selective in its choice of target proteins and does not octanoylate the KGDH-E2 protein.
Both AtLPLA and AtLIP2 Are Essential for Embryo Development
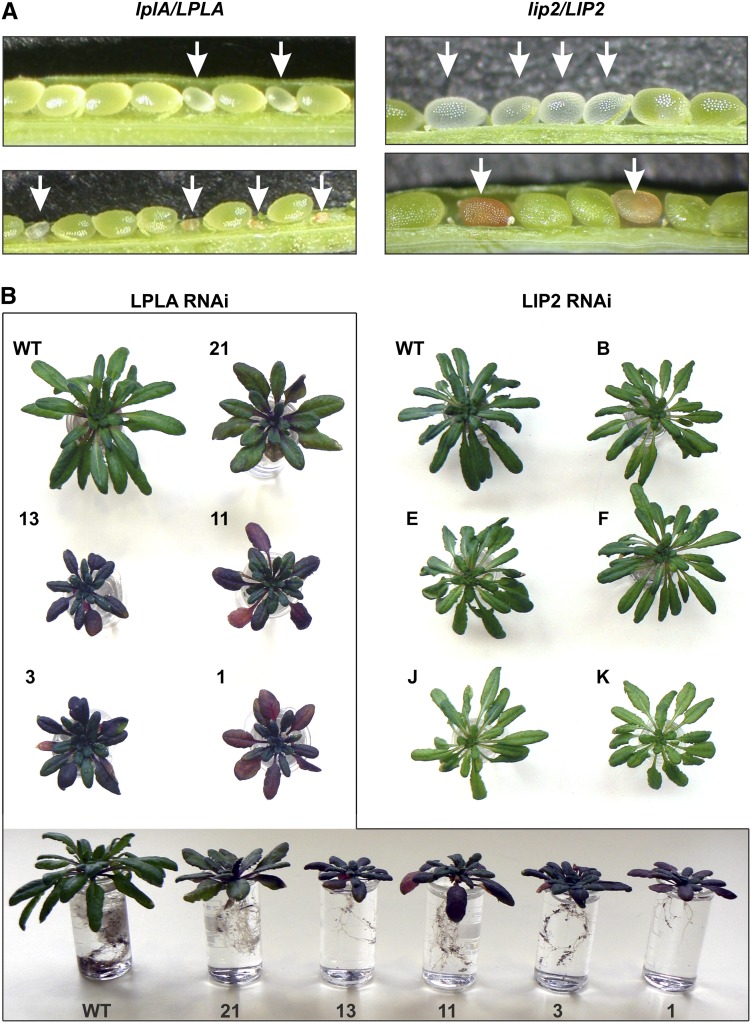
LplA and LipB (LPLA and LIP2 in Arabidopsis) participate in redundant lipoylation pathways in E. coli (Morris et al., 1995), but the simultaneous presence of both enzymes is necessary for protein lipoylation in yeast (Schonauer et al., 2009) and an LPLA-related enzyme is required for E2 protein lipoylation in human (Soreze et al., 2013). To better understand the role of LPLA and LIP2 in plants, we attempted to isolate homozygous transfer DNA (T-DNA) insertion lines (Supplemental Fig. S6). After selfing of more than 60 individual heterozygous plants for each of the two examined T-DNA insertion lines, however, homozygous progeny could not be detected. This was also the case if the plants were grown in air containing 1% CO2 to prevent negative effects from the accumulation of photorespiratory intermediates, which would occur if a significant fraction of leaf H-protein remains unlipoylated. Siliques from heterozygous individuals from both lines displayed atypically large numbers of aborted seeds (Fig. 3A), which were homozygous, as confirmed by PCR with genomic DNA from young aberrant mutant seeds and wild-type seeds as a control (data not shown).
Figure 3.
LPLA and LIP2 are essential for normal seed and plant development of Arabidopsis. A, Young (top) and fully developed (bottom) siliques of selfed heterozygous lplA/LPLA and lip2/LIP2 plants. Arrows point to aberrant seeds. Segregation ratios are shown in Table I. B, Phenotyping of LPLA RNAi (boxed) and LIP2 RNAi lines. Plants were grown for 7 weeks in air containing 1% CO2 to exclude overlap from possibly impaired photorespiratory metabolism. Blue-red discoloration, particularly of the older leaves of LPLA RNAi plants, begins within 1 week after the start of bolting. It was not observed in LIP2 RNAi plants at any developmental stage. WT, Wild type.
Table I shows significant 3:1 segregation ratios. Wild-type plants outcrossed from lplA and lip2 heterozygotes showed very few aborted seeds (less than 1%), corresponding to typical numbers for the Columbia-0 (Col-0) wild type. More potential knockout lines for these two genes were not available from seed stocks, and none for the lipoyl synthase gene AtLIP1. LPLA and LIP2 apparently do not participate in redundant pathways but are essential for mitochondrial protein lipoylation already during the seed development of Arabidopsis. This conclusion implies that the protein lipoylation network in Arabidopsis mitochondria is more similar to that in eukaryotes than in E. coli.
Table I. Segregation of normal-looking and aborted seeds of LPLA and LIP2 T-DNA insertion lines (seven plants each) compared with Col-0 wild-type plants (3:1; P < 0.05, χ2 = 3.84).
| Genotype | Total Seeds | Normal Seeds | Aborted Seeds | Percentage of Aborted Seeds | χ2 |
|---|---|---|---|---|---|
| lplA/LPLA | 835 | 646 | 189 | 22.6 | 2.49 |
| lip2/LIP2 | 748 | 580 | 168 | 22.5 | 2.57 |
| LPLA/LPLA | 996 | 991 | 5 | 0.5 | – |
| LIP2/LIP2 | 825 | 820 | 5 | 0.6 | – |
| Col-0 | 866 | 863 | 3 | 0.3 | – |
LPLA Is Necessary for the Lipoylation of PDH-E2 Proteins in Leaves and Roots
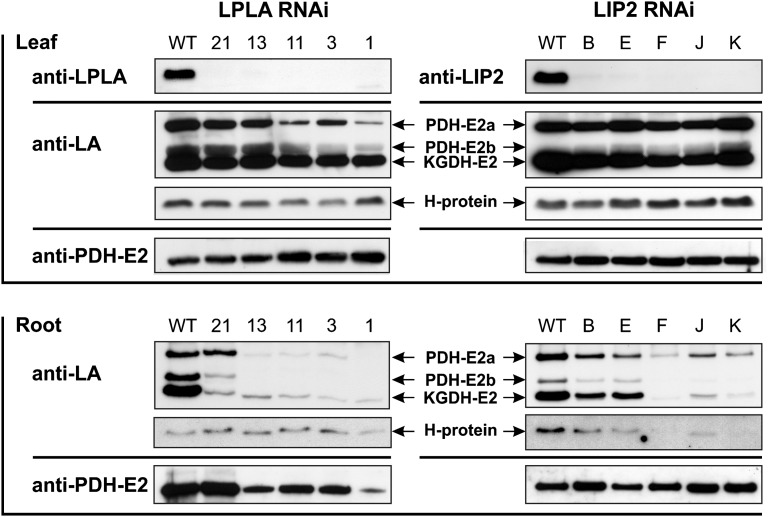
Since AtLPLA and AtLIP2 null mutants were not viable, we generated RNA interference (RNAi) lines to examine knockdown effects. Individual plants were examined by immunoblotting using specific antibodies directed against the two impaired enzymes, LA epitopes and apoPDH-E2 protein. Lines with reduced LPLA and LIP2 levels were selected under high-CO2 conditions (1%) to avoid potential negative effects caused by H-protein dysfunction in the photorespiratory pathway (Fig. 3B). The LPLA RNAi lines showed growth retardation of rosettes and roots. Within 1 week after the start of bolting, these plants displayed a strong blue-red discoloration, particularly of older leaves, indicating that LPLA deficiency caused severe metabolic stress. This can also be seen in the considerable increases of leaf Pro contents in all lines: for example, a significant 35-fold increase in the RNAi line LPLA-11 (35.4 ± 11.1-fold) or an 80-fold increase in the RNAi line LPLA-3 (82.3 ± 5.4-fold). In leaves, there was a very distinct reduction of PDH-E2 lipoylation, whereas the levels of apoPDH-E2, lipoylated KGDH-E2, and H-protein were barely affected (Fig. 4). Due to the different flux modes of the tricarboxylic acid cycle, as discussed by Sweetlove et al. (2010), corresponding changes in the in vivo activities of these enzymes are difficult to quantify. As a suitable proxy, we examined leaf metabolite levels of intermediates of the photorespiratory and tricarboxylic acid cycles (Table II). The changes observable in Gly, Ser, and glycerate indicated that the perturbation of photorespiratory metabolism was relatively small. Similarly, there were also no clear changes in the levels of pyruvate, citrate, and α-ketoglutarate. Notably, however, the levels of malate, succinate, and fumarate were significantly increased in several lines. The levels of γ-amino butyric acid (GABA), which is the key intermediate of the GABA shunt, which can bypass the reactions catalyzed by KGDH and succinyl-CoA ligase (Studart-Guimarães et al., 2007), were also significantly increased (5- to 8-fold) in several lines. Additional metabolite data are shown in Supplemental Table S2. In roots, PDH-E2 lipoylation levels were heavily reduced in most lines. Moreover, there was also a distinct reduction in KGDH-E2 lipoylation, whereas H-protein again was essentially unaltered.
Figure 4.
Immunoblots of RNAi lines with reduced contents of LPLA and LIP2. Ten micrograms of leaf and 5 µg of root proteins were separated on SDS-polyacrylamide gels and blotted onto polyvinylidene difluoride membranes. Antibodies used were directed against Arabidopsis LPLA (anti-LPLA), Arabidopsis mitochondrial octanoyltransferase (anti-LIP2), LA epitopes of lipoylated E2 proteins and H-protein (anti-LA), and a fragment of Arabidopsis mitochondrial apoPDH-E2a (At3g52200; anti-PDH-E2a). PDH-E2 variants were named as described by Taylor et al. (2002). Numbers and letters represent individual RNAi lines. WT, Wild type.
Table II. Leaf metabolite profiling of wild-type and LPLA RNAi plants.
Values are presented as nmol g−1 fresh weight (means ± se from four individual plants). Values in boldface are significantly different from wild-type values according to a two-tailed Student’s t test (P < 0.05). n.d., Not determined.
| Metabolite | Wild Type | LPLA-1 | LPLA-3 | LPLA-11 | LPLA-13 | LPLA-21 |
|---|---|---|---|---|---|---|
| Gly | 0.11 ± 0.02 | 0.29 ± 0.09 | 0.33 ± 0.03 | 0.13 ± 0.03 | 0.23 ± 0.06 | 0.12 ± 0.01 |
| Ser | 0.84 ± 0.09 | 1.49 ± 0.42 | 1.90 ± 0.13 | 1.20 ± 0.22 | 1.21 ± 0.17 | 1.27 ± 0.35 |
| Glycerate | 0.18 ± 0.02 | 0.40 ± 0.04 | 0.54 ± 0.01 | 0.37 ± 0.03 | 0.32 ± 0.02 | 0.26 ± 0.05 |
| Pyruvate | 0.11 ± 0.02 | 0.11 ± 0.02 | 0.18 ± 0.02 | 0.16 ± 0.01 | 0.14 ± 0.03 | 0.20 ± 0.03 |
| Citrate | 0.23 ± 0.04 | 0.24 ± 0.09 | 0.07 ± 0.02 | 0.08 ± 0.04 | 0.14 ± 0.08 | 0.60 ± 0.21 |
| α-Ketoglutarate | n.d. | 0.07 ± 0.05 | 0.07 ± 0.01 | 0.04 ± 0.01 | 0.07 ± 0.05 | 0.28 ± 0.11 |
| Succinate | 0.19 ± 0.04 | 0.70 ± 0.05 | 0.71 ± 0.07 | 0.54 ± 0.08 | 0.59 ± 0.12 | 0.41 ± 0.12 |
| Fumarate | 8.03 ± 1.68 | 20.34 ± 4.67 | 13.89 ± 0.92 | 13.63 ± 1.01 | 16.72 ± 1.15 | 12.78 ± 3.53 |
| Malate | 1.35 ± 0.34 | 7.81 ± 1.71 | 13.90 ± 0.68 | 6.19 ± 0.65 | 9.74 ± 2.31 | 4.56 ± 1.22 |
| GABA | 0.03 ± 0.01 | 0.11 ± 0.03 | 0.26 ± 0.01 | 0.16 ± 0.02 | 0.17 ± 0.04 | 0.13 ± 0.04 |
By contrast, the phenotypes of LIP2 RNAi plants were not distinctly different from those of wild-type plants despite their very low LIP2 contents. Leaves of these lines showed somewhat reduced KGDH-E2 lipoylation, particularly in lines F and J, with possibly some minor changes in PDH-E2 lipoylation, but no alterations were visible for H-protein. Again, apoPDH-E2 levels were unchanged and similar to those in the wild type. For roots, the RNAi effects were more distinct, with a strong reduction in KGDH-E2, PDH-E2, and H-protein lipoylation in lines F, J, and K. Notably, roots of LIP2 RNAi plants were the only case where not only PDH-E2 and/or KGDH-E2 lipoylation was reduced but also an impaired H-protein lipoylation could be observed.
Arabidopsis Leaf GLDH Protein Is Fully Lipoylated in Vivo But Not PDH-E2 and KGDH-E2
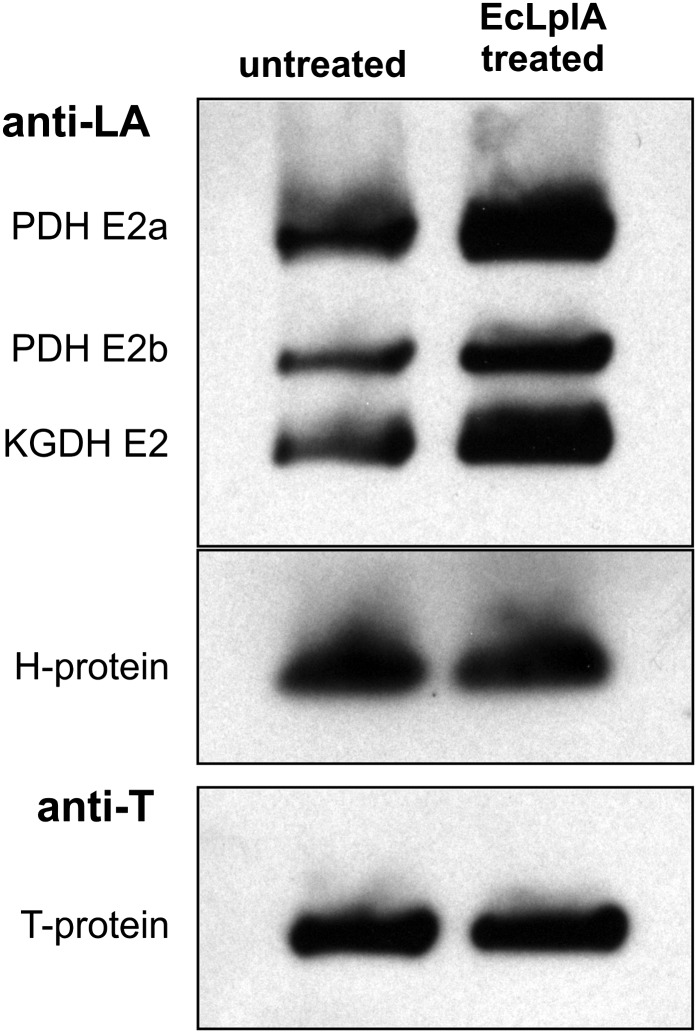
Finally, we were interested to find out whether the E2 proteins and the H-protein are fully lipoylated in organello. We examined whether the addition of E. coli LPLA, LA, and ATP would enhance the levels of lipoylation of matrix proteins prepared from purified leaf mitochondria (Fig. 5). In this experiment, we used an excess of recombinant EcLplA to exclude discrimination between the individual target proteins. An antibody against the GDC T-protein, which is an unlipoylated protein, provided an internal control for equal loading. We observed that a considerable fraction of both PDH-E2 and KGDH-E2 is present in the unlipoylated form in organello but can be fully lipoylated by EcLplA in vitro. Notably, this treatment did not enhance the lipoylation of the H-protein, indicating that this target protein is fully lipoylated in wild-type Arabidopsis leaf mitochondria. While this result does not provide conclusive evidence that the H-protein could act as an intermediate octanoyl acceptor in plants, it is in line with the sequence of events discussed for protein lipoylation in yeast (Hermes and Cronan, 2013).
Figure 5.
Arabidopsis GDC H-protein is fully lipoylated but PDH-E2 and KGDH-E2 are incompletely lipoylated in wild-type mitochondria. Immunoblotting of 10 µg of matrix protein prepared from purified wild-type Arabidopsis mitochondria was done without and with incubation with E. coli LPLA. ATP and LA were added to all samples, and lipoylation levels were examined using an anti-LA antibody. As an internal control, the blot was reincubated with antibodies specific for GDC T-protein.
DISCUSSION
There are two known sites of protein lipoylation in plant cells, plastids and mitochondria. Plastids vitally depend on a partially redundant de novo lipoylation pathway (Wada et al., 2001b; Yasuno and Wada, 2002; Ewald et al., 2014). By contrast, the inactivation of mitochondrial fatty acid synthesis, which triggers the de novo synthesis of lipoyl groups, can be well tolerated, particularly under nonphotorespiratory conditions (Ewald et al., 2007). This indicates that at least two routes exist in plants for the lipoylation of mitochondrial proteins. One of these pathways is well known and starts with the synthesis of octanoyl-ACP (Wada et al., 1997, 2001a; Yasuno and Wada, 1998). The other one is not exactly known but likely relies on the usage of compounds such as free lipoate or octanoate by LPLAs similar to that in other organisms.
To better characterize this second pathway in plants, we used the as yet only reported plant lipoate-protein ligase as a query sequence (Kang et al., 2007) and identified Arabidopsis protein At3g29010 as the homologous enzyme. We also confirmed the in silico prediction of Kang et al. (2007) that plant LPLAs are located in the mitochondrion. The AtLPLA gene is expressed to similar levels in all examined plant organs. By contrast, the AtLIP2 gene is preferentially expressed in leaves and other photosynthesizing organs and shows little expression in roots and flowers. Given that about 95% of leaf H-protein is lipoylated by the mtKAS-dependent de novo lipoylation pathway (Ewald et al., 2007), our study underlines the major contribution of de novo lipoylation in the supply of photorespiring leaf mitochondria with the needed massive amounts of lipoylated H-protein for high GDC activity (Fig. 4, leaf side). This also means that some basal protein lipoylation, including about 5% of H-protein lipoylation, occurs in an mtKAS-independent manner in the mitochondria of all plant organs. It is reasonable to suppose that this process likely requires lipoate-protein ligase. As mentioned, a candidate protein had already been identified (Kang et al., 2007), but functional data were not available.
To find out whether AtLPLA shows lipoate-protein ligase activity, we transformed an overexpression construct into the LplA− LipB− E. coli strain TM137 to test for complementation of the metabolic defects. Uncomplemented TM137 would require, in addition to Glc, acetate to bypass the inactive PDH and succinate to bypass the inactive KGDH; growth on Glc as the only carbon source would indicate the restoration of PDH and KGDH activity by the Arabidopsis protein (Morris et al., 1995). This experiment was set up exactly the same way as reported for the OsLPLA complementation of TM137 (Kang et al., 2007). Unexpectedly, however, transformation of AtLPLA into TM137 resulted in only some very minor growth enhancement on Glc as the only carbon source. At present, we cannot explain these different outcomes in our experiments and those of Kang et al. (2007). The very similar amino acid sequences of OsLPLA and AtLPLA suggest that they have similar catalytic features.
Lipoate-protein ligases are a diverse group of enzymes. Mammalian, yeast, and some prokaryotic LPLAs require lipoyl- or octanoyl-NMP (Christensen and Cronan, 2009) or octanoyl-CoA (Hermes and Cronan, 2013), which are provided by separate enzymes, such as the so-called lipoate-activating enzyme of bovine animals (Fujiwara et al., 2001). In the archaeon T. acidophilum, the two activities are sequestered on two proteins, LPLA and LPLB, encoded by adjacent genes, which form a bipartite hybrid LPLA (Christensen and Cronan, 2009). Other LPLAs, such as E. coli LPLA, carry an extra C-terminal domain (CTD) of about 80 to 100 amino acids that catalyzes the activation of lipoate/octanoate to lipoyl/octanoyl-AMP in these bifunctional enzymes. Plant LPLAs show adequate sequence identity to prokaryotic LPLAs only over the N-terminal range of about 200 to 210 amino acids but lack the CTD of bifunctional lipoate-protein ligases. This structural feature strongly suggested that plant LPLAs are not bifunctional but require a separate enzyme for substrate activation, explaining the very poor complementation of TM137 by AtLPLA.
This hypothesis fits well with our observation that apoKGDH-E2 is lipoylated with an extremely low efficiency in AtLPLA-expressing TM137, requiring distinctly more than 12 h for a detectable signal. On the other hand, the expression of AtLPLA resulted in wild-type-like levels of PDH-E2 lipoylation already after 12 h. While these data suggest different routes for the lipoylation of the two E2 proteins in TM137 cells expressing AtLPLA, they do not provide clear evidence in favor of or against our hypothesis that plant LPLAs depend on substrate activation by a separate enzyme. The donor substrate for apoPDH2 lipoylation by AtLPLA in TM137 is also somewhat elusive, because it cannot be provided by the inactivated EcLPLA in this strain. Similar to our results, a small complementation effect was observed when T. acidophilum LPLA, without the lipoate-activating partner enzyme LPLB, was expressed in the LipA− LplA− E. coli strain TM131 (Christensen and Cronan, 2009). These authors discussed that E. coli in addition to EcLplA possibly harbors another, yet unknown enzyme for lipoate/octanoate activation. Alternatively, the substrate specificity of AtLPLA could be relaxed and allow the usage of other donor substrates than octanoyl-NMP or lipoyl-NMP.
To exclude bias by unknown E. coli enzymes, we tested in vitro how efficiently AtLPLA can attach 1-14C-labeled octanoic acid to purified recombinant plant target proteins (apoH-protein and apoPDH-E2) and to extracted bacterial target proteins, all produced in the lipoylation-deficient strain TM137 to ensure that they are unmodified apoproteins. Because it was not possible to identify CTD, LPLB, or LAE homologs in plant protein databases, we produced recombinant bovine LAE (Fujiwara et al., 2001; Hasse et al., 2009) in TM137 to avoid contamination by E. coli LplA and LipB for use as the octanoate/lipoate-activating helper enzyme, which has highest activity with GTP and lower activity with ATP. This experiment clearly demonstrated that AtLPLA is an octanoyl-protein ligase, but its activity depends on the supply of octanoyl-GMP or octanoyl-AMP (ATP signals were visible but much weaker). The control with the bifunctional E. coli LplA produced signals for both bacterial apoE2 proteins, whereas LAE alone produced only some likely unspecific minor signals. It is also clear that AtLPLA cannot octanoylate bacterial apoKGDH-E2, at least with octanoyl-NMP, which is in line with our observation that succinate supplementation results in a higher growth enhancement than the addition of acetate to AtLPLA-expressing TM137. In the context of target protein specificity, it is interesting that the smaller of two lipoate ligases of apicomplexan parasites, such as the malaria parasite Plasmodium falciparum, PfLipL2, is more similar to plant LPLAs than the somewhat larger PfLIP1 and also shows a distinct if not exclusive preference for E. coli PDH-E2 (Allary et al., 2007).
Collectively, these data demonstrate that LPLA from Arabidopsis and other plants are octanoate/lipoate protein ligases that require a separate enzyme for substrate activation and have a more restricted target protein spectrum in comparison with EcLplA. We tested whether a chimeric protein comprising the entire AtLPLA sequence fused to the C-terminal domain of E. coli LPLA (the last 85 amino acids) would show lipoylation or octanoylation activity without LAE, but these experiments were unsuccessful so far (data not shown). Bovine LAE has similarities to members of the superfamily of plant acyl-activating enzymes (Shockey et al., 2003; Souza et al., 2008), and it remains to be seen whether plant LAE can be identified in this group.
The role of LPLA in plants has not been examined before. Plants can get along very well without the mtKAS pathway of mitochondrial fatty acid biosynthesis (Ewald et al., 2007), at least in nonphotorespiratory conditions. Hence, it was sensible to assume that, in the presence of LPLA, mitochondrial octanoyltransferase LIP2 would also be dispensable; however, this was not the case. We first attempted to select T-DNA insertion knockout lines for LPLA and LIP2. Only a few such lines are available in public seed stocks for LPLA and LIP2 and, to our knowledge, none for LIP1. In repeated trials, we failed to produce homozygous knockout lines for both enzymes but observed aberrant seeds in a 1:3 segregation ratio in siliques of selfed heterozygous individuals. These seeds were homozygous, which demonstrates that plants cannot tolerate the deletion of either enzyme, at least during seed development. Similarly, RNAi plants with lower levels of LPLA were heavily impaired in normal air (data not shown), and their performance was not improved in high-CO2 conditions. Slow leaf and root growth and purple leaves indicated heavy metabolic damage and severe stress. The considerably elevated leaf contents of Pro, which is a stress response indicator (Kavi Kishor and Sreenivasulu, 2014), also point in that direction. Lower LIP2 activity in a corresponding set of LIP2 RNAi lines was distinctly better tolerated, even with LIP2 contents that were below the detection limit by immunoblotting.
Concerning the lipoylation of target proteins in planta, less LPLA caused progressively reduced levels of PDH-E2 lipoylation in leaves and roots of Arabidopsis, whereas KGDH-E2 lipoylation was barely affected in leaves but clearly reduced in roots. H-protein lipoylation was essentially unaltered in leaves and roots of LPLA RNAi plants. We also examined the levels of total PDH-E2; the observed alterations (slightly more in leaves and less in roots) were uncorrelated to the changes in the lipoylation profiles. As is seen in the changes of leaf metabolite levels shown in Table II, altered lipoylation profiles translate into altered metabolism, with minor adjustments of the photorespiratory cycle and more distinct perturbations in the tricarboxylic acid cycle. Given the possible operation of different flux modes of the tricarboxylic acid cycle, it is quite difficult to discriminate which of the observed changes are due to an effect on PDH and which on an effect on KGDH. That said, it seems likely that the changes in α-ketoglutarate and GABA levels are consequences of the partial KGDH knockdown, while the changes in fumarate, malate, and succinate could well be due to reduced activity of the PDH and the operation of the tricarboxylic acid cycle in the reverse direction (Rocha et al., 2010; Sweetlove et al., 2010).
The lower total PDH-E2 content in roots indicates higher protein degradation in these highly stressed plants, which to some extent could bias our immunoblotting assessment of protein lipoylation in this particular case. In summary, the changes in protein lipoylation in LPLA RNAi plants show that LPLA is crucial not only for seed but also general plant development. Additionally, they confirm our view that LPLA activity is essential for apoPDH-E2 lipoylation and cannot be replaced by the mtKAS-LIP2 route. By contrast, apoKGDH-E2 lipoylation is not directly dependent on LPLA in leaf mitochondria but is clearly related to LPLA activity in roots. In combination, the high effects of the AtLPLA knockdown on apoPDH-E2 lipoylation in leaves and roots in comparison with minor (leaves) and distinct (roots) effects on apoKGDH lipoylation suggest two conclusions (Fig. 6). First, apoPDH-E2 is exclusively lipoylated by AtLPLA in leaves and roots. Second, apoKGDH-E2 is exclusively lipoylated by AtLPLA in roots but can become lipoylated via the mtKAS-LIP2 pathway in leaves. Additionally, the large knockdown effects on the E2 proteins in roots of LPLA RNAi plants are not accompanied by corresponding changes in H-protein lipoylation. This observation suggests that H-protein may be lipoylated via mtKAS-LIP2-LIP1 also in roots, with some degree of LIP2-mediated octanoyl transfer to the E2 proteins.
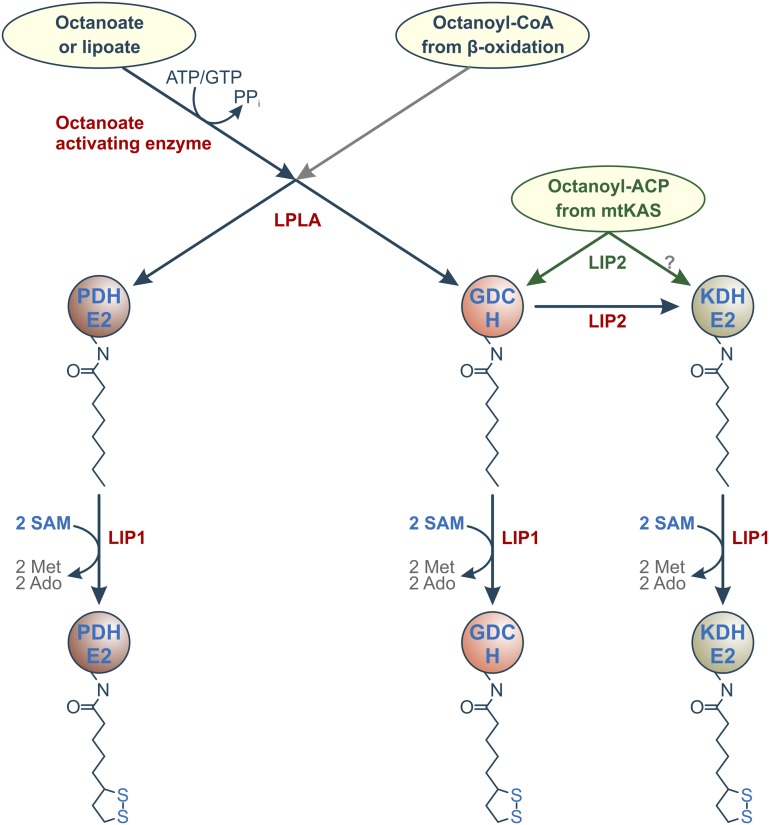
Figure 6.
Model of protein lipoylation in plant mitochondria. Given that protein lipoylation in roots is fully independent on fatty acid biosynthesis (Ewald et al., 2007), it is likely that H-protein acts as an octanoyl-relay protein, as in other organisms (Schonauer et al., 2009). The basal protein lipoylation network is supplemented by the mtKAS route in photorespiring organs (drawn in green). It will require in vitro studies to decide whether LIP2 can octanoylate KGDH-E2 with octanoyl-ACP as a donor. As yet, there is no evidence that the gray route starting from octanoyl-CoA operates in plants as it does in yeast (Hermes and Cronan, 2013).
We have yet no explanation for why leaves of LPLA RNAi plants displayed a very characteristic discoloration from green to purple within 1 week after bolting. However, this striking phenotypic difference between the two sets of RNAi plants supports our view that AtLPLA and AtLIP2 contribute to different routes of mitochondrial protein lipoylation. Corresponding to the better health of LIP2 RNAi plants, the total PDH-E2 control showed no significant alteration relative to the wild type in leaves or roots. Also, alterations in protein lipoylation in plants were less pronounced in leaves, in which apoPDH-E2 and H-protein lipoylation were at wild-type levels despite a very considerable reduction in LIP2. The effects of low LIP2 contents on the lipoylation of root proteins were much stronger than in leaves and, notably, included H-protein. This result was somewhat unexpected, because H-protein is known to be lipoylated primarily via the de novo pathway, at least in leaf mitochondria, with their very high H-protein requirement of the photorespiratory pathway (Wada et al., 1997; Ewald et al., 2007). It appears that very low activities of LPLA or LIP2 mainly impair the lipoylation of leaf PDH-E2 or KGDH-E2, respectively, but are not limiting for H-protein lipoylation in leaves. This preferential lipoylation of H-protein in plant leaves is similar to protein lipoylation in yeast, where LIP2 preferentially octanoylates the H-protein, which then acts as an intermediate octanoyl donor to E2 proteins (Schonauer et al., 2009). A recent addition to this concept for yeast is that octanoyl-ACP accumulates once all H-protein is octanoylated, triggering octanoyl-CoA synthesis by acyl-ACP:CoA transferase with subsequent octanoyl-CoA-dependent octanoylation of E2 proteins by LIP3 (Hermes and Cronan, 2013). Recent data on the orthologous human lipoyltransferase LIPT1 point in the same direction by the demonstration that LIPT1 mutations cause fatal PDH and KGDH deficiencies (Soreze et al., 2013). At present, some features of mtKAS knockout plants (Ewald et al., 2007) and the fact that AtLPLA requires octanoyl-NMP (this study) suggest that the lipoylation network at least of leaf mitochondria, with their very high H-protein content, may operate in a similar but still somewhat different manner. The major difference seems to be that mitochondrial fatty acid synthesis, although it also occurs in other organisms (Witkowski et al., 2007), has a very high capacity in photosynthesizing plant cells that can be used for KGDH-E2 lipoylation. It remains to be shown whether this occurs directly from octanoyl-ACP or indirectly via H-protein. The unimpaired lipoylation of root proteins in mtKAS knockout plants (Ewald et al., 2007), however, shows that the de novo pathway is not crucial but an adaptation to the specific lipoate requirements of the high-flux photorespiratory pathway in leaves, as suggested by Wada et al. (1997). LPLA together with LIP2 would then provide a more basal essential protein lipoylation network that operates independently in all plant cells.
As in other organisms, the substrate specificities of LIP2, LPLA, and obviously the elusive lipoate/octanoate-activating enzyme in plants are not exactly known. In accordance with the models proposed for yeast (Schonauer et al., 2009; Hermes and Cronan, 2013), several of our observations suggest that the significance of H-protein in plants could also go beyond its function during Gly decarboxylation and include functioning as an intermediate octanoyl donor to apoE2 proteins, particularly apoKGDH-E2. This hypothesis receives some support from our demonstration that only H-protein is fully lipoylated in wild-type leaf mitochondria but not in the E2 proteins (Fig. 5), but this provides no conclusive evidence. Roots of LIP2 RNAi plants were the only observed case were the levels of not only the examined holo-E2 proteins but also the lipoylated H-protein decreased progressively, which could be cautiously interpreted as an H-protein:apoKGDH-E2 donor:acceptor relationship. It is difficult to test models of yeast protein lipoylation in plants, because they are unable to survive without functional GDC (Engel et al., 2007). However, the perception of H-protein as an octanoyl/lipoyl donor protein in many organisms is quite sensible also in light of the fact that this protein is present even in organisms that have lost other GDC components (Spalding and Prigge, 2010). It is also interesting that a recently discovered novel kind of amidotransferase (LIPL) transfers octanoyl moieties from octanoyl-H-protein to E2 apoproteins in Bacillus subtilis (Christensen et al., 2011; Martin et al., 2011). Again, LIPL homologs are difficult to identify in plant protein databases, but this does not exclude that such enzymes could be present in plants. In light of these studies with other organisms and our own observation of fully lipoylated H-protein in combination with incomplete lipoylated E2 proteins in Arabidopsis RNAi plants, it is well possible but still needs to be proven by in vitro studies that H-protein could act as an intermediate donor of octanoyl or lipoyl groups in plants, too. This would require future evidence that AtLPLA cannot directly react with plant KGDH-E2, which would be different from LIP3 in yeast (Hermes and Cronan, 2013) and possibly LIPT1 in mammals (Soreze et al., 2013).
In conclusion, we have identified Arabidopsis LPLA as an essential mitochondrial enzyme that can use octanoyl-NMPs as the donor substrate to octanoylate apoPDH-E2 and apoH-proteins but shows no reactivity with bacterial and likely plant KGDH-E2. While octanoyl-GMP and octanoyl-AMP are substrates for LPLA in vitro, the octanoate-activating enzyme of plants is not known, and usage of other substrates is possible. In planta, LPLA is crucial for the lipoylation of apoPDH-E2, and this function cannot be replaced by the mtKAS route. H-protein and KGDH-E2 are mostly lipoylated via the mtKAS-LIP2-LIP1 route in leaves. Given that protein lipoylation is mtKAS independent in roots (Ewald et al., 2007), these two proteins are also lipoylated via the LPLA-LIP2-LIP1 sequence of reactions, in which H-protein likely acts as an octanoyl donor to apoKGDH-E2. We assume that the required octanoyl chains are provided by the β-oxidation of imported of acyl-CoA entities (Masterson and Wood, 2000, 2009). The likely events on a basal lipoylation route in plants would then be octanoylation of PDH-E2 and H-protein by LPLA followed by octanoyl transfer from H-protein to KGDH-E2, as illustrated in Figure 6.
MATERIALS AND METHODS
Plants and Growth Conditions
Wild-type Arabidopsis (Arabidopsis thaliana) ecotype Col-0 and T-DNA insertion lines (Sessions et al., 2002; Alonso et al., 2003) for AtLPLA (SALK_003427) and AtLIP2 (SAIL_662_H11 and SAIL_1229_F08) were obtained from the Nottingham Arabidopsis Stock Centre (http://nasc.nott.ac.uk). Plants were grown under a 10-h-light/14-h-dark (22°C/18°C) cycle at 100 to 150 μmol photons m−2 s−1 in controlled environment chambers.
Reverse Transcription-PCR Studies
Complementary DNA (cDNA) was obtained from total RNA purified from different Arabidopsis organs harvested in the middle of the light period (Nucleospin RNA Plant Kit [Macherey-Nagel] and RevertAid H minus cDNA synthesis kit [MBI Fermentas]). AtLPLA transcripts were amplified by PCR using primers AtLPLA-S2 (R1062) and AtLPLA-A (R1068). All primer sequences used in this work are shown in Supplemental Table S1. For AtLIP2 transcripts, primers AtLIP2-S (R982) and AtLIP2-A (R983) were used. Transcripts from the constitutively expressed gene At2g09990 encoding the 40S ribosomal protein S16 (primers R176 and R177) were used for calibration.
Genotyping of T-DNA Mutants for AtLPLA and AtLIP2
Genomic DNA from leaves of the T-DNA lines SALK_003427 (lplA-1) and SAIL_662_H11 (lip2-1) was subjected to standard PCR (Master Mix; Qiagen) with primers specific for the left border (R741 for SALK T-DNA and R1044 for SAIL T-DNA) and gene-specific primers for AtLPLA (P189) and AtLIP2 (R982). The obtained fragments were directly sequenced to verify the insertion sites. Zygosity of the mutations was tested with gene-specific primer pairs (AtLPLA, P189/P190; AtLIP2, R982/R983) encompassing the respective T-DNA insertion. Genotyping of SAIL_1229_F08 revealed the absence of a T-DNA insertion in the AtLIP2 gene. Genomic DNA from seeds was isolated by grinding three young aborted or wild-type seeds in 10 µL of 10 mm Tris-HCl (pH 8) followed by incubation at 95°C for 10 min. One microliter of the supernatant obtained after centrifugation was used as a template for PCR as above.
RNAi Lines
PCR amplificates of the AtLIP2 cDNA comprising nucleotides 533 to 844 flanked by BamHI and SalI sites (sense orientation; primers P129 and P130) or XhoI and BglII sites (antisense orientation; primers P132 and P131), respectively, were ligated into corresponding cloning sites downstream (sense fragment) and upstream (antisense fragment) of the GA 20-oxidase intron in vector pUC-RNAi (Chen et al., 2003). The same procedure was used to prepare the AtLPLA RNAi construct by using primers R1062 and P223 (sense) and primers P222 and P224 (antisense). The entire RNAi cassettes were excised and inserted into vector pGREEN0029 via the PstI site just to add flanking XbaI and HindIII sites for subsequent ligation into the pGREEN cauliflower mosaic virus 35S cassette (http://www.pgreen.ac.uk). These new cassettes, now including left and right borders, were PCR amplified with primers R1069 and R1070 and inserted into the NotI site of pGREEN0179 (Supplemental Fig. S7). This vector was used to transform Arabidopsis Col-0 (Clough and Bent, 1998) by using Agrobacterium tumefaciens strain GV3101psoup. Transgenic seeds were selected on 20 µg mL−1 hygromycin plates for several generations. T3 or T4 lines were used for experiments.
Heterologous Overexpression and Antibody Generation
cDNA was prepared from Arabidopsis leaf RNA as described above. PCR amplificates produced with primers R1062/R1068 for full-length LPLA (At3g29010) and with primers R1058/R1102 for full-length AtLIP2 (At1g04640) were ligated into BamHI and HindIII sites of the expression vector pASK-IBA7plus (IBA). Expression in Escherichia coli BL21 was induced by the addition of 200 µg L−1 anhydrotetracycline. After 12 h of shaking at 20°C, cells were collected (10 min, 6,000g, and 4°C), resuspended in 20 mm sodium phosphate (pH 7.8 and 4°C), and disrupted by sonification (Braun Labsonic U, 50 W; four 15-s bursts). After centrifugation (20,000g, 30 min, and 4°C), the recombinant proteins were affinity purified on Strep-Tactin matrix columns following standard protocols (http://www.iba-go.com), concentrated by using Vivaspin columns with a 5-kD exclusion limit (Sartorius), and rebuffered by passage through PD-10 columns (GE Healthcare) equilibrated in 10/10 mm Tris/MOPS (pH 7.5), 0.5 mm MgSO4, and 5 mm 2-mercaptoethanol.
For immunization, proteins were separated by SDS-PAGE followed by electroelution of the AtLPLA or AtLIP2 band (Electro Eluter model 422; Bio-Rad). About 1 mg of lyophilized protein was used to immunize rabbits (Seqlab). The antiserum was affinity purified by overnight incubation of a 1:8 dilution in Tris-buffered saline (4°C) with the respective electrophoretically purified nitrocellulose-bound recombinant protein. After multiple washes with Tris-buffered saline, immunoglobulins were eluted with 100 mm Gly-HCl (pH 2.5), immediately brought to pH 7 with 1 m Tris-HCl (pH 8), stabilized by the addition of 1 mg mL−1 bovine serum albumin, and stored at −20°C.
Complementation of the LplA− LipB− E. coli Strain TM137
For unknown reasons, the above recombinant vector pASK-IBA7plus did not express detectable amounts of AtLPLA in the lipoylation-deficient E. coli strain TM137 (rspl lipB182::Tn1000dKn::lplA329::Tn10dTc; Morris et al., 1995). Therefore, the entire coding sequence of AtLPLA was amplified by reverse transcription (RT)-PCR (primers R1063 and R1068) and ligated into pBAD-HisA (Invitrogen) via XhoI and HindIII sites, yielding pBAD-HisA/LPLA. Wild-type E. coli K12, untransformed TM137, and TM137 transformed with nonrecombinant pBAD-HisA served as controls in the complementation test with pBAD-HisA/LPLA. Cells were streaked on M9 plates (1.5% [w/v] agar, 34 mm Na2HPO4, 22 mm KH2PO4, 8.5 mm NaCl, 19 mm NH4Cl, 2 mm MgSO4, 0.1 mm CaCl2, and 0.001% [w/v] thiamine) supplemented with 0.4% (w/v) Glc as a carbon source and 0.2% (w/v) Ara to induce expression. Plates containing different supplements (5 mm sodium acetate to bypass PDH and 5 mm sodium succinate to bypass KGDH), 500 ng mL−1 LA, and antibiotics (100 µg mL−1 kanamycin for the LIPB− mutation, 1.7 µg mL−1 tetracycline for the LPLA− mutation, and 100 µg mL−1 ampicillin for pBAD-HisA selection) were incubated for 48 h at 28°C.
Liquid cultures were grown in 50 mL of M9 medium containing 0.4% (w/v) Glc, 0.2% (w/v) Ara, 40 µg mL−1 casamino acids (Difco), 500 ng mL−1 LA, 5 mm sodium acetate and/or sodium succinate, and antibiotics as above and inoculated with 500 µL (optical density at 600 nm of about 0.75) of the respective E. coli strains. Optical density at 600 nm values were measured after 24 and 48 h at 28°C (200 rpm). Ten-milliliter fractions were set aside as a control before induction with 0.2% (w/v) Ara (TM137/pBAD-HisA and TM137/pBAD-HisA/AtLPLA). More 10-mL fractions were harvested 12 and 24 h after induction. Cells were collected by centrifugation (10 min, 6,000g, and 4°C), resuspended in 20 mm sodium phosphate buffer (pH 7.8 and 4°C), 1 mm phenylmethanesulfonyl fluoride, and 0.2 mm Pefabloc (Roth), and disrupted by sonication (Braun Labsonic U, 50 W; four 15-s bursts). The extracts were cleared by centrifugation (20,000g, 30 min, and 4°C), and proteins were examined by SDS-PAGE and immunoblotting using antibodies against the LA epitopes or the His tag as described below.
Production of AtLPLA, LAE, apoPDH-E2, and apoH-Protein for Enzymatic Studies
Bovine liver cDNA (obtained from Dr. Thorsten Viergutz) was used to amplify the mature LAE open reading frame by PCR (primers P474 and P475, including BamHI and HindIII restriction sites). A 222-amino acid fragment of the PDH-E2a subunit (At3g52200) was amplified by RT-PCR (primers AtPDHE2a-SacI-S2, P463; AtPDHE2a-SaII-A2, P464; introducing terminal SacI and SalI restriction sites). The coding sequence for the mature Arabidopsis H-protein (At2g35370; GLDH1) was amplified by RT-PCR (primers AtGLDH1-BamHI-S, P213; AtGLDH1-Sall-A, P214; introducing terminal BamHI and SalI restriction sites). The amplificates were ligated into pASK-IBA7plus. These three overexpression vectors and the pBAD-HisA/LPLA vector above were transformed into TM137. Transformed TM137 cells were grown in Luria-Bertani medium supplemented with 5 mm each of sodium acetate and sodium succinate and antibiotics as above (ampicillin, kanamycin, and tetracycline). Expression was induced by the addition of 200 µg L−1 anhydrotetracycline (pASK/IBA7plus/LAE) or 0.2% (w/v) Ara (pBAD-HisA/LPLA) followed by growth for 12 h at 20°C. Recombinant proteins were collected from the Strept-Actin matrix as described above.
Lipoylation Activity of LPLA
Lipoylation activity was tested in 100-µL assays containing 10/10 mm Tris/MOPS (pH 7.5), 5 mm MgSO4, 5 mm GTP (the preferred substrate of LAE; Fujiwara et al., 2001) or 5 mm ATP, 0.4 mm dithiothreitol, and 0.19 mm [1-14C]octanoic acid (0.1 mCi mL−1, 55 mCi mmol−1; Sigma-Aldrich). Target apoproteins were 10 µg of Arabidopsis H-protein (GLDH1), 5 µg of Arabidopsis PDH-E2a polypeptide, or 50 µg of E. coli TM137 soluble protein. A total of 2.5 µg of AtLPLA plus or minus 0.5 µg of LAE or 2.5 µg of E. coli LplA (Hasse et al., 2009) was added as indicated, and the reaction mixture was incubated for 12 h at 28°C. After stopping the reaction after 10 min at 95°C, 20-µL aliquots were separated on 12% (w/v) denaturing polyacrylamide gels. Gels were dried onto Whatman filter paper, and 14C-labeled proteins were detected by autoradiography using Hyperfilm ECL (GE Healthcare, Amersham).
For in vitro lipoylation of Arabidopsis mitochondrial proteins, 20 µg of matrix protein (Keech et al., 2005) was incubated with 5 µg of recombinant E. coli LPLA in 100 µL of buffer containing 100 mm MOPS and 100 mm Tris (pH 7.5), 50 mm MgCl2, 5 mm ATP, and 400 µm LA acid (dl-racemic mixture, saturating for E. coli LPLA) for 16 h (Hasse et al., 2009). As the control, 20 µg of mitochondrial matrix proteins was incubated without LA and E. coli LPLA. Assays were stopped by heating for 10 min at 95°C. Twenty-microliter aliquots of untreated and E. coli LPLA-treated samples were analyzed by SDS-PAGE followed by immunoblotting as described below.
Protein Extraction and Immunoblotting
Arabidopsis leaf and root extracts were prepared by grinding 100 mg of tissue in 200 μL of ice-cold 75 mm Tris-HCl (pH 7.5) containing 1.5 mm EDTA, 1 mm phenylmethanesulfonyl fluoride, 0.2 mm Pefabloc (Roth), and 2 mm NaHSO3. Homogenates were cleared by centrifugation (20,000g, 10 min, and 4°C). Mitochondria were isolated from wild-type Arabidopsis rosette leaves according to Keech et al. (2005) and resuspended in a buffer containing 0.3 m Suc, 10 mm TES-KOH (pH 7.5), 10 mm potassium phosphate, and 2 mm EDTA. Matrix extracts were prepared by exposure of purified mitochondria to four freeze-thaw cycles, followed by centrifugation at 44,000g for 30 min.
Ten micrograms of total protein per lane (Bradford, 1976) was separated on 12% (w/v) denaturing polyacrylamide gels (Laemmli, 1970; Wittig et al., 2006) and electrotransferred onto polyvinylidene difluoride membrane. For immunodetection, we used affinity-purified antibodies against LA (monoclonal anti-lipoate IgG at 1:4,000 dilution; Calbiochem), Arabidopsis LIP2 (1:500), Arabidopsis LPLA (1:500), the T-protein subunit of potato (Solanum tuberosum) GDC (1:2,000), the H-protein subunit of Flaveria trinervia GDC (GDC-H; 1:2,000), and the large subunit of tobacco (Nicotiana tabacum) Rubisco (1:5,000 dilution) in combination with a horseradish peroxidase-conjugated anti-rabbit immunoglobulin (Bio-Rad). An anti-His horseradish peroxidase conjugate (Miltenyi Biotec) was used to immunodetect His-tagged proteins.
Transient Expression in Tobacco Protoplasts
The 5′ end of AtLPLA cDNA encoding the first 102 amino acids was PCR amplified by using the primer pair P496/P505. The fragment was ligated into the pGFP-2 vector (Wendt et al., 2000) via the introduced XbaI and XhoI restriction sites to generate plasmid pGFP2-LPLA for transient transformation of tobacco protoplasts. Isolation of tobacco protoplasts was done according to Wendt et al. (2000). Plasmid DNA was introduced and fluorescence was detected as described recently (Ewald et al., 2014).
Metabolite Analysis
For metabolite analysis by gas chromatography coupled to mass spectrometry, LPLA RNAi plants were grown as described above. Samples were taken from fully expanded rosette leaves in four biological replicates at the inflorescence emergence stage at the middle of the day (growth stage 5.1 according to Boyes et al. [2001]). Samples were immediately frozen in liquid nitrogen and stored at −80°C as recommended by Fernie et al. (2011). A total of 50 mg was used for metabolite extraction, derivatization, and analysis as described by Lisec et al. (2006).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression and transcriptional coregulation by electronic northern data.
Supplemental Figure S2. Fluorescence signals of an AtLPLA-GFP fusion expressed in tobacco protoplasts.
Supplemental Figure S3. Alignment of LPLAs including bifunctional LPLAs with C-terminal extensions.
Supplemental Figure S4. Phylogenetic relations between enzymes involved in protein lipoylation.
Supplemental Figure S5. Lack of complementation of TM137 cells transformed with pBAD-HisA-AtLPLA on M9 minimal medium.
Supplemental Figure S6. Arabidopsis T-DNA insertion knockout lines for LIP2 and LPLA.
Supplemental Figure S7. Strategy for setting up the RNAi constructs.
Supplemental Table S1. Primers used in this work (introduced restriction sites are underlined).
Supplemental Table S2. Leaf metabolite profiling of wild-type and LPLA RNAi plants (fold change).
Supplementary Material
Acknowledgments
We thank Ursula Bauwe (University of Rostock) for technical assistance, Dr. Dirk Hasse (Swedish University of Agricultural Sciences) for discussions, Dr. Thorsten Viergutz (Leibniz Institute for Farm Animal Biology) for bovine liver cDNA, and Elfriede K. Pistorius (University Bielefeld) for anti-RBCL antibody.
Glossary
- LA
lipoic acid
- LAE
lipoate-activating enzyme
- T-DNA
transfer DNA
- Col-0
Columbia-0
- RNAi
RNA interference
- GABA
γ-amino butyric acid
- CTD
C-terminal domain
- RT
reverse transcription
- cDNA
complementary DNA
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. BA 1177/9) in close cooperation with FOR1186.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Allary M, Lu JZQ, Zhu L, Prigge ST. (2007) Scavenging of the cofactor lipoate is essential for the survival of the malaria parasite Plasmodium falciparum. Mol Microbiol 63: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bauwe H, Hagemann M, Fernie AR. (2010) Photorespiration: players, partners and origin. Trends Plant Sci 15: 330–336 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J. (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Chen S, Hofius D, Sonnewald U, Börnke F. (2003) Temporal and spatial control of gene silencing in transgenic plants by inducible expression of double-stranded RNA. Plant J 36: 731–740 [DOI] [PubMed] [Google Scholar]
- Christensen QH, Cronan JE. (2009) The Thermoplasma acidophilum LplA-LplB complex defines a new class of bipartite lipoate-protein ligases. J Biol Chem 284: 21317–21326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen QH, Martin N, Mansilla MC, de Mendoza D, Cronan JE. (2011) A novel amidotransferase required for lipoic acid cofactor assembly in Bacillus subtilis. Mol Microbiol 80: 350–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rébeillé F. (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6: 167–176 [DOI] [PubMed] [Google Scholar]
- Engel N, van den Daele K, Kolukisaoglu U, Morgenthal K, Weckwerth W, Pärnik T, Keerberg O, Bauwe H. (2007) Deletion of glycine decarboxylase in Arabidopsis is lethal under nonphotorespiratory conditions. Plant Physiol 144: 1328–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald R, Hoffmann C, Neuhaus E, Bauwe H. (2014) Two redundant octanoyltransferases and one obligatory lipoyl synthase provide protein-lipoylation autonomy to plastids of Arabidopsis. Plant Biol (Stuttg) 16: 35–42 [DOI] [PubMed] [Google Scholar]
- Ewald R, Kolukisaoglu U, Bauwe U, Mikkat S, Bauwe H. (2007) Mitochondrial protein lipoylation does not exclusively depend on the mtKAS pathway of de novo fatty acid synthesis in Arabidopsis. Plant Physiol 145: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Aharoni A, Willmitzer L, Stitt M, Tohge T, Kopka J, Carroll AJ, Saito K, Fraser PD, DeLuca V. (2011) Recommendations for reporting metabolite data. Plant Cell 23: 2477–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Takeuchi S, Okamura-Ikeda K, Motokawa Y. (2001) Purification, characterization, and cDNA cloning of lipoate-activating enzyme from bovine liver. J Biol Chem 276: 28819–28823 [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Toma S, Okamura-Ikeda K, Motokawa Y, Nakagawa A, Taniguchi H. (2005) Crystal structure of lipoate-protein ligase A from Escherichia coli: Determination of the lipoic acid-binding site. J Biol Chem 280: 33645–33651 [DOI] [PubMed] [Google Scholar]
- Gueguen V, Macherel D, Jaquinod M, Douce R, Bourguignon J. (2000) Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J Biol Chem 275: 5016–5025 [DOI] [PubMed] [Google Scholar]
- Hasse D, Mikkat S, Hagemann M, Bauwe H. (2009) Alternative splicing produces an H-protein with better substrate properties for the P-protein of glycine decarboxylase. FEBS J 276: 6985–6991 [DOI] [PubMed] [Google Scholar]
- Hermes FA, Cronan JE. (2013) The role of the Saccharomyces cerevisiae lipoate protein ligase homologue, Lip3, in lipoic acid synthesis. Yeast 30: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan SW, Cronan JE., Jr (2003) The Escherichia coli lipB gene encodes lipoyl (octanoyl)-acyl carrier protein:protein transferase. J Bacteriol 185: 1582–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Jeong HK, Lee E, Natarajan S. (2007) Characterization of a lipoate-protein ligase A gene of rice (Oryza sativa L.). Gene 393: 53–61 [DOI] [PubMed] [Google Scholar]
- Kavi Kishor PB, Sreenivasulu N. (2014) Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ 37: 300–311 [DOI] [PubMed] [Google Scholar]
- Keech O, Dizengremel P, Gardeström P. (2005) Preparation of leaf mitochondria from Arabidopsis thaliana. Physiol Plant 124: 403–409 [Google Scholar]
- Kim DJ, Kim KH, Lee HH, Lee SJ, Ha JY, Yoon HJ, Suh SW. (2005) Crystal structure of lipoate-protein ligase A bound with the activated intermediate: insights into interaction with lipoyl domains. J Biol Chem 280: 38081–38089 [DOI] [PubMed] [Google Scholar]
- Laemmli UK. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lernmark U, Gardeström P. (1994) Distribution of pyruvate dehydrogenase complex activities between chloroplasts and mitochondria from leaves of different species. Plant Physiol 106: 1633–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Behal R, Oliver DJ. (2003) Disruption of plE2, the gene for the E2 subunit of the plastid pyruvate dehydrogenase complex, in Arabidopsis causes an early embryo lethal phenotype. Plant Mol Biol 52: 865–872 [DOI] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Martin N, Christensen QH, Mansilla MC, Cronan JE, de Mendoza D. (2011) A novel two-gene requirement for the octanoyltransfer reaction of Bacillus subtilis lipoic acid biosynthesis. Mol Microbiol 80: 335–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson C, Wood C. (2000) Mitochondrial β-oxidation of fatty acids in higher plants. Physiol Plant 109: 217–224 [Google Scholar]
- Masterson C, Wood C. (2009) Influence of mitochondrial β-oxidation on early pea seedling development. New Phytol 181: 832–842 [DOI] [PubMed] [Google Scholar]
- Mooney BP, Miernyk JA, Randall DD. (2002) The complex fate of α-ketoacids. Annu Rev Plant Biol 53: 357–375 [DOI] [PubMed] [Google Scholar]
- Morikawa T, Yasuno R, Wada H. (2001) Do mammalian cells synthesize lipoic acid? Identification of a mouse cDNA encoding a lipoic acid synthase located in mitochondria. FEBS Lett 498: 16–21 [DOI] [PubMed] [Google Scholar]
- Morris TW, Reed KE, Cronan JE., Jr (1995) Lipoic acid metabolism in Escherichia coli: the lplA and lipB genes define redundant pathways for ligation of lipoyl groups to apoprotein. J Bacteriol 177: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J. (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge JB, Kuhn DN, Stumpf PK. (1979) Subcellular localization of acyl carrier protein in leaf protoplasts of Spinacia oleracea. Proc Natl Acad Sci USA 76: 1194–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perham RN. (2000) Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem 69: 961–1004 [DOI] [PubMed] [Google Scholar]
- Posner MG, Upadhyay A, Bagby S, Hough DW, Danson MJ. (2009) A unique lipoylation system in the Archaea: lipoylation in Thermoplasma acidophilum requires two proteins. FEBS J 276: 4012–4022 [DOI] [PubMed] [Google Scholar]
- Rocha M, Licausi F, Araújo WL, Nunes-Nesi A, Sodek L, Fernie AR, van Dongen JT. (2010) Glycolysis and the tricarboxylic acid cycle are linked by alanine aminotransferase during hypoxia induced by waterlogging of Lotus japonicus. Plant Physiol 152: 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonauer MS, Kastaniotis AJ, Kursu VAS, Hiltunen JK, Dieckmann CL. (2009) Lipoic acid synthesis and attachment in yeast mitochondria. J Biol Chem 284: 23234–23242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, Aux G, McElver J, Patton D, Dietrich B, Ho P, Bacwaden J, Ko C, et al. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14: 2985–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimakata T, Stumpf PK. (1982) Isolation and function of spinach leaf beta-ketoacyl-[acyl-carrier-protein] synthases. Proc Natl Acad Sci USA 79: 5808–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockey JM, Fulda MS, Browse J. (2003) Arabidopsis contains a large superfamily of acyl-activating enzymes: phylogenetic and biochemical analysis reveals a new class of acyl-coenzyme A synthetases. Plant Physiol 132: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreze Y, Boutron A, Habarou F, Barnerias C, Nonnenmacher L, Delpech H, Mamoune A, Chrétien D, Hubert L, Bole-Feysot C, et al. (2013) Mutations in human lipoyltransferase gene LIPT1 cause a Leigh disease with secondary deficiency for pyruvate and alpha-ketoglutarate dehydrogenase. Orphanet J Rare Dis 8: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza CdeA, Barbazuk B, Ralph SG, Bohlmann J, Hamberger B, Douglas CJ. (2008) Genome-wide analysis of a land plant-specific acyl:coenzyme A synthetase (ACS) gene family in Arabidopsis, poplar, rice and Physcomitrella. New Phytol 179: 987–1003 [DOI] [PubMed] [Google Scholar]
- Spalding MD, Prigge ST. (2010) Lipoic acid metabolism in microbial pathogens. Microbiol Mol Biol Rev 74: 200–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studart-Guimarães C, Fait A, Nunes-Nesi A, Carrari F, Usadel B, Fernie AR. (2007) Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the γ-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol 145: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetlove LJ, Beard KFM, Nunes-Nesi A, Fernie AR, Ratcliffe RG. (2010) Not just a circle: flux modes in the plant TCA cycle. Trends Plant Sci 15: 462–470 [DOI] [PubMed] [Google Scholar]
- Taylor NL, Day DA, Millar AH. (2002) Environmental stress causes oxidative damage to plant mitochondria leading to inhibition of glycine decarboxylase. J Biol Chem 277: 42663–42668 [DOI] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. (2005) The Botany Array Resource: e-Northerns, expression angling, and promoter analyses. Plant J 43: 153–163 [DOI] [PubMed] [Google Scholar]
- Tsunoda JN, Yasunobu KT. (1967) Mammalian lipoic acid activating enzyme. Arch Biochem Biophys 118: 395–401 [DOI] [PubMed] [Google Scholar]
- Wada H, Shintani D, Ohlrogge J. (1997) Why do mitochondria synthesize fatty acids? Evidence for involvement in lipoic acid production. Proc Natl Acad Sci USA 94: 1591–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M, Yasuno R, Jordan SW, Cronan JE, Jr, Wada H. (2001a) Lipoic acid metabolism in Arabidopsis thaliana: cloning and characterization of a cDNA encoding lipoyltransferase. Plant Cell Physiol 42: 650–656 [DOI] [PubMed] [Google Scholar]
- Wada M, Yasuno R, Wada H. (2001b) Identification of an Arabidopsis cDNA encoding a lipoyltransferase located in plastids. FEBS Lett 506: 286–290 [DOI] [PubMed] [Google Scholar]
- Wendt UK, Wenderoth I, Tegeler A, Von Schaewen A. (2000) Molecular characterization of a novel glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.). Plant J 23: 723–733 [DOI] [PubMed] [Google Scholar]
- Witkowski A, Joshi AK, Smith S. (2007) Coupling of the de novo fatty acid biosynthesis and lipoylation pathways in mammalian mitochondria. J Biol Chem 282: 14178–14185 [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schägger H. (2006) Blue native PAGE. Nat Protoc 1: 418–428 [DOI] [PubMed] [Google Scholar]
- Yasuno R, von Wettstein-Knowles P, Wada H. (2004) Identification and molecular characterization of the β-ketoacyl-[acyl carrier protein] synthase component of the Arabidopsis mitochondrial fatty acid synthase. J Biol Chem 279: 8242–8251 [DOI] [PubMed] [Google Scholar]
- Yasuno R, Wada H. (1998) Biosynthesis of lipoic acid in Arabidopsis: cloning and characterization of the cDNA for lipoic acid synthase. Plant Physiol 118: 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuno R, Wada H. (2002) The biosynthetic pathway for lipoic acid is present in plastids and mitochondria in Arabidopsis thaliana. FEBS Lett 517: 110–114 [DOI] [PubMed] [Google Scholar]
- Zhao X, Miller JR, Jiang Y, Marletta MA, Cronan JE. (2003) Assembly of the covalent linkage between lipoic acid and its cognate enzymes. Chem Biol 10: 1293–1302 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.