Abstract
In a broad attempt to improve the understanding of the genetic regulation of serum IgA levels, the heritability was estimated in over 12 000 Swedish twins, and a genome-wide association study was conducted in a subsample of 9617. Using the classical twin model the heritability was found to be significantly larger among females (61%) compared with males (21%), while contribution from shared environment (20%) was only seen for males. By modeling the genetic relationship matrix with IgA levels, we estimate that a substantial proportion (31%) of variance in IgA levels can ultimately be explained by the investigated SNPs. The genome-wide association study revealed significant association to two loci: (i) rs6928791 located on chromosome 6, 22 kb upstream of the gene SAM and SH3 domain containing 1 (SASH1) and (ii) rs13300483 on chromosome 9, situated 12 kb downstream the CD30 ligand (CD30L) encoding gene. The association to rs13300483 was replicated in two additional independent Swedish materials. The heritability of IgA levels is moderate and can partly be attributable to common variation in the CD30L locus.
INTRODUCTION
Immunoglobulin A (IgA) is one of the three major immunoglobulins in humans, and it is the most abundant antibody isotype in secretions. While a dimeric form is dominant at mucosal sites, a monomeric form is observed in serum (1). Many studies have been conducted to investigate the genetic contribution to the biological variation of serum IgA concentrations (2–17). Di Franco et al. suggested that IgA concentrations are under genetic and environmental influence (2). Earlier studies on familial resemblance suggested IgA concentrations to be under control of a single gene (3,4). However, given what we now know about the multifactorial nature of the regulation of most molecules in blood, the genetic influences are likely to be complex. The cause of higher IgA concentrations (up to 20% on average) in males compared with females is currently not known, but has been suggested to be due to both genetic as well as environmental influences (8,14,16). Despite the higher average levels in males, they also have higher prevalence of selective IgA deficiency (IgAD) and subnormal levels of IgA than women (16). Moreover, IgA concentrations increase with age (5–16,18). The factors underlying this phenomenon are not understood, although Gonzalez-Quintela et al. proposed an accumulation of chronic inflammation with age as one possible explanation (8).
Heritability estimations of IgA levels from twin studies are scarce. The few we are aware of have utilized small number of twins, profoundly limiting the precision of the estimates (19–21). In the present genome-wide association study (GWAS), we utilized a large population-based Swedish cohort of twins measured for serum IgA levels. Data on almost 5000 complete twin pairs were available within the cohort, enabling for the first time a precise partitioning of the variance of IgA level into additive genetic, shared environment and non-shared environmental components. Furthermore, thanks to recent developments of ways to utilize the genome-wide dense single-nucleotide polymorphism (SNP) marker data generated by GWAS, the present sample also enables estimation of the ‘chip heritability’ (22). With this approach, the proportion of the trait variance captured by the investigated markers as a whole can be estimated.
GWASes have become the standard method of interrogating molecular genetic influences to complex traits. Many of these studies have focused on quantitative traits in blood serum, which have resulted in the mapping of hundreds of significantly associated loci (23–26). Also serum immunoglobulin levels have been object of GWA studies. In 2012, Yang et al. identified in a two-stage analysis a SNP in the TNFSF13 gene that showed genome-wide significant association to IgA (P = 2 × 10−8) in the discovery sample that dropped to borderline significance in the combined discovery and replication data set consisting of 1999 plus 1496 subjects (27). To date, there have also been three GWA studies published investigating the regulation of serum Immunoglobulin E levels (28–30). These studies found novel associated SNPs in the FCER1A, the STAT6 and the IL13 genes.
To identify loci involved in regulating normal serum IgA concentrations we used the IgA measurements of almost 10 000 of the Swedish twins participating in the TwinGene study that also had available genome-wide SNP array data. Replication of the top genome-wide significant finding were sought in two independent samples (i) EIRA, which is a study of Swedish cases of rheumatoid arthritis (RA) and controls, and (ii) BAMSE, a Swedish study of allergy in children.
RESULTS
Twin heritability and GCTA
The classical twin design examines the proportion of variance in a trait that is due to additive genetic factors (A), against the proportions that are due to shared (C) or non-shared environment (E), from here on referred to as the ACE model. ACE models of IgA level were fit on square root-transformed IgA values adjusted for age at sampling and batch. The resulting distribution was normal with similar mean and variance between the zygosity classes. The heritability (A) was found to be 0.50 (95% CI 0.40–0.54), shared environment (C) 0.00 (95% CI 0.00–0.00) and non-shared environment (E) 0.50 (95% CI 0.46–0.54; see Table 1). We also investigated a sex-limited model in which the sex specific parameters were allowed to differ from each other. This revealed that there was a significant quantitative difference in variance components between the sexes (P < 0.01). The heritability was observed to be larger among the females (0.61, 95% CI 0.53–0.66) compared with males (0.21, 95% CI 0.06–0.38). Furthermore, for males, a significant contribution of shared environment (C = 0.20, 95% CI 0.06–0.32) was detected, while this component was close to null and non-significant among females (Table 1). As indicated by similar covariances in opposite and same-sex DZ twins no evidence for qualitative sex differences were found (P = 0.99), suggesting the same genetic factors to be involved in males and females.
Table 1.
Sex-combined and sex-limited twin ACE model on genetic and environmental influences on IgA levela
| Sex | A% (95% CI) | C% (95% CI) | E% (95% CI) |
|---|---|---|---|
| All | 0.503 | 0.000 | 0.497 |
| (0.402–0.540) | (0.000–0.000) | (0.459–0.541) | |
| Male | 0.221 | 0.200 | 0.588 |
| (0.058–0.379) | (0.064–0.322) | (0.526–0.654) | |
| Female | 0.613 | 0.003 | 0.382 |
| (0.532–0.622) | (0.000–0.062) | (0.459–0.541) |
Test for Qualitative sex difference P > 0.99. Test for quantitative sex difference P < 0.01.
aSquare root-transformed and adjusted for age at sampling, batch, and three principal components displaying the strongest association to IgA levels.
By using the genome-wide complex trait analysis (GCTA) approach that estimates the proportion of phenotypic variance explained by genome-wide SNPs, we found a highly significant estimate of the proportion of genetic variance to total phenotypic variance, V(g)/V(p), of IgA level captured by all investigated markers, V(g)/V(p) = 0.31 (SE 0.06, P = 9e−10; Table 2). Inspired by the sex differences in heritability observed by the twin model, we also tested a GCTA model, including gene by sex interaction, but found the interaction term to be insignificant (P = 0.08).
Table 2.
Partitioning of IgA level variance by GCTA methoda
| Source | Variance | SE |
|---|---|---|
| V genotypic (g) | 0.408 | 0.0785 |
| V environmental (e) | 0.928 | 0.0779 |
| V phenotypic (p) | 1.337 | 0.0250 |
| V (g)/V (p) | 0.306 | 0.0580 |
aAdjusted for three principal components displaying the strongest association to IgA levels.
Discovery GWAS
The discovery sample consisted of 9617 unique genomes whereof 1110 belonged to monozygous twin pairs. Thus for 1110 genomes, we had dual IgA measurements (one from each twin). By using the within twin pair average as the phenotypic value for such genomes the statistical power increases (by reducing environmental influences). IgA levels displayed weak but significant correlation with age at sampling, sex and zygosity. Further, when investigating the association between IgA level and the main genetic principal components (PCs), we observed a significant association with three of these PCs. Thus, we subsequently included these parameters as covariates in the GWAS.
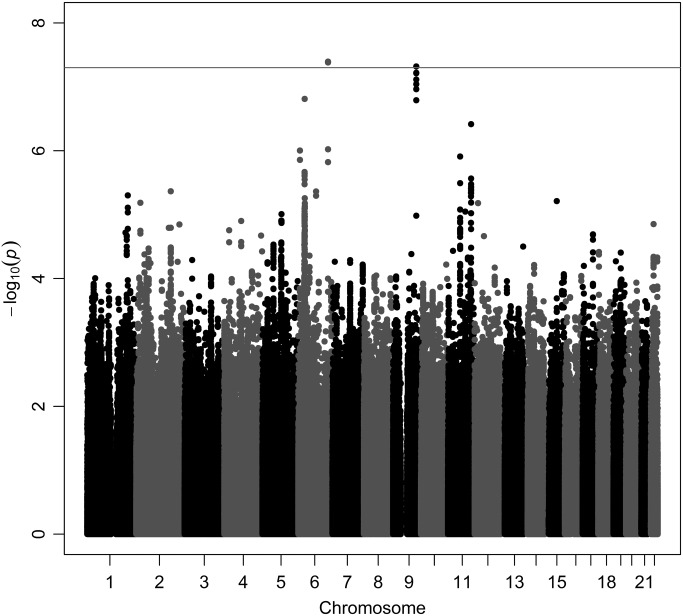
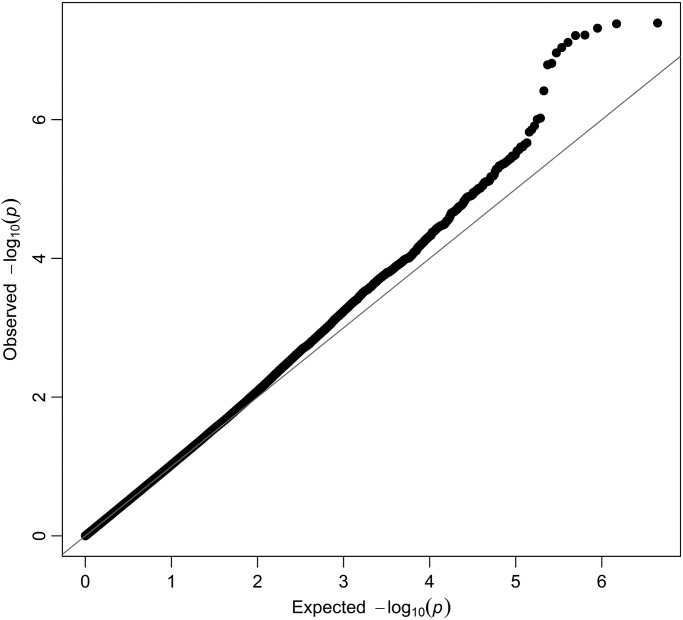
The GWAS result for IgA levels are displayed as a Manhattan plot in Figure 1, and the distribution of observed P-values are depicted in the quantile–quantile plot (Q–Q plot) (Fig. 2). Two loci surpass genome-wide significance level of 5 × 10−8. On chromosome 6q the association is displayed by two imputed SNPs, rs6928791 and rs12665468 (both P = 4 × 10−8). The directly genotyped SNP showing strongest association within this locus is rs10484601 (P = 1.5 × 10−6). The lead SNP, rs6928791, is located 22 kb upstream of the gene SASH1 (see Table 3).
Figure 1.
Manhattan plot of SNPs for serum IgA levela. aSNPs are plotted on the x-axis according to their position on each chromosome against association with serum IgA level on the y-axis (shown as –log10 P-value). The solid line indicates the threshold for genome-wide significance (P < 5 × 10−8).
Figure 2.
Quantile–quantile plot of SNPs for serum IgA level.
Table 3.
Top associations in the discovery set (chr 6q and chr 9q) after including adjustments for covariates
| Chromosome | SNP | Position | Coding allele | Non-coding allele | Frequency of coding allele | INFOa | Beta | Standard error | P-value |
|---|---|---|---|---|---|---|---|---|---|
| 6 | rs6928791 | 148937045 | A | G | 0.1209 | 0.9587 | −0.1383 | 0.0252 | 4.041e−08 |
| 6 | rs12665468 | 148938309 | A | C | 0.1203 | 0.9283 | −0.1408 | 0.0256 | 4.152e−08 |
| 6 | rs12614 | 32022158 | C | T | 0.9226 | 1.0081 | 0.1621 | 0.0309 | 1.545e−07 |
| 9 | rs13300483 | 116683183 | C | T | 0.7661 | 0.9835 | −0.1167 | 0.0214 | 4.801e−08 |
| 9 | rs7860414 | 116581271 | C | T | 0.2163 | 0.9788 | 0.1144 | 0.0211 | 6.03e−08 |
| 9 | rs7853287 | 116589148 | A | G | 0.2174 | 0.9630 | 0.1150 | 0.0212 | 6.133e−08 |
aProper_info from IMPUTE2.
The second locus surpassing the genome-wide significance level is located on chromosome 9q, with the lead SNP being rs13300483 (P = 5 × 10−8). rs13300483 is directly genotyped and tags a 100 kb haplotype block represented by four directly genotyped and four imputed SNPs, all showing associations below P < 10−5 level. The top signal is situated 12 kb downstream the gene CD30L, also known as tumor necrosis factor (ligand) superfamily, member 8 (TNFSF8) or CD153. No other loci show genome-wide significance to IgA level, but a list of top-100 SNPs can be found in Supplementary Material, Table S1.
It is not with great margin our two loci pass the threshold for genome-wide significance, despite the relatively large discovery sample. In order to gain confidence in the findings we therefore sought replication in independent materials. We were able to identify two independently collected Swedish samples for which genotypic information and IgA level measurements were available. Because the size of both these replication samples (EIRA n = 2365, BAMSE n = 420) were substantially smaller than the initial discovery sample, we decided only to replicate one of the loci to limit the burden of multiple tests in the replication phase. With the rationale of having most confidence in the chromosome 9q finding due to associations displayed by several, both directly genotyped as well as imputed markers, the replications were restricted to this locus. The lead SNP (rs13300483) plus one imputed SNP (rs7853287) in LD were selected for further replication association analysis. For a complete set of summary association statistics, see Supplementary Material section.
Replication analysis of top locus
Both rs13300483 and rs7853287 displayed allele frequencies in both replication samples that were close to identical to what was seen among the TwinGene participants, supporting the notion that potential biases arising from population or methodological differences in genotyping assays were limited. The association between IgA level and the selected variants at the CD30L locus on 9q replicated in the EIRA material with P = 0.006 for rs13300483 and P = 0.003 for rs7853287, with direction of effects consistent with the TwinGene sample and similar, albeit somewhat smaller effect sizes (Tables 4 and 5). In the considerable smaller BAMSE material, the effects for both SNPs were again consistent in direction and magnitude and significant for both markers, P = 0.044 for rs13300483 and P = 0.038 for rs7853287 (Table 4).
Table 4.
Associations to the chromosome 9q top finding, in each of the three samplesa
| Cohort | SNP | Coding allele | Non-coding allele | Frequency of coded allele | Beta | P-value (one-sided) | N |
|---|---|---|---|---|---|---|---|
| TwinGene | rs13300483 | C | T | 0.7661 | −0.1167 | 4.801e−08 | 9,586 |
| rs7853287 | A | G | 0.2174 | 0.1150 | 6.133e−08 | 9,586 | |
| EIRA | rs13300483 | C | T | 0.7676 | −0.0710 | 0.0124 (0.0062) | 2,365 |
| rs7853287 | A | G | 0.2131 | 0.0653 | 0.0064 (0.0032) | 2,365 | |
| BAMSE | rs13300483 | C | T | 0.7594 | −0.1005 | 0.087 (0.0435) | 420 |
| rs7853287 | A | G | 0.2076 | 0.1089 | 0.076 (0.0380) | 420 |
aReported P-values are two-sided, and one-sided within parentheses for EIRA and BAMSE cohorts (replication). Adjustments for three principal components displaying the strongest association to IgA levels were undertaken in each of the three samples.
Table 5.
Meta-analysis estimates for the three combined samplesa
| SNP | Coding allele | Non-coding allele | Weight | Z-score | P-value | Direction |
|---|---|---|---|---|---|---|
| rs13300483 | C | T | 12371 | 6.214 | 5.177e−10 | +++ |
| rs7853287 | A | G | 12371 | 6.285 | 3.276e−10 | +++ |
aAll P-values going in to the analysis were two-sided.
Meta-analysis
When combining evidence for association from the TwinGene, EIRA and BAMSE studies (Table 5) jointly in meta-analysis the P-values reached 5.2 × 10−10 for rs13300483 and 3.3 × 10−10 for rs8753287. Thus, all three studies lend support to a robust association between genetic variation close to the CD30L gene and serum IgA levels, with a consistent beta-coefficient in the range of 0.07 to 0.12 g/l per minor allele.
DISCUSSION
For GWAS to be successful, there needs to be genetic variation segregating in the population that impacts the trait. Familial aggregation in nuclear families is consistent with such genetic influences, but may also arise from shared environmental influences. The classic way to disentangle genetic from environmental contributions to familial trait resemblance is by twin modeling of MZ and DZ twins. More pronounced similarity among MZ twins indicate genetic influences. We here show that the relative importance of inherited genes is 50% for IgA level and that there are important shared environmental influences specifically among males. What underlies this male environmental component is not known, but could, for example, reflect differences in social acceptance of exposures to alcohol. Heavy alcohol consumption has been reported to be affecting the IgA levels markedly (8,12). Given that habit of heavy drinking is more socially accepted among males than among females, it may contribute to the male-specific shared environmental component. The GCTA analysis confirmed the substantial heritability found by the twin model and gave a ‘chip heritability’ V(g)/V(p) of 0.31. Previous papers using the GCTA method for various traits and diseases have found that the ‘chip heritability’ tends to be in the order of 1/4 to 1/2 of the twin-based heritability (31). Here, in contrast, we found the Vg/Vp to be over 70% of the twin-based sex averaged estimate, indicating an unusually large proportion of the genetic variability is captured by the investigated chip markers.
The IgA discovery GWAS found variation close to CD30L to be significantly associated with IgA. The locus was replicated in two independent materials from the Swedish population lending strong support to this locus. CD30L constitutes an excellent candidate gene for IgA regulation, with results from several in vitro as well as animal models indicating complex involvement in B-cell proliferation, differentiation and antibody production (32).
Despite that our discovery study found two genome-wide significant loci, we chose to restrict our replication attempt to the one single locus (the CD30L), as we believed it displayed the most compelling evidence. This decision was taken on the basis of the limited size and power of the replication materials in combination with the notion that the 6q locus only was supported by imputed SNPs at the genome-wide significance level. The CD30L locus was in contrast supported by directly genotyped as well as imputed SNPs. Even so, we think that the 6q locus merits particular attention in future independent studies.
A previous, smaller GWAS on Chinese subjects reported borderline significance to one single locus, rs3803800, chr 17, P = 2.97e−07 (27). Interestingly, rs3803800 is located within the gene TNFSF13, another member of the tumor necrosis factor ligand superfamily. TNFSF13 encodes a protein known as APRIL, a ligand involved in regulation of B-cell proliferation, just like our hit TNFSF8 on chromosome 9. Mice deficient in TNFSF13 have been reported to show a reduced plasma cell survival (33). Despite that our study comes up with a different significant top finding, we think it is remarkable and informative that these two completely independent GWAS studies, performed in two very different populations, both find variation close to tumor necrosis factor ligand genes as single significant hits. In our main discovery sample, rs3803800 was associated with consistent direction of effect and a P-value of 0.0008, thus lending additional support to the previous finding by Yang et al. (27).
The serum IgA level is of immediate clinical importance for subjects with pronounced deficiency. This condition is relatively rare in Western populations with a prevalence reported to be in the range of about 1/600 (34), and even lower in China with reported prevalence of 1/1615 and 1/4100 (35,36). This could suggest a different genetic background of IgAD in Chinese populations, but a recent study by Wang et al. (37) found that most of the investigated Chinese IgAD subjects carried the same IgAD-associated risk haplotypes found in Caucasians. Given that the present study and the previously performed IgA level GWAS study both indicates associated loci that are distinct from known IgAD haplotypes, it appears to be different loci that are responsible for pronounced deficiency compared with deviances in the normal range. Previously, the idea of one gene regulating IgA levels in a Mendelian manner has been proposed (3,4). Based on familial aggregation, it was estimated that this gene could account for ∼12% of the phenotypic variation (4). The remaining variation was thought to be due to environmental effects. The heritability of 50% we find in this study does not lend support to this viewpoint. Instead, a larger and more complex influence of genetic variation to IgA level is indicated.
Genome-wide searches for genetic association to normal variation in quantitative traits have been applied successfully for a large number of immunological traits, including serum levels of fibrinogen (23), beta-2-microblogulin (38), high-density lipoprotein cholesterol (39), immunoglobulin M (40), Immunoglobulin G (41), Immunoglobulin E (42) and complement component 3 and 4 (43). Given the multifactorial, complex nature of most quantitative traits investigated to date, it is likely that the overlap between genetic factors predisposing to extreme values and concomitant clinical manifestations strongly overlap with variants contributing to normal variation. For body mass index (with clinical manifestations of obesity and anorexia), this hypothesis has previously been suggested based on population studies on correlations among relatives (44), and recently also confirmed using mega-analyses of genetic data on >250K subjects (45).
The main finding of this paper is the identification and replication of the associated locus close to the CD30L gene. The lead SNP is located in between CD30L and TNFSF15. We note that previous GWAS on different forms of inflammatory bowel disease (IBD), especially Crohn's disease and ulcerative colitis have found strong associations to the same genes (46–48). Because there are also documented associations between IBDs and IgA levels (49–53), this colocalization of GWAS findings reinforces the potential clinical relevance of our finding and encourages more detailed investigations of the role of genetic influences to IgA in inflammatory disease.
In conclusion, our study found the heritability of IgA levels to be 0.50 (95% CI 0.40–0.54), with a significant difference between females (0.61, 95% CI 0.53–0.66) and males (0.21, 95% CI 0.06–0.38). In a GWAS of Swedish 9617 genome samples, we found association between IgA levels and markers close to the CD30L gene, and further replicated these findings in two separately collected samples. This gene is a new good candidate for further functional immunological studies.
MATERIALS AND METHODS
Ethics statement
The study was approved by the local ethics committee at Karolinska Institutet (DNR: 2007-664-31, 2011-463-32, 02-420, 2011-2037-32), and all participants gave informed consent.
Discovery cohort TwinGene
The TwinGene study (54), conducted between 2004 and 2008, is a population-based Swedish study of twins born between 1911 and 1958. The study participants have previously responded to a telephone interview called Screening Across the Lifespan of Twins cohort (SALT) (55,56). In total, 22 390 subjects were invited to TwinGene. In total, 12 591 (56%) individuals participated by donating blood to the study, and by answering questionnaires about life style and health.
The participants were asked to make an appointment at their local health care facility on Monday to Thursday mornings (not the day before a national holiday), to ensure that the sample would reach the Karolinska Institutet Bio bank in Stockholm the following day by overnight mail. The participants were instructed to fast from 8 pm (20 : 00) the previous night. A total volume of 50 ml of blood was drawn from each individual by venipuncture.
Iga serum measurement
After arrival to the biobank the serum was stored in liquid nitrogen. Aliquots were withdrawn for each sample and kept at –20°C until IgA measurements were performed. The samples were sorted on to 96-well plates based on zygosity, and 96-well plates with different zygosities were used to prepare the 384-well plates used for spotting. IgA concentration was measured by a reverse-phase protein microarray. In brief, 1 : 10 diluted serum samples were spotted on epoxy-coated microarray slides (Corning, USA) using the 2470 microarray spotter (Aushon, USA). Up to 4600 different samples (including reference samples with known IgA concentration and negative control) were spotted on one slide with five replicates each. The following adjustments were made: spot diameter of 260 µm, temperature of 25°C and humidity of 50%. After spotting, the slides were kept in a highly humid environment overnight, followed by air-drying.
Spotted slides were blocked in PBS/BSA (2%). For detection of IgA rabbit antihuman IgA (Dako Cytomation, Denmark) and Alexa Fluor 555 conjugated goat antirabbit antibodies (Molecular Probes, USA) were used diluted in PBST (1 : 100 000 resp. 1 : 60 000). Images were generated with high-resolution microarray scanner (Agilent Technologies, USA) and analyzed using GenePix Pro 7 (Molecular Devices, USA). Owing to the high reproducibility of the replicates, a replicate-specific scaling could be abstained. The unit of measurement is g/l. Measurement of IgA level was successful for almost all TwinGene participants, n = 12 530 (99.5%). After IgA measurement, the whole sample was tested for effects based on the position on the array and the processing date. No such effects were detected.
DNA extraction and genotyping
One 7 ml EDTA tube of blood is used for DNA extraction using Puregene extraction kit (Gentra Systems, Minneapolis, USA). After extraction, the DNA was subsequently stored at −20°C. After excluding subjects in whom the DNA concentration in the stock solution was <20 ng/µl, as well as subset of 302 female monozygous twin pairs participating in a previous genome-wide effort, DNA from all available dizygous twins + one twin from each available MZ twin pair (n = 9896) was sent to Uppsala, Sweden for genome-wide genotyping using the Illumina OmniExpress bead chip. Genotyping results for 9836 subjects and 731 442 autosomal SNPs passed the initial lab-based quality control (QC). In further QC, SNPs with >3% missing information (GENO > 0.03) (n = 3922), a minor allele frequency <1% (n = 79 893), or a Hardy–Weinberg equilibrium (HWE) test P ≤ 1e−7 (n = 3071), were excluded. Individuals with low genotyping success (MIND > 0.03) (n = 10), male heterozygosity of X-chromosomes (n = 36), deviations in heterozygosity of >5 SD from the population mean (n = 49), or detection of unknown (cryptic) relatedness (n = 124), were excluded. After the QC there were 9617 individuals and 644 556 autosomal SNPs remaining.
Imputation
Imputation of missing SNPs was performed using IMPUTE 2 (57) on build 36 and release 22 of HapMap. The data set was split in groups of thousand individuals, imputed separately and then merged, resulting in 2 585 290 reference SNPs. SNPs with INFO < 0.9 were filtered which removed 358 310 SNPs, leaving the final data set to consist of 2 226 980 SNP markers.
Adjustment for covariates
IgA levels for the TwinGene individuals were available for all genotyped subjects passing genotype QC. Prior association analysis the levels were adjusted for sex, zygosity, batch, age at sampling and the three genetic PCs displaying the strongest association to IgA levels. For complete monozygotic (MZ) twin pairs, one twin was randomly selected to be genotyped. When IgA level was measured for both members of such MZ pairs the average IgA value was calculated and used as phenotypic value for that genome.
Heritability
The classical twin design is used to estimate the relative importance of genetic and environmental effects. The sample consisted of the whole TwinGene cohort, including 12 530 twins from 7568 pairs. Among them, 3128 were monozygotic twins, 4795 were same-sex dizygotic (DZ) twins, and 4607 were opposite-sex DZ twins. Twin methodology relies on the different genetic relatedness between MZ and DZ twins. The MZ twins are genetically identical, whereas DZ twins, just like any full siblings, on average share 50% of their segregating alleles. If genes influence a trait, there will be more pronounced twin similarity within MZ twin pairs than within DZ twin pairs. By modeling of twin variance and covariance structures in MZ and DZ pairs the variation in a phenotype is decomposed into additive genetic (A), shared environmental (C), and non-shared environmental (E) factors. We used OpenMx in R (58), a structural equation modeling software, to perform maximum-likelihood model-fitting analyses with raw data. To improve normality the IgA level was square root-transformed. Because of a small but significant association to age and batch, IgA values were further adjusted for these potential confounders in males and females separately. IgA levels were also adjusted for the three genetic PCs displaying the strongest association to IgA levels.
Genome-wide complex trait analysis
Variance explained by all SNPs was estimated by restricted maximum likelihood (REML) modeling of the genetic relationship matrix (GRM) with IgA levels as implemented in the GCTA, version 1.11, software package (22). Since GCTA relies on comparisons between subjects not closely related to each other, the sample was filtered for close relations. For complete DZ twin-pairs one member of each pair was therefore randomly selected rendering the sample reduced to 6902 participants. A further restriction was implemented by only considering pairs with relatedness <0.025 which led to exclusion of 1098, leaving n = 5804. Of these, 5784 had IgA levels measured and thus constituting the final sample on which GCTA analysis was conducted. The analysis was adjusted for the three genetic PCs displaying the strongest association to IgA levels.
GWAS replication samples
EIRA
The epidemiological investigation of rheumatoid arthritis (EIRA) is a population-based case–control study of incident cases of RA. Controls were randomly selected from the Swedish national population registry and were matched to the patients for age, sex and residential area. More details about this study population have been described elsewhere (59). In the current study 2365 individuals (70.9% females, median age 54.0 years) recruited between 1996 and 2007 were included and used as a replication cohort. The analysis was adjusted for the three genetic PCs displaying the strongest association to IgA levels.
BAMSE
BAMSE is a Swedish birth cohort study. A total number of 4089 newborn infants were recruited between 1994 and 1996 in the Stockholm area (60). Questionnaires with a focus on lifestyle factors and the children's symptoms related to asthma and allergic diseases were answered by the parents when the children were ∼1, 2, 4 and 8 years old. At 4 years of age, serum IgA levels were determined in all children, using enzyme-linked immunosorbent assay as previously described (18). At 8 years of age, DNA was extracted from 2033 samples after exclusion of samples with too little blood, lack of questionnaire data, or if parental consent to genetic analysis of the sample was not obtained. From these samples, all children with a doctor's diagnosis of asthma (ever) and children with no history of allergic diseases (controls) underwent GWAS genotyping (n = 485). Those that also had PCs available (n = 420) were included in the present study (61). The analysis was adjusted for the three genetic PCs displaying the strongest association to IgA levels.
Statistical analysis of genetic effect
A total of 9617 genotyped individuals were investigated in the discovery GWAS for association to SNP markers in an additive linear model using Plink v1.07 (62) on imputed dosages. Correlated data within dizygotic twin pairs was taken into account by specifying a cluster variable. Associations in the BAMSE replication data set were analyzed using ProbABEL (63), while EIRA analyses were performed using Plink v1.07 (62).
The meta-analysis was performed using METAL (64).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the SwediCsh Research Council (M-2005-1112); GenomeEUtwin (EU/QLRT-2001-01254, QLG2-CT-2002-01254); National Institutes of Health DK (U01-066134); the Heart and Lung Foundation (20070481); and the Swedish Foundation for Strategic Research.
Supplementary Material
REFERENCES
- 1.Woof J.M., Russell M.W. Structure and function relationships in IgA. Mucosal Immunol. 2011;4:590–597. doi: 10.1038/mi.2011.39. [DOI] [PubMed] [Google Scholar]
- 2.Di Franco P., Brai M., Misiano G., Piazza A.M., Giorgi G., Cossarizza A., Franceschi C. Genetic and environmental influences on serum levels of immunoglobulins and complement components in monozygotic and dizygotic twins. J. Clin. Lab. Immunol. 1988;27:5–10. [PubMed] [Google Scholar]
- 3.McGue M., Gerrard J.W., Lebowitz M.D., Rao D.C. Commingling in the distributions of immunoglobulin levels. Hum. Hered. 1989;39:196–201. doi: 10.1159/000153860. [DOI] [PubMed] [Google Scholar]
- 4.Borecki I.B., McGue M., Gerrard J.W., Lebowitz M.D., Rao D.C. Familial resemblance for immunoglobulin levels. Hum. Genet. 1994;94:179–185. doi: 10.1007/BF00202866. [DOI] [PubMed] [Google Scholar]
- 5.Buckley C.E., III, Buckley E.G., Dorsey F.C. Longitudinal changes in serum immunoglobulin levels in older humans. Fed. Proc. 1974;33:2036–2039. [PubMed] [Google Scholar]
- 6.Cassidy J.T., Nordby G.L., Dodge H.J. Biologic variation of human serum immunoglobulin concentrations: sex-age specific effects. J. Chronic Dis. 1974;27:507–516. doi: 10.1016/0021-9681(74)90026-5. [DOI] [PubMed] [Google Scholar]
- 7.Crisp H.C., Quinn J.M. Quantitative immunoglobulins in adulthood. Allergy Asthma Proc. 2009;30:649–654. doi: 10.2500/aap.2009.30.3292. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez-Quintela A., Alende R., Gude F., Campos J., Rey J., Meijide L.M., Fernandez-Merino C., Vidal C. Serum levels of immunoglobulins (IgG, IgA, IgM) in a general adult population and their relationship with alcohol consumption, smoking and common metabolic abnormalities. Clin. Exp. Immunol. 2008;151:42–50. doi: 10.1111/j.1365-2249.2007.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grundbacher F.J., Shreffler D.C. Changes in human serum immunoglobulin levels with age and sex. Z. Immunitatsforsch. Allerg. Klin. Immunol. 1970;141:20–26. [PubMed] [Google Scholar]
- 10.Kalff M.W. A population study on serum immunoglobulin levels. Clin. Chim. Acta. 1970;28:277–289. doi: 10.1016/0009-8981(70)90092-6. [DOI] [PubMed] [Google Scholar]
- 11.Listi F., Candore G., Modica M.A., Russo M., Di Lorenzo G., Esposito-Pellitteri M., Colonna-Romano G., Aquino A., Bulati M., Lio D., et al. A study of serum immunoglobulin levels in elderly persons that provides new insights into B cell immunosenescence. Ann. N. Y. Acad. Sci. 2006;1089:487–495. doi: 10.1196/annals.1386.013. [DOI] [PubMed] [Google Scholar]
- 12.McMillan S.A., Douglas J.P., Archbold G.P., McCrum E.E., Evans A.E. Effect of low to moderate levels of smoking and alcohol consumption on serum immunoglobulin concentrations. J. Clin. Pathol. 1997;50:819–822. doi: 10.1136/jcp.50.10.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Radl J., Sepers J.M., Skvaril F., Morell A., Hijmans W. Immunoglobulin patterns in humans over 95 years of age. Clin. Exp. Immunol. 1975;22:84–90. [PMC free article] [PubMed] [Google Scholar]
- 14.Stoica G., Macarie E., Michiu V., Stoica R.C. Biologic variation of human immunoglobulin concentration. I. Sex-age specific effects on serum levels of IgG, IgA, IgM and IgD. Med. Interne. 1980;18:323–332. [PubMed] [Google Scholar]
- 15.Stoica G.H., Samborschi C., Michiu V. Influence of sex and age on serum immunoglobulin concentrations in healthy subjects. Med. Interne. 1978;16:23–31. [PubMed] [Google Scholar]
- 16.Weber-Mzell D., Kotanko P., Hauer A.C., Goriup U., Haas J., Lanner N., Erwa W., Ahmaida I.A., Haitchi-Petnehazy S., Stenzel M., et al. Gender, age and seasonal effects on IgA deficiency: a study of 7293 Caucasians. Eur. J. Clin. Invest. 2004;34:224–228. doi: 10.1111/j.1365-2362.2004.01311.x. [DOI] [PubMed] [Google Scholar]
- 17.Janzi M., Melen E., Kull I., Wickman M., Hammarstrom L. Rare mutations in TNFRSF13B increase the risk of asthma symptoms in Swedish children. Genes Immun. 2012;13:59–65. doi: 10.1038/gene.2011.55. [DOI] [PubMed] [Google Scholar]
- 18.Janzi M., Kull I., Sjoberg R., Wan J., Melen E., Bayat N., Ostblom E., Pan-Hammarstrom Q., Nilsson P., Hammarstrom L. Selective IgA deficiency in early life: association to infections and allergic diseases during childhood. Clin. Immunol. 2009;133:78–85. doi: 10.1016/j.clim.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 19.Hatagima A., Cabello P.H., Krieger H. Causal analysis of the variability of IgA, IgG, and IgM immunoglobulin levels. Hum. Biol. 1999;71:219–229. [PubMed] [Google Scholar]
- 20.Rowe D.S., Boyle J.A., Buchanan W.W. Plasma immunoglobulin concentrations in twins. Clin. Exp. Immunol. 1968;3:233–244. [PMC free article] [PubMed] [Google Scholar]
- 21.Grundbacher F.J. Heritability estimates and genetic and environmental correlations for the human immunoglobulins G, M, and A. Am. J. Hum. Genet. 1974;26:1–12. [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dehghan A., Yang Q., Peters A., Basu S., Bis J.C., Rudnicka A.R., Kavousi M., Chen M.H., Baumert J., Lowe G.D., et al. Association of novel genetic Loci with circulating fibrinogen levels: a genome-wide association study in 6 population-based cohorts. Circ. Cardiovasc. Genet. 2009;2:125–133. doi: 10.1161/CIRCGENETICS.108.825224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer T.E., Verwoert G.C., Hwang S.J., Glazer N.L., Smith A.V., van Rooij F.J., Ehret G.B., Boerwinkle E., Felix J.F., Leak T.S., et al. Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six Loci influencing serum magnesium levels. PLoS Genet. 2010;6:e1001045. doi: 10.1371/journal.pgen.1001045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka T., Roy C.N., Yao W., Matteini A., Semba R.D., Arking D., Walston J.D., Fried L.P., Singleton A., Guralnik J., et al. A genome-wide association analysis of serum iron concentrations. Blood. 2010;115:94–96. doi: 10.1182/blood-2009-07-232496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igl W., Johansson A., Wilson J.F., Wild S.H., Polasek O., Hayward C., Vitart V., Hastie N., Rudan P., Gnewuch C., et al. Modeling of environmental effects in genome-wide association studies identifies SLC2A2 and HP as novel loci influencing serum cholesterol levels. PLoS Genet. 2010;6:e1000798. doi: 10.1371/journal.pgen.1000798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang C., Jie W., Yanlong Y., Xuefeng G., Aihua T., Yong G., Zheng L., Youjie Z., Haiying Z., Xue Q., et al. Genome-wide association study identifies TNFSF13 as a susceptibility gene for IgA in a South Chinese population in smokers. Immunogenetics. 2012;64:747–753. doi: 10.1007/s00251-012-0636-y. [DOI] [PubMed] [Google Scholar]
- 28.Granada M., Wilk J.B., Tuzova M., Strachan D.P., Weidinger S., Albrecht E., Gieger C., Heinrich J., Himes B.E., Hunninghake G.M., et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J. Allergy Clin. Immunol. 2012;129:840–845. doi: 10.1016/j.jaci.2011.09.029. e821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moffatt M.F., Gut I.G., Demenais F., Strachan D.P., Bouzigon E., Heath S., von Mutius E., Farrall M., Lathrop M., Cookson W.O., et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weidinger S., Gieger C., Rodriguez E., Baurecht H., Mempel M., Klopp N., Gohlke H., Wagenpfeil S., Ollert M., Ring J., et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4:e1000166. doi: 10.1371/journal.pgen.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamin D.J., Cesarini D., van der Loos M.J., Dawes C.T., Koellinger P.D., Magnusson P.K., Chabris C.F., Conley D., Laibson D., Johannesson M., et al. The genetic architecture of economic and political preferences. Proc. Natl Acad. Sci. USA. 2012;109:8026–8031. doi: 10.1073/pnas.1120666109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kennedy M.K., Willis C.R., Armitage R.J. Deciphering CD30 ligand biology and its role in humoral immunity. Immunology. 2006;118:143–152. doi: 10.1111/j.1365-2567.2006.02354.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belnoue E., Pihlgren M., McGaha T.L., Tougne C., Rochat A.F., Bossen C., Schneider P., Huard B., Lambert P.H., Siegrist C.A. APRIL is critical for plasmablast survival in the bone marrow and poorly expressed by early-life bone marrow stromal cells. Blood. 2008;111:2755–2764. doi: 10.1182/blood-2007-09-110858. [DOI] [PubMed] [Google Scholar]
- 34.Hammarström L., Smith C. Genetic approach to common variable immunodeficiency and IgA deficiency. In: Ochs H.D., Smith C.I.E., Puck J.M., editors. Primary Immunodeficiency Diseases: A Molecular and Genetic Approach. 2nd ed. New York: Oxford University Press; 2007. pp. 313–325. [Google Scholar]
- 35.Feng M.L., Zhao Y.L., Shen T., Huang H., Yin B., Liu R.Z., Qian K.C., Liu D.Z. Prevalence of immunoglobulin A deficiency in Chinese blood donors and evaluation of anaphylactic transfusion reaction risk. Transfus. Med. 2011;21:338–343. doi: 10.1111/j.1365-3148.2011.01082.x. [DOI] [PubMed] [Google Scholar]
- 36.Feng L. [Epidemiological study of selective IgA deficiency among 6 nationalities in China] Zhonghua yi xue za zhi, 72. 1992;88–90:128. [PubMed] [Google Scholar]
- 37.Wang N., Lu P., Ling B., Zhu Z., Hammarstrom L. Caucasian origin of disease associated HLA haplotypes in Chinese blood donors with IgA deficiency. J. Clin. Immunol. 2014;34:157–162. doi: 10.1007/s10875-013-9983-1. [DOI] [PubMed] [Google Scholar]
- 38.Tin A., Astor B.C., Boerwinkle E., Hoogeveen R.C., Coresh J., Kao W.H.L. Genome-wide association study identified the human leukocyte antigen region as a novel locus for plasma beta-2 microglobulin. Human Genet. 2013;132:619–627. doi: 10.1007/s00439-013-1274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Surakka I., Whitfield J.B., Perola M., Visscher P.M., Montgomery G.W., Falchi M., Willemsen G., de Geus E.J.C., Magnusson P.K.E., Christensen K., et al. A genome-wide association study of monozygotic twin-pairs suggests a locus related to variability of serum high-density lipoprotein cholesterol. Twin Res. Hum. Genet. 2012;15:691–699. doi: 10.1017/thg.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang M., Wu Y., Lu Y., Liu C., Sun J., Liao M., Qin M., Mo L., Gao Y., Lu Z., et al. Genome-wide scan identifies variant in TNFSF13 associated with serum IgM in a healthy Chinese male population. PloS One. 2012;7:e47990. doi: 10.1371/journal.pone.0047990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao M., Ye F., Zhang B., Huang L., Xiao Q., Qin M., Mo L., Tan A., Gao Y., Lu Z., et al. Genome-wide association study identifies common variants at TNFRSF13B associated with IgG level in a healthy Chinese male population. Genes Immun. 2012;13:509–513. doi: 10.1038/gene.2012.26. [DOI] [PubMed] [Google Scholar]
- 42.Levin A.M., Mathias R.A., Huang L., Roth L.A., Daley D., Myers R.A., Himes B.E., Romieu I., Yang M., Eng C., et al. A meta-analysis of genome-wide association studies for serum total IgE in diverse study populations. J. Allergy Clin. Immunol. 2012;131:1176–1184. doi: 10.1016/j.jaci.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang X., Sun J., Gao Y., Tan A., Zhang H., Hu Y., Feng J., Qin X., Tao S., Chen Z., et al. Genome-wide association study for serum complement C3 and C4 levels in healthy Chinese subjects. PLoS Genet. 2012;8:e1002916. doi: 10.1371/journal.pgen.1002916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magnusson P.K., Rasmussen F. Familial resemblance of body mass index and familial risk of high and low body mass index. A study of young men in Sweden. Int. J. Obes. Relat. Metab. Disord. 2002;26:1225–1231. doi: 10.1038/sj.ijo.0802041. [DOI] [PubMed] [Google Scholar]
- 45.Berndt S.I., Gustafsson S., Magi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L., Croteau-Chonka D.C., Day F.R., et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franke A., McGovern D.P.B., Barrett J.C., Wang K., Radford-Smith G.L., Ahmad T., Lees C.W., Balschun T., Lee J., Roberts R., et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat. Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson C.A., Boucher G., Lees C.W., Franke A., D'Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A., et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ammann A.J., Hong R. Selective IgA deficiency: presentation of 30 cases and a review of the literature. Medicine. 1971;50:223–236. [PubMed] [Google Scholar]
- 50.Strober W., Sneller M.C. IgA deficiency. Ann. Allergy. 1991;66:363–375. [PubMed] [Google Scholar]
- 51.Schaffer F.M., Monteiro R.C., Volanakis J.E., Cooper M.D. IgA deficiency. Immunodefic. Rev. 1991;3:15–44. [PubMed] [Google Scholar]
- 52.Heneghan M.A., Stevens F.M., Cryan E.M., Warner R.H., McCarthy C.F. Celiac sprue and immunodeficiency states: a 25-year review. J. Clin. Gastroenterol. 1997;25:421–425. doi: 10.1097/00004836-199709000-00004. [DOI] [PubMed] [Google Scholar]
- 53.Cunningham-Rundles C. Physiology of IgA and IgA deficiency. J. Clin. Immunol. 2001;21:303–309. doi: 10.1023/a:1012241117984. [DOI] [PubMed] [Google Scholar]
- 54.Magnusson P.K., Almqvist C., Rahman I., Ganna A., Viktorin A., Walum H., Halldner L., Lundstrom S., Ullen F., Langstrom N., et al. The Swedish twin registry: establishment of a biobank and other recent developments. Twin Res. Hum. Genet. 2013;16:317–329. doi: 10.1017/thg.2012.104. [DOI] [PubMed] [Google Scholar]
- 55.Lichtenstein P., De Faire U., Floderus B., Svartengren M., Svedberg P., Pedersen N.L. The Swedish Twin Registry: a unique resource for clinical, epidemiological and genetic studies. J. Intern. Med. 2002;252:184–205. doi: 10.1046/j.1365-2796.2002.01032.x. [DOI] [PubMed] [Google Scholar]
- 56.Pedersen N.L., Lichtenstein P., Svedberg P. The Swedish Twin Registry in the third millennium. Twin Res. 2002;5:427–432. doi: 10.1375/136905202320906219. [DOI] [PubMed] [Google Scholar]
- 57.Howie B.N., Donnelly P., Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boker S., Neale M., Maes H., Wilde M., Spiegel M., Brick T., Spies J., Estabrook R., Kenny S., Bates T., et al. OpenMx: an open source extended structural equation modeling framework. Psychometrika. 2011;76:306–317. doi: 10.1007/s11336-010-9200-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holmqvist M.E., Wedren S., Jacobsson L.T., Klareskog L., Nyberg F., Rantapaa-Dahlqvist S., Alfredsson L., Askling J. No increased occurrence of ischemic heart disease prior to the onset of rheumatoid arthritis: results from two Swedish population-based rheumatoid arthritis cohorts. Arthritis Rheum. 2009;60:2861–2869. doi: 10.1002/art.24855. [DOI] [PubMed] [Google Scholar]
- 60.Kull I., Melen E., Alm J., Hallberg J., Svartengren M., van Hage M., Pershagen G., Wickman M., Bergstrom A. Breast-feeding in relation to asthma, lung function, and sensitization in young schoolchildren. J. Allergy Clin. Immunol. 2010;125:1013–1019. doi: 10.1016/j.jaci.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 61.Melen E., Granell R., Kogevinas M., Strachan D., Gonzalez J.R., Wjst M., Jarvis D., Ege M., Braun-Fahrlander C., Genuneit J., et al. Genome-wide association study of body mass index in 23 000 individuals with and without asthma. Clin. Exp. Allergy. 2013;43:463–474. doi: 10.1111/cea.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aulchenko Y.S., Struchalin M.V., van Duijn C.M. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.