Abstract
The oxidative-stress-responsive kinase 1 (OSR1) and the STE20/SPS1-related proline/alanine-rich kinase (SPAK) are key enzymes in a signaling cascade regulating the activity of Na+-K+-2Cl− cotransporters (NKCC1-2) and Na+-Cl− cotransporter (NCC). Both kinases have a conserved carboxyl-terminal (CCT) domain, which recognizes a unique peptide motif present in OSR1- and SPAK-activating kinases (with-no-lysine kinase 1 (WNK1) and WNK4) as well as their substrates (NKCC1, NKCC2, and NCC). Utilizing various modalities of the Rosetta Molecular Modeling Software Suite including flexible peptide docking and protein design, we comprehensively explored the sequence space recognized by the CCT domain. Specifically, we studied single residue mutations as well as complete unbiased designs of a hexapeptide substrate. The computational study started from a crystal structure of the CCT domain of OSR1 in complex with a hexapeptide derived from WNK4. Point mutations predicted to be favorable include Arg to His or Trp substitutions at position 2 and a Phe to Tyr substitution at position 3 of the hexapeptide. In addition, de novo design yielded two peptides predicted to bind to the CCT domain: FRFQVT and TRFDVT. These results, which indicate a little bit more freedom in the composition of the peptide, were confirmed through the use of yeast two-hybrid screening.
Keywords: Computer modeling, Protein design, Peptide conformation, Ste20 kinases, Rosetta Modeling Suite, Chloride transport
INTRODUCTION
SPAK (SPS1-related Proline/Alanine-rich Kinase) and OSR1 (Oxidative Stress Response 1 kinase) are members of the Ste20-related family of protein kinases (Dan, et al., 2001; Delpire, 2009). They modulate the activity of cation-chloride cotransporters (Gagnon, et al., 2006; Grimm, et al., 2012; Lin, et al., 2011; McCormick, et al., 2011; Rafiqi, et al., 2010), which are involved ion secretion (Kurihara, et al., 2002; Matthews, et al., 1993) and reabsorption (Gimenez and Forbush, 2005; Pacheco-Alvarez, et al., 2006) across a variety of epithelia, Cl− homeostasis in neurons (Austin and Delpire, 2011; Blaesse, et al., 2009; Delpire, 2000; Delpire and Austin, 2010), and cell volume maintenance and regulation in many cells (Gagnon and Delpire, 2012). Molecular studies have demonstrated that kinase binding to the substrate is a pre-requisite for the function of SPAK and OSR1 (Gagnon, et al., 2006; Piechotta, et al., 2003; Piechotta, et al., 2002). The interaction between SPAK/OSR1 and substrates involves a ~90 residue domain located at the C-terminal tail of the kinase and a short conserved peptide located within the substrate (Piechotta, et al., 2002). Yeast 2-hybrid analyses indicated that the peptide needed to be nine residues long, at least when located at the extreme C-terminus of the bait protein. Amino acid alignment between the cytosolic N-terminal tails of membrane transporters (related targets) revealed the preliminary conserved sequence: Arg-Phe-Xaa-Val (Piechotta, et al., 2002). Following a large yeast 2-hybrid screen that used the conserved carboxyl-terminal domain of SPAK as bait (Piechotta, et al., 2003), the motif was expanded to [Val/Ser/Gly]-Arg-Phe-Xaa-[Val/Iso]-Xaa-Xaa-[Thre/Ser/Val/Iso]. A whole mouse proteome search identified some 170 proteins containing this expanded motif (Delpire and Gagnon, 2007).
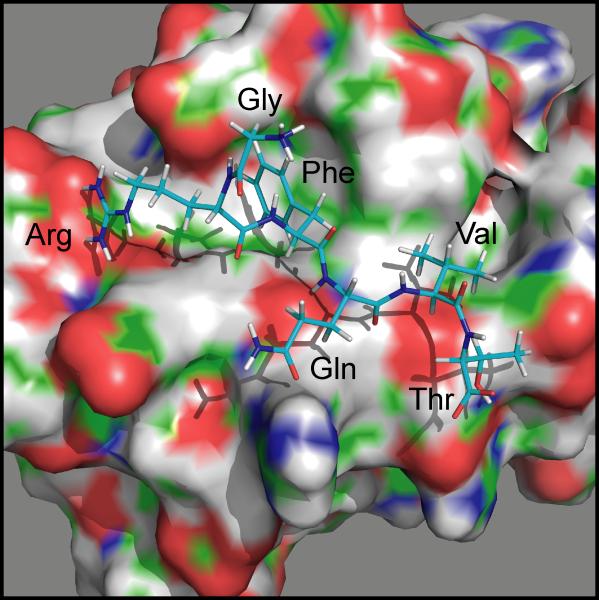
The crystal structure of the ~90 amino acid human OSR1 domain, which includes an embedded GRFQVT hexapeptide from human WNK4, was resolved at 1.95 Å (PDB ID: 2v3s, (Villa, et al., 2007)). This domain, which was termed Conserved Carboxyl-Terminal (CCT) (Villa, et al., 2007) or Protein Fold 2 (PF2) (Lee, et al., 2009), is represented in Figure 1. The most salient feature is a hydrophobic groove that accommodates the Phe and Val residues of the peptide. While the protein fold was originally thought to be unique to SPAK and OSR1 (Villa, et al., 2007), we later showed that it is also present, at least partially, downstream of the catalytic domain of WNK4 (Delpire and Gagnon, 2008; Gagnon and Delpire, 2012). Availability of this domain – peptide structure allowed us to use the docking and design applications of Rosetta, a protein structure prediction and functional design software package, to assess the binding of various hexapeptides in the pocket and estimate binding energies. Thus, this analysis allows us to better understand the amino acid requirements for the interaction by ranking peptides with favorable to unfavorable energies. It also comprehensively determines the peptide sequences consistent with OSR1 and SPAK interaction thereby identifying potential sequences that have not yet been implied but can now be tested experimentally.
Figure 1. PyMol rendering of the hydrophobic pocket of OSR1 with the GRFQVT peptide of WNK4.
The surface representation of the OSR1 domain highlights negative (red), positive (blue), and polar (green) moieties.
MATERIAL AND METHODS
Computational Modeling
We started with a three-dimensional representation of the crystal structure of the CCT/PF2 domain of OSR1 kinase in complex with a hexapeptide (GRFQVT) derived from WNK4 (Figure 1). This file was obtained from the Protein Data Bank (http://www.rcsb.org/) as 2v3s, representing the work performed by Villa and collaborators (Villa, et al., 2007). Crystallographic coordinates for extraneous molecules and fragments were removed, leaving a lone CCT-peptide complex. The resulting structure was then energy minimized using the Rosetta 3.4 relax application (Raman, et al., 2009; Verma and Wenzel, 2007), according to the score12 energy function. This protocol adjusts the protein backbone and side chain torsion angles as a means of correcting local crystallographic bias, minimizing internal clashes, and moving the structure into an energy minimum on the Rosetta score12 energy function. As Rosetta employs a stochastic Monte Carlo Metropolis sampling strategy, multiple trajectories are needed to search the conformational (and sequence) space comprehensively. Here one hundred relaxed CCT-peptide complexes were produced and the top five models were chosen based on lowest Rosetta total energy score.
The native hexapeptides of the five relaxed complexes were then copied into separate files. Utilizing a simple python script, each residue of the hexapeptides was mutated into the other 19 canonical amino acids at all six positions, resulting in a total of 114 mutated peptides for each relaxed structure. The backbone atoms of all of these mutants had similar three-dimensional coordinates as their starting hexapeptides with the alterations only occurring at the side chains. These engineered peptides were then recombined with their corresponding unbound CCT domains (CCT-peptide complexes with peptides removed).
The mutated hexapeptides, along with the native forms, were then docked into the CCT domains by utilizing the FlexPepDock application of Rosetta 3.4 (London, et al., 2011; Raveh, et al., 2010) with the following flags: -use_input_sc, -ex1, -ex2, -pep_refine, and -unboundrot. Two hundred models were produced for each relaxed structure with a total of one thousand models created for each mutated hexapeptide. A low-resolution pre-optimization flag (-lowres_preoptimize) was employed in half of the docking runs in order to sample a larger peptide conformational space. A Rosetta binding energy (ddG) was calculated to assess the stability of the docked protein-peptide complex. The top ten models for each mutation, regardless of initial relaxed structure, were determined based on ddG and reweighted total energy score. The reweighted score is a combination of interface score, peptide score, and total score and is a better scoring function than score12 in the case of flexible peptides docking onto their receptors (Raveh, et al., 2011). After averaging the scores of the ten models, each mutant was compared to the native CCT-peptide and other mutated peptide complexes.
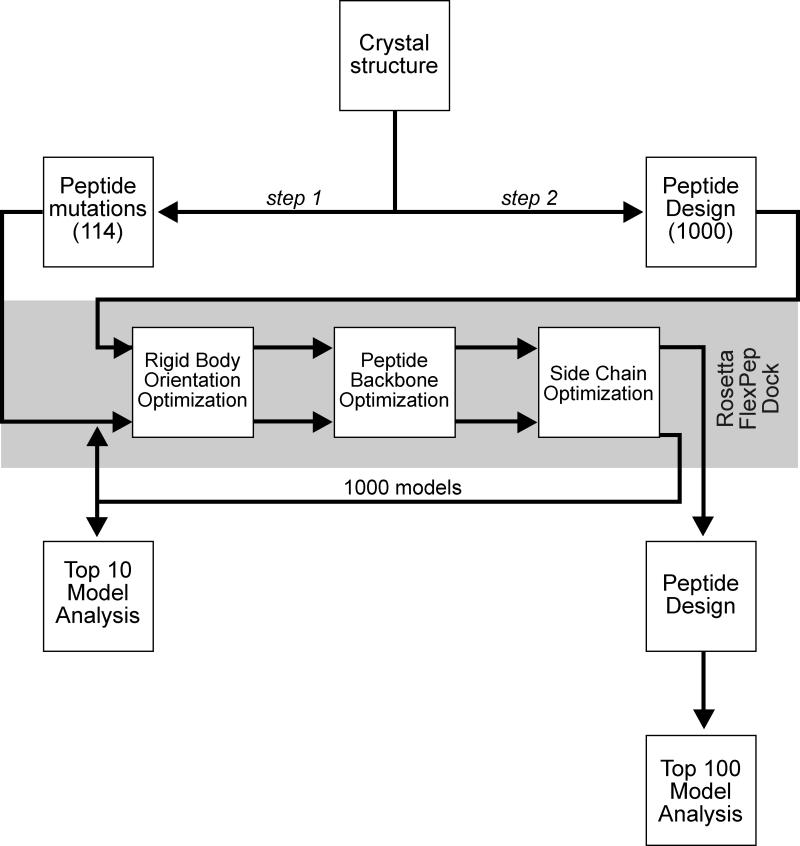
Separately, the design module (Jha, et al., 2010; Kuhlman and Baker, 2000) of Rosetta 3.4.1 was applied to the native hexapeptide in order to sample the sequence space consistent with CCT binding in a more unbiased fashion. In our procedure, only the hexapeptide was targeted for redesign while the CCT domain was left untouched. High resolution docking of these designed peptides into the binding pocket of the CCT domain was achieved through the use of FlexPepDock with similar options as previously described. After docking, another iterative round of design was performed on the hexapeptide to further refine the list of mutable residues. Again, two hundred decoys were produced for each relaxed structure with a total of one thousand models being created. The top one hundred models based on ddG were analyzed and compared to the native structure using PyMOL (PyMOL Molecular Graphics System, Version 1.5.0.4, Schrödinger, LLC). This entire process is outlined in Figure 2.
Figure 2. Scheme utilized in our modeling study.
Step 1: The GRFQVT peptide complexed in WNK4 was initially removed from the crystal structure (2v3s) and docked back into the hydrophobic pocket of the OSR1 CCT/PF2 domain along with 114 hexapeptide variants (point mutations at every position with the other 19 canonical amino acid residues). One thousand models were created using Rosetta FlexPepDock for each peptide and the top 10 models from each of the 1000 runs were averaged for analysis. Step 2: The Rosetta design application was used to produce 1000 whole hexapeptide mutants based on binding of the individual residues of the wild-type hexapeptide to the CCT/PF2 domain. FlexPepDock was then employed and binding energies of the complexes were calculated. The top 100 models were analyzed.
Yeast-2-Hybrid Analysis
The regulatory domain of mouse SPAK (residues 353 to 556) fused to the GAL4 activating domain in pACT2 was originally isolated from a Clontech mouse brain library (Piechotta, et al., 2002). The clone was re-transformed into PJ69-4A cells (James, et al., 1996)) according to standard yeast handling procedures (Yeast Handbook, Clontech) and plated on –LEU plates. Sense and anti-sense oligonucleotides were purchased from Sigma Genosys. Upon annealing, the oligonucleotides create overhanging 5’ EcoRI and 3’ BamHI sites that are directly used for ligation. The annealed adaptors encode 14 amino acid peptides that includes EF (EcoRI site), QLVG (linker), RFQVT or mutant (PF2 target peptides), and SSK followed by a stop codon. The QLVGRFQVTSSK sequence is original to the SPAK binding site in WNK4 ((Piechotta, et al., 2003; Villa, et al., 2007). The adaptors are ligated downstream of the Gal4 binding domain in the pGBDUc2 vector. Yeast cells containing the regulatory domain of SPAK in pACT2 were then transformed with individual peptide clones in pGBDUc2 and plated on double dropout -LEU, -URA plates. Yeast clones were then re-streaked on double-dropout plates as controls and triple dropout –LEU, -URA, -His, 2 mM 3-amino-1,2,4-triazole plates.
Analysis of Motif Frequency
The National Center for Biotechnology Information protein database (http://www.ncbi.nlm.nih.gov/) was then searched for Mus musculus AND “RecName” to capture known full-length proteins. The search was performed on 10/25/1013 and yielded 17,049 protein sequences. All proteins were saved in FASTA format in a single file which was opened with WordPerfect to find and remove all hard returns (HRt) codes and subsequently saved in ASCII DOS (Delimited Text) format. Using a small routine written in Visual Basic (Microsoft), the entire text file was searched for specific sequences allowing multiple residues per position as described in Delpire and Gagnon (Delpire and Gagnon, 2007). At the end of the search, the number of proteins with motifs, the total number of motifs, the protein names, and the motif sequences are copied into a single text file.
RESULTS
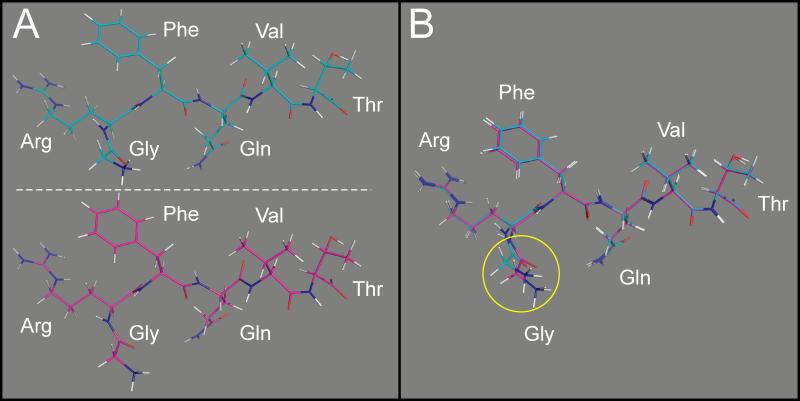
The first objective of our study was to model the binding of the native GRFQVT hexapeptide from WNK4 into the hydrophobic pocket of the OSR1 kinase's CCT domain, using the Rosetta computational suite. For this purpose, the peptide was first extracted out of the CCT domain and docked back into the pocket in 1000 configurations. For each configuration, the ddG binding energy was calculated. A lower, more negative binding energy is indicative of an energetically stable complex. Using the best (lowest energy) model, the position of each amino acid in the hexapeptide was compared to the native position of the peptide in the crystal structure. Figure 3 and Table 1 show that there is only a root mean square deviation (RMSD) of 0.38 Å between the two hexapeptides, indicating that Rosetta can appropriately model the interaction between the hexapeptide and the CCT domain of OSR1.
Figure 3. Position of the WNK4 GRFQVT peptides in the OSR1 CCT/PF2 domain.
Structure of the domain is hidden to highlight the structure and position of each amino acid. A, top: position of the peptide in the native crystal structure. A, bottom: position of the peptide after Rosetta FlexPepDock modeling. B: Super-imposition of the two peptides. There is only a sub-angstrom all-atom difference between the model and the crystal structure.
Table 1.
GRFQVT residue-specific parameters.
| Hexamer (or Total) | Glycine (−1) | Arginine (+1) | Phenylalanine (+2) | Glutamine (+3) | Valine (+4) | Threonine (+5) | |
|---|---|---|---|---|---|---|---|
| RMSD (Å) | 0.384 | 1.175 | 0.257 | 0.144 | 0.1 | 0.081 | 0.101 |
| ddG | −17.59 | 0.002 | −1.504 | −2.356 | −2.553 | −0.831 | −0.523 |
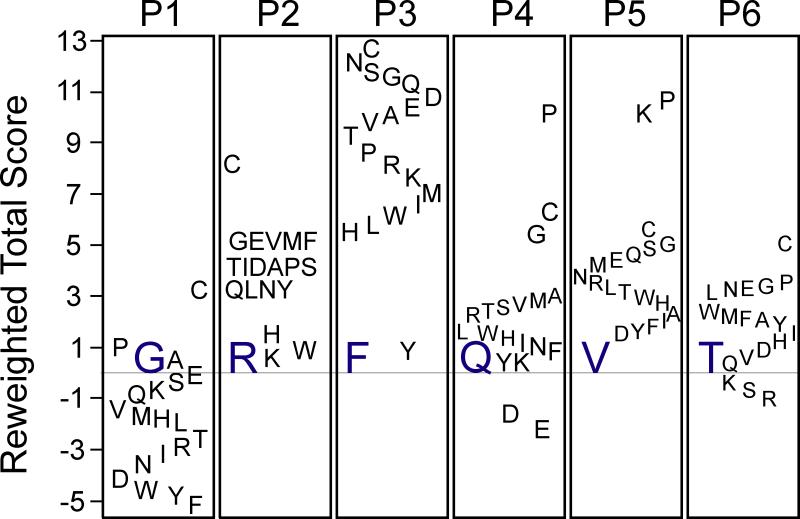
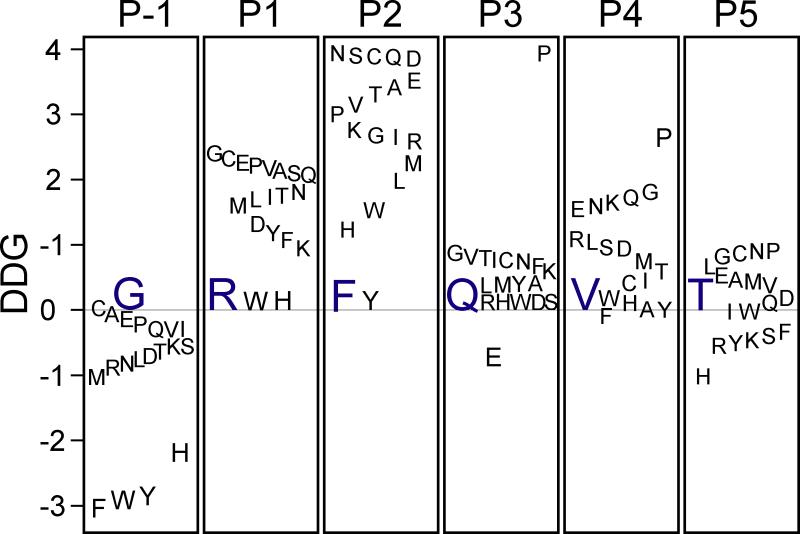
Amino acid substitutions were then systematically created at each of the six positions in the peptide and each mutant peptide modeled for binding using FlexPepDock. It is shown that the reweighted total scores of native Arg, Phe, and Val at positions 2, 3, and 5, respectively, were lower than any of the mutant scores (Figure 4). However, almost all of the position 1 mutants have a more favorable energy than the native Gly. Also, only Asp and Glu possessed lower reweighted energies than Gln at position 4 while Lys, Ser, and Arg were the only residues less energetic than Thr at position 6. The reweighted total score was linearly related to the total score and could be used interchangeably to produce similar results. The binding energies (ddGs) of the point mutants followed comparable trends to their reweighted total scores (Figure 5). The native Arg at position 2 had the lowest ddG, corresponding to the most stable complex. Phe at position 3 and Val at position 5 each only had one mutant residue with greater stability, Tyr and Phe, respectively. Similar to its reweighted total score, all of the hexapeptides with point mutations at the first position bound more tightly than the native peptide. Only Glu had a lower ddG than Gln at position 4 and almost half of the residue 6 mutants possess lower ddGs.
Figure 4. Relative energy scores (reweighted total scores) of hexapeptide mutants.
The difference in reweighted total scores between every possible canonical amino acid and the original WNK4 residues at each of the six positions (P1-P6) is indicated. Negative values indicate better scores (more stable complexes) whereas positive values indicate less stability than wild-type amino acids. Original amino acids are indicated in blue. Scores are given in Rosetta Energy Units (REU).
Figure 5. Relative binding energies (ddG) of hexapeptide mutants.
The difference in binding energy (ddG) between every possible canonical amino acid and the original WNK4 residue at each of the six positions (P1-P6) is indicated. Negative values indicate better energy whereas positive values indicate worse score than wild-type amino acids. Original amino acids are indicated in blue. Binding energies are given in Rosetta Energy Units (REU).
Rosetta design yielded similar results. In this case, rather than manually assigning all 20 amino acids at each position, the Rosetta design, guided by the Rosetta energy function, mutated each position in a Monte Carlo Metropolis search of the full sequence space to identify the optimal hexapeptide sequence. The Arg, Phe, and Val in positions 2, 3, and 5, respectively, were recovered after design (Figure 6). Gly at position 1 was replaced with a Thr while Gln at position 4 was replaced with an Asp. Lastly, the position 6 Thr was mostly mutated to Lys, Arg, or Val. The dominant residues at each position corresponded to favorable hexapeptides with low reweighted total scores and ddGs in the manual point mutation analysis.
Figure 6. Sequence logo of designed hexapeptides.
The original peptide appears below the x-axis and the relative sizes of the letters indicate their frequency in the sequences. Arg 2, Phe 3, and Val 5 were conserved in this experiment.
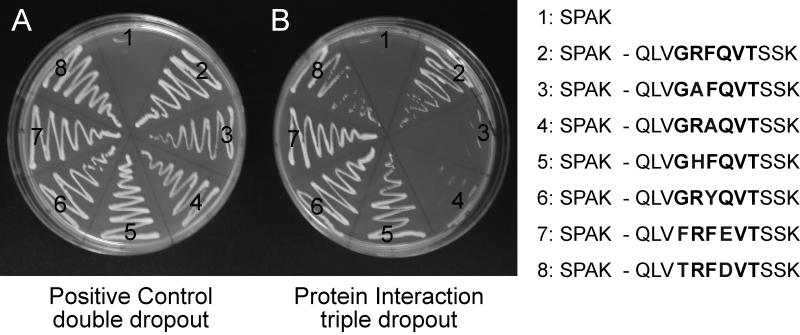
Based on the information gained from the computer modeling, we created peptides to test for protein-protein interaction using yeast-2 hybrid analysis. Yeast cells were first transfected with the PF2 domain of SPAK fused downstream of the GAL4 activating domain in the pACT2 vector. As the vector confers yeast survival on plates lacking leucine, transfected yeast cells were selected on -LEU plates. These cells were then transfected with different peptide sequences fused to the GAL4 binding domain in the vector pGBDUc2. As this vector confers survival on plates lacking uracil, doubly transfected yeast cells were selected on -LEU, -URA plates. As seen in Figure 7A, in the absence of pGBDUC2 transfection, there is no growth of SPAK-PF2_pACT2 containing yeast cells in positive control double dropout plates. In contrast, all fully transfected yeast cells grew under these -LEU, -URA conditions. As yeast cells will survive in the absence of histidine upon positive protein-protein interaction, the cells were re-plated on triple dropout -LEU, -URA, -HIS plates (Figure 7B). Consistent with previous studies, the wild-type peptide GRFQV (condition 2) interacted with the PF2 domain of SPAK promoting yeast survival, whereas the GAFQV (condition 3) and GRAQV (condition 4) mutants who served as negative controls demonstrated absence of interaction. These mutations were not computationally favored (Figures 4 & 5). Because the Rosetta modeling indicated that His and Tyr residues could substitute for Arg and Phe residues, respectively, we tested interaction of peptides GHFQV (condition 5) and GRYQV (condition 6) with the PF2 domain of SPAK. Both peptides demonstrated positive yeast 2-hybrid interaction. Similarly, sequences that Rosetta found optimal through systematic single amino acid substitution (FRFEVT, condition 7) or through peptide design (TRFDVT, condition 8) also interacted with the PF2 domain of SPAK in our yeast-2 hybrid assay.
Figure 7. Yeast 2-hybrid analysis.
Yeast transformed with different cDNA clones were plated onto agar media lacking essential nutrients: uracil, leucine, and/or histidine. For each pair, the regulatory domain of SPAK was fused to the GAL4 activating domain in pACT2, and a 12 amino acid peptide (sequence on the right inset) was fused to the GAL4 binding domain in pGBDUc2. (A): control plate lacking two essential nutrients (uracil and leucine) to demonstrate transformation efficiency. (B) Yeast survival on plate lacking three essential nutrients (uracil, leucine, and histidine) exists only upon interaction between the GAL4 fusion proteins translated from each clone.
DISCUSSION
The purpose of this study was to better understand the amino acid requirements for binding to the CCT domain of OSR1 by expanding our knowledge from specific point mutations in the Arg-Phe-Xaa-Val binding motif to mutations that include every canonical amino acid at all motif positions plus the two residues surrounding this motif. Since performing all of these experiments in a laboratory setting would be both cumbersome and expensive, we decided to perform these analyses in silico. We utilized the Rosetta protein modeling suite, a unified software package that is routinely acknowledged for its protein design and structure prediction accuracy. Starting from a monomer of the CCT domain bound to a hexapeptide derived from one of its substrates, our initial procedure of remodeling this complex using the FlexPepDock application produced an all-atom model of the hexapeptide which had a sub-angstrom RMSD compared to the crystal structure. This modeling accuracy is on par with previous experiments utilizing this application (Raveh, et al., 2010). Upon further analysis, all hexapeptide residues in this model had a RMSD of < 0.3Å with the exception of glycine (Figure 3B, Table 1), which is most likely a result of its lack of affinity to the binding pocket as represented by its trivial contribution to the peptide binding energy (Table 1).
We then mutated the hexapeptide by two different computational algorithms and complexed these novel substrates with the CCT domain in order to access their binding energies. In the hexapeptides that were altered using the Rosetta design protocol, the Arg, Phe, and Val at positions 2, 3, and 5, respectively, were conserved 97%, 98%, and 98% of the time, respectively (Figure 6). These conserved residues correspond to the amino acids with the lowest reweighted total scores in the point mutation portion of this experiment (Figure 4). This is an important correlation since the design protocol evaluates the relative favorability of the transformed sequence based on its energy score compared to the previous model (Kuhlman and Baker, 2000). Since, on average, the total energies of the Arg, Phe, and Val at positions 2, 3, and 5, respectively, are more favorable than the other canonical amino acids, there is a low probability of replacement during design. The conservation of these positions is perhaps unsurprising given their molecular-level interactions. As noted by Villa et al (Villa, et al., 2007), Arg at position 2 of the designed hexapeptides forms salt bridges with Asp at position 459 and Glu acid at position 467. Similarly, Phe 3 in the designed substrates hydrophobically interacts with Phe 452, Leu 468, Ala 471, and Leu 473 while Val 5 hydrophobically stacks with Ile 450.
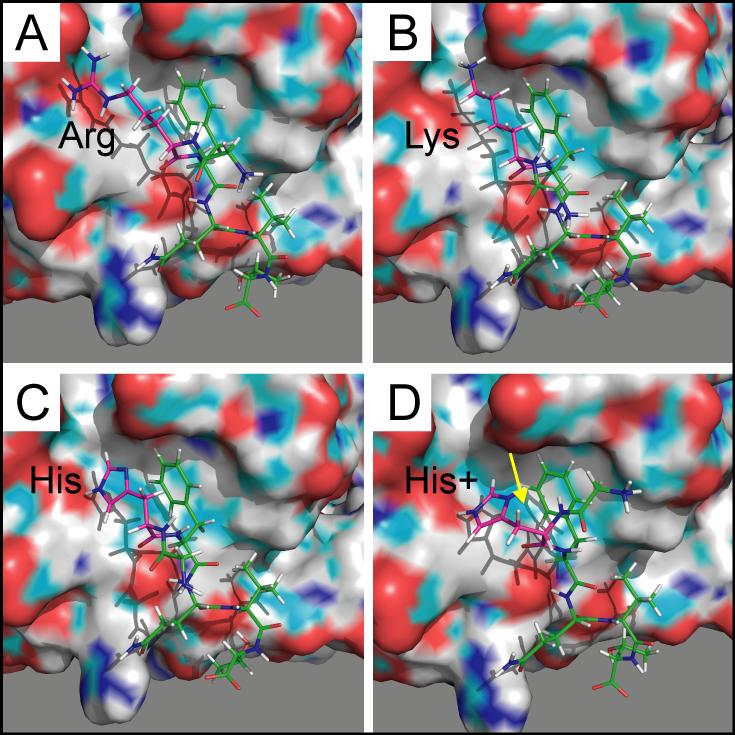
Various mutations of Arg at position 2 disrupt key interactions within the CCT binding pocket. Although replacement of this native residue with a Lys confers a similar positively charged side chain, the discrepancy in its side chain length mitigates its interactions with Asp 459 (distance 3.4 Å) and Glu 467 (distance 2.9 Å), as represented in Figure 8B. This mutation increases both the overall Rosetta energy (Figure 4) and the hexapeptide binding energy (Figure 5). Likewise, altering the native Arg to a His destabilizes the complex through disruption of these important interactions, which is a consequence of increased distances between the side groups and decreased polarity of histidine's substituent (Figure 8C). Although these two point mutants result in a lower receptor affinity, these differences are small relative to the wild-type peptide (Figure 5) such that binding most likely would not be precluded. In fact, the His variant promoted yeast survival in the yeast two-hybrid screen in this experiment (Figure 7) while the Lys mutant has already been shown to produce a positive yeast two-hybrid interaction (Piechotta, et al., 2002). Of note, protonating histidine's side chain as a means of increasing its positive charge slightly worsens its predicted binding energy, most likely resulting from a conformational change within the binding pocket (Figure 8D). Lastly, the Trp variant also appears to favorably bind to the receptor (Figure 5). Although the salt bridges with Asp 459 and Glu 467 are completely lost, this is mostly compensated by increased hydrophobic interactions with Phe 452 and Val 464 (not shown).
Figure 8. PyMol rendering of peptides and OSR1/CCT binding pocket.
A. GRFQVT wild-type peptide with arginine side chains extending towards negative charges within the pocket. B. GKFQVT mutant peptide with shorter side chain of lysine. C & D. GHFQVT mutant peptides are shown with or without protonation of the imidazole ring. Proton is highlighted with yellow arrow. The CCT/PF2 domains are rendered in surface mode, whereas the peptides are rendered in stick mode. The surface drawing of the domains highlights negative (red), positive (blue), and polar (green) moieties.
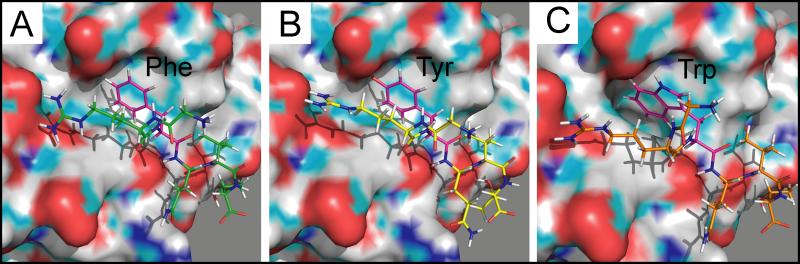
As with the native Arg, mutations in Phe at position 3 lead to destabilized complexes. The one clear exception is Tyr, whose mutant possesses a lower binding energy (Figure 5) while its total Rosetta energy is slightly higher than the native hexapeptide (Figure 4). In addition to analogous hydrophobic effects from the binding pocket residues, the tyrosine mutant forms a hydrogen bond with Asp 459 (distance 2.9 Å), further stabilizing the complex (Figure 9B). This is reflected in tyrosine's individual ddG of −3.0 Rosetta Energy Units (REU), significantly less than the native Phe (Table 1). As expected, this point mutation promoted yeast survival in the yeast two-hybrid screen (Figure 7). Due to the aromatic phenyl ring in its side chain, we expected similar hydrophobic packing (and stability) with Trp as with both the native Phe and the mutant Tyr. Although there are some comparable nonpolar interactions, steric effects due to the bulky indole group alter the conformation of the binding pocket (Figure 9C). This leads to a change in the position of the substrate's Arg and Val relative to their cognate receptor residues, thereby destabilizing the complex.
Figure 9. PyMol rendering of peptides and OSR1/CCT binding pocket.
A. GRFQVT wild-type peptide with arginine side chains extending towards negative charges within the pocket. B. Tyrosine residue substitutes well for the phenylalanine (GKYQVT). C. Tryptophan residue modifies the shape (side chains) of the binding pocket and disrupts the position of the arginine and valine. The CCT/PF2 domains are rendered in surface mode, whereas the peptides are rendered in stick mode. The surface drawing of the domains highlights negative (red), positive (blue), and polar (green) moieties.
Although the almost complete conservation of Val at position 5 in the design (Figure 6) can be attributed to its low total energy score (Figure 4), it is surprising that the Ile variant at this position did not emerge in the design. Apart from Val, Ile is the only known amino acid that exists in the SPAK/OSR1 RFx[V/I] binding motif (Piechotta, et al., 2002). Upon further inspection, Ile 5 also forms hydrophobic stacking with Ile 450, similar to the native Val. However, the discrepancies in their total scores (~1 REU) may have precluded Ile in the design at this position.
There is complete replacement of two of the native hexapeptide residues during the design protocol: Gly at position 1 and Gln at position 4. Gly is typically replaced by Thr (91%, Figure 6), which is predicted to form a more stable complex (Figure 4) with superior binding (Figure 5) when compared to the native hexapeptide. In our models, threonine's side chain forms internal hydrogen bonds with Arg 2 and Phe 3, thereby stabilizing the substrate's secondary structure. In contrast, the native Gln residue is predominantly substituted to an Asp (77%, Figure 6) to form interactions across the interface. Although the side chains of the native Gln and the mutant Asp interact with Arg 451 in our models (both distances 2.9 Å), the mutant's ionic interaction is much stronger than the hydrogen bond formed with the Gln, as represented by glutamine's lower individual ddG of −3.2 REU. This salt bridge stabilizes the complex (Figure 4), leading to residue replacement in the hexapeptide design. These computationally favorable variants were confirmed via yeast two-hybrid screening. The most prevalent designed hexapeptide, TRFDVT (Figure 6), promoted yeast survival (Figure 7). In addition, the mutant with the highest binding affinities at positions 1 and 4, FRFEVT (Figure 5), also produced a positive yeast interaction (Figure 7).
In order to assess its effects on binding, the Thr at position 6 was mutated into a phosphorylated Thr. As expected, this destabilized the complex (total energy increased by ~1.5 REU) and decreased the hexapeptide binding energy (ddG increased by ~ 1 REU). In fact, phosphothreonine's individual ddG was slightly positive (0.1 REU) compared to the negative individual binding energy in the native Thr (Table 1). This may be partially explained by the phosphate's effects on the hydrogen bond formed between Thr 6 and Asp 449 (distance 3.0 Å). With phosphorylation, this interaction weakens and the distance between the atoms increases (3.2 Å). We reported in a 2007 study that Ser and Thr residues were over-represented in protein motifs at this position (Delpire and Gagnon, 2007). The prospect of phosphorylating a residue within the binding motif raises the possibility that phosphorylation might regulate protein interaction between the kinase and its targets.
In summary, our study showed that the Rosetta Molecular Modeling Software Suite can very precisely model the binding of the GRFQVT peptide to the CCT/PF2 domain of OSR1. As the modeled binding is exceedingly similar to the one observed in the crystal structure, the software suite is highly likely to be suitable for predicting the binding of chemical structures in the binding pocket. Our in silico mutagenesis study shows that the nature of the residues required for the binding in the CCT/PF2 pocket might be more flexible than previously recognized. This includes several substitutions of the Arg residue and the possible substitution of the Phe residue to a Tyr, for which we also provide experimental evidence. Although a large number of proteins have been identified within the mouse proteome as containing such alternative sequences (Table 2), to date there is no data associating these proteins to SPAK and OSR1 function
Table 2.
Alternative sequences in mouse proteome.
| Motif Searched | Number of Proteins | Number of Motifs | Motif | Protein | |
|---|---|---|---|---|---|
| 1 | [FWYH] [RWH] [FY] [QED] V | 54 | 57 | ||
| 2 | T R F [DVE] V [TKR] | 1 | 1 | TRFEVTGLM | Titin |
| 3 | R F x [VI] | 1305 | 1446 | ||
| 4 | H F x [VI] | 717 | 777 | ||
| 5 | R Y x [VI] | 1071 | 1167 | ||
| 6 | H Y x [VI] | 658 | 730 | ||
| 7 | W F x [VI] | 333 | 340 | ||
| 8 | W Y x [VI] | 257 | 260 |
Supplementary Material
ACKNOWLEGEMENTS
This work was supported by NIH grant RO1 GM074771 to ED. Thomas Austin is supported by a Foundation for Anesthesia Education and Research (FAER) grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Austin TM, Delpire E. Inhibition of KCC2 in mouse spinal cord neurons leads to hyper-sensitivity to thermal stimulation. Anesth. Analg. 2011;113:1509–1515. doi: 10.1213/ANE.0b013e31822e0a5d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaesse P, Airaksinen MS, Rivera C, Kaila K. Cation-chloride cotransporters and neuronal function. Neuron. 2009;61:820–838. doi: 10.1016/j.neuron.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Dan I, Watanabe NM, Kusumi A. The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell. Biol. 2001;11:220–230. doi: 10.1016/s0962-8924(01)01980-8. [DOI] [PubMed] [Google Scholar]
- 4.Delpire E. Cation-chloride cotransporters in neuronal communication. NIPS. 2000;15:309–312. doi: 10.1152/physiologyonline.2000.15.6.309. [DOI] [PubMed] [Google Scholar]
- 5.Delpire E. The Mammalian family of Sterile20p-like protein kinases. Pflügers Arch. 2009;458:953–967. doi: 10.1007/s00424-009-0674-y. [DOI] [PubMed] [Google Scholar]
- 6.Delpire E, Austin TM. Kinase regulation of Na+-K+-2Cl− cotransport in Primary afferent neurons. J Physiol. 2010;588:3365–3373. doi: 10.1113/jphysiol.2010.190769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delpire E, Gagnon KB. Genome-wide analysis of SPAK/OSR1 binding motifs. Physiol. Genomics. 2007;28:223–231. doi: 10.1152/physiolgenomics.00173.2006. [DOI] [PubMed] [Google Scholar]
- 8.Delpire E, Gagnon KB. SPAK and OSR1: STE20 kinases involved in the regulation of ion homoeostasis and volume control in mammalian cells. Biochem. J. 2008;409:321–331. doi: 10.1042/BJ20071324. [DOI] [PubMed] [Google Scholar]
- 9.Gagnon KB, Delpire E. Molecular Physiology of SPAK and OSR1: Two Ste20-Related Protein Kinases Regulating Ion Transport. Physiol. Rev. 2012;92:1577–1617. doi: 10.1152/physrev.00009.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagnon KB, England R, Delpire E. Volume sensitivity of cation-chloride cotransporters is modulated by the interaction of two kinases: SPAK and WNK4. Am. J. Physiol. Cell Physiol. 2006;290:C134–C142. doi: 10.1152/ajpcell.00037.2005. [DOI] [PubMed] [Google Scholar]
- 11.Gimenez I, Forbush B. Regulatory phosphorylation sites in the NH2 terminus of the renal Na-K-Cl cotransporter (NKCC2). Am. J. Physiol. Renal Physiol. 2005;289:F1341–F1345. doi: 10.1152/ajprenal.00214.2005. [DOI] [PubMed] [Google Scholar]
- 12.Grimm PR, Taneja TK, Liu J, Coleman R, Chen YY, Delpire E, Wade JB, Welling PA. SPAK Isoforms and OSR1 Regulate Sodium-Chloride Co-Transporters In a Nephron-Specific Manner. J. Biol. Chem. 2012;287:37673–37690. doi: 10.1074/jbc.M112.402800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James P, Halladay J, Craig EA. Genomic libraries and host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jha RK, Leaver-Fay A, Yin S, Wu Y, Butterfoss GL, Szyperski T, Dokholyan NV, Kuhlman B. Computational design of a PAK1 binding protein. J. Mol. Biol. 2010;400:257–270. doi: 10.1016/j.jmb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhlman B, Baker D. Native protein sequences are close to optimal for their structures. Proc. Natl. Acad Sci. U.S.A. 2000;97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurihara K, Nakanishi N, Moore-Hoon ML, Turner RJ. Phosphorylation of the salivary Na(+)-K(+)-2Cl(−) cotransporter. Am. J. Physiol. Cell Physiol. 2002;282:C817–C823. doi: 10.1152/ajpcell.00352.2001. [DOI] [PubMed] [Google Scholar]
- 17.Lee SJ, Cobb MH, Goldsmith EJ. Crystal structure of domain-swapped STE20 OSR1 kinase domain. Protein Sci. 2009;18:304–313. doi: 10.1002/pro.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin SH, Yu IS, Jiang ST, Lin SW, Chu P, Chen A, Sytwu HK, Sohara E, Uchida S, Sasaki S, Yang SS. Impaired phosphorylation of Na+-K+-2Cl− cotransporter by oxidative stress-responsive kinase-1 deficiency manifests hypotension and Bartter-like syndrome. Proc Natl Acad Sci U S A. 2011;108:17538–17543. doi: 10.1073/pnas.1107452108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.London N, Raveh B, Cohen E, Fathi G, Schueler-Furman O. Rosetta FlexPepDock web server--high resolution modeling of peptide-protein interactions. Nucl. Acids Res. 2011;39:W249–253. doi: 10.1093/nar/gkr431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matthews JB, Awtrey CS, Thompson R, Hung T, Tally KJ, Madara JL. Na+-K+-2Cl− cotransport and Cl− secretion evoked by heat-stable enterotoxin is microfilament dependent in T84 cells. Am. J. Physiol. 1993;265:G370–G378. doi: 10.1152/ajpgi.1993.265.2.G370. [DOI] [PubMed] [Google Scholar]
- 21.McCormick JA, Mutig K, Nelson JH, Saritas T, Hoorn EJ, Yang C-L, Rogers S, Curry J, Delpire E, Bachmann S, Ellison DH. A SPAK Isoform Switch Modulates Renal Salt Transport and Blood Pressure. Cell Metabolism. 2011;14:352–364. doi: 10.1016/j.cmet.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacheco-Alvarez D, Cristóbal PS, Meade P, Moreno E, Vazquez N, Muñoz E, Díaz A, Juárez ME, Giménez I, Gamba G. The Na+:Cl− cotransporter is activated and phosphorylated at the amino-terminal domain upon intracellular chloride depletion. J. Biol. Chem. 2006;281:28755–28763. doi: 10.1074/jbc.M603773200. [DOI] [PubMed] [Google Scholar]
- 23.Piechotta K, Garbarini NJ, England R, Delpire E. Characterization of the interaction of the stress kinase SPAK with the Na+-K+-2Cl− cotransporter in the nervous system: Evidence for a scaffolding role of the kinase. J. Biol. Chem. 2003;278:52848–52856. doi: 10.1074/jbc.M309436200. [DOI] [PubMed] [Google Scholar]
- 24.Piechotta K, Lu J, Delpire E. Cation-chloride cotransporters interact with the stress-related kinases SPAK and OSR1. J. Biol. Chem. 2002;277:50812–50819. doi: 10.1074/jbc.M208108200. [DOI] [PubMed] [Google Scholar]
- 25.Rafiqi FH, Zuber AM, Glo ver M, Richardson C, Fleming S, Jovanovic S, Jovanovic A, O'Shaughnessy KM, Alessi DR. Role of the WNK-activated SPAK kinase in regulating blood pressure. EMBO Mol Med. 2010;2:63–75. doi: 10.1002/emmm.200900058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raman S, Vernon R, Thompson J, Tyka M, Sadreyev R, Pei J, Kim D, Kellogg E, DiMaio F, Lange O, Kinch L, Sheffler W, Kim BH, Das R, Grishin NV, Baker D. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins. 2009;77:89–99. doi: 10.1002/prot.22540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raveh B, London N, Schueler-Furman O. Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins. 2010;78:2029–2040. doi: 10.1002/prot.22716. [DOI] [PubMed] [Google Scholar]
- 28.Raveh B, London N, Zimmerman L, Schueler-Furman O. Rosetta FlexPepDock ab-initio: simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One. 2011;6:e18934. doi: 10.1371/journal.pone.0018934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma A, Wenzel W. Protein structure prediction by all-atom free-energy refinement. BMC Struct. Biol. 2007;7:12. doi: 10.1186/1472-6807-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villa F, Goebel J, Rafiqi FH, Deak M, Thastrup J, Alessi DR, van Aalten DMF. Structural insights into the recognition of substrates and activators by the OSR1 kinase. EMBO Rep. 2007;8:839–845. doi: 10.1038/sj.embor.7401048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.