Abstract
This article describes the construction and characterization of a mariner-based transposon vector designed for use in oral streptococci, but with a potential use in other Gram-positive bacteria. The new transposon vector, termed pMN100, contains the temperature-sensitive origin of replication repATs-pWV01, a selectable kanamycin resistance gene, a Himar1 transposase gene regulated by a xylose-inducible promoter, and an erythromycin resistance gene flanked by himar inverted repeats. The pMN100 plasmid was transformed into Streptococcus mutans UA159 and transposon mutagenesis was performed via a protocol established to perform high numbers of separate transpositions despite a low frequency of transposition. The distribution of transposon inserts in 30 randomly picked mutants suggested that mariner transposon mutagenesis is unbiased in S. mutans. A generated transposon mutant library containing 5000 mutants was used in a screen to identify genes involved in the production of sucrose-dependent extracellular matrix components. Mutants with transposon inserts in genes encoding glycosyltransferases and the competence-related secretory locus were predominantly found in this screen.
Keywords: Biofilm, extracellular matrix, glycosyltransferases, mariner, mutagenesis, Streptococcus mutans, transposon.
Introduction
Oral streptococci are Gram-positive, facultative anaerobes that prevail on tooth surfaces in complex multi-species biofilms called dental plaque. Occasionally, due to environmental factors, the biofilm becomes cariogenic. There is a strong correlation between a prolonged acidic environment at the enamel and an increased population of Streptococcus mutans, which in the end leads to an accelerated progress of caries (Takahashi and Nyvad 2008). S. mutans has developed a variety of mechanisms to colonize the tooth surfaces and to outnumber other bacteria in a cariogenic biofilm (Lemos et al. 2005; Ahn et al. 2006). Oral streptococci may also enter the blood stream, not only through dental procedures but also during daily activities like eating and tooth brushing. In the blood circulation, they can potentially bind to for example laminin, fibrin, collagen, and platelets, and subsequently adhere to damaged heart valves and cause endocarditis (Beg et al. 2002; Sato et al. 2004).
Mutant library screens have provided significant data with regard to identification of genes responsible for specific phenotypes in bacteria. A number of different transposon-based gene delivery systems have been used for creating mutant libraries in bacteria. In the case of oral streptococci, transposon mutagenesis has been carried out with Tn916, Tn4001, Tn917 and ISS1 (Spatafora et al. 1995; Gutierrez et al. 1996; Boyd et al. 2000a,b). Tn916 harbors a preferred insertion site consisting of an A-rich sequence separated by six bases from a T-rich sequence. Tn10 needs a 6-bp unique target sequence, and Tn5 is dependent on host factors and not adapted to some bacteria (Choi and Jung, 2009). The Tn917 transposon has been shown not to be inserted randomly in the chromosome of various bacteria (Slater et al. 2003; Shi et al. 2009). Moreover, transposon mutagenesis with ISS1 may be problematic as some Streptococcus species contains endogenous ISS1 elements that can recombine with the inserted ISS1, resulting in chromosomal deletions (Thibessard et al. 2002). We sought to develop a system for unbiased transposon mutagenesis in oral streptococci. To this end we employed the mariner transposon, which has been shown to insert at random positions in the bacterial chromosomes that have been analyzed so far, and which does not have endogenous counterparts in bacteria (Lampe 2010). In addition, the recognition signal for insertion of the mariner transposon is only two basepairs (TA dinucleotides) (Lampe et al. 1999). Among streptococci, the mariner system was first used in Streptococcus pneumonia using an in vitro mutagenesis protocol, and later developed as a signature-tagged mutagenesis system in Streptococcus sanguinis (Akerley et al. 1998; Paik et al. 2005).
The plasmid pBTn contains a xylose-inducible Himar1-transposase gene that mediates transposition of an erythromycin resistance gene flanked with himar inverted repeats (Li et al. 2009). pBTn contains a temperature-sensitive origin of replication that is functional in Staphylococcus aureus and Bacillus subtilis, but not in streptococci. The plasmid pTV1-OK contains genes required for transposon Tn917 mutagenesis (Gutierrez et al. 1996), as well as the temperature-sensitive origin of replication repATs-pWV01 which is functional in streptococci. In this study, we have fused parts of the pBTn and pTV1-OK plasmids to obtain a new plasmid, pMN100, which contains the temperature-sensitive origin of replication repATs-pWV01, the selectable kanamycin resistance gene aphA3, the Himar1 transposase gene regulated by a xylose-inducible promoter, and an erythromycin resistance gene flanked by himar inverted repeats. We transformed S. mutans UA159 with pMN100 and have provided evidence that transpositions occur with an unbiased distribution in the genome. In addition, as a proof of concept, we created a mariner transposon mutant library containing 5000 mutants, and performed a screen to identify genes involved in the production of sucrose-dependent extracellular matrix components.
Materials and Methods
Bacterial strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 1. For routine cultivation S. mutans UA159 (Ajdic et al. 2002), belonging to Bratthall serotype c, was grown in Tryptone soya broth (TSB) at 37°C aerobically. Tryptone soya agar (TSA) with and without sucrose were used for plating and anaerobic incubation (10% H2, 10% CO2 and 80% N2). Escherichia coli strains were grown in Luria-Bertani medium at 37°C. Where appropriate, antibiotics were used for bacterial cultures at the following concentrations (if nothing else is stated): for S. mutans strains, erythromycin (Sigma-Aldrich, St. Louis, MO) at 5 μg/mL, kanamycin (AppliChem GmbH, Darmstadt, Germany) at 300 μg/mL; for E. coli strains, kanamycin at 50 μg/mL.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics or sequence | Source orreference |
|---|---|---|
| Streptococcus mutans | ||
| UA159 | American type culture collection (ATCC 700610) | Ajdic et al. (2002) |
| E. coli | ||
| HB101 | recA thi pro leu hsdRM, Smr; strain used for maintenance and proliferation of plasmids | Kessler et al. (1992) |
| DH5α | F−, ϕ80dlacZΔM15, Δ(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rk−, mk+), phoA, supE44, λ−, thi-1, gyrA96, relA1 | Invitrogen |
| Plasmids | ||
| pBTn | 1.45 kbp ermB fragment of Tn551, xylose-inducible promoter and xylR, Himar1 transpose gene | Li et al. (2009) |
| pTV1-OK | repATs-pWV01Ts aphA3 Tn917(erm) | Gutierrez et al. (1996) |
| pMN100 | repATs-pWV01Ts aphA3 1.45 kbp ermB fragment of Tn551, xylose-inducible promoter and xylR, Himar1 transpose gene | This study |
Construction of the pMN100 transposon vector
The pMN100 transposon vector (see Fig. 1) was constructed as follows. Purified pTV1-OK (Oiaprep® Spin Miniprep kit; Qiagen GmbH, Hilden, Germany) was digested with EcoRI and PstI. The 3.5 kb backbone fragment, containing the temperature-sensitive repATs replicon and the aphA3 gene encoding kanamycin resistance, was agarose gel extracted, blunt ended with T4 DNA polymerase and dephosphorylated with shrimp alkaline phosphatase. pBTn was digested with KpnI and PstI, and the 4.4 kb fragment, containing the Himar1 transposase gene, the xylose-inducible promoter, the xylR regulator gene and the erythromycin resistance gene flanked by himar inverted repeats, was agarose purified and blunt ended as described above. The fragments were ligated and transformed into E. coli HB101 (Kessler et al. 1992). The new transposon vector, named pMN100, was confirmed by restriction analyses and partial sequencing.
Figure 1.
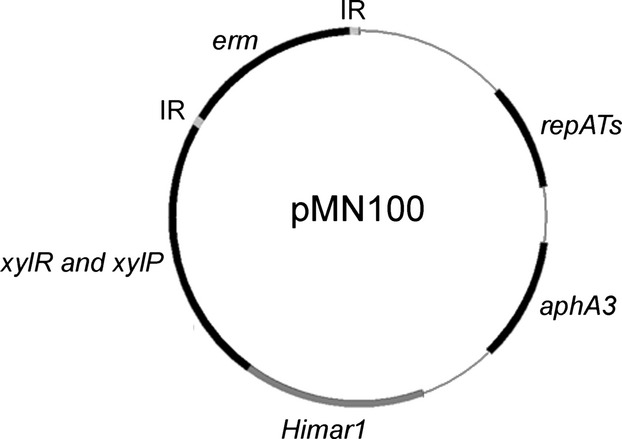
Schematic of the Himar1-based transposon vector pMN100. Gene designations: repATs-pWVO1, temperature-sensitive origin of replication; aphA3, kanamycin resistance gene; erm, erythromycin resistance gene; xylR and xylP, xylose regulator gene and promoter; Himar1, transposase gene; IR, mariner inverted repeats.
Transformation of S. mutans
The transformation method was based on previously published procedures (Cvitkovitch et al. 1998). A volume of 250 μL from an S. mutans overnight culture grown in Todd-Hewitt medium (THB) was added to 12 mL THB containing 25% heat-inactivated horse serum (PAA Laboratories GmbH, Pasching, Austria). The cultures were incubated at 37°C anaerobically, and when they reached an OD600 between 0.15 and 0.25 they were left at room temperature for 10 min. The pMN100 plasmid was added (∼1 μg) and the tubes were incubated at 30°C for 1 h. Cultures were diluted with 5 mL THB supplemented with horse serum and incubated for an additional hour. Cells were harvested by centrifugation and, resuspended in THB and spread on TSA plates containing erythromycin and 2 mg/mL kanamycin. The plates were incubated at 30°C for 3 days.
Transposon mutagenesis in S. mutans, and mutant library preparation
An overnight culture of S. mutans/pMN100, grown at 30°C in TSB supplemented with kanamycin and erythromycin, was diluted 1:100 into tubes containing 1 mL TSB (without dextrose) supplemented with 0.5% xylose and kanamycin and erythromycin. The cultures (usually 300) were incubated for 24 h at 30°C. Ten μL from each culture was subsequently transferred to tubes containing 1 mL TSB supplemented with erythromycin and incubated at the restrictive temperature of 40°C for 24 h. Then, 100 μL of each culture was plated on TSA plates containing 10 μg/mL erythromycin, and the plates were incubated at 37°C for 48–72 h. Five colonies per transposition culture were picked separately to a kanamycin and an erythromycin agar plate, and the plates were incubated for 48 h. Generally, three clones per transposition culture, with an Ermr and Kms phenotype, were picked to microtiter plates containing 100 μL of TSB and grown for 24 h at 37°C. Glycerol was added to an end concentration of 15% and the plates were preserved at −80°C for future screening. All cultivations, except for the start overnight culture, were done in 2 mL eppendorf tubes. To create a library of 5000 clones, ∼1700 separate transposition cultures were grown.
Identification of interrupted genes by arbitrary primed PCR
In order to identify the location of the mariner transposon insertions, the arbitrary primed PCR protocol outlined by Li et al. (2009) was followed. Briefly, in the first round of PCR one of the arbitrary primers was paired with a primer that reads out from the erythromycin resistance gene. Approximately 100 ng of purified chromosomal DNA, obtained using Qiagen's DNeasy® Blood & Tissue kit, was used as template. In the second PCR a part of the product from the first PCR was PCR amplified with an arb-3 primer and a nested erm primer. Purified second-round products were used for DNA sequence analyses performed by Macrogen by the use of the nested erm primer. Insertion sites were identified with BLASTN searches, against the annotated sequence of S. mutans UA159, using software provided by NCBI.
Southern blot analysis
Southern blot analysis was performed with a DIG labeling kit and NBT/BCIP tablets were used for detection according to the manufacturer's (Roche Diagnostics GmbH, Mannheim, Germany) protocols. The amplified 1.3 kb product using primer IRXbaI and IRPstI and comprising the erythromycin gene was used as a probe (Li et al. 2009). Chromosomal DNA obtained from mutants was digested with EcoRI.
Colony morphology assay
Primary screening was made by plating spots directly from transposon library microtiter plate glycerol stocks onto TSA plates (14 cm diameter petri dishes) containing 1% sucrose, using a replicator with 3-mm pins. The plates were incubated at 37°C for 2 days. The colony morphology of collected semi-rough colony morphology mutants from the primary screening was confirmed in a second assay where 5 μL from newly made glycerol stocks were spot-inoculated on TSA with and without 1% sucrose. The plates were incubated at 37°C for 3 days. Pictures of colonies from the second plate assay were taken with a Canon EOS 400D camera (Canon Inc.,Tokyo, Japan).
Results
Construction of a mariner transposon vector suitable for use in oral streptococci
A mariner transposon vector was constructed with the aim to perform unbiased transposon mutagenesis in oral streptococci. The components of the new transposon vector were taken from the plasmids pTV1-OK and pBTn (Gutierrez et al. 1996; Li et al. 2009). The backbone of pTV1-OK, containing the temperature-sensitive origin of replication repATs-pWV01 and the selectable kanamycin resistance gene aphA3, was fused with the pBTn part comprising an erythromycin gene flanked with himar inverted repeats and a Himar1 transposase gene regulated by a xylose-inducible promoter. The new transposon vector, named pMN100, is schematically shown in Figure 1. The pMN100 plasmid can replicate in streptococci grown at 30°C. Expression of the Himar1 transposase can be induced with xylose, and it promotes transposition of the erythromycin resistance gene flanked with himar inverted repeats. The pMN100 plasmid cannot replicate at 40°C and the bacteria will, therefore, be cured from the plasmid if they are grown at 40°C. Bacteria in which the erythromycin resistance gene is inserted into the chromosome will display erythromycin resistance at 40°C.
Establishment of a mariner transposition protocol for S. mutans
The mariner transposon vector pMN100 was introduced into S. mutans UA159. Restriction analysis of plasmids purified from S. mutans/pMN100 transformants grown at permissive temperature indicated that the plasmid is structurally stable in S. mutans. The ability of the S. mutans/pMN100 strain to mediate transposition was investigated, using basically the transposition protocol described by Li et al. (2009) for pBTn-mediated transposition in S. aureus. The procedure resulted in integration of the erythromycin gene at TA dinucleotides in the chromosome of S. mutans UA159 (data not shown), confirming that the pMN100 plasmid functions as a transposon vector as anticipated. However, the procedure was not optimal since the number of unique clones per transposition culture was low (data not shown).
In order to be able to create comprehensive transposon libraries, we set up a novel transposition protocol. In the protocol described by Li et al. (2009) cultures were grown at permissive temperature, and were then cultivated for 24 h at restrictive temperature three times before plating. With S. mutans harboring pMN100, it was possible to obtain a high frequency (above 90%) of cured (i.e., plasmid-free) bacteria after only one overnight cultivation at restrictive temperature. This improvement, besides preparing transposition cultures in small volumes, that is, eppendorf tube scale, made it possible to perform a high number of transpositions in a short time. S. mutans/pMN100 was grown to stationary phase in TSB supplemented with erythromycin and kanamycin at permissive temperature, and the culture was then diluted into several tubes containing TSB (without dextrose) supplemented with xylose, erythromycin and kanamycin and these cultures were grown at permissive temperature overnight. The resulting stationary phase cultures were diluted into TSB containing erythromycin, and were then incubated overnight at restrictive temperature. Thereafter a part of these cultures were spread on TSA plates with erythromycin, and the plates were incubated at restrictive temperature. Colonies on the plates appeared after 2–3 days of incubation. Thereafter the putative transposon mutants were picked and analyzed separately on erythromycin and kanamycin plates. The frequency of temperature resistant Ermr and Kms clones, after a transposition procedure, was generally above 90%. When we applied our new transposition protocol, the sizes of the mutant colonies were more heterogeneous compared with the outcome when we followed the protocol described by Li et al. (2009). This was expected as our protocol involves only one cultivation at the restrictive temperature whereas the protocol of Li et al. (2009) contains three consecutive cultivations at the restrictive temperature. Throughout mutant library preparation we chose not to pick the smallest colonies in order to avoid mutants with slow growth. For library preparation, three Ermr and Kms clones were chosen per transposition culture, and thereby 60% of the mutants had unique inserts (i.e., were non siblings), as judged by DNA sequencing of 30 randomly picked mutants. Southern blot analysis of 10 mutants showed that only one transposon integration per mutant had occurred (data not shown), which is in agreement with the observed low frequency of transposon integration.
Evidence for unbiased mariner transposon mutagenesis in S. mutans
Mariner tranposons have been shown to insert at random positions in the bacterial chromosomes that have been analyzed so far. To investigate if mariner transposon insertion occurs at random sites also in the S. mutans UA159 chromosome, the insertion sites in 30 randomly picked transposon mutants were determined by arbitrary primed PCR followed by sequencing of the PCR product. As shown in Figure 2, the transposon inserts are well distributed over the entire S. mutans UA159 genome, indicating that mariner transposon mutagenesis is unbiased in S. mutants. There are a few larger gaps, for example, between 1.14 and 1.46-Mbp, but this is likely attributed to the presence of clusters of essential genes.
Figure 2.
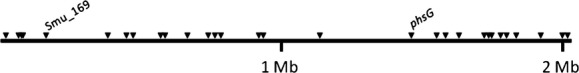
Mapping of unique mariner transposon insertion sites in 30 randomly picked S. mutansUA159 transposon mutants. The 2.03 Mbp genome of S. mutansUA159 is shown linearized. Arrow heads indicate approximate transposon insertion sites. Two gene names are included to indicate genome orientation.
Isolation of mutants defective in the production of sucrose-induced extracellular matrix components
We created a S. mutans mariner transposon mutant library containing 5000 mutants, and performed a screen to identify genes involved in the production of sucrose-dependent extracellular matrix components in S. mutans. The formation by bacteria of wrinkled or rough colonies on solid medium is indicative of the production of extracellular matrix components such as exopolysaccharides, large adhesive proteins and aggregative fimbriae (Rainey and Travisano 1998; Friedman and Kolter 2004; Simm et al. 2004; Fazli et al. 2013). The fact that S. mutans forms rough colonies on solid medium containing sucrose, whereas it forms smooth colonies on solid medium without sucrose (Fig. 3) indicates that it produces sucrose-dependent extracellular matrix components. This provided a basis for screening of the mutant library for mutants that are defective in the production of sucrose-dependent extracellular matrix components. However, we were not able to isolate any mutant that gave rise to a smooth colony morphology on TSA sucrose plates similar to the colonies of the S. mutans wild type growing on TSA agar without sucrose. Instead, we isolated mutants with semi-rough colony morphology. The isolated mutants had a flatter colony structure with a more transparent appearance compared to the wild type colony morphology (Fig. 3). Sequence analyses of the regions flanking the transposon inserts in the mutants that displayed semi-rough colony morphologies on TSA sucrose plates revealed 14 unique transposon mutants. Six of the transposon inserts were located in gtf genes encoding glycosyltransferases (Table 2), which are capable of synthesizing polymers of glucans from sucrose, and of importance for sucrose-dependent biofilm formation. Four of the isolated transposon inserts were located in the comAB operon (Table 2), also referred to as competence-related secretory locus (cslAB) (Petersen and Scheie 2000). The other transposon insertions were located in an alcohol-acetalaldehyde dehydrogenase gene, a putative oligopeptide transporter and in two different ORFs encoding putative proteins with unknown functions (Table 2).
Figure 3.
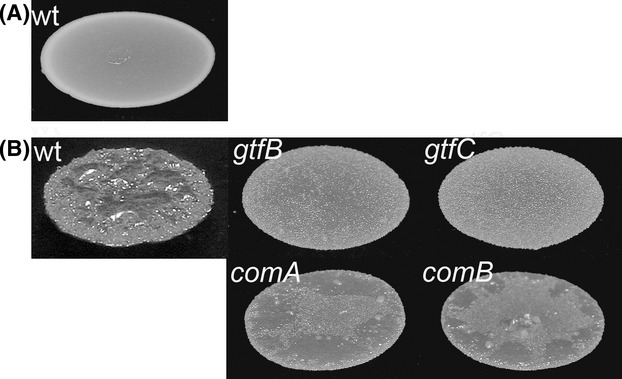
Colony morphology on TSA agar medium without sucrose (A) and with sucrose (B) of S. mutansUA159 wild type and the selected transposon mutants gtfB, gtfC, comA, and comB.
Table 2.
Transposon insertion sites in S. mutans UA159 mutants isolated in a screen for smooth or semi-rough colony morphology on TSA sucrose agar plates
| Mutant designation | Transposon insertion site | Affected gene | Putative function of gene product |
|---|---|---|---|
| 1B6 | 958790 | gtfC | Glycosyltransferase |
| 12H5 | 150205 | adhE | Alcohol-acetalaldehyde dehydrogenase |
| 14A8 | 956319 | gtfC | Glycosyltransferase |
| 30C6 | 953840 | gtfB | Glycosyltransferase |
| 30F6 | 578023 | Smu_616 | Hypothetical protein |
| 36C7 | 959225 | gtfC | Glycosyltransferase |
| 41D1 | 272044 | comA | CSP transport |
| 42B6 | 956088 | gtfC | Glycosyltransferase |
| 42G11 | 892647 | Smu_941c | Hypothetical protein |
| 43G8 | 244904 | oppA | Putative oligopeptide ABC transporter |
| 46C7 | 273621 | comB | CSP transport |
| 47H9 | 271530 | comA | CSP transport |
| 48B3 | 952972 | gtfB | Glycosyltransferase |
| 50B1 | 271269 | comA | CSP transport |
Discussion
We considered several mutagenesis strategies and transposition vectors, before we chose to employ the temperature-sensitive repATs origin of replication and the mariner transposon with inducible transposase. Plasmid pTV1-OK (Gutierrez et al. 1996), which served as the source of the repATs replicon, is a temperature-sensitive derivative of pWVO1, a broad-host-range plasmid originally isolated from Lactococcus lactis but with the ability to replicate in various streptococcal species. Thus, the feature that the repATs can initiate replication in streptococci at a permissive temperature of 30°C, but not at temperatures above 37°C is well established. Plasmid pTV1-OK is a delivery vector for Tn917 mutagenesis, however, the Tn917 transposon has been reported to have insertion hot spots in the chromosome of various bacteria, making library construction inefficient. For example, in Streptococcus equi, 60% of transposon inserts had occurred within a 15 kb region of the genome (Slater et al. 2003). An extreme regional preference for Tn917 mutagenesis has also been shown in Enterococcus faecalis and Bacillus subtilis (Shi et al. 2009). On the contrary, the mariner transposon system has been used in many bacterial species without any apparent insertion bias in the chromosome (Rubin et al. 1999; Bae et al. 2004; Wong and Akerley 2008; Beare et al. 2009).
Other systems than the mariner-based one have previously been used to create mutants in streptococci. A non-transposon-mediated mutagenesis strategy employed Sau3AI digested S. mutans GS-5 chromosomal DNA that was ligated to a linearized vector and used to transform S. mutans GS-5 (Yoshida and Kuramitsu 2002). This suicide plasmid-mediated insertion-mutagenesis procedure resulted in ∼12.000 S. mutans mutants, which were screened for genes that mediated sucrose independent biofilm formation. However, this procedure requires a high frequency of transformation, which is not displayed by all streptococcal strains. Cvitkovitch et al. (1998) generated a transposon mutant library using pTV1-OK, but this system is based on Tn917 and subject to extreme insertion bias as described above. Recently, ISS1 transposition mutagenesis, with a temperature-sensitive vector, has been used successfully in S. mutans (Thibessard et al. 2002). ISS1 appears to integrate randomly in the genome of streptococci (Thibessard et al. 2002). However, some Streptococcus species contain endogenous ISS1 elements which may mediate homologous recombination events between the ISS1 originating from the transposon vector and the endogenous ISS1 sequences (Thibessard et al. 2002).
We used our new mariner transposon vector to create an S. mutans mutant library, and performed a screen for mutants deficient in producing sucrose-dependent extracellular matrix components. S. mutans forms rough colonies on TSA sucrose agar plates but smooth colonies on agar plates without sucrose, and mutants that display less rough colony morphology on TSA sucrose agar plates may be impaired in the synthesis of sucrose-dependent extracellular matrix components. Six of the S. mutans tranposon mutants that displayed semi-rough colony morphology on TSA sucrose plates had transposon inserts in gtf genes encoding glycosyltransferases. Sucrose is known to be a strong inducer of S. mutans biofilm formation, dependent on the three glycosyltransferases GtfB, GtfC and GtfD that from sucrose synthesize different variants of glucans (Bowen and Koo 2011). Collectively, the Gtf's play a major role in the development and establishment of the extracellular matrix in sucrose-induced S. mutans biofilm. The gtfB and gtfC single mutants that were isolated in this study displayed semi-rough colony morphology on TSA sucrose plates, which seem to be an intermediate phenotype between the colony morphology displayed by the wild type on TSA sucrose plates and on plates without sucrose. It is possible that knockout of more than one gtf gene in S. mutans is necessary to obtain a smooth colony morphology on TSA sucrose plates (Hanada and Kuramitsu 1988). However, we found that several other genes, including the comAB genes (as addressed below), are also required for the rough S. mutans colony morphology on TSA sucrose plates. We did not isolate gtfD transposon mutants in this study, which is in agreement with earlier studies showing that gtfD mutants display growth defects (Koo et al. 2010), as during mutant library preparation we consistently avoided picking mutants which formed smaller colonies than the wild type. Four of the S. mutans transposon mutants that displayed a semi-rough colony morphology on TSA sucrose plates had transposon inserts in the competence-stimulating peptide (CSP) exporter genes comAB. ComA is an ABC transporter and ComB an accessory protein, which together constitute the secretory apparatus that is involved in processing and export of CSP. A role of CSP signaling in the production of sucrose-induced matrix components is interesting and warrants further investigations.
In our screen for S. mutans mutants that form semi-rough colonies on sucrose plates, we also found transposon insertions in an alcohol-acetaldehyde dehydrogenase gene, a putative oligopeptide transporter gene, and two different ORFs encoding putative proteins with unknown functions. Mutants with transposon insertion in these genes were only isolated once, unlike the case with the gft and com genes described above. For the mutants that were only isolated once, there is a risk that a secondary mutation in the genome causes the semi-rough colony morphology instead of the transposon insertion. For instance, a relatively rare event involving endogenous recombination of gtfB and gtfC in S. mutans can result in smooth colony morphology on sucrose plates (Narisawa et al. 2011). Therefore, the genes inactivated by transposon insertion in the four mutants that were only isolated once should only be regarded as candidates for being involved in the production of sucrose-dependent extracellular matrix components. Further analysis is required to confirm this.
After an era with an enormous development regarding gene discovery and publication of genome sequences, there is still a need of various systems to study gene function (van Opijnen and Camilli 2013). We present a new mariner transposon vector which should be a valuable tool for producing mutant libraries in streptococcal species, as it enables unbiased transposon mutagenesis. Because the transposon vector can replicate in streptococci and subsequently, after inducible transposition, can be cured by a temperature shift, it is useful for streptococcal strains that display a low plasmid transformation frequency.
Acknowledgments
We thank Paula J. Crowley for plasmid pTV1-OK, Michael Otto for plasmid pBTn, and Joseph J. Ferretti for providing us with S. mutans UA159. This study was supported by a grant from Tandlægefonden.
Conflict of Interest
None declared.
References
- Ahn SJ, Wen ZT, Burne RA. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect. Immun. 2006;74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Rubin EJ, Camilli A, Lampe DJ, Robertson HM, Mekalanos JJ. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, et al. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc. Natl. Acad. Sci. USA. 2004;101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J. Bacteriol. 2009;191:1369–1381. doi: 10.1128/JB.01580-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beg AM, Jones MN, Miller-Torbert T, Holt RG. Binding of Streptococcus mutans to extracellular matrix molecules and fibrinogen. Biochem. Biophys. Res. Commun. 2002;298:75–79. doi: 10.1016/s0006-291x(02)02390-2. [DOI] [PubMed] [Google Scholar]
- Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45:69–86. doi: 10.1159/000324598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd DA, Cvitkovitch DG, Bleiweis AS, Kiriukhin MY, Debabov DV, Neuhaus FC, et al. Defects in D-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 2000a;182:6055–6065. doi: 10.1128/jb.182.21.6055-6065.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd DA, Thevenot T, Gumbmann M, Honeyman AL, Hamilton IR. Identification of the operon for the sorbitol (Glucitol) Phosphoenolpyruvate:sugar phosphotransferase system in Streptococcus mutans. Infect. Immun. 2000b;68:925–930. doi: 10.1128/iai.68.2.925-930.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Jung K. Applications of transposon-based gene delivery system in bacteria. J. Microbiol. Biotechnol. 2009;19:217–228. doi: 10.4014/jmb.0811.669. [DOI] [PubMed] [Google Scholar]
- Cvitkovitch D, Gutierrez J, Crowley P, Wojciechowski L, Hillman J, Bleiweis A. Tn917 transposon mutagenesis and marker rescue of interrupted genes of Streptococcus mutans. Methods Cell Sci. 1998;20:1–12. [Google Scholar]
- Fazli M, McCarthy Y, Givskov M, Ryan RP, Tolker-Nielsen T. The exopolysaccharide gene cluster Bcam1330-Bcam1341 is involved in Burkholderia cenocepacia biofilm formation, and its expression is regulated by c-di-GMP and Bcam1349. Microbiologyopen. 2013;2:105–122. doi: 10.1002/mbo3.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 2004;51:675–690. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- Gutierrez JA, Crowley PJ, Brown DP, Hillman JD, Youngman P, Bleiweis AS. Insertional mutagenesis and recovery of interrupted genes of Streptococcus mutans by using transposon Tn917: preliminary characterization of mutants displaying acid sensitivity and nutritional requirements. J. Bacteriol. 1996;178:4166–4175. doi: 10.1128/jb.178.14.4166-4175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada N, Kuramitsu HK. Isolation and characterization of the Streptococcus mutans gtfC gene, coding for synthesis of both soluble and insoluble glucans. Infect. Immun. 1988;56:1999–2005. doi: 10.1128/iai.56.8.1999-2005.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler B, de Lorenzo V, Timmis KN. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 1992;233:293–301. doi: 10.1007/BF00587591. [DOI] [PubMed] [Google Scholar]
- Koo H, Xiao J, Klein MI, Jeon JG. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J. Bacteriol. 2010;192:3024–3032. doi: 10.1128/JB.01649-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampe DJ. Bacterial genetic methods to explore the biology of mariner transposons. Genetica. 2010;138:499–508. doi: 10.1007/s10709-009-9401-z. [DOI] [PubMed] [Google Scholar]
- Lampe DJ, Akerley BJ, Rubin EJ, Mekalanos JJ, Robertson HM. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Abranches J, Burne RA. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 2005;7:95–107. [PubMed] [Google Scholar]
- Li M, Rigby K, Lai Y, Nair V, Peschel A, Schittek B, et al. Staphylococcus aureus mutant screen reveals interaction of the human antimicrobial peptide dermcidin with membrane phospholipids. Antimicrob. Agents Chemother. 2009;53:4200–4210. doi: 10.1128/AAC.00428-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa N, Kawarai T, Suzuki N, Sato Y, Ochiai K, Ohnishi M, et al. Competence-dependent endogenous DNA rearrangement and uptake of extracellular DNA give a natural variant of Streptococcus mutans without biofilm formation. J. Bacteriol. 2011;193:5147–5154. doi: 10.1128/JB.05240-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. 2013;11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik S, Senty L, Das S, Noe JC, Munro CL, Kitten T. Identification of virulence determinants for endocarditis in Streptococcus sanguinis by signature-tagged mutagenesis. Infect. Immun. 2005;73:6064–6074. doi: 10.1128/IAI.73.9.6064-6074.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen FC, Scheie AA. Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral Microbiol. Immunol. 2000;15:329–334. doi: 10.1034/j.1399-302x.2000.150511.x. [DOI] [PubMed] [Google Scholar]
- Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- Rubin EJ, Akerley BJ, Novik VN, Lampe DJ, Husson RN, Mekalanos JJ. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Okamoto K, Kagami A, Yamamoto Y, Igarashi T, Kizaki H. Streptococcus mutans strains harboring collagen-binding adhesin. J. Dent. Res. 2004;83:534–539. doi: 10.1177/154405910408300705. [DOI] [PubMed] [Google Scholar]
- Shi Q, Huguet-Tapia JC, Peters JE. Tn917 targets the region where DNA replication terminates in Bacillus subtilis, highlighting a difference in chromosome processing in the firmicutes. J. Bacteriol. 2009;191:7623–7627. doi: 10.1128/JB.01023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simm R, Morr M, Kader A, Nimtz M, Romling U. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 2004;53:1123–1134. doi: 10.1111/j.1365-2958.2004.04206.x. [DOI] [PubMed] [Google Scholar]
- Slater JD, Allen AG, May JP, Bolitho S, Lindsay H, Maskell DJ. Mutagenesis of Streptococcus equi and Streptococcus suis by transposon Tn917. Vet. Microbiol. 2003;93:197–206. doi: 10.1016/s0378-1135(03)00030-0. [DOI] [PubMed] [Google Scholar]
- Spatafora G, Rohrer K, Barnard D, Michalek S. A Streptococcus mutans mutant that synthesizes elevated levels of intracellular polysaccharide is hypercariogenic in vivo. Infect. Immun. 1995;63:2556–2563. doi: 10.1128/iai.63.7.2556-2563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 2008;42:409–418. doi: 10.1159/000159604. [DOI] [PubMed] [Google Scholar]
- Thibessard A, Fernandez A, Gintz B, Decaris B, Leblond-Bourget N. Transposition of pGh9:ISS1 is random and efficient in Streptococcus thermophilus CNRZ368. Can. J. Microbiol. 2002;48:473–478. doi: 10.1139/w02-038. [DOI] [PubMed] [Google Scholar]
- Wong SM, Akerley BJ. Identification and analysis of essential genes in Haemophilus influenzae. Methods Mol. Biol. 2008;416:27–44. doi: 10.1007/978-1-59745-321-9_3. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Kuramitsu HK. Multiple Streptococcus mutans genes are involved in biofilm formation. Appl. Environ. Microbiol. 2002;68:6283–6291. doi: 10.1128/AEM.68.12.6283-6291.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


