Muliciliate cells (MCCs) play critical roles in producing luminal fluid flow in a number of organ systems. To differentiate, MCCs undergo a unique program of centriole assembly initiated by the small coiled-coil protein multicilin. Kintner and colleagues show that multicilin forms a ternary complex with e2f4, e2f5, and their dimerization partner, dp1, which activates centriole biogenesis genes. Multicilin thus coopts the e2f regulation of cell cycle genes to drive centriole assembly. Furthermore, this complex is disabled by mutations in human multicilin that cause a severe congenital mucociliary clearance disorder.
Keywords: centrioles, e2f4, multiciliate cells
Abstract
Multiciliate cells employ hundreds of motile cilia to produce fluid flow, which they nucleate and extend by first assembling hundreds of centrioles. In most cells, entry into the cell cycle allows centrioles to undergo a single round of duplication, but in differentiating multiciliate cells, massive centriole assembly occurs in G0 by a process initiated by a small coiled-coil protein, Multicilin. Here we show that Multicilin acts by forming a ternary complex with E2f4 or E2f5 and Dp1 that binds and activates most of the genes required for centriole biogenesis, while other cell cycle genes remain off. This complex also promotes the deuterosome pathway of centriole biogenesis by activating the expression of deup1 but not its paralog, cep63. Finally, we show that this complex is disabled by mutations in human Multicilin that cause a severe congenital mucociliary clearance disorder due to reduced generation of multiple cilia. By coopting the E2f regulation of cell cycle genes, Multicilin drives massive centriole assembly in epithelial progenitors in a manner required for multiciliate cell differentiation.
The centriole, a cylindrical organelle made up of triplet microtubules, both organizes the mitotic spindle during cell division and, in G0, converts into a basal body, docks at the plasma membrane, and organizes the outgrowth of the ciliary axoneme. Since supernumerary centrioles disrupt the bipolar mitotic spindle and cause abnormal chromosomal segregation, the number of centrioles is invariant and only increases during the centriolar cycle, a highly regulated process initiated at the G1–S transition that leads to centriole duplication in preparation for chromosomal segregation. Mitosis places strict limits on centriole duplication and maturation, and these limitations generally limit cells to a single cilium during G0.
In contrast, the multiciliate cell (MCC) is a specialized epithelial cell type that extends anywhere from 150 to 200 motile cilia per cell in order to produce a vigorous fluid flow critical to human health in several organ systems (Fliegauf et al. 2007). To extend these cilia, differentiating MCCs undergo an unusual form of centriole assembly and maturation that yields hundreds of mother centrioles, thus generating the basal bodies required for multiple cilia extension (Sorokin 1968). Unlike dividing cells, MCCs generate these centrioles in G0 after progenitors leave the cell cycle, and much of this assembly occurs on structures called deuterosomes that recently have come into focus molecularly (Tang 2013). The mechanisms that initiate this massive centriole assembly during MCC differentiation are largely unknown.
Previous work identified Multicilin, a small coiled-coil protein encoded by the mcidas gene, as a critical regulator of MCC differentiation (Stubbs et al. 2012). Mcidas lies within a conserved chromosomal region that appears dedicated to MCC differentiation, flanked on one side by ccno, a gene recently shown to be mutated in a human syndrome called “congenital mucociliary clearance disorder with reduced generation of multiple motile cilia” (RGMC) (Wallmeier et al. 2014), and on the other side by cdc20B, a gene containing three microRNAs, miR449a–c, which are also required for MCC differentiation (Marcet et al. 2011). Among these genes, Multicilin appears to be a critical, upstream, transcriptional coregulator, since it is both necessary and sufficient to induce gene expression required for MCC differentiation within the epithelial progenitors (Stubbs et al. 2012). Human mutations in MCIDAS also cause RGMC and reduce the expression of target genes up-regulated by Multicilin, indicating that its function in MCC differentiation is conserved and relevant to human disease (Boon et al. 2014). However, because Multicilin lacks a DNA-binding domain, its mechanisms of action, presumably via a transcriptional complex, is unknown.
To identify this complex, we turned to the E2F family of transcription factors, which are the key activators and repressors of gene expression involved in cell cycle progression (Trimarchi and Lees 2002). One member of this family, E2f4, acts as a repressor of cell cycle genes and is mainly thought to promote cycle exit and quiescence in G0 (Sadasivam and DeCaprio 2013). In E2f4-null mice, however, the cell cycle was largely unaffected, reflecting extensive genetic overlap with E2f5, but then the mice surprisingly died of respiratory failure due to an early loss of MCCs in the airways (Danielian et al. 2007). Since E2f4 can regulate genes required for centriole assembly during cell division (Fischer et al. 2014), this observation raised the possibility that Multicilin could act via E2f4 during MCC differentiation.
Results
Multicilin binds to E2f4 or E2f5 in a ternary complex with Dp1
We used RNA injection to express tagged proteins in MCC progenitors in the developing larval skin (Stubbs et al. 2012) in order to test the ability of Multicilin to bind to E2f4 alone and in the presence of its obligatory dimerization binding partner, TfDp1 (referred to here as Dp1) (Trimarchi and Lees 2002). Pull-down of Flag-tagged Multicilin from extracts readily recovered both GFP-E2f4 and GFP-Dp1, indicating that the three form a complex (Fig. 1A, lane1). Flag-Multicilin did not efficiently recover GFP-E2f4 from extracts in the absence of GFP-Dp1, suggesting that the complex was ternary in nature (Fig. 1A, lane 2). The same complex was detected using different tag combinations (Fig. 1B, lane1; data not shown) and also formed with E2f5, a “repressor E2f” closely related to E2f4 (Fig. 1A, lane 3). However, Multicilin did not bind detectably to an “activator E2f,” E2f1, expressed alone or with Dp1 (Fig. 1A, lanes 5,6), or to E2f7 (data not shown), a member of the E2f family that acts as a constitutive repressor (Trimarchi and Lees 2002). We examined whether endogenous proteins also form a similar complex by preparing extracts from animal caps where MCC differentiation was either blocked or increased by changing Notch activity (Deblandre et al. 1999). Pull-down of endogenous Multicilin also recovers endogenous E2f4 from extracts with increased MCC differentiation but not from those where mcidas expression and MCC differentiation were blocked (Supplemental Fig. 1). Finally, as a negative control, we tested the related coiled-coil protein geminin (Gmnn), the only previously known binding partner for Multicilin (Pefani et al. 2011). Gmnn does not detectably bind to E2f4 and Dp1 alone (Fig. 1B, lane 2; Supplemental Fig. 2A, lane 2) but will form a quaternary complex with E2f4, Dp1, and Multicilin (Fig. 1B, lane 3), whose significance is explored below. In sum, these results indicate that a ternary complex, called EDM, can form between E2f4 or E2f5, Dp1, and Multicilin but not between Multicilin and other E2f family members either alone or in a heterodimer with Dp1.
Figure 1.
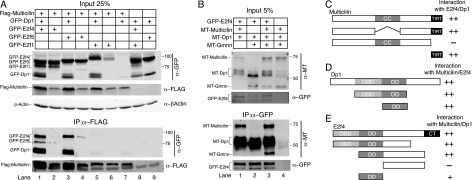
Multicilin interactions with E2f family members and Gmnn. (A,B) Extracts of stage 12 animal caps were prepared from Xenopus embryos that were injected with the indicated RNAs and subjected to Western analysis with the indicated antibody before (input) and after immunoprecipitation (IP). (C–E) The domain structures of Multicilin (C), Dp1 (D), and E2f4 (E) are diagrammed along with deletion mutants that were tested for their ability to form the EDM complex, summarizing data shown in Supplemental Figure 2.
We mapped the regions of Multicilin, E2f4, and Dp1 required to form the EDM complex using deletion mutants. For Multicilin, complex formation is mediated through a 45-amino-acid region at the C terminus, called the TIRT domain (Fig. 1C; Supplemental Fig. 2A), which has previously been shown to act as a strong dominant-negative mutant of MCC differentiation (Stubbs et al. 2012). Complex formation requires the dimerization domain of Dp1 (Fig. 1D; Supplemental Fig. 2B) and that of E2f4 (Fig. 1E; Supplemental Fig. 2C). The regions required and the results above are consistent with a model in which Multicilin binds via the TIRT domain to the region where E2f4 and Dp1 dimerize, leaving the coiled-coil domain of Multicilin to mediate additional interactions. Further structural work with isolated proteins will be required to determine the exact composition of this complex and how it forms.
E2f4ΔCT blocks basal body formation
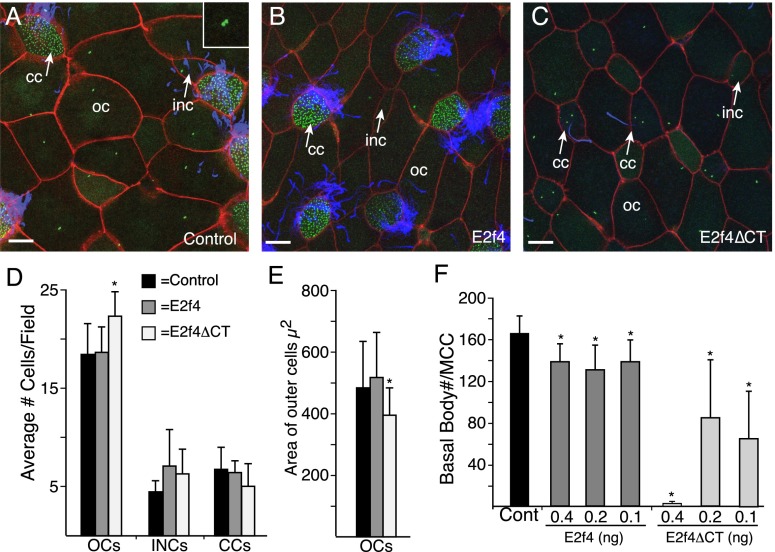
Because a complete loss of the repressor E2f proteins would likely cause severe cell cycle defects, we took a dominant-negative approach to test whether the EDM complex is critical for MCC differentiation. A form of E2f4 was generated with a myc epitope tag and nuclear localization signal (NLS) but lacking 140 amino acids from the C terminus (E2f4ΔCT) (Supplemental Fig. 2C). When E2f4ΔCT was expressed in early Xenopus embryos by RNA injection, the cell cycle appeared largely normal, based on cell size, but MCCs failed to form, and in their stead were cells with a MCC-like morphology that stained internally for acetylated tubulin and/or extended a single or sometimes two cilia (Fig. 2C–E). This phenotype was titratable in terms of basal body formation (Fig. 2F), indicating that E2f4ΔCT is not blocking a switch to undergo MCC differentiation but rather their ability to activate basal body assembly. Wild-type E2f4 misexpression produced very little in terms of a phenotype, and E2f4ΔCT misexpression appeared not to alter centriole number in other skin cell types (Supplemental Fig. 3A) or their differentiation, based on the analysis of ionocytes, a cell type that differentiates along with the MCCs in the skin (Supplemental Fig. 3B–F; Quigley et al. 2011). Finally, a similar truncated form of E2f5 gave the same phenotype as E2f4ΔCT, suggesting that the two inhibitory E2fs are likely to act redundantly in the EDM complex (data not shown). Thus, E2f4ΔCT specifically inhibits MCC differentiation by blocking a process of centriole assembly unique to these cells.
Figure 2.
MCC differentiation is blocked in the skin of E2f4ΔCT-expressing embryos. (A–C) Shown are confocal images of embryonic skin (stage 28) of control embryos (A) or embryos injected with RNA encoding E2f4 (B) or E2f4ΔCT (C). Embryos were also injected with RNA encoding mRFP (red) and Hyls1-GFP (green) to mark membranes and basal bodies/centrioles, respectively, and stained for cilia (blue). Cells extending one or two cilia are marked with an arrow in C. Different cell types are identified based on morphology and cilia staining as outer cells (OCs), MCCs, small intercalated cells (INCs), and ciliated cells (CCs). Bar, 10 μm. (D) Representation of different skin cell types in control or E2f4- or E2f4ΔCT-expressing embryos based on 12 fields (98 μm2) from six embryos. (E) Effects on the cell cycle can be indirectly read out by the size of outer cells (Stubbs et al. 2012). E2f4ΔCT significantly (P < 0.05) decreases average cell size based on 24 cells from six embryos, indicating that it weakly promotes cell division. (F) Basal body number in MCCs in control or E2f4- or E2f4ΔCT-expressing embryos. Basal body counts based on 10 to 15 MCCs taken from six embryos. In all graphs, error bars indicate SD, and values significantly different (P < 0.05) from controls based on a two-tailed t-test are marked (asterisks).
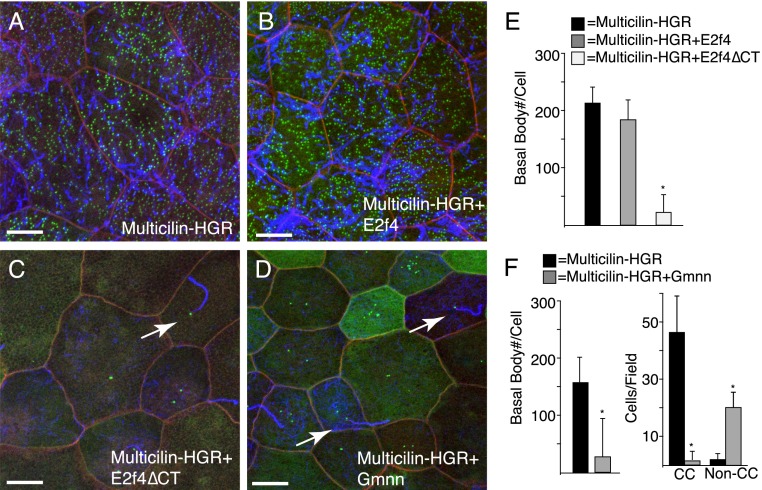
If E2f4ΔCT disables the EDM complex, one would predict that it should act in parallel with Multicilin. To test this prediction, E2f4ΔCT was expressed along with an inducible form of Multicilin, called Multicilin-HGR, which will induce most if not all of the cells in the skin to undergo MCC differentiation following activation with dexamethasone (DEX) (Fig. 3A,F; Stubbs et al. 2012). Coexpression of e2f4ΔCT but not e2f4 RNA strongly blocked the ability of Multicilin-HGR to induce centriole assembly (Fig. 3C,E). In a similar manner, we found that Gmnn, which binds to the EDM complex, also severely disrupts MCC differentiation when misexpressed: Fewer MCCs formed, and many extended just one or two cilia (Supplemental Fig. 4). When Gmnn was coexpressed with Multicilin-HGR, it also blocked the induction of centriole assembly, producing the same phenotype as E2f4ΔCT (Fig. 3D,F). In sum, disruption of the EDM complex with E2f4ΔCT or Gmnn disables the ability of Multicilin to drive centriole assembly. In contrast, the ability of Multicilin to induce motile cilia formation, presumably by inducing foxj1 expression, is less affected.
Figure 3.
E2f4ΔCT and Gmnn block Multicilin-induced basal body assembly during MCC differentiation. (A–D) Shown are confocal images of the embryonic skin expressing Multicilin-HGR alone (A) or with E2f4 (B), E2f4ΔCT (C), or Gmnn (D). Multicilin-HGR activity was induced with DEX at stage 12 and embryos fixed at stage 28. Membranes are marked with mRFP (red), basal bodies with Hyls1-GFP (green), and cilia in blue. Arrows denote monociliated cells. Bar, 10 μm. (E) Basal body number induced by ectopic expression of Multicilin-HGR activity in the presence of E2f4 and E2f4ΔCT. (F) Basal body number or the number of ciliated cells per field induced by Multicilin-HGR alone or in the presence of Gmnn. Basal body counts based on 10 to 15 cells taken from six embryos. Error bars indicate SD, and values significantly different (P < 0.05) from controls based on a two-tailed t-test are marked (asterisks)
Genes regulated by the EDM complex during MCC differentiation
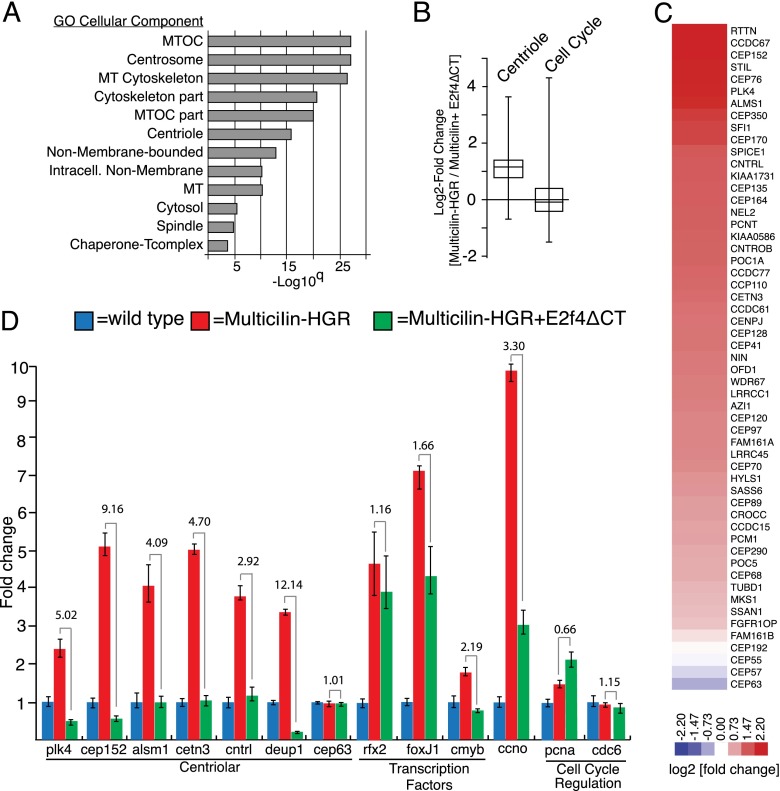
To assess the range of gene expression potentially induced by the EDM complex, we used RNA sequencing (RNA-seq) to assay skin progenitors induced to undergo MCC differentiation using Multicilin-HGR in the presence or absence of E2f4ΔCT. The most significant gene ontology (GO) terms of the 700 genes significantly down-regulated by E2f4ΔCT in this experiment are those associated with centrioles or related structures (Fig. 4A). Indeed, essentially all of the genes currently associated with centriole biogenesis (Azimzadeh et al. 2012) are down-regulated by E2f4ΔCT (Fig. 4C; Supplemental Table 1), including those encoding structural components such as Cetn3 (<2.4-fold), Cep135 (<2.6-fold), and Cep76 (<4.3-fold); the proximal regulators of centriole assembly, such as Plk4 (<4.2-fold) (Bettencourt-Dias et al. 2005; Habedanck et al. 2005), Cep152 (<4.7-fold) (Cizmecioglu et al. 2010; Dzhindzhev et al. 2010; Hatch et al. 2010), or Sass6 (<1.8-fold); or more peripheral assembly components such as Stil (<4.3-fold), Cenpj (<2.3-fold) (Tang et al. 2011), Ccp110 (<2.45-fold) (Chen et al. 2002), and KIAA0753 (<3.7-fold) (Firat-Karalar et al. 2014). Strikingly, E2f4ΔCT also strongly down-regulated the expression of deup1 (ccdc67; <4.8-fold), a key component of the deuterosome-mediated pathway of centriole assembly during MCC differentiation, but not its paralog, cep63, which is required for centriole assembly via the centriolar pathway (Fig. 4C; Supplemental Table 1; Zhao et al. 2013). Finally, E2f4ΔCT strongly down-regulated ccno, a gene mutated in human RGMC and up-regulated by Multicilin (Supplemental Table 1; Wallmeier et al. 2014). In contrast, the cell cycle genes classically thought to be regulated by E2f4 were not significantly changed or increased (Fig. 4B; Supplemental Table 3), with the notable exception of ccna1, a cyclin previously shown to be markedly up-regulated in differentiating MCCs (Hoh et al. 2012); rbl2, which paradoxically is normally required by E2f4 to act as a repressor in G0; and plk1, another member of the polo-like kinase family that is known to contribute to centriole assembly. These findings were validated using quantitative RT–PCR for a select group of centriolar assembly genes, cilia transcription factors, and cell cycle genes (Fig. 4D). E2f4ΔCT was less effective at inhibiting rfx2 and foxj1 expression induced by Multicilin-HGR, in line with the observations that it is also less effective at blocking cilia formation during MCC differentiation (Fig. 2C, arrows) or is induced by Multicilin ectopically (Fig. 3C, arrow). Together, these results indicate that Multicilin, by forming a complex with E2f4/Dp1, activates the expression of genes required for a massive expansion of centriole assembly while largely not affecting the ability of E2f4/Dp1 to repress genes involved in other aspects of the cell cycle.
Figure 4.
E2f4ΔCT inhibits centriole gene expression during MCC differentiation. (A) RNA-seq analysis of skin progenitors isolated from Xenopus embryos expressing Multicilin-HGR alone or with E2f4ΔCT. Skin progenitors were isolated at stage 10, treated with DEX at stage 11 to induce MCC differentiation, and then extracted for RNA 9 h later. GO terms (P < 0.01) for genes down-regulated by E2f4ΔCT with the highest significance for microtubule (MT)-associated structures, including centrioles. GO term analysis for genes up-regulated by E2f4ΔCT failed to find a significant term. (B) Tukey box plot showing fold changes in response to E2f4ΔCT in the background of Multicilin-HGR-induced differentiation for both centriole components based on the supplemented list generated by Azimzadeh et al. (2012) or for cell cycle genes (Kegg Pathway) (Supplemental Tables 2, 3) (C) Heat map showing log2 fold change in response to E2f4ΔCT in Multicilin-HGR samples. (D) RNA samples were generated as in A but then analyzed in triplicate for the expression of the indicated gene using quantitative RT–PCR. RNA levels are shown after normalization to ubiquitously expressed ornithine decarboxylase (odc) RNA and are set relative to a value of 1 for uninjected controls. Fold change in the presence and absence of E2f4ΔCT is indicated.
Chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) reveals E2f4-binding sites in MCC progenitors
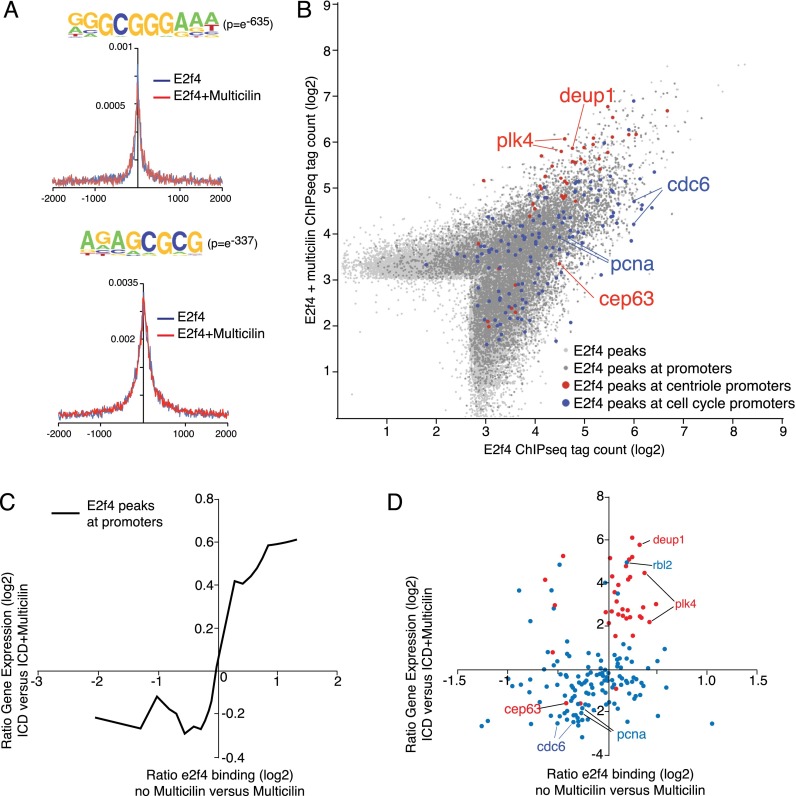
We next asked whether the EDM complex regulates genes involved in basal body assembly during MCC differentiation directly or indirectly. ChIP-seq was carried out on skin progenitors expressing a GFP-tagged form of E2f4 either alone or in the presence of Multicilin-HGR to induce MCC differentiation. Genomic fragments recovered with ChIP were sequenced and mapped to the Xenopus laevis genome, and positions with a high density of reads were called as peaks, identifying E2f4-binding sites under these two conditions. The most enriched sequence motifs within these fragments, based on a de novo search, correspond to E2f consensus motifs in other species (Fig. 5A; Supplemental Fig. 5) and are located at the peak center, suggesting that we efficiently captured binding sites for E2f4 in skin progenitors (Fig. 5A). When E2f4-bound peaks were mapped relative to features in the X. laevis genome, they were enriched at promoters, consistent with experiments performed in mammals (Supplemental Fig. 5; Lee et al. 2011), and overlapped significantly with the promoters of the orthologs bound in mouse B-cell lymphoma (CH12) cells by E2f4, as determined by ChIP-seq experiments as part of the ENCODE project (Mouse ENCODE Consortium et al. 2012), with strikingly similar consensus motifs (Supplemental Fig. 5B–H). Xenopus and mouse E2f4 also occupied a significant fraction of the promoters for cell cycle and centriolar biogenesis genes, suggesting functional and regulatory conservation (Supplemental Fig. 5H–J).
Figure 5.
E2f4-binding sites in skin progenitors and differentiating MCCs. ChIP-seq analysis was carried out on skin progenitors expressing E2f4-GFP in the presence or absence of Multicilin-HGR. Recovered DNA was sequence-aligned to the X. laevis genome, peaks were called, and tags within those peaks were counted. (A) Top two motifs enriched in E2f4 ChIP-seq and the positions of those motifs in immunoprecipitated genomic DNA from both experiments. Motif statistics are from E2f4 + Multicilin; for other conditions, see Supplemental Figure 6. (B) Tag densities for all E2f4 peaks, E2f4 peaks in promoters (±1 kb from the transcription start site), E2f4 peaks in the promoters of centriole genes, and E2f4 peaks in the promoters of cell cycle genes in the presence or absence of Multicilin. Centriole genes are strongly bound by E2f4 and even more so in the presence of Multicilin. (C) Moving average of the ratio of expression of all genes with promoters bound by E2f4 in the presence or absence of Multicilin. Note the general increase in transcription of genes with promoters more strongly bound in the presence of Multicilin. (D) Ratio of expression of centriole and cell cycle genes and binding of E2f4-GFP in the presence or absence of Multicilin.
Change of E2f4-binding targets and their expression by Multicilin
We next asked whether the genomic sites bound by E2f4-GFP are different in the presence or absence of Multicilin. While we found substantial overlap between the two conditions, there was increased E2f4-GFP binding (>1.5-fold) in genes required for centriole assembly (Azimzadeh et al. 2012) in the presence of Multicilin and decreased E2f4-GFP binding (>1.5-fold) in genes that are critical to cell cycle progression (Fig. 5B; Supplemental Tables 4, 5; Kanehisa and Goto 2000; Kanehisa et al. 2014), including members of the prereplication complex. We also observed that genes bound more tightly by E2f4 in the presence of Multicilin tended to increase in expression under those conditions (Fig. 5C), and this applied in particular to cell cycle and centriole assembly genes (Fig. 5D), suggesting a tight correlation between differences in E2f4 binding under the influence of Multicilin and the expression of genes downstream from those binding sites (Supplemental Fig. 6).
Together, these results indicate that the EDM complex likely regulates the expression of centriolar assembly genes directly. Furthermore, these results suggest that Multicilin affects E2f4/Dp1 activity differentially at genes associated with centriole assembly versus those involved in other aspects of cell cycle progression.
Mutations in human Multicilin disrupt the EDM complex
Recently, exome sequencing of patients with a severe RGMC identified missense mutations in the human MCIDAS gene that change conserved residues within the TIRT domain; e.g., Arg381 into histidine (R381H) and Gly366 into asparagine (G366D) (Boon et al. 2014). When forms of Xenopus Multicilin with equivalent mutations (Multicilin-R370H and Multicilin-G355D) are expressed in embryos, they fail to induce ectopic MCC differentiation (Boon et al. 2014), suggesting that the mutations affect EDM formation or activity. To examine this possibility, we first asked whether these mutations affect binding of Multicilin to E2f4 and Dp1. Both Multicilin-G355D and Multicilin-R370H appear stable based on their steady-state levels of expression (Fig. 6A) and both still localize to the nucleus (data not shown) but then differ dramatically in their ability to form the EDM complex: G355D disrupts EDM complex formation, thus explaining its mutant phenotype, while R370H still forms a complex, apparently as well as wild-type Multicilin (Fig. 6A).
Figure 6.
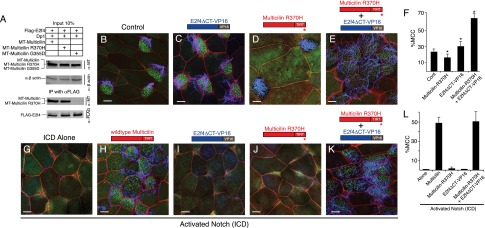
Mutations that cause human RGMC affect EDM complex formation and function. (A) Extracts of skin progenitors (stage 12) isolated from embryos injected with the indicated RNAs were subjected to Western blot analysis prior to (input) or after immunoprecipitation (IP) using the indicated antibodies. (B–E) Shown are confocal images of the skin in embryos expressing Multicilin-R370H and/or E2f4ΔCT-VP16 as indicated. Membranes are marked with mRFP (red), basal bodies are marked with Chibby-GFP (green), and cilia are labeled in blue. (F) The percentage of MCCs in the skin for each RNA injection. Values that differ significantly from the control based on a two-tailed t-test are marked ([*] P < 0.01). (G–K) Shown are confocal images of the skin in embryos expressing activated Notch (ICD) alone or with Multicilin-R370H and/or E2f4ΔCT-VP16 as indicated. Membranes are marked with mRFP (red), basal bodies are marked with Chibby-GFP (green), and cilia are labeled in blue. (L) The percentage of cells that are MCCs in the skin for each RNA injection. Values for Multicilin- and Multicilin-R370H/E2f4ΔCT-VP16-expressing embryos are not significantly different. Data were obtained from 10 fields from five embryos. Error bars indicate SD. Bars, 10 μm.
To account for the R370H phenotype, we reasoned that the EDM complex still forms but now lacks the ability to transcriptionally activate its target. To test this idea, we exploited a construct in which the potent activation domain from VP16 was fused to the C terminus of E2f4ΔCT, called E2f4ΔCT-VP16. While E2f4ΔCT inhibits centriole assembly in MCCs (Fig. 2), expression of E2f4ΔCT-VP16 causes MCCs to form more basal bodies than normal (195 ± 31 vs. 159 ± 19, P = 0.0002) (Supplemental Figs. 7C,D,F, 8M). Expression of E2f4ΔCT-VP16 also potentiates basal body formation induced ectopically by Multicilin (Supplemental Fig. 7F), in line with the idea that the EDM complex acts as a transcriptional activator to drive centriole gene expression. However, the number of differentiating MCCs or the centriole number in outer cells was not dramatically or consistently changed in response to the expression of E2f4ΔCT-VP16 (Fig. 6F; Supplemental Fig. 7E,G), suggesting that E2f4ΔCT-VP16 activity in terms of centriole assembly depends on Multicilin in the context of MCC differentiation.
Based on these observations, we reasoned that E2f4ΔCT-VP16 might synthetically rescue the defect observed with Multicilin-R370H. As shown above, the density of MCCs that form in the skin was not reproducibly altered by E2f4ΔCT-VP16 and was reduced by Multicilin-R370H (Fig. 6C,D,F; Supplemental Figs. 7, 8). In contrast, in embryos injected with both RNAs, ectopic MCCs were induced at a frequency similar to that of wild-type Multicilin (Fig. 6E,F; data not shown) and, strikingly, even when carried out in the presence of activated Notch (ICD) to inhibit endogenous Multicilin expression (Fig. 6G–L; Stubbs et al. 2012). Synthetic rescue did not occur between E2f4ΔCT-VP16 and Multicilin that lacks a TIRT domain or with the G355D mutant, both of which cannot form the EDM complex (Supplemental Fig. 8G–J). Only weak synthetic rescue occurred between the Multicilin-R370H and wild-type E2f4 (Supplemental Fig. 8D,E). Finally, we assayed the response of select target genes in the centriole assembly pathway to Multicilin-R370H in the presence and absence of E2f4ΔCT-VP16 in an ICD background. In comparison with wild-type Multicilin, Multicilin-R370H was less active in inducing Plk4, Cep152, and deup1 expression, but E2f4ΔCT-VP16 substantially rescued its activity (Supplemental Fig. 9). Together, these results indicate that Multicilin binding via the TIRT domain to the EDM complex creates an activation domain required to promote full gene expression underlying basal body amplification during MCC differentiation. Both the G355D and R370H mutations in human Multicilin underscore the importance of the EDM complex in the activation of gene expression required for MCC differentiation in both the Xenopus skin and the human airways.
Discussion
Here we show that Multicilin forms a complex with E2f4 or E2f5 along with Dp1 and that this complex binds and activates gene expression required for basal body assembly during MCC differentiation. Our findings indicate that Multicilin coopts the inhibitory E2fs to activate key components that drive centriole assembly during the centriolar cycle, such as Plk4, Cep152, and Stil, while also activating the expression of deup1, the paralog of cep63 required for centriole assembly via the deuterosome rather than the centriolar pathway (Zhao et al. 2013). This complex also activates the expression of ccno, a gene required for centriole assembly in MCCs and mutated in human RGMC (Wallmeier et al. 2014). Thus, the EDM complex accounts for how differentiating MCCs assemble a large number of centrioles despite the fact that this assembly occurs in G0. These results also predict that E2f4, E2f5, and Dp1 are candidate genes for loci mutated in RGMC.
How does Multicilin selectively modulate E2f transcription, thereby activating centriole assembly genes but not other components of the cell cycle? The data obtained by ChIP-seq analysis suggest that EDM complex binding is selectively enhanced at activated promoters, presumably based on additional interactions with other factors, some of which could be gene-specific, depending on its promoter or enhancer architecture. These interactions presumably recruit new factors to the EDM complex that binds to genes in the centriole biogenesis pathway to activate their expression while not precluding repression that occurs at genes involved in other aspects of cell cycle progression. The EDM complex could recruit positive factors via the C-terminal domain of E2f4/5, a region required for the EDM complex to function. The EDM complex could also recruit factors through the TIRT domain, which is required not only for the EDM complex to form but also for its transcriptional activity, based on the R381H mutation in human MULTICILIN. Finally, the EDM complex could recruit positive regulators through the coiled-coil domain of Multicilin, based on the ability of Gmnn to bind here and inhibit EDM activity. Identification of the factors that interact with the EDM complex through these sites will be essential for determining how centriole and cell cycle genes are differentially regulated during MCC differentiation.
Dividing cells tightly regulate the process of centriole assembly to maintain proper centriole number and avoid chromosomal segregation defects associated with multipolar mitotic spindles. In light of this, both the expression and activity of Multicilin are likely to be under tight negative control in dividing cells, given its potent ability to coordinately regulate a large number of genes that drive centriole assembly. In epithelial progenitors, Multicilin activity, in terms of centriole assembly, could be gated by Gmnn, which is maintained at high levels in dividing cells but is degraded as cells transit into G0 (McGarry and Kirschner 1998). Such inhibitory interactions may be critical to prevent Multicilin activity from prematurely promoting centriole assembly in MCC progenitors with potentially catastrophic consequences.
Multicilin induces not only centriole assembly via the deuterosome-mediated pathway but also motile extension via its ability to up-regulate additional transcriptional regulators; namely, Foxj1 and the RFX transcription factors (Yu et al. 2008; Stubbs et al. 2012; Chung et al. 2014). Since the former pathway is much more sensitive to inhibition by E2f4ΔCT and Gmnn than the latter, Multicilin could activate motile cilium formation by forming additional complexes other than the one with E2f proteins. Nonetheless, prominent E2f4-binding sites are present in the proximal promoters of both foxj1 and rfx2, and the human mutations that disable the EDM complex also disable the ability of Multicilin to activate motile cilia extension. Thus, the difference between the two pathways is more likely to be due to a different cast of cofactors that interact with the EDM complex to activate different target genes during MCC differentiation.
Like Deup1, Multicilin is present in only a subset of vertebrate species, suggesting that it recently evolved to drive a program of multiciliogenesis (Zhao et al. 2013). While the appearance of new cell type features is often associated with regulatory changes in enhancer elements (Carroll 2008), Multicilin could be an example of a transcriptional coregulator that evolved to coopt an existing transcriptional factor complex to coordinately regulate a large number of genes required for multiciliogenesis. The coordinated transcriptional regulation of the centriole assembly pathway by the E2f proteins is likely to have pre-existed as a component of cell cycle regulation. The appearance of deup1 by duplication of cep63, along with its cell cycle regulatory machinery, coupled with Multicilin to selectively act on this machinery, puts in place critical components used in the centriolar cycle to drive a massive expansion of centrioles during MCC differentiation. Further dissection of the centriole assembly pathway and why it selectively responds transcriptionally to Multicilin is thus key to understanding the origins and development of this novel ciliated cell type.
Materials and methods
X. laevis embryos, RNA synthesis, and microinjections
X. laevis embryos were prepared by in vitro fertilization using standard protocols (Sive et al. 1998). Synthetic, capped mRNA was generated in vitro as described previously using DNA templates described below (Stubbs et al. 2008). In vitro synthesized RNA was injected into animal blastomeres of embryo at the two-cell or four-cell stage (typically 0.1–5.0 ng of RNA per embryo).
DNA constructs
Templates used to generate tracer RNAs encoding Hyls1-GFP (centriole/basal body marker), Chibby-GFP (basal body marker), Centrin4-RFP (centriole/basal body marker), and membrane-localized RFP (mRFP) have been described previously (Stubbs et al. 2012; Wallmeier et al. 2014). Templates for RNAs encoding Multicilin-HGR, myc, or Flag-tagged Multicilin or for different deleted myc-tagged forms of Multicilin were described previously (Stubbs et al. 2012), as were templates for RNAs encoding activated Notch (ICD) or for the DNA-binding mutant of Su(H) (Deblandre et al. 1999). Missense mutations were introduced into Multicilin using PCR or site-directed mutagenesis using PCR primers listed in Supplemental Table 6, sequenced, and cloned into the CS2-MT vector. Templates used to generate RNA encoding tagged forms of X. laevis E2f4, E2f5, E2f1, E2f7, Dp1, and Gmnn were obtained using PCR amplification of cDNA from a stage 17 cDNA library using primers listed in Supplemental Table 6, sequenced, and then cloned into CS2 vectors that add 6-myc epitope tags, a NLS, a Flag tag, or a GFP tag, all at the N terminus. Deleted forms of Dp1, E2f4, or E2f5 were generated using restriction enzyme cleavage or by PCR amplification followed by sequencing. The transactivation domain (amino acids 413–490) from HSV1 was fused onto the E2f4ΔCT to create E2f4ΔCT-VP16.
RNA preparation, RNA-seq, and quantitative RT–PCR
RNA was generated from animal caps isolated at stage 10 from embryos that were injected at the two-cell stage with RNAs, targeting all four animal quadrants. Animal caps were cultured in 0.5× MMR, treated with 1 μM DEX at stage 11 to induce Multicilin-HGR, and harvested at 9 h (through stage 18) with the proteinase K method followed by phenol-chloroform extractions, lithium precipitation, and treatment with RNase-free DNase and a second series of phenol-chloroform extractions and ethanol precipitation. RNA-seq libraries were constructed with Illumina TruSeq RNA sample preparation kit version 2 according to the manufacturer’s instructions and sequenced on a HiSeq 2000 or 2500 at 1 × 50 or 1 × 100 base pairs to a depth of 20 million to 40 million reads. Each RNA-seq condition was performed in triplicate using animal caps isolated from different females.
RNA samples as prepared above were also used to measure gene expression using quantitative PCR as described previously (Stubbs et al. 2012). Briefly, RNA was converted in cDNA using reverse transcription and then assayed by quantitative PCR using the appropriate primer pairs (Supplemental Table 7) in triplicate with the ABI Prism 7900HT thermal cycler (Life Technology). Samples were normalized to the levels of ornithine decarboxylase (odc) RNA as an internal control.
ChIPs and library construction
Animal caps were isolated at stage 10 from embryos injected with RNAs encoding GFP-E2f4 alone or with Multicilin-HGR, treated with DEX at stage 11 to induce Multicilin activity, and harvested 6 h later (roughly stage 16). Samples were prepared for ChIP using methods described in Blythe et al. (2009) with the following modifications: About 220 animal caps were fixed for 30 min in 1% formaldehyde, and chromatin was sheared on a BioRuptor (30 min; 30 sec on and 2 min off at highest power setting). Tagged proteins with associated chromatin were immunoprecipitated with a rabbit anti-GFP (Invitrogen catalog no. A11122, lot no. 1296649). DNA fragments were then end-repaired (New England Biolabs, end repair module), adenylated (New England Biolabs, Klenow fragment 3′–5′ exo- and da-tailing buffer), ligated to standard Illumina indexed adapters (TruSeq version 2), and PCR-amplified (New England Biolabs, Phusion, 16 cycles). Libraries were then sequenced on a HiSeq 2500 at 1 × 50 or 1 × 100 to a depth of 10 million to 30 million reads.
All raw sequencing data were deposited at NCBI Gene Expression Omnibus (accession number is pending).
Bioinformatics
RNA-seq
FASTQ reads were aligned to the X. laevis transcriptome, MayBall version (Dobin et al. 2013; Chung et al 2014; http://daudin.icmb.utexas.edu) with RNA-STAR. Aligned reads were then counted with eXpress (Roberts and Pachter 2011), and, for interspecies comparisons, counts of A and B forms of X. laevis transcripts were combined. DESeq was then used to normalize, estimate dispersion, and test differential expression using rounded raw collapsed counts from eXpress. For visualization purposes, RNA-seq reads were also mapped to version 7.1 with Bowtie2 (Langmead and Salzberg 2012) and loaded as tracks into the Integrative Genomics Viewer browser (Thorvaldsdottir et al. 2013).
ChIP-seq
To identify the positions of features in the X. laevis genome, the longest cDNA in the transcriptome (MayBall version) were aligned to X. laevis genome build version 7.1 (JGI, Xenbase) with BLAT (Kent 2002), keeping only the highest-scoring hit. FASTQ reads from ChIP-seq were then mapped to version 7.1 with Bowtie2 (Langmead and Salzberg 2012), peaks were called with HOMER (Heinz et al. 2010), their positions were annotated relative to known exons, and sequences in these peaks were then interrogated for motif enrichment with HOMER (Heinz et al. 2010; Lin et al. 2012).
Phenotypic analysis of MCC differentiation
MCC differentiation was assessed in embryos using confocal microscopy to visualize cell boundaries marked with mRFP, Hyls1-GFP, or Chibby-GFP to label basal bodies and staining with a mouse antibody directed against acetylated tubulin (1:1000; Sigma) followed by a cy5 secondary to label cilia. Embryos were processed by a short 10-min fix in 3.7% formaldehyde and 0.25% glutaraldehyde in PBT (PBS with 0.1% TritonX-100) followed by antibody staining. PSC differentiation was analyzed in embryos injected with mGFP RNA followed by staining with a mouse monoclonal anti-AE1 (1:250; Iowa Developmental Studies Hybridoma Bank [IDSHB] clone IVF-12) and rabbit anti-Atp6V1B1/2 (1:100; Santa Cruz Biotechnology) as described previously (Quigley et al. 2011). Embryos were mounted in PVA with DABCO and imaged on a Bio-Rad Radiance 2100 or Zeiss LSM710 confocal microscope.
Data for basal body counting typically were collected from two to three randomly chosen fields from five to 10 embryos per sample. Cell type number, basal body number, and cell size were quantified using ImageJ software. Statistical significance was performed in all experiments using two-tailed t-tests.
Coimmunoprecipitation and Western analysis
Xenopus embryos were injected with RNAs at the two- or four-cell stage, targeting all four animal quadrants. Animal caps were cut at stage 10, and protein was extracted with lysis buffer (25 mM Tris-HCl at pH 7.5, 300 mM NaCl, 1%Triton-100, supplemented with a cocktail of protein inhibitors) at the equivalent of stage 13. GFP-tagged proteins were recovered from extracts following overnight incubation at 4°C with rabbit anti-GFP antibody (Invitrogen, no. A11122) using protein A agarose beads (Millipore, 16-157) for at least 2 h at 4°C. For Flag-tagged proteins, extracts were incubated with anti-Flag M2 beads (Sigma, A2220) for 2 h at 4°C. Beads were washed at least six times with lysis buffer and boiled in Laemmli loading buffer. Proteins were separated by 10% SDS-PAGE and transferred to PVDF membrane (Millipore). Membranes were blocked using 0.1% Casein buffer (Bio-Rad, no. 161-0782) and incubated in primary antibody diluted in 0.1% Casein buffer: mouse monoclonal anti-Myc (1:125; 9E10), rabbit anti-GFP (1:500; Cell Signaling, 2956P), rabbit anti-Flag (1:500; Cell Signaling, 2368P), rabbit anti-Multicilin 5226 (1:1000), and rabbit anti-E2f4 (1:200; Santa Cruz Biotechnology, sc-1082). Primary antibodies were detected using LI-COR Odyssey infrared imaging following incubation in goat anti-rabbit Alexa Fluor 680 (1:10,000; Invitrogen, A21076) or goat anti-mouse Alexa Fluor 680 (1:10,000; Invitrogen, A21058). Light chain-specific secondary antibody (Alexa Fluor 680 goat anti-mouse IgG, light chain-specific, 1:50,000; Jackson ImmunoResearch Laboratories, 115-625-174) was used for Western blots when needed to eliminate background due to immunoglobulin heavy chains. The endogenous interaction between Multicilin and E2f4 was analyzed using a Western blot analysis based on HRP goat anti-rabbit IgG, light chain-specific (1:15,000; Jackson ImmunoResearch Laboratories), and ECL (Amersham, GE)
Acknowledgments
We thank The International Xenopus laevis Genome Project Consortium, especially the Harland, Rokhsar, and Taira laboratories, as well as all data contributors to the consortium. We thank members of the Kintner laboratory and Dr. Tony Hunter for comments on the manuscript. This work was supported by National Institutes of Health grant GM96021 to C.K. We thank Dr. Bing Li (University of California at Los Angeles) for bioinformatics assistance. C.K. and L.M. were responsible for the design of the experiments and writing of the manuscript. L.M., I.Q., and C.K. were responsible for the experiments and the assembly of the figures, while H.O. provided information about the human Multicilin mutants.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.243832.114.
References
- Azimzadeh J, Wong ML, Downhour DM, Sanchez Alvarado A, Marshall WF 2012. Centrosome loss in the evolution of planarians. Science 335: 461–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol 15: 2199–2207 [DOI] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kessler DS, Klein PS 2009. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev Dyn 238: 1422–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon M, Wallmeier J, Ma L, Loges NT, Jaspers M, Olbrich H, Dougherty G, Werner C, Amirav I, Hevroni A, et al. 2014. Mutations in MCIDAS result in a congenital mucociliary clearance disorder with reduced generation of multiple motile cilia (RGMC). Nat Commun (in press) [DOI] [PubMed] [Google Scholar]
- Carroll SB 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Chen Z, Indjeian VB, McManus M, Wang L, Dynlacht BD 2002. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev Cell 3: 339–350 [DOI] [PubMed] [Google Scholar]
- Chung MI, Kwon T, Tu F, Brooks ER, Gupta R, Meyer M, Baker JC, Marcotte EM, Wallingford JB 2014. Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife 3: e01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu O, Arnold M, Bahtz R, Settele F, Ehret L, Haselmann-Weiss U, Antony C, Hoffmann I 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J Cell Biol 191: 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Bender Kim CF, Caron AM, Vasile E, Bronson RT, Lees JA 2007. E2f4 is required for normal development of the airway epithelium. Dev Biol 305: 564–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblandre GA, Wettstein DA, Koyano-Nakagawa N, Kintner C 1999. A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development 126: 4715–4728 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev NS, Yu QD, Weiskopf K, Tzolovsky G, Cunha-Ferreira I, Riparbelli M, Rodrigues-Martins A, Bettencourt-Dias M, Callaini G, Glover DM 2010. Asterless is a scaffold for the onset of centriole assembly. Nature 467: 714–718 [DOI] [PubMed] [Google Scholar]
- Firat-Karalar EN, Rauniyar N, Yates JR 3rd, Stearns T 2014. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr Biol 24: 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Quaas M, Wintsche A, Muller GA, Engeland K 2014. Polo-like kinase 4 transcription is activated via CRE and NRF1 elements, repressed by DREAM through CDE/CHR sites and deregulated by HPV E7 protein. Nucleic Acids Res 42: 163–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H 2007. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8: 880–893 [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA 2005. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–1146 [DOI] [PubMed] [Google Scholar]
- Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J Cell Biol 191: 721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh RA, Stowe TR, Turk E, Stearns T 2012. Transcriptional program of ciliated epithelial cells reveals new cilium and centrosome components and links to human disease. PLoS ONE 7: e52166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42: D199–D205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BK, Bhinge AA, Iyer VR 2011. Wide-ranging functions of E2F4 in transcriptional activation and repression revealed by genome-wide analysis. Nucleic Acids Res 39: 3558–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Benner C, Mansson R, Heinz S, Miyazaki K, Miyazaki M, Chandra V, Bossen C, Glass CK, Murre C 2012. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol 13: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcet B, Chevalier B, Luxardi G, Coraux C, Zaragosi LE, Cibois M, Robbe-Sermesant K, Jolly T, Cardinaud B, Moreilhon C, et al. 2011. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat Cell Biol 13: 693–699 [DOI] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93: 1043–1053 [DOI] [PubMed] [Google Scholar]
- Mouse ENCODE Consortium, Stamatoyannopoulos JA, Snyder M, Hardison R, Ren B, Gingeras T, Gilbert DM, Groudine M, Bender M, Kaul R, et al. 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pefani DE, Dimaki M, Spella M, Karantzelis N, Mitsiki E, Kyrousi C, Symeonidou IE, Perrakis A, Taraviras S, Lygerou Z 2011. Idas, a novel phylogenetically conserved geminin-related protein, binds to geminin and is required for cell cycle progression. J Biol Chem 286: 23234–23246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley IK, Stubbs JL, Kintner C 2011. Specification of ion transport cells in the Xenopus larval skin. Development 138: 705–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L 2011. RNA-seq and find: entering the RNA deep field. Genome medicine 3: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadasivam S, DeCaprio JA 2013. The DREAM complex: master coordinator of cell cycle-dependent gene expression. Nat Rev Cancer 13: 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H, Grainger RM, Harland RM 1998. The early development of Xenopus laevis: a laboratory manual. Cold Spring Harbor Press, Cold Spring Harbor, NY [Google Scholar]
- Sorokin SP 1968. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci 3: 207–230 [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C 2008. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet 40: 1454–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, Kintner C 2012. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol 14: 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TK 2013. Centriole biogenesis in multiciliated cells. Nat Cell Biol 15: 1400–1402 [DOI] [PubMed] [Google Scholar]
- Tang CJ, Lin SY, Hsu WB, Lin YN, Wu CT, Lin YC, Chang CW, Wu KS, Tang TK 2011. The human microcephaly protein STIL interacts with CPAP and is required for procentriole formation. EMBO J 30: 4790–4804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdottir H, Robinson JT, Mesirov JP 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA 2002. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 3: 11–20 [DOI] [PubMed] [Google Scholar]
- Wallmeier J, Al-Mutairi D, Chen C, Loges N, Petra P, Menchen T, Ma L, Shamseldin HE, Olbrich H, Dougherty G, et al. 2014. Mutations in CCNO result in congenital mucociliary clearance disorder with reduced generation for multiple motile cilia. Nat Genet 46: 646–651 [DOI] [PubMed] [Google Scholar]
- Yu X, Ng CP, Habacher H, Roy S 2008. Foxj1 transcription factors are master regulators of the motile ciliogenic program. Nat Genet 40: 1445–1453 [DOI] [PubMed] [Google Scholar]
- Zhao H, Zhu L, Zhu Y, Cao J, Li S, Huang Q, Xu T, Huang X, Yan X, Zhu X 2013. The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat Cell Biol 15: 1434–1444 [DOI] [PubMed] [Google Scholar]