Abstract
The exploitation of catalytic promiscuity and the application of de novo design have recently opened the access to novel, non-natural enzymatic activities. Here we describe a structural bioinformatic method for predicting catalytic activities of enzymes based on three-dimensional constellations of functional groups in active sites (‘catalophores’). As a proof-of-concept we identify two enzymes with predicted promiscuous ene-reductase activity (reduction of activated C–C double bonds) and compare them with known ene-reductases, that is, members of the Old Yellow Enzyme family. Despite completely different amino acid sequences, overall structures and protein folds, high-resolution crystal structures reveal equivalent binding modes of typical Old Yellow Enzyme substrates and ligands. Biochemical and biocatalytic data show that the two enzymes indeed possess ene-reductase activity and reveal an inverted stereopreference compared with Old Yellow Enzymes for some substrates. This method could thus be a tool for the identification of viable starting points for the development and engineering of novel biocatalysts.
 Enzymes are very efficient reaction catalysts, though taking advantage of this synthetically is hampered by their notorious specificity. Here, the authors identify important arrangements of active site residues and use structural bioinformatics to successfully predict enzyme activity.
Enzymes are very efficient reaction catalysts, though taking advantage of this synthetically is hampered by their notorious specificity. Here, the authors identify important arrangements of active site residues and use structural bioinformatics to successfully predict enzyme activity.
Enzymes are increasingly used in biotechnological and biocatalytic processes for the ‘green’ production of chemical compounds1 and in medical applications2. Natural enzymes, however, are generally not fully optimized for these purposes. Consequently, not all chemical reactions potentially interesting for biotechnological applications are represented in the currently available enzyme portfolio. Thus, the identification and design of novel enzyme functionalities is of utmost importance.
Current approaches include classic wet-lab screening and protein engineering efforts3, as well as bioinformatic techniques. The latter often rely on similarities of amino acid sequences (for example, using BLAST or the Catalytic Site Atlas4) or on the conservation of the overall structures and folds of proteins (for example, using PDBeFold5). These computational approaches are mainly used to identify homologous enzymes. By de novo enzyme design, on the other hand, it has recently been possible to build biocatalysts from scratch6,7,8 and these methods were successfully applied to generate a variety of protein functions9,10. However, the designed enzymes also had some drawbacks, such as modest catalytic efficiency or decreased enzyme stability9,11.
In this context, the reutilization of existing enzymes could be beneficial. It has been observed that some natural enzymes—apart from their original catalytic activity—also catalyse completely unrelated chemical reactions as a side activity, known as ‘catalytic promiscuity’12. Examples include the catalysis of C–C bond formation by hydrolases13 or the nitroaldolase activity of hydroxynitrile lyases14. Catalytic promiscuity is generally also seen as the starting point for the evolution of novel enzyme activities in nature15. Thus, it could also be a viable basis for engineering novel enzymes3 provided that a method for the detection of potential catalytic promiscuity is available. Earlier work on three-dimensional (3D) active site templates and database mining, such as TESS16 and JESS17, has shown the potential of structure-based approaches for predicting catalytic activity.
Here we describe a method to identify enzymes with specific active site geometries and thus to predict (promiscuous) activities by screening the Protein Data Bank (PDB) using specific 3D templates, which mimic a ‘minimal’ catalytic active site constellation (‘catalophore’) in the style of the theozyme concept18. As a test case for our approach, we choose ene-reductase (Old Yellow Enzyme, OYE) activity, that is, the reduction of activated C–C double bonds. This class of enzymes is structurally and mechanistically very well characterized and the reaction has a high potential in biocatalysis, because it allows the simultaneous introduction of two new stereocentres19. We identify two enzymes for which we predict promiscuous ene-reductase activity. These enzymes are compared with known ene-reductases regarding their 3D structures (including substrate binding), their biochemical properties, as well as their biocatalytic substrate scope and stereospecificities. These data clearly show that the two enzymes—despite completely different sequences and folds—share salient features of typical OYEs, especially their ene-reductase activity. This example indicates that search methods using structural active site templates may be useful tools for the identification of viable starting points for the development and engineering of novel biocatalysts.
Results
Database search
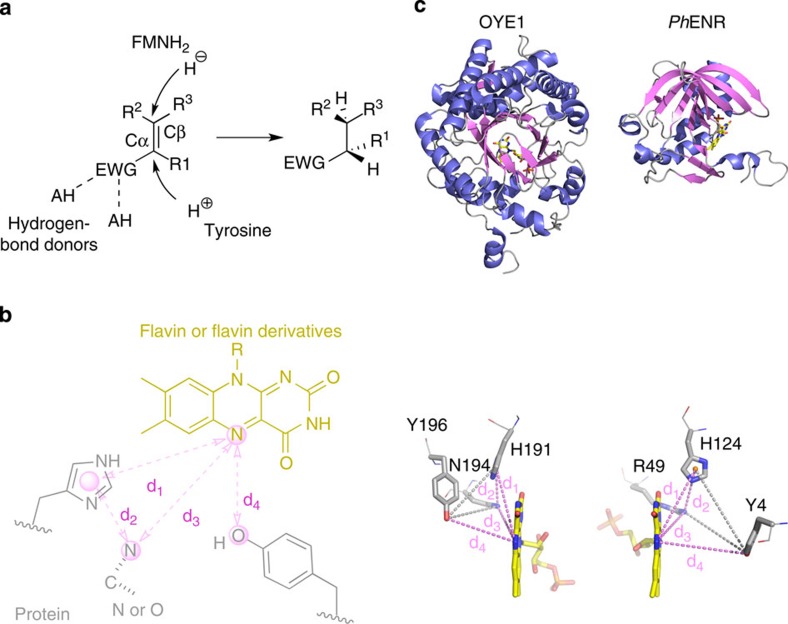
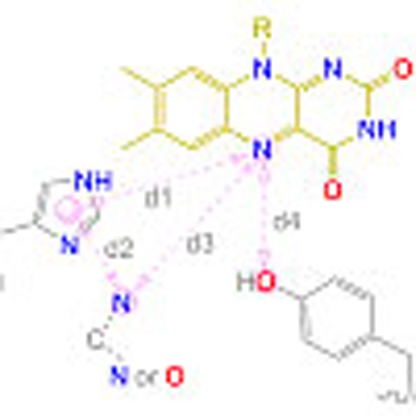
The database search motif was derived through an analysis of active site constellations in available OYE structures (Supplementary Table 1 and Supplementary Fig. 1). The catalytic site of OYEs harbours a flavin mononucleotide (FMN) cofactor, which donates a hydride to the Cβ atom of the substrate. In addition, it usually comprises a pair of amino acid residues (typically histidine/histidine or asparagine/histidine) that act as H-bonding donors to the electron-withdrawing group of the substrate, and a conserved tyrosine residue (in some cases a cysteine), which is necessary to deliver a proton onto the Cα atom during turnover (Fig. 1a). We defined four distances to characterize the relative position of the required functional groups (Fig. 1b): from N5 of the flavin cofactor to the geometric centre of the imidazole ring of histidine (d1), between the histidine and the second hydrogen bonding donor (d2), between N5 and the second hydrogen bonding donor (d3) and between N5 and the OH group of the tyrosine (d4).
Figure 1. Overview of search template and comparison between OYEs and the identified hits.
(a) Hydrogenation of activated C–C double bonds catalysed by OYEs (EWG, electron withdrawing group; HA, hydrogen bonding donor). (b) Search template with distance descriptors derived from OYE structures. Amino acids of the protein and the flavin cofactor are represented in grey and yellow, respectively. Spheres represent atoms/pseudo-atoms used for defining distance ranges (d1=7.5–7.9 Å, d2=4.6–5.4 Å, d3=6.1–6.8 Å, d4=5.0–8.9 Å). Undefined bond valences are indicated by dotted lines. (c) Comparison of the overall and active site structures of OYE1 (PDB entry: 1OYB) from Saccharomyces pastorianus as a typical member of the OYE family and one protomer of PhENR (PDB entry: 2R6V) identified by the catalophore search. The lower panel shows the positions of the catalytic residues relative to FMN. Distance descriptors d1–d4 are shown as magenta dotted lines. Additional grey dotted lines complete a tetrahedron to better show the approximate mirror symmetry.
This motif was then applied in searches in the commercial Relibase+ database20. In addition, we implemented an in-house database of protein structures and a script-based search environment using the programme YASARA Structure21. Searches using both database platforms yielded mostly equivalent lists of hits, which not surprisingly contained mainly OYE structures (Supplementary Table 2). The latter were excluded based on sequence similarity as well as on the occurrence of OYE-specific sequence patterns22. Two oxidoreductases (PDB entries: 2I0K and 3FBS) were discarded, because the tyrosine residue was located behind the flavin (relative to the other components of the catalophore) and would therefore be unable to act as the proton donor. A flavocytochrome C (PDB entry: 1JRZ) was discarded because of the additional haem cofactor bound to this protein.
The two remaining hits, annotated as ‘putative styrene monooxygenase reductase component’ (PDB entries: 1USC and 1USF) from Thermus thermophilus (subsequently named TtENR) and as ‘FMN-binding protein’ (PDB entry: 2R6V) from Pyrococcus horikoshii (subsequently named PhENR), were further analysed. These two enzymes from extremophiles are homodimers showing a split barrel-like fold23,24 with anti-parallel β-sheets and with one, non-covalently bound FMN molecule per subunit (Fig. 1c). This fold is completely different to the (α,β)8- or TIM-barrel fold of OYEs. Despite the different fold types, protein sizes and the overall low sequence identities of <10%, both groups of enzymes exhibit very similar arrangements of active site functional groups. Interestingly, however, the active site constellations in PhENR and TtENR show approximate mirror symmetry to OYE structures (Fig. 1c) with the re-face of the isoalloxazine ring (instead of the si-face) being accessible to the substrate, indicating an inverted stereochemical preference25 of the two enzymes compared with OYEs.
X-ray crystallography
We determined X-ray crystal structures of ligand complexes of PhENR and TtENR to analyse the respective binding modes in comparison with OYEs. Although both enzymes crystallized easily and fast, obtaining structures of ligand complexes by soaking and co-crystallization experiments turned out to be more difficult. We managed, however, to obtain diffracting crystals soaked with 2-cyclohex-1-enone, p-hydroxybenzaldehyde (pHB) and 1,4-hydroquinone, and to determine the structures of the corresponding complexes (Supplementary Table 3). When soaking the crystals with these three ligands, a pronounced colour change from yellow to deep orange was observed, already indicating ligand binding in the vicinity of the FMN cofactor. Indeed, clear residual electron density near the isoalloxazine ring of the flavin was observed in all cases (Supplementary Figs 2 and 3).
An analysis using the PISA server26 confirmed that both proteins are present as dimers in the crystal (and very likely to be in solution). The previously deposited structures of both enzymes, which were identified in the database search (PDB entries: 1USC, 1USF and 2R6V), are very similar to the newly determined complex structures. The root-mean-square deviations after superposition were 0.1–0.2 Å for the comparison of the PhENR structures with PDB entry 2R6V and 0.2–0.3 Å for the comparison of the TtENR structures with PDB entries 1USC and 1USF.
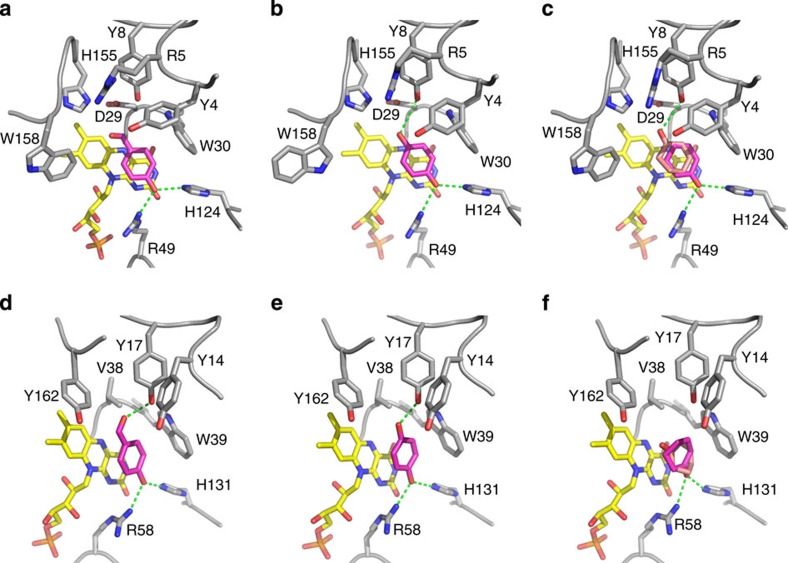
pHB is known as a high-affinity ligand of OYEs27,28 and its binding mode has been elucidated for several members of this enzyme family (Supplementary Fig. 4). In PhENR and TtENR, an equivalent binding mode was observed. Specifically, the phenolate oxygen atom is hydrogen bonded by His-124 and Arg-49 in PhENR or by His-131 and Arg-58 in TtENR (Fig. 2a,d). Structurally, these residues correspond to the conserved His and Asn residues in OYEs (for example, His-191 and Asn-194 in OYE1 from Saccharomyces pastorianus29, Supplementary Fig. 4), which act as H-bonding donors to the electron-withdrawing group. The aldehyde group of the ligand is H-bonded to Tyr-17 in TtENR (again similar to the binding to OYEs), while a clear interaction partner is missing in the active site of PhENR. The third important residue in the active site of OYEs is a tyrosine, which is supposed to act as a proton donor. The residues Tyr-4 in PhENR and Tyr-14 in TtENR (Fig. 2) correspond to this proton donor in OYEs.
Figure 2. X-ray crystal structures of ligand complexes of PhENR and TtENR.
Complexes of PhENR with p-hydroxybenzaldehyde (a), 1,4-hydroquinone (b) and 2-cyclohexen-1-one (c). Complexes of TtENR with p-hydroxybenzaldehyde (d), 1,4-hydroquinone (e) and 2-cyclohexen-1-one (f). Amino acid residues are shown in grey, the flavin cofactor in yellow and the bound ligands in magenta. The second, alternate binding mode of 2-cyclohexen-1-one is shown in pink. Hydrogen bonding interactions are depicted as green, dashed lines. The figure was prepared using PyMOL (Schrodinger Inc.).
The same binding partners and interactions were also observed in the structures of the two complexes with 1,4-hydroquinone, another phenolic ligand of OYEs (Fig. 2b,e). In the structures of the complexes with 2-cyclohexene-1-one, on the other hand, we observed two alternate binding modes in both cases (Fig. 2c,f). 2-Cyclohexen-1-one is a typical OYE substrate, which is reduced to cyclohexanone. In the first binding mode the carbonyl group is again H-bonded to His-124 and Arg-49 in PhENR or to His-131 and Arg-58 in TtENR, respectively. In these structures the Cβ atoms of the α,β-unsaturated carbonyl moiety align well with the N5 atom of the flavin cofactor with distances of 3.4 and 4.5 Å. The relative positions of the predicted proton donors (Tyr-4 in PhENR and Tyr-14 in TtENR), however, appear to be less optimal with 6–7 Å distances between the OH groups of the tyrosine residues and the Cα atom of the potential substrate. In the second, presumably non-productive binding mode, the ligand is flipped by ∼180°. In PhENR the carbonyl group is then H-bonded to Tyr-8, but it does not form any specific polar interactions in TtENR.
Biochemical characterization
OYE was the first flavin-dependent enzyme isolated from the brewers’ bottom yeast by Warburg and Christian30, and since then has been recognized as a paradigm of flavoproteins. Therefore, a large body of biochemical information is available for the yeast enzyme as well as for other members of the OYE family identified in eubacteria, fungi and plants31. We were curious to see to what extent PhENR and TtENR share properties of the yeast enzyme despite the completely different protein fold.
Ligand binding
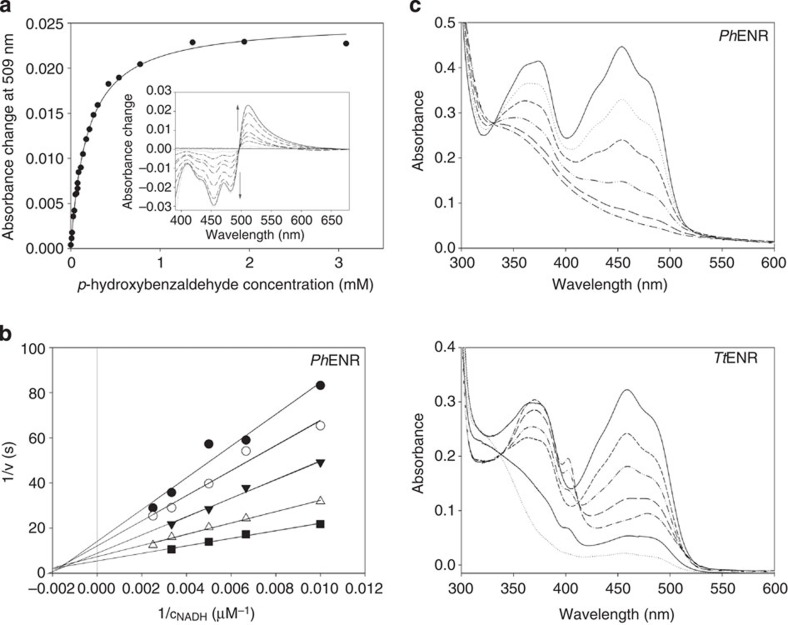
We investigated the binding of different potential ligands using ultraviolet–visible spectroscopic titration experiments. As observed in the soaking experiments, addition of pHB to PhENR and TtENR gave rise to spectral perturbations in the 550 to 750 nm range, indicating the formation of a charge-transfer complex28 in a concentration-dependent manner (Fig. 3a). The resulting dissociation constants (Kd) of 36 and 25 μM for PhENR and TtENR, respectively, are higher than for yeast OYE (0.1 μM)32 but comparable to the Bacillus subtilis homologue YqjM (24 μM)33. Furthermore, binding of the OYE model substrate 2-cyclohexen-1-one was demonstrated for both enzymes, but the determined Kd-values of 76 and 40 mM for PhENR and TtENR, respectively, were substantially higher than for yeast OYE (32 μM)34. In addition, we found that 1,4-benzoquinone also binds to both enzymes. However, the Kd-values (1.8 mM for PhENR and 0.58 mM for TtENR) were much lower than in the case of 2-cyclohexen-1-one. This finding suggested that quinones may also act as substrates in the oxidative half reaction (oxidation of the enzyme-bound flavin cofactor).
Figure 3. Biochemical and spectroscopic analysis.
(a) Difference titration of TtENR with p-hydroxybenzaldehyde. The determined Kd value was 25 μM. The insert shows the difference ultraviolet–visible spectra. (b) Double-reciprocal plot of initial rate measurements with PhENR as a function of NADH at 20 (filled circles), 30 (open circles), 50 (filled triangles), 90 (open triangles) and 150 (filled squares) μM of 1,4-benzoquinone. (c) Ultraviolet–visible spectra of anaerobic photoreduction of PhENR (upper panel) and TtENR (lower panel), where the peak at 402 nm indicates the presence of the red radical species.
Enzyme kinetics
We tested the reoxidation of reduced FMN in PhENR and TtENR by different, typical OYE substrates and by 1,4-benzoquinone using stopped-flow kinetic measurements under anoxic conditions. Indeed, reoxidation of the reduced cofactor in PhENR by 1,4-benzoquinone was complete within the dead time of the stopped-flow instrument, which means that the oxidative rate kox is >1,000 s−1. 2-Cyclohexen-1-one, on the other hand, reoxidized the enzyme at a rate of kox=18 s−1 comparable to the rate determined for yeast OYE (kox=102 s−1) (ref. 34). Interestingly, although 2-methyl-2-pentenal did not generate noticeable spectral perturbations in titration experiments (see above), we were able to determine the reoxidation rate (kox=48 s−1) and the dissociation constant (Kd=1.26 mM) in stopped-flow experiments.
Yeast OYE is efficiently reduced by NADPH showing biphasic kinetics for the reduction of the flavin cofactor35 with rates of 1.18 and 0.3 s−1. In contrast, reduction of PhENR and TtENR with NADPH showed monophasic kinetics with reductive rates (kred) of 0.06 and 0.15 s−1, respectively. An analysis of the concentration dependence of the reduction yielded dissociation constants (Kd) of 114 μM (PhENR) and 17 μM (TtENR). Interestingly, TtENR but not PhENR was reduced also by NADH with a kred of 0.25 s−1, but with a markedly higher dissociation constant (Kd=300 μM).
Steady-state turnover experiments with TtENR using NADPH and dioxygen as substrates yielded turnover rates of kcat=0.36 s−1 and a KM(NADPH) of 50 μM. This implies that the reductive half reaction is rate-limiting for turnover, because the reoxidation of the cofactor in an air-saturated buffer is two orders of magnitude faster (kox=57 s−1). The presence of 2-cyclohexen-1-one, 2-methyl-2-pentenal, 4-ketoisophorone, 2-methylcyclo-2-pentenone or pHB had no effect on the rate of NADPH or NADH oxidation. However, 1,4-benzoquinone significantly increased the rate of NADPH and NADH oxidation in a concentration-dependent manner, giving rise to a family of intersecting lines (Fig. 3b). This is in contrast to the kinetic behaviour of yeast OYE (OYE2 and OYE3), where the reaction proceeds via a ping-pong bi–bi mechanism involving binary complexes36. Although the particular concentration dependence (Fig. 3b) may be explained by the formation of a ternary complex, based on the structural constraints of the active site (see Fig. 2) it appears more likely to be that the leading substrate (NADPH or NADH) binds to the enzyme in a rapid-equilibrium manner37.
Photoreduction and redox potential
To further investigate the redox properties of the two enzymes, we monitored the photoreduction of the FMN cofactor under anoxic conditions. In both cases the cofactor was fully reduced to the hydroquinone species, accompanied by a loss of the absorbance peaks at 450 and 370 nm (Fig. 3c). Only in the case of TtENR, small traces of the red anionic semiquinone appeared during photoirradation, but this radical species is clearly not stabilized thermodynamically. This behaviour is similar to yeast OYE, where photoreduction also proceeds to the fully reduced species with only a minor occurrence of the red anionic radical38. However, the determination of the redox potentials using NADH or NADPH as reducing agents revealed a major difference to yeast OYE. The measured potentials (PhENR: −325/−320 mV; TtENR: −322/−317 mV, with NADPH and NADH, respectively) were ∼90 mV lower than that of yeast OYE (−230 mV)38. Hence, PhENR and TtENR substantially alter the redox potential of the FMN cofactor (−210 mV in solution) leading to an ∼1,000-fold energetic destabilization of the reduced hydroquinone species, that is, the reduced FMN is a much more powerful reducing agent in both cases.
Biocatalytic experiments
Several typical OYE substrates were tested to assess the substrate scope and the stereospecificity of PhENR and TtENR (Table 1). The activation of the C–C double bond necessary to be accepted as a substrate appears to be higher compared with OYEs, but it follows the same general trend19,39. Therefore, α- and/or β-alkylated ketones (Table 1 6a–9a), which are only weak substrates for other ene-reductases, were not accepted by either of the two enzymes or only converted to a very low extent. On the other hand, 2-methyl-2-pentenal and citral (3a and 4a) as well as compounds bearing two activating groups (4-ketoisophorone 2a and N-phenyl-2-methylsuccinimide 5b) were good substrates. The unsubstituted ketone 2-cyclohexen-1-one (1a) was accepted but at lower conversion rates. In most cases, PhENR turned out to be the more efficient biocatalyst of the two enzymes and we even obtained full conversion in the case of 2-methyl-2-pentenal.
Table 1. Biocatalytic transformations.
As indicated by the mirror symmetry of the active sites of PhENR and TtENR compared with typical OYEs (Fig. 1c), both enzymes indeed showed an inverted stereopreference for interesting substrates (that is, 4-ketoisophorone 2a and N-phenyl-2-methylsuccinimide 5bb), thereby yielding the opposite enantiomer of the reduced product.
Discussion
Our crystallographic, biochemical and biocatalytic studies show that the two enzymes PhENR and TtENR which were discovered through the catalophore approach share—despite a completely different sequence and fold—some salient features of typical OYEs. These common properties include the binding modes of phenolic ligands and substrates, the reduction of activated C–C double bonds and the utilization of NADPH as the preferred reducing agent. However, the data also unveiled some crucial differences, such as an altered substrate spectrum (for example, quinones), a different kinetic mechanism and a drastically more negative redox potential. Interestingly, the measured redox potentials were also significantly more negative than those obtained for typical reductase components of styrene monooxygenases40. This suggests that the putative annotation of TtENR (see above) might have to be reconsidered. Taken together, PhENR and TtENR could be described as NADPH-dependent quinone reductases with significant OYE-like side activities. Such apparent catalytic promiscuity was exactly what we set out to detect with our motif-based catalophore approach.
Regarding the potential for biocatalytic applications, the two enzymes turned out to be less efficient catalysts than typical OYEs (Supplementary Table 4), although high conversions were observed for more activated substrates (Table 1). The lower conversion of 2-cyclohexen-1-one could be explained by the occurrence of two alternate binding modes (one productive and one possibly non-productive), which were observed in the X-ray crystal structures of the complexes of PhENR and TtENR (Fig. 2c,f). A remarkable result was the computationally predicted and experimentally confirmed inverted stereopreference of PhENR and TtENR as in the case of 4-ketoisophorone and N-phenyl-2-methylsuccinimide. Although there are several ways known to switch the enantiopreference of ene-reductases (either by substrate- or enzyme-engineering)41,42,43, up to now the (S)-enantiomers of these compounds were not accessible by asymmetric bioreduction19,39. The lower conversions obtained with PhENR and TtENR are not entirely unexpected, given that the reduction of activated double bonds appears to be only a side activity of these enzymes. However, both enzymes could very well serve as starting points, especially for the development and engineering of stereocomplementary ene-reductases.
In summary, this successful test case exemplifies the workflow of identifying (promiscuous) enzyme activity starting from mechanistic and structural considerations. It is a legitimate question, whether the required presence of a cofactor in this specific case is challenging the general applicability of the method, because the presence of a flavin derivative in an enzyme might already imply oxidoreductase activity. Technically, the flavin only contributed another set of geometric restraints to the search template. Of course, increasing the number of these restraints decreases the number of hits, which have to be evaluated. From a functional point of view, it is true that a flavin present in the active site increases the chances of the particular enzyme being an oxidoreductase. In fact, however, only few flavo-enzymes actually show ene-reductase activity and there are also examples of non-redox flavo-enzymes44.
Search methods using 3D catalytic site constellations are independent of annotations, for example, based on sequence and overall structure similarity. In fact, an increasing number of protein structures are deposited in the PDB, for which no functional annotation is available to date. Another benefit of the method lies in the fact that each hit corresponds to a protein, which has previously been expressed and was stable enough for purification and structure determination. We think that this approach can more generally be used to predict catalytic activities based on 3D structures and to identify novel, non-natural or promiscuous enzyme activities. Thus, it provides an alternative for and complement to existing computational search and design methods, as well as classic wet-lab strategies.
Methods
Database searches
Database searches were carried out in December 2011 against Relibase+ (v2.2)20 and an in-house database of protein structures, both containing roughly 78,000 PDB entries at that time.
For the search in the Relibase+ database the Relibase+ 2D/3D Sketcher was used to create the template with distance cut-offs derived from known OYE structures (Supplementary Table 1 and Supplementary Fig. 5). Because of hard-coded time limits of the Relibase+ (v.2.2) search engine, a subset of structures containing flavin or flavin derivatives was defined before the catalophore search.
A slightly modified version of the YANACONDA script (available as a Supplementary File) was executed in the programme YASARA Structure21 to search for constellations of functional groups in structures stored in our in-house database consisting of protein structures obtained from the PDB. Code required for the database API is removed for easier reading and stand-alone execution. The script was executed against each available database entry on a Linux-based high-performance computing cluster. It starts by loading the first model record of each PDB file into memory for further processing. The distance ranges d1–d4 can be adapted by changing variables d11–d42 in lines 52 to 62. Structures not containing flavin-like cofactors are discarded (lines 65 to 74). Lines 75 to 163 contain the routines for the distance descriptors. If all distance requirements are fulfilled, a scene file containing the identified constellation is produced and saved, as well as a comma-separated values (CSV) file containing the measured distances, residues and residue numbers for further processing. The obtained results were collected using a Structured Query Language (SQL) backend for data storage and analysis. Owing to slightly different database stocks and search algorithms between Relibase+ and the in-house system, PDB entries 3FBS, 3HGO and 2Q3R were not identified as hits in the in-house database system, whereas PDB entry 3KFT was not identified in the Relibase search.
Protein expression and purification
PhENR
The plasmid pSpeedET-Pho containing a gene (GI:14590716) from Pyrococcus horikoshii in a pSpeedET vector was provided by the JCSG (Joint Center for Structural Genomics, CA). The vector pSpeedET is based on a modified pBAD-Thio vector (Invitrogen Life Technologies, Carlsbad, CA) with an additional tobacco-etch-virus (TEV) protease cleavage site. Escherichia coli TOP10F’ cells (Invitrogen Life Technologies) transformed with pSpeedET-Pho were grown in lysogeny broth (LB) medium supplemented with kanamycin (50 μg ml−1) at 37 °C. Expression of recombinant protein was initiated by addition of 0.02% arabinose to A600∼0.8 cultures, and cultivation was continued for 20 h at 25 °C.
TtENR
A gene (GI:75345313) encoding a putative monooxygenase reductase component from Thermus thermophilus was purchased from Epoch Biolabs Inc. (Sugar Land, TX, USA). No codon optimization was performed. The gene was amplified without stop codon by PCR (forward primer: 5′-AATCACATATGAGGTCCTACCGGGCGC-3′, reverse primer: 5′-AATCAAAGCTTCGGCGCAAAGGTCTCCTCG-3′) from the obtained plasmid. The PCR product was ligated into the NdeI/HindIII restriction site of pET26b(+), thereby fusing it to a carboxy-terminal His-tag. The construct was confirmed by sequencing (LGC Genomics, Berlin, Germany) and transformed into the expression host E. coli BL21 (DE3) Gold (Agilent Technologies-Stratagene, La Jolla, CA). The transformed cells were grown in lysogeny broth (LB) medium supplemented with kanamycin (50 μg ml−1) at 37 °C. Expression of recombinant protein was initiated by addition of IPTG (isopropyl-β-D-thiogalactoside; final concentration 0.5 mM) to A600∼0.8 cultures and cultivation was continued for 20 h at 25 °C.
Expression and solubility was monitored by SDS–PAGE. The harvested cells were resuspended in cold buffer A (20 mM sodium phosphate buffer pH 7.4 containing 0.5 M NaCl and 10 mM imidazole) and disrupted by sonication (6 min, 80% duty cycle, 70% output; Branson Sonifier S-250, Danbury, CT, USA). The suspension was centrifuged for an hour at 50,000g. The supernatant was filtered through a 0.45-μm syringe filter and incubated with Ni Sepharose 6 Fast Flow (GE Healthcare, Uppsala, Sweden) for 20 min. The Ni Sepharose was then filled into an empty PD-10 column. After washing with 30 mM imidazole, the bound protein was eluted with 300 mM imidazole in buffer A. Fractions were analysed by SDS–PAGE, pooled and concentrated using Vivaspin 20 Centrifugal Filter Units (10,000 Da cut-off; Sartorius) and desalted on PD-10 columns (GE Healthcare) into 100 mM Tris/HCl, pH 7 (PhENR) or into 20 mM Tris/HCl, pH 7 containing 100 mM NaCl (TtENR).
The last purification step for both enzymes was performed on the Äkta Avant (GE Healthcare) using a prepacked Superdex 16/60 column. The eluent was 10 mM Tris/HCl (pH 7.0) and 100 mM NaCl, with an isocratic flow rate of 1 ml min−1. The protein samples were dialysed against 10 mM Tris/HCl pH 7.5, concentrated to 14 (PhENR) and 7 mg ml−1 (TtENR), and stored at −20 °C.
X-ray crystallography
Initial crystallization trials employing Morpheus and JCSG+ screens (Molecular Dimensions, Newmarket, UK) were set up using an Oryx8 robot (Douglas Instruments, Hungerford, UK). Crystals of PhENR were obtained by vapour diffusion at 20 °C in conditions containing PEG 3350 or 6000 (10–20% w/v) as precipitation agent and different salts (50–200 mM magnesium chloride, potassium formate, sodium formate, ammonium formate, magnesium formate). The crystallization drops were prepared by mixing 0.3–0.5 μl of protein solution (14 mg ml−1) with an equal volume of the reservoir solution.
Initial crystals of TtENR were obtained in crystallization screens using the microbatch method. Several conditions from the Morpheus screen yielded diffraction quality crystals, although growth defects were regularly observed. The best diffracting crystals were obtained with conditions containing 12.5% PEG 1000, 12.5% w/v PEG 3350 and 12.5% v/v 2-methyl-2,4-pentanediol. Diffraction data were collected from crystals obtained in acetate buffer pH 4.5, in 0.1 M sodium HEPES/MOPS buffer system pH 7.5 and in 0.1 M Bicine/Trizma base buffer system pH 8.5. Crystals obtained at pH 7.5 were orthorhombic, the monoclinic crystal used for determining the structure of the 2-cyclo-hexen-1-one complex was obtained at pH 8.5.
Crystals of PhENR and TtENR were soaked for at least 5 min and up to 24 h with potential ligands, including 2-cyclopenten-1-one, 2-cyclohexen-1-one, 2-methyl-2-cyclohexen-1-one, 2-methyl-2-cyclopenten-1-one, 3-methyl-2-cyclopenten-1-one, (E)-2-methylpent-2-enal, 4-ketoisophorone, pHB, 1,4-hydroquinone, NADP+ and NAD+. Co-crystallizations trials with putative ligands were set up using the microbatch method and employing the Morpheus screen. All soaking and co-crystallization experiments were performed under aerobic conditions.
Diffraction data were collected from flash-cooled crystals (at 100 K) at the synchrotrons ESRF (beamlines ID14-4, wavelengths 1.004 and 0.980 Å, and BM-14, wavelength 0.976 Å) and Elletra (beamline 5.2R, wavelength 1.0 Å). The data were indexed and scaled using the programmes XDS45 and SCALA46. Structures of PhENR and TtENR were determined by molecular replacement using structures of both proteins deposited in the PDB entries (2R6V and 1USC) as search models. Molecular replacement was performed using the programme Phaser47 followed by rounds of refinement using Refmac5 (ref. 48) interspersed with manual model rebuilding in Coot49. The refinement was only continued if clear difference electron density close to the FMN cofactor indicative of a bound ligand was observed after rigid body refinement. In all cases, Rfree-values50 were computed from 5% randomly chosen reflections not used for the refinement. Twin refinement, as included in Refmac5, had to be applied for the refinement of the structure of TtENR in complex with 2-cyclohexen-1-one (twinning operator h, -k, -l; twinning fraction 0.48). The stereochemical quality of the models was checked using the programme Molprobity51. In all structures, more than 97% of the residues had their main-chain torsion angles in the favoured regions of the Ramachandran plot and none in the disallowed regions. Data collection and refinement statistics are summarized in Supplementary Table 3.
Spectrophotometric methods
To determine the extinction coefficient of enzyme-bound FMN, SDS was added to a concentration of 2% and the solution was incubated at 80 °C for 5 min to release the cofactor. Ultraviolet–visible absorption spectra were recorded before and after denaturation of the respective enzyme with a photometer (model Specord 200 plus from Analytik Jena AS, Jena, Germany). All measurements were performed in 20 mM Tris-HCl, pH 8.0, using 1 cm quartz cuvettes.
Difference titrations were carried out at 25 °C in tandem cuvettes by the addition of the putative binding partner [2-cyclohexen-1-one, 2-methyl-2-pentenal, 4-ketoisophorone (2,6,6-trimethylcyclohex-2-ene-1,4-dione), 2-methylcyclo-2-penten-1-one, pHB, 1,4-benzoquinone] to a 60-μM enzyme solution in the sample cell and to buffer (50 mM Tris-HCl, pH 7.5) in the reference cell.
For photoreduction, the protein was diluted in 50 mM Tris-HCl, pH 7.5, containing 1 mM EDTA and catalytic amounts of 5-deazaflavin. Quartz cuvettes were rendered anaerobic in the nitrogen atmosphere of a glove box. Reduction of the flavin cofactor was achieved by illumination of the sample with a 250-W halogen lamp. Spectra from 300 to 600 nm were recorded at 25 °C until no further changes were observed. Next, the cuvette was opened to air and the absorbance spectrum was recorded.
Steady state and stopped-flow kinetics
Steady-state turnover of the enzymes was determined by monitoring the oxidation of NADPH and NADH in the presence of molecular oxygen as substrate. Initial velocities were measured by monitoring the decrease of absorbance at 340 nm. All reactions were carried out in 50 mM potassium phosphate (pH 7.0) at 75 °C. The reaction mixture contained 2–5 μM of the respective enzyme, and NADPH or NADH in a concentration range of 50–350 μM. The enzyme activity was calculated using an extinction coefficient of 6,220 M−1 cm−1 for NADPH and NADH. Steady-state turnover rates were also measured by monitoring the oxidation of NADPH or NADH in the presence of 1,4-benzoquinone (20–100 μM) and other putative substrates (2-cyclohexen-1-one, 2-methyl-2-pentenal, 4-ketoisophorone, 2-methyl-2-cyclopenten-1-one, pHB).
Stopped flow measurements were carried out with a Hi-Tech (SF-61DX2) stopped flow device (TgK Scientific Limited, Bradford-on-Avon, UK) positioned in a glove box from Belle Technology (Weymouth, UK) at 37 °C. Two reactant solutions were joined in single-mixing mode. FMN oxidation and reduction were measured by monitoring changes in A458 with a KinetaScanT diode array detector (MG-6560, TgK Scientific Limited). Initial rates were calculated by fitting the curves with Specfit 32 (Spectrum Software Associates, Chapel Hill, NC, USA) using a function of two exponentials.
The reductive half reaction was carried out by mixing 40 μM enzyme with NADPH or NADH at a concentration of 0.4–10.0 mM in 100 mM Tris-HCl (pH 7.4). Rate constants for the oxidative half reaction were determined by reduction of 40 μM enzyme in 50 mM potassium phosphate (pH 7.0) by the addition of sodium dithionite. Excess sodium dithionite was removed using a Sephadex PD-10 desalting column inside the glove box. After mixing with the respective substrate at various concentrations in the range of 50 μM to 50 mM, or with air-saturated buffer, the reoxidation of the FMN cofactor was monitored. The used buffer was prepared by flushing with nitrogen followed by incubation in the glove box. The enzyme and substrate solutions were prepared by incubation in the anoxic glove box and dilution with the described buffer.
Redox potential measurement
Redox potentials of PhENR and TtENR were measured by titration of the oxidized enzyme in an anaerobic cuvette with NADPH or NADH (end concentration between 20 and 120 μM) as described previously34. The solutions were supplemented with 5 μM methylviologen. The titration was recorded by following the ultraviolet–visible spectrum. At the chemical equilibrium of the reaction, the redox potential of NADP+/NADPH equals that of the enzyme. The relative amount of reduced and oxidized enzyme was calculated from the absorbance at 458 nm (TtENR) or 453 nm (PhENR), and the relative amount of NADP+ and NADPH was calculated from the absorbance at 337 nm (TtENR) or 330 nm (PhENR), the wavelength where the oxidized and reduced form of each enzyme have the same extinction coefficient. The redox potential of the flavin cofactor was calculated according to the Nernst equation.
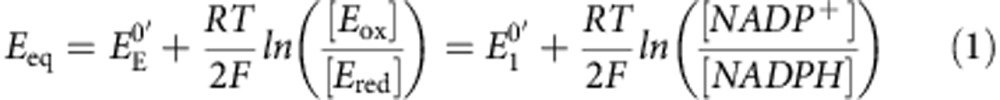 |
In this equation, Eeq represents the redox potential of both couples at the equilibrium, 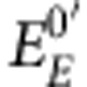 refers to the midpoint redox potential of the enzyme, and
refers to the midpoint redox potential of the enzyme, and 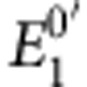 refers to the redox potential of NADP+/NADPH, the value of which was taken as −320 mV. The possibility that the enzyme may have different affinities for NADPH and NADP+ might influence the measured value of the redox potential. Therefore, NADH was also used instead of NADPH for titration.
refers to the redox potential of NADP+/NADPH, the value of which was taken as −320 mV. The possibility that the enzyme may have different affinities for NADPH and NADP+ might influence the measured value of the redox potential. Therefore, NADH was also used instead of NADPH for titration.
Substrates and reference compounds
2-Cyclohexen-1-one 1a, cyclohexanone 1b, (E)-2-methylpent-2-enal 3a, rac-2-methylpentanal rac-3b, citral (mixture of E/Z-isomers) 4a, rac-citronellal rac-4b, (S)-citronellal (S)-4b, N-phenyl-2-methylmaleimide 5a, 3-methyl-2-cyclohexen-1-one 7a, rac-3-methylcyclohexan-1-one rac-7b, 2-methyl-2-cyclopenten-1-one 8a, rac-2-methylcyclopentan-1-one rac-8b and rac-3-methylcyclopentan-1-one rac-9b were purchased from Sigma Aldrich (St Louis, MO, USA). 3-Methyl-2-cyclopenten-1-one 9a was purchased from Fluka (Buchs, Switzerland). 4-Ketoisophorone 2a was purchased from ABCR (Karlsruhe, Germany). rac-2-Methylcyclohexan-1-one rac-6b was purchased from Alfa Aesar (Karlsruhe, Germany). (R)-Levodione (R)-2b was obtained by bioreduction of 4-ketoisophorone using baker’s yeast (62% e.e.); rac-N-phenyl-2-methylsuccinimide rac-5b and 2-methyl-2-cyclohexen-1-one 6a were synthesized as previously reported52. rac-2,3-Epoxy-1-cyclohexanone rac-1c and rac-2,3-epoxy-3,5,5-trimethyl-1,4-cyclohexanedione rac-2c were synthesized as previously reported53. Chemical formulas of substrates and products are shown in Supplementary Fig. 6.
Biocatalytic transformations
In a first step, PhENR and TtENR were screened for the reduction of the C=C bond of 1a, 2a and 3a by adding an aliquot of the enzyme (100 μg ml−1) to a 1.5-ml microcentrifuge tube containing Tris-HCl buffer (0.8 ml, 50 mM, pH 7, 8 or 9), the substrate (10 mM) and the reducing agent (NADH or NADPH, 15 mM). In case of 1a and 2a, the major observed reaction was a Weitz–Scheffer epoxidation (Supplementary Fig. 7) most likely to be due to the FMN-hydroperoxy intermediate or possibly due to free H2O2, which are produced by the reaction of the reduced cofactor with molecular oxygen53 (1a: TtENR: <2% 1b, 9–28% 1c; PhENR: <3% 1b, 4–7% 1c; 2a: TtENR: <0% 2b, 29–83% 2c, PhENR: <0% 2b, 7–29% 2c). Compound 3a showed no epoxidation (Supplementary Table 5, entries 1–3) and was therefore selected for further optimization of the reaction conditions.
In addition, the optimization was carried out in the absence of oxygen (Supplementary Table 5, entries 4–15) to suppress any NAD(P)H oxidase reaction. In procedure, an aliquot of the enzyme (100 μg ml−1) was added to a 2-ml screw top glass vial containing degassed buffer (0.8 ml, 50 mM either Tris-HCl buffer, pH 7 or 8; or phosphate buffer, pH 6.2, 7, 7.5 or 8), the substrate (10 mM) and the reducing agent (15 mM). The vial was sealed with a Teflon septum and a screw cap with a hole and flushed with 5 ml Ar. Optimal conditions were 50 mM phosphate buffer for TtENR and 50 mM Tris-HCl buffer for PhENR both at pH 7, as well as NADPH as reducing agent for both enzymes.
In both procedures, the mixture was shaken for 24 h at 30 °C and 120 r.p.m. The products were extracted with EtOAc (2 × 0.5 ml) and the combined organic phases were dried over Na2SO4. Achiral GC was used to determine conversions and chiral GC or HPLC were used to determine the enantiomeric excess. In case of 5a, the substrate was dissolved in DMF (1% final DMF conc.) for all biotransformations. The reaction products were analysed using achiral GC, as well as chiral GC and HPLC. For details see Supplementary Methods.
Author contributions
G.S., C.C.G. and K.G. planned and designed the project; G.S., C.C.G. and K.G. generated the catalophore search templates, built up an in-house structural database, developed software tools, performed the database searches and analysed the data; K.S., O.S., M.O. and H.S. expressed and purified the enzymes; A.B., T.P.K. and P.M. performed the biochemical characterization of the enzymes; C.W. and K.F. performed the biocatalytic transformations. T.P.K. and A.L. determined the crystal structures. All co-authors discussed the results and contributed to the manuscript. G.S., C.C.G. and K.G. wrote the paper.
Additional information
Accession codes: Atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 3ZOC, 3ZOD, 3ZOE, 3ZOF, 3ZOG and 3ZOH.
How to cite this article: Steinkellner, G. et al. Identification of promiscuous ene-reductase activity by mining structural databases using active site constellations. Nat. Commun. 5:4150 doi: 10.1038/ncomms5150 (2014).
Supplementary Material
Supplementary Figures 1-7, Supplementary Tables 1-5, Supplementary Methods and Supplementary References
Acknowledgments
This work has been supported by the Federal Ministry of Science, Research and Economy (BMWFW), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol and ZIT—Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG, as well as by the Austrian Science Funds (FWF) through projects L428 (to K.G.) and P22722 (to K.F.). We acknowledge the ESRF (Grenoble, France), SLS (Villigen, Switzerland), Elletra (Trieste, Italy) and DESY (Hamburg, Germany) for providing synchrotron radiation facilities at the respective storage rings and for the support of the beamline staff during diffraction data collection.
References
- Huisman G. W. & Collier S. J. On the development of new biocatalytic processes for practical pharmaceutical synthesis. Curr. Opin. Chem. Biol. 17, 284–292 (2013). [DOI] [PubMed] [Google Scholar]
- McGrath B. M. & Walsh G. Directory of Therapeutic Enzymes CRC Press (2006). [Google Scholar]
- Turner N. J. Directed evolution drives the next generation of biocatalysts. Nat. Chem. Biol. 5, 567–573 (2009). [DOI] [PubMed] [Google Scholar]
- Furnham N. et al. The Catalytic Site Atlas 2.0: Cataloging catalytic sites and residues identified in enzymes. Nucleic Acids Res. 42, D485–D489 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E. & Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- Röthlisberger D. et al. Kemp elimination catalysts by computational enzyme design. Nature 453, 190–195 (2008). [DOI] [PubMed] [Google Scholar]
- Jiang L. et al. De novo computational design of retro-aldol enzymes. Science 319, 1387–1391 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel J. B. et al. Computational design of an enzyme catalyst for a stereoselective bimolecular diels-alder reaction. Science 329, 309–313 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss G., Çelebi-Ölçüm N., Moretti R., Baker D. & Houk K. N. Computational enzyme design. Angew. Chem. Int. Ed. 52, 5700–5725 (2013). [DOI] [PubMed] [Google Scholar]
- Kries H., Blomberg R. & Hilvert D. De novo enzymes by computational design. Curr. Opin. Chem. Biol. 17, 221–228 (2013). [DOI] [PubMed] [Google Scholar]
- Richter F., Leaver-Fay A., Khare S. D., Bjelic S. & Baker D. De novo enzyme design using Rosetta3. PLoS One 6, e19230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornscheuer U. T. & Kazlauskas R. J. Catalytic promiscuity in biocatalysis: Using old enzymes to form new bonds and follow new pathways. Angew. Chem. Int. Ed. 43, 6032–6040 (2004). [DOI] [PubMed] [Google Scholar]
- Branneby C. et al. Carbon-carbon bonds by hydrolytic enzymes. J. Am. Chem. Soc. 125, 874–875 (2003). [DOI] [PubMed] [Google Scholar]
- Purkarthofer T. et al. A biocatalytic Henry reaction—The hydroxynitrile lyase from Hevea brasiliensis also catalyzes nitroaldol reactions. Angew. Chem. Int. Ed. 45, 3454–3456 (2006). [DOI] [PubMed] [Google Scholar]
- O’Brien P. J. & Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6, R91–R105 (1999). [DOI] [PubMed] [Google Scholar]
- Wallace A. C., Borkakoti N. & Thornton J. M. Tess: A geometric hashing algorithm for deriving 3D coordinate templates for searching structural databases. Application to enzyme active sites. Protein Sci. 6, 2308–2323 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker J. A. & Thornton J. M. An algorithm for constraint-based structural template matching: Application to 3D templates with statistical analysis. Bioinformatics 19, 1644–1649 (2003). [DOI] [PubMed] [Google Scholar]
- Tantillo D. J., Chen J. & Houk K. N. Theozymes and compuzymes: Theoretical models for biological catalysis. Curr. Opin. Chem. Biol. 2, 743–750 (1998). [DOI] [PubMed] [Google Scholar]
- Stuermer R., Hauer B., Hall M. & Faber K. Asymmetric bioreduction of activated C:C bonds using enoate reductases from the old yellow enzyme family. Curr. Opin. Chem. Biol. 11, 203–213 (2007). [DOI] [PubMed] [Google Scholar]
- Bergner A., Günther J., Hendlich M., Klebe G. & Verdonk M. Use of relibase for retrieving complex three-dimensional interaction patterns including crystallographic packing effects. Biopolymers 61, 99–110 (2001). [DOI] [PubMed] [Google Scholar]
- Krieger E., Koraimann G. & Vriend G. Increasing the precision of comparative models with YASARA NOVA—A self-parameterizing force field. Proteins 47, 393–402 (2002). [DOI] [PubMed] [Google Scholar]
- Oberdorfer G., Steinkellner G., Stueckler C., Faber K. & Gruber K. Stereopreferences of Old Yellow Enzymes: Structure correlations and sequence patterns in enoate reductases. ChemCatChem 3, 1562–1566 (2011). [Google Scholar]
- Murzin A. G., Brenner S. E., Hubbard T. & Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247, 536–540 (1995). [DOI] [PubMed] [Google Scholar]
- Andreeva A. et al. Data growth and its impact on the SCOP database: New developments. Nucleic Acids Res. 36, D419–D425 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford P. F., Wagner U. G., Jiang Y., Faber K. & Kazlauskas R. J. Enantiocomplementary enzymes: Classification, molecular basis for their enantiopreference, and prospects for mirror-image biotransformations. Angew. Chem. Int. Ed. 47, 8782–8793 (2008). [DOI] [PubMed] [Google Scholar]
- Krissinel E. & Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007). [DOI] [PubMed] [Google Scholar]
- Matthews R. G., Massey V. & Sweeley C. C. Identification of p-hydroxybenzaldehyde as the ligand in the green form of old yellow enzyme. J. Biol. Chem. 250, 9294–9298 (1975). [PubMed] [Google Scholar]
- Abramovitz A. S. & Massey V. Interaction of phenols with Old Yellow Enzyme. Physical evidence for charge-transfer complexes. J. Biol. Chem. 251, 5327–5336 (1976). [PubMed] [Google Scholar]
- Fox K. M. & Karplus P. A. Old yellow enzyme at 2 Å resolution: Overall structure, ligand binding, and comparison with related flavoproteins. Structure 2, 1089–1105 (1994). [PubMed] [Google Scholar]
- Warburg O. & Christian W. Über das gelbe Ferment und seine Wirkungen. Biochem. Z. 266, 377–341 (1933). [Google Scholar]
- Gudipati V., Koch K., Lienhart W. D. & Macheroux P. The flavoproteome of the yeast Saccharomyces cerevisiae. BBA Proteins Proteom 1844, 535–544 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitz A. S. & Massey V. Purification of intact Old Yellow Enzyme using an affinity matrix for the sole chromatographic step. J. Biol. Chem. 251, 5321–5326 (1976). [PubMed] [Google Scholar]
- Fitzpatrick T. B., Amrhein N. & Macheroux P. Characterization of YqjM, an Old Yellow Enzyme homolog from Bacillus subtilis involved in the oxidative stress response. J. Biol. Chem. 278, 19891–19897 (2003). [DOI] [PubMed] [Google Scholar]
- Xu D., Kohli R. M. & Massey V. The role of threonine 37 in flavin reactivity of the old yellow enzyme. Proc. Natl Acad. Sci. USA 96, 3556–3561 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey V. & Schopfer L. M. Reactivity of Old Yellow Enzyme with alpha-NADPH and other pyridine nucleotide derivatives. J. Biol. Chem. 261, 1215–1222 (1986). [PubMed] [Google Scholar]
- Niino Y. S., Chakraborty S., Brown B. J. & Massey V. A new Old Yellow Enzyme of Saccharomyces cerevisiae. J. Biol. Chem. 270, 1983–1991 (1995). [DOI] [PubMed] [Google Scholar]
- Taylor K. B. Enzyme Kinetics and Mechanisms Kluwer Academic Publishers (2002). [Google Scholar]
- Stewart R. C. & Massey V. Potentiometric studies of native and flavin-substituted old yellow enzyme. J. Biol. Chem. 260, 13639–13647 (1985). [PubMed] [Google Scholar]
- Toogood H. S., Gardiner J. M. & Scrutton N. S. Biocatalytic reductions and chemical versatility of the Old Yellow Enzyme family of flavoprotein oxidoreductases. ChemCatChem 2, 892–914 (2010). [Google Scholar]
- Morrison E., Kantz A., Gassner G. T. & Sazinsky M. H. Structure and mechanism of styrene monooxygenase reductase: New insight into the FAD-transfer reaction. Biochemistry 52, 6063–6075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhi S. K., Bougioukou D. J. & Stewart J. D. Site-saturation mutagenesis of tryptophan 116 of Saccharomyces pastorianus old yellow enzyme uncovers stereocomplementary variants. J. Am. Chem. Soc. 131, 3271–3280 (2009). [DOI] [PubMed] [Google Scholar]
- Bougioukou J. D., Kille S., Taglieber A. & Reetz M. Directed evolution of an enantioselective enoate-reductase: Testing the utility of iterative saturation mutagenesis. Adv. Synth. Catal. 351, 3287–3305 (2009). [Google Scholar]
- Oberdorfer G., Gruber K., Faber K. & Hall M. Stereocontrol strategies in the asymmetric bioreduction of alkenes. Synlett 23, 1857–1864 (2012). [Google Scholar]
- Macheroux P., Kappes B. & Ealick S. E. Flavogenomics—A genomic and structural view of flavin-dependent proteins. FEBS J. 278, 2625–2634 (2011). [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006). [DOI] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N., Vagin A. A. & Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleywegt G. J. & Brunger A. T. Checking your imagination—Applications of the free R-Value. Structure 4, 897–904 (1996). [DOI] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. et al. Asymmetric bioreduction of C=C bonds using enoate reductases OPR1, OPR3 and YqjM: Enzyme-based stereocontrol. Adv. Synth. Catal. 350, 411–418 (2008). [Google Scholar]
- Mueller N. J., Stueckler C., Hall M., Macheroux P. & Faber K. Epoxidation of conjugated C=C-bonds and sulfur-oxidation of thioethers mediated by NADH:FMN-dependent oxidoreductases. Org. Biomol. Chem. 7, 1115–1119 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-7, Supplementary Tables 1-5, Supplementary Methods and Supplementary References