Abstract
In breast cancer (BrCa), overexpression of the nuclear co-activator NCOA1 (SRC-1) is associated with disease recurrence and resistance to endocrine therapy. To examine the impact of NCOA1 overexpression on morphogenesis and carcinogenesis in the mammary gland (MG), we generated MMTV-hNCOA1 transgenic [Tg(NCOA1)] mice. In the context of two distinct transgenic models of breast cancer, NCOA1 overexpression did not affect the morphology or tumor forming capability of MG epithelial cells. However, NCOA1 overexpression increased the number of circulating BrCa cells and the efficiency of lung metastasis. Mechanistic investigations showed that NCOA1 and c-Fos were recruited to a functional AP-1 site in the macrophage attractant CSF1 promoter, directly upregulating CSF1 expression to enhance macrophage recruitment and metastasis. Conversely, silencing NCOA1 reduced CSF1 expression and decreased macrophage recruitment and BrCa cell metastasis. In a cohort of 453 human breast tumors, NCOA1 and CSF1 levels correlated positively with disease recurrence, higher tumor grade and poor prognosis. Together, our results define an NCOA1/AP-1/CSF1 regulatory axis that promotes BrCa metastasis, offering a novel therapeutic target for impeding this process.
Introduction
NCOA1 is a member of the p160 SRC family that also contains NCOA2 (GRIP1/TIF2/SRC-2) and NCOA3 (AIB1/ACTR/SRC-3) (1). These NCOAs interact with nuclear hormone receptors and other transcription factors (TFs) to facilitate the assembly of transcriptional protein complexes for chromatin remodeling and activation of gene expression (1). Since these coactivators are strong boosters of gene expression, these proteins are usually unstable and present at low concentrations in normal cells (2) and changes in either their concentration or activity significantly impact their target gene expression (3). Accordingly, overexpression of these coactivators is often linked with human diseases such as cancer. Specifically, NCOA3 is amplified and overexpressed in subsets of breast, prostate, ovarian, hepatocellular and pancreatic cancers (4–8). Forced overexpression of NCOA3 in the mouse mammary gland (MG) epithelium induces tumorigenesis, while knockout of NCOA3 suppresses oncogene- or chemical carcinogen-induced MG and prostate tumorigenesis (9–13). Furthermore, NCOA2 is a commonly amplified oncogene that is associated with an enhanced androgen receptor function in prostate cancer (14). Moreover, NCOA1 is also overexpressed in a subset of breast tumors that express HER2 and give poor prognosis (15). However, the in vivo role of NCOA1 overexpression in breast cancer (BrCa) remains to be defined.
Recent studies have suggested that NCOA1 is required for BrCa metastasis. Knockout of Ncoa1 significantly inhibits mammary tumor metastasis to the lung in transgenic MMTV-polyoma middle T [Tg(PyMT)] or Tg(Neu) BrCa mouse models (16, 17). Knockdown of NCOA1 in human BrCa cells also suppresses their invasion and metastasis (18–20). At the molecular level, NCOA1 serves as a coactivator for different TFs to upregulate the expression of several genes that promote the epithelial-mesenchymal transition (EMT), migration, invasion and metastasis of BrCa cells. The known NCOA1-regulated genes include Twist1, integrin α5, SDF1, HER2 and c-Myc (16, 18, 19, 21–23). Since NCOA1 is a critical coactivator that may control BrCa metastasis through interaction with multiple TFs important for the metastatic process, further characterization of the TF partners of NCOA1 and their target genes will aid in elucidating the regulatory gene networks of cancer metastasis and identifying potential targets for inhibiting cancer metastasis.
CSF1 is expressed in multiple cell types such as osteoblasts, uterine epithelial cells and different types of cancer cells, and it plays important roles in organ development and physiological functions such as MG and placental development (24–27). CSF1 regulates the proliferation, differentiation and survival of mononuclear phagocytic cells and their bone marrow progenitors (26). CSF1 secreted from BrCa cells recruits cancer-associated macrophages (CAMs) to promote metastasis (28). CSF1 is overexpressed in 70% of breast tumors and its overexpression is associated with macrophage infiltration, tumor cell invasion, advanced tumor grades and poor prognosis (28, 29). Knockout of CSF1 inhibits lung metastasis from MG tumors, while transgenic expression of CSF1 in both CSF1 knockout and WT mammary epithelium restores or enhances macrophage recruitment and lung metastasis in the Tg(PyMT) mouse model (30). A paracrine loop between tumor cells and macrophages has been shown to be required for BrCa cell migration (31). In this regulatory loop, cancer cells secrete CSF1 to recruit and stimulate macrophages. In turn, macrophages secrete epidermal growth factor (EGF) to stimulate tumor cells to migrate and metastasize. However, the TFs and coactivators that regulate CSF1 expression in BrCa cells are still unknown.
In this study, we generated both BrCa mouse models and cell lines with overexpression or knockout/knockdown of NCOA1 or CSF1 to investigate whether NCOA1 directly regulates CSF1 expression to promote BrCa metastasis.
Materials and Methods
Transgenic mice
The MMTV-hNCOA1 transgene was constructed (Fig 1A). Tg(NCOA1) mice were generated as described in Supplementary Methods. Tg(NCOA1) mice were crossed with Tg(Neu) mice (32) and Tg(TVA) mice (33) respectively to generate female Tg(Neu), Tg(NCOA1)×Tg(Neu), Tg(TVA) and Tg(NCOA1)×Tg(TVA) mice for experiments. Mouse genotypes were determined by PCR using transgene-specific primers listed in Supplementary Table S1. All mice have a FVB strain background. Animal protocols were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine.
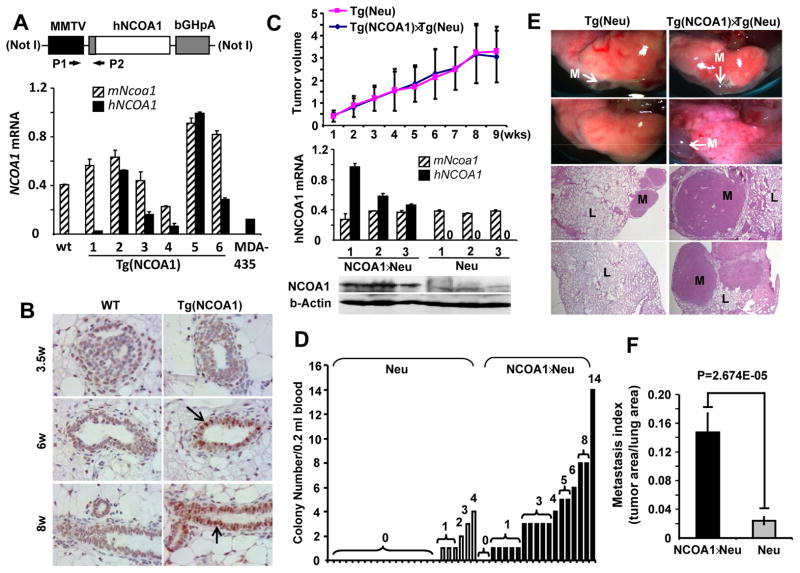
Fig. 1. Generation of Tg(NCOA1) mice and mammary tumor metastasis in Tg(Neu) (Neu) and Tg(NCOA1)×Tg(Neu) (NCOA1×Neu) mice.
A. The MMTV-hNCOA1 transgene and qPCR analysis of hNCOA1 mRNA expression. RNA was extracted from MGs of WT and Tg(NCOA1) mice and MDA-MB-435 cells. qPCR was performed using hNCOA1 cDNA-specific P1/P2 primers. Human (hNCOA1) and mouse (mNcoa1) are indicated. bGHpA, bovine growth hormone polyadenylation sequence. B. IHC was performed using an antibody that detects both mNcoa1 and hNCOA1. Arrows indicate NCOA1-overexpressing mammary epithelial cells. MG sections were prepared from female WT and Tg(NCOA1) mice with indicated weeks of ages. C. Tumor growth curves were obtained from 27 Tg(Neu) and 26 Tg(NCOA1)×Tg(Neu) mice by measuring tumor volumes (cm3). Tumor specimens were collected at 9 weeks after mammary tumors were palpated and subjected to Western blotting using a NCOA1 antibody that recognizes both hNCOA1 and mNcoa1. β-actin served as a loading control. Three tumors from each genotype group were assayed. D. The number of tumor cell colonies derived from circulating tumor cells in the blood samples of Tg(Neu) and Tg(NCOA1)×Tg(Neu) mice that had developed mammary tumors for 9 weeks. Student’s T test indicates a significant difference (p=0.001) between the two groups. E. Metastatic tumor foci on the lung surfaces and H&E-stained lung (L) sections with metastatic (M) tumors in Tg(Neu) and Tg(NCOA1)×Tg(Neu) mice. F. Lung metastasis indexes are presented as the ratio of tumor area to the total tumor and lung area in Tg(Neu) (n=15) and Tg(NCOA1)×Tg(Neu) (n=10) mice.
Cell culture
MDA-MB-231 and MCF-7 cell lines were obtained from the Tissue Culture Core in Baylor College of Medicine. The two Ncoa1 Knockout Tg(PyMT) (PyMT×Ncoa1-K1/K2) and the two Ncoa1 WT Tg(PyMT) (PyMT×Ncoa1-W1/W2) cell lines were developed from mouse MG tumors as previously described (19). The MDA-231-LM3.3 BrCa cell line was developed from a lung metastatic focus derived from a xenograft tumor of MDA-MB-231-LM2 cells (34) in the MG of a SCID mouse. These cells were authenticated by STR DNA fingerprinting in The University of Texas MD Anderson Cancer Center Characterized Cell Line Core. Cells were cultured as described previously (19).
Promoter-reporter constructs, cell transfection and luciferase assay
Two DNA fragments spanning base pairs −1956–284 (F1) or −790–284 (F2) of the human CSF1 promoter region were amplified by high fidelity PCR and subcloned into pGL3 plasmid with a luciferase reporter. The primer sequences used for PCR are listed in Supplementary Table S1. Another set of the mutant CSF1 promoter-reporter plasmids were constructed based on the pGL3-F2 plasmid that contains the F2 DNA fragment. In this set, one, two or three AP-1-binding sites at bp −614, −300 or −106 positions were deleted individually or in combination by PCR-assisted mutagenesis. HeLa cells in 24-well plates were co-transfected with one of the pGL3-based promoter-reporter plasmids, pCR3.1-NCOA1 plasmid, and one of the plasmids for PEA3, TCF-4, NF-κB and AP-1 (c-Jun/c-Fos) expression as described (18, 19). Mock plasmids were used to compensate for total DNA in each transfection. After 48 hours, cells were lysed for measuring luciferase activity, which were then normalized to total protein used in each assay as described (19, 35).
Macrophage recruitment and macrophage-induced tumor cell invasion assays in a transwell co-culture system
Tg(Neu) and Tg(NCOA1)×Tg(Neu) tumor cells isolated from individual mouse MG tumors were cultured in 24-well plate and transfected with non-targeting siRNAs (control) or siRNAs targeting NCOA1 or CSF1. The transfected cells were cultured in DMEM medium with 10% serum for 48 hours and then changed to the serum-free DMEM medium. Matrigel-coated invasion chambers were mounted to the top of the 24-well plate and 5×104 of RAW-264.7 mouse macrophages suspended in the same serum-free medium were loaded to each upper chamber. After culturing for 20 hours, the non-invading macrophages above the transwell membrane were removed, and the macrophages adhered to the bottom surface of this membrane were fixed, stained and counted as described previously (18, 19, 35). The number of recruited macrophages was normalized to the number of total tumor cells in the lower chamber.
Detection of NCOA1 and CSF1 proteins by IHC in human breast tumors
The tissue microarrays (TMAs) were prepared from archived human breast tumor specimens (n=560) of a patient cohort as described [(15, 35) and Supplementary Methods]. IHC was performed using NCOA1 and CSF1 antibodies as described in Supplementary Methods. The immunostaining intensities for NCOA1 and CSF1 were independently scored by a pathologist (Z.Y.) and an investigator (L.Q.) according to the Allred scoring system (36), and the average score was used for each sample. Pearson Chi-square test was used for categorical variables to compare two proportions. Kaplan Meier estimates of recurrence-free functions were computed and Logrank test was applied to compare the difference of the recurrence-free curves among different groups. A p-value of less than 0.05 was considered to be statistically significant.
Other methods
Examination of MG morphology, epithelial proliferation, MG tumor growth and lung metastasis were performed as described previously (10, 11, 16, 33, 37, 38). IHC, Western blotting and ELISA were performed as described previously (16, 18, 19). Quantitative real time RT-PCR (qPCR), knockdown and expression of NCOA1, and chromatin immunoprecipitation (ChIP) assay were also performed as described previously (18, 19). Oligonucleotide primers used in qPCR and ChIP assays are listed in Supplementary Table S1. Please refer to Supplementary Information for detailed description of these methods.
Results
Generation of Tg(NCOA1) mice for NCOA1 overexpression in MECs
To overexpress hNCOA1 in the mouse MECs, we constructed the MMTV-hNCOA1 transgene (Fig. 1A). Microinjection of the transgene DNA into the fertilized oocytes generated 41 pups, 13 of which harbored the transgene. From these founders with the transgene, we developed 6 Tg(NCOA1) transgenic lines that expressed different levels of the transgene as measured by hNCOA1-specific qPCR. Two of the lines expressed higher hNCOA1, which was similar to the level of endogenous mNcoa1 mRNA (Fig. 1A). The different levels of mNcoa1 expression in different mice might be due to the different phases of estrus cycle. Since total MG RNA was used in the assay, hNCOA1 was only expressed in MECs, and mNcoa1 was expressed in both MECs and other MG cells, the hNCOA1 mRNA should be much higher than mNcoa1 mRNA in MECs of these Tg(NCOA1) mice. Indeed, IHC revealed a significant increase of NCOA1 protein in MECs of Tg(NCOA1) mice as examined at ages of 6 and 8 weeks compared with age-matched WT mice (Fig. 1B). Based on these results, we kept two transgenic lines that displayed higher levels of hNCOA1 expression for further experiments, and these two lines showed similar features in all experiments described below.
WT and Tg(NCOA1) mice showed no significant changes in MG ductal morphogenesis, MEC proliferation index, and the number of macrophages around MG ducts when examined at 3.5, 6 and 8 weeks of ages (Supplementary Fig. S1A–E). Tg(NCOA1) mice also exhibited normal lactation function and developed no MG tumors during the examining period from newborn to 14-month-old. These results demonstrate that NCOA1 is successfully overexpressed in MECs of Tg(NCOA1) mice, and this overexpression does not cause any obvious abnormal phenotypes.
NCOA1 overexpression promotes BrCa metastasis in Tg(Neu) and Tg(TVA)+RCAS-PyMT mouse models
Palpable solid MG tumors comparably developed in Tg(Neu) and Tg(NCOA1)×Tg(Neu) mice during 6–12 months of ages and showed similar growth speeds. These tumors exhibited histopathological morphologies of poorly differentiated adenocarcinomas (data not shown). Total NCOA1 mRNA and protein were increased in Tg(NCOA1)×Tg(Neu) tumors versus Tg(Neu) tumors as measured by qPCR and Western blot (Fig. 1C). Interestingly, after examining the circulating tumor cells by culturing blood samples collected from mice borne mammary tumors for 9 weeks, we found that both the frequency and number of tumor cell colonies formed from the blood samples of Tg(NCOA1)×Tg(Neu) mice were much higher than those from Tg(Neu) mice (Fig. 1D). The metastatic foci in the lung were also more common and greater in area in Tg(NCOA1)×Tg(Neu) mice when compared with Tg(Neu) mice. Statistical analysis of tumor area in lungs revealed a significant increase of metastatic index in Tg(NCOA1)×Tg(Neu) mice compared with that of Tg(Neu) mice (Fig. 1E and F).
We also induced MG tumorigenesis in Tg(TVA) and Tg(NCOA1)×Tg(TVA) mice by intra-ductal injection of RCAS-PyMT avian virus as described (33). In these mice, the TVA receptor for RCAS virus is expressed in MECs, so that the injected RCAS-PyMT virus specifically infects some of these TVA-expressing cells to express PyMT for tumorigenic transformation (33). Again, no significant differences in MG tumorigenesis and tumor growth were observed in Tg(TVA)+RCAS-PyMT and Tg(NCOA1)×Tg(TVA)+RCAS-PyMT mice (Supplementary Fig. S2A and data not shown). However, the number of circulating tumor cells, the tumor foci in the lung and the metastatic index were significantly increased in Tg(NCOA1)×Tg(TVA)+RCAS-PyMT mice versus Tg(TVA)+RCAS-PyMT mice (Supplementary Fig. S2B–D). Together, the results from both Tg(NCOA1)×Tg(Neu) and Tg(NCOA1)×Tg(TVA)+RCAS-PyMT mouse models indicate that NCOA1 overexpression significantly enhances spontaneous BrCa metastasis.
NCOA1 overexpression promotes CSF1 expression in MG tumor cells
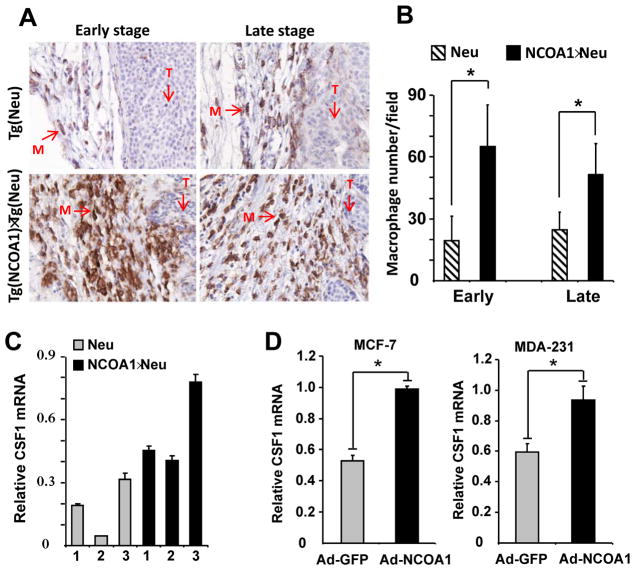
Macrophage recruitment to the primary tumor site is important for tumor progression to advanced malignant stages (28–31). To examine the effect of NCOA1 overexpression on the recruitment of macrophages, immunostaining against F4/80, a specific macrophage marker, was performed on MG tumor sections from Tg(Neu) and Tg(NCOA1)×Tg(Neu) mice at 2–3 weeks (early stage) and 9 weeks (late stage) after tumors were first detected. More macrophages were observed on the sections of Tg(NCOA1)×Tg(Neu) tumors versus the sections of Tg(Neu) tumors at both stages. Quantitative analysis confirmed that the average number of macrophages was significantly increased in Tg(NCOA1)×Tg(Neu) tumors versus Tg(Neu) tumors (Fig. 2A and B). Consistently, qPCR analysis revealed that the expression levels of CSF1 mRNA were higher in all three examined NCOA1-overexpressing tumors compared with NCOA1 WT tumors (Fig. 2C). Furthermore, adenovirus-mediated overexpression of hNCOA1 in MCF-7 and MDA-MB-231 human BrCa cells significantly upregulated CSF1 expression (Fig. 2D). These results demonstrate that NCOA1 overexpression in MG tumor cells results in up-regulation of CSF1 expression.
Fig. 2. NCOA1 overexpression in mammary tumor cells enhances macrophage recruitment and CSF1 expression.
A. Immunostaining of macrophages (brown color) with F4/80 antibody in Tg(Neu) (Neu) and Tg(NCOA1)×Tg(Neu) (NCOA1×Neu) mammary tumors. T, tumor area; M, macrophage. B. Macrophage numbers in Tg(Neu) and Tg(NCOA1)×Tg(Neu) tumors at early and late stages. Macrophages in 10-viewing fields at 400× magnification were counted on each tumor section. Five tumors were analyzed for each group. Data are presented as mean ± standard deviation (SD). *, p<0.05. C. qPCR measurement of CSF1 mRNA levels in individual Tg(Neu) and Tg(NCOA1)×Tg(Neu) mammary tumors. D. Adenovirus-mediated NCOA1 expression (Ad-NCOA1) in MCF-7 and MDA-MB-231 cells induced CSF1 expression as assayed by qPCR. Adenovirus-mediated GFP expression (Ad-GFP) served as a control.
NCOA1 regulates CSF1 expression in BrCa cells
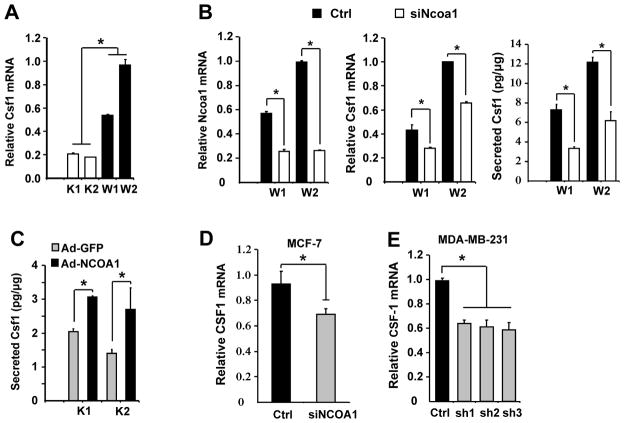
To determine whether NCOA1 regulates CSF1 expression, we measured Csf1 mRNA levels in Ncoa1-negative PyMT×Ncoa1-K1/K2 and Ncoa1-positive PyMT×Ncoa1-W1/W2 mouse MG tumor cells (19). We found that Csf1 mRNA levels decreased 3–5 fold in PyMT×Ncoa1-K1/K2 versus PyMT×Ncoa1-W1/W2 cells (Fig. 3A). Knockdown of NCOA1 by siRNA significantly reduced Csf1 mRNA levels and secreted Csf1 protein in both PyMT×Ncoa1-W1/W2 cell lines and their conditioned media (Fig. 3B). Conversely, adenovirus-mediated re-expression of NCOA1 in both PyMT×Ncoa1-K1/K2 cell lines significantly increased CSF1 protein concentration in their conditioned media (Fig. 3C). More importantly, knockdown of NCOA1 in MCF-7 and MDA-MB-231 human BrCa cells also significantly reduced CSF1 mRNA expression (Fig. 3D and E). These results, together with those shown in Fig. 2C and D, indicate that NCOA1 expression levels are tightly associated with CSF1 expression levels in both mouse and human BrCa cells.
Fig. 3. NCOA1 regulates CSF1 expression in BrCa cells.
A. Relative Csf1 mRNA levels measured by qPCR in PyMT×Ncoa1-K1/K2 and PyMT×Ncoa1-W1/W2 mouse mammary tumor cell lines. *, p<0.05. B. Knockdown of Ncoa1 expression in W1 and W2 cells by siRNA (left panel) reduced Csf1 mRNA expression (central panel) and secreted Csf1 protein in the culture medium (right panel). *, p<0.05. C. Adenovirus-mediated expression of NCOA1 (Ad-NCOA1) in K1 and K2 cells increased the concentration of secreted Csf1 compared with adenovirus-mediated expression of GFP (Ad-GFP). *, p<0.05. D and E. Knockdown of NCOA1 by siRNA in MCF-7 cells or by three different shRNAs in MDA-MB-231 cells decreased CSF1 mRNA expression compared with normal NCOA1 expression in non-targeting siRNA and shRNA-treated cells. *, p<0.05.
NCOA1 associates with the proximal region of the CSF1 promoter in BrCa cells
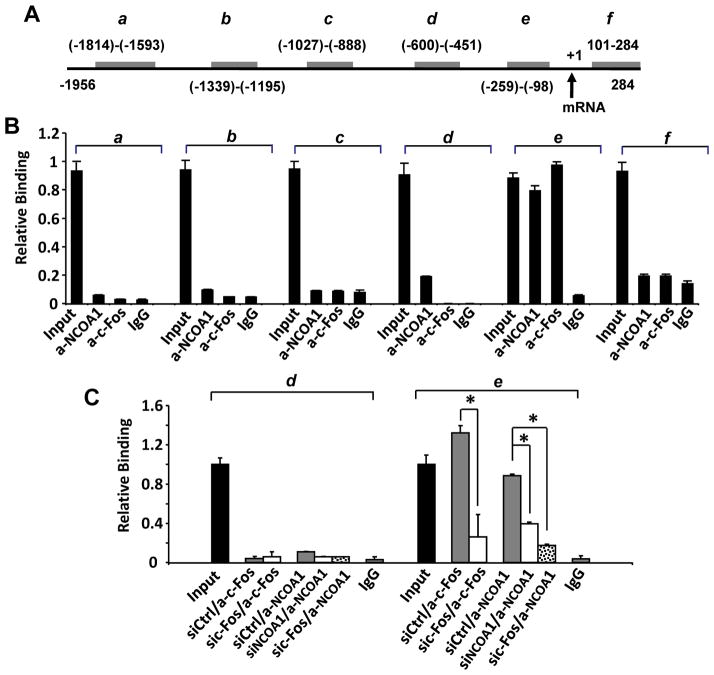
To examine the association of NCOA1 with CSF1 promoter in BrCa cells, we designed six pairs of PCR primers for amplifying DNA fragments a–f and performed ChIP assays to identify specific NCOA1-associating chromatin regions near the promoter from bp −1956 to 284 (Fig. 4A). We performed qPCR to measure the eluted DNA immunoprecipitated by NCOA1 or c-Fos antibody from the protein-DNA complexes extracted from MDA-MB-231 cells. These analyses revealed that NCOA1 associated strongly with region e and very weakly with region d, while c-Fos mainly associated with region e (Fig. 4B). As expected, knockdown of NCOA1 diminished its association with region e and abolished its weak association with region d, and knockdown of c-Fos diminished its association with region e. More importantly, knockdown of c-Fos also abolished and diminished NCOA1 recruitment to region d and region e, respectively (Fig. 4C). These results indicate that both NCOA1 and c-Fos are mainly associated with region e and NCOA1 is recruited to this region through c-Fos.
Fig. 4. NCOA1 and c-Fos are associated with a region proximate to the CSF1 promoter.
A. The 5′ regulatory sequence of the hCSF1 gene. Gray bars indicate genomic DNA fragments a–f amplified by PCR in ChIP assays. The locations of each fragment are numbered in reference to the transcriptional initiation site (NCBI NT_032977), which is indicated by an arrow. B. Representative ChIP assay results. DNA-protein complexes extracted from MDA-MB-231 cells were subjected to immunoprecipitation with NCOA1 antibody. 5′ DNA fragments (a–f) of the CSF1 gene in the eluted ChIP DNA were measured by real time PCR. The non-immune IgG served as a control, and 3% of the DNA-protein complexes was used as the input control. C. NCOA1 or c-Fos expression was knocked down by siRNAs in MDA-MB-231 cells and ChIP assays were performed using antibodies against c-Fos, NCOA1 or non-immune IgG. DNA fragments d and e in the eluted ChIP DNA were measured by real time PCR. *, p < 0.05.
NCOA1 serves as a coactivator for AP-1 to promote CSF1 promoter activity
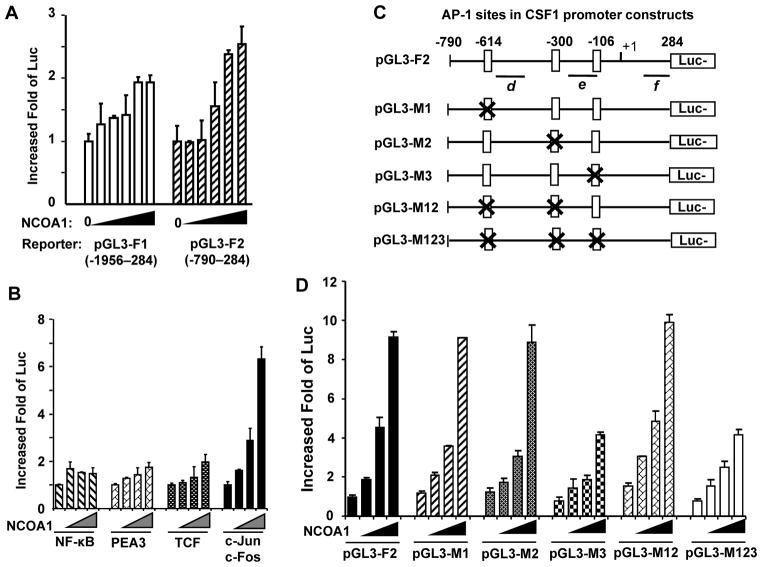
To determine the role of NCOA1 in regulation of CSF1 transcription, two luciferase reporters with 5′ CSF1 DNA sequences from bp −1956 to 284 (pGL3-F1) and from bp −790 to 284 (pGL3-F2) were constructed. In agreement with the presence of NCOA1-associating regions in both reporters, the luciferase activities derived from both reporters in transfected HeLa cells were significantly enhanced by NCOA1 expression in a dose-dependent manner (Fig.5A). These results suggest that NCOA1 overexpression potentiates the transcriptional activity of the 5′ CSF1 promoter region from bp −790 to 284 in pGL3-F2 reporter, which contains the NCOA1-associated regions d and e (bp −600 to −98 in Fig. 4A). Thus, we proposed that NCOA1 promotes CSF1 promoter activity mainly through its recruitment to the region from bp −600 to −98.
Fig. 5. NCOA1 co-activates AP-1 to promote the transcriptional activity of the CSF1 promoter.
A. HeLa cells in 24-well plates were transfected with pGL3-F1 or pGL3-F2 reporters (0.2 μg/well) and pCR3.1-NCOA1 plasmid (0, 0.15, 0.3, 0.6, 0.9 and 1.2 μg/well). B. HeLa cells were transfected with pGL3-F2 reporter (0.2 μg/well), one of the indicated TF expression plasmids (0.05 μg/well) and NCOA1 expression plasmid (0, 0.15, 0.3 and 0.6 μg/well). C. The NCOA1-associated regions proximate to the CSF1 promoter contain three AP-1 sites. The specific sites are individually or combinatorially deleted in different reporters as indicated. Luc, luciferase. D. HeLa cells were transfected with an indicated pGL3 reporter (0.2 μg/well), c-Jun/c-Fos expression plasmids (0.1 μg/well each) and NCOA1 expression plasmid (0, 0.015, 0.3 and 0.6 μg/well). In all transfection assays, luciferase activity was assayed 48 hours post transfection and normalized to total cellular protein.
NCOA1 and NCOA1-interacting proteins have been reported to associate with multiple TFs including NF-κB, PEA3, TCF-4 and AP-1 (c-Jun/c-Fos) and coactivate their transcriptional activities in cancer cells (18, 19, 39, 40). Therefore, we tested whether NCOA1 overexpression could coactivate any of these TFs to enhance the CSF1 promoter activity as reflected by luciferase activity from the pGL3-F2 reporter. Transfection with NF-κB, PEA3 or TCF-4 expression plasmid respectively activated their cognate responsive reporters, indicating these TFs expressed well in these transfected cells (Supplementary Fig. S3). However, co-expression of NCOA1 with NF-κB, PEA3 or TCF-4 did not or only slightly increase the luciferase activity of the pGL3-F2 reporter containing the 5′ CSF1 regulatory sequence associated with NCOA1. Interestingly, co-expression of NCOA1 with c-Jun and c-Fos dramatically increased the luciferase activity of the pGL3-F2 reporter in a NCOA1 dose-dependent manner, showing a six-fold induction at the highest level of NCOA1 expression (Fig. 5B). There are three putative AP-1-binding sites at base pairs −614 (TGATTAATCA), −300 (TGACTCA) and −106 (TGAATCA) (Fig. 5C) of the 5′ CSF1 promoter region tested in the pGL3-F2 reporter. Deletion of the −614 site (pGL3-M1), the −300 site (pGL3-M2), or both of these sites (pGL3-M12) in the pGL3-F2 reporter did not significantly affect NCOA1 and c-Jun/c-Fos-induced reporter activity. However, deletion of the −106 site (pGL3-M3) or all three putative AP-1 binding sites (pGL3-M123) in the pGL3-F2 reporter not only decreased the basal reporter activities induced by AP-1, but also significantly compromised NCOA1 and AP-1-induced reporter activities (Fig. 5D). These results indicate that the −106 AP-1-binding site is the major one responsible for NCOA1 and c-Jun/c-Fos to enhance the transcriptional activity of the CSF1 promoter. NCOA1 expression still showed some promoting activity to activate the pGL3-M123 reporter that lacks all AP-1 sites, suggesting that NCOA1 may also weakly coactivate some other TFs in the cells to enhance the reporter activity.
Knockdown of either NCOA1 or CSF1 in Tg(NCOA1)×Tg(Neu) mouse MG tumor cells reduces macrophage recruitment and tumor cell invasion
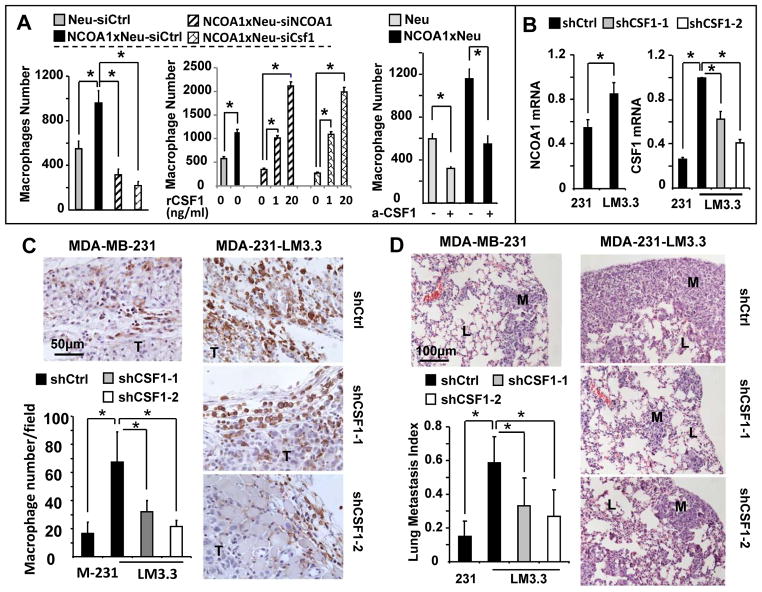
To determine whether NCOA1 overexpression in mouse mammary tumor cells is responsible for the increased macrophage recruitment, we assessed macrophage invasion attracted by NCOA1-overexpressed mammary tumor cells in a Matrigel layer-based transwell assay. We found that Tg(NCOA1)×Tg(Neu) tumor cells seeded in the lower chambers were able to attract nearly two times more macrophages from the upper chamber than the same number of wild type Tg(Neu) tumor cells could. Knockdown of either NCOA1 or CSF1 in Tg(NCOA1)×Tg(Neu) tumor cells by siRNAs reduced more than 65% and 75% of their macrophage recruitment capability, respectively (Fig. 6A, left panel). Addition of recombinant CSF1 protein to the lower chambers abolished the effect of either NCOA1 or CSF1 knockdown in Tg(NCOA1)×Tg(Neu) cells on macrophage recruitment (Fig. 6A, middle panel). Furthermore, addition of neutralizing CSF1 antibody to the lower chambers with wild type Tg(Neu) or Tg(NCOA1)×Tg(Neu) cells also significantly reduced the number of recruited macrophages (Fig. 6A, right panel). These results indicate that NCOA1-upregulated CSF1 expression in breast tumor cells plays a crucial role in macrophage recruitment.
Fig. 6. Knockdown of CSF1 blocks NCOA1-promoted macrophage recruitment in culture and in mice and inhibits metastasis.
A. The numbers of macrophages recruited by non-targeting siRNA-transfected Tg(Neu) (Neu-siCtrl) and Tg(NCOA1)×Tg(Neu) (NCOA1×Neu-siCtrl) tumor cells, hNCOA1 and Csf1 siRNA-transfected Tg(NCOA1)×Tg(Neu) (NCOA1×Neu-siNCOA1 and NCOA1×Neu-siCsf1) tumor cells, and untransfected Tg(Neu) (Neu) and Tg(NCOA1)×Tg(Neu) (NCOA1×Neu) tumor cells. Cells were treated with or without recombinant Csf1 (rCsf1) protein or CSF1 antibody (a-CSF1) as indicated. The assay was performed using a transwell co-culture system with the indicated tumor cells in the lower chambers and macrophages above a matrigel layer in the upper chambers. The knockdown efficiency of hNCOA1 and Csf1 mRNAs were evaluated by qPCR (Supplementary Fig. S4). B. qPCR measurement of relative NCOA1 and CSF1 mRNA levels in MDA-MB-231shCtrl, MDA-231-LM3.3shCtrl, MDA-231-LM3.3shCSF1-1 and/or MDA-231-LM3.3shCSF1-2 cells. These cells were generated by lentiviral expression of control shRNA (shCtrl) and CSF1 shRNAs (shCSF1-1 and shCSF1-2). The expression levels of these mRNAs were normalized to 18S rRNA. C and D. MDA-MB-231shCtrl, MDA-231-LM3.3shCtrl, MDA-231-LM3.3shCSF1-1 and MDA-231-LM3.3shCSF1-2 cells (2×106) were injected into the 4th pair of mammary fat pads of SCID mice (n=5). Mice were sacrificed 30 days after injection. F4/80 immunostaining was performed to identify macrophages (brown) on the xenograft tumor (T) sections. F4/80-positive cells were counted as described in Fig. 2B. Data are presented as mean ± SD (Panel C). H&E staining of non-adjacent lung sections was performed to determine metastasis index, which was presented as the average ratio of lung metastasis (M) area to the total metastasis and lung (L) area (Panel D). * in all panels, p < 0.05 by Student’s t-test.
Knockdown of CSF1 expression in human BrCa cells with NCOA1 overexpression reduces macrophages in the xenograft tumors and decreases lung metastases in SCID mice
The MDA-231-LM3.3 human BrCa cell line has a much stronger lung metastasis potential than its original MDA-MB-231 parent cell line. We found that the expression levels of both NCOA1 and its target CSF1 are significantly increased in MDA-231-LM3.3shCtrl cells versus MDA-MB-231shCtrl cells harboring a non-targeting control shRNA (Fig. 6B). We also generated MDA-231-LM3.3shCSF1-1 and MDA-231-LM3.3shCSF1-2 cells with stable knockdown of CSF1 mRNA using two different lentiviral shRNAs (Fig. 6B). We injected these cells into the mammary fat pads of SCID mice, and compared their tumor growth rates and metastases. SCID mice are devoid of functional T and B cells but still retain macrophages which could be activated by a T-cell-independent mechanism (41). The xenograft tumors derived from all four groups of cells became palpable within a week after injection and these tumors also grew comparably (data not shown). However, the average numbers of F4/80-positive macrophages in MDA-MB-231shCtrl, MDA-231-LM3.3shCSF1-1 and MDA-231-LM3.3shCSF1-2 tumors were significantly less than the number of macrophages in MDA-231-LM3.3shCtrl tumors (Fig. 6C). Accordingly, the extent of lung metastases derived from MDA-MB-231shCtrl, MDA-231-LM3.3shCSF1-1 and MDA-231-LM3.3shCSF1-2 tumors was much smaller than that derived from MDA-231-LM3.3shCtrl tumors (Fig. 6D). These results suggest that the upregulated CSF1 expression in MDA-231-LM3.3 cells with NCOA1 overexpression mediates, at least in part, the NCOA1-promoted BrCa metastasis.
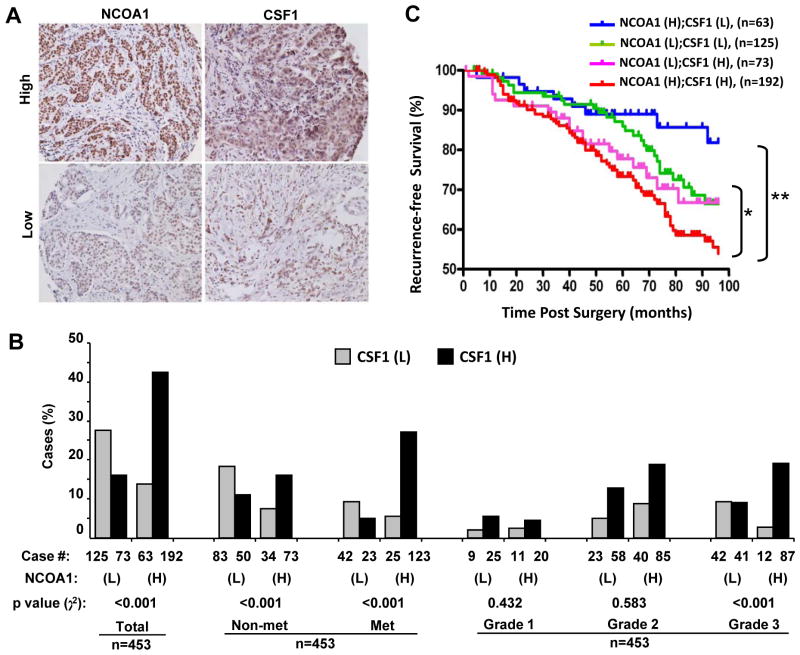
The coupled high expression of both NCOA1 and CSF1 in human breast tumors positively correlates with lymph node metastasis, high tumor grade and poor prognosis
To examine the expression association between NCOA1 and CSF1 in human BrCa and to address the role of their co-expression in metastasis development, we examined the expression of NCOA1 and CSF1 in 453 breast tumors by IHC. In positively stained tumor cells, NCOA1 immunoreactivity is mainly located in the nucleus, while CSF1 immunoreactivity is distributed both intra- as well as extracellularly (Fig. 7A). Medium to high levels (3< scores ≤8, designated as high expression groups) of NCOA1 and CSF-1 proteins were detected in 56.3% and 58.5% tumor samples, respectively. Importantly, as many as 42.4% of tumors showed high expression of both NCOA1 and CSF1, but only 13.9% and 16.1% of tumors expressed high NCOA1 with low CSF1 (0≤ scores ≤3) and low NCOA1 with high CSF1, respectively. This indicates a positive association of NCOA1 expression with CSF1 expression in these breast tumors (Fig. 7B). Furthermore, only 16.1% (73 out of 240) of lymph node-negative tumors showed high NCOA1 and CSF1 expression, while as many as 27.2% (123 out of 213) of lymph node-positive tumors had high NCOA1 and CSF1 expression. However, node-positive status was not associated with either NCOA1 or CSF1 high expression alone (Fig. 7B). These results suggest that tumors with high expression levels of both NCOA1 and CSF1 have greater metastatic potentials. Moreover, a significantly higher number of grade 3 tumors expressed both NCOA1 and CSF1 at higher levels versus grades 1 and 2 tumors, suggesting a positive correlation between high tumor grade and high NCOA1 and CSF1 expression (Fig. 7B). Finally, patients with high expression of both NCOA1 and CSF1 demonstrated a significantly worse disease-free survival than patients with low expression of both NCOA1 and CSF1 or with high NCOA1 expression alone. There were no significant differences in disease recurrence among other patient groups (Fig. 7C). These results suggest that the coupled overexpression of NCOA1 and CSF1 in BrCa plays a crucial role in disease recurrence and therefore serves as a marker of poor prognosis.
Fig. 7. The expression of NCOA1 and CSF1 proteins in human breast tumors and their association with lymph node metastasis, tumor grade and recurrence-free survival.
A. Representative images for high and low IHC signals of NCOA1 and CSF1 in human breast tumors. The original images were taken at 100×. B. The case numbers and percentages of NCOA1(L);CSF1(L), NCOA1(L);CSF1(H), NCOA1(H);CSF1(L) and NCOA1(H);CSF1(H) breast tumors in the total cohort and subgroups divided by negative (Non-met) and positive (Met) lymph node metastases and tumor grades. C. Kaplan Meier recurrence-free survival curves for patient groups as indicated. H, high expression group (3< IHC scores ≤8); L, low expression group (0≤ IHC scores ≤3); n, number of patients; * and **, p < 0.05 and 0.01 by Logrank test.
Discussion
Recent studies have suggested many detrimental roles of altered expression of transcriptional coactivators in development of human diseases including cancers (1, 42, 43). Thus, there is an urgent need to define the exact roles of coactivators in carcinogenesis and their underlying molecular mechanisms using animal models and molecular approaches. In this study, we investigated mammary tumorigenesis and metastasis in Tg(NCOA1)×Tg(Neu) mice and RCAS-PyMT retrovirus-infected Tg(NCOA1)×Tg(TVA) mice with hNCOA1 overexpression in their MECs. NCOA1 overexpression itself did not affect MG morphogenesis in wild type and Tg(NCOA1) mice. Nor did its overexpression affect oncogene-induced mammary tumor initiation and growth. Moreover, our previous study showed NCOA1 knockout did not affect mammary tumor formation and growth in Tg(PyMT) mice (16). These results suggest that deregulated NCOA1 expression itself is not oncogenic, which is different from the oncogenic capability of overexpressed NCOA3 (9–11). Therefore, the p160 coactivator family members may have distinct contributions to BrCa initiation and growth in addition to their possible redundant functions.
NCOA1 expression is low in normal human MECs, but high in metastatic BrCa exhibiting early recurrence, resistance to endocrine therapy and poor disease-free survival (15, 44). Our study further demonstrate that NCOA1 overexpression significantly promotes mammary tumor cell dissemination into the blood circulation, followed by a significantly increased incidence of lung metastasis in both Tg(NCOA1)×Tg(Neu) and Tg(NCOA1)×Tg(TVA)+RCAS-PyMT BrCa mouse models, indicating NCOA1 overexpression indeed drives BrCa metastasis in vivo. On the other hand, it has been shown that knockout of NCOA1 drastically reduced BrCa metastasis in Tg(PyMT) and Tg(Neu) mice (16, 17). Together, these clinical and experimental findings suggest NCOA1 as a potential target for controlling BrCa metastasis.
We found that NCOA1 overexpression in mouse mammary tumors and human BrCa cells positively correlates with CSF1 expression, and NCOA1 knockout or knockdown in these cells reduces CSF1 expression. We further showed that both NCOA1 and c-Fos associate with a previously known AP-1 site (45), and this association activates the CSF1 promoter. These results clearly identified CSF1 as a direct novel target gene of NCOA1 and c-Jun/c-Fos in BrCa cells. Recent studies have also demonstrated that NCOA1 serves as a coactivator for PEA3, c-Jun/c-Fos, Ets-2 and HOXC11 to upregulate Twist1, integrin α5 (ITGA5), c-Myc and S100β expression, respectively, which in turn promotes BrCa cell EMT, migration, invasion and/or resistance to endocrine therapies (18, 19, 46, 47). Together, these findings indicate that NCOA1 can coactivate different TFs to regulate multiple target genes important for BrCa.
The BrCa cell-produced CSF1 plays a crucial role to stimulate cancer cell invasion and metastasis through both paracrine and autocrine pathways (48). The paracrine pathway consists of CSF1 from BrCa cells, CAMs recruited by CSF1, EGF from CAMs and EGF-stimulated BrCa cell invasion and metastasis (28–31). The recruited CAMs can also induce immunosuppression against tumors to accelerate cancer progression (49), produce more uPA (urokinase-type plasminogen activator) to augment tumor cell invasion (50), and secrete pro-angiogenic factors such as IL-6, VEGF, MCP-1 and TGFβ to stimulate angiogenesis (28). The autocrine pathway also stimulates BrCa cell invasion and this is mediated by the CSF1 receptor, a transmembrane tyrosine kinase receptor in BrCa cells (48). Our current study further revealed an important role of NCOA1-upregulated CSF1 in promotion of BrCa metastasis through its paracrine pathway. Specifically, we demonstrated that Tg(NCOA1)×Tg(Neu) tumor cells with overexpression of hNCOA1 and hNCOA1-induced CSF1 are much more capable versus Tg(Neu) tumor cells to recruit macrophages in culture. Knockdown of either NCOA1 or NCOA1-induced CSF1 in these cells compromised their ability to recruit macrophages in culture, suggesting CSF1 mediates macrophage recruitment induced by NCOA1 overexpression. We also demonstrated that the MDA-MB-231-derived highly metastatic MDA-231-LM3.3 cells had much higher expression of both NCOA1 and CSF1 versus MDA-MB-231 cells, while knockdown of either NCOA1 or CSF1 could block CAM recruitment to the xenograft tumors and tumor cell metastasis to the lung in mice. Again, this suggests NCOA1 overexpression promotes macrophage recruitment and BrCa metastasis through the up-regulation of CSF1 expression. Taken together, our novel findings indicate that NCOA1 serves as a coactivator for AP-1 to upregulate CSF1 in BrCa cells, which forms a regulatory axis to promote BrCa cell invasion and metastasis through enhancing paracrine pathway of the CSF1 signaling.
In human breast tumors, NCOA1 expression has been shown to be an independent marker for predicting disease recurrence and endocrine therapy resistance (15, 44). However, the expressional and functional relationship between NCOA1 and CSF1 in human BrCa has not been previously investigated. In the current study, we identified a positive correlation between NCOA1 and CSF1 protein expression in human breast tumors. More importantly, we found this correlation is associated with lymph node metastasis, tumor grade and recurrence. These results suggest that NCOA1-induced CSF1 expression in human BrCa also plays an important role in driving BrCa progression and metastasis. Therefore, targeting NCOA1 could inhibit CSF1 expression and CSF1 expression-induced BrCa cell invasion and metastasis. Given that NCOA1 also upregulates other genes important for BrCa cell survival, invasion and metastasis, such as Twist1, ITGA5, c-Myc and S100β (18, 19, 46, 47), NCOA1 may be a potential molecular target for inhibiting BrCa progression and metastasis.
Supplementary Material
Acknowledgments
We thank Dr. Hongwu Chen for providing NCOA1-expressing adenovirus, Dr. Joan Massagué for providing MDA-MB-231-LM2 cells, and the Baylor College of Medicine Transgenic Mouse Core (partially supported by) for microinjecting the transgene DNA. This study is supported by R01CA112403, R01DK058242, P01DK059820 and R01CA124820 NIH grants, 2012SZ0073 Sichuan Science and Technology Agency grant, and RP120732-P5 Cancer Prevention and Research Institute of Texas grant.
Footnotes
The authors have no conflict of financial interests.
References
- 1.Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9:615–30. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li X, Lonard DM, Jung SY, Malovannaya A, Feng Q, Qin J, et al. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell. 2006;124:381–92. doi: 10.1016/j.cell.2005.11.037. [DOI] [PubMed] [Google Scholar]
- 3.O’Malley BW. Masters of the genome. Nat Rev Mol Cell Biol. 2010;11:311. doi: 10.1038/nrm2886. [DOI] [PubMed] [Google Scholar]
- 4.Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–8. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 5.Zhou HJ, Yan J, Luo W, Ayala G, Lin SH, Erdem H, et al. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65:7976–83. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]
- 6.Tanner MM, Grenman S, Koul A, Johannsson O, Meltzer P, Pejovic T, et al. Frequent amplification of chromosomal region 20q12-q13 in ovarian cancer. Clin Cancer Res. 2000;6:1833–9. [PubMed] [Google Scholar]
- 7.Xu Y, Chen Q, Li W, Su X, Chen T, Liu Y, et al. Overexpression of transcriptional coactivator AIB1 promotes hepatocellular carcinoma progression by enhancing cell proliferation and invasiveness. Oncogene. 2010;29:3386–97. doi: 10.1038/onc.2010.90. [DOI] [PubMed] [Google Scholar]
- 8.Henke RT, Haddad BR, Kim SE, Rone JD, Mani A, Jessup JM, et al. Overexpression of the nuclear receptor coactivator AIB1 (SRC-3) during progression of pancreatic adenocarcinoma. Clin Cancer Res. 2004;10:6134–42. doi: 10.1158/1078-0432.CCR-04-0561. [DOI] [PubMed] [Google Scholar]
- 9.Torres-Arzayus MI, Font de Mora J, Yuan J, Vazquez F, Bronson R, Rue M, et al. High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell. 2004;6:263–74. doi: 10.1016/j.ccr.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 10.Kuang SQ, Liao L, Wang S, Medina D, O’Malley BW, Xu J. Mice lacking the amplified in breast cancer 1/steroid receptor coactivator-3 are resistant to chemical carcinogen-induced mammary tumorigenesis. Cancer Res. 2005;65:7993–8002. doi: 10.1158/0008-5472.CAN-05-1179. [DOI] [PubMed] [Google Scholar]
- 11.Kuang SQ, Liao L, Zhang H, Lee AV, O’Malley BW, Xu J. AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res. 2004;64:1875–85. doi: 10.1158/0008-5472.can-03-3745. [DOI] [PubMed] [Google Scholar]
- 12.Chung AC, Zhou S, Liao L, Tien JC, Greenberg NM, Xu J. Genetic ablation of the amplified-in-breast cancer 1 inhibits spontaneous prostate cancer progression in mice. Cancer Res. 2007;67:5965–75. doi: 10.1158/0008-5472.CAN-06-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tien JC, Liu Z, Liao L, Wang F, Xu Y, Wu YL, et al. The steroid receptor coactivator-3 is required for the development of castration-resistant prostate cancer. Cancer Res. 2013;73:3997–4008. doi: 10.1158/0008-5472.CAN-12-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redmond AM, Bane FT, Stafford AT, McIlroy M, Dillon MF, Crotty TB, et al. Coassociation of estrogen receptor and p160 proteins predicts resistance to endocrine treatment; SRC-1 is an independent predictor of breast cancer recurrence. Clin Cancer Res. 2009;15:2098–106. doi: 10.1158/1078-0432.CCR-08-1649. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, Yuan Y, Liao L, Kuang SQ, Tien JC, O’Malley BW, et al. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc Natl Acad Sci U S A. 2009;106:151–6. doi: 10.1073/pnas.0808703105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han JS, Crowe DL. Steroid receptor coactivator 1 deficiency increases MMTV-neu mediated tumor latency and differentiation specific gene expression, decreases metastasis, and inhibits response to PPAR ligands. BMC Cancer. 2010;10:629. doi: 10.1186/1471-2407-10-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin L, Chen X, Wu Y, Feng Z, He T, Wang L, et al. Steroid receptor coactivator-1 upregulates integrin alpha expression to promote breast cancer cell adhesion and migration. Cancer Res. 2011;71:1742–51. doi: 10.1158/0008-5472.CAN-10-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator-1 regulates twist expression and promotes breast cancer metastasis. Cancer Res. 2009;69:3819–27. doi: 10.1158/0008-5472.CAN-08-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McBryan J, Theissen SM, Byrne C, Hughes E, Cocchiglia S, Sande S, et al. Metastatic progression with resistance to aromatase inhibitors is driven by the steroid receptor coactivator SRC-1. Cancer Res. 2012;72:548–59. doi: 10.1158/0008-5472.CAN-11-2073. [DOI] [PubMed] [Google Scholar]
- 21.Kishimoto H, Wang Z, Bhat-Nakshatri P, Chang D, Clarke R, Nakshatri H. The p160 family coactivators regulate breast cancer cell proliferation and invasion through autocrine/paracrine activity of SDF-1alpha/CXCL12. Carcinogenesis. 2005;26:1706–15. doi: 10.1093/carcin/bgi137. [DOI] [PubMed] [Google Scholar]
- 22.Myers E, Hill AD, Kelly G, McDermott EW, O’Higgins NJ, Buggy Y, et al. Associations and interactions between Ets-1 and Ets-2 and coregulatory proteins, SRC-1, AIB1, and NCoR in breast cancer. Clin Cancer Res. 2005;11:2111–22. doi: 10.1158/1078-0432.CCR-04-1192. [DOI] [PubMed] [Google Scholar]
- 23.McCartan D, Bolger JC, Fagan A, Byrne C, Hao Y, Qin L, et al. Global characterization of the SRC-1 transcriptome identifies ADAM22 as an ER-independent mediator of endocrine-resistant breast cancer. Cancer Res. 2012;72:220–9. doi: 10.1158/0008-5472.CAN-11-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kacinski BM. CSF-1 and its receptor in ovarian, endometrial and breast cancer. Ann Med. 1995;27:79–85. doi: 10.3109/07853899509031941. [DOI] [PubMed] [Google Scholar]
- 25.Pollard JW, Bartocci A, Arceci R, Orlofsky A, Ladner MB, Stanley ER. Apparent role of the macrophage growth factor, CSF-1, in placental development. Nature. 1987;330:484–6. doi: 10.1038/330484a0. [DOI] [PubMed] [Google Scholar]
- 26.Wiktor-Jedrzejczak W, Gordon S. Cytokine regulation of the macrophage (M phi) system studied using the colony stimulating factor-1-deficient op/op mouse. Physiol Rev. 1996;76:927–47. doi: 10.1152/physrev.1996.76.4.927. [DOI] [PubMed] [Google Scholar]
- 27.Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127:2269–82. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- 28.Lin EY, Gouon-Evans V, Nguyen AV, Pollard JW. The macrophage growth factor CSF-1 in mammary gland development and tumor progression. J Mammary Gland Biol Neoplasia. 2002;7:147–62. doi: 10.1023/a:1020399802795. [DOI] [PubMed] [Google Scholar]
- 29.Tang R, Beuvon F, Ojeda M, Mosseri V, Pouillart P, Scholl S. M-CSF (monocyte colony stimulating factor) and M-CSF receptor expression by breast tumour cells: M-CSF mediated recruitment of tumour infiltrating monocytes? J Cell Biochem. 1992;50:350–6. doi: 10.1002/jcb.240500403. [DOI] [PubMed] [Google Scholar]
- 30.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–40. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–9. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 32.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992;89:10578–82. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du Z, Podsypanina K, Huang S, McGrath A, Toneff MJ, Bogoslovskaia E, et al. Introduction of oncogenes into mammary glands in vivo with an avian retroviral vector initiates and promotes carcinogenesis in mouse models. Proc Natl Acad Sci U S A. 2006;103:17396–401. doi: 10.1073/pnas.0608607103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin L, Liao L, Redmond A, Young L, Yuan Y, Chen H, et al. The AIB1 oncogene promotes breast cancer metastasis by activation of PEA3-mediated matrix metalloproteinase 2 (MMP2) and MMP9 expression. Mol Cell Biol. 2008;28:5937–50. doi: 10.1128/MCB.00579-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harvey JM, Clark GM, Osborne CK, Allred DC. Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol. 1999;17:1474–81. doi: 10.1200/JCO.1999.17.5.1474. [DOI] [PubMed] [Google Scholar]
- 37.Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O’Malley BW. The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc Natl Acad Sci U S A. 2000;97:6379–84. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ, O’Malley BW. Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science. 1998;279:1922–5. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 39.Koh SS, Li H, Lee YH, Widelitz RB, Chuong CM, Stallcup MR. Synergistic coactivator function by coactivator-associated arginine methyltransferase (CARM) 1 and beta-catenin with two different classes of DNA-binding transcriptional activators. J Biol Chem. 2002;277:26031–5. doi: 10.1074/jbc.M110865200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Na SY, Lee SK, Han SJ, Choi HS, Im SY, Lee JW. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor kappaB-mediated transactivations. J Biol Chem. 1998;273:10831–4. doi: 10.1074/jbc.273.18.10831. [DOI] [PubMed] [Google Scholar]
- 41.Bancroft GJ, Kelly JP. Macrophage activation and innate resistance to infection in SCID mice. Immunobiology. 1994;191:424–31. doi: 10.1016/S0171-2985(11)80448-1. [DOI] [PubMed] [Google Scholar]
- 42.Lonard DM, Lanz RB, O’Malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–87. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- 43.Walsh CA, Qin L, Tien JC, Young LS, Xu J. The function of steroid receptor coactivator-1 in normal tissues and cancer. Int J Biol Sci. 2012;8:470–85. doi: 10.7150/ijbs.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fleming FJ, Myers E, Kelly G, Crotty TB, McDermott EW, O’Higgins NJ, et al. Expression of SRC-1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC-1. J Clin Pathol. 2004;57:1069–74. doi: 10.1136/jcp.2004.016733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Konicek BW, Xia X, Rajavashisth T, Harrington MA. Regulation of mouse colony-stimulating factor-1 gene promoter activity by AP1 and cellular nucleic acid-binding protein. DNA Cell Biol. 1998;17:799–809. doi: 10.1089/dna.1998.17.799. [DOI] [PubMed] [Google Scholar]
- 46.Al-azawi D, Ilroy MM, Kelly G, Redmond AM, Bane FT, Cocchiglia S, et al. Ets-2 and p160 proteins collaborate to regulate c-Myc in endocrine resistant breast cancer. Oncogene. 2008;27:3021–31. doi: 10.1038/sj.onc.1210964. [DOI] [PubMed] [Google Scholar]
- 47.deBlacam C, Byrne C, Hughes E, McIlroy M, Bane F, Hill ADK, et al. HOXC11-SRC-1 regulation of S100beta in cutaneous melanoma: new targets for the kinase inhibitor dasatinib. Br J Cancer. 2011;105:118–23. doi: 10.1038/bjc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patsialou A, Wyckoff J, Wang Y, Goswami S, Stanley ER, Condeelis JS. Invasion of human breast cancer cells in vivo requires both paracrine and autocrine loops involving the colony-stimulating factor-1 receptor. Cancer Res. 2009;69:9498–506. doi: 10.1158/0008-5472.CAN-09-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.al-Sarireh B, Eremin O. Tumour-associated macrophages (TAMS): disordered function, immune suppression and progressive tumour growth. J R Coll Surg Edinb. 2000;45:1–16. [PubMed] [Google Scholar]
- 50.Hildenbrand R, Wolf G, Bohme B, Bleyl U, Steinborn A. Urokinase plasminogen activator receptor (CD87) expression of tumor-associated macrophages in ductal carcinoma in situ, breast cancer, and resident macrophages of normal breast tissue. J Leukoc Biol. 1999;66:40–9. doi: 10.1002/jlb.66.1.40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.