Abstract
The bitter taste serves as an important natural defence against the ingestion of poisonous foods and is thus believed to be indispensable in animals. However, vampire bats are obligate blood feeders that show a reduced behavioural response towards bitter-tasting compounds. To test whether bitter taste receptor genes (T2Rs) have been relaxed from selective constraint in vampire bats, we sampled all three vampire bat species and 11 non-vampire bats, and sequenced nine one-to-one orthologous T2Rs that are assumed to be functionally conserved in all bats. We generated 85 T2R sequences and found that vampire bats have a significantly greater percentage of pseudogenes than other bats. These results strongly suggest a relaxation of selective constraint and a reduction of bitter taste function in vampire bats. We also found that vampire bats retain many intact T2Rs, and that the taste signalling pathway gene Calhm1 remains complete and intact with strong functional constraint. These results suggest the presence of some bitter taste function in vampire bats, although it is not likely to play a major role in food selection. Together, our study suggests that the evolutionary reduction of bitter taste function in animals is more pervasive than previously believed, and highlights the importance of extra-oral functions of taste receptor genes.
Keywords: vampire bats, bitter, taste receptor, pseudogenization, diet
1. Introduction
Mammals typically have five primary taste modalities dedicated to the evaluation of diets, of which the bitter taste serves as an important natural defence against the ingestion of poisonous foods and is thus believed to be indispensable in animals [1]. Although vertebrate bitter taste receptor genes (T2Rs or Tas2rs) diverge tremendously in number from 0 in the bottlenose dolphin to 51 in the African clawed frog [2], multiple intact T2Rs are maintained to ensure the functionality of detecting toxins in food sources for these animals, with the exception of the bottlenose dolphin [2,3]. The dolphin represents the first mammal to lack functional bitter taste receptors, probably because they swallow food whole, rendering the taste dispensable [3]. The great reduction of bitter taste function in the dolphin is surprising because natural toxins typically taste bitter, so the bitter taste represents an important natural defence against the ingestion of poisonous chemicals such as plant alkaloids and insect toxins [4–6].
Vampire bats are the only mammals that feed exclusively on blood [7] and the extreme narrowness of their diets may have rendered these bats poor tasters [8,9]. Indeed, all extant vampire bats (three species: common vampire bat, Desmodus rotundus; white-winged vampire bat, Diaemus youngi and hairy-legged vampire bat, Diphylla ecaudata) have lost sweet and umami tastes [9–11]; the common vampire bat behaviourally showed a reduction towards bitter-tasting compounds [11]. Furthermore, vampire bats use odour cues for prey detection [12] and use infrared sensors to locate capillary-rich areas of skin [7,13]. These capabilities may have further reduced their taste sensitivity [9]. To test whether bitter taste receptor genes (T2Rs) have been relaxed from selective constraint in vampire bats, we sampled all three vampire bats and 11 non-vampire bats across the phylogeny and examined nine one-to-one orthologous T2Rs that are shared in four bats representing two major groups of bats (Yangochiroptera and Yinpterochiroptera) and thus are assumed to be functionally conserved in all bats. We found that, of these functionally conserved T2Rs common to other bats, vampire bats have a significantly greater percentage of pseudogenized T2Rs than other bats. We also found that vampire bats retain many intact and putatively functional T2Rs.
2. Material and methods
(a). Gene identification and taxon coverage
We identified T2Rs from the draft genome sequences of the four bats in the Ensembl genome database (Pteropus vampyrus and Myotis lucifigus) and an earlier study (Pteropus alecto and Myotis davidii) [14]. Because vertebrate T2Rs are intronless and approximately 300 codons in length, the gene identification approach was straightforward. We used all T2Rs from human, rat, dog and chicken as queries to TblastN against the four bat genomes following a previous study [15], and confirmed the presence of seven transmembrane domains using the TMHMM method [16]. All candidate T2Rs were verified by the best hits with known T2Rs using BlastN searches against the entire GenBank [15].
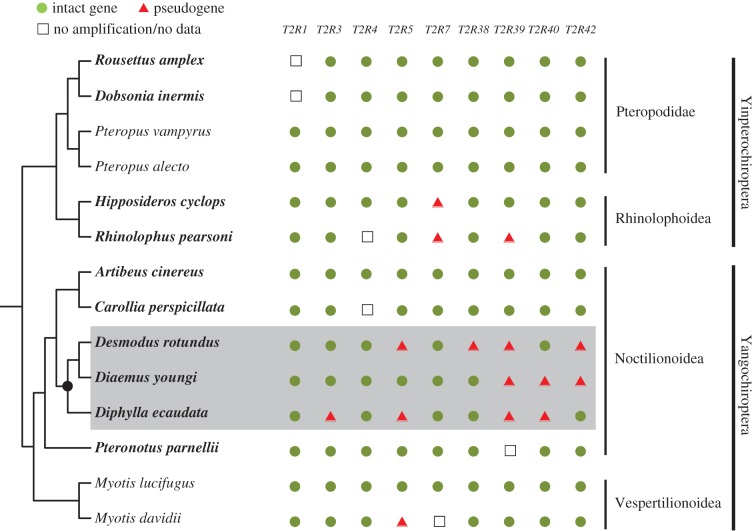
Our dataset of bats contained all three species of vampire bats and 11 species of non-vampire bats (figure 1). We attempted to include bat species that are both closely and distantly related to vampire bats. Specifically, two bats are affiliated with the same family Phyllostomidae as the three vampire bats; one belongs to Mormoopidae, a bat family that is most closely related to Phyllostomidae; two are from the other family in the same suborder Yangochirotera; the remaining six bats are from more distantly related families in the other suborder Yinpterochiroptera (figure 1; electronic supplementary material, table S1). The bat order Chiroptera is divided into two suborders: Yinpterochiroptera and Yangochiroptera, which comprise two and three superfamilies, respectively [17]. We sequenced T2Rs from 10 bats and identified T2Rs from the draft genome sequences of four additional bats. These species represent four of the five superfamilies of bats (figure 1).
Figure 1.
The species tree of the 14 bats studied, with intact and pseudogenized T2Rs being indicated. Intact genes are characterized by an intact open reading frame (ORF), while pseudogenes are characterized by a disrupted ORF resulting from nonsense and/or frame-shifting mutations. Tree topology follows a previous study [17]. The ten species in bold are those sequenced in this study, whereas the four remaining species are those with available genome sequences. All three vampire bats are shaded in grey. The common ancestor of vampire bats discussed in the text is indicated as a black circle. (Online version in colour.)
(b). Polymerase chain reaction amplification and DNA sequencing
Based on the sequence alignments of T2Rs from the four bats with available genome sequences, we designed a suite of primers (electronic supplementary material, table S2) to amplify the nine one-to-one orthologous T2Rs in 10 bats (figure 1). All bat tissues were loaned from the American Museum of Natural History, and the identity of each bat was confirmed by sequencing the complete coding sequences of the mitochondrial cytochrome b (Cytb) gene (electronic supplementary material, figure S1). Genomic DNAs were isolated using Qiagen DNeasy kits. Polymerase chain reactions (PCRs) were performed following our previously described methods [9,10]. PCR products were sequenced directly with the same primer sets as for PCR amplifications. When the direct sequencing did not work, PCR products were cloned into the pMD19-T vector (Takara) and sequenced from both strands. We additionally amplified T1R3 and Calhm1 using the primer sequences listed in the electronic supplementary material, table S2. All sequences newly generated by PCRs were deposited in GenBank under accession numbers KJ55725–KJ557347.
(c). Sequence alignment and phylogenetic reconstruction
The resulting sequences were aligned with MEGA v. 5.2 [18], and checked by eye. Nucleotide sequence alignments were generated according to protein sequence alignments and were subsequently used to reconstruct phylogenetic trees. Phylogenetic reconstruction for each dataset was conducted using a Bayesian approach, implemented in MrBayes v. 3.2 [19]. Six Markov chains were run simultaneously with as many generations as needed to ensure that the standard deviation of split frequencies was less than 0.01. We discarded the first 400 000 generations as burn-in and sampled the chains every 1000 generations. The best-fitting model of sequence evolution for each dataset was estimated by Modeltest v. 3.7 [20].
(d). Construction of ancestral sequences and tests for selection
Ancestral sequences of vampire bats were reconstructed using the Bayesian method [21] implemented in the baseml program in PAML [22] and the parsimony method [23]. To determine whether vampire bats have undergone differential selective pressures as compared to other bats, we estimated the ratio of non-synonymous to synonymous substitution rates (termed ω), which is an indicator of natural selection, with ω being less than 1, equal to 1 and more than 1 indicating purifying selection, neutral evolution and positive selection, respectively. We next undertook likelihood ratio tests of selection using branch models in the codeml program in PAML. For each gene, we conducted three tests (table 1). First, we tested whether the overall ω is significantly smaller than 1 in non-vampire bats. Second, we tested whether there is a significant difference in ω between the common ancestor of vampire bats and all other bats. Third, we tested whether there is a variation in ω before and after the divergence of vampire bats.
Table 1.
Likelihood ratio tests of selective pressures on bat T2Rs and Calhm1. p-values for each likelihood ratio test are given, and significant p-values (<0.05) are indicated in bold. See the electronic supplementary material, table S4 for details of parameter estimates.
| models | model compared | genes |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| T2R1 | T2R3 | T2R4 | T2R5 | T2R7 | T2R38 | T2R39 | T2R42 | T2R40 | Calhm1 | ||
| Test 1: all non-vampire bats | |||||||||||
| A. All branches have the same ω
B. All branches have the same ω = 1 |
B versus A | 0.0651 | 1.9 × 10−6 | 3.0 × 10−8 | 9.5 × 10−8 | 0.002 | 1.1 × 10−24 | 5.7 × 10−6 | 0.75 | 1.1 × 10−22 | 1.9 × 10−154 |
| Test 2: all non-vampire bats plus ancestral sequence of vampire bats | |||||||||||
| C. All branches have the same ω
D. Ancestral branch of vampire bats has ω2 and other branches have ω1 |
D versus C | 0.189 | 0.202 | 0.295 | 0.502 | 0.997 | 0.927 | 0.463 | 0.742 | 0.873 | 0.985 |
| Test 3: all bats after removing nonsense mutations in pseudogenes | |||||||||||
| E. All vampire bats branch have ω2, the other branches have ω1
F. Ancestral branch to vampire bats has ω3, branches connecting three vampire bats have ω2, other branches have ω1 |
F versus E | 0.262 | 0.105 | 0.42 | 1 | 0.362 | 0.327 | 0.869 | 0.633 | 0.117 | 0.119 |
3. Results
(a). Survey of bitter taste receptor genes in four bat genomes
A total of 39, 34, 26 and 24 T2Rs were identified from M. davidii, M. lucifigus, P. alecto and P. vampyrus, respectively (electronic supplementary material, table S3). Among them, 79 T2Rs are intact with at least 270 codons, start codon, stop codon and seven transmembrane domains (electronic supplementary material, table S3); the nomenclature of bat T2Rs followed that for human T2Rs [24]. The deduced protein sequences of these intact genes were aligned and translated back to nucleotides and the resulting alignment was used to reconstruct a phylogenetic tree using the Bayesian approach. We found three Myotis specific clusters in the tree (electronic supplementary material, figure S2), suggestive of functional innovation of bitter taste in these insect-eating bats [25] because many insects rely on chemical defence against predators [6,26,27]. Notably, we identified seven clades containing four genes from each of the four bats (electronic supplementary material, figure S2), showing a one-to-one orthologous relationship. We also included two additional genes as one-to-one orthologues despite their absence in one of the four bats due to pseudogenization (T2R5) or incomplete sequencing (T2R7) (figure 1; electronic supplementary material, figure S2). The four bats analysed above belong to the two major groups of bats (Yinpterochiroptera and Yangochiroptera; electronic supplementary material, table S1) and these one-to-one orthologous T2Rs are assumed to be functionally conserved in all bats because same bitter taste receptors tend to recognize certain bitter-tasting compounds better than other bitter-tasting chemicals, and thus tend to have the same functions [28]. We did not examine other T2Rs because those genes are specific to certain species (electronic supplementary material, figure S2) that do not represent the conserved taste function in bats.
(b). Pseudogenization of bitter taste receptor genes in vampire bats
To test whether bitter taste function is reduced in vampire bats, we examined the nine T2Rs in all three vampire bats, seven non-vampire bats and the four additional non-vampire bats with available genome sequences (figure 1; electronic supplementary material, table S1). We sequenced 85 T2R gene segments from the 10 bats, including the three vampire bats, which ranged from 528 to 872 bp in length. Phylogenetic trees reconstructed using each T2R gene generally agree with the established species tree [17] (electronic supplementary material, figure S3), suggesting that each T2R gene newly generated in various bats is orthologous. After aligning with 35 T2Rs of the four bats with genome sequences, our analysis of 120 genes discovered 105 T2Rs that retain intact open reading frames (ORFs), of which 89 intact ones were identified from a total of 93 genes in 11 non-vampire bats. These results strongly support the assumption that these T2Rs are of functional importance across all non-vampire bats. By contrast, the remaining 15 T2Rs contain ORF-disrupting mutations such as nonsense mutations and frame-shifting deletions (figure 2). In 12 of the 15 T2Rs, the first nonsense mutations are located near the 5′ end, resulting in the loss of multiple transmembrane domains of the proteins (figure 2). The remaining three genes (T2R5 of Diphylla ecaudata, T2R7 of R. pearsonii and T2R38 of Desmodus rotundus) contain the first nonsense mutations near the 3′ end (figure 2), which would lead to the loss of at least one transmembrane domain of the receptors because the final transmembrane domains of the bitter taste receptors are located at the very end of the coding region [29]. Therefore, none of the 15 truncated receptors is functional. Of the 15 pseudogenized T2Rs, 11 were amplified from vampire bats, while four were identified from non-vampire bats (figure 1). For these functionally conserved T2Rs common to non-vampire bats, the percentage of pseudogenes is significantly greater for vampire bats (11/27 = 40.7%) than for non-vampire bats (4/93 = 4.3%) (p < 0.001, Fisher's exact test), indicative of substantial reduction of bitter taste function in vampire bats.
Figure 2.
Alignments of T2Rs with the first ORF-disrupting mutations boxed. Dashes indicate alignment gaps and numbers in parentheses indicate nucleotide positions following the reference sequences. Reference sequences were from Myotis lucifugus or Pteropus vampyrus depending on the phylogenetic positions.
Among vampire bats, T2R39 is pseudogenized in all three vampire bats, two of which (Diaemus youngi and Diphylla ecaudata) share multiple frame-shifting deletions and nonsense mutations that are unshared in the third vampire bat (Desmodus rotundus) (figure 2). Because Diaemus youngi and Diphylla ecaudata diverged at the origin of vampire bats, the common ORF-disrupting mutations between them suggest that the pseudogenization of T2R39 occurred in the common ancestor of vampire bats following additional mutations in Desmodus rotundus (figure 1). However, we cannot rule out the possibility that T2R39 was pseudogenized independently in the three bats. Moreover, we observed T2R42 to have one 2-bp deletion and one premature stop codon shared between Desmodus rotundus and Diaemus youngi, although this gene remains intact in Diphylla ecaudata. This result suggests that the pseudogenization of T2R42 arose in the common ancestor of Desmodus rotundus and Diaemus youngi after its separation from Diphylla ecaudata. Additionally, T2R5 and T2R40 are pseudogenized in two vampire bats with no shared ORF-disrupting mutations, and T2R3 and T2R38 contain disruptive mutations in one of the three vampire bats, suggesting that the four genes were pseudogenized independently. Therefore, we found extensive losses of T2Rs in vampire bats, but the common disruptive mutations that cause pseudogenization among all three vampire bats are absent, despite them sharing a common ancestry of blood-feeding [9,30,31].
(c). Likelihood ratio tests of selective pressures on bat bitter taste receptors and taste signalling pathway
To examine the functional implications of T2Rs in non-vampire bats and to explore when the functional constraint on T2Rs became relaxed in vampire bats, we estimated the ω ratio for each of the nine T2Rs using a likelihood approach [22]. We undertook three tests for each gene, respectively. First, we analysed all non-vampire bats in this study, and estimated the same ω (model A in table 1) for all branches of the species tree (figure 1). The ω ratio is significantly smaller than 1 in each of the seven T2Rs (see the comparison with model B in table 1), suggesting that these genes are under purifying selection and thus functionally important. By contrast, the remaining two genes (T2R1 and T2R42) have an elevated ω ratio close to 1 (table 1), indicative of a relaxation of functional constraint on the two genes. Second, we inferred the sequence of the common ancestor of vampire bats (black circle in figure 1) for each of the nine T2Rs using both Bayesian and parsimony approaches [21,23], and estimated ω ratios of T2Rs for the common ancestor and other bats. We found that a model (model D in table 1) that allows a variation in ω between the common ancestor of vampire bats and all other bats is not significantly better than a simpler model (model C in table 1) that assumes the same ω across the tree for any gene (see table 1 for p-values). Third, we removed the nonsense mutations of pseudogenized T2Rs in the three vampire bats and compared them with sequences from other bats. For each gene, we examined a model (model F in table 1) allowing a variation in ω between the ancestral branch of vampire bats and four branches connecting the three vampire bats. We found that the ω ratio of each gene for the ancestral branch is not significantly different from that of the four branches (see the p-values in table 1), after comparing with a simpler model (model E in table 1) assuming that the same ω ratio for the five branches. For details of parameter estimates for selection tests on bat T2Rs, see the electronic supplementary material, table S4. Collectively, these results suggest that seven of nine T2Rs are under strong functional constraint and evolutionarily conserved, and that relaxation of functional constraint resulting in pseudogenized T2Rs may have arisen recently.
In addition to taste receptors, taste signalling pathways downstream of taste receptors are also essential for taste function. For example, CALHM1 (calcium homeostasis modulator 1) contributes to neurotransmission of taste stimuli; the loss of CALHM1 has rendered severely impaired responses to sweet, umami and bitter tastants [32]. We sequenced the complete coding sequences of Calhm1 from all three vampire bats and seven other bats in this study (figure 1) and found these genes to be complete and intact in all bats. Likelihood ratio tests of selective pressures suggest that Calhm1 is under strong purifying selection in bats (table 1). For details of these selection tests, see the electronic supplementary material, text S1 and table S4. Coupled with the observations of many intact and evolutionarily conserved T2Rs in vampire bats (figure 1 and table 1), our genetic data unambigously suggest that vampire bats still retain some bitter taste function, despite the losses of sweet and umami tastes [9,10].
4. Discussion
Behavioural tests have demonstrated that vampire bats possess poorly developed taste ability because they showed indifference to sweet and detected bitter, sour and salty tastants in high concentrations [11], and they even lost taste-aversion learning for poison avoidance [33]. Our genetic data are fully consistent with the behavioural tests. First, the sweet taste receptor gene (T1R2) was pseudogenized in each of the three vampire bats [9], which appeared consistent with the behavioural study [11]. In addition, we found the T1R3 to have a common 26 bp deletion in Desmodus rotundus [10] and Diphylla ecaudata, which would shift the ORF and result in loss of most transmembrane domains of the receptor in their common ancestor (electronic supplementary material, figure S4). Because T1R3 encodes the shared subunit of sweet and umami taste receptors [34], this finding strongly suggests that both sweet and umami tastes were lost in the common ancestor of vampire bats approximately 26 Ma [9], although the umami taste sensitivity has not been examined behaviourally [11]. Second, many pseudogenized T2Rs in vampire bats suggest that their bitter taste is greatly reduced and the reduction of bitter taste was also observed behaviourally in Desmodus rotundus [11]. Third, the evolutionary conservation of several T2Rs and taste signaling pathway strongly support the behavioural finding in which vampire bats still retain some bitter taste ability, evidenced by the detection of bitter tastants in relatively higher concentrations [11]. Consistent with the genetic data supporting the view of retaining some bitter taste in vampire bats, anatomical studies discovered normal taste buds in the canonical taste structures [35] and electrophysiological recordings identified functional taste receptors in these bats [36].
In addition to the bottlenose dolphin [3] and other whales [37], vampire bats also showed the evolutionary reduction of bitter taste function, suggesting that the reduction or major loss of bitter taste in animals is more pervasive than previously believed. All three vampire bats are obligate feeders on mammalian or bird blood [30], a food type that is unlikely ever to be bitter or toxic to these animals. This highly specialized diet with extremely narrow components would result in extensive reduction of bitter taste function in vampire bats, which would never encounter toxic foods in nature, despite many natural toxins tasting bitter [4,5]. Furthermore, instead of just taste, vampire bats use a combination of smell, echolocation and heat to find their prey and locate the skin with rich capillaries [7]. The utilization of various sensory systems may have further rendered the sense of taste less important [9]. Nonetheless, in view of the residual bitter taste conferring avoidance to higher concentrations of bitter tastants in vampire bats [11], it is not unexpected to observe many putatively functional T2Rs in these animals. Although the functional T2Rs are unlikely to play a major role in food selection for vampire bats, they could function in several extragustatory tissues [38]. For example, T2Rs are expressed in the gastrointestinal and tracheal tracts [38–40]; T2Rs are also involved in additional functions apart from bitter taste, such as regulation of glucose homeostasis [41] and delay of gastric emptying [42]. Analogous to these findings, the intact T2Rs in vampire bats may function in extra-oral tissues. An alternative hypothesis to explain our finding of many intact T2Rs in vampire bats is that the ancestors of vampire bats did not originally feed on blood and the specific dietary changes may have arisen recently, although these animals share a common ancestry of blood-feeding [9,30,31]. Regardless, future scrutiny of expression profiling and functional characterization of T2Rs in vampire bats will provide a better understanding of the evolution of bitter taste in animals.
Supplementary Material
Acknowledgements
We thank Julie Feistein and Neil Duncan from the American Museum of Natural History for access to bat tissues. We are grateful to Zubing Zhan and Liang Ao for technical assistance in the laboratory.
Data accessibility
Funding statement
This work was supported by National Natural Science Foundation of China (31300313) and a start-up fund from Wuhan University awarded to H.Z.
References
- 1.Yarmolinsky DA, Zuker CS, Ryba NJP. 2009. Common sense about taste: from mammals to insects. Cell 139, 234–244. ( 10.1016/j.cell.2009.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li D, Zhang J. 2014. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol. Biol. Evol. 31, 303–309. ( 10.1093/molbev/mst219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang P, Josue J, Li X, Glaserb D, Li W, Branda JG, Margolskeea RF, Reeda DR, Beauchamp GK. 2012. Major taste loss in carnivorous mammals. Proc. Natl Acad. Sci. USA 109, 4956–4961. ( 10.1073/pnas.1118360109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glendinning JI. 1994. Is the bitter rejection response always adaptive? Physiol. Behav. 56, 1217–1227. ( 10.1016/0031-9384(94)90369-7) [DOI] [PubMed] [Google Scholar]
- 5.Garica J, Hankins WG. 1975. The evolution of bitter and the acquisition of toxiphobia. In Olfaction and Taste V, Proc. of the 5th Int. Symp. in Melbourne, Australia (eds. Denton DA, Coghlan JP.), pp. 39–45. New York, NY: Academic Press. [Google Scholar]
- 6.de Jong P, Holloway G, Brakefield PM, de Vos H. 1991. Chemical defence in ladybird beetles (Coccinellidae) II. Amount of reflex fluid, the alka-loid adaline and individual variation in defence in 2-spot ladybirds (Adalia bipunctata). Chemoecology 2, 15–19. ( 10.1093/beheco/art013) [DOI] [Google Scholar]
- 7.Altringham JD. 1996. Bats: biology and behaviour. Oxford, UK: Oxford University Press. [Google Scholar]
- 8.Zhao H, Zhang J. 2012. Mismatches between feeding ecology and taste receptor evolution: an inconvenient truth. Proc. Natl Acad. Sci. USA 109, E1464 ( 10.1073/pnas.1205205109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao H, Zhou Y, Pinto CM, Charles-Dominique P, Galindo-González J, Zhang S, Zhang J. 2010. Evolution of the sweet taste receptor gene Tas1r2 in bats. Mol. Biol. Evol. 27, 2642–2650. ( 10.1093/molbev/msq152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao H, Xu D, Zhang S, Zhang J. 2012. Genomic and genetic evidence for the loss of umami taste in bats. Genome Biol. Evol. 4, 73–79. ( 10.1093/gbe/evr126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thompson RD, Elias DJ, Shumake SA, Gaddis SE. 1982. Taste preferences of the common vampire bat (Desmodus rotundus). J. Chem. Ecol. 8, 715–721. ( 10.1007/BF00988313) [DOI] [PubMed] [Google Scholar]
- 12.Bahlman JW, Kelt DA. 2007. Use of olfaction during prey location by the common vampire bat (Desmodus rotundus). Biotropica 39, 147–149. ( 10.1111/j.1744-7429.2006.00218.x) [DOI] [Google Scholar]
- 13.Kurten L, Schmidt U. 1982. Thermoperception in the common vampire bat (Desmodus rotundus). J. Comp. Physiol. A 146, 223–228. ( 10.1007/BF00610241) [DOI] [Google Scholar]
- 14.Zhang G, et al. 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science 339, 456–460. ( 10.1126/science.1230835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi P, Zhang J. 2006. Contrasting modes of evolution between vertebrate sweet/umami receptor genes and bitter receptor genes. Mol. Biol. Evol. 23, 292–300. ( 10.1093/molbev/msj028) [DOI] [PubMed] [Google Scholar]
- 16.Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182. ( 10.1007/978-1-59745-574-9_7) [DOI] [PubMed] [Google Scholar]
- 17.Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307, 580–584. ( 10.1126/science.1105113) [DOI] [PubMed] [Google Scholar]
- 18.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. ( 10.1093/molbev/msr121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. ( 10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 20.Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. ( 10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 21.Yang Z, Kumar S, Nei M. 1995. A new method of inference of ancestral nucleotide and amino acid sequences. Genetics 141, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang ZH. 2007. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. ( 10.1093/molbev/msm088) [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Nei M. 1997. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J. Mol. Evol. 44(Suppl. 1), S139–S146. ( 10.1007/PL00000067) [DOI] [PubMed] [Google Scholar]
- 24.Shi P, Zhang J, Yang H, Zhang YP. 2003. Adaptive diversification of bitter taste receptor genes in mammalian evolution. Mol. Biol. Evol. 20, 805–814. ( 10.1093/molbev/msg083) [DOI] [PubMed] [Google Scholar]
- 25.Zhou Y, Dong D, Zhang S, Zhao H. 2009. Positive selection drives the evolution of bat bitter taste receptor genes. Biochem. Genet. 47, 207–215. ( 10.1007/s10528-008-9218-y) [DOI] [PubMed] [Google Scholar]
- 26.Blum M. 1981. Chemical defense of arthropods. New York, NY: Academic Press. [Google Scholar]
- 27.Weatherston J, Percy JE. 1970. Arthropod defensive secretions. New York, NY: Academic Press. [Google Scholar]
- 28.Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. 2000. T2Rs function as bitter taste receptors. Cell 100, 703–711. ( 10.1016/S0092-8674(00)80706-0) [DOI] [PubMed] [Google Scholar]
- 29.Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. 2000. A novel family of mammalian taste receptors. Cell 100, 693–702. ( 10.1016/S0092-8674(00)80705-9) [DOI] [PubMed] [Google Scholar]
- 30.Fenton MB. 1992. Wounds and the origin of blood-feeding in bats. Biol. J. Linnean Soc. 47, 161–171. ( 10.1111/j.1095-8312.1992.tb00662.x) [DOI] [Google Scholar]
- 31.Wetterer AL, Rockman MV, Simmons NB. 2000. Phylogeny of phyllostomid bats (Mammalia : Chiroptera): data from diverse morphological systems, sex chromosomes, and restriction sites. Bull. Am. Mus. Nat. Hist. 248, 1–200. () [DOI] [Google Scholar]
- 32.Taruno A, et al. 2013. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature 495, 223–226. ( 10.1038/nature11906) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ratcliffe JM, Fenton MB, Galef BG., Jr 2003. An exception to the rule: common vampire bats do not learn taste aversions. Anim. Behav. 65, 385–389. ( 10.1006/anbe.2003.2059) [DOI] [Google Scholar]
- 34.Xu H, Staszewski L, Tang H, Adler E, Zoller M, Li X. 2004. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc. Natl Acad. Sci. USA 101, 14 258–14 263. ( 10.1073/pnas.0404384101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park H, Hall ER. 1951. The gross anatomy of the tongues and stomachs of eight New World bats. Trans. Kans. Acad. Sci. 54, 64–72. [Google Scholar]
- 36.Suthers RA. 1970. Vision, olfaction, taste. In Biology of bats (ed. Wimsatt WA.). New York, NY: Academic Press. [Google Scholar]
- 37.Feng P, Zheng JS, Rossiter SJ, Wang D, Zhao H. 2014. Massive losses of taste receptor genes in toothed and baleen whales. Genome Biol. Evol. 6, 1254–1265. ( 10.1093/gbe/evu095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behrens M, Meyerhof W. 2011. Gustatory and extragustatory functions of mammalian taste receptors. Physiol. Behav. 105, 4–13. ( 10.1016/j.physbeh.2011.02.010) [DOI] [PubMed] [Google Scholar]
- 39.Rozengurt E, Sternini C. 2007. Taste receptor signaling in the mammalian gut. Curr. Opin. Pharmacol. 7, 557–562. ( 10.1016/j.coph.2007.10.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu SV, Rozengurt N, Yang M, Young SH, Sinnett-Smith J, Rozengurt E. 2002. Expression of bitter taste receptors of the T2R family in the gastrointestinal tract and enteroendocrine STC-1 cells. Proc. Natl Acad. Sci. USA 99, 2392–2397. ( 10.1073/pnas.042617699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dotson CD, et al. 2008. Bitter taste receptors influence glucose homeostasis. PLoS ONE 3, e3974 ( 10.1371/journal.pone.0003974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glendinning JI, Yiin YM, Ackroff K, Sclafani A. 2008. Intragastric infusion of denatonium conditions flavor aversions and delays gastric emptying in rodents. Physiol. Behav. 93, 757–765. ( 10.1016/j.physbeh.2007.11.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.