Abstract
The human leukocyte antigen (HLA) restriction of the IgE response to different allergens in humans has been a subject of numerous published studies. However, the role and contribution of specific HLA class II molecules in the pathogenesis of allergic airway inflammation are unknown and difficult to assess. HLA-DQ6 and HLA-DQ8 transgenic mice lacking endogenous mouse class II gene expression were actively immunized and later challenged intranasally with short ragweed (SRW) allergenic extract. The HLA-DQ transgenic mice developed pulmonary eosinophilia and lung tissue damage. We also found an increase in total protein (TP) level and IL-5 production in bronchoalveolar lavage (BAL) fluid and an increase in SRW-specific Th2-type immunoglobulins (IgG1, IgG2b) and total serum IgE levels. Under similar treatment, DQ-negative full-sib control mice were normal. The allergic response could be significantly inhibited or abrogated in HLA-DQ mice by systemic treatment with anti-DQ mAb. The in vivo responses of HLA-DQ6 and HLA-DQ8 mice showed differences in terms of levels of eosinophilia, BAL protein, IL-5 concentration, and lung hyperreactivity to inhaled methacholine. These findings demonstrate the crucial role for specific HLA-DQ molecules in SRW-specific CD4+ T-cell activation and resulting recruitment of eosinophils into the airways.
J. Clin. Invest. 103:1707–1717 (1999).
Introduction
The familial incidences of allergy and asthma suggest a genetic basis for these diseases. Recently, genome-wide searches and candidate gene approaches have been used to examine the possible involvement of a number of genes in the development of atopy and asthma. The regions of potential linkage to markers were detected with one or more asthma phenotypes on chromosomes 5q, 6p, 11q, 12q, 13q, and 14q, respectively (1–3).
Human leukocyte antigen (HLA) molecules are encoded by highly polymorphic gene families located on chromosome 6p. They play a pivotal role in regulating the T-cell immune response and have been implicated in susceptibility to a wide range of diseases with an immunologic basis. The association of HLA haplotypes and ragweed allergy was the first human Ir (immune response) gene to be recognized (4), and HLA-DR2 and HLA-DQ6 restriction of IgE reactions to antigen 5 is well documented (5–7). Recently, strong relationships between the immune response to several highly purified allergens and specific HLA-DR and DQ haplotypes have been published (reviewed in ref. 8). In addition, specific HLA-DR and DQ molecules have been shown to be implicated in house dust mite asthma (9, 10), aspirin-induced asthma (11), soybean epidemic asthma (12), and occupational asthma and atopy involving isocyanates (13) or acid anhydrides (14). However, the role and contribution of specific HLA class II molecules in the development of allergic asthma are unknown and are difficult to assess in humans because of their genetic heterogeneity.
Eosinophils play an important role in the pathogenesis of allergic airway inflammation. The infiltration of the airway wall with a large number of activated eosinophils and release of their toxic granule proteins and lipid metabolites cause damage to bronchial epithelial cells (15–19). It is believed that an airway hyperresponsiveness, the hallmark of asthma, occurs as a result of this eosinophil-mediated damage. In the majority of clinical studies, pulmonary eosinophilia has been shown to correlate with disease severity (15, 17), and the resolution of airway eosinophilia correlates with the remission of asthma symptoms (16).
Recently, a number of murine models of antigen-induced pulmonary eosinophilia and airway hyperreactivity (AHR) have been developed to study the role of B cells (20, 21), T cells (20, 22–24), mast cells (25), NK cells (20), dendritic cells (26), IgE and its receptor (27, 28), cytokines (29–32), leukotrienes (33), and costimulatory molecules (34, 35) in the mechanisms of immune responses in asthma. However, in all of these animal models, MHC class II molecules are mouse derived. We have generated transgenic mice expressing human DQ6 (HLA-DQA1*0103 and HLA-DQB1*0601) and DQ8 (HLA-DQA1*0301 and HLA-DQB1*0302) genes (36–38). These mice lack endogenous class II molecules, and the only functional class II molecules on antigen-presenting cells (APCs) are human DQ6 or DQ8.
We previously reported that our HLA-DQ transgenic mice vigorously responded to short ragweed (SRW) in vitro, with Th2-type cytokine production (39). The in vitro response was mediated by CD4+ HLA-DQ–restricted T cells. The current study shows that HLA-DQ mice develop significant lung eosinophilia, epithelial damage, and AHR in response to intranasally administered SRW allergen. The disease is allergen specific, restricted by the HLA-DQ molecules, and mediated by CD4+ T cells. The humanized mouse model of SRW-induced allergic asthma will be valuable in identifying allergenic epitopes critical for disease and development of potential immunotherapy.
Methods
Mice.
The production and characterization of transgenic mice expressing human DQ6 (HLA-DQA1*0103 and HLA-DQB1*0601) and DQ8 (HLA-DQA1*0301 and HLA-DQB1*0302) genes in mice deficient in endogenous class II molecules (H-2Aβ0) have been described previously (36–38). Expression of HLA class II molecules in H-2Aβ0 mice induces the selection of CD4+ Vβ TCR+ cells and restores CD4+ T-cell population in the periphery to a substantial level (5.0–9.3%) (37). There is no surface expression of H-2Aαb, H-2Eβb, or H-2Aα/HLA-DQβ hybrid molecules on peripheral blood lymphocyte in HLA-DQ transgenic mice (36–38). The DQ-negative, Aβ0 full sibs were used as controls. Mice were bred and maintained in the pathogen-free Immunogenetics Mouse Colony at the Mayo Clinic.
Flow cytometry.
The levels of expression of HLA-DQ molecules were monitored on freshly prepared spleen mononuclear cells (MNCs) and bronchoalveolar lavage (BAL) cells using two-color flow cytometry (37, 38). Commercial anti-murine conjugated mAb’s (PharMingen, San Diego, California, USA) used for cell staining included anti–CD3-PE (17A2), anti–B220-PE (RA3-6B2), anti–Mac1α-PE (M1/70), and anti–CD11c-FITC (HL3). Anti–HLA-DQ mAb IVD12 was obtained from a cell line prepared in our laboratory. DTAF goat anti-mouse IgG Fc (Accurate Chemical & Scientific Corp., Westbury, New York, USA) was used as a secondary antibody. Cells treated with biotin-conjugated anti-DQ (IVD12) were then cultured with PE-labeled streptavidin (PharMingen). Both single- and two-color flow cytometry were performed using a FACS Vantage cytometer and analyzed with CellQuest software (both from Becton Dickinson Immunocytometry Systems, Mountain View, California, USA).
Antigen.
SRW (Ambrosia artemisiifolia) extract was purchased from Bayer Corp. (Elkhart, Illinois, USA).
Anesthetic agent.
Stock avertin solution (10 g of 99% 2,2,2-tribromoethanol [Aldrich Chemical Co., Milwaukee, Wisconsin, USA]) was dissolved in 10 mL TERT-amyl alcohol (catalog no. A730-1; Fisher Scientific Co., Fairlawn, New Jersey, USA). One hundred microliters of stock solution was diluted in 5 mL PBS. An intraperitoneal injection of 1 mL per mouse was used for euthanizing, and 0.2–0.3 mL was administered for anesthesia.
Immunizations with SRW.
Age-matched H-2Aβ0, HLA-DQ6, HLA-DQ8, and, as a control for mouse genetic background, C57BL/6 (B6/J) mice were used. Each experimental group consisted of 4–5 animals. Mice were sensitized by intraperitoneal injection of 40 μg SRW extract adsorbed to 1 mg aluminum hydroxide in 0.5 mL sterile endotoxin-free PBS (pH 7.2; GIBCO BRL, Grand Island, New York, USA) A booster injection was given 7 days later. On day 14, mice were challenged intranasally 2 times 6 hours apart by application of 250 μg/100 μL of dialyzed extract in the nostrils under light anesthesia. Intranasal application was performed as described previously (33, 40) using a sterile 1-cc plastic syringe with a sterile plastic tip for micropipette. Vehicle-sensitized control mice were injected with an equal volume of aluminum hydroxide suspension in PBS and were challenged intranasally with PBS alone. Another group of mice was injected and boosted with SRW extract as already described here, but they were challenged intranasally with PBS only. Forty-eight hours after challenge, mice were euthanized and BAL fluids were withdrawn from mouse lungs.
In vivo antibody administration.
Purified non–complement-fixing anti–HLA-DQ mAb IVD12 or isotype-matched IgG1 antibody MOPC-21 (Sigma Chemical Co., St. Louis, Missouri, USA) was used. Five hundred micrograms of mAb was injected intraperitoneally 24 hours before, simultaneous with, and after each allergen application. All reagents for in vivo applications were routinely tested for endotoxin contamination using the Timed Gel Formation Endotoxin Kit (Sigma Chemical Co.) per manufacturer’s instructions. No contamination was observed.
Diffusion of nasally administered solutions into the mouse respiratory tract.
Ethidium bromide in PBS (4%) was applied intranasally to determine which parts of the mouse lungs came in contact with solutions given nasally (41).
BAL cell composition.
Mice were euthanized at 48 hours after the last intranasal challenge. BAL fluids were collected as described earlier (40). BAL cells were washed once with PBS by centrifugation at 200 g for 5 minutes. The cell pellets were resuspended in 1 mL PBS. Total leukocytes in BAL fluids were determined for each sample with standard hemocytometer. Cytospin preparations were made with 200 μL of BAL fluid (Cytospin 2; Shandon Inc., Pittsburgh, Pennsylvania) and stained by Giemsa. The differential cell counts were determined from 4 high-power fields using a ×10 wide-field eyepiece (Leeds Precision Instruments Inc., Minneapolis, Minnesota, USA) and were evaluated in a double-blind manner. The absolute macrophage, neutrophil, lymphocyte, and eosinophil counts were ascertained (32).
Total protein concentration in BAL fluids.
The level of total protein (TP) was measured using a commercial kit (Bio-Rad Laboratories Inc., Hercules, California, USA) using the Bradford method.
Cytokine protein levels in BAL fluids.
IL-4, IL-5, IFN-γ, and TNF-α were measured by sandwich ELISA. Mini-kits for IL-4, IFN-γ, and TNF-α from Genzyme Diagnostics (Cambridge, Massachusetts, USA) were used according to instructions provided. IL-5 content was measured using a mini-kit from Endogen Inc. (Cambridge, Massachusetts, USA). BAL samples were diluted 1:1 with dilution buffer before applying to plates. All ELISA assays were performed as described previously (39). The sensitivity was as follows: IL-4, 15 pg/mL; IL-5, 5 pg/mL; IFN-γ, 10 pg/mL; TNF-α, 10 pg/mL.
Histologic examination.
In separate experiments, the trachea and lungs were removed from euthanized mice (n = 4–5 per group) 48 hours after allergen challenge. Sections 4-mm thick were prepared as described (42), using 10% formalin for fixation and paraffin for embedding, and then were stained with hematoxylin and eosin. Evaluation of histologic alterations was done with 3 samples for each mouse in a double-blind study.
Immunohistochemistry.
The sections from formalin-fixed, paraffin-embedded lung tissue were deparaffinized using a xylene/ethanol/water gradient. To view and count eosinophils, immunohistochemical staining using a rabbit anti-mouse major basic protein (MBP), kindly provided by G.J. Gleich (Mayo Clinic), was performed as described (30). The sections were examined using fluorescence microscopy.
The specific staining for HLA-DQ expression was performed using a culture supernatant from the cell line producing mAb specific for HLA-DQ α-chain (IVD 12) as a primary antibody, and a biotinylated anti-mouse F(ab) (DAKO Corp., Carpinteria, California, USA) as a secondary antibody. The avidin-biotin complex (Vector Laboratories, Burlingame, California, USA) was used for detection and Hanker-Yates reagent (Polysciences Inc., Warrington, Pennsylvania, USA) for viewing (43). Slides were counterstained for 30 seconds with hematoxylin. The sections were examined using light microscopy.
Cell proliferation assays.
Mice were sensitized and challenged as already described here. Spleens were harvested 48 hours after intranasal challenge, and MNCs were prepared for in vitro cultures as described previously (37, 39). The cells were cocultured with medium alone, 10 μg/mL of ConA, or 200 μg/mL of SRW. Cultures were incubated for 24 hours and were then pulsed with 1 μCi of [3H]thymidine per well during the final 24-hour incubation. Triplicate wells were harvested and radioactivity was measured in a beta counter (Beckman Instruments, Palo Alto, California, USA).
The BAL cells were obtained from SRW-treated H-2Aβ0 and HLA-DQ6 mice on day 5 after challenge, pooled within the experimental group (n = 10 and n = 5, respectively), and prepared for the in vitro cultures. Briefly, 500,000 cells per well were cocultured with SRW for 48 hours, labeled with [3H]thymidine, and harvested on day 3 of incubation.
For in vitro mAb blocking study, culture supernatants from the cell lines producing mAb’s specific for HLA-DQ α-chain (IVD 12), HLA-DQ6/8 (TB1), HLA-DR (L227, cross-reacts with HLA-DQ6β), H-2Aαb (7-16-17), H-2Aβb (25-5-16), H-2Eα/βb (Y- 17), CD4 (GK1.5), and CD8 (53-6.72) were prepared in our laboratory (38, 39).
Airway hyperresponsiveness.
AHR in PBS- or SRW-sensitized mice was measured 48 hours after the last intranasal challenge by recording respiratory pressure curves by whole-body plethysmography (model PLY 3211; Buxco Electronics Inc., Sharon, Connecticut, USA) in response to the increasing doses of inhaled methacholine (Sigma Chemical Co.) as described previously (32, 44). This method allows measurements of spontaneous breathing in an unrestrained mouse. Values of PenH (enhanced pause, index of pulmonary airflow obstruction) were calculated by BioSystem XA software (Buxco Electronics, Inc., Sharon, Connecticut, USA) using the following equation: PenH = (Te/RT-1) × (Pef/Pif), where Te is expiration time, RT is relaxation time, Pef is peak expiratory flow, and Pif is peak inspiratory flow.
Allergen-specific antibody levels and serum total IgE.
H-2Aβ0, HLA-DQ6, and HLA-DQ8 mice were bled before the first immunization with SRW, on days 6 and 13 during the sensitization protocol, and again 48 hours and 96 hours after intranasal challenge. Sera were prepared and stored at –70°C until assayed for SRW-specific antibody and total IgE levels.
The amount of antigen-specific IgM, IgG1, IgG2a, IgG2b, IgG3, and IgA in the sera was measured by ELISA. Briefly, 10 μg/mL of SRW extract in PBS (pH 7.2) was used to coat plates (Immulon 1; Dynatech Laboratories Inc., Chantilly, Virginia, USA). After incubating with sera diluted 1:50 or 1:100 in Tris-buffered saline (pH 7.2) for 2 hours at 37°C, plates were washed, and appropriate alkaline-phosphatase–conjugated mAb (Southern Biotechnology Associates Inc., Birmingham, Alabama, USA) was added to the wells. After 1 hour of incubation at 37°C, and following washes, an optimal concentration of phosphatase substrate (p-NPP; Southern Biotechnology Associates Inc.) was added. Plates were incubated for 30 minutes at room temperature and then read in a microtiter autoreader (Bio-Rad Inc., Pleasanton, California, USA) at 415 nm, with a reference wavelength of 490 nm. Serum antibody levels were quantified by comparison to purified isotype standards (Southern Biotechnology Associates Inc.) added to each plate, using the Microplate Manager software for the Macintosh computer (Bio-Rad Inc.). The sensitivity was as follows: IgM, 5 ng/mL; IgG1, 10 ng/mL; IgG2a, 40 ng/mL; IgG2b, 20 ng/mL; IgA, 2.5 ng/mL; IgG3, 40 ng/mL.
An IgE-specific ELISA was used to quantitate total IgE antibody levels in serum using matching antibody pairs (R35-72 and R35-92) obtained from PharMingen. As an internal positive control, pooled serum from SRW-immunized BALB/c mice was included in each assay.
Statistics.
Data are summarized as mean ± SEM. To calculate significance levels between treatment groups, Student’s t test (Sigma Plot) was used. Differences between values were considered to be significant at P < 0.05.
Results
Expression of HLA-DQ molecule in the BAL and lymphoid tissue of transgenic mice.
Flow cytometry analysis of spleen MNCs revealed that DQ transgene is expressed on 7.38–15.41% of CD3+ cells, 2.71–8.15% of Mac 1α+ cells, 25.68–32.51% of B220+ cells, and 0.9–1.53% of CD11c+ cells. In BAL, 80.66–92.92% of Mac 1α+ cells and 79.68–90.33% of B220+ cells coexpressed DQ antigen. There was no significant difference in transgene expression between HLA-DQ6 and HLA-DQ8 mice.
Distribution in the respiratory tract of solutions administered nasally.
Using 100 μL of ethidium bromide for intranasal administration, bright signals were found in the mouse trachea, bronchi, and bronchioli, whereas staining was weak in the lung parenchyma (data not shown). Therefore, even if intranasal delivery of liquid allergen does not completely reflect the natural way of human exposure to airborne allergens, this volume allows the antigen deposition in small airway vessels.
SRW-induced BAL eosinophilia is present in HLA-DQ transgenic mice after airway challenge.
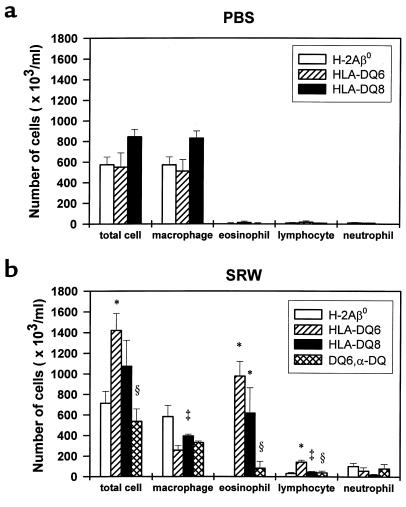
In the unimmunized H-2Aβ0 and HLA-DQ6 mice, BAL fluids yielded 515,000 ± 84,606 and 490,000 ± 61,913 cells/mL, respectively. The majority of cells (>98%) were small resting macrophages (not shown). The total number of cells and cellular distribution in the BAL fluids of PBS-treated mice was similar to that of naive mice (Figures 1a and 2a). In contrast, there was a significant increase in total number of BAL cells at 48 hours after SRW intranasal challenge in sensitized HLA-DQ6 transgenic mice (Figure 2, a and b; P < 0.01 compared with PBS-treated counterparts and SRW-treated H-2Aβ0 mice). The HLA-DQ mice, but not MHC class II knockout mice, were able to develop eosinophilic infiltration into BAL (Figure 1, c and b; and Figure 2b), which amounted to 69.00 ± 5.14% and 51 ± 14.30% in HLA-DQ6 and HLA-DQ8 mice, respectively. BAL lymphocyte differential was greatly increased in HLA-DQ6 mice (Figure 2b; P < 0.01 vs. H-2Aβ0 mice). Thus, eosinophils were the primary cell type in BAL of SRW-treated HLA-DQ mice, whereas macrophages predominated in H-Aβ0 mice. The genetic background control group of B6/J mice showed only 10 ± 5% of eosinophilia in response to SRW. These data indicate that the prevalence of eosinophilia in BAL fluids after allergen inoculation depends on the presence of specific HLA-DQ molecules.
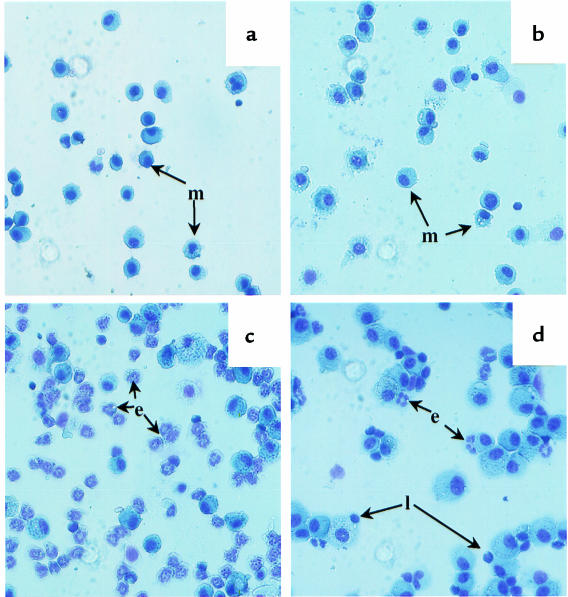
Figure 1.
Representative examples of BAL fluid cells isolated from PBS-treated HLA-DQ or H-2Aβ0 mice (a), SRW-sensitized and -challenged H-2Aβ0 (b) and HLA-DQ6 (c) mice, and anti-DQ–treated SRW-immunized HLA-DQ6 mice (d) 48 hours after intranasal challenge with allergenic extract. No eosinophils were recovered in BAL from any groups of PBS-treated mice (a) or SRW-sensitized and -challenged MHC class II knockout mice (b). Eosinophils were found to dominate the cellular infiltrate into BAL of HLA-DQ6 and HLA-DQ8 transgenic mice (c). Only a few eosinophils can be seen in anti-DQ–treated SRW-immunized mice (d). e, eosinophils; l, lymphocytes; m, macrophages.
Figure 2.
Cellular composition of BAL fluid of PBS-treated (a) and SRW-sensitized and -challenged (b) mice. Each experimental group consisted of 4–5 animals. Mice were immunized as described in Methods. BAL fluids were obtained at 48 hours after appropriate (PBS or SRW) intranasal challenge. The results for each group of mice are expressed as mean ± SEM from 1 of 3 independent experiments. *Significant differences (P < 0.04) between SRW-treated H-2Aβ0 and HLA-DQ mice. ‡P < 0.05 HLA-DQ6 vs. HLA-DQ8 mice. §P < 0.05 anti-DQ–treated HLA-DQ6 vs. mAb-untreated SRW-immunized counterparts.
In vivo treatment of HLA-DQ6 mice with anti-DQ mAb during sensitization inhibits airway eosinophilia.
To evaluate the importance of HLA-DQ molecules on APCs in the development of allergic inflammation in our model, HLA-DQ6 mice were treated with anti-DQ mAb as described in Methods. The number of total leukocytes, macrophages, eosinophils, and lymphocytes recovered from BAL of anti-DQ–treated mice was significantly lower than that in nontreated mice (Figure 2b). Eosinophils represented only 10.9 ± 8.1% of the total cells recovered. At the same time, mice treated with control isotype antibody (mouse IgG1) did not show significant changes in either total number of lavage cells or in cell differentials compared with SRW-sensitized and -challenged counterparts (not shown). These results suggest that specific HLA-DQ molecules are essential for the development of allergen-induced BAL eosinophilia, as anti-DQ treatment of SRW-challenged HLA-DQ mice “normalized” the profile of the infiltrating cells.
Increased protein concentrations were detected in BAL fluids of SRW-treated HLA-DQ mice.
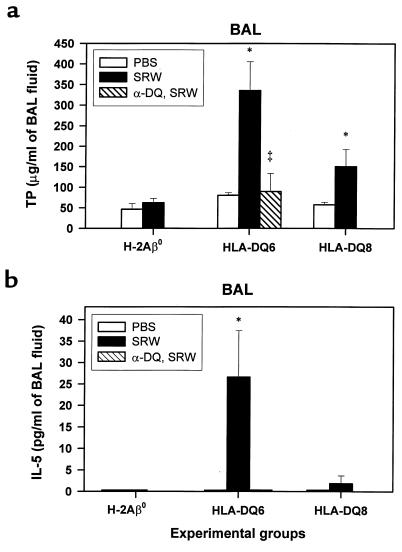
The changes in lung permeability elicited by SRW instillation were measured by the change in TP contents of BAL fluids. At 48 hours after intranasal challenge with SRW, a significant increase in TP was observed in both HLA-DQ6 and HLA-DQ8 mice compared with PBS-treated counterparts (Figure 3a; P < 0.01 and P < 0.05, respectively). TP level in BAL fluid of SRW-sensitized and -challenged B6/J mice (49.47 (8.72 μg/mL) was not different from other control groups (not shown). There was a more than 3-fold decrease in TP level in anti-DQ–treated HLA-DQ6 mice compared with mAb-untreated SRW-sensitized HLA-DQ6 mice (89.68 ± 43.79 and 335.54 ± 69.88 μg/mL, respectively) (Figure 3a). Thus, the leakage of plasma proteins due to increased vascular permeability or increased local protein production in the lung was one of the characteristics of allergic response to SRW in HLA-DQ transgenic mice, and it could be reduced by anti-DQ mAb treatment.
Figure 3.
TP levels (a) and IL-5 concentration (b) measured in BAL fluid supernatants. Mice were treated with PBS or sensitized to SRW as described in Methods. BAL fluids were obtained from each group of mice (n = 4–5) at 48 hours after last intranasal challenge with either PBS or SRW. TP levels (a) were detected by the Bradford method. A significant increase in TP was present in both HLA-DQ6 and HLA-DQ8 mice compared with PBS-treated counterparts (P < 0.01 and P < 0.05, respectively). Levels of IL-5 were determined by ELISA (b). There was no detectable IL-5 in BAL from PBS-treated mice. SRW sensitization and challenge resulted in the increase of IL-5 content in BAL of HLA-DQ mice. There was no detectable IL-5 in BAL of SRW-sensitized H-2Aβ0 mice or anti-DQ–treated SRW-sensitized HLA-DQ6 mice. Data are expressed as mean ± SEM from 1 of 3 independent experiments.
Increased levels of IL-5 in BAL fluids of SRW-treated HLA-DQ mice.
To examine the profile of cytokines involved in generating and perpetuating the lung inflammatory response in our models, BAL fluids obtained from PBS-treated or SRW-sensitized mice at 48 hours after appropriate challenge were analyzed for the presence of IL-4, IL-5, IFN-γ, and TNF-α cytokines. No IL-4, IFN-γ, or TNF-α was detected in any samples. No IL-5 was found in PBS-treated mice (Figure 3b). SRW sensitization and challenge resulted in the significant increase of IL-5 content in BAL of HLA-DQ6 mice (26.61 ± 10.81 pg/mL) but was lower in HLA-DQ8 mice (Figure 3b). There was no detectable IL-5 cytokine in BAL of SRW-sensitized H-2Aβ0 mice or anti-DQ–treated SRW-sensitized HLA-DQ6 mice. Thus, BAL eosinophilia is associated with an increase of the IL-5 secretion into airways of HLA-DQ transgenic mice and may be abrogated by systemic anti-DQ treatment.
Lung eosinophilia and epithelial damage in SRW-sensitized and -challenged HLA-DQ mice.
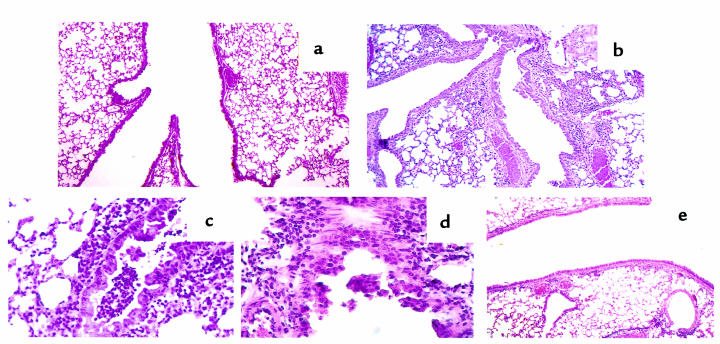
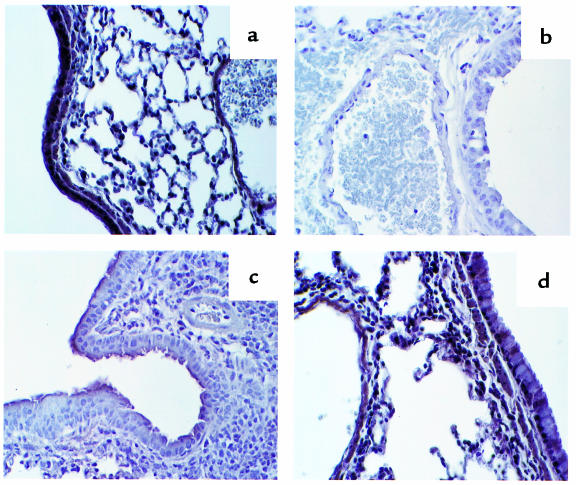
The extent of the allergic airway inflammation induced by SRW exposure was further studied by histologic analysis. The lungs of unimmunized or PBS-treated H-2Aβ0 and HLA-DQ6 transgenic mice appeared normal with no sign of inflammation (Figure 4a). The lungs of SRW-treated HLA-DQ6 and HLA-DQ8 mice contained multifocal dense inflammatory infiltrates (Figure 4b), and the infiltrates consisted primarily of eosinophils (Figure 4, c and d). Epithelial damage, including hypertrophy, sloughing off epithelial cells, and increased spaces between them (Figure 4, b–d), was observed in HLA-DQ mice. The lungs of SRW-sensitized and -challenged H-2Aβ0 mice or SRW-sensitized transgenic mice challenged intranasally with PBS were not significantly different from the PBS control (not shown). Systemic anti-DQ mAb treatment of HLA-DQ6 mice during sensitization to SRW significantly decreased the lung inflammation (Figure 4e).
Figure 4.
Histopathology of lung tissue from PBS (a) and SRW-sensitized and -challenged (b–e) mice. Tissue section were prepared as described in Methods and were stained with hematoxylin and eosin. The photomicrograph in a shows that there is no inflammation in lung tissue from PBS-treated mice (×10). This is contrasted with multiple sites of inflammation in tissue taken 48 hours after allergen challenge of SRW-sensitized HLA-DQ6 (b) transgenic mice (×10). (c and d) Lung sections of an HLA-DQ6 mouse treated with SRW show a diffuse eosinophilic infiltration around and in the bronchioli, with involvement of the bronchiolar epithelium and associated epithelial damage. (e) Reduction in peribronchial and perivascular inflammation in HLA-DQ6 mice after treatment with anti-DQ mAb.
Immunostaining for mouse MBP.
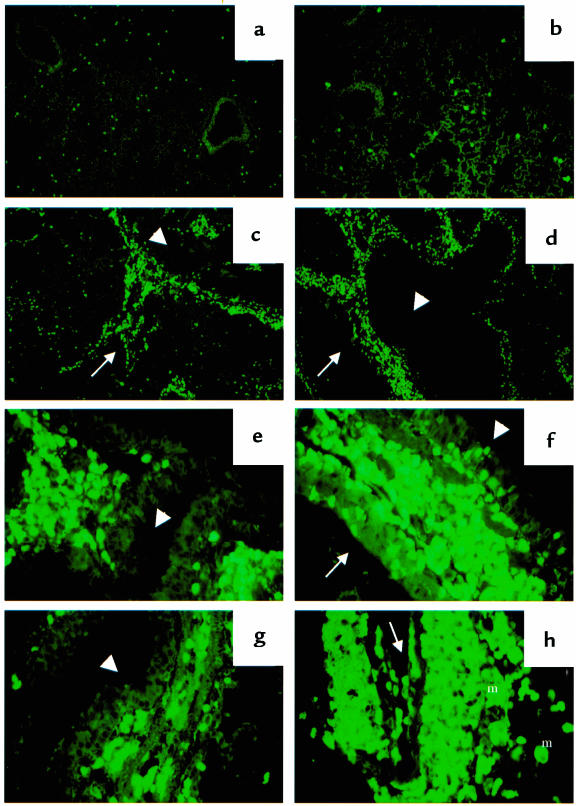
To view and examine the level of eosinophilia in the lung tissue and identify their granule release, we performed immunostaining of eosinophil MBP using anti-MBP antibody. In PBS-treated mice, a few mature eosinophils were distributed throughout the lung parenchyma (Figure 5a). A similar picture was seen in lungs of SRW-sensitized and -challenged H-2Aβ0 mice (Figure 5b). In contrast, the massive perivascular and peribronchial eosinophilic infiltration was found in both lines of HLA-DQ transgenic mice (Figure 5, c and d). Using higher magnification, margination and migration of eosinophils through the blood vessel wall to the epithelium, and their granule release in airway mucosa and epithelium, were detected (Figure 5, e–g). Also, as supportive data for eosinophil activation and granule release, positive staining of infiltrating macrophages for MBP as a result of its uptake was found (Figure 5h). However, no extensive areas of extracellular MBP were observed. In anti-DQ–treated HLA-DQ6 mice, eosinophils were primarily seen in the lung parenchyma and around some blood vessels, and only a few were noticed in the mucosal layer or the lumen of the airways (not shown).
Figure 5.
Immunostaining of lung tissue for mouse eosinophil MBP. H-2Aβ0, HLA-DQ6, or HLA-DQ8 mice were immunized, boosted, and challenged intranasally with SRW allergenic extract as described in Methods. PBS-treated, PBS-challenged mice served as a control for this study. (a) Lung of PSB-treated HLA-DQ6 mouse with a few eosinophils in lung tissue (×10). (b) Lung of SRW-sensitized and -challenged H-2Aβ0 mouse (×10). Severe perivascular and peribronchial concentration of eosinophils in SRW-treated HLA-DQ6 (c) and HLA-DQ8 (d) mice (×10). (e–h) High-power magnification of lungs from HLA-DQ mice (×40). Eosinophil migration from blood vessel to airway mucosa and epithelium in HLA-DQ6 (e) and HLA-DQ8 (f) transgenic mice, and their granule release (f and g). Positive staining of infiltrating macrophages for MBP (h) as a result of its uptake from degranulating eosinophils. Arrow, blood vessel; arrowhead, bronchiole; m, macrophages.
Decrease of HLA-DQ expression in lungs of transgenic mice by SRW treatment.
To study the lung tissue distribution of the HLA-DQ transgene products in mice, specific immunostaining was performed, rendering DQ-expressing sites brown and cell nuclei blue. Examination of lung tissue from PBS-treated HLA-DQ6 transgenic mice revealed that all epithelial cells lining the air spaces, and vascular endothelial cells expressed HLA-DQ (Figure 6a). This was not observed in lung sections of transgene-negative animals treated with PBS or SRW (Figure 6b). SRW sensitization and challenge of HLA-DQ transgenic mice resulted in a significant loss of transgene expression in lung tissue (Figure 6c). The infiltrating inflammatory cells that were primarily eosinophils did not express HLA-DQ. Low-intensity expression was detected only on the apical part of bronchial epithelium (Figure 6c) and on endothelium of some large blood vessels (not shown). Anti-DQ treatment during protocol of sensitization to SRW reconstitutes the expression of HLA-DQ in transgenic mice (Figure 6d).
Figure 6.
Immunohistochemical localization of HLA-DQ molecules in lungs, as detected by the brown avidin-biotin peroxidase reaction product. Representative areas of the airway tissue from PBS-treated HLA-DQ6 mice revealed transgene expression on epithelium and endothelium (a), which was not observed in the lungs of transgene-negative H-2Aβ0 mice (b). (c) SRW sensitization and challenge of HLA-DQ mice markedly reduce the antigen expression and lead to the prominent infiltration with cells not having HLA-DQ marker on their surface. (d) Anti-DQ treatment reconstitutes transgene expression in allergen-sensitized HLA-DQ6 mice.
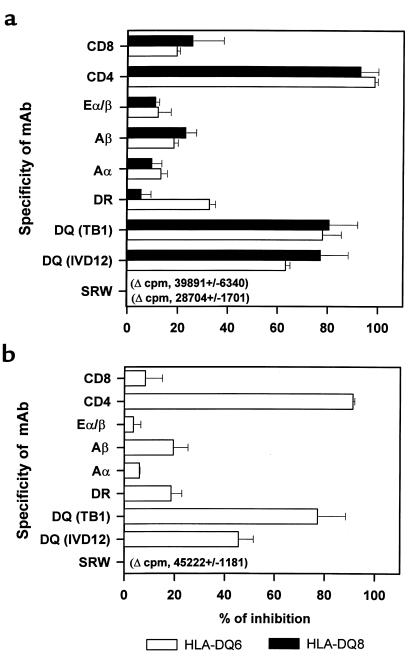
Inhibition of the immune response to SRW in transgenic mice by anti-DQ and anti-CD4 mAb’s.
To identify the subpopulation of T cells necessary for the response to SRW in transgenic mice, different monoclonal antibodies were added to the wells containing cultures of spleen MNCs with SRW (Figure 7a). Proliferation was inhibited in cultures with anti-CD4 mAb by 98.55 ± 1.27% for HLA-DQ6 mice and by 92.92 ± 7.21% for HLA-DQ8 mice. The use of anti-DQ mAb (TB1) resulted in the inhibition of in vitro response by 77.96 ± 7.52% (HLA-DQ6) and 80.5 ± 11.49% (HLA-DQ8). Other mAb’s used showed no significant effect. No proliferation was observed in cultures of spleen MNCs obtained from PBS-treated animals or SRW-treated H-2Aβ0 mice.
Figure 7.
Inhibition of SRW-specific HLA-DQ spleen MNC (a) and BAL cell (b) proliferative response. (a) Spleens were harvested from mice at 48 hours after intranasal challenge with SRW, and spleen MNCs were cocultured in vitro with allergenic extract in the presence of the indicated mAb’s. The data represent the percentage of inhibition of [3H]thymidine incorporation in triplicate cultures with SRW for 3 mice per experimental group from 1 of 2 independent blocking experiments. (b) BAL fluids were obtained from mice on day 5 after challenge, and BAL cells were cocultured with SRW and mAb for 3 days. The values displayed are the result of pooled BAL cells from 5 mice per strain and are representative of 2 analyses. Changes in cpm values for cultures with SRW alone (0% inhibition) are indicated.
BAL cells from HLA-DQ6 mice proliferated strongly during in vitro culture with SRW, as determined by 3[H]thymidine uptake (Figure 7b). Blocking of either DQ or CD4 molecules inhibited SRW-specific proliferation, whereas other mAb’s had no effect on SRW-induced response. The H-2Aβ0 BAL cells did not respond to SRW (change in cpm, 187 ± 138). Thus, the immune response to SRW in both the lymphoid tissue and BAL cellular infiltrates of HLA-DQ transgenic mice is mediated by CD4-positive HLA-DQ–restricted T cells.
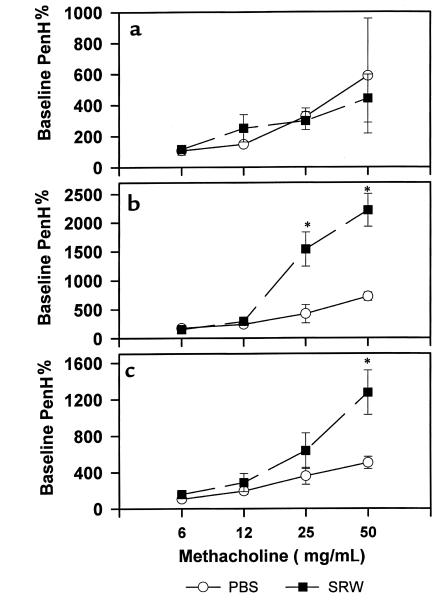
Increase of airway reactivity in SRW-treated HLA-DQ transgenic mice.
To study whether HLA-DQ6 and HLA-DQ8 molecules contribute to the development of airway hyperresponsiveness, we challenged PBS- or SRW-treated mice with increasing concentrations of methacholine in a whole-body plethysmograph. Figure 8 shows that SRW-treated HLA-DQ mice developed a strong AHR 48 hours after last intranasal challenge with allergenic extract, compared with PBS exposed counterparts. In contrast, SRW treatment of H-2Aβ0 mice had no significant effect on AHR, and the methacholine response was comparable to that observed in PBS control. Thus, specific HLA-DQ molecules are required for the induction of AHR in vivo.
Figure 8.
Assessment of lung physiology. Airway responsiveness in SRW-sensitized H-2Aβ0 mice (a) and transgenic mice expressing HLA-DQ6 (b) or HLA-DQ8 (c) molecules was measured 48 hours after intranasal challenge with SRW (filled squares). Control mice (open circles) were treated with PBS as described in Methods. Results are expressed for each methacholine concentration as percent of PenH (index of airway obstruction) values after PBS exposure (n = 3 for each experimental group). *P < 0.04 SRW sensitized/challenged vs. PBS sensitized/challenged HLA-DQ mice.
HLA-DQ6 mice are capable of producing Th2-type SRW-specific antibodies.
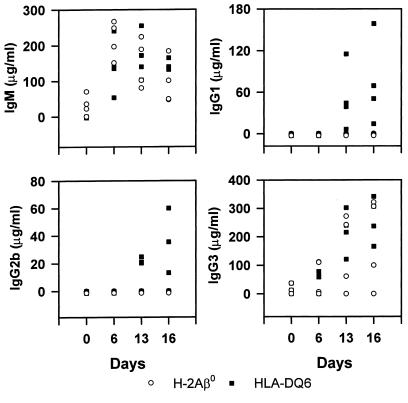
Mice were bled, and sera were collected at various times and assayed for the presence of SRW-specific antibodies by ELISA. The immune sera did not react with a control antigen, human thyroglobulin (OD, 0.01). The immunization of H-2Aβ0 and HLA-DQ6 mice with ragweed extract induced a high level of production of IgM-specific antibodies that showed gradual decline after immunizations (Figure 9). The IgG response was significantly increased after a booster injection and challenge and was composed mainly of IgG1, IgG2b, and IgG3 antibodies in HLA-DQ6 mice, and only IgG3 antibodies in H-2Aβ0 mice, and did not involve the IgG2a subclass (Figure 9). Serum levels of specific IgA were below the limit of detection (not shown). This confirms the ability of the HLA-DQ6 transgenic mice to generate Th2-dependent antibody responses to SRW extract (IgG1 and IgG2b); however, anti-ragweed IgM and IgG3 antibodies predominate. As expected, the levels of SRW-specific antibody isotypes in unimmunized mice were below the level of detection. However, trace amounts of SRW-specific IgG3 antibodies were detected in preimmune sera of both HLA-DQ6 and H-2Aβ0 mice, and IgM antibodies only in H-2Aβ0 mice.
Figure 9.
SRW-specific antibody. Mice were bled before the first immunization, immunized, and challenged intranasally with SRW as described in Methods, and then bled on days 6 and 13 during the protocol of sensitization, and again 48 hours (day 16) and 96 hours (day 18) after intranasal challenge. Levels of SRW-specific antibody were determined by ELISA. The points represent the concentration of antibody for individual mouse (n = 4 per group). Note the increase in IgG1 and IgG2b content in SRW-treated HLA-DQ6, but not H-2Aβ0, mice.
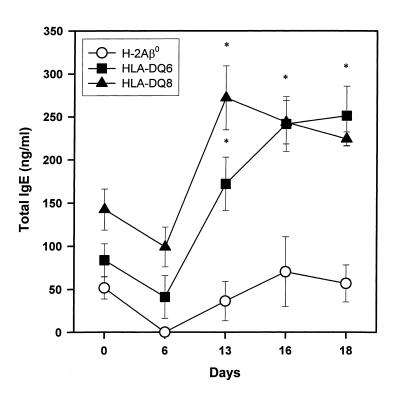
Increase in total IgE level in the immune sera of SRW-sensitized HLA-DQ mice.
In view of the association of allergic disorders with elevated serum IgE levels, we measured total IgE at different time points during sensitization to SRW. At day 6 after first injection with SRW, a decrease in serum IgE was found in all groups of mice (Figure 10). In the sera of SRW-boosted HLA-DQ6 and HLA-DQ8 mice, the significant increase in total IgE (241.35 ± 32.07 and 243.44 ± 25.23 ng/mL, compared with preimmune control 83.77 ± 18.95 and 142.35 ± 23.89 ng/mL, respectively; P < 0.01 and P < 0.02, respectively) was found, which was even more pronounced 48 hours after challenge in HLA-DQ6 mice. In contrast, H-2Aβ0 mice did not exhibit an increase of total serum IgE during sensitization and challenge.
Figure 10.
Changes in serum concentration of total IgE induced by SRW treatment in H-2Aβ0, HLA-DQ6, and HLA-DQ8 mice. Serum samples were analyzed for IgE content by ELISA. Results shown here are from a single experiment representative of 3 experiments and are expressed as mean ± SEM (n = 4 for each experimental group). *P < 0.02 compared with IgE concentration in preimmune sera (day 0).
Discussion
To our knowledge, the data presented here comprise the first report demonstrating the role and contribution of specific HLA class II molecules in the pathogenic mechanism(s) in atopic diseases and asthma. We have generated an HLA-DQ transgenic mouse model of SRW-induced asthma to demonstrate that (a) there is a preferential perivascular and peribronchial accumulation of eosinophils 48 hours after allergen challenge; (b) the local increase of IL-5 parallels infiltration of lungs with eosinophils; (c) there is a strong increase in airway reactivity to nonspecific bronchoconstrictor methacholine; (d) there is an induction of SRW-specific immunoglobulins corresponding to Th2-type helper function to B cells, and an increased total IgE level; (e) the in vivo response to SRW is mediated by CD4-positive HLA-DQ–restricted T cells; and (f) systemic treatment with anti-DQ mAb reduces airway eosinophilia and lymphocyte infiltration and effectively abrogates tissue damage and IL-5 release. Other important findings here include (a) the absence of allergic response in MHC class II knockout mice, and (b) significantly lower (6- to 5-fold) SRW-induced eosinophilic inflammation in the genetic background control C57BL/6 mice.
This study demonstrates the expression of HLA class II molecules in normal lung tissue in transgenic mice, similar to that in humans.Recent studies have shown that human lung alveolar (45, 46) and bronchial (45, 47) epithelial cells and vascular endothelial cells (47) express MHC class II molecules. Chronic bronchitis (48), transplant rejection (47), and chronic asthmatic conditions (48, 49) were accompanied by overexpression of class II MHC antigens on lung epithelial cells. Our studies shows that an acute attack of asthma may decrease HLA class II expression on bronchial epithelium (Figure 6). In a chronic stage, it may be enhanced.
We used SRW extract as a natural airborne allergen instead of ovalbumin, which has been used in most studies involving murine models of asthma (22, 25–27, 29, 31, 33–35, 42). Although it is believed that asthma can be induced mostly by house dust mite, cockroach, mold, cat and dog allergens, and birch pollen (50), several recent studies have shown the involvement of SRW pollen in atopic asthma in humans (51–54).
The availability of well-characterized inbred mice enabled the creation of murine models of antigen-induced bronchopulmonary eosinophilia to address selected pathogenetic issues (20, 33, 42, 55–61). In allergen-sensitized inbred mice, allergen challenge is noted to induce the same changes in immune profile that take place in human subjects with allergic asthma. The use of transgenic and knockout mice for the multitude of mouse cytokines (29, 31, 32, 62), cells (20–22, 25, 60, 62), and adhesion molecules (22) has shown new evidence for the role and significance of the immune system in the mechanisms of allergic tissue inflammation and AHR. Our HLA-DQ transgenic mice express individual functional human class II molecules in the absence of endogenous mouse class II genes, and they provide a new approach to study the genetics of human diseases (37, 38, 63). The severe inflammatory reaction, involving predominantly eosinophils, and strong AHR to nebulized methacholine measured in the lungs of our transgenic animals are most likely due to HLA-DQ–restricted immune responses. Observed variances in the in vivo responses of HLA-DQ6 and HLA-DQ8 mice to SRW (levels of eosinophilia, BAL protein, and IL-5 content, AHR) may be explained by previously demonstrated differences in epitope recognition (39) and their immunogeneity.
The HLA molecules have been implicated in several measures of the allergic phenotype and asthma in a number of studies (reviewed in ref. 1). The multicenter Collaborative Study on the Genetics of Asthma has shown an evidence for increased allele sharing for markers on 6p21.3-23 (P = 0.01) in Caucasian sibling pairs with asthma. Because 75% of the family members with asthma had at least 1 positive skin test, it was proposed that this finding may be similar to previously reported association between atopy and the HLA loci, or may be related to function of TNF-α gene located in this region (1). We did not find any measurable amount of TNF-α cytokine in BAL of HLA-DQ transgenic mice developing strong lung eosinophilia at 48 hours after allergen challenge. However, RT-PCR analysis of total RNA extracted from lung tissue showed the presence of TNF-α transcripts in all groups of mice sensitized to SRW including H-2Aβ0 mice (not shown in results).
Histologic findings in SRW-sensitized and -challenged HLA-DQ transgenic mice (Figure 4, b–d) are different from results obtained using the conventional laboratory mice (21, 22, 40, 42, 55, 64) and are more reminiscent of human asthmatic pathologies (17, 18, 65). A characteristic of human disease is the occurrence of eosinophil lysis with free eosinophil granules in the airway mucosa (18, 66). Immunostaining with polyclonal antibodies to mouse MBP enabled us to see these distributions in HLA-DQ transgenic mice (Figure 5, e–g) illustrative for eosinophil activation. These data indicate that eosinophil infiltration of lung tissue and epithelial damage depend on the presence of APCs bearing specific HLA class II molecules, as well as CD4+ T cells, because active sensitization of H-2Aβ0 mice with SRW allergen cannot induce eosinophil accumulation. Treatment with anti-DQ mAb greatly diminished SRW-induced airway eosinophilia and normalized the profile of infiltrating cells. Importantly, no detectable IL-5 was found in BAL of these treated animals. These findings support the possibility that allergen-specific CD4+ T cells can be abrogated for therapeutic purposes by specific manipulation of HLA class II antigen presentation. Thus, HLA class II transgenic mice offer the opportunity to investigate the role of distinct HLA molecules in allergen sensitivity and may be useful for devising new immune-based therapies for allergic asthma.
Acknowledgments
This research was supported by National Institutes of Health grant AI-14764 and by funds from Boehringer Ingelheim Pharmaceuticals Inc. The immunostaining for mouse MBP was done in the Dermatology Research Laboratory at Mayo Clinic with the technical assistance of Ellen Peterson and Terry George, under the supervision of Kristin M. Leiferman. The lung function measurements were performed in the Allergic Diseases Research Laboratory with generous help from Koji Iigima and Mickey Valyasevi. We thank Julie Hanson and her staff (Immunogenetics Mouse Colony) for outstanding mouse production; P. Zhou, S. Chen, and J. Baich for generating the transgenic mice; Michele Smart for tissue typing of transgenic mice and FACS analysis; Gerald J. Gleich (Allergic Diseases Research Laboratory) for the generous gift of the anti-mouse MBP antibody; Teresa Ryan, Carol Swanson, and Marcy Kulawski (General Hematology Laboratory) for hematoxylin/eosin staining of the cytospin slides; Linda McGee and her staff (Pathology Specimen Processing Laboratory) for help in preparing mouse lung histology slides; Tomas Beito for preparing the cell hybridomas; and Kristen M. Drescher for help with immunostaining for HLA-DQ. Also, we thank Gerald J. Gleich and H. Kita for providing helpful advice and supportive interest in our work. We are grateful to K.M. Leiferman, H. Kita, and Ellen Peterson for critically reviewing the manuscript.
References
- 1.Bleecker ER, Postma DS, Meyers DA. Evidence for multiple genetic susceptibility loci for asthma [review] Am J Respir Crit Care Med. 1997;156:S113–S116. doi: 10.1164/ajrccm.156.4.12tac7. [DOI] [PubMed] [Google Scholar]
- 2.Barnes KC, Marsh DG. The genetics and complexity of allergy and asthma. Immunol Today. 1998;19:325–332. doi: 10.1016/s0167-5699(97)01241-3. [DOI] [PubMed] [Google Scholar]
- 3.Marone J. Asthma: recent advances. Immunol Today. 1998;19:5–9. doi: 10.1016/s0167-5699(97)01187-0. [DOI] [PubMed] [Google Scholar]
- 4.Levine BB, Stember RH, Fontino M. Ragweed hayfever: genetic control and linkage to HLA haplotypes. Science. 1972;178:1201–1203. doi: 10.1126/science.178.4066.1201. [DOI] [PubMed] [Google Scholar]
- 5.Marsh DG, et al. HLA-Dw2: a genetic marker for human immune response to short ragweed pollen allergen Ra5. I. Response resulting primarily from natural antigenic exposure. J Exp Med. 1982;155:1439–1451. doi: 10.1084/jem.155.5.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang S-K, Zwollo P, Marsh DG. Class II major histocompatibility complex restriction of human T cell responses to short ragweed allergen, Amb a V. Eur J Immunol. 1991;21:1469–1473. doi: 10.1002/eji.1830210621. [DOI] [PubMed] [Google Scholar]
- 7.Goldstein R, Yang WH, Drouin MA, Karsh J. Studies of the HLA class II alleles involved in human responses to ragweed allergens Ambrosia artemisiifolia V (Ra5S) and Ambrosia trifida V (Ra5G) Tissue Antigens. 1992;39:122–127. doi: 10.1111/j.1399-0039.1992.tb01921.x. [DOI] [PubMed] [Google Scholar]
- 8.Joost van Neerven RJ, Ebner C, Yssel H, Kapsenberg ML, Lamb JR. T-cell responses to allergens: epitope-specificity and clinical relevance [review] Immunol Today. 1996;17:526–532. doi: 10.1016/0167-5699(96)10058-x. [DOI] [PubMed] [Google Scholar]
- 9.Stephan V, et al. Mite allergy, clinical atopy, and restriction by HLA class II immune response genes. Pediatr Allergy Immunol. 1996;7:28–34. doi: 10.1111/j.1399-3038.1996.tb00102.x. [DOI] [PubMed] [Google Scholar]
- 10.Gerbase-DeLima M, Gallo CA, Daher S, Sole D, Naspitz CK. HLA antigens in asthmatic children. Pediatr Allergy Immunol. 1997;8:150–152. doi: 10.1111/j.1399-3038.1997.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 11.Dekker JW, et al. Aspirin-induced asthma and HLA-DRB1 and HLA-DPB1 genotypes. Clin Exp Allergy. 1997;27:574–577. [PubMed] [Google Scholar]
- 12.Soriano JB, et al. HLA class II genes in soybean epidemic asthma patients. Am J Respir Crit Care Med. 1997;156:1394–1398. doi: 10.1164/ajrccm.156.5.9701064. [DOI] [PubMed] [Google Scholar]
- 13.Mapp CE, Balboni A, Baricordi R, Fabbri LM. Human leukocyte antigen association in occupational asthma induced by isocyanates. Am J Respir Crit Care Med. 1997;156:S139–S143. doi: 10.1164/ajrccm.156.4.12-t-12. [DOI] [PubMed] [Google Scholar]
- 14.Young RP, Barker RD, Pile KD, Cookson WOCM, Newman-Taylor AJ. The association of HLA-DR3 with specific IgE to inhaled acid anhydrides. Am J Respir Crit Care Med. 1995;151:219–221. doi: 10.1164/ajrccm.151.1.7812558. [DOI] [PubMed] [Google Scholar]
- 15.Azzawi M, et al. Identification of activated T lymphocytes and eosinophils in bronchial biopsies in stable atopic asthma. Am Rev Respir Dis. 1990;142:1407–1413. doi: 10.1164/ajrccm/142.6_Pt_1.1407. [DOI] [PubMed] [Google Scholar]
- 16.Bousquet J, et al. Eosinophilic inflammation in asthma. N Engl J Med. 1990;323:1033–1039. doi: 10.1056/NEJM199010113231505. [DOI] [PubMed] [Google Scholar]
- 17.Ohashi Y, Motojima S, Fukuda T, Makino S. Airway hyperresponsiveness, increased intracellular spaces of bronchial epithelium, and increased infiltration of eosinophils and lymphocytes in bronchial mucosa in asthma. Am Rev Respir Dis. 1992;145:1469–1476. doi: 10.1164/ajrccm/145.6.1469. [DOI] [PubMed] [Google Scholar]
- 18.Reed CE. The importance of eosinophils in the immunology of asthma and allergic disease. Ann Allergy. 1994;72:376–380. [PubMed] [Google Scholar]
- 19.Kita H, Gleich GJ. Chemokines active on eosinophils: potential roles in allergic inflammation. J Exp Med. 1996;183:2421–2426. doi: 10.1084/jem.183.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hamelmann E, et al. Allergic airway sensitization induces T cell activation but not airway hyper-responsiveness in B cell–deficient mice. Proc Natl Acad Sci USA. 1997;94:1350–1355. doi: 10.1073/pnas.94.4.1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korsgren M, Erjefalt JS, Korsgren O, Sundler F, Persson CGA. Allergic eosinophil-rich inflammation develops in lungs and airways of B cell–deficient mice. J Exp Med. 1997;185:885–892. doi: 10.1084/jem.185.5.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gonzalo JA, et al. Eosinophil recruitment to the lung in a murine model of allergic inflammation. The role of T cells, chemokines, and adhesion receptors. J Clin Invest. 1996;98:2332–2345. doi: 10.1172/JCI119045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Sanctis GT, et al. T-lymphocytes regulate genetically determined airway hyperresponsiveness in mice. Nat Med. 1997;3:460–462. doi: 10.1038/nm0497-460. [DOI] [PubMed] [Google Scholar]
- 24.Li X-M, Schofield B, Wang Q-F, Kim K-H, Huang S-H. Induction of pulmonary allergic responses by antigen-specific Th2 cells. J Immunol. 1998;160:1378–1384. [PubMed] [Google Scholar]
- 25.Takeda K, et al. Development of eosinophilic airway inflammation and airway hyperresponsiveness in mast cell–deficient mice. J Exp Med. 1997;186:449–454. doi: 10.1084/jem.186.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambrecht BN, Salomon B, Klatzmann D, Pauwels RA. Dendritic cells are required for the development of chronic eosinophilic airway inflammation in response to inhaled antigen in sensitized mice. J Immunol. 1998;160:4090–4097. [PubMed] [Google Scholar]
- 27.Haczku A, et al. CD23 deficient mice develop allergic airway hyperresponsiveness following sensitization with ovalbumin. Am J Respir Crit Care Med. 1997;156:1945–1955. doi: 10.1164/ajrccm.156.6.9701087. [DOI] [PubMed] [Google Scholar]
- 28.Mehlhop PD, et al. Allergen-induced bronchial hyperreactivity and eosinophilic inflammation occur in the absence of IgE in a mouse model of asthma. Proc Natl Acad Sci USA. 1997;94:1344–1349. doi: 10.1073/pnas.94.4.1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foster PS, Hogan SP, Ramsay AJ, Matthaei KI, Young IG. Interleukin 5 deficiency abolished eosinophilia, airway hyperreactivity, and lung damage in a mouse asthma model. J Exp Med. 1996;183:195–201. doi: 10.1084/jem.183.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamelmann E, et al. Antiinterleukin-5 antibody prevents airway hyperresponsiveness in a murine model of airway sensitization. Am J Respir Crit Care Med. 1997;155:819–825. doi: 10.1164/ajrccm.155.3.9117011. [DOI] [PubMed] [Google Scholar]
- 31.Hogan SP, Mould A, Kikutani H, Ramsay AJ, Foster PS. Aeroallergen-induced eosinophilic inflammation, lung damage, and airway hyperreactivity in mice can occur independently of IL-4 and allergen-specific immunoglobulins. J Clin Invest. 1997;99:1329–1339. doi: 10.1172/JCI119292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JJ, et al. Interleukin-5 expression in the lung epithelium of transgenic mice leads to pulmonary changes pathognomonic of asthma. J Exp Med. 1997;185:2143–2156. doi: 10.1084/jem.185.12.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henderson WR, Jr, et al. The importance of leukotrienes in airway inflammation in a mouse model of asthma. J Exp Med. 1996;184:1483–1494. doi: 10.1084/jem.184.4.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keane-Myers A, Gause WC, Linsley PS, Chen SJ, Wills-Karp M. B7-CD28/CTLA-4 costimulatory pathways are required for the development of T helper cell 2-mediated allergic airway responses to inhaled antigens. J Immunol. 1997;158:2042–2049. [PubMed] [Google Scholar]
- 35.Tsuyuki S, Tsuyuki J, Einsle K, Kopf M, Coyle AJ. Costimulation through B7-2 (CD86) is required for the induction of lung mucosal T helper cell 2 (Th2) immune response and altered airway responsiveness. J Exp Med. 1997;185:1671–1679. doi: 10.1084/jem.185.9.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng S, et al. Expression and function of HLA-DQ8 (DQA1*0301/ DQB1*0302) genes in transgenic mice. Eur J Immunogenet. 1996;23:15–20. doi: 10.1111/j.1744-313x.1996.tb00260.x. [DOI] [PubMed] [Google Scholar]
- 37.Nabozny GH, et al. HLA-DQ8 transgenic mice are highly susceptible to collagen-induced arthritis. A novel model for human polyarthritis. J Exp Med. 1996;183:27–37. doi: 10.1084/jem.183.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bradley DS, et al. HLA-DQB1 polymorphism determine incidence, onset, and severity of collagen-induced arthritis (CIA) in transgenic mice: implications in human rheumatoid arthritis. J Clin Invest. 1997;100:2227–2234. doi: 10.1172/JCI119760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapoval SP, et al. HLA-DQ6 and HLA-DQ8 transgenic mice respond to ragweed allergen and recognize distinct set of epitopes on short and giant ragweed group 5 antigens. J Immunol. 1998;161:2032–2037. [PubMed] [Google Scholar]
- 40.Yu C-K, Lee S-C, Wang J-Y, Hsiue T-R, Lei H-Y. Early-type hypersensitivity-associated airway inflammation and eosinophilia induced by Dermatophagoides farinae in sensitized mice. J Immunol. 1996;156:1923–1930. [PubMed] [Google Scholar]
- 41.Karachunski PI, Ostlie NS, Okita DK, Conti-Fine BM. Prevention of experimental myasthenia gravis by nasal administration of synthetic acetylcholine receptor T epitope sequences. J Clin Invest. 1997;100:3027–3035. doi: 10.1172/JCI119857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacLean JA, Ownbey R, Laster AD. T cell–dependent regulation of eotaxin in antigen-induced pulmonary eosinophilia. J Exp Med. 1996;184:1461–1469. doi: 10.1084/jem.184.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drescher KM, Murray PD, David CS, Pease LR, Rodrigues M. CNS cell populations are protected from virus-induced pathology by distinct arms of the immune system. Brain Pathol. 1999;9:21–31. doi: 10.1111/j.1750-3639.1999.tb00206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamelmann E, et al. Noninvasive measurement of airway responsiveness in allergic mice using barometric plethysmography. Am J Respir Crit Care Med. 1997;156:766–775. doi: 10.1164/ajrccm.156.3.9606031. [DOI] [PubMed] [Google Scholar]
- 45.Glanville AR, et al. The distribution of MHC class I and II antigens on bronchial epithelium. Am Rev Respir Dis. 1989;139:330–334. doi: 10.1164/ajrccm/139.2.330. [DOI] [PubMed] [Google Scholar]
- 46.Cunningham AC, Kirby JA. Regulation and function of adhesion molecule expression by human alveolar epithelial cells. Immunology. 1995;86:279–286. [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor PM, Rose ML, Yacoub MH. Expression of MHC antigens in normal human lungs and transplanted lungs with obliterative bronchiolitis. Transplantation. 1989;48:506–510. doi: 10.1097/00007890-198909000-00030. [DOI] [PubMed] [Google Scholar]
- 48.Vignola AM, et al. HLA-DR and ICAM-1 expression on bronchial epithelial cells in asthma and chronic bronchitis. Am Rev Respir Dis. 1993;148:689–694. doi: 10.1164/ajrccm/148.3.689. [DOI] [PubMed] [Google Scholar]
- 49.Vignola AM, et al. Functional and phenotypic characteristics of bronchial epithelial cells obtained by brushing from asthmatic and normal subjects. Allergy. 1993;48:32–38. doi: 10.1111/j.1398-9995.1993.tb04696.x. [DOI] [PubMed] [Google Scholar]
- 50.Moffat MF, Cookson WOCM. The genetics of specific allergy. Monogr Allergy. 1996;33:71–96. [PubMed] [Google Scholar]
- 51.Liu MC, et al. Immediate and late inflammatory responses to ragweed antigen challenge of the peripheral airways in allergic asthmatics. Cellular, mediator, and permeability changes. Am Rev Respir Dis. 1991;144:51–58. doi: 10.1164/ajrccm/144.1.51. [DOI] [PubMed] [Google Scholar]
- 52.Pin I, et al. Changes in the cellular profile of induced sputum after allergen-induced asthmatic responses. Am Rev Respir Dis. 1992;145:1265–1269. doi: 10.1164/ajrccm/145.6.1265. [DOI] [PubMed] [Google Scholar]
- 53.Kang BC, Johnson J, Veres-Thorner C. Atopic profile of inner-city asthma with a comparative analysis on the cockroach-sensitive and ragweed-sensitive subgroups. J Allergy Clin Immunol. 1993;92:802–811. doi: 10.1016/0091-6749(93)90057-m. [DOI] [PubMed] [Google Scholar]
- 54.Yurovsky VV, et al. T-cell repertoire in the blood and lungs of atopic asthmatics before and after ragweed challenge. Am J Respir Cell Mol Biol. 1998;18:370–383. doi: 10.1165/ajrcmb.18.3.2935. [DOI] [PubMed] [Google Scholar]
- 55.Renz H, et al. Aerosolized antigen exposure without adjuvant causes increased IgE production and increased airway responsiveness in the mouse. J Allergy Clin Immunol. 1992;89:1127–1138. doi: 10.1016/0091-6749(92)90296-e. [DOI] [PubMed] [Google Scholar]
- 56.Kung TT, et al. Characterization of a murine model of allergic pulmonary inflammation. Int Arch Allergy Immunol. 1994;105:83–90. doi: 10.1159/000236807. [DOI] [PubMed] [Google Scholar]
- 57.Garlisi CG, et al. T cells are necessary for Th2 cytokine production and eosinophil accumulation in airways of antigen-challenged allergic mice. Clin Immunol Immunopathol. 1995;75:75–83. doi: 10.1006/clin.1995.1055. [DOI] [PubMed] [Google Scholar]
- 58.Kips JC, et al. Importance of interleukin-4 and interleukin-12 in allergen-induced airway changes in mice. Int Arch Allergy Immunol. 1995;107:115–118. doi: 10.1159/000236947. [DOI] [PubMed] [Google Scholar]
- 59.Bell SJD, Metzger WJ, Welch CA, Gilmour MI. A role for Th2 T-memory cells in early airway obstruction. Cell Immunol. 1996;170:185–194. doi: 10.1006/cimm.1996.0151. [DOI] [PubMed] [Google Scholar]
- 60.Coyle AJ, et al. Central role of immunoglobulin (Ig) E in the induction of lung eosinophil infiltration and T helper 2 cell cytokine production: inhibition by a non-anaphylactogenic anti-IgE antibody. J Exp Med. 1996;183:1303–1310. doi: 10.1084/jem.183.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harris N, Campbell C, Le Gros G, Ronchese F. Blockade of CD28/B7 co-stimulation by mCTLA4-Hgamma1 inhibits antigen-induced lung eosinophilia but not Th2 cell development or recruitment in the lung. Eur J Immunol. 1997;27:155–161. doi: 10.1002/eji.1830270123. [DOI] [PubMed] [Google Scholar]
- 62.Zhang Y, Rogers KH, Lewis DB. β2-Microglobulin-dependent T cells are dispensable for allergen-induced T helper 2 responses. J Exp Med. 1996;184:1507–1512. doi: 10.1084/jem.184.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taneja V, David CS. HLA transgenic mice as humanized mouse models of disease and immunity. J Clin Invest. 1998;101:921–926. doi: 10.1172/JCI2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y, et al. Influence of the route of allergen administration and genetic background on the murine allergic pulmonary response. Am J Respir Crit Care Med. 1997;155:661–669. doi: 10.1164/ajrccm.155.2.9032210. [DOI] [PubMed] [Google Scholar]
- 65.Frigas E, Loegering DA, Solley GO, Farrow GM, Gleich GJ. Elevated levels of the eosinophil granule major basic protein in the sputum of patients with bronchial asthma. Mayo Clin Proc. 1981;56:345–353. [PubMed] [Google Scholar]
- 66.Coyle AJ, Uchida D, Ackerman SJ, Mitzner W, Irvin CG. Role of cationic proteins in the airway. Hyperresponsiveness due to airway inflammation. Am J Respir Crit Care Med. 1994;150:S63–S71. doi: 10.1164/ajrccm/150.5_Pt_2.S63. [DOI] [PubMed] [Google Scholar]