Significance
Bromodomain and extraterminal (BET) proteins bind acetylated proteins, including histones, and regulate transcription. Interestingly, inhibitors of BET proteins (BETi) can block cancer cell proliferation and induce apoptosis in a wide range of tumor types. To date many of the effects of BETi have been attributed to transcriptional suppression of genes like the MYC oncogene. We show that genetically-engineered Myc-induced lymphoma mouse models are highly sensitive to BETi without MYC transcription being suppressed. Our data suggest broad effects on transcription by BETi including a set of genes being induced. Here a genetic and functional link between BET proteins and histone deacetylases is unraveled that opens up avenues for combination therapies against cancer.
Keywords: mouse models, JQ1, vorinostat, Brd2, Brd4
Abstract
The bromodomain and extraterminal (BET) domain family of proteins binds to acetylated lysines on histones and regulates gene transcription. Recently, BET inhibitors (BETi) have been developed that show promise as potent anticancer drugs against various solid and hematological malignancies. Here we show that the structurally novel and orally bioavailable BET inhibitor RVX2135 inhibits proliferation and induces apoptosis of lymphoma cells arising in Myc-transgenic mice in vitro and in vivo. We find that BET inhibition exhibits broad transcriptional effects in Myc-transgenic lymphoma cells affecting many transcription factor networks. By examining the genes induced by BETi, which have largely been ignored to date, we discovered that these were similar to those induced by histone deacetylase inhibitors (HDACi). HDACi also induced cell-cycle arrest and cell death of Myc-induced murine lymphoma cells and synergized with BETi. Our data suggest that BETi sensitize Myc-overexpressing lymphoma cells partly by inducing HDAC-silenced genes, and suggest synergistic and therapeutic combinations by targeting the genetic link between BETi and HDACi.
The bromodomain and extraterminal (BET) domain family of proteins Brd2, Brd3, Brd4, and BrdT bind via their tandem bromodomains (BD1 and BD2) to acetylated lysines in histones and other proteins (1). On binding, they regulate the transcription of genes critical for cell-cycle progression and apoptosis. Therefore, BET proteins have emerged as interesting proteins for targeted intervention of cancer.
Recently, the small-molecule BET inhibitor (+)-JQ-1 (hereafter JQ1) was found to be a potent and specific suppressor of B cell-lineage malignancies (2, 3). In acute myelogenous leukemia, BRD4 is essential for tumor maintenance, and JQ1 recapitulates the effects of RNA interference of BRD4 (4, 5). JQ1 was subsequently shown to have an antiproliferative effect in other hematological malignancies and solid organ tumors including glioblastoma, prostate cancer, and neuroblastoma (6–10). The current model of how BET inhibitors (BETi) inhibit tumor cell proliferation places inhibition of MYC as mediating activity in lymphoid tumors, with Myc-independent activity in some solid tumor types such as lung adenocarcinoma (11). However, it has not been clear in hematopoietic tumor types whether the antiproliferative effects of BETi are mediated by suppression of MYC expression or whether effects on MYC are a correlative bystander of the mechanism, perhaps useful as a biomarker but not necessarily mechanistic (12).
We have assessed the effect of RVX2135, a novel and orally bioavailable selective inhibitor of Brd2, Brd3, Brd4, and BrdT, in in vitro and in vivo models of Myc-induced lymphoma. We find that the effects are mediated by broad transcriptional changes and that these are genetically and functionally linked to histone deacetylase inhibitors.
Results
RVX2135 Blocks Proliferation of Myc-Induced Mouse Lymphoma Cells and Induces Caspase-Dependent Apoptosis.
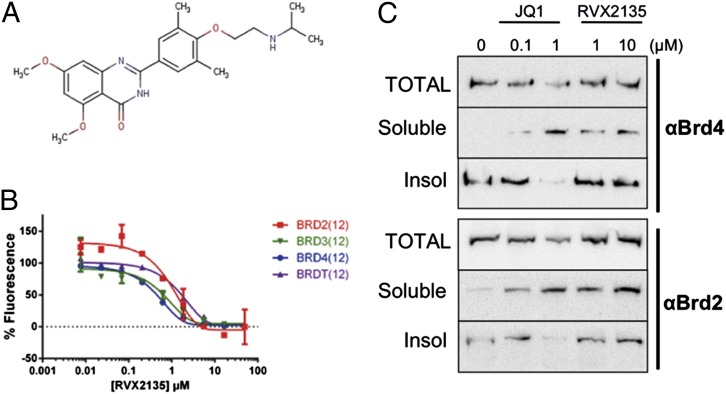
RVX2135 is a novel small-molecule BET bromodomain inhibitor that is structurally unrelated to the benzodiazepine derivative compounds but is in the same chemical scaffold group as RVX-208 developed by Zenith Epigenetics Corp. (Fig. 1A) (13–15). In a fluorescence resonance energy transfer assay in vitro, RVX2135 and the prototype BET inhibitor JQ1 displaced all four BET family members from tetraacetylated histone peptide (Brd2, Brd3, Brd4, and BrdT) with low or submicromolar potency (Fig. 1B and Fig. S1A). Because the antiproliferative effects of BETi in hematological malignancies correlate with suppression of MYC transcription, we investigated the effects of RVX2135 and JQ1 in transgenic models where c-Myc drives lymphomagenesis. In λ-Myc and Eµ-Myc mice, mouse (Eµ-Myc) or human (λ-Myc) Myc genes are placed under ectopic control of IgH or IgL enhancers, respectively. Mice carrying these transgenes invariably develop B-cell lymphomas of varying maturity with onset ranging from 3 to 12 mo (median survival 100 d) (16, 17). These lymphomas are transplantable, and we have also established cell lines that grow readily in vitro as well as in C57BL/6 mice following transplantation. Treating two of the cell lines with RVX2135 and JQ1 confirmed that the BET proteins Brd2 and Brd4 can be displaced from chromatin (Fig. 1C and Fig. S1B) in a dose-dependent manner.
Fig. 1.
Features of RVX2135, a novel BET inhibitor. (A) Structure of the chemical scaffold from which RVX2135 was developed. (B) FRET assay dose–response curves showing displacement of BET proteins from an acetylated histone-derived peptide by RVX2135. Data are mean values ± SD (error bars). (C) λ663 cells were treated for 24 h with the indicated concentrations of BET inhibitors JQ1 and RVX2135. Total protein lysates, the soluble fraction, and the insoluble fraction of detergent-generated lysates were loaded on a gel for Western blot analysis using the indicated antibodies. The method is described in greater detail in Fig. S1.
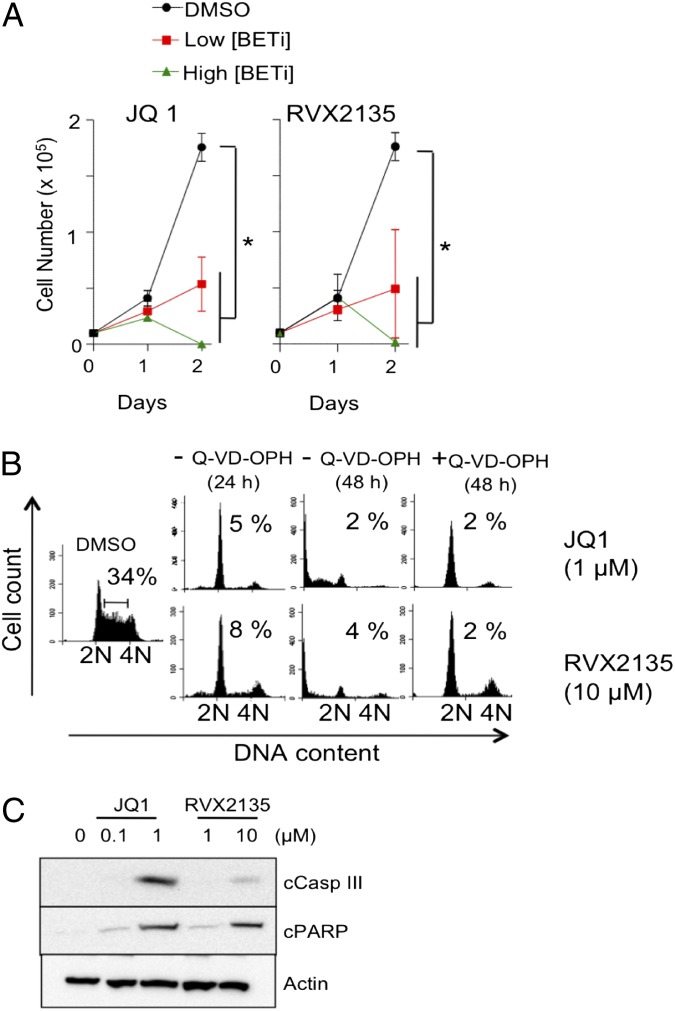
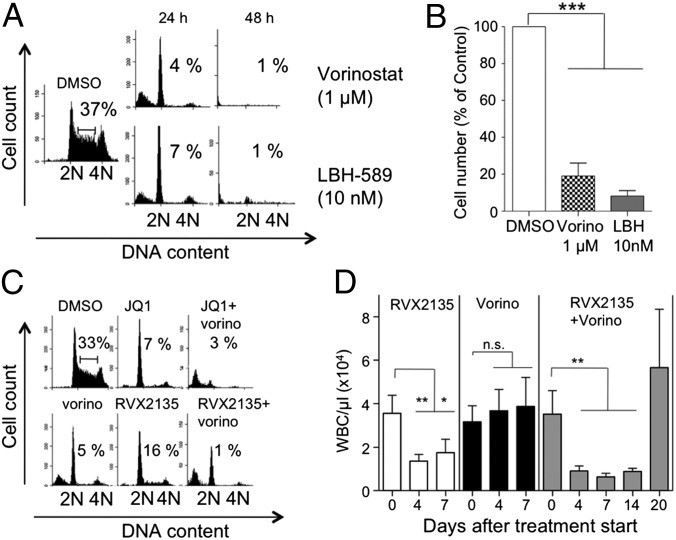
To investigate the antiproliferative effects of RVX2135 and JQ1 on lymphoma cells from Myc-transgenic mice, we performed cell count and flow cytometric analysis of in vitro propagated cell lines. Both inhibitors potently inhibited growth, with JQ1 showing ∼10-fold higher potency than RVX2135 in λ820 cells. At lower concentrations of BETi—100 nM JQ1 or 1 µM RVX2135—cell proliferation was severely restricted but the cells remained viable (Fig. 2A). At 10 times higher concentrations, cells underwent cell-cycle arrest by 24 h and died by 48 h (Fig. 2 A and B and Fig. S2 A and B). Similar data were obtained in Eµ239 cells (Fig. S2 C–E). The presence of a sub-G1 population in the DNA histogram suggested that the cells died by apoptosis (Fig. 2B and Fig. S2 B, C, and E); indeed, when cells were pretreated with the pan-caspase inhibitor Q-VD-OPH, sub-G1 accumulation was abolished but G1 arrest was maintained and enhanced. Both cleaved caspase 3 and PARP were detected by Western blotting upon treatment with high concentrations of BETi (Fig. 2C), suggesting involvement of the mitochondrial apoptotic pathway.
Fig. 2.
BET inhibitors induce cell-cycle arrest and apoptosis of Myc-transgenic mouse lymphoma cells. (A) λ820 cells were treated for 48 h with different concentrations of BETi and counted by trypan blue exclusion. Low concentrations of BETi were defined as 100 nM JQ1 and 1 µM RVX2135, whereas high concentrations were defined as 1 µM JQ1 and 10 µM RVX2135. Errors bars represent the standard deviation from three independent experiments. *P < 0.05. (B) λ820 cells were treated for 24 and 48 h with the indicated concentrations of BETi in the presence or absence of the pan-caspase inhibitor Q-VD-OPH. Cell-cycle distribution was analyzed by flow cytometry of 7-AAD–stained cells. Numbers show the proportion of cells in S phase. Quantifications of S phase and apoptosis (sub-G1) in three independent experiments are presented in Fig. S1 A and B. (C) λ820 cells were treated for 24 h with the indicated concentrations of BETi. Cells were lysed and analyzed by Western blotting for cleaved caspase 3 and cleaved PARP, markers of apoptosis. β-Actin was used as a loading control.
BETi such as JQ1 have previously been shown to exhibit broad antiproliferative effects in many cancer cell lines of both solid and hematological origin. To assess whether RVX2135 affects cell types other than Myc-induced lymphoma, we cultured five melanoma cell lines in the absence or presence of 10 µM RVX2135. Similar to previous studies on a different BET inhibitor (18), RVX2135 suppressed cell-cycle progression (Fig. S3A). Moreover, neither JQ1 nor RVX2135 was cytotoxic in normal cells, as they did not overtly affect viability or proliferation of low-passage mouse embryo fibroblasts (Fig. S3B).
RVX2135 Is an Orally Bioavailable Compound with Potent Effect Against Lymphoma in Vivo.
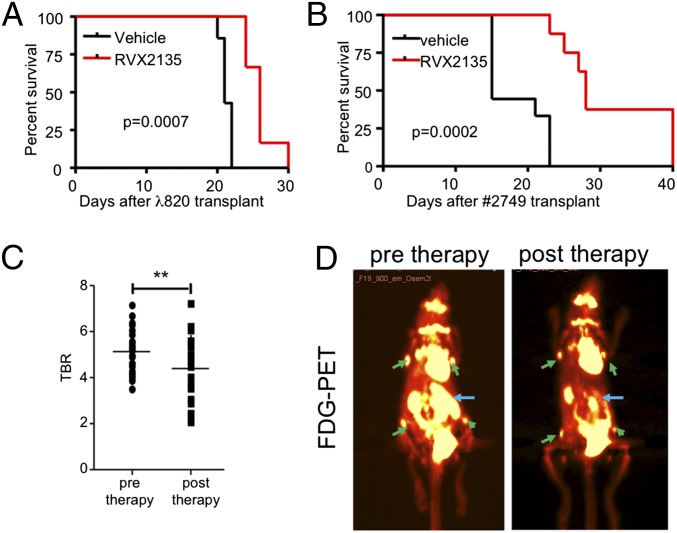
To establish whether RVX2135 was effective in immunocompetent mouse models of lethal lymphoma, we used two transplant models. First, in vitro cultured λ820 cells were transplanted into C57BL/6 mice. Exome sequencing of this cell line revealed two inactivating Trp53 frame-shift mutations in exons 4 and 8 (Fig. S4). Because it has been in culture for several years, we therefore also serially transplanted cells from a dispersed lymphoma of a λ-Myc mouse (ID 2749) into B6 mice without subjecting the cells to culture. When these two models were established, we allowed the cells to home for 4 d. The mice were then randomly divided into two groups receiving 75 mg/kg RVX2135 or vehicle bidaily by oral gavage. The vehicle-treated mice carrying λ820 cells started to show signs of disease approximately 3 wk after transplantation, whereas RVX2135-treated mice succumbed to lymphoma approximately 1 wk later (Fig. 3A), similar to the survival benefit seen for JQ1 administered by i.p. injection in other hematological cancer models (2, 4, 5, 10). However, RVX2135 doubled both the median and overall survival of mice carrying 2749 lymphoma (Fig. 3B), with disseminated disease and splenomegaly eventually developing. The compound was well-tolerated throughout the course of the two experiments.
Fig. 3.
RVX2135 causes potent therapeutic responses in mouse models of aggressive Myc-induced lymphoma. (A) λ820 cells were transplanted into syngenic B6 mice via tail vein injection. Four days after injection, mice were dosed with 75 mg/kg [twice a day (b.i.d.); 5 d/wk) RVX2135 (n = 6) or vehicle (n = 7). Mice were monitored daily for signs of lymphoma (visible palpable lymphomas, panting suggesting thymic lymphoma, or overall health appearance), and four of the vehicle-treated mice and all of RVX2135-treated mice were killed when they showed signs of disease. The three remaining vehicle-treated mice were used in an experiment shown in Fig. S5C. (B) A lymphoma arising in a λ-Myc mouse (ID 2749) was transplanted into recipient B6 mice via tail vein injection accompanied by treatment with either vehicle or RVX2135. Four days after injection, mice were dosed with 75 mg/kg b.i.d. RVX2135 (n = 8) or vehicle (n = 9). Mice were monitored daily for signs of lymphoma and were killed when they showed signs of disease. (C) Four mice were transplanted with lymphoma cells from a λ-Myc mouse (2749). Twelve days after transplantation, when mice were yet to show manifest disease, they were injected with [18F]FDG and scanned with a PET/computed tomography imager. All mice had a strong signal in the spleen and in several lymph nodes. The signal ratio comparing normal tissue (skeletal muscle) and lymphoma was calculated. Mice were treated with four doses of 75 mg/kg b.i.d. RVX2135 and imaged posttherapy. The tumor PET signal-to-background ratio (TBR) of all measured PET-positive sites (n = 23), representing the spleens of all mice and four or five lymph nodes, is shown. All point (Fig. 3C) are individual data point. **P < 0.01. (D) Representative PET image of one mouse before and after treatment with RVX2135. PET-positive lymphoid tissues used in the TBR calculation in C are shown with green (lymph nodes) and blue (spleen arrows).
To reduce the likelihood of selection of resistant clones or the confounding effects of systemic illness, we assessed the efficacy of acute dosing of RVX2135 on a separate group of mice transplanted with primary lymphoma cells (from mouse 2749). Twelve days after transplantation the mice were injected with [18F]fluorodeoxyglucose (FDG), and a baseline positron emission tomography (PET) signal was obtained to image tumor cell metabolism. Mice were then treated with bidaily doses of RVX2135 for 2 d followed by PET imaging. RVX2135 treatment significantly reduced the metabolic activity of lymphoma growing in the lymph nodes and spleen of the animal (Fig. 3C), with a particularly marked reduction in tumor load in the spleen (Fig. 3D).
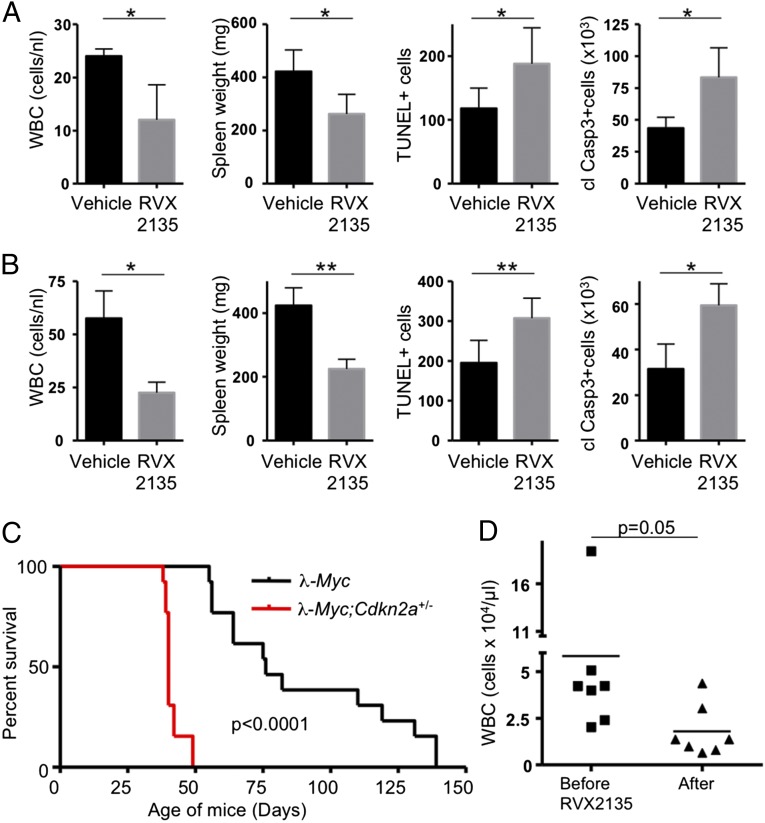
Although the in vitro data suggested that BETi resulted in apoptosis, we also investigated whether this occurred in vivo. We treated mice with an increased number of leukocytes in blood samples before they had developed palpable lymphomas requiring their sacrifice. Acute treatment of mice carrying either λ820 cells or 2749 cells resulted in a decrease in white blood cell counts and spleen size and an increase in cleaved caspase 3 and TUNEL-positive cells at 36 and 48 h posttreatment initiation (Fig. 4 A and B and Fig. S5 A and B), respectively. Moreover, Western blot analysis of cleaved PARP in lymph node lymphomas from acutely and long term-treated mice carrying λ820 cells also provided evidence that many RVX2135-treated lymphoma cells died by apoptosis (Fig. S5C). Taken together, these results suggest that BETi can induce rapid apoptosis of Myc-induced lymphoma cells in vitro as well as in vivo.
Fig. 4.
RVX2135 causes apoptosis and rapid therapeutic responses in mouse models of aggressive Myc-induced lymphoma. (A and B) Mice transplanted with λ820 cells (A) or serially transplanted with lymphoma from mouse 2749 (B) were monitored for elevated leukocyte count (white blood cells; WBCs) in the blood drawn from a hind leg vein (vena saphena). When counts had reached significantly above normal mouse blood WBCs (5–15 cells per nL), the mice were randomized to treatment with either vehicle or 75 mg/kg RVX2135 (b.i.d.). Mice in groups of three were killed 2 h after one to five bidaily doses; shown here are the experiments where significant reductions in spleen size had occurred (four doses during 36 h for λ820 cells and five doses during 48 h for 2749). After sacrifice, spleens were weighed and subsequently fixed in formalin and processed for TUNEL staining or immunohistochemistry to detect cleaved caspase 3, markers of apoptosis. Counts are either from whole sections (caspase 3; n = 6) or representative fields of view (TUNEL; n = 6). Representative images are shown in Fig. S5. (C) λ-Myc mice were interbred to Cdkn2a knockout mice to generate λ-Myc;Cdkn2a+/− mice. These were followed for signs of lymphoma. Typically at 40–50 d of age the mice developed lymphoma with an associated leukocytosis, which is an acceleration of a similar rate to that seen in Eµ-Myc;Cdkn2a+/− mice and λ-Myc;p53+/− mice (50–52). (D) Seven λ-Myc;Cdkn2a+/− mice exhibiting leukocytosis (>20 WBCs per nL), as assessed by WBC count of peripheral blood, were treated with 75 mg/kg b.i.d. RVX2135 for 3 d. Six hours after the last dose, blood samples were drawn and a WBC count was performed. Six out of seven mice responded, and data were analyzed by a paired t test. Errors bars represent the standard deviation. *P < 0.05, **P < 0.01.
So far all experiments had been performed in mice whose tumors had developed via transplantation. To investigate the efficacy of RVX2135 in a model of spontaneously developing lymphoma, λ-Myc mice were interbred to Cdkn2a knockout mice to generate λ-Myc;Cdkn2a+/− mice. Instead of developing lymphoma at a mean latency of 100 d as λ-Myc mice do, λ-Myc;Cdkn2a+/− mice developed lymphoma at around 40 d (Fig. 4C). Treating mice with palpable lymphoma with RVX2135 suppressed lymphoma-associated leukocytosis in 6/7 mice (Fig. 4D). RVX2135 is therefore an orally bioavailable BET inhibitor with potent antilymphoma activity in both transplant and genetically engineered mouse models of Myc-induced lymphoma.
BET Inhibitor Induces a Complex Transcriptional Program Without Specifically Inactivating Transgenic Myc Transcription.
The cell-cycle arrest and apoptosis observed in our B-cell lymphoma models were similar to the effects described by others in multiple myeloma, pre-B acute lymphoblastic leukemia, and Burkitt lymphoma, all malignancies in which inhibition of MYC transcription correlates with BET inhibitor activity (2, 19, 20). To investigate whether BET inhibition correlated with Myc suppression in our murine Myc-induced lymphoma lines, we analyzed Myc transcript levels. We performed quantitative (q)RT-PCR and Western blot analyses of various murine lymphoma cell lines derived from Eµ-Myc or λ-Myc mice treated with various concentrations of JQ1 and RVX2135. Unexpectedly, down-regulation of c-Myc transcription (Fig. S6A) was not observed in any of the cell lines, and a reduced c-Myc protein level was only observed in λ820 cells at the highest concentrations but not in three other cell lines or in lymphomas harvested from treated mice (Fig. S6 B and C). Treating λ820 cells with a GSK3β inhibitor rescued the protein levels (Fig. S6D), suggesting the possibility that c-Myc protein was destabilized by BETi via activation of the GSK3β/Fbw7/proteasome pathway in this cell line (21). We therefore conclude that the biological effects observed with BETi in Myc-transgenic mice are independent of transcriptional repression of c-Myc.
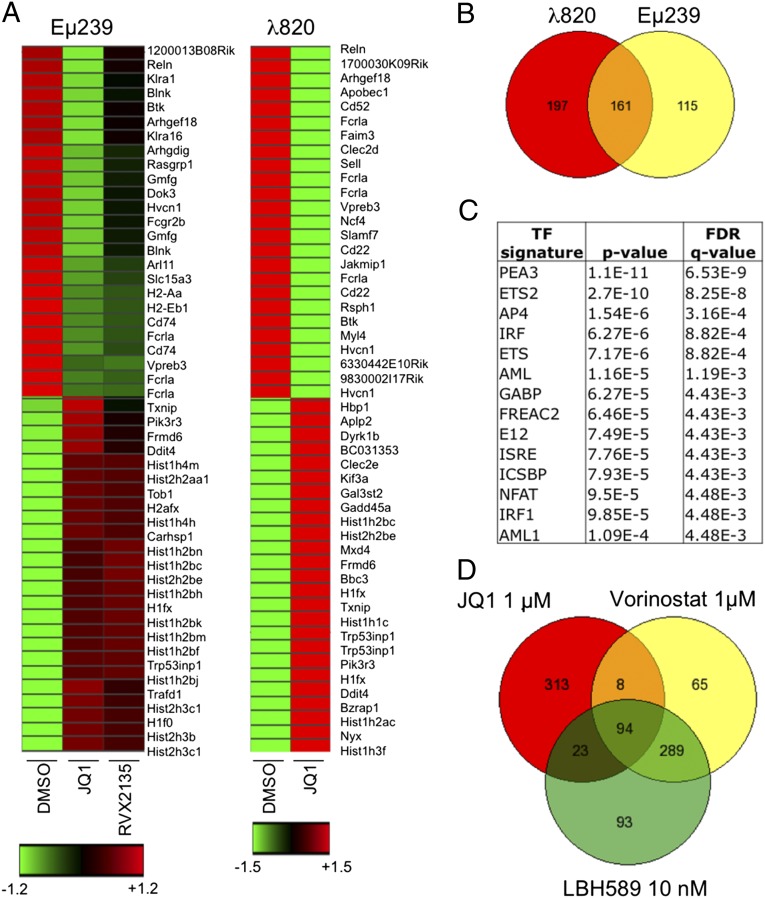
To gain further insights into the transcriptional effects of BETi, we next performed transcriptional profiling of λ820 and Eµ239 cells treated with BETi. In cells treated with JQ1, 358 genes were down-regulated more than twofold in λ820 cells and 276 genes in Eµ239 cells (Datasets S1 and S2). Hierarchical clustering analysis of the 50 most up- or down-regulated genes visualized the abundance of cell-surface markers of B cells (Il7r, Vpreb3, Fcrla, and Cd74) among the down-regulated genes, and p53-regulated genes (Bbc3, Trp53inp1, Ddit4, and Gadd45a) among the up-regulated genes (Fig. 5A). There was an overlap of 161 genes down-regulated by JQ1 in both cell lines (Fig. 5B). We then used the gene set enrichment analysis (GSEA) application at the Broad Institute (www.broadinstitute.org/gsea/msigdb/annotate.jsp) to compute overlaps between the Molecular Signatures Database (MSigDB; Broad Institute) signatures and the genes down-regulated in both cell lines by JQ1. This analysis revealed an enrichment of genes regulated by several transcription factors, including members of the ETS family (Fig. 5C and Dataset S3). Myc-regulated genes were only down-regulated in λ820 cells, not surprisingly, as Myc protein levels were decreased in these cells (Fig. S6B). Our data indicate that BETi affect several transcriptional programs and that similar biological effects can be observed whether Myc target genes are suppressed or not.
Fig. 5.
BETi induce broad transcriptional effects affecting several growth-promoting gene signatures. (A) Supervised hierarchical clustering of Illumina BeadChip microarray data in λ820 and Eµ239 cells treated with BETi. Shown are the 50 most down- and up-regulated genes (fold change). Red color indicates induced expression, black means unchanged expression, and green indicates down-regulated expression. Some of the genes up-regulated by BET inhibitor treatment were confirmed by qRT-PCR (Fig. S7). (B) Venn diagram of the genes down-regulated by treatment with λ820 and Eµ239 cells with 1 µM JQ1 for 24 h. Data of average expression levels (n = 2) in Eµ239 cells are present in Dataset S2. (C) Gene set enrichment analysis of genes coregulated (down) by JQ1 in both λ820 and Eµ239 cells. Shown are top transcription factors (TFs) associated with the gene signatures. GSEA of genes shared and uniquely regulated in λ820 and Eµ239 cell lines is shown in Dataset S3. FDR, false discovery rate; q-value, the statistical significance of an FDR test. (D) Venn diagram showing the similarities and differences in the expression profiles of λ820 cells treated with the indicated concentrations of BETi or HDACi for 24 h. Data of average expression levels are presented in Datasets S1 and S4.
A next possible avenue of investigation would be to try to genetically rescue genes down-regulated by BETi. However, previous similar experiments have failed to rescue the effects of BETi by overexpressing MYC or FOSL1, for example (2, 11). Instead, we further analyzed genes that are up-regulated by BETi, because these have largely been ignored in previous studies. Besides p53-regulated genes, several histone mRNAs were among the most highly induced genes (Fig. 5A, Fig. S7A, and Datasets S1 and S2). Both λ820 and Eµ239 cells have Trp53 mutations, and the p53 family members p73 and p63 are only expressed at low levels. On the other hand, some of these induced genes are targets of the tumor suppressor and transcription factor Egr1 (22, 23), which was up-regulated by BETi in both lymphoma lines (Datasets S1 and S2). Egr1 has recently been shown to mediate p53-independent apoptosis in Myc-overexpressing cells (24, 25), suggesting a possible component of BET inhibitor-induced cell death.
BETi Induce Genes and Biological Effects Reminiscent of Histone Deacetylase Inhibitors.
Further GSEA and MSigDB overlap analyses revealed a high degree of similarity between our BET inhibitor-induced transcriptional changes and those seen in human multiple myeloma (MM) cell lines treated with the histone deacetylase inhibitor trichostatin A (TSA) and the DNA methyltransferase inhibitor decitabine (26). About a third of the genes reported to be induced by TSA and decitabine in MM cell lines are also induced by BETi in λ820 and Eµ239 cells (Datasets S1 and S2). To investigate whether BETi and histone deacetylase inhibitors (HDACi) share a similar transcriptional signature, λ820 cells were treated with the HDAC inhibitor vorinostat or LBH-589 and analyzed by expression profiling. Microarray analysis of λ820 cells treated with vorinostat or LBH-589 showed that 25% of all genes induced by BETi were also induced by vorinostat or LBH-589 (Fig. 5D and Dataset S4). This is a fourfold higher overlap than what is to be expected by chance if 8,000 genes are assumed to be possible to regulate. The similarity was not observed among genes suppressed by HDACi and BETi. Interestingly, genes with possible links to the apoptotic phenotype that were induced by both BETi and HDACi included genes such as Egr1, Trp53inp1, Gadd45a, and Bbc3. In the qRT-PCR validation of select histone mRNA, the similarity between HDACi and BETi was again confirmed (Fig. S7 A and B). Myc transcription was not decreased by HDACi—at variance with the case in human MM (26)—but c-Myc protein levels were slightly down-regulated at the higher concentration, again phenocopying the effect of BETi (Figs. S6 and S8 A and B). HDACi induced a marker of DNA damage response signaling, phosphorylation of the histone variant H2Ax (γH2Ax; Fig. S8B). This is a known consequence of HDAC inhibition (27), and recently a similar finding has been described in human cells treated with JQ1 (28). We confirmed that treatment with RVX2135 or JQ1 results in a γH2Ax signal, even in 2749 lymphomas treated in vivo with RVX2135, again pointing to the similarity in effects between BETi and HDACi (Figs. S6C and S8C).
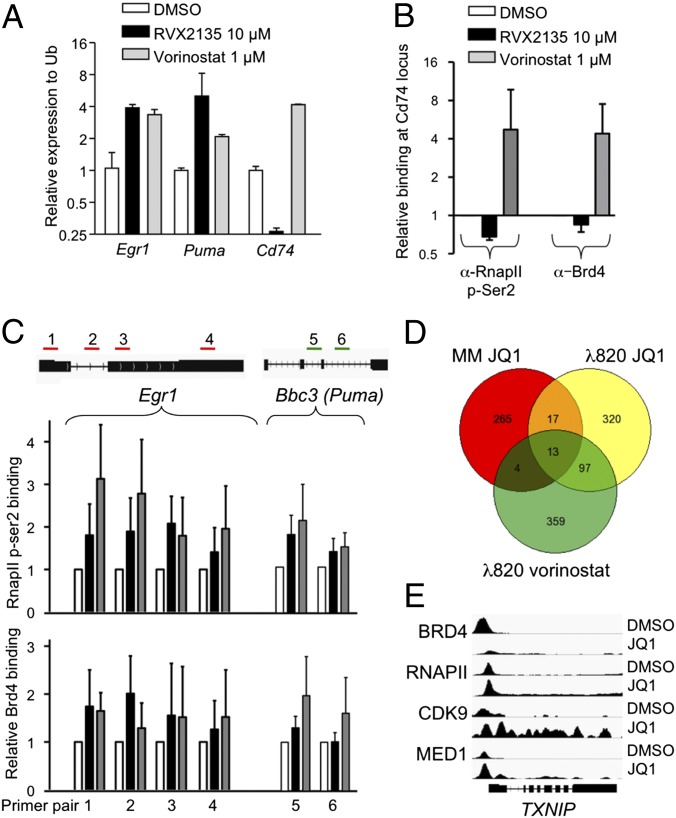
The similarity between BET inhibitor- and HDAC inhibitor-induced genes suggested a common mechanism. BETi have been shown to induce transcription in other pathological settings such as HIV, inflammation, and arteriosclerosis, but this has largely been ignored in the cancer field. In HIV, the mechanism is believed to be mediated by a transient inactivation of the HEXIM1–7S small nuclear ribonucleoprotein (snRNP) by BETi, resulting in a release of the positive transcription elongation factor p-TEFb (29–33). p-TEFb can then phosphorylate the C-terminal domain of RNA polymerase II (34), ensuring transcription elongation. Because Brd4 also recruits p-TEFb to acetylated histones (35, 36), thereby promoting transcription pause release of growth-promoting genes, it is possible that p-TEFb dynamics is altered in BET inhibitor-treated cells, favoring stress-induced genes such as p53 targets, Egr1, and others. Interestingly, treatment with the HDAC inhibitor vorinostat also results in transient release of p-TEFb, favoring HIV transcription (30). To investigate this at the molecular level, we studied gene occupancy of Brd4 and RNA polymerase II phosphorylated at the p-TEFb site of serine 2 by chromatin immunoprecipitation (ChIP) assay. We focused on three genes regulated by both BETi and HDACi in the expression profiling. The Egr1 and Bbc3 (Puma) genes are both induced by BETi and HDACi. Cd74 was also analyzed, because it is repressed by BETi but induced by HDACi (Fig. 6A and Datasets S1, S2, and S4) and thus functions as a control for inhibitor-specific effects. Indeed, ChIP assay showed that the promoter region of Cd74 was occupied by both transcriptionally active RNA polymerase II and Brd4. Treatment with RVX2135 resulted in decreased binding of Brd4 and p-Ser2-RNA polymerase II on this gene but, in contrast, an increased binding was observed in vorinostat-treated cells (Fig. 6B). We also analyzed occupancy of Brd4 and p-Ser2-RNA polymerase II on four sites along the Egr1 gene and two sites in the Bbc3 and found that binding of Ser2-phosphorylated RNA polymerase II increased by BET inhibitor and HDAC inhibitor treatment on both genes, whereas Brd4 only increased binding to Bbc3 upon vorinostat binding (Fig. 6C). This suggests that Brd4 itself could be involved in recruiting RNA polymerase to certain (e.g., Egr1), but not all (e.g., Bbc3), nontranscribed genes when it cannot bind acetylated histones. To challenge this notion, we downloaded publicly available datasets of microarrays and sequencing data from ChIP assays (ChIP-seq) of MM1.S cells treated with JQ1 (37). We first compared the datasets to investigate whether there were any genes that were induced by JQ1 in both MM cells and λ820 cells, as well as in λ820 cells treated with vorinostat. Although the overlap only contained around 10% of the genes, some recurrently seen genes emerged such as histone genes and Txnip (Figs. 5A and 6D). Interestingly, ChIP-seq data revealed that JQ1 increased the binding of BRD4, RNA polymerase II, the active kinase CDK9 of the p-TEFb elongation factor, and Mediator downstream of the transcription initiation site. ChIP-seq analyses of the dynamics of these factors on other genes coregulated by BETi and HDACi in λ820 cells showed that induced genes were only uniformly regulated with respect to RNA polymerase II, and not BRD4 (Fig. S9).
Fig. 6.
BETi and HDACi induce transcription of stress-regulated genes. (A) Quantitative RT-PCR analyses of Egr1, Bbc3 (Puma), and Cd74 expression in λ820 cells treated with the indicated compounds. (B and C) ChIP assay experiment of cells treated with the indicated compounds. Following ChIP, qPCR analyses were performed using primers directed against the promoter region of Cd74 (B) or the indicated regions of Egr1 (green) or Bbc3 (red). Shown are the mean values from three independent experiments of relative binding compared in DMSO-treated cells. Errors bars represent the standard deviation from three independent experiments. (D) Venn diagram showing the similarities between genes induced in multiple myeloma MM1.S cells treated with 500 nM JQ1 for 6 h (GEO dataset GSE44931) and λ820 cells treated with 1 µM JQ1 or 1 µM vorinostat for 24 h. (E) IGV screenshot showing gene occupancy of the indicated proteins in ChIP-seq data of the TXNIP gene.
To investigate whether BETi and HDACi share both a similar transcriptional signature and biological effects, we treated λ820 cells with additional HDACi. Many of the second-generation HDACi were very cytotoxic, sometimes exhibiting nanomolar activity against this cell line (Fig. 7A and Fig. S10A). Interestingly, select concentrations of all HDACi phenocopied the effects of BETi by initially inducing a cell-cycle arrest at 24 h and later an apoptotic phenotype at 48 h (Fig. 7 A and B). Because BETi and HDACi induce similar genes but also exhibit unique transcriptional signatures, we hypothesized that combination therapies might be useful. To test this notion, we treated cultured λ820 cells with BETi and HDACi in combination. Interestingly, combined BET inhibitor and HDAC inhibitor treatment resulted in apoptosis at time points and concentrations where the respective monotherapies only caused cell-cycle arrest (Fig. 7C and Fig. S10 A and B). To test whether this synergy could be visualized in an animal model, we transplanted B6 mice with the 2749 serially transplanted lymphoma line. When white blood cell counts had increased above those of healthy mice, we treated mice with the Food and Drug Administration (FDA)-approved HDAC inhibitor vorinostat, RVX2135, or both in combination. Previous studies in lymphoma-bearing mice had shown monotherapy efficacy of HDAC inhibitors including vorinostat, but we used a much lower concentration, 40 mg⋅kg−1⋅d−1, instead of 100–200 mg⋅kg−1⋅d−1 (38, 39). RVX2135 and the combination treatment efficiently reduced the leukocytosis in just a few days, whereas vorinostat did not (Fig. 7D). After a week of treatment, the blood counts remained low but single-treated mice had developed a steady increase in leukocytosis, or even palpable lymphoma, and were thus killed. At this time the combination group appeared healthy and the mice were treated for an additional week before stopping the treatment. One week after treatment cessation, mice developed lymphoma, demonstrating that the mice were not cured by the 2-wk combination treatment. Nevertheless, our data suggest that HDACi and BETi synergize to kill Myc-induced lymphoma in mice, which results in improved overall survival.
Fig. 7.
Effects of BETi are similar to effects of HDAC inhibitors. (A) λ820 cells were treated for 24 and 48 h with the indicated concentrations of HDACi vorinostat or LBH-589. Cell-cycle distribution was analyzed by flow cytometry of 7-AAD–stained cells. Numbers show the proportion of cells in S phase. Additional HDACi used are shown in Fig. S11A. (B) λ820 cells were treated for 24 h with the indicated concentrations of HDACi and counted by using the gate of the viable cells in the flow cytometer. (C) λ820 cells were treated for 24 h with 1 µM JQ1 or 10 µM RVX2135 monotherapy or in combination with 1 µM HDAC inhibitor vorinostat. Cell-cycle distribution was analyzed by flow cytometry of 7-AAD–stained cells. Numbers show the proportion of cells in S phase. Data of S phase and apoptosis (sub-G1) from three independent experiments are shown in Fig. S11B. (D) Mice carrying 2749 lymphoma cells were acutely treated with bidaily oral doses of 75 mg/kg RVX2135, daily i.p. injections of 40 mg/kg vorinostat, or a combination of both treatments when showing WBC counts suggestive of leukocytosis. Single-treated mice were treated until WBC counts started to increase again and/or palpable lymphomas appeared. Combination-treated mice were maintained on treatment for an additional week, at which time they were scored as disease-free by palpation and WBC count. They were then taken off treatment until they showed signs of lymphoma and were killed. Shown in the graphs is the average ± SD, five mice per treatment group. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
We have characterized the novel BET bromodomain inhibitor RVX2135 and shown that it displaces all BET proteins from acetylated histones (Fig. 1), similar to the previously described BETi JQ1 (3). RVX2135 belongs to a different scaffold, which has a higher selectivity for BD2 over BD1 of the BET proteins. This property is shared by RVX-208, a BET inhibitor with the same scaffold that is undergoing clinical trial for arteriosclerosis (13–15). Nevertheless, despite the BD2 selectivity, RVX2135 blocks cell-cycle progression and induces cell death in Myc-induced mouse lymphoma cells in vitro and in mice (Figs. 2–4). Cell death could be blocked by a pan-caspase inhibitor, suggesting apoptosis as the mode of cell death. RVX2135 not only suppresses transcription but also induces several histone- and apoptosis-related genes (Figs. 5 and 6). These features are shared by the structurally unrelated BET inhibitor JQ1, suggesting that BET proteins are the relevant molecular targets to exert potent antilymphoma activity.
Previous studies in hematological malignancies and some solid tumors have suggested that the main target of inhibition by BETi is MYC (2–10). By using genetically engineered mouse models of lymphoma, it has here been possible to uncouple BET inhibitor effects on Myc transcription from the effects of BETi on other transcription factor networks. Interestingly, despite maintaining c-Myc expression, the murine B-cell lymphoma cells are as sensitive to BETi as the human lymphoma cells previously studied. Therefore, the fact that there is no genetic rescue in the mouse model demonstrates that BETi block lymphoma cell proliferation and induce apoptosis independent of inhibiting Myc transcription in B-cell lymphoma. This finding underscores the difficulty in identifying monogenic mechanisms that explain effects of inhibition of epigenetic regulators.
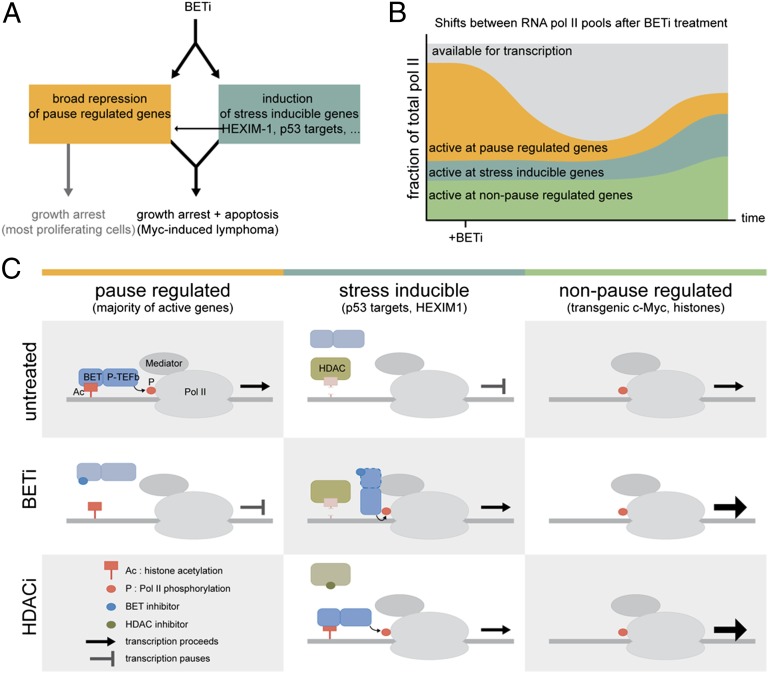
From the analyses of our microarray experiments shown in Figs. 5 and 6, we can conclude that there are broad effects on the transcriptome in BET inhibitor-treated cells. In the absence of a unifying mechanism involving one or a few transcription factors as the “targets” of BETi, we propose a model where it is regulation of transcriptional pausing, and not initiation, that is mostly affected by BETi (Fig. 8A). In many growth-regulated genes, RNA polymerase pauses after the initiation of transcription. The transcriptional pause release factor p-TEFb phosphorylates the C-terminal domain of paused RNA polymerase II, causing it to resume transcription (34). Brd4 recruits p-TEFb to active genes (Fig. 8C) (35, 36). Because histone marks are not identically placed in all cancer cells, we speculate that Brd4–p-TEFb and transcriptional elongation (40, 41) will be displaced differentially by BETi in different cell types as well. By expression profiling it will appear as though a specific transcriptional program has been turned off, when in fact it is any transcriptionally active gene that is no longer pause-released. It is tempting to speculate that this explains why overexpression of the suspected target gene of BETi, for example MYC or FOSL1, fails to rescue all phenotypes of BET (2, 11).
Fig. 8.
BET inhibitors cause major effects on transcription of growth-promoting genes. (A) Summary of effects of BETi in cancer cells. Depending on which genes get suppressed and/or activated, the cells either undergo cell-cycle arrest and apoptosis (Myc-induced lymphoma) or only undergo cell-cycle arrest (e.g., melanoma). (B) BETi cause major repression of transcription by blocking Brd4–p-TEFb–mediated pause release. Furthermore, some genes are induced, likely because of repositioning of p-TEFb and RNA pol II to stress-induced genes such as p53 target genes, immediate early genes (Egr1, Fos, Jun), histone genes, and HEXIM1, a negative regulator of p-TEFb. (C) Schematic model, based on data presented herein (Fig. 6), publicly available ChIP-seq data (Fig. S9), and conclusions from published studies in the cancer and HIV fields. (Left) In untreated cells, Brd4–p-TEFb mediate the phosphorylation of Ser2 on the RNA pol II C-terminal domain. This results in elongation of paused transcription. Upon BET inhibitor treatment, Brd4 can no longer bind acetylated histones, resulting in transcriptional pausing of many genes, including those needed for cell proliferation. HDAC inhibitor treatment does not result in suppression of the same genes, as judged by microarray. (Center) Stress-induced genes are stimulated by both BETi and HDACi via Brd4-dependent (e.g., Egr1 and TXNIP) and Brd4-independent (Bbc3 and JUN) mechanisms. The Brd4-independent mechanism may involve a transient release of p-TEFb from HEXIM1–7S RNP complexes, as seen in induction of HIV transcription by BETi and HDACi and another BET protein. (Right) Genes that are not pause-regulated in a BET-dependent manner can continue transcription or even enhance it (Fig. S9, histone genes). Whether or not this represents an active event or simply reflects enhanced availability of p-TEFb and RNA pol II as suggested in B remains to be elucidated.
BETi have been shown to also induce transcription in other pathological settings such as HIV, inflammation, and arteriosclerosis (1). The mechanisms involved are not entirely elucidated, but in HIV transcription it again points to regulation of the transcription elongation factor p-TEFb (42). A lot of p-TEFb is bound and inactivated by a complex containing HEXIM1 and the 7S snRNP. Treatments such as JQ1, vorinostat, and UV have been shown to cause a transient release of p-TEFb from this inactive complex, enabling the recruitment of p-TEFb to the promoters of, for example, the HIV genome, resulting in transcriptional pause release of HIV genes and HEXIM1 itself (30, 33). In HIV, the Tat protein (HIV-tat) can take over and stimulate transcription pause release by blocking the reformation of the inactive p-TEFb–HEXIM1–7S snRNP complex (32). This is an attractive model (Fig. 8) to explain the data obtained with BETi and HDACi here as well. It would be interesting to investigate whether there exists any factor similar to HIV-tat also in BET inhibitor and HDAC inhibitor-treated cancer cells. It is possible that this factor could be Brd4 or any of the other BET proteins—in HIV, Brd2 has been implicated as well (31)—because upon BET inhibitor treatment they cannot bind acetylated histones and stimulate the normally active genes when bound by BETi. They could then relocate to other transcriptional complexes containing, for example, Mediator and Cdk8, to which Brd4 has been shown to bind independent of acetylated histones (43). We hypothesize that HDACi in turn would recruit BET proteins by generating ectopic acetylated groups on histones or p-TEFb itself. Evidence for Brd4-dependent (Egr1) and -independent (Puma) transcriptional stimulation by BETi can be seen in Fig. 6 and in some genes that overlap between our dataset and publicly available ChIP-seq data of multiple myeloma cells treated with JQ1 (compare BRD4 occupancy on the TXNIP gene in Fig. 6 with that on the CDKN1A, JUN, or CDKN1C genes in Fig. S9). The number of genes induced by JQ1 in the public dataset was smaller, and not many genes overlapped with our gene list. This could be due to data collection after a 6-h treatment with JQ1, instead of 24 h as in our dataset. Because of these differences, the accuracy and potential magnitude of our proposed model (Fig. 8) must await future ChIP-seq data on BET inhibitor- and HDAC inhibitor-treated cells.
The similarity in response and transcriptional output between HDACi and BETi seen in Figs. 5–7 is of high clinical significance. Several HDAC inhibitors have already proven to have efficacy in mouse models of Myc-induced lymphoma (38, 39), but the molecular mechanism is not fully understood. The model put forward here correctly predicts that only induced genes would be shared between HDAC inhibitor and BET inhibitor transcriptional output. However, we cannot rule out that part of the observed synergy is indeed due to the different genes whose transcription is suppressed by HDACi and BETi. Nevertheless, the demonstrated synergy between BETi and HDACi in vivo shown in Fig. 7 may help limit potential side effects, because lower therapeutic concentrations may be feasible.
The in vivo data presented here and previous studies using other BETi have shown that inhibition of BET proteins is tolerated in mice (3, 18, 44, 45). BETi exhibit broad transcriptional effects by targeting the general transcriptional elongation factor p-TEFb’s interplay with Brd4. It is possible that systemic features of cancer such as inflammation (46) and muscle wasting by glucocorticosteroid treatment (47), known to be mediated by transcriptional elongation, could be targeted by BETi too for the benefit of cancer patients. Importantly, however, early trials will have to determine whether a combination treatment using BETi and HDACi will be tolerated in cancer patients. Because the HDAC inhibitor vorinostat (Zolinza) is FDA-approved, these types of clinical trials are tractable in a not-too-distant future.
Experimental Procedures
BET Inhibitors.
The (+)-enantiomer of JQ1 was purchased from Cayman Chemicals or synthesized. RVX2135 was synthesized by Zenith Epigenetics Corp. Fluorescence resonance energy transfer was measured between purified bromodomains and tetraacetylated H4 peptide in a Synergy H4 (BioTek) in the presence of increasing concentrations of RVX2135 or JQ1.
Mice and Tumor Analysis.
All animal experiments were performed in accordance with the regional animal ethics committee of Gothenburg approval (approval nos. 287/2011 and 288/2011). λ-Myc mice have been previously described (17), and Cdkn2a knockout mice were obtained from Jackson Laboratories. C57BL/6-Tyr (albino) mice were transplanted with lymphoma cells. When showing signs of morbidity and tumor development, mice were either acutely treated or killed. RVX2135 was formulated to a final composition including 10% PEG300, 2.5% (vol/vol) Tween 80 (pH 4).
Cell Culture and Treatments.
Mouse B-lymphoma cells were established from single-cell suspensions of tumors arising in Eµ-Myc– and λ-Myc–transgenic mice by serial passage in culture. All cells were grown in RPMI 1640 with stable glutamin (E15-885, PAA Laboratories) supplemented with 10% (vol/vol) FBS, 50 µM β-mercaptoethanol, and antibiotics. Sequencing data of λ820 cells were analyzed as described before (48). All treatments were performed using BETi or compounds from Selleck Chemicals dissolved in DMSO. DMSO concentrations never reached above 0.1%.
Cell-Cycle Analysis.
One million cells per milliliter were lysed and stained for 30 min at 37 °C in modified Vindelöv’s solution [20 mM Tris, 100 mM NaCl, 1 µg/mL 7-aminoactinomycin D (7-AAD), 20 μg/mL RNase, and 0.1% Nonidet P-40 adjusted to pH 8.0] followed by analysis of DNA content using the FL3 channel (linear mode, cell cycle) or FL3 channel (logarithmic mode, apoptosis).
RNA Preparation and Analyses.
λ820 (in biological triplicates or duplicates) or Eµ239 cells (in biological duplicates) were cultured in the presence of the indicated concentrations of JQ1, RVX2135, vorinostat, or LBH-589 at the indicated time points. RNA was prepared using the NucleoSpin RNA II Kit (Macherey-Nagel). The analysis of gene expression was performed using the Illumina BeadChip System at the Genomics Core Facility, University of Gothenburg. Mouse Ref-8 v2 BeadChip Arrays (Illumina) were used following the manufacturer’s protocol. Primary data were collected from the BeadChips using the Illumina BeadArray Reader (HiScan) and analyzed using the supplied software. Data normalization was performed by cubic spline normalization using Illumina’s GenomeStudio software equipped with the necessary modules. Before further analysis, forcePos adjustment was performed to avoid negative data arising during normalization. Clustering and visualization of genes were performed using Spotfire. For qRT-PCR, total RNA was prepared from cultured cells or tumors. cDNA was prepared from 500 ng total RNA using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative RT-PCR was performed using KAPA SYBR FAST ABI Prism 2X qPCR Master Mix (Kapa Biosystems). Data analyses were performed by comparing ΔΔCt values, using Ubiquitin (Ub) as the reference gene and with a control sample set as relative expression 1.
Chromatin Immunoprecipitation Assay and Analyses.
ChIP was carried out using the manufacturer’s protocol (SimpleChIP Plus Enzymatic Chromatin IP Kit; Cell Signaling Technology). Briefly, cells treated with vehicle, RVX2135, or vorinostat were fixed with formaldehyde and lysed; chromatin was partially digested by micrococcal nuclease and then sonicated. Lysates were quantified and ∼7.5 μg of chromatin was loaded per IP and the rest was used as input. ChIP was carried out using anti-Brd4 or anti-phosphorylated RNA polymerase II (S2) antibodies, where anti-rabbit IgG was used as an isotype control. Purified and reverse–cross-linked chromatin were analyzed by quantitative PCR (RNA analysis) using primers designed to Egr1 or Bbc3. Gene occupancy was calculated using the formula % total = 2(Ct input − Ct ChIP) × % input used for ChIP. Data were then normalized to the vehicle-treated samples. Data from three independent experiments were plotted. For analyses of publicly available ChIP-seq and Affymetrix expression profiling data, Gene Expression Omnibus (GEO) dataset GSE44931 was downloaded. Genes coregulated upon BET inhibitor or HDAC inhibitor treatment in our murine lymphoma cell line and in the multiple myeloma cell line MM1.S were inspected for occupancy of BRD4, RNA polymerase II, CDK9, and Mediator using IGV software (Broad Institute).
Immunoblotting.
Cells were lysed as previously described (49). Protein extracts (20–50 μg per lane) were then electrophoretically separated on ClearPAGE or Criterion Precast SDS/PAGE gels (C.B.S. Scientific; Bio-Rad) and transferred to membranes (Protran; GE Healthcare Bio-Sciences) before being blotted with specific antibodies (Dataset S5). In all experiments, successful transfer was confirmed by Ponceau red (0.2% solution; SERVA Electrophoresis) staining of the nitrocellulose membranes following transfer. For analyses of chromatin-bound proteins, an alternative lysis procedure was used, detailed in Fig. S1.
Positron Emission Tomography.
Before administration of the radiotracer [18F]FDG, all mice were anesthetized with 4% isoflurane (Abbott) using a veterinary anesthesia system (Vetland Medical). During PET imaging, the dose was reduced to 1.5% isoflurane. Dynamic imaging with [18F]FDG-PET was performed using a small-animal PET system (MicroPET Focus 120; Siemens Preclinical Solutions); 3.7–7.4 MBq [18F]FDG per mouse was injected intravenously, and data were acquired for 60 min. All acquisitions were performed in list-mode format and histogrammed into a framed sinogram. The sinogram was reconstructed into a 128 × 128 × 95 voxel image using the filtered back-projection method with a cutoff at the Nyquist frequency. Voxel size equals 0.433 × 0.433 × 0.796 mm3. The data were normalized and corrected for random coincidences, dead time, and decay.
Analyses of in Vivo Apoptosis.
Spleens from treated mice were fixed in formalin and embedded in paraffin. Thin sections (4-µm) were generated, and the sections were subjected to immunohistochemical analyses according to standard methodology using an antibody directed against cleaved caspase 3 (Dataset S5). Stained sections were analyzed by light microscopy (Fig. S5), and slides were scanned using a Mirax automated slide scanner (Zeiss) using the default setting for bright-field microscopy. The virtual slides were then imported to BioPix iQ, and the number of cleaved caspase 3-positive cells in the whole section was quantified and analyzed using the HSB tool to mark and quantify the 3,3′-diaminobenzidine stain. For TUNEL staining, other sections from the same spleens were processed for immunofluorescence using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Millipore). The slides were analyzed under a fluorescence microscope (Fig. S5), and the images of representative fields of view were imported into ImageJ software (National Institutes of Health). Here the amount of FITC-stained cells was counted using the cell-count function.
Statistical Analysis.
The bars shown represent the mean ± SD. Student’s t test or Kaplan–Meier (log-rank) analyses were performed using Prism (GraphPad Software). Statistically significant differences are indicated as *P < 0.05, **P < 0.01, or ***P < 0.001.
Supplementary Material
Acknowledgments
We thank Sofia Nordstrand at the animal facility (Department of Experimental Biomedicine, University of Gothenburg) for animal care, Susanne Mükusch at VisSciCom (Germany) for science graphics, Histocenter (Gothenburg, Sweden) for immunohistochemistry services, and Filip Culev Stern at the Genomics Core Facility of University of Gothenburg for microarray services. This work was supported by grants from the Swedish Cancer Society, the Swedish Research Council, the Sahlgrenska Academy, and BioCARE, a National Strategic Cancer Research Program at University of Gothenburg (to J.A.N.), Deutsche Forschungsgemeinschaft, Jose Carreras Leukämie Stiftung and Deutsches Konsortium für translationale Krebsforschung (to U.B.K.), and from the Assar Gabrielsson Foundation and Lundgrenska stiftelserna (to J.B. and S.V.M.). Clinical validation of the current work is being carried out using a grant from the Region Västra Götaland (Sahlgrenska University Hospital, Gothenburg).
Footnotes
Conflict of interest statement: E.M.G., H.C.H., and K.G.M. are employees of Zenith Epigenetics Corp.
This article is a PNAS Direct Submission. E.V.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406722111/-/DCSupplemental.
References
- 1.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12(7):465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delmore JE, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrmann H, et al. Small-molecule inhibition of BRD4 as a new potent approach to eliminate leukemic stem- and progenitor cells in acute myeloid leukemia AML. Oncotarget. 2012;3(12):1588–1599. doi: 10.18632/oncotarget.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao X, et al. Disruption of the MYC-miRNA-EZH2 loop to suppress aggressive B-cell lymphoma survival and clonogenicity. Leukemia. 2013;27(12):2341–2350. doi: 10.1038/leu.2013.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tolani B, Gopalakrishnan R, Punj V, Matta H, Chaudhary PM. Targeting Myc in KSHV-associated primary effusion lymphoma with BET bromodomain inhibitors. Oncogene. June 24, 2013 doi: 10.1038/onc.2013.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao L, et al. Androgen receptor promotes ligand-independent prostate cancer progression through c-Myc upregulation. PLoS ONE. 2013;8(5):e63563. doi: 10.1371/journal.pone.0063563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Z, et al. Inhibition of BET bromodomain targets genetically diverse glioblastoma. Clin Cancer Res. 2013;19(7):1748–1759. doi: 10.1158/1078-0432.CCR-12-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ott CJ, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120(14):2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockwood WW, Zejnullahu K, Bradner JE, Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109(47):19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu A, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154(5):1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLure KG, et al. RVX-208, an inducer of ApoA-I in humans, is a BET bromodomain antagonist. PLoS ONE. 2013;8(12):e83190. doi: 10.1371/journal.pone.0083190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholls SJ, et al. Efficacy and safety of a novel oral inducer of apolipoprotein A-I synthesis in statin-treated patients with stable coronary artery disease: A randomized controlled trial. J Am Coll Cardiol. 2011;57(9):1111–1119. doi: 10.1016/j.jacc.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Picaud S, et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci USA. 2013;110(49):19754–19759. doi: 10.1073/pnas.1310658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams JM, et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 17.Kovalchuk AL, et al. Burkitt lymphoma in the mouse. J Exp Med. 2000;192(8):1183–1190. doi: 10.1084/jem.192.8.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segura MF, et al. BRD4 sustains proliferation and represents a new target for epigenetic therapy in melanoma. Cancer Res. 2013;73(20):6264–6276. doi: 10.1158/0008-5472.CAN-13-0122-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Da Costa D, et al. BET inhibition as a single or combined therapeutic approach in primary paediatric B-precursor acute lymphoblastic leukaemia. Blood Cancer J. 2013;3:e126. doi: 10.1038/bcj.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mertz JA, et al. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci USA. 2011;108(40):16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sears R, et al. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 2000;14(19):2501–2514. doi: 10.1101/gad.836800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krones-Herzig A, et al. Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53. Cancer Res. 2005;65(12):5133–5143. doi: 10.1158/0008-5472.CAN-04-3742. [DOI] [PubMed] [Google Scholar]
- 23.Zhan Q, Chen IT, Antinore MJ, Fornace AJ., Jr Tumor suppressor p53 can participate in transcriptional induction of the GADD45 promoter in the absence of direct DNA binding. Mol Cell Biol. 1998;18(5):2768–2778. doi: 10.1128/mcb.18.5.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, et al. Domain-specific c-Myc ubiquitylation controls c-Myc transcriptional and apoptotic activity. Proc Natl Acad Sci USA. 2013;110(3):978–983. doi: 10.1073/pnas.1208334110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boone DN, Qi Y, Li Z, Hann SR. Egr1 mediates p53-independent c-Myc-induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci USA. 2011;108(2):632–637. doi: 10.1073/pnas.1008848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heller G, et al. Genome-wide transcriptional response to 5-aza-2′-deoxycytidine and trichostatin A in multiple myeloma cells. Cancer Res. 2008;68(1):44–54. doi: 10.1158/0008-5472.CAN-07-2531. [DOI] [PubMed] [Google Scholar]
- 27.Bhaskara S, et al. Deletion of histone deacetylase 3 reveals critical roles in S phase progression and DNA damage control. Mol Cell. 2008;30(1):61–72. doi: 10.1016/j.molcel.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Floyd SR, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498(7453):246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee C, et al. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol. 2012;92(6):1147–1154. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem. 2012;287(43):36609–36616. doi: 10.1074/jbc.M112.410746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boehm D, et al. BET bromodomain-targeting compounds reactivate HIV from latency via a Tat-independent mechanism. Cell Cycle. 2013;12(3):452–462. doi: 10.4161/cc.23309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, Guo J, Wu Y, Zhou Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013;41(1):277–287. doi: 10.1093/nar/gks976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J, et al. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Reports. 2012;2(4):807–816. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271(43):27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 35.Jang MK, et al. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19(4):523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, et al. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19(4):535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 37.Lovén J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153(2):320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellis L, et al. The histone deacetylase inhibitors LAQ824 and LBH589 do not require death receptor signaling or a functional apoptosome to mediate tumor cell death or therapeutic efficacy. Blood. 2009;114(2):380–393. doi: 10.1182/blood-2008-10-182758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindemann RK, et al. Analysis of the apoptotic and therapeutic activities of histone deacetylase inhibitors by using a mouse model of B cell lymphoma. Proc Natl Acad Sci USA. 2007;104(19):8071–8076. doi: 10.1073/pnas.0702294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138(1):129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zippo A, et al. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell. 2009;138(6):1122–1136. doi: 10.1016/j.cell.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 42.Rice AP. P-TEFb as a target to reactivate latent HIV: Two Brds are now in hand. Cell Cycle. 2013;12(3):392–393. doi: 10.4161/cc.23556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17(2):194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picaud S, et al. PFI-1, a highly selective protein interaction inhibitor, targeting BET bromodomains. Cancer Res. 2013;73(11):3336–3346. doi: 10.1158/0008-5472.CAN-12-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dawson MA, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Belkina AC, Nikolajczyk BS, Denis GV. BET protein function is required for inflammation: Brd2 genetic disruption and BET inhibitor JQ1 impair mouse macrophage inflammatory responses. J Immunol. 2013;190(7):3670–3678. doi: 10.4049/jimmunol.1202838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Proserpio V, Fittipaldi R, Ryall JG, Sartorelli V, Caretti G. The methyltransferase SMYD3 mediates the recruitment of transcriptional cofactors at the myostatin and c-Met genes and regulates skeletal muscle atrophy. Genes Dev. 2013;27(11):1299–1312. doi: 10.1101/gad.217240.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bhadury J, López MD, Muralidharan SV, Nilsson LM, Nilsson JA. Identification of tumorigenic and therapeutically actionable mutations in transplantable mouse tumor cells by exome sequencing. Oncogenesis. 2013;2:e44. doi: 10.1038/oncsis.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Höglund A, Nilsson LM, Forshell LP, Maclean KH, Nilsson JA. Myc sensitizes p53-deficient cancer cells to the DNA-damaging effects of the DNA methyltransferase inhibitor decitabine. Blood. 2009;113(18):4281–4288. doi: 10.1182/blood-2008-10-183475. [DOI] [PubMed] [Google Scholar]
- 50.Schmitt CA, McCurrach ME, de Stanchina E, Wallace-Brodeur RR, Lowe SW. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13(20):2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nilsson LM, et al. Mouse genetics suggests cell-context dependency for Myc-regulated metabolic enzymes during tumorigenesis. PLoS Genet. 2012;8(3):e1002573. doi: 10.1371/journal.pgen.1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13(20):2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.