Significance
Regulatory T cells (Tregs) are crucial for preventing autoimmunity, and thus discovering an efficient means of generating antigen-specific Tregs is a medical priority. To this end, we demonstrate that transcription factor Krüppel-like factor 2 (KLF2) is necessary for the generation of antigen-induced Tregs and their in vivo counterpart, peripheral Tregs. Moreover, pharmaceutical drugs that stabilize KLF2 protein levels during the transition from CD4+CD25− T cell to CD4+CD25+FoxP3+ Treg augment production of these tolerizing lymphocytes. Results from this study indicate that KLF2 is a viable target for altering Treg development, which may significantly impact patients prescribed statins.
Abstract
Regulatory T cells (Tregs) are a specialized subset of CD4+ T cells that maintain self-tolerance by functionally suppressing autoreactive lymphocytes. The Treg compartment is composed of thymus-derived Tregs (tTregs) and peripheral Tregs (pTregs) that are generated in secondary lymphoid organs after exposure to antigen and specific cytokines, such as TGF-β. With regard to this latter lineage, pTregs [and their ex vivo generated counterparts, induced Tregs (iTregs)] offer particular therapeutic potential because these cells can be raised against specific antigens to limit autoimmunity. We now report that transcription factor Krüppel-like factor 2 (KLF2) is necessary for the generation of iTregs but not tTregs. Moreover, drugs that limit KLF2 proteolysis during T-cell activation enhance iTreg development. To the authors’ knowledge, this study identifies the first transcription factor to distinguish between i/pTreg and tTreg ontogeny and demonstrates that KLF2 is a therapeutic target for the production of regulatory T cells.
Regulatory T cells (Tregs) are a vital component of self-tolerance (1, 2); however, the signaling events that govern Treg development are not well defined. Treg development and function is dependent upon the forkhead-winged helix transcription factor FoxP3, which is coregulated by signaling molecules downstream of T-cell receptors and cytokine receptors. At the same time, robust antigen receptor and cytokine receptor stimulation activates PI3K, which negatively regulates FoxP3 expression (3, 4). This occurs when PI3K activates protein kinase B (PKB), which in turn phosphorylates and inactivates forkhead-box O (Foxo) transcription factors that promote FoxP3 expression (5–7). Consistent with this model, conditional deletion of Foxo1 and Foxo3 results in reduced numbers of thymic- and peripheral-derived Tregs (7, 8). Moreover, the Tregs that eventually populate Foxo1/3-deficient animals are hyperproliferative, skew toward differentiated effector lineages, and lack suppressive functions. Within the context of CD4+ T-cell biology, a critical molecule regulated by Foxo1 is Krüppel-like factor 2 (KLF2) (9, 10), a zinc-finger transcription factor that is also inactivated in a PI3K-dependent manner (11). KLF2 maintains T-cell homeostasis, in part by promoting expression of sphingosine-1-phosphate receptor 1 (S1P1) (12–14). Surprisingly, S1P1 suppresses expression of FoxP3, as evidenced by increased numbers of thymus-derived Treg (tTregs) and induced Treg (iTreg) cells after T cell-specific excision in S1P1 gene-targeted animals (15, 16). On the one hand, studies using Foxo1/3-deficient mice suggest that KLF2 promotes Treg development and/or function, whereas reports using S1P1-deficient animals predict an opposing outcome. To clarify this apparent contradiction and gain further insight into Treg biology, we compared gene-targeted mouse models that excised Klf2 within the T-cell vs. Treg compartments. We now report that (i) KLF2 is selectively required for the generation of iTregs but not tTregs, (ii) KLF2 is necessary during the inductive phase of iTreg development to promote FoxP3 transcription, and (iii) iTreg production can be augmented by stabilizing KLF2 protein levels with pharmaceutical drugs.
Results
Treg Frequencies Are Maintained in Klf2 Gene-Targeted Mice.
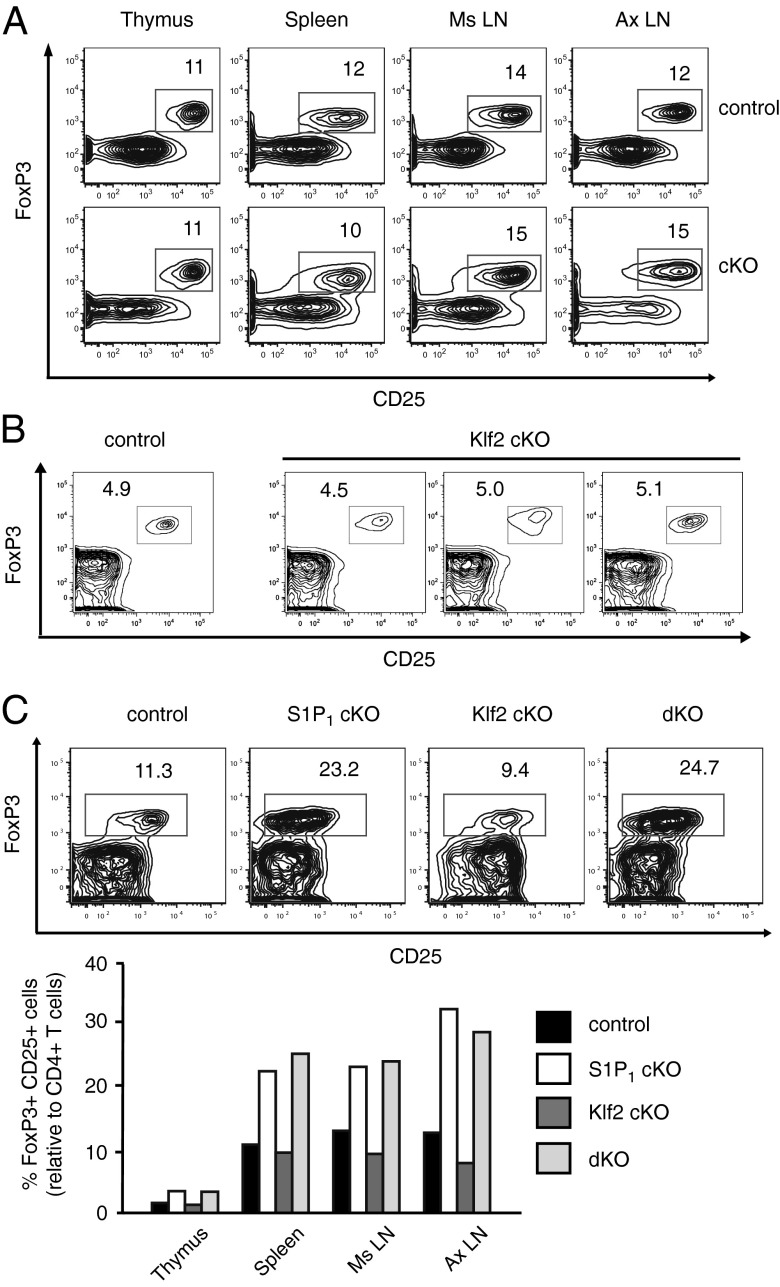
KLF2 is a zinc-finger transcription factor that has a documented role in controlling naive T-cell migration patterns (12–14). Its expression is promoted by Foxo1, a PKB-regulated transcription factor that has also been reported to control T-cell circulation (9, 10). Moreover, Foxo1 and Foxo3 promote Treg development and function; mice lacking these transcription factors have reduced percentages of FoxP3+ T cells (8), especially at 3 wk of age (7), and fail to develop functional Tregs. To determine whether Foxo1 and Foxo3 were acting through Klf2 to control Treg biology, we examined regulatory T cells in Klf2 gene-targeted animals. Using Lck-cre transgenic mice that specifically excised floxed alleles of Klf2 (Klf2fl/fl) within the T-cell compartment (Lck-cre; Klf2fl/fl), we found no obvious Treg defects, as determined by CD4+CD25+FoxP3+ T-cell frequencies in primary and secondary lymphoid tissues (Fig. 1A). Likewise, Treg frequencies in the thymus of neonates seemed normal (Fig. 1B), which would suggest that Foxo-mediated Treg development was not dependent upon KLF2.
Fig. 1.
Treg frequencies in Klf2 gene-targeted mice. (A) Percent FoxP3+CD25+ Tregs relative to CD4+ T cells in tissues harvested from Klf2fl/fl (control) vs. Lck-cre; Klf2fl/fl (cKO) littermates (6 wk of age). This experiment was repeated four times. Ms LN, mesenteric lymph nodes; Ax LN, axillary lymph nodes. (B) Percent Tregs relative to CD4+ thymocytes in 3-wk-old Klf2fl/fl (control) or Lck-cre; Klf2fl/fl (cKO) littermates. This experiment was performed three times. (C) Proportion of Tregs present in Klf2fl/fl (control), Vav-cre; S1P1fl/fl (S1P1 cKO), Vav-cre; Klf2fl/fl (Klf2 cKO), and Vav-cre; S1P1fl/fl; Klf2fl/fl (dKO) littermates (n = 2 mice per group). FACS plots: flow cytometric analysis of CD4+ T cells in the spleen. % FoxP3+CD25+ Tregs are indicated. Bar graph: average frequency of Tregs relative to CD4+ T cells in lymphoid organs. Mice were 7 wk old. This experiment was repeated twice (excluding S1P1 cKO littermates, which were analyzed once).
Recent reports have demonstrated that S1P1 suppresses generation of Tregs (15, 16), and because KLF2 promotes S1P1 expression in the CD4+ T-cell lineage (12–14), we decided to examine the relationship between KLF2 and S1P1 relative to Treg development. Using Vav-cre transgenic mice to thoroughly excise floxed alleles within the T-cell compartment, including early stages of thymocyte development, we found that S1P1 gene-targeted mice (Vav-cre; S1P1fl/fl or Vav-cre; S1P1fl/fl; Klf2fl/fl) expressed increased frequencies of Tregs, whereas this phenotype was not present in animals lacking KLF2 alone (Vav-cre; Klf2fl/fl) (Fig. 1C). From these results we conclude that KLF2 does not contribute to the overt Treg homeostatic defects observed in Foxo1/3 or S1P1 gene-targeted mice.
KLF2 Is Required for the Ex Vivo Generation of iTregs.
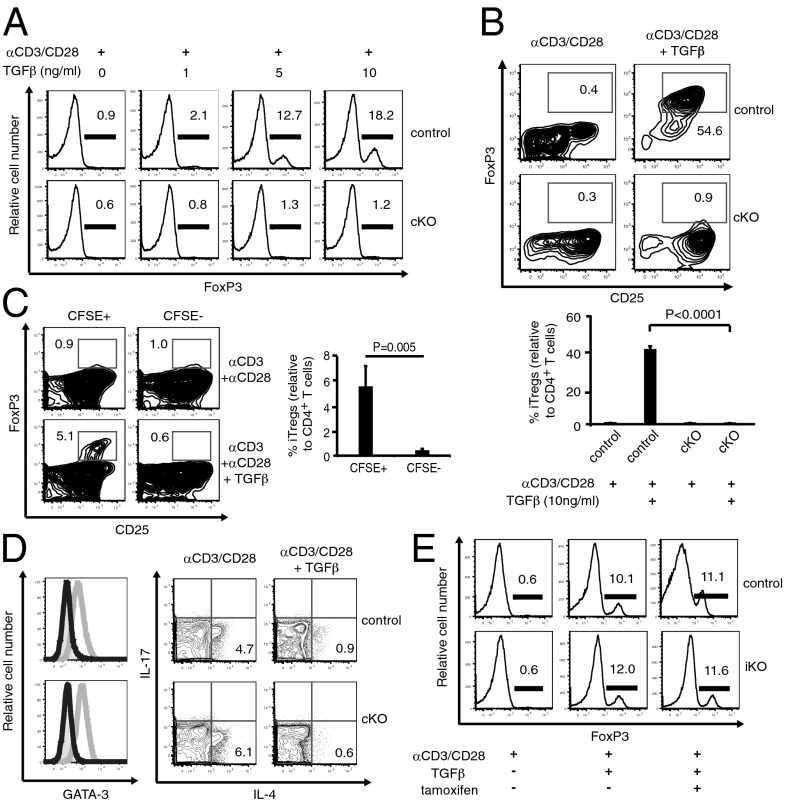
We have previously demonstrated that KLF2 differentially impacts lymphocyte homeostasis in a lineage-specific manner (17). Therefore, we decided to examine iTreg and tTreg development independent of one another. Strikingly, we found that KLF2 was necessary for the ex vivo generation of iTregs. Antigen stimulation in the presence of TGF-β induced regulatory T-cell production; however, CD4+ T cells from Lck-cre; Klf2fl/fl mice did not differentiate into FoxP3-expressing iTregs under these conditions (Fig. 2A). Similarly, KLF2-deficient CD4+CD25− (single-positive) thymocytes were unable to differentiate into FoxP3+CD25+ cells, demonstrating that defective iTreg development was not secondary to aberrant T-cell migration (Fig. 2B). A published report indicated that KLF2-deficient CD4+ T cells secrete elevated levels of IL-4 after T-cell receptor (TCR) stimulation (18), raising the possibility that IL-4 was suppressing TGF-β–mediated iTreg generation (19). To address this issue, KLF2-sufficient and KLF2-deficient CD4+ T cells were cocultured under iTreg differentiating conditions. As shown in Fig. 2C, only KLF2-sufficient T cells were capable of producing iTregs, demonstrating the cell-intrinsic nature of this defect. Moreover, we did not detect increased expression of the TH2-associated transcription factor, GATA3, or elevated production of IL-4 by KLF2-deficient splenic CD4+ T cells after antigen stimulation (Fig. 2D), indicating that this cytokine was not responsible for the iTreg defect in the present study. To determine when KLF2 was necessary during iTreg generation, we used a tamoxifen-inducible cre system (T2-cre; Klf2fl/fl) to excise Klf2 after TGF-β–induced FoxP3 expression. As shown in Fig. 2E, Klf2 excision did not affect the stability of FoxP3 expression, indicating that KLF2 was required solely at the induction stage of iTreg generation.
Fig. 2.
KLF2 is required for the ex vivo generation of iTregs. (A) Ex vivo generation of iTregs from splenic CD4+CD25− T cells derived from Klf2fl/fl (control) or Lck-cre; Klf2fl/fl (cKO) mice. Histograms display FoxP3+ T-cell frequencies. This experiment was repeated four times. (B) iTreg development using CD4+CD8−CD25− thymocytes from Klf2fl/fl (control) vs. Lck-cre; Klf2fl/fl (cKO) mice. Representative contour plots (Upper) and a chart of accumulated data (Lower) from four mice per cohort are shown. This experiment was repeated three times. (C) Ex vivo generation of iTregs using cocultured CD4+CD25− T cells harvested from Klf2fl/fl (CFSE+) and Lck-cre; Klf2fl/fl (CFSE−) mice. Contour plots and pooled results for Treg induction (αCD3+αCD28+TGFβ) are shown. This experiment was repeated three times. (D) CD4+ T cells from Klf2fl/fl (control) vs. Lck-cre; Klf2fl/fl (cKO) mice were evaluated for their propensity to skew toward the TH2 lineage and secrete IL-4. (Left) Histogram overlays of GATA-3 expression after culturing conditions that favor CD4+ T-cell differentiation toward a neutral (solid gray), TH2 (gray line), or iTreg lineage (black line). (Right) Generation of intracellular cytokines after CD4+ T-cell activation (Left) vs. iTreg differentiation (Right). This experiment was performed twice. (E) Tamoxifen-induced excision of Klf2 after ex vivo generation of iTregs using CD4+CD25− T cells from T2cre vs. T2cre; Klf2fl/fl animals. T cells from T2cre; Klf2fl/fl mice excised >95% Klf2 as measured by RT-PCR. This experiment was repeated twice.
In Vivo Development of iTregs Is KLF2-Dependent.
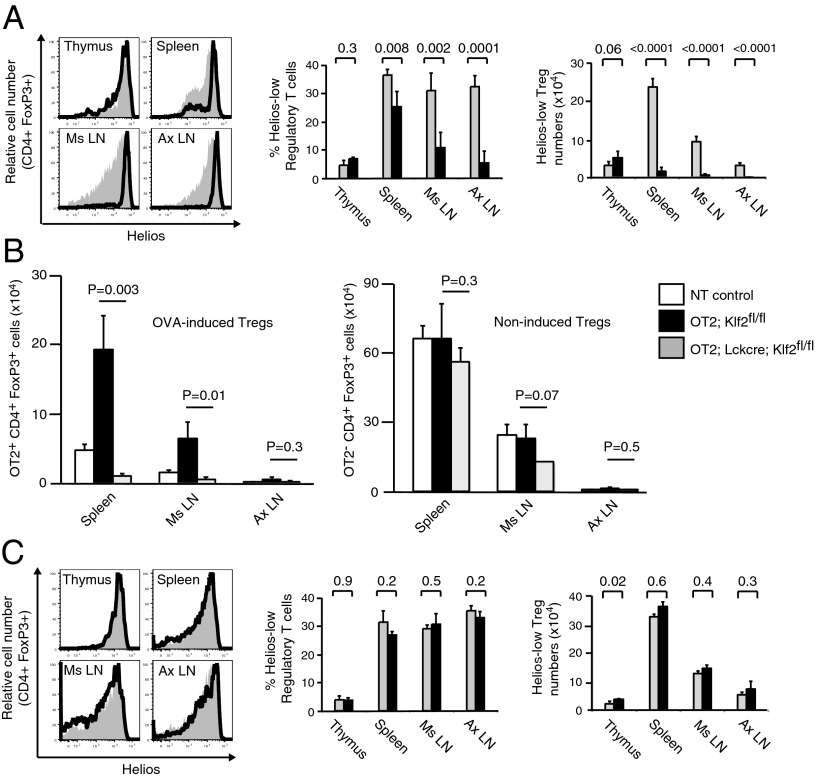
We next addressed the physiological relevance of KLF2 by examining peripheral Treg (pTreg) populations under homeostatic conditions. Reports indicate that Helios (an Ikaros transcription factor family member) and Neuropilin 1 (Nrp1, a membrane-bound surface receptor) are preferentially but not exclusively expressed by tTregs relative to pTregs (20–25). To determine whether either of these molecules were appropriate surrogates for pTreg identification, we initially characterized expression patterns within the entire regulatory T-cell compartment. Most FoxP3+CD4+ thymocytes belong to the tTreg lineage, whereas peripheral FoxP3+CD4+ T cells are composed of both tTregs and pTregs, with a partial preference for pTregs in mucosal-associated lymph nodes. Consistent with this phenotype, few HelioslowFoxP3+CD25+ T cells were detected in the thymus, whereas increased frequencies were found in secondary lymphoid organs (SLO) and after ex vivo Treg induction (Fig. S1). Although Nrp1low stains also identified iTregs in culture, expression did not strictly correlate with known pTreg tissue distribution patterns found under steady-state conditions. For this reason we focused on Helios expression as a proxy to distinguish between tTreg (Helioshigh) and pTreg (Helioslow) populations. As shown in Fig. 3A, most FoxP3+CD4+ thymocytes expressed high levels of Helios, whereas FoxP3+CD4+ T cells in SLO of wild-type animals contained a spectrum of Helios expression. In contrast, the majority of FoxP3+CD4+ T cells in Lck-cre; Klf2fl/fl mice were Helioshigh, both in primary and secondary lymphoid tissues. These Helioshigh FoxP3+CD25+ T cells were phenotypically quiescent (Fig. S1D), thus excluding the possibility that KLF2-deficient Tregs were transiently expressing Helios owing to aberrant activation (22). Significantly fewer Helioslow Tregs were present in the SLO of Lck-cre; Klf2fl/fl mice relative to littermate controls, consistent with an in vivo defect in pTreg generation. To directly assess de novo generation of pTregs, KLF2 gene-targeted mice were crossed onto a TCR transgenic background (OT2) that recognizes ovalbumin peptide (p323-339, OVA) in the context of H2-IAb. OVA-specific (Vα2+/Vβ5+) CD4+FoxP3+ Tregs are absent in OT2 animals; however, OT2 mice placed on OVA-infused drinking water develop transgenic pTregs in gut-associated lymphoid tissues (GALT; e.g., mesenteric lymph nodes, spleen) (26, 27). In agreement with surrogate Helios staining results, OT2; Lck-cre; Klf2fl/fl mice were unable to generate OVA-specific FoxP3+ Tregs (Fig. 3B), although this defect did not significantly impact nontransgenic Treg numbers. Moreover, low frequencies of CD4+ T cells expressing a TH2-like profile were identified in the GALT of Lck-cre; Klf2fl/fl mice (Fig. S2), a phenotype that has previously been associated with a lack of pTregs in vivo (28). To confirm that KLF2 was necessary for induction but not maintenance of FoxP3 expression within the pTreg compartment, we next examined Helios expression in Klf2fl/fl mice bred onto a FoxP3-cre transgenic background (FoxP3-cre; Klf2fl/fl). Consistent with our ex vivo results, we found similar frequencies and numbers of Helioslow cells in SLO harvested from FoxP3-cre; Klf2fl/fl mice and littermate controls (Fig. 3C). These data indicate that under normal physiological conditions, KLF2 is necessary during the inductive stage of pTreg development. Despite the lack of pTregs, Lck-cre; Klf2fl/fl mice were healthy and did not exhibit signs of autoimmunity, including spontaneous T-cell activation or pathogenic TH1/TH17 differentiation (Fig. S3A). Moreover, KLF2-deficient T cells maintained their effector functions (Fig. S3 B–E), suggesting that autoimmunity was actively repressed by a functional Treg compartment. Direct analysis of Tregs from Lck-cre; Klf2fl/fl mice confirmed that KLF2-deficient Tregs retained their suppressive properties (Fig. S3F). Therefore, we conclude that tTregs are sufficient to prevent autoimmunity under homeostatic conditions and posit that pTregs are necessary to limit tissue damage associated with an inflammatory environment.
Fig. 3.
KLF2 is required to establish a pTreg compartment in vivo. (A) Expression of Helios within the Treg compartment (CD4+FoxP3+CD25+) of Klf2fl/fl (gray histograms) vs. Lck-cre; Klf2fl/fl (open histograms) mice (n = 4 mice per group). Representative histogram overlays are displayed. Percentage and absolute number of Helioslow regulatory T cells from Klf2fl/fl (gray bars) vs. Lck-cre; Klf2fl/fl (black bars) mice. Error bars (SD) and P values are displayed. This experiment was repeated four times. (B) De novo generation of pTregs in non-TCR transgenic control (white bars), OT2; Klf2fl/fl (black bars), and OT2; Lck-cre; Klf2fl/fl (gray) mice placed on ovalbumin-infused drinking water. (Left) Absolute number of OT2+ CD4+FoxP3+ Tregs isolated from tissues associated with pTreg generation [spleen, mesenteric lymph nodes (Ms LN)] as well as control tissue [axillary lymph nodes (Ax LN)]. (Right) Absolute number of non-TCR transgenic OT2− CD4+FoxP3+ Tregs isolated from same animals. This experiment was conducted twice, n = 3 mice per cohort. Error bars (SD) and P values (Student t test) are indicated. (C) Comparison of Helios expression within the Treg compartment of Klf2fl/fl (gray histograms, gray bars) vs. FoxP3-cre; Klf2fl/fl (open black histograms, black bars) mice (n = 3 animals per group). Bar graphs display percentage and absolute number of Helioslow Tregs, including error bars (SD) and P values. This experiment was repeated four times.
KLF2 Directly Regulates FoxP3 Expression Within the iTreg Lineage.
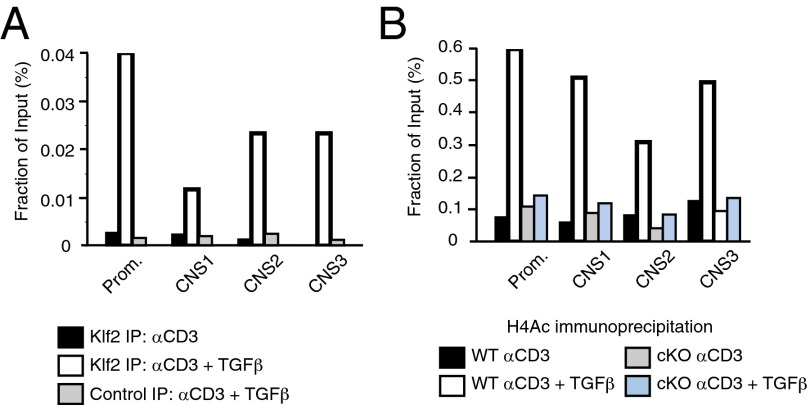
Foxp3 expression is orchestrated by transcription factors that selectively bind to key genetic elements, including the promoter region and three conserved noncoding sequences (CNS1–3) downstream of the transcriptional start site (29). These enhancer elements are differentially used by iTregs and tTregs; for instance, CNS1 is necessary for the generation of iTregs, whereas it is dispensable for tTreg production. To determine whether KLF2 bound to these regulatory regions, ChIP assays were performed using DNA from CD4+ T cells that were TCR-stimulated in the presence (iTregs) or absence (mock control) of TGF-β. As shown in Fig. 4A, KLF2 bound to the promoter and all three enhancer elements of TGF-β–stimulated T cells. Acetylated histone H4 (H4Ac) epigenetically marks transcriptionally permissive areas of chromatin (30), as demonstrated by H4Ac association with FoxP3 regulatory sites after iTreg induction of wild-type CD4+ T cells (Fig. 4B). In contrast, minimal H4Ac was recovered from ChIP assays using KLF2-deficient nuclear extracts. From these assays we contend that KLF2 is necessary to open compacted chromatin associated with the FoxP3 locus, thus permitting iTreg development under conditions that favor KLF2 expression.
Fig. 4.
KLF2 directly promotes FoxP3 transcription within the iTreg lineage. (A) ChIP assays using antibody directed against KLF2 after lysis of wild-type CD4+CD25− T cells cultured under conditions that promote T-cell activation (black) vs. iTreg differentiation (white). Isotype control antibody (gray) is included for iTreg ChIP assays. DNA primers are specific for the promoter or enhancer elements (CNS1-3) of FoxP3. KLF2 binding to the regulatory regions of FoxP3 is graphed relative to input amounts of genomic DNA. These experiments were performed three times. (B) Wild-type (WT) and KLF2-deficient (cKO) CD4+CD25− T cells were cultured under activating (black, gray) vs. iTreg-inducing conditions (white, blue) before performing ChIP assays using antibody directed against acetylated histone H4. This experiment was conducted twice.
Drugs That Stabilize KLF2 Expression Enhance iTreg Production.
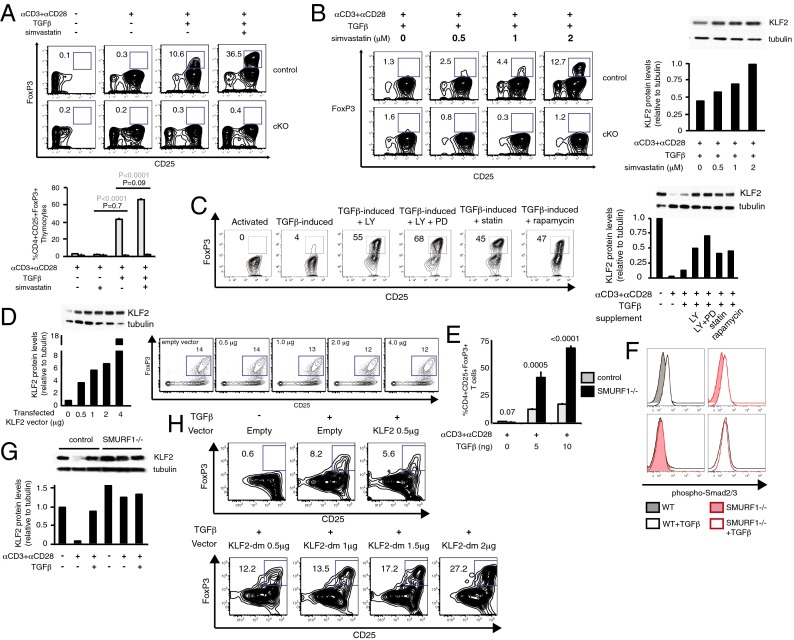
KLF2 is necessary for the generation of iTregs, raising the possibility that this transcription factor can be manipulated to enhance i/pTreg development for clinical purposes. The 3-hydroxy-3-methylglutaryl CoA reductase inhibitor simvastatin has previously been shown to enhance Klf2 expression in endothelium (31, 32) and activated T cells (33), which led us to test this US Food and Drug Administration-approved drug on T cells undergoing iTreg induction. Consistent with our previous experiments, simvastatin increased production of iTregs using cultured CD4+CD25− T cells or thymocytes (Fig. 5A) in a manner that paralleled KLF2 expression (Fig. 5B). Furthermore, this direct correlation extended to other inhibitory compounds that suppressed TCR-mediated down-regulation of KLF2 (Fig. 5C and Fig. S4). Analysis of simvastatin-treated Tregs indicated that this drug enhanced/stabilized KLF2 at the mRNA and protein level (Fig. S5), raising the possibility that one of these factors was responsible for augmented iTreg development. Overexpression of KLF2 using an established mammalian vector did not amplify FoxP3+ T-cell frequencies (Fig. 5D), which suggests that KLF2 stability rather than transcription was the primary means of regulating KLF2-mediated events after T-cell activation. To investigate the potential relationship between KLF2 stability and regulatory T-cell development, iTreg production was analyzed using CD4+CD25− T cells from Sma and Mad homologue (SMAD) specific E3 ubiquitin protein ligase 1 (SMURF1) gene-targeted animals. In keeping with a previous report that demonstrated that SMURF1 targets KLF2 for ubiquitin-mediated degradation (34), we found that ex vivo iTreg production was enhanced in the absence of SMURF1 (Fig. 5E). Importantly, SMURF1 deficiency did not uncouple iTreg’s requirement for TGF-β (Fig. 5E) or affect the activation status of SMAD molecules that act directly downstream of TGF-βR1 (Fig. 5F). Instead, SMURF1-deficient cells exhibited a defect in KLF2 stability that was most evident after T-cell activation (Fig. 5G). All of these results are consistent with KLF2 stability being a key rate-determining step during Treg induction, and for that reason we repeated transfection/induction experiments using a mutant form of KLF2 that is no longer recognized by the E3 ubiquitin ligase complex, SCFFBW7 (35). SCFFBW7 is a multisubunit RING-finger ligase that targets proteins for 26S proteasomal destruction, and in this context KLF2 has recently been identified as a substrate in endothelial cells (35). Despite the fact that several E3 ubiquitin ligases have a documented role in promoting KLF2 degradation (34–36), we found that CD4+CD25+FoxP3+ frequencies were enhanced when CD4+CD25− T cells were transfected with the mutant form of KLF2 before ex vivo Treg induction (Fig. 5H). From this we conclude that KLF2 degradation is a key facet of CD4+ T-cell conversion that can be modified to enhance the generation of i/pTregs. Of note, overexpression of KLF2 using pharmaceutical drugs during the inductive phase of iTreg production did not impair suppressive activity or cell viability (Fig. S6 A and B). Likewise, elevated levels of KLF2 did not affect ex vivo Treg suppression (Fig. S6C), which suggests that KLF2 and the proteins that interface with this transcription factor are potential immunotherapeutic targets.
Fig. 5.
Increased KLF2 expression correlates with enhanced generation of iTregs. (A) Ex vivo generation of iTregs using CD4+CD25− T cells from Lck-cre (control) vs. Lck-cre; Klf2fl/fl (cKO) littermates in the presence of simvastatin. (Upper) Contour plots of iTregs (FoxP3+CD25+) after induction of splenic T cells. (Lower) Same assay performed in triplicate using CD4+CD8−CD25− thymocytes from control (gray) vs. cKO (black) mice. Statistical differences between control groups (gray numbers) and KLF2-deficient populations (black numbers) are shown. Both sets of experiments were repeated three times. (B and C) A comparison between iTreg induction and KLF2 protein levels. (B) (Left) FACS analysis of KLF2-replete (control) vs. KLF2-deficient (cKO) CD4+CD25− T cells cultured under suboptimal iTreg conditions with or without increasing concentrations of simvastatin. (Right) Immunoblots of KLF2/tubulin and corresponding densitometry plots using total cell lysate from CD4+CD25− T cells cultured (12 h) as specified. Both sets of experiments were repeated three times. (C) (Left) Contour plots of iTregs using wild-type CD4+CD25− T cells cultured with the indicated compounds (LY, LY294002; PD, PD98059; statin, simvastatin). Activated, αCD3 + αCD28; TGFβ-induced, αCD3 + αCD28 + TGFβ. (Right) Western blots of KLF2/tubulin and corresponding densitometry data from CD4+CD25− T cells cultured for 12 h under the specified conditions. These experiments were repeated four times. (D) iTreg production after transfection of wild-type CD4+CD25− T cells with KLF2 vector. Left panel: densitometry plot of KLF2 protein levels (relative to tubulin) 24 h after transfection with pcDNA-KLF2. (Right) Contour plots of T cells after KLF2 transfection (24 h, vector concentrations indicated) and 48 h iTreg induction. This experiment was repeated twice. (E) Quantification of ex vivo iTreg production using splenic CD4+CD25− T cells harvested from wild-type (gray bars) vs. SMURF1-deficient (black bars) animals. Error bars (SD) and P values are shown. n = 2 experiments, performed in triplicate. (F) Histogram overlays displaying Smad2/3 phosphorylation after TGF-β stimulation of CD4+CD25− T cells harvested from wild-type vs. SMURF1-deficient mice. This experiment was performed once, in triplicate. (G) KLF2/tubulin immunoblot and corresponding densitometry plot of 12-h cultured CD4+ T cells harvested from wild-type vs. SMURF1-deficient mice. This experiment was repeated twice. (H) Contour plots of CD4+ T cells after transfection [24 h with pcDNA3 (empty), pcDNA3-KLF2 (KLF2), or pcDNA3-KLF2-dm (KLF2-dm)] and 48 h iTreg induction. Percentages of iTregs are included. This experiment was repeated twice.
Discussion
Numerous reports have demonstrated that Foxo1 promotes expression of KLF2, which in turn drives expression of S1P1 within the T-cell compartment. For this reason, it has been difficult to reconcile the dichotomy in Treg development that occurs in Foxo1 vs. S1P1 gene-targeted animals; in the former, tTreg and pTreg populations are decreased, whereas in the latter, both Treg lineages are increased. Because KLF2 bridges these two signaling components, we were surprised to discover that KLF2 is not required for the generation of tTregs and is only necessary for pTreg (and ex vivo iTreg) development. tTregs arise from self-reactive CD4+CD8+ thymocytes (37), and because these cells do not yet express KLF2 (KLF2 is initially expressed at the single-positive thymocyte stage), it is evident that other Foxo1/3-transcribed factors are necessary to establish CD4+CD25+FoxP3+ thymocytes. Interestingly, S1P1 thymic expression patterns are similar to those of KLF2, which leads us to speculate that S1P1 modifies FoxP3 expression after the initial generation of tTregs. If true, this would suggest that KLF2 and its transcriptional target, S1P1, regulate pTreg levels by initiating FoxP3 transcription and limiting its continued expression, respectively. This temporal separation in FoxP3 regulation may explain the counterintuitive Treg phenotypes displayed by Foxo1 and S1P1 gene-targeted animals.
With regard to the molecular mechanisms that underpin iTreg and pTreg development, current results indicate that KLF2 is absolutely necessary during the initial stages of Treg induction but is redundant once FoxP3 is stably expressed. We propose that KLF2 is incorporated into a signaling complex that provides transcriptional accessibility to the entire FoxP3 locus, after which additional nuclear factors stabilize the open conformation in a KLF2-independent manner. Consistent with a previous report (38), this larger signaling complex may include histone acetyltransferases that reverse p300/CBP-associated factor-mediated FoxP3 gene silencing in naïve CD4+ T cells. Importantly, we demonstrate that KLF2 is a rate-limiting factor during this inductive process that can be targeted with drugs to enhance iTreg production via increased KLF2 stability. Perhaps because KLF2 is a transcription factor, its regulation is typically described in terms of mRNA levels and Foxo1-mediated transcription; however, current results indicate that KLF2 protein levels are far more relevant when describing biological events in postactivated lymphocytes. Case in point, concurrent with TCR stimulation that promotes KLF2 degradation, KLF2 proteolysis is partially inhibited by TGF-β–mediated signaling events that favor iTreg production over other CD4+ T-cell differentiation outcomes. Moreover, this block in TCR-mediated KLF2 degradation is a bottleneck in iTreg production that can be amplified using a diverse array of chemical inhibitors. Future work will determine whether drugs that stabilize KLF2 protein levels expand pTreg populations in vivo, especially during inflammatory outbreaks encountered in autoimmune diseases. In this regard, it is worth noting that statins, which are currently prescribed to approximately one-quarter of all Americans over the age of 45 y (www.cdc.gov/nchs/hus.htm), have a documented role in alleviating autoimmune symptoms that are independent of their lipid-reducing properties (39). At the same time, there are numerous situations in which Tregs hinder desired immune responses, such as those directed against tumors; under these circumstances pharmaceuticals that stabilize KLF2 protein levels would be predicted to harm patient outcome. Instead, it may prove advantageous to treat the T-cell compartment with drugs that promote KLF2 degradation via E3 ubiquitin ligases such as WWP1 (36), SMURF1 (34), or FBW7 (35).
Materials and Methods
Mice.
Klf2fl/fl mice were generated as previously described (40). SMURF1 gene-targeted mice (41) were obtained from the laboratory of Ying E. Zhang (Center for Cancer Research, National Cancer Institute, Bethesda, MD). Lck-cre mice were purchased from Taconic, and all other mice were purchased from Jackson Laboratories. All KLF2 gene-targeted animals were back-crossed a minimum of 10 generations onto a C57BL/6 background. Mice were housed in pathogen-free conditions in accordance with the Institutional Animal Care and Use Committee at Vanderbilt University.
Antibodies.
Anti-Thy1.2 (53-2.1), -CD3ε (145-2C11), -CD4 (GK1.5), -CD8α (53-6.7), -CD25 (PC61.5), -CD28 (37.51), -CD44 (IM7), -CD69 (H1.2F3), -CD71 (C2), -IFN-γ (XMG1.2), -IL4 (11b11), -IL17 (TC11-18H10), -GATA3 (L50-823), and –phospho Smad2/3 (072-670) were purchased from BD Biosciences. Anti-FoxP3 (FJK-16s) and -CD152 (UC10) were obtained from eBioscience USA. Anti-Helios (22F6) was purchased from Biolegend. Anti-Neuropilin was purchased from R&D Systems. Anti-KLF2 and -Acetyl–Histone H4 (H4Ac) were purchased from Millipore. Anti-α-tubulin was obtained from Cell Signaling Technologies.
Flow Cytometry and Cell Sorting.
Intracellular and surface staining was performed using standard procedures. For cell quantification, CaliBRITE Beads (BD Biosciences) were added to individual tubes before acquisition. CD4+CD25+ T cells were isolated using a Treg isolation kit (Miltenyi Biotec) according to the manufacturer’s instructions.
Ex Vivo Induction of Tregs.
FACS- or MACS-sorted naïve CD4+CD25− cells (2 × 106 cell/mL) were preincubated with or without LY294002 (10 µM), PD98059 (10 µM), simvastatin (2 µM), or rapamycin (100 nM), activated with plate-bound anti-CD3ε (2 µg/mL) and soluble -CD28 (2 µg/mL) antibody, then cultured with TGF-β (1–10 ng/mL) in complete medium containing IL-2 (5 ng/mL). After 2–4 d of culture cells were collected and stained for FoxP3.
In Vivo Induction of Tregs.
Klf2fl/fl, OT2; Klf2fl/fl, and OT2; Lck-cre; Klf2fl/fl mice were placed on chicken ovalbumin (p323-339)-infused drinking water (20 mg/mL) for 7 d, after which animals were killed and analyzed for pTreg development. Antibodies directed against Vα2 and Vβ5 (BD Biosciences) were used to identify OVA-specific T cells.
Ex Vivo Excision of KLF2.
A tamoxifen-inducible cre transgenic mouse model (T2-cre) (42) was used to excise KLF2 after ex vivo generation of Tregs. After 4 d of iTreg culture, cells were treated with 100 nM 4-hydroxy-tamoxifen for an additional 72 h. Klf2 excision was assessed by RT-PCR, and cells were analyzed by flow cytometry.
ChIP Assays.
FACS-sorted naïve CD4+CD25− T cells were activated and cultured for 4 d. ChIP assays were performed using the EZ-ChIP kit (Millipore) according to the manufacturer’s protocol. Immunoprecipitations were performed using anti-Klf2 antibody, anti-H4Ac or rabbit IgG control antibody. The following primer sets were used to amplify the DNA from KLF2 pull-downs: Foxp3 Promoter F 5′-CTC TGT GGT GAG GGG AAG AA-3′; Foxp3Promoter R 5′CGC AGA CCT CGC TCT TCT AA-3′; Foxp3 CNS1 F 5′- ACG TAT CTC TCT AGT GGG TCT GGA-3′; Foxp3 CNS1 R 5′- TGA GGA ACA GTG CAG GAC AG-3′; Foxp3 CNS2 F 5′-CTA GCC ACT TCT CGG AAC GA-3′; Foxp3 CNS2 R 5′-CAG CTT CCT GCA CTG TCT GTT-3′; Foxp3 CNS3 F 5′- GCC CAC ACC TCT TCT TCC TT-3′; Foxp3 CNS3 R 5′- GGG ACC CAT AAA CCA CTT CC-3′.
Nucleofection of Quiescent CD4+CD25− T Cells.
Transient nucleofection was performed using the Lonza/Amaxa Mouse T-cell Nucleofector Kit (VPA-1006) according to the manufacturer’s instructions. Nucleofection of purified splenic CD4+CD25− T cells (1 × 106) from C57BL/6J mice was performed with the indicated amounts of pcDNA3 (empty vector), pcDNA3-KLF2 (40), or pcDNA3-dmKLF2 plasmid (35). Cells were subsequently cultured under iTreg-inducing conditions before FACS analysis.
Statistical Analysis.
Data were analyzed using a two-tailed Student t test. P values ≤ 0.05 were considered statistically significant.
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323493111/-/DCSupplemental.
References
- 1.Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol. 2010;10(7):490–500. doi: 10.1038/nri2785. [DOI] [PubMed] [Google Scholar]
- 2.Josefowicz SZ, Lu LF, Rudensky AY. Regulatory T cells: Mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–564. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205(3):565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauer S, et al. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. Proc Natl Acad Sci USA. 2008;105(22):7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada Y, et al. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. J Exp Med. 2010;207(7):1381–1391. doi: 10.1084/jem.20100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma J, Ding Y, Fang X, Wang R, Sun Z. Protein kinase C-θ inhibits inducible regulatory T cell differentiation via an AKT-Foxo1/3a-dependent pathway. J Immunol. 2012;188(11):5337–5347. doi: 10.4049/jimmunol.1102979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouyang W, et al. Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat Immunol. 2010;11(7):618–627. doi: 10.1038/ni.1884. [DOI] [PubMed] [Google Scholar]
- 8.Kerdiles YM, et al. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33(6):890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fabre S, et al. FOXO1 regulates L-Selectin and a network of human T cell homing molecules downstream of phosphatidylinositol 3-kinase. J Immunol. 2008;181(5):2980–2989. doi: 10.4049/jimmunol.181.5.2980. [DOI] [PubMed] [Google Scholar]
- 10.Kerdiles YM, et al. Foxo1 links homing and survival of naive T cells by regulating L-selectin, CCR7 and interleukin 7 receptor. Nat Immunol. 2009;10(2):176–184. doi: 10.1038/ni.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waugh C, Sinclair L, Finlay D, Bayascas JR, Cantrell D. Phosphoinositide (3,4,5)-triphosphate binding to phosphoinositide-dependent kinase 1 regulates a protein kinase B/Akt signaling threshold that dictates T-cell migration, not proliferation. Mol Cell Biol. 2009;29(21):5952–5962. doi: 10.1128/MCB.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai A, Hu H, Yeung M, Chen J. Kruppel-like factor 2 controls T cell trafficking by activating L-selectin (CD62L) and sphingosine-1-phosphate receptor 1 transcription. J Immunol. 2007;178(12):7632–7639. doi: 10.4049/jimmunol.178.12.7632. [DOI] [PubMed] [Google Scholar]
- 13.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442(7100):299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 14.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9(3):292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 15.Liu G, et al. The receptor S1P1 overrides regulatory T cell-mediated immune suppression through Akt-mTOR. Nat Immunol. 2009;10(7):769–777. doi: 10.1038/ni.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu G, Yang K, Burns S, Shrestha S, Chi H. The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nat Immunol. 2010;11(11):1047–1056. doi: 10.1038/ni.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoek KL, et al. Follicular B cell trafficking within the spleen actively restricts humoral immune responses. Immunity. 2010;33(2):254–265. doi: 10.1016/j.immuni.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinreich MA, et al. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31(1):122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dardalhon V, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(-) effector T cells. Nat Immunol. 2008;9(12):1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto N, et al. Foxp3-dependent and -independent molecules specific for CD25+CD4+ natural regulatory T cells revealed by DNA microarray analysis. Int Immunol. 2006;18(8):1197–1209. doi: 10.1093/intimm/dxl060. [DOI] [PubMed] [Google Scholar]
- 21.Thornton AM, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol. 2010;184(7):3433–3441. doi: 10.4049/jimmunol.0904028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottschalk RA, Corse E, Allison JP. Expression of Helios in peripherally induced Foxp3+ regulatory T cells. J Immunol. 2012;188(3):976–980. doi: 10.4049/jimmunol.1102964. [DOI] [PubMed] [Google Scholar]
- 23.Zabransky DJ, et al. Phenotypic and functional properties of Helios+ regulatory T cells. PLoS ONE. 2012;7(3):e34547. doi: 10.1371/journal.pone.0034547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weiss JM, et al. Neuropilin 1 is expressed on thymus-derived natural regulatory T cells, but not mucosa-generated induced Foxp3+ T reg cells. J Exp Med. 2012;209(10):1723–1742, S1. doi: 10.1084/jem.20120914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yadav M, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. 2012;209(10):1713–1722, S1–S19. doi: 10.1084/jem.20120822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mucida D, et al. Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest. 2005;115(7):1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun CM, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204(8):1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Josefowicz SZ, et al. Extrathymically generated regulatory T cells control mucosal TH2 inflammation. Nature. 2012;482(7385):395–399. doi: 10.1038/nature10772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463(7282):808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sen-Banerjee S, et al. Kruppel-like factor 2 as a novel mediator of statin effects in endothelial cells. Circulation. 2005;112(5):720–726. doi: 10.1161/CIRCULATIONAHA.104.525774. [DOI] [PubMed] [Google Scholar]
- 32.Parmar KM, et al. Statins exert endothelial atheroprotective effects via the KLF2 transcription factor. J Biol Chem. 2005;280(29):26714–26719. doi: 10.1074/jbc.C500144200. [DOI] [PubMed] [Google Scholar]
- 33.Bu DX, et al. Statin-induced Krüppel-like factor 2 expression in human and mouse T cells reduces inflammatory and pathogenic responses. J Clin Invest. 2010;120(6):1961–1970. doi: 10.1172/JCI41384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie P, et al. Smurf1 ubiquitin ligase targets Kruppel-like factor KLF2 for ubiquitination and degradation in human lung cancer H1299 cells. Biochem Biophys Res Commun. 2011;407(1):254–259. doi: 10.1016/j.bbrc.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 35.Wang R, et al. FBW7 regulates endothelial functions by targeting KLF2 for ubiquitination and degradation. Cell Res. 2013;23(6):803–819. doi: 10.1038/cr.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Srinivasan SV, Lingrel JB. WWP1-dependent ubiquitination and degradation of the lung Krüppel-like factor, KLF2. Biochem Biophys Res Commun. 2004;316(1):139–148. doi: 10.1016/j.bbrc.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 37.Lio CW, Hsieh CS. Becoming self-aware: The thymic education of regulatory T cells. Curr Opin Immunol. 2011;23(2):213–219. doi: 10.1016/j.coi.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong Y, et al. Polycomb antagonizes p300/CREB-binding protein-associated factor to silence FOXP3 in a Kruppel-like factor-dependent manner. J Biol Chem. 2012;287(41):34372–34385. doi: 10.1074/jbc.M111.325332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khattri S, Zandman-Goddard G. Statins and autoimmunity. Immunol Res. 2013;56(2-3):348–357. doi: 10.1007/s12026-013-8409-8. [DOI] [PubMed] [Google Scholar]
- 40.Lee JS, et al. Klf2 is an essential regulator of vascular hemodynamic forces in vivo. Dev Cell. 2006;11(6):845–857. doi: 10.1016/j.devcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 41.Yamashita M, et al. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell. 2005;121(1):101–113. doi: 10.1016/j.cell.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1(1):113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.