Significance
Seasonality in disease incidence is ubiquitous among infectious diseases. Seasonal drivers include weather variables, such as temperature and humidity, and social factors (e.g., contact patterns). Attempts to make long-term predictions of infectious diseases are hampered by the inability to understand the complex interplay of these factors. Here, we model the dynamics of seasonal influenza based on a high-quality 12-year Israeli dataset. The dynamics are completely explainable by the time evolution of the model equations, the antigenic jumps of the virus, and the climatic forcing, yielding high-correlation fits to the data (r = 0.94). The forecasting ability is greatly increased through the predictability of the system’s transient dynamics, resulting in accurate predictions (r = 0.93) that have not yet been found elsewhere.
Keywords: model forecasting, infectious disease, climate, Bayesian epidemic model, predictive model
Abstract
Human influenza occurs annually in most temperate climatic zones of the world, with epidemics peaking in the cold winter months. Considerable debate surrounds the relative role of epidemic dynamics, viral evolution, and climatic drivers in driving year-to-year variability of outbreaks. The ultimate test of understanding is prediction; however, existing influenza models rarely forecast beyond a single year at best. Here, we use a simple epidemiological model to reveal multiannual predictability based on high-quality influenza surveillance data for Israel; the model fit is corroborated by simple metapopulation comparisons within Israel. Successful forecasts are driven by temperature, humidity, antigenic drift, and immunity loss. Essentially, influenza dynamics are a balance between large perturbations following significant antigenic jumps, interspersed with nonlinear epidemic dynamics tuned by climatic forcing.
Influenza outbreaks have been documented in the scientific literature in records that extend back to at least 1650 (1), making it an exceptional example of a persisting, recurrent disease. Being a respiratory infection, influenza spreads rapidly from person to person through a population in the form of virus particles airborne as respiratory droplets or aerosols. Depending on the circumstances, influenza typically infects between 10% and 50% of a given population and has become a source of considerable human morbidity and mortality (2). There is much controversy in identifying the seasonal drivers that generate annual influenza epidemics and the processes that give rise to their large variability (3–12). This is an outstanding problem of influenza research today. Using long-term modeling, a recent study (9) gave support to the possibility that absolute humidity is the predominant determinant of influenza seasonality in temperate zones, driving disease transmission and controlling the timing of individual wintertime outbreaks. Another study investigated the physical properties of absolute humidity on influenza virus transmission and influenza virus survival (3). However, a general understanding of the mechanisms underlying influenza seasonal variation remains quite limited (8). Here, we use a simple mathematical model to unravel the interplay between climate and evolution to predict long-term influenza dynamics correctly for the years since June 2010.
A requirement for the generation of recurrent epidemics is a sufficient and continuous source of new susceptible individuals arising in the population, enough to fuel each new outbreak (13). In the case of influenza, infected individuals recover with immunity but eventually become susceptible again because of the rapidly evolving nature of the influenza virus (7, 14). Positive selection exerted by the host immune system leads to a continual antigenic drift of the influenza virus’s glycoproteins, particularly the main antigen, hemagglutinin, thus allowing the virus to eventually evade the immune system (15). The process of antigenic drift thereby creates an important renewed source of susceptible individuals. Hence, evolutionary forces are considered tremendously important in shaping complex recurrent patterns of infectious diseases and explain why influenza is regarded as “an invariable disease caused by a variable virus” (1).
The changing rate of antigenic drift also has a significant impact on the timing and amplitude of influenza outbreaks (16). Recent studies reveal that the evolution of influenza A H3N2’s main antigen is punctuated in character such that the drift occurs within discrete antigenic clusters (neutral periods), but with jumps to newly arising clusters after irregular periods (17, 18). A significant jump for the A H3N2 lineage last occurred during the 2003–4 season with the appearance of the A/Fujian virus strain coinciding with a sharp influenza outbreak approximately 2 mo earlier than usual, with a normal attack rate. Nevertheless, it is difficult to demonstrate a consistent and conclusive direct link between the size of antigenic jumps and changes in influenza dynamics at the population level (6, 19–21).
Results
Our modeling is based on a high-quality 11.5-y dataset from June 2001 to January 2013 of daily influenza-like illness (ILI) cases reported in Israel’s largest city, Tel Aviv, as plotted in Fig. 1. The data were obtained from the Maccabi Health Maintenance Organization (HMO), whose medical surveillance covers ∼45% of the local Tel Aviv population. An analysis of laboratory tests of ILI cases from sentinel clinics showed the ILI dataset to be highly correlated with the incidence of confirmed influenza cases (SI Appendix). Its high level of coverage and the data quality make this one of the finest available influenza surveillance datasets by world standards. We make use of the classical susceptible–infected–recovered–susceptible (SIRS) epidemic model (Materials and Methods) in which all individuals in the population are assumed to be in only one of three classes: susceptible (S), infected (I), and recovered (R). S individuals move to the I class after contact with an infective and transmission of the disease. Infective individuals eventually recover and move to the R class, whereas R individuals eventually become susceptible once more, closing the SIRS loop (S → I → R → S) in a manner that mimics the effects of antigenic drift (14).
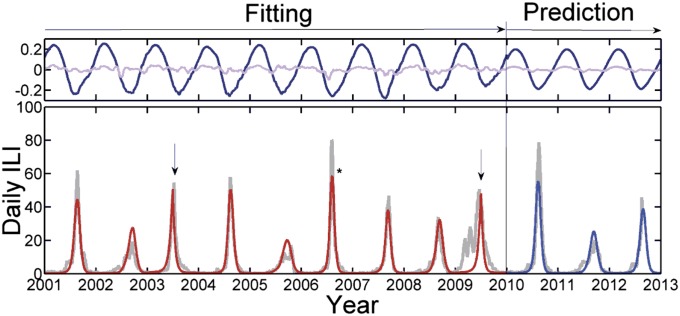
Fig. 1.
(Lower) The smoothed daily ILI cases reported in Tel Aviv (light gray), with June 1 indicated on the x-axis for each year. Time series from the climate-driven deterministic SIRS model fitted to the ILI data (2001–2010) is shown (red), with fit correlation r = 0.94 to the period leading up to the pandemic in 2009. (Upper) The climate data consist of centered, normalized daily temperature (dark blue) and relative humidity readings (light violet), both scaled according to estimated weight. The pure prediction from June 1, 2010 is driven by the average climate of all years with fit r = 0.93. The outbreaks dominated by influenza B are indicated, and the asterisk highlights the abnormally high amplitude of 2006–2007. The epochal jumps in antigenic drift are indicated with arrows.
The model requires six basic parameters, which we provide with the best-fitting estimates (and ∼95% credible intervals) found from the modeling procedure: (i) The parameter Λ = 0.17 ± 0.01 describes the rate of immunity loss due to antigenic drift and the rate of new susceptible individuals entering the population per year. (ii) The reproduction number R0 represents the number of infections transmitted by a typical individual over the lifetime of the disease in a wholly susceptible population. The model assumes that the basic reproduction rate, estimated as R0 = 2.9 ± 0.1, is constant for all years, as discussed further below. (iii and iv) Seasonal forcing is included by modulating R0 with the locally observed temperature and relative humidity time series seen in Fig. 1 (Upper). The forcing δ(t) is specified by weights that control the influence of these climatic variables: temperature weight wT = 0.13 ± 0.02 and relative humidity weight wRH = 0.028 ± 0.003. Although for generality we include relative humidity and temperature as climate variables (22), similar but slightly inferior results also are obtained by using only absolute humidity as a single driver (3). (v and vi) Two separate parameters are used to take into account the special characteristics of years with epochal antigenic jumps. One parameter enhances the loss-of-immunity rate Λ′ = Λ + Λ* over the entire year (Materials and Methods and SI Appendix). The other parameter makes it possible to accommodate the early arrival of outbreaks observed in epochal jump years, e.g., the A/Fujian outbreak in 2003–4. This may be achieved by enhancing Reff over the first 6 mo of the season when the epidemic grows, by δ′ = δ + Δ. We fit the two parameters Λ* and Δ over the epochal jump years 2003–2004 and 2009–2010 only. Finally, the model requires a single boundary condition, S(t0) = 0.31 ± 0.02 (fitted value), which represents the fractional size of the susceptible population on the first of June 2001, at the beginning of the modeling period. All model runs start out of season on the first of June; thus, we always have I(t0), = 0 although there is a small continuous migration influx (Materials and Methods).
The simplicity of this elementary SIRS model would deem it unlikely to mimic Israel’s complex influenza dynamics, as seen in Fig. 1. However, the model fit of the first 8 y of the data (June 2001 to June 2009) correlates with the observed data to an unusual degree, with correlation r = 0.94 (and r = 0.90 for the full fitting period, including the unusual pandemic year 2009/10). Note that all model time series correlations with observed data reported here are highly significant (P < 0.001).
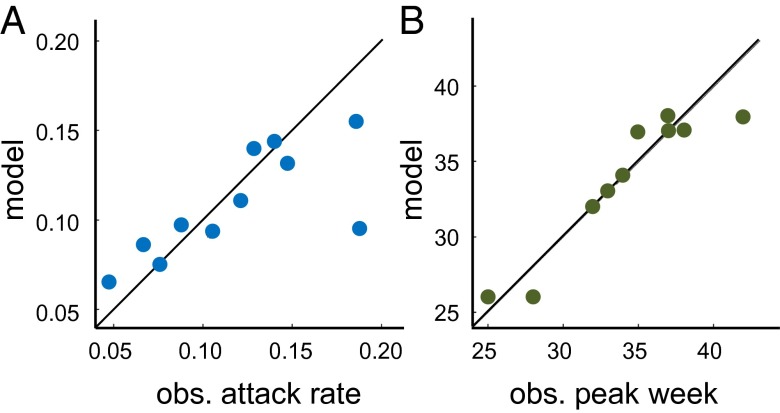
To assess the quality of the fit further, we calculated the attack rates and the week of peak incidence in the data and the fit for each of the seasons (not including the last season, for which we have only partial data). We then computed the Pearson correlation between the data and the fit of these attributes. Fig. 2 shows the result of this analysis. For the attack rates, we obtained a correlation of r = 0.74 (P = 0.01) The correlation was driven down because of the pandemic season of 2009–2010, in which there were several unusual waves in a single year. Without this season, the correlation in the attack rates would exceed r = 0.9. For the peak epidemic week, we obtained a correlation of r = 0.94 (P < 0.001).
Fig. 2.
Quality of model fit as assessed by correspondence of (A) attack rate at r = 0.55, P = 0.01. The main outlier in the match of attack rates is the H1N1 outbreak in 2009, without which the fit exceeds r = 0.9. (B) Peak epidemic week vs. model fit correspondence is r = 0.94, P < 0.001.
The excellent fit is a feature that also holds with the fully stochastic version of the model (SI Appendix). Because there are only six degrees of freedom and one boundary condition, this is not even one parameter per year. Also note that the large year-to-year variability in epidemic size and timing appears to be random in character, making it surprising that the mechanistic model can capture the intricacies of the data despite the major qualitative differences in the shape of the epidemic from year to year. For example, compare the large-scale symmetrically shaped epidemic in 2004–5 with the small, curtailed asymmetric outbreak in 2005–6. The model nevertheless captures both epidemics accurately. This accuracy is a result of the complex interaction among the changing supply of new susceptible individuals arising due to loss of immunity in the population through antigenic drift, the strong transient dynamics following the appearance of a new strain, and the timing of the climate signal each year (Fig. 1). Note that the years dominated by influenza B fall effortlessly within the model dynamics, providing possible population-level observational support for the notion of broader cross-reactivity of the immune response to influenza, as previously reported (23–25).
The recent global H1N1 pandemic in 2009–10 deviates the most from the normal influenza outbreak dynamics. Given that influenza epidemics almost always are triggered at the beginning of winter, one cannot expect our model to fit an outbreak beginning as early as April, as was the case for the H1N1 pandemic. The model can accommodate only the main peak of the outbreak, which occurred at the same time as the 2003–4 outbreak. Here, we are fitting δ′ and Λ′ to both the 2003–4 and the 2009–10 pandemics. When attempting to forecast such outbreaks, we note that the possibility of an antigenic jump year sometimes is known in advance, during the previous season, and methods are being refined to predict the possibility of a forthcoming jump year. Once expected, we can forecast the epidemic trajectory using our estimated jump parameters.
Having obtained parameter estimates via the fitting procedure, we can use the model to predict the ILI dynamics following the severe 2009–10 pandemic. Below, we further discuss the probability of consecutive antigenic jumps, and we therefore assume that the seasons following 2009–10 were normal. As Fig. 1B demonstrates, the model successfully predicts the trajectories of the subsequent three normal outbreaks from June 2010 through March 2013 with a high correlation of r = 0.93, although these outbreaks are still clearly different in timing, amplitude, and length. When making pure predictions into the future, it is assumed that prevailing local climate conditions are unknowable, and hence the model is driven by daily average climate data determined from the entire previous period.
To identify the significant components of our model, we apply a bottom-up approach by first considering the dynamics that arise with a hypothetical climate that is purely periodic. This is accomplished by assuming R0 is periodic and sinusoidal of the standard form (13, 26). However, sinusoidal forcing leaves the model dynamics periodic and uniform, as shown in Fig. 3A, and therefore incapable of capturing any year-to-year variability seen in the real data. This very regular behavior represents the limit cycle of the outbreak dynamics. Using a simple square-pulse function for the climate driver, we arrive at the following analytical approximation for the effective reproduction rate R0S0 ≡ Reff, where S0 is the susceptible population at outbreak (SI Appendix, section 5):
| [1] |
When inserting realistic parameter values (e.g., R0 = 3.5, δ = 0.2, Λ = 0.15; for details and references, see SI Appendix), the right-hand-side term is Reff ∼ 1.22. Note that previous findings from a multiyear analysis (6) of seasonal data from Australia, France, and the United States yielded in general Reff ∼ 1.3. Even for wide ranges of realistic influenza parameters, the approximation bounds the overall Reff within the interval [1.1, 1.4] (SI Appendix). The formula also shows how antigenic drift via Λ provides a contribution to raising Reff above unity.
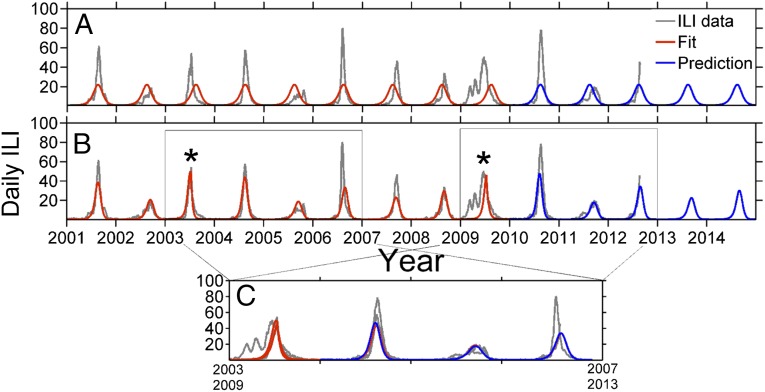
Fig. 3.
(A) Model outcome of fitting (red) a pure sinusoidal seasonality driver: r = 0.65. The prediction (blue) is the continuation of the model after the fitting period ends. (B) Model outcome by including only antigenic jumps (*) together with the pure seasonality driver: r = 0.84. (C) Data collapse showing the HHL(H) motif (gray shade) also exhibited by the model (red and blue).
The key factors responsible for the success of the model can be understood from Fig. 3. The basic sinusoidally forced model always has an outbreak in the high season (winter) and thus is endowed with a baseline average correlation of r = 0.65 against the data at any time, as seen in Fig. 3A. Upon inclusion of antigenic jumps in the years 2003 and 2009, this simple model and the data appear to lock in synchrony to a much higher degree, with the correlation jumping to r = 0.84, as shown in Fig. 3B. Inclusion of the real climate signal, rather than sinusoidal forcing, finally raises the correlation up to r = 0.94 for the model fit in Fig. 1.
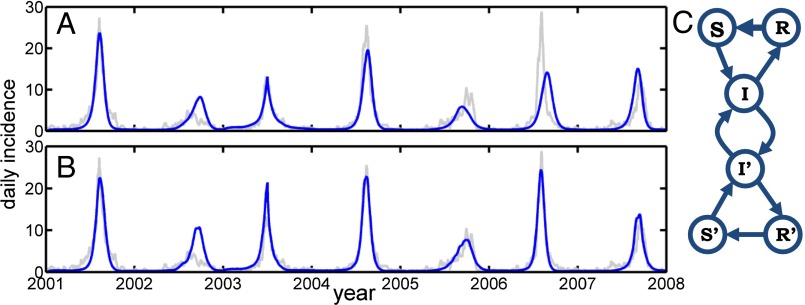
As a further test of the method, we applied the same fitting/prediction scheme to ILI data from Jerusalem, which has a 10% health insurance coverage rate. The returned fit correlated to the data with r = 0.84 (compare with Tel Aviv, where r = 0.94); the lower correlation is a result of the poor fit of the June 1, 2004–2007 period (see Fig. 4A). Because thousands of Jerusalem residents commute daily to Tel Aviv and surrounding areas, we included a fitted fraction, ψ = 0.30 ± 0.05, of the infected population in Tel Aviv to infect the susceptible population of Jerusalem, and vice versa, to make an interacting two-city model. The model then could fit the observed Jerusalem ILI data with a much higher correlation of r = 0.94 (Tel Aviv slightly diminished at r = 0.93), largely because of the June 1, 2004–2007 period (see Fig. 4B). The results thus demonstrate a metapopulation in action and further corroborate that the SIRS model is working effectively, even for two geographically separated cities.
Fig. 4.
Jerusalem ILI. (A) Data (gray) and model (blue) of a fit to Jerusalem influenza epidemics, r = 0.84. We fit Jerusalem and Tel Aviv simultaneously, although uncoupled, with the same universal R0. (B) The metapopulation model with mutual exchange of infectives between Tel Aviv and Jerusalem, r = 0.94. (C) Network diagram of the coupled populations.
Discussion
The model’s transient dynamics after large perturbations (e.g., the 2003–4 A/Fujian antigenic jump) have an important impact on the epidemic system (Fig. 3). After large antigenic jumps (asterisks in Fig. 3), the trajectory of the infective population follows a damped double-period oscillation (27) that corresponds well to the observed disease dynamics for at least 3 y. This overall dynamical motif may be characterized as “high–high–low–(med/high),” or simply HHL(H), in terms of coarse-grained epidemic peak amplitudes. During such a transient, the climate driver variability seems to be a second-order modulating effect. Upon exiting the transient, however, the model returns to seasonal outbreaks in which the detailed variability is dominated by climate dynamics. Long-term predictions in this latter regime depend on access to high-accuracy prediction of local climate, which currently is not available.
In Fig. 3C, we compare the influenza dynamics following the appearance of new strains by plotting the 4 y beginning with the A/Fujian year in 2003–4 together with the 3.5 y beginning with the A/H1N1 pandemic in 2009–10. Also included are model runs (red and blue) based on a pure sinusoidal climatic driver. The data seemingly collapse in the sense that the observed data in both windows sit closely on top of each other, except for the fourth year. In other words, they form the same HHL(H) motif. It appears more than just coincidence that on the two occasions an epochal jump year appears, the double-period damping transient or the motif in the time series ensues, as this data collapse identifies. To explore the randomness of the underlying motif idea, we can assume that years have random independent peak heights (H, M, or L). The probability of the motif occurring by chance alone is significantly small, P < 1% (SI Appendix). Unsurprisingly, the major deviance of the data collapse—and the worst correspondence to the model—is the early part of the unusual 2009 H1N1 pandemic, during summer/fall of 2009 (visible as two “humps” before the main winter outbreak). In Fig. 3C, the fourth-year predictions also are degraded, as the transient oscillations will have died down to a level at which the climate starts to dominate again. This explains the prediction errors in Fig. 3B, in which sinusoidal rather than true climatic forcing is used. The true climatic signal provides a superior prediction of the 2006–7 outbreak (compare Fig. 1 with Fig. 3B and SI Appendix). Similarly, by using average climate data, we successfully trace the 2012–13 outbreak, predicting a normal outbreak initiation time and matching the amplitude peak closely (Fig. 1, time series end).
That the simple model fits the data well raises many questions. Most plausibly, the population mixing inside Israel conforms closely to the basic SIRS model assumptions so that the mean field dynamics provide a faithful caricature of influenza transmission and immunity loss in a large population. Historically, the success of the SIR model for single epidemics derives from this feature, but what is new here is the unusual long-term year-to-year predictability reproduced in addition. It would appear that the dynamics are completely explainable by the combined time evolution of the model equations, the antigenic jumps, and the climatic forcing. The hypothesis outlined here prescribes not only that climate is a key seasonal driver of influenza in semi-isolated coastal Mediterranean populations (Tel Aviv) with hot and humid climates, but also that one city may act as a hub for disease and drive satellites. The latter of these statements we derive from the fact that the model fit to Jerusalem influenza dynamics improves considerably when coupled with Tel Aviv, but not vice versa. The forecasting ability is increased strongly through the predictability of the system’s transient dynamics. These characteristics serve to enhance the effect of climate on influenza spread in a truly unique and predictable fashion, and they help explain why the quality of our model fits has not yet been found elsewhere.
Materials and Methods
The ILI data were obtained from the Maccabi HMO for the period June 1, 2001 to January 2013. The period from June 2001 marked a fundamental switch in surveillance and coding used by the HMO. We smoothed the data by interpolating the missing weekend reports and subsequently using a wavelet approximation to minimize noise (SI Appendix, section 3). All ILI and climate datasets may be obtained from the following link: www.tau.ac.il/~lewi/Data_flu_climate.txt.
Climate data of relative humidity and temperature in Tel Aviv were obtained from monitoring stations of the Israeli Meteorological Society and the Ministry of Environmental Protection of Israel. The time series were smoothed analogously to the ILI time series above. The resulting time series then were centered to zero mean and normalized to unit variance, which is why there are no units in Fig. 1, Upper.
The dynamics of the epidemic outbreaks were modeled based on a deterministic age-of-infection SIR model (28), which we extended to an SIRS model by adding a simple feedback from the recovered population to the susceptible population via loss of immunity caused by steady antigenic drift. In this model, the number of newly infected at time t is
| [2] |
where S is the number of susceptible, R0 is the basic reproduction rate of a fully susceptible population, P is the 7-d infectivity profile (d = 7) chosen as a gamma distribution with a mean of 2.7 d and variance of 1.8 d (11), and N is the reporting population at a reporting rate of 10%. The average period of immunity of a typical individual immunity, here 1/Λ, usually is 2–10 y (Λ ∈ [0.1: 0.5]). For a model with a daily time step, we set λ = Λ/365. The infectives pool follows: I(t) = I(t − 1) + i(t) − i(t − d); the dynamics of the susceptible pool then is
| [3] |
The infected individuals recover after d days; thus, the recovered population growth is
| [4] |
Finally, there also is a constant influx of influenza arriving in Tel Aviv from any destination in the world, which was set at one person per day all year round. The influenza influx cannot be higher than 10 per day, as this gives a model baseline ILI above one influenza case per day in the low season (10 × 10% = 1, taking into account a 10% reporting rate). Conversely, an influx rate much below 0.1 sick persons per day eventually will render the model unable to reproduce the “small” outbreaks because the infected and susceptible populations may never reach the epidemic threshold Reff = 1 in those years.
In this model, the disease dynamics are modeled neutrally for all strains and compositions thereof between punctuations, i.e., the ILI cases are treated as one disease only. Although this is a nontrivial approximation, it is not uncommon in the seasonal flu-modeling literature (4, 10, 11, 29); furthermore, recent findings indicate the possibility of broad cross-reactivity, further justifying the approximation. These recent studies seem to find in vivo and in vitro evidence of functional cross-reactivity between human monoclonal antibodies and epitopes from hemagglutinin of both influenza A and influenza B (30).
The simple climate driver is modeled as . The real climate driver is expressed as a linear combination; thus, δ(t) = wTT(t) + wRHRH(t), where T is daily average temperatures and RH is relative humidity, both normalized to the interval [−0.5, 0.5]. The antigenic jump is a modification first to δ′(t) = δ(t) + Δ, Δ = 0.26(0.02), for the period June 1 to December 1. A modification in immunity-loss rate λ′ = λ + λ*, λ* = 0.04(0.01) is modeled throughout the year.
Inference is performed by Bayesian Monte Carlo sampling of the a priori likelihood function; see SI Appendix for details and analysis.
Supplementary Material
Acknowledgments
We thank Drs. Jodie McVernon, James McCaw, Haggai Katriel, and Amit Huppert. We are grateful for the support of the European Union FP7 EPIWORK grant, the Israel Science Foundation, and the Israel Ministry of Health. J.B.A. is supported by the Carlsberg Foundation. R.Y. is supported by the Israel National Institute for Health Policy and Health Services Research. We appreciate the support of Maccabi Health Care Services for providing the ILI data. B.T.G. was supported by the Bill and Melinda Gates Foundation, the Science and Technology Directorate, Department of Homeland Security Contract HSHQDC-12-C-00058, and the Research and Policy in Infectious Disease Dynamics program of the Science and Technology Directorate, Department of Homeland Security, and the Fogarty International Center, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321656111/-/DCSupplemental.
References
- 1.Potter CW. A history of influenza. J Appl Microbiol. 2001;91(4):572–579. doi: 10.1046/j.1365-2672.2001.01492.x. [DOI] [PubMed] [Google Scholar]
- 2.Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med. 2000;51:407–421. doi: 10.1146/annurev.med.51.1.407. [DOI] [PubMed] [Google Scholar]
- 3.Shaman J, Kohn M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc Natl Acad Sci USA. 2009;106(9):3243–3248. doi: 10.1073/pnas.0806852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Finkenstädt BF, Morton A, Rand DA. Modelling antigenic drift in weekly flu incidence. Stat Med. 2005;24(22):3447–3461. doi: 10.1002/sim.2196. [DOI] [PubMed] [Google Scholar]
- 5.Finkelman BS, et al. Global patterns in seasonal activity of influenza A/H3N2, A/H1N1, and B from 1997 to 2005: Viral coexistence and latitudinal gradients. PLoS One. 2007;2(12):e1296. doi: 10.1371/journal.pone.0001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowell G, Miller MA, Viboud C. Seasonal influenza in the United States, France, and Australia: Transmission and prospects for control. Epidemiol Infect. 2008;136(6):852–864. doi: 10.1017/S0950268807009144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earn DJD, Dushoff J, Levin SA. Ecology and evolution of the flu. Trends in Ecology and Evolution. 2002;17(7):334–340. [Google Scholar]
- 8.Lipsitch M, Viboud C. Influenza seasonality: Lifting the fog. Proc Natl Acad Sci USA. 2009;106(10):3645–3646. doi: 10.1073/pnas.0900933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaman J, Pitzer VE, Viboud C, Grenfell BT, Lipsitch M. Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biol. 2010;8(2):e1000316. doi: 10.1371/journal.pbio.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Noort S, Águas R, Ballesteros S, Gomes M. The role of weather on the relation between influenza and influenza-like illness. J Theor Biol. 2011;298:131–137. doi: 10.1016/j.jtbi.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 11.Truscott J, et al. Essential epidemiological mechanisms underpinning the transmission dynamics of seasonal influenza. J R Soc Interface. 2012;9(67):304–312. doi: 10.1098/rsif.2011.0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yaari R, Katriel G, Huppert A, Axelsen JB, Stone L. Modelling seasonal influenza: The role of weather and punctuated antigenic drift. J R Soc Interface. 2013;10(84):20130298. doi: 10.1098/rsif.2013.0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stone L, Olinky R, Huppert A. Seasonal dynamics of recurrent epidemics. Nature. 2007;446(7135):533–536. doi: 10.1038/nature05638. [DOI] [PubMed] [Google Scholar]
- 14.Pease CM. An evolutionary epidemiological mechanism, with applications to type A influenza. Theor Popul Biol. 1987;31(3):422–452. doi: 10.1016/0040-5809(87)90014-1. [DOI] [PubMed] [Google Scholar]
- 15.Nelson MI, Holmes EC. The evolution of epidemic influenza. Nat Rev Genet. 2007;8(3):196–205. doi: 10.1038/nrg2053. [DOI] [PubMed] [Google Scholar]
- 16.Koelle K, Cobey S, Grenfell B, Pascual M. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science. 2006;314(5807):1898–1903. doi: 10.1126/science.1132745. [DOI] [PubMed] [Google Scholar]
- 17.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 18.Rambaut A, et al. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453(7195):615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu A, Peng Y, Du X, Shu Y, Jiang T. Correlation of influenza virus excess mortality with antigenic variation: application to rapid estimation of influenza mortality burden. PLOS Comput Biol. 2010;6(8):e1000882. doi: 10.1371/journal.pcbi.1000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf YI, et al. Projection of seasonal influenza severity from sequence and serological data. PLoS Curr. 2010;2:RRN1200. doi: 10.1371/currents.RRN1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viboud C, et al. Transmissibility and mortality impact of epidemic and pandemic influenza, with emphasis on the unusually deadly 1951 epidemic. Vaccine. 2006;24(44-46):6701–6707. doi: 10.1016/j.vaccine.2006.05.067. [DOI] [PubMed] [Google Scholar]
- 22.Earn DJ, et al. Effects of school closure on incidence of pandemic influenza in Alberta, Canada. Ann Intern Med. 2012;156(3):173–181. doi: 10.7326/0003-4819-156-3-201202070-00005. [DOI] [PubMed] [Google Scholar]
- 23.Lavenu A, Valleron AJ, Carrat F. Exploring cross-protection between influenza strains by an epidemiological model. Virus Res. 2004;103(1-2):101–105. doi: 10.1016/j.virusres.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 24.Cobey SE. 2009. Ecological and evolutionary dynamics of influenza viruses. PhD thesis (Univ of Michigan, Ann Arbor)
- 25.Towers S, Feng Z, Hupert N. 2010. Short-term heterologous immunity after severe influenza A outbreaks. arXiv:1007.3017.
- 26.Earn DJ, Rohani P, Bolker BM, Grenfell BT. A simple model for complex dynamical transitions in epidemics. Science. 2000;287(5453):667–670. doi: 10.1126/science.287.5453.667. [DOI] [PubMed] [Google Scholar]
- 27.Aron JL, Schwartz IB. Seasonality and period-doubling bifurcations in an epidemic model. J Theor Biol. 1984;110(4):665–679. doi: 10.1016/s0022-5193(84)80150-2. [DOI] [PubMed] [Google Scholar]
- 28.Katriel G, Yaari R, Huppert A, Roll U, Stone L. Modelling the initial phase of an epidemic using incidence and infection network data: 2009 H1N1 pandemic in Israel as a case study. J R Soc Interface. 2011;8(59):856–867. doi: 10.1098/rsif.2010.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia Y, Gog JR, Grenfell BT. Semiparametric estimation of the duration of immunity from infectious disease time series: Influenza as a case-study. Journal of the Royal Statistical Society: Series C (Applied Statistics) 2005;54:659–672. [Google Scholar]
- 30.Dreyfus C, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337(6100):1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.