Abstract
Genetically Engineered Mouse (GEM) models are a pillar of functional cancer research. Here we developed RapidCaP, a GEM modeling system that uses surgical injection for viral gene delivery to prostate. We show that in Pten-deficiency, loss of p53 suffices to trigger metastasis to distant sites at greater than 50% penetrance by 4 months, consistent with results from human prostate cancer genome analysis. Live bioluminescence tracking showed that endogenous primary and metastatic disease responds to castration before developing lethal castration resistance. To our surprise, the resulting lesions showed no activation of Akt but activation of the Myc oncogene., Using RapidCaP, we show that Myc drives local prostate metastasis and is critical for maintenance of metastasis, as shown by using the Brd4 inhibitor JQ1.
Taken together, our data suggest that a ‘MYC-switch’ away from AKT forms a critical and druggable event in PTEN-mutant prostate cancer metastasis and castration resistance.
Introduction
Prostate cancer is the most common malignancy in men and the second-leading cause of cancer-related deaths in the Western world. Similar to other solid tumors, metastasis is the major cause of mortality and morbidity of patients and since the discovery of androgen ablation therapy in the 1930's (1), therapeutic options have only marginally changed (2). The current breakthroughs in analysis of the prostate cancer genome, have revealed that the gradual accumulation of genome alterations is intimately associated with metastatic disease progression and the derived genome landscape revealed a critical role for copy number changes (3-6). In the past, making sense of putative oncogenic events was made possible due to functional validation of candidate genes by mouse genetic engineering of prostate cancer (7-15). Today however, two major problems have arisen: first, the GEM models have not so far reconstituted highly penetrant metastatic prostate cancer. Second, the time associated with de novo generation of GEM models is far beyond today's speed of candidate gene discovery (16). As a result, the use of GEM models has so far presented hardly any options for functional validation of genes that may form hallmarks of metastasis. As a corollary, the discovery of biology and mechanisms of the reverse process, therapy of metastatic disease, has also eluded prostate GEM models.
Today's ideal prostate GEM model should thus combine: highly penetrant metastatic disease progression, emergence of castration resistant metastasis, simple visualization for therapy, fully preserved architecture of naturally evolved lesions, which are embedded in their intact (micro-) environment and immune system as judged by histology analysis (17, 18).
While metastasis is indeed sometimes seen in post-mortem analysis, the reported penetrance is too low for pre-clinical studies (19). Furthermore, promoters that drive transgenes in prostate are typically androgen dependent (e.g., the probasin promoter), thus making them incompatible with hormone ablation therapy. Finally, a major drawback of classic genetic engineering lies in the time, cost and effort needed for GEM generation. Projects carry typically a high risk as scientists become ‘locked in’ with a few selected candidate gene alterations, the combination of which requires further lengthy breeding. Furthermore, state of the art imaging systems like ultrasound or magnetic resonance imaging are expensive and require dedicated expert staff. The above major shortcomings of classic GEM models have unfortunately put them out of sync with today's speed of human cancer genome analysis and the resulting need for fast validation of candidate cancer genes (20). As a consequence, animal modelers of cancer are actively exploring new approaches (16).
Here, we developed a new mouse model that is designed for analysis and therapy of metastatic prostate cancer, termed RapidCaP. Using a surgical process to deliver viral transgenes into prostate, we are able to achieve tissue specific single or multiple gene alternations, such as knockout (KO), knockdown and over-expression without need for cross-breeding of animals that harbor multiple engineered alleles. Inclusion of a luciferase marker with target genes enables live monitoring of metastasis, therapy induced regression and relapse. Histology analysis reveals new biology of metastasis and delivers lead candidate genes, which can be functionally validated using the RapidCaP system.
Results
Stable transgene delivery to epithelial prostate cells by virus injection
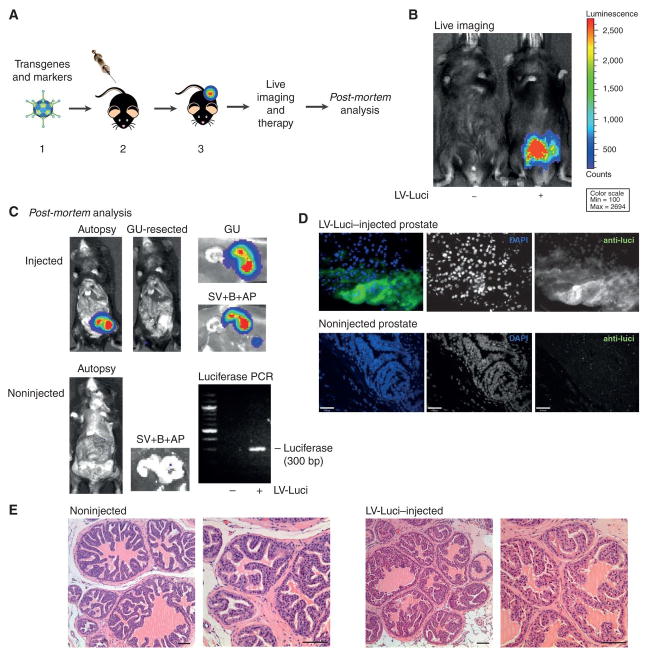
To overcome the limitations of germline based GEM-models for prostate cancer, we pursued a strategy, depicted in Fig. 1A, where transgenes are delivered by direct injection of lentivirus (LV) into an anterior prostate gland (see Methods). Infected prostate cells are designed to express oncogenic transgenes and a marker gene, luciferase, for bioluminescence (BL) imaging to allow tracking of disease progression or the results of therapy, and to guide post-mortem autopsy analysis to tissues of interest. As shown, this approach allowed successful monitoring of mouse with injected prostate by live imaging (Fig. 1B), and post-mortem analysis (Fig. 1C) 60 days post injection revealed luciferase signal only in the injected anterior prostate and adjacent seminal vesicle (SV, see below for discussion of SV signal, Fig. 2A). PCR analysis revealed the presence of the luciferase transgene in the animal with injected anterior prostate (Fig. 1C, bottom right panel), while immunofluorescence (IF) based histology using anti-luciferase antibodies revealed expression of luciferase in the prostatic epithelium. Although infection of non-epithelial cells can by no means be excluded, epithelial IF signal typically clearly dominated over stromal signal (Fig. S1A, see also Fig. 1D). Based on FACS analysis with fluorescent marker transgenes our technique infects some 0.3% of the ∼100 million anterior prostate cells (not shown). Histology comparison of injected and non-injected glands revealed no morphological alterations in the injected glands and immunohistochemistry (IHC) analysis of the PTEN pathway and the Ki-67 proliferation marker did not reveal any anomalies (Fig. S1B). Successfully injected/ infected prostates stained negative for the CD3 T-cells marker and no signs of inflammatory responses were observed (Fig. S1C, top and middle panels). These results demonstrated that viral transgene delivery and stable integration into genomic DNA in the anterior prostate epithelium is feasible with our technique.
Figure 1. Stable transgene delivery to epithelial prostate cells by virus injection.
A. The RapidCaP system. 1- Design and production of lentivirus harboring candidate cancer genes and marker genes. 2- Surgical injection of virus directly into anterior prostate gland. 3- Non-invasive bioluminescence imaging to monitor disease progression and therapeutic effects. Note that Adenovirus is depicted for purely aesthetic reasons. B. Live imaging of lentiviral luciferase (LV-Luci) injected Bl6 mice and non-injected control at 60 days post injection suggests persistent transgene expression. C. Post-mortem autopsy imaging of LV-Luci and non-injected control mice at 98 days post injection. The genito-urinary (GU) tract comprises bladder (B), seminal vesicle (SV) and anterior prostate (AP). Luminescence signal is found in the (injected) right anterior prostate lobe and also in the seminal vesicle. Note that FACS sorting experiments fail to identify infected (tomato-FP) seminal vesicle cells, potentially suggesting SV signal to be extracellular. PCR of prostate genomic DNA reveals the luciferase transgene (300 bp) only in the LV-Luci injected prostate lobe. D. Anti-luciferase immunofluorescence on LV-Luci- and non-injected control prostate reveals luciferase expression in prostate luminal epithelium. Scale bar, 47μm. E. H&E staining of LV-Luci injected and non-injected control prostate reveals retention of normal glandular architecture. Scale bar, 100μm.
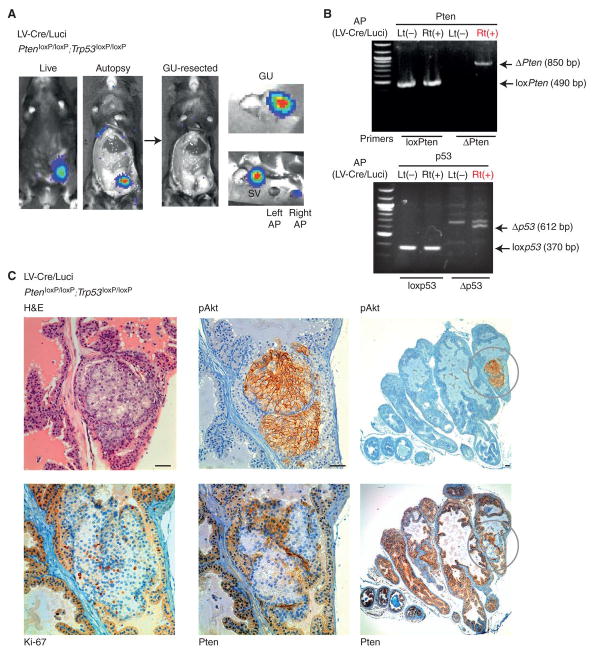
Figure 2. Prostate specific LV-Cre/ Luci delivery results in focal disease.
A. Live and post-mortem images of LV-Cre/Luci injected PtenloxP/loxP; Trp53loxP/loxP mouse at 69 days post injection. Dissection reveals similar distribution to that shown in Fig. 1C. Luminescence signal was seen in LV-Cre/Luci injected prostate and seminal vesicle. B. PCR analysis confirms recombination of Pten and Trp53 in LV-Cre/luci injected PtenloxP/loxP; Trp53loxP/loxP specifically in the injected right (Rt) anterior prostate. C. Immunohistochemistry analysis of LV-Cre/luci injected PtenloxP/loxP; Trp53loxP/loxP prostate reveals a focal lesion in the luminal epithelium of one gland (H&E), which also displays increased proliferation, specific to epithelial cells (Ki-67). Loss of Pten expression, activation of Akt (pAkt), and increased proliferation are also found in this region (middle panels), which is a unique focus of disease in the cross section of the entire AP (right panels). Note that surrounding stroma remains unaffected in all panels. Scale bars, 100μm (top right), 50μm (all other panels).
Prostate specific delivery of Cre recombinase results in focal Pten/ Trp53 deficient disease
Analysis of mouse models of prostate cancer, revealed that loss of Trp53 function is a critical step for disease progression in Pten-pathway mutant animals (21, 22). Thus, we next injected Lentiviral Cre-luciferase double-transgenic virus (LV-Cre/Luci) into prostates of Ptenloxp/loxp; Trp53loxp/loxp. As shown in Fig. 2A, LV-Cre/Luci injected mice typically show a strong luciferase signal in the genitourinary (GU) region. Post-mortem analysis (69 days post injection) reveals that the injected anterior prostate and associated seminal vesicle are the source- discussed below. In contrast, 62% of LV-Luci virus injected mice demonstrated loss of signal within 78 days (not shown). PCR analysis of genomic DNA from anterior prostate revealed recombination of the tumor suppressors, Pten and Trp53, specifically in the LV-Cre/Luci injected right (Rt) anterior prostate (AP, Fig. 2B, ΔPten, Δp53). Strikingly, anterior prostate IHC analysis (see Fig. 2C) revealed the highly focal nature of disease initiation in this model: only a small region within one gland showed increased proliferation (Ki-67), loss of Pten and strong Akt activation, while the vast majority of glands in the prostate retained completely normal morphology and staining for these proteins (Fig. 2C, right panels, pAkt, Pten). Note that increased proliferation and cell expansion were not seen in stromal cells. Indeed, after histopathology analysis of more than 60 prostate injected cases, the only cell type of which we observed expansion were epithelial prostate cells. Furthermore, we again observed no immunogenicity of the viral injection procedure as shown by CD3 staining (see Fig. S1C). These data demonstrated that the RapidCaP system triggered focal disease initiation, which is thought to be a feature of the human disease initiation process. Note that in contrast, the probasin-driven Cre conditional knockout (cKO) systems of Pten and Trp53 or Smad4 delete their target genes in every cell of the prostate epithelium, which results in massive expansion of prostate size to several centimeters in diameter (19, 22, 23) and is accompanied by a non-epithelial sarcoma phenotype.
Deletion of Pten and Trp53 triggers disease dissemination
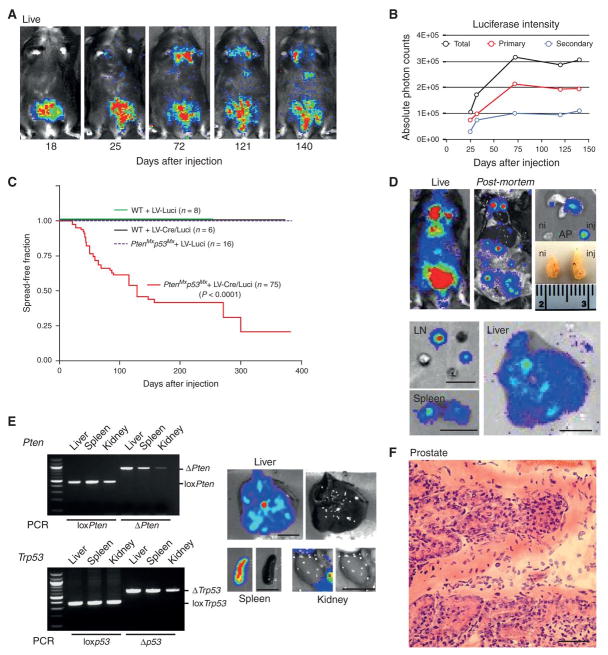
Analysis of the human prostate cancer genome has revealed that PTEN and TP53 are co-deleted in half of all metastatic samples (3, 21). In mouse however, the complete Pten and Trp53 deletion in prostate resulted in massive and lethal tumor growth (22, 23). In order to investigate disease progression in the above RapidCaP system (LV-Cre/Luci injection into Ptenloxp/loxp; Trp53loxp/loxp prostate), mice were imaged for a period of several months. As shown in Fig. 3A, the luminescence signal was initially observed in the lower abdominal region (up to 25 days post injection, p.i.). Later however, strong signal spread was detected to the upper and mid body (72d to 140 d p.i.). Typically, the signal first disseminated to below the neck and later to the mid-abdomen, as shown. Quantification of whole body, primary and secondary signal (Fig. 3B), revealed how both primary and secondary signals increased over time. Most importantly, no spreading of luminescence signal was observed in any of the control injection cohorts (see Fig. 3C). As mentioned above, 62%, or 18 of 29 non-gene modifying control injected mice instead showed a decrease of the primary luciferase signal over time (see Fig. S2 for typical examples of a RapidCaP time course). Overall (See Fig. 3C), 50% of animals showed disease dissemination 4 months post injection. In post-mortem analysis, secondary signals were confirmed in lymph nodes (mediastinal, lumbar and caudal), spleen and liver (shown in Fig. 3D), while pancreas and lung, as well as organs near the GU tract (excluding bladder) were also often signal positive. Importantly, LV-Cre/Luci injected prostate was macroscopically indistinguishable from the non-injected prostate (Fig. 3D, ni vs. inj), revealing a stark contrast to the probasin-Cre driven Pten/ Trp53 cKO model. To date, after analysis of more than 300 injected animals, those without gene alteration never showed luciferase signal spread in post-mortem analysis. Note that the seminal vesicle on the side of the injected prostate was invariably luminescence positive from the day of injection, also in luciferase only control injections. This argues against positive biological selection, and in agreement, we never found phenotypic changes by IHC analysis of seminal vesicles (not shown). In fact, seminal vesicles have never shown any phenotypes in the many whole body KO models driven by heterozygosity of Pten (11, 15, 21). Recombination of Pten and Trp53 in the signal positive secondary organ sites was confirmed by PCR (Fig. 3E). Histopathological examination of prostates revealed focal invasion as shown by the frequent focal budding of epithelial cells (Fig. 3F).
Figure 3. Pten/ Trp53 deletion triggers disease dissemination.
A. Time course of a typical RapidCaP experiment (showing one LV-Cre/Luci injected PtenloxP/ loxP; Trp53loxP/loxP mouse) reveals disease spreading to distant sites starting at 72 days post injection. B. Quantification of signal intensity from mouse shown in Fig. 3A. Total body, secondary and primary signal measurements are shown. C. Kaplan-Meier analysis of disease dissemination reveals early onset and high penetrance, specifically in PtenloxP/loxP; Trp53loxP/loxP mice (p<0.0001). D. Live (219 days post injection) and post-mortem autopsy of an LV-Cre/Luci injected PtenloxP/loxP; Trp53loxP/loxP mouse reveals luciferase positive secondary organ sites (lymph-nodes, LN, spleen and liver) and a minor difference in size of prostate lobes (injected, inj. vs. non-injected, ni). Scale bar, 1cm. E. PCR analysis reveals Pten/ Trp53 recombination in the secondary tissues (weakest in kidney). Scale bar, 1cm. F. H&E staining of an LV-Cre/Luci injected PtenloxP/loxP; Trp53loxP/loxP prostate lesion (192 days post injection) reveals a typical budding gland with features of focal invasion. Scale bar, 50μm.
Histology analysis confirms metastatic prostate cancer in RapidCaP
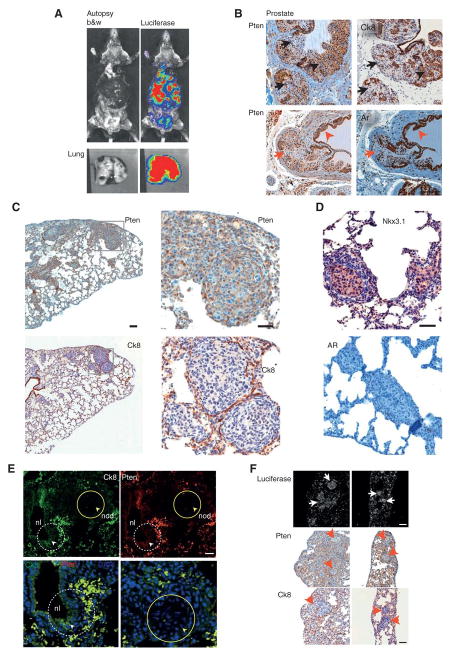
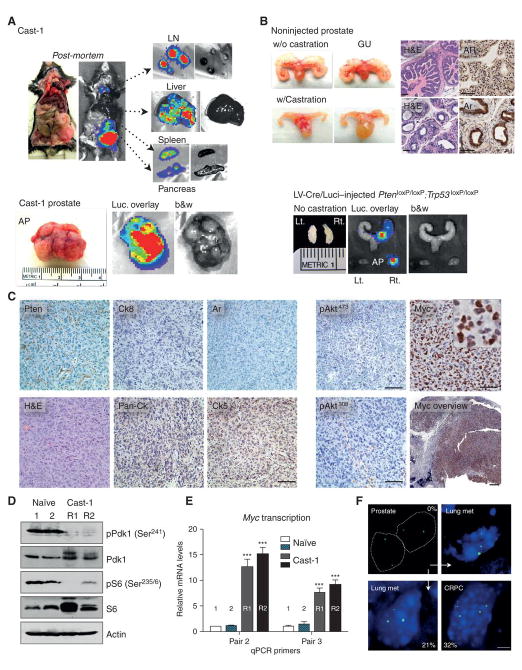
To investigate if the signal dissemination in RapidCaP is prostate metastasis, histopathological analysis was performed on the animal shown in Fig. 4A. First, we defined markers for mutant cancer cells by analyzing prostate. As expected, focal regions in prostate lost Pten expression (Fig. 4B). In addition, the epithelial markers cytokeratin-8 (Ck8) and androgen receptor (Ar), were absent specifically in regions where Pten (and p53) was no longer expressed (Fig. 4B, arrows), in full agreement with previous results (24), and in contrast to normal prostate (Fig. S3C, left panels). Inspecting the lungs of this animal (Figs. 4C-D), we found many small tumor nodules, some of which demonstrated tumor within intravascular spaces and others within lung parenchyma. The nodules were composed of proliferating atypical cells with large, pleomorphic, hyperchromatic nuclei and scant to moderate amounts of amphophilic cytoplasm. These cells were immunohistochemically characterized by loss of Pten as well as loss of CK8 and AR, thus matching the features of the mutant prostate epithelial cells. The metastatic nodules showed focal, faint positivity for the prostatic differentiation marker Nkx3.1, further supporting prostatic origin. The loss of the low molecular weight Ck8 was particularly informative because the normal alveolar and bronchiolar lung epithelium displayed high expression of this cytokeratin (see also Fig. S3C, right panels), while the metastatic nodules stood out as negative mutant epithelial islands. In order to confirm that the metastatic cells were truly double negative for Pten and CK8, double immunofluorescence (IF) labeling was performed on lung sections. Indeed (see Fig. 4E), metastatic nodules in lung (nod) were negative for both Pten and Ck8 (yellow circles and arrowheads) while normal bronchiolar epithelium (nl) showed high expression of both proteins (white, dashed circles and arrowheads). Next, we used anti-luciferase IF to further study the metastatic nodules. Only injected prostate was stained, thus establishing our positive control for the procedure (Fig. S3A). As shown in Figure S3B, lungs with metastatic nodules (H&E) showed readily detectable luciferin-positive cells and nodules (see Luci). These sites corresponded to tumor sites with loss of Pten and Ck8 staining, as shown in Fig. 4F. Importantly, Ki-67 IHC analysis revealed that metastatic nodules contained cells with a high proliferation rate (Fig. S3D). We next confirmed that Nkx3.1 levels were high in prostate, the lung metastatic nodules but markedly reduced in the castration resistant prostate cancer (CRPC, Fig. S3E), thus inversely correlating with the expression levels of Myc, as published previously (25). Additional cases of lung metastasis confirming the immunohistological correlations are shown in Fig. S4. Taken together, our results are consistent with extensive prostate cancer metastasis to lung in RapidCaP.
Figure 4. Histogenic analysis confirms prostate cancer metastasis to lung.
A. Autopsy imaging of the mock-castrated RapidCaP mouse (#3) from Fig. 4 reveals disease dissemination to lung, lymph nodes, spleen and liver at the trial endpoint (10 months post injection). B. Identification of histogenic markers that define prostate cancer cells by immunohistochemistry (IHC) analysis of prostate in animal from A: Pten & Ck8 (black arrows, see black arrowheads for region with normal protein levels) and Pten & Ar (red arrows, see red arrowheads for region with normal protein levels). Note that ‘loss’ of AR-staining is a hallmark of Pten/p53-null prostate (see text). Scale bar, 100μm. C, D. IHC analysis of lung from above mouse reveals metastatic nodules that are Pten-, Ck8- and AR-negative and positive for the prostate epithelial marker Nkx3.1. Boxes indicate areas of magnification. Scale bars, 100μm (top & bottom left panels), 50μm (all other panels). E. Double-immunofluorescence (IF) labeling confirms double-negativity for Pten & Ck8 in a metastatic lung nodule (nod) shown in (C-D) (yellow circles & arrowheads). Note that in contrast, co-labeling of Pten and Ck8 in adjacent normal lung epithelia (nl, white dashed circles and arrowheads) shows double positive staining. Scale bar, 50μm. F. Anti-luciferase antibody staining and IF analysis of lung from (C-F) confirms that metastatic nodules are luciferase positive (white arrows) and Pten / Ck8 deficient (red arrowheads). Scale bar, 100μm.
Pten/ Trp53-deficient RapidCaP metastasis responds to castration
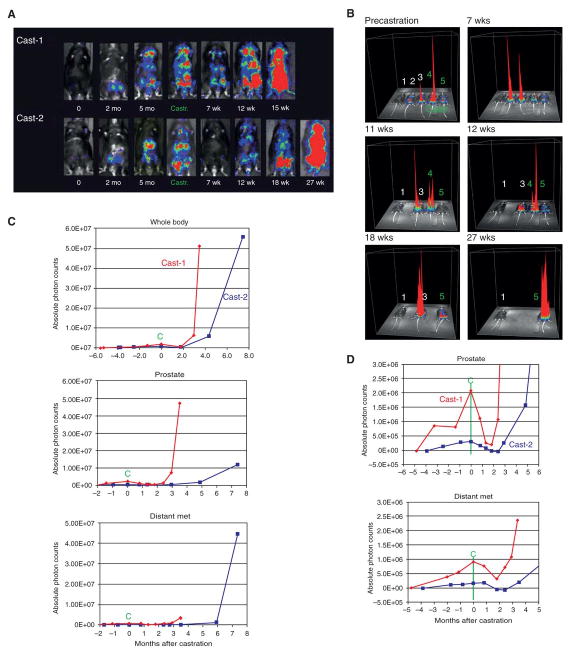
Androgen ablation therapy is the standard of care for advanced prostate cancer causing widespread atrophy of prostate cells and the derived metastases. Surgical castration was conducted on mice with secondary signals and bioluminescence imaging was continued once per week on a total of 16 animals as summarized in Fig. S5A-C: 7 castrated Pten/ Trp53 mutant RapidCaP animals, 7 mock-castrated Pten/ Trp53 mutant RapidCaP animals, and two castrated LV-luci prostate-injected normal controls. Figure 5 illustrates the typical results obtained from a group of 5 trial (Figure 5A-B), consisting of two castrated animals (Fig. 5A, cast-1, cast-2 and Fig. 5B, #4, #5,), and two non-castrated controls (Fig. 5B, #2, #3), one of which underwent a mock castration surgery (#3) and a non-treated wt mouse (#1). The castrated animals showed partial (cast-1) or complete (cast-2) therapy response within 7 weeks post castration, and importantly both the primary and secondary signals regressed (Fig. 5A, 7 wk, see also Fig. S5A, low). However, regression was followed by aggressive relapse of both primary and secondary signals, now growing much more rapidly than prior to castration (see Fig. 5A, 7wk - 27 wk, Fig. S5A), a hallmark of human castration resistance. In contrast (see Figs. 5B, S5B), both control animals showed steadily increasing signal intensity. Quantification (Figs. 5C, S5C) confirmed this sharp increase in intensity for both primary and secondary disease, resulting in a typical ‘hockey stick’ growth behavior (compare pre-castration to post-castration growth) just before these animals needed to be sacrificed to prevent excessive tumor burden. Clear signs of morbidity, such as palpable tumors, low activity, hunched backs, and a rough hair coat were observed in these animals. Fig. 5D shows the quantitative analysis of regression and relapse, and breakdown of the data into primary and secondary disease. This analysis revealed the kinetics of how prostate and metastatic signals in both animals responded to castration. Post-mortem analysis of animal #4, from Fig. 5B illustrates the typical metastasis to organs including lymph nodes, liver, spleen and pancreas (some of which were palpable). The prostate itself was massively enlarged spanning close to 3 cm in diameter, in stark contrast to the 5-7 mm diameter of non-castrated Pten/Trp53 deficient RapidCaP prostate (Fig. 6A, bottom, see also Fig. S6, slide overviews).
Figure 5. Metastatic signals respond to castration.
A. Live imaging time course of two castrated RapidCaP mice (LV-Cre/Luci injected PtenloxP/loxP; Trp53loxP/loxP mouse). Surgical castration was performed at 5 months post injection on mice harboring distant secondary disease (see Castr.). Imaging analysis reveals differential response and recurrence of disease after castration. B. 3D-plot of signal intensity time course from (A), including control mice: 1, wt untreated mouse- 2, RapidCaP mouse- 3, mock-castrated RapidCaP mouse- 4&5, castrated RapidCaP mice. C. Quantification of luminescence signals in castrated animals reveals a sharp increase in the rate of disease progression after relapse. “C” denotes castration. D. Close-up analysis of graphs from (C) shows that both primary and secondary disease respond to castration, confirming their hormone dependence.
Figure 6. Analysis of castration resistant prostate cancer.
A. Post-mortem analysis of Cast-1 animal after relapse (see Fig. 4A) shows metastasis to distant organs and massive prostate enlargement. B. Castration results in prostate epithelium atrophy and diffuse cytoplasmic Ar staining (upper panels). Note that Pten/ Trp53-loss in RapidCaP does not cause a significant increase in prostate size (lower panels). Scale bar, 100μm. C. IHC analysis of Cast-1 prostate shows loss of Akt-activation, lost/ low cytokeratin epithelial and basal cell markers Ck8 and CK5, respectively. In contrast, there is strong activation of Myc in tumor cells in the therapy resistant prostate tumor. Scale bars, 1mm (Myc overview), 50μm (all other panels). D. Western blotting analysis of PI 3-Kinase reveals no feedback (p-S6) or parallel pathway (p-Pdk1) activation of castration resistant tumor shown in (C). E. A strong increase in Myc mRNA expression is detected in the castration resistant tumor from. Error bars are s.d., ***p = 0.0001 (C). F. FISH analysis reveals Myc gene amplification in disease progression from prostate to lung metastasis of the same animal. Myc gene amplification is also observed in a castration resistant tumor (CRPC). Percentage of nuclei with > 2 copies is indicated, scale bar 5μm.
Taken together, the castration experiments confirmed that RapidCaP mice developed prostate metastatic disease, which initially responds to androgen deprivation and eventually relapses to develop castration resistant prostate cancer. It is likely that residual disease after castration as shown in the Cast-1 animal (Fig. 5A, Cast-1, 7wk) is due to pre-existing AR low/ negative cells such as the ones shown in Fig. 4B, D, which could expand upon castration to form AR-negative CRPC tumors (see Fig. 6C). To study the molecular makeup of castration resistant disease we turned to IHC analysis.
Myc expression can drive metastasis in the absence of Akt activation
IHC analysis of the relapsed prostate tumor (see Fig. 6C and S6) confirmed the consistent emergence of anaplastic cells with loss/ background levels of Pten, Ar, and CK8, consistent with mutant prostate of non-castrated mice (shown in Fig. 4B-F). However Akt was not activated in spite of Pten-loss (Fig. 6C and S6C, right panels). We then analyzed components of the PI3Kinase pathway in order to understand if feedback mechanisms (26, 27) or parallel Pdk1 kinase activation (28) was suppressing pAkt. As shown (Fig. 6D), Pdk1 was not phosphorylated in the castration resistant tumor, where pAkt levels were low. Furthermore, we found lower ribosomal protein S6 phosphorylation after castration than in wt prostate (see also Fig. S7C for IHC analysis of phospho-S6). These results suggested that PI 3-kinase signaling is low in metastasis and after castration. Instead, there was strong Myc staining seen in both nuclei and cytoplasm throughout the tumor mass (Fig. 6C and Fig. S6, Myc overview). Note that the mock castrated Pten/ Trp53- deficient RapidCaP prostate showed weak or absent and exclusively nuclear Myc (Fig. S3F), similar to normal prostate (not shown). Myc RNA expression sharply increased in castration resistant cancer (Fig. 6E) and using FISH analysis (Fig. 6F), we found that the Myc gene was amplified in 32% of CRPC nuclei. We also observed Myc amplification in metastasis (21% frequency), while the corresponding primary prostate showed no gene amplification. These data suggested that Myc and not phospho-Akt (pAkt) may be driving metastasis and castration resistant cancer, in contrast to disease initiation, where we observed strong Akt activation (see pAkt, Fig. 2C). We frequently saw mixed prostate glands with p-Akt high and low staining regions (Fig. S7A), which also showed low pS6 and Ck8 staining but were positive for Myc and Ar. These findings suggest that the low pAkt phenotype first emerges in prostate and is selected for during metastasis and castration resistance, along with partial amplification of the gene and strongly increased transcription.
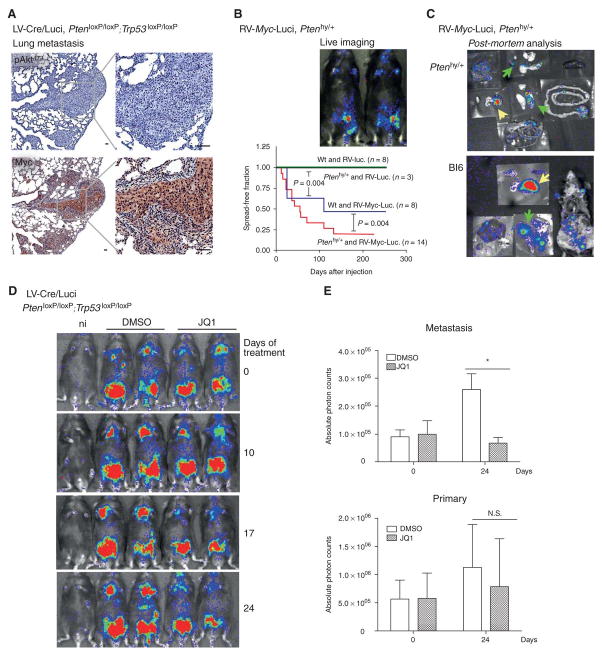
Next, we made full use of the RapidCaP system for discovery and validation of a metastasis driver gene. First, we studied Akt and Myc status in prostate metastasis to lung. As shown (Fig. 7A) metastasis to lung in the mock-castrated animal showed absence of Akt activation but high Myc levels in cytoplasm and nuclei of all metastatic nodules. This situation was comparable to the IHC staining observed in the castrated animal prostate (Fig. 6C), but very different from the mock-castrated animal's own prostate (see Fig. S3F). Therefore, our results suggested that increased and/ or mislocalized Myc may contribute specifically to the metastatic process (see discussion). To test this hypothesis we generated a cohort of Myc-driven RapidCaP mice.
Figure 7. Myc succeeds Akt activation and is required for growth of metastatic prostate cancer.
A. IHC analysis of phospho-Akt (Ser473) in metastatic lung nodules of LV-Cre/Luci injected PtenloxP/loxP; Trp53loxP/loxP mice reveals absence of pAkt activation. In contrast, Myc levels are strongly increased, especially in the cytoplasm (compared to prostate, Fig. S3F). Note the apparent further Myc increase in castration resistant disease (Fig. 6C) compared to this mock-castrated metastasis. Scale bar, 50μm. B. Top, typical live imaging of RV-Myc-luci injected Ptenhy/+ mice shows lower abdominal signals and absence of distant metastasis (in contrast to LV-Cre/Luci injected PtenloxP/loxP; Trp53loxP/loxP mice, Fig. 3A). Note that Ptenhy/+ mice express only 75% Pten throughout their body. Bottom, Kaplan-Meier analysis of disease spread in indicated RapidCaP models reveals that Myc expression can drive local dissemination of prostate cancer and cooperate with Pten-suppression. Log-rank (Mantel-Cox) testing shows that wt, Ptenhy/+, as well as negative control study arms are significantly different (p = 0.004). C. Autopsy analysis of RV-Myc-luci injected mice reveals local, mostly lower abdominal disease dissemination from prostate (yellow arrows) to secondary sites (green arrows), including local lymph nodes. See also Supplementary Fig. S7C. D. Treatment of metastasis in LV-Cre/Luci injected PtenloxP/loxP; Trp53loxP/loxP mice using the Myc antagonizing Brd4 inhibitor JQ1 (at 50 mg/kg/day) reveals regression of metastatic disease compared to the disease progression in DMSO treated mice. E. Quantification of disease regression from (D) shows significant effect on metastasis (*p = 0.026) but not primary disease. Error bars are s.d.
A retroviral construct harboring Myc and luciferase was used (see Fig. S7B and Methods) and validated in vitro prior to injection into mice (not shown). Two animal genotypes were selected for injection: Ptenhy/+ (“Pten hyper”) mice with 25% reduced Pten levels (11), to test if Myc cooperates with activation of Akt in driving metastasis, and WT mice, to test if Myc can drive metastasis on its own. As shown in Fig. 7B (top), Ptenhy/+ mice with Myc-luciferase retrovirus (RV-Myc-luci) injected prostates retained their luminescence signals until they were sacrificed for analysis, and so did the WT mice (not shown). There were however no signals detected in live imaging beyond the lower abdominal region in both genotypes, suggesting that no strong dissemination to distant organs had occurred. Typical post-mortem imaging (Fig. 7C, S7C, 54 days p.i.) revealed luciferase positivity of the injected (right) anterior prostate lobes in both genotypes (yellow arrows) and Myc overexpression was confirmed by RT-qPCR (Fig. S7D). Importantly, post-mortem visualization revealed that the majority of Myc-virus injected animals had secondary signals (green arrows). These were limited to organs close to prostate, such as local lymph nodes and epididymal fat pads confirming the lack of distant dissemination observed in live imaging. Statistical cohort analysis (Fig. 7B, bottom) showed significant frequencies of local disease dissemination in both the Pten-mutant and in the WT mice, even if a significant increase in local metastasis was observed in the mutant cohort. Thus, Myc over-expression suffices to induce local metastasis in normal prostate, but the process can be accelerated through even only modest suppression of Pten.
Finally, to probe if Myc was needed to drive growth or maintenance of the metastatic cells we used the Myc suppressing Brd4 inhibitor JQ1 (29-31) on mice with established metastases. We confirmed that JQ1 administration (daily at 50 mg kg−1) suppressed Myc transcription in prostate disease tissues already after 4 days (Fig. S8). As shown (Fig. 7D), JQ1 inhibited growth of metastatic cells within 17 days and had little effect on primary disease signal (quantified in Fig. 7E). Taken together, our data suggest that the spontaneously increased Myc expression observed in metastasis or castration resistance is a key driver and potential drug target against Pten-mutant metastasis.
Discussion
Virus-based in vivo gene transfer has been used to study various cancer types (32, 33). The RapidCaP system presents us with new technology for the exploration of metastatic prostate cancer and its therapy. This approach has delivered two unexpected results: First, we showed that deletion of Pten and Trp53 tumor suppressors suffices to trigger metastatic prostate cancer (CaP) at high penetrance within a few months. Second, Pten/ Trp53-deficient metastasis shows no Akt activation, but Myc activation instead.
The first result validates our previous analysis of mouse genetics and human CaP genomes, where we found that p53 is a gatekeeper for metastatic progression of PTEN mutant CaP (21). The result is corroborated by our finding that p53 is haplosufficient for preventing distant metastasis as seen by live imaging of PtenloxP/loxP; Trp53 +/loxP RapidCaP mice (not shown). These findings are in line with a critical role of the p53-mediated senescence response after Pten-loss (22, 34, 35) and its potential in preventing metastatic CaP (36). These results could not readily be obtained with the classic Pten-mutant GEM models where metastatic disease is not, or only rarely observed, as primary prostate cancers grow to lethal size (22, 23). In patients, this genetic setting occurs in half of the recently analyzed metastatic cancer genomes (3, 37, 38).
The second result demonstrates the utility of the RapidCaP approach, which allows for identification and fast validation of metastasis mechanisms. The loss of Akt activation in Pten-deficient metastatic disease and castration resistant tumors is unexpected. This could suggest that these tissues no longer respond to PI 3-Kinase pathway inhibition, in spite of Pten-loss, a hypothesis that can be tested using RapidCaP. The metastatic Pten/ Trp53-mutant disease shows clearly less pAkt signal than the primary disease initiating counterpart (compare Fig. 6C, right panels and 2C, right panels). While the mechanism of pAkt-suppression remains to be determined, it is apparent that Myc is over-expressed in this setting, suggesting the existence of a Myc-switch in Pten mutant metastatic disease progression. Intriguingly, in this setting Myc is also strongly expressed in the cytoplasm in contrast to the nuclear, weak Myc expression in normal prostate. Recently, a cytoplasmic cleavage product of Myc, termed Myc-Nick, has been shown to control muscle cell differentiation (39). Thus, it will be interesting to see if Myc-Nick is present in the metastatic or castration resistant lesion.
The MYC gene is frequently amplified, especially in metastatic human CaP as part of the signature chr. 8p amplification (3) and has previously been functionally validated as driver of CaP (14, 40). It is known to cooperate with PI 3-Kinase pathway activation in mouse models of mainly primary disease (41, 42); (reviewed in (40)). The non-epithelial (Ck8-deficient) histology of Pten/ Trp53-mutant metastasis remains to be validated in PTEN/ TP53-mutant human CaP, which represents between 54% and 65% of metastatic CaP cases (3, 37, 38). The RapidCaP system however, revealed a specific role for Myc as a druggable driver of Pten-mutant metastasis. These results suggest that a quantitative and qualitative change in Myc expression succeeds Akt-activation in driving prostate metastasis. Furthermore, we have seen that Pten/ Trp53-mutant metastasis is still androgen dependent, and that disease relapse is characterized by absence of pAkt and high Myc levels. It remains to be determined if and how Myc function suppresses Akt activity and how these two cancer pathways can best be targeted coordinately to prevent disease relapse.
Thanks to its versatility, accelerated time frame and amenability to genome sequencing, the RapidCaP system is very well positioned to explore these questions and thus may help end death and suffering from today's incurable prostate cancer.
Methods
Mice
PtenloxP/loxP; Trp53loxP/loxP, Ptenhy/+ transgenic and C57BL/6 or 129SV / C57BL/6 wt mice were used in this study. All protocols for mouse experiments were in accordance with institutional guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC). PtenloxP/loxP; Trp53loxP/loxP transgenic mice were generated by crossing PtenloxP/loxP with Trp53loxP/ loxP (43). For genotyping, tail DNA was subjected to polymerase chain reaction analysis with the following primers. For PtenloxP/loxP, primer 1 (5′-TGTTTTTGACCAATTAAAGTAGGCTG TG-3′) and primer 2 (5′-AAAAGTTCCCCTGCTGATGATTTGT-3′) were used. For Trp53loxP/loxP, primer 1 (5′-CACAAAAACAGGTTAAACCCAG-3′) and primer 2 (5′-AGCACATAGGAGGCAGAGA C-3′) were used.
Viral constructs
Lentiviral constructs - The Luc.Cre lentiviral plasmid (Tyler Jacks, Addgene plasmid 20905), pMD2.G and psPAX2 were purchased. The luciferase lentiviral plasmid was generated by eliminating CreNLS from the above construct. The retroviral Myc-luciferase construct was a kind gift from Dr. Scott W. Lowe.
Retrovirus productions and infections
Retrovirus was produced by calcium phosphate transfection, 6x106 phoenix cells were plated in 10 cm plates 6 to 12 hours prior to transfection with 15 μg of target construct and 5 μg of ecotropic helper plasmid. Fresh media was added 12 hours after transfection and viral supernatant was collected four times at 24, 36, 48 and 60 hours post-transfection. Viral supernatant was filtered through a 0.45 μm filter, then concentrated by ultracentrifugation (2 h at 20,000 × g). Prior to the viral injection to prostate, in vitro infection test for each batch of virus was conducted.
Lentivirus productions, infections and injections
Lentiviruses were produced by calcium phosphate transfection. 293FT cells were plated for transfection density 8×106 cells per 10 cm plate. 10 μg of target plasmid was combined with helper constructs, 8.5 μg of pMD2.G and 3.5 μg of psPAX2, for transfection. For retroviruses, ecotropic phoenix cells were plated in 10 cm dishes 6 to 12 hours prior to transfection with 15 μg of target construct and 5 μg of ecotropic helper plasmid. Both viruses were harvested at 24, 36, 48 and 60 h post-transfection and centrifuged (4500 rpm, 15 min) prior to filtering through 0.45-μm-pore cellulose acetate filters. Viral supernatant was concentrated by ultracentrifugation (2 h at 20,000 × g), then an in vitro infection test for each batch of virus was conducted in advance.
Cell lines
293FT and ecotropic Phoenix cells were a kind gift from Dr. Scott Lowe. They were authenticated for propagation of lentivirus and retrovirus, respectively.
Intra-prostate injection
After exposure to anesthesia (Isoflurane, 2%), the lower half of the abdomen was shaved and the mouse was placed in a surgery hood. The mouse was constantly exposed to Isoflurane via a nose cone for the entire duration of the 10-minute surgery. The shaved region was cleaned with betadine, followed by sterile PBS 3 times. A 0.5 inch incision in both the skin and peritoneum was made along the lower abdominal midline to allow the right anterior prostate to be positioned for injection on a sterile support. Typically 30 μl of concentrated virus was injected into the right anterior prostate. The incision was then sutured and the skin was stapled shut using 2 to 3 stainless steel EZ Clip wound closures. After animals were observed for complete recovery from anesthesia, they were warmed under a heating lamp to regain the ability to maintain sternal recumbence and given DietGel.
Surgical castration
An anesthetized and surgically prepared animal was placed in dorsal recumbency. Both testes were then pushed down into the scrotal sacs by gently applying pressure to the abdomen. A 1-2 cm ventral midline incision was made in the scrotum and the skin was retracted to expose the tunica. The tunica was pierced and the testes were pushed out one at a time. The testes were then raised to expose the underlying blood vessels and tubules. The fat pad, which adheres to testis, was then grasped with blunt forceps to locate the vas deferens with the prominent blood vessel running along it. The testis is dissected away from the fat pad and removed. The fat pad is then pushed back into the scrotal sac. All deferential vessels and ducts were replaced back into the tunica. Skin incisions were closed with stainless steel wound closures and removed 7-10 days post-operation.
Bioluminescence Imaging
In vitro, in vivo and ex vivo Bioluminescence imaging was performed using an Xenogen IVIS Spectrum imager, which utilizes a highly sensitive, cooled CCD camera mounted in a light-tight camera box.
For in vitro imaging, MEFs (1× 105 cells/well) were seeded in 12-well plate and infected with serial volume of virus. At 24 hrs post infection, the medium was replaced with fresh medium and infection efficiency was measured by bioluminescence imaging 5 days post infection. For in vitro imaging, luciferin (D-luciferin, Potassium Salt, Gold Biotechnology) was added to each well at 150 mg/ml of final concentration in PBS and photons were collected for 3 min. For in vivo imaging, animals received luciferin at 200 mg/kg by intra-peritoneal injection 5 min prior to imaging. The animals were then anesthetized using 2% isoflurane and placed onto the warmed stage inside the camera box. The animals received continuous exposure to 2% isoflurane to sustain sedation for 3 min of imaging. For quantification, regions of interest (ROI) were measured with standardized rectangular regions covering the mouse trunk and extremities. The measured signal was quantified as photons/second (ph/sec) using the Living Image software v.4.2 (Xenogen). Background bioluminescence in vivo was in the range 3∼6X104 ph/sec. For ex vivo imaging, animals were humanely euthanized, tissues of interest excised, and placed individually on paraffin film and imaged for 3 min after 3 mg of D-luciferin (200 μl of 15 mg/ml in PBS) were dropped in each organ. Tissues were subsequently fixed in 10% neutral-buffered formalin (Sigma) overnight and prepared for standard histopathology evaluation. For JQ1 treatment trials, JQ1 stock (100 mg ml−1 ) in DMSO was diluted by dropwise addition of a 10% 2-hydroxypropyl-β-cyclodextrin carrier (Sigma) under vortexing, resulting in a 5 mg ml−1 final solution. Mice were intraperitoneally injected daily with freshly diluted JQ1 (50 mg kg−1) or the same volume of carrier containing 5% DMSO.
PCR analysis to confirm injection
PCR analysis of Cre-mediated recombination in PtenloxP/loxP; Trp53loxP/loxP transgenic mice was performed on genomic DNA extracted from Cre virus injected or non-injected prostates. For Pten recombination, primer 2 (5′-AAAAGTTCCCCTGCTGATGATTTGT-3′) and primer 3 (5′-CCCCCAAGTCAATTGTTAGGTCTGT-3′)(11) were used. For Trp53 recombination, primer 1 (5′-CACAAAAACAGGTTAAACCCAG-3′) and primer 3 (5′-GAAG ACAGAAAAGGGGAGGG-3′)(44) were used. For detecting luciferase, primer 1 (5′-GAGGTTCCATCTGCCAGGTA-3′) and primer 2 (5′-CACACAGTTCGCCTCTT TGA-3′) were used.
Histology and Immunohistochemistry analysis
Tissues were fixed in 10% buffered formalin for 24 hrs, followed by gentle wash and transfer to PBS. Tissues were fixed in 10% buffered formalin for 24 hrs, followed by gentle wash and transfer to PBS. Then, paraffin-embedded tissues were sectioned 6 μm thick, placed on charged glass slides and stained with hematoxylin & eosin, or the appropriate immunohistochemical stains. Antigen retrieval was performed by incubating the slides in 0.01 M citric acid buffer (pH 6.0) at 95°C for 15 min. Slides were then allowed to cool to room temperature for 20 min in a citric acid buffer. After washing with deionized water, the slides were transferred to TBS (pH 7.4) for 5 min. The following detection and visualization procedures were performed according to manufacturer's protocol. Slides were counterstained in Mayer's hematoxylin, dehydrated, cleared, and cover slipped. Negative control slides were run without primary antibody. Control slides known to be positive for each antibody were incorporated. For androgen receptor (AR;D6F11, 1:500, Cell Signaling) and cytokeratin 8 (CK8; EP1628Y, 1:250, Novus Biologicals), cytokeratin 5 (CK5; AF138, 1:4000; Covance), luciferase (NB100-1677, 1:500, Novus Biologicals), pAKT(S473) (D9E, 1:50, Cell Signaling), pAKT(Thr308) (9275, 1:100, Cell Signaling), PTEN (138G6, 1:50, Cell Signaling), Pan-CK (sc-15367, 1:1000, Santa Cruz Biotechnology), Ki67(VP-K452, 1:2000, Vector Laboratories), pS6 Ser235/236 (4856, 1:200, Cell Signaling), Nkx3.1(1:6000, a kind gift from Dr. Chuck Bieberich), cMyc (1472-1, 1:50, Epitomics), GFP (ab290, 1:500 Abcam) CD3 (ab5690, 1:1000, Abcam) antibodies were used using the Discovery XT processor system (Ventana Medical Systems) as previously described (21) or sections were first blocked with 5% normal horse serum and 1% BSA (in TBS) for 1 hour at room temperature, and then the primary antibodies were diluted, as suggested by the manufacturer, and incubated overnight at 4°C. Following three 10 min. washes with TBS, sections were incubated with biotinylated secondary antibody for 30 min. at room temperature and rinsed with TBS three times for 10 min. Finally, the sections were treated with diaminobenzidine for 3 min and rinsed with distilled water to end the reaction, mounted on gelatin-coated slides, air-dried, dehydrated with 70%–100% alcohol, cleared with xylene and cover-slipped for microscopic observation. After examination of all immunohistochemical and special stains, stained slides were digitally scanned using the Aperio ScanScope software (Vista, California).
Double immunofluorescence staining was done with rabbit monoclonal cytokeratin 8 (CK8; EP1628Y, 1:250, Novus Biologicals) and mouse monoclonal PTEN antibody (138G6, 1:50, Cell Signaling), followed by incubation with Alexa Fluor 488 donkey anti-rabbit (1:500, Life Technologies) and Alexa Fluor 546 goat anti-mouse (1:500, Life Technologies) antibodies for 1 hour at room temperature for visualization. To stain cell nuclei, sections were incubated with a 10 μg/mL solution of 4′,6-diamidino-2-phenylindole (DAPI) for 1 hour at room temperature. Sections were rinsed in TBS and distilled water serially, and finally mounted with Mounting Medium (H-1000, Vector Laboratories, Burlingame, CA). Stained slides were imaged and analyzed using both the Ultraview VoX Spinning disk confocal microscope (PerkinElmer) and the Aperio ScanScope software (Vista, California).
Western blotting analysis
Fresh frozen tissues were lysed with RIPA buffer (50 mM NaCl, 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM Tris, pH 8.0, 10 μg/mL aprotinin, 1 mM phenylmethanesulfonyl fluoride (PMSF), 1 mM Na2VO4, 10 μg/mL pepstatin, 10 μg/mL leupeptin). Tissue lysates were separated by electrophoresis in SDS-polyacrylamide gels and transferred to nitrocellulose membranes. After blocking, membranes were hybridized with primary antibodies. Subsequently, membranes were incubated with horseradish peroxidase conjugated secondary antibodies and signals were detected with enhanced chemiluminescence. The following antibodies were used : anti-pPDK1 Ser241 (Cell Signaling, #3061), anti-PDK1 (Cell Signaling, #3062), anti-pS6 Ser235/236 (Cell Signaling, #4856), anti-S6 (Cell Signaling, #2217), beta actin-HRP (Sigma, #A3854).
RNA Extraction, Reverse Transcription, and Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from prostate or lymph node tissues using the Trizol reagent (Life Technologies) according to the manufacturer's instructions. 2 μg RNA were used for first strand synthesis and cDNA produced using random primers and SuperScript II (Invitrogen). RNA expression was measured by real-time quantitative reverse transcription-PCR, using the Roche LightCycler 480 (Roche Applied Science) based on the SYBR Green method. Each assay was done in triplicate and the expression of each gene was calculated relative to expression of β-actin cDNA. Quantification was based on a standard curve obtained by serial dilution of the indicated control RT reaction.
DNA Fluorescence in situ hybridization (FISH)
To detect cMyc, the probe was made using nick-translated kit (Abbot Molecular, 07J00-001) as the manufacturer's protocol. The paraffin embedded tissue sections were treated with 5% pepsin for 20 min at 37°C after de-paraffinization. Tissues were permeabilized with 0.5% TX-100/PBST for 5 min and RNase A was treated for 1 hr at RT. For hybridization, cellular DNA was denatured in 70% deionized formamide/2X SSC for 7 min at 80 °C. Hybridization of oligo probes was performed in 50 % deionized formamide, 10% dextran sulfate, 2× SSC, yeast tRNA, salmon sperm DNA, mouse CotI DNA and 50 ng nick translated probe in a humidified chamber for overnight at 37°C. Tissues were washed with 50% deionized formamide/2X SSC at 37°C and then 15 min in 2X SSC and 1X SSC, respectively. Tissues were mounted and images obtained using a Perkin Elmer Spinning Disk Confocal Microscope. For quantification, the percentage of cells with Myc copies >2 were counted for more between 200-300 nuclei per tissue.
Supplementary Material
Significance.
The RapidCaP system introduces fast and flexible genetics for functional analysis and therapy of endogenous metastatic prostate cancer. The approach introduces targeting of MYC as a critical strategy against PTEN-deficient lethal prostate cancer.
Acknowledgments
We would like to thank Scott Lowe, Alea Mills, Leemor Joshua-Tor, Scott Powers, Chris Vakoc, Junwei Shi, Charles Bieberich and the members of the Trotman lab for valuable discussion and help with experimental protocols and reagents, the CSHL Animal Resources team, Lisa Bianco, Lotus Altholtz, Jodi Coblentz, Michael Cahn for help with RapidCaP modeling, maintenance and trials, Aigoul Nourjanova, Raisa Puzis, Denise Hoppe, K. Manova, and A. Barlas for help with histopathology procedures, Scott Lowe, Nilgun Tasdemir and Luke Dowe for help with reagents, and D. Tsang for help with the manuscript. This work was supported by grants to L.C.T. from the NIH (CA137050), the Department of the Army (W81XWH-09-1-0557), the STARR foundation, the Robertson Research Fund of Cold Spring Harbor Laboratory and by support from the CSHL CTD2 Cancer Target Discovery and Development Network Grant (U01CA168409 S. Powers, PI).
Footnotes
Conflict of Interest Statement: The authors declare that they have no competing financial interests.
References
- 1.Huggins C, Stevens RC, Hodges CV. Studies on prostatic cancer: ii. The effects of castration on advanced carcinoma of the prostate gland. Arch Surg. 1941;43:209–23. [Google Scholar]
- 2.Chen Y, Scher HI. Prostate cancer in 2011: Hitting old targets better and identifying new targets. Nat Rev Clin Oncol. 2012;9:70–2. doi: 10.1038/nrclinonc.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baca SC, Garraway LA. The genomic landscape of prostate cancer. Front Endocrinol (Lausanne) 2012;3:69. doi: 10.3389/fendo.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, et al. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nature genetics. 2012;44:685–9. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irshad S, Abate-Shen C. Modeling prostate cancer in mice: something old, something new, something premalignant, something metastatic. Cancer Metastasis Rev. 2013;32:109–22. doi: 10.1007/s10555-012-9409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Podsypanina K, Ellenson LH, Nemes A, Gu J, Tamura M, Yamada KM, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci U S A. 1999;96:1563–8. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–55. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 10.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27:222–4. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 11.Trotman LC, Niki M, Dotan ZA, Koutcher JA, Di Cristofano A, Xiao A, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–21. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 13.King JC, Xu J, Wongvipat J, Hieronymus H, Carver BS, Leung DH, et al. Cooperativity of TMPRSS2-ERG with PI3-kinase pathway activation in prostate oncogenesis. Nat Genet. 2009;41:524–6. doi: 10.1038/ng.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, et al. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–38. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- 15.Trotman LC, Alimonti A, Scaglioni PP, Koutcher JA, Cordon-Cardo C, Pandolfi PP. Identification of a tumour suppressor network opposing nuclear Akt function. Nature. 2006;441:523–7. doi: 10.1038/nature04809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heyer J, Kwong LN, Lowe SW, Chin L. Non-germline genetically engineered mouse models for translational cancer research. Nature reviews Cancer. 2010;10:470–80. doi: 10.1038/nrc2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh M, Ferrara N. Modeling and predicting clinical efficacy for drugs targeting the tumor milieu. Nature biotechnology. 2012;30:648–57. doi: 10.1038/nbt.2286. [DOI] [PubMed] [Google Scholar]
- 18.Singh M, Murriel CL, Johnson L. Genetically engineered mouse models: closing the gap between preclinical data and trial outcomes. Cancer research. 2012;72:2695–700. doi: 10.1158/0008-5472.CAN-11-2786. [DOI] [PubMed] [Google Scholar]
- 19.Ittmann M, Huang J, Radaelli E, Martin P, Signoretti S, Sullivan R, et al. Animal models of human prostate cancer: the consensus report of the New York meeting of the Mouse Models of Human Cancers Consortium Prostate Pathology Committee. Cancer research. 2013;73:2718–36. doi: 10.1158/0008-5472.CAN-12-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen M, Pratt CP, Zeeman ME, Schultz N, Taylor BS, O'Neill A, et al. Identification of PHLPP1 as a Tumor Suppressor Reveals the Role of Feedback Activation in PTEN-Mutant Prostate Cancer Progression. Cancer Cell. 2011;20:173–86. doi: 10.1016/j.ccr.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470:269–73. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwata T, Schultz D, Hicks J, Hubbard GK, Mutton LN, Lotan TL, et al. MYC overexpression induces prostatic intraepithelial neoplasia and loss of Nkx3.1 in mouse luminal epithelial cells. PLoS One. 2010;5:e9427. doi: 10.1371/journal.pone.0009427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrington LS, Findlay GM, Lamb RF. Restraining PI3K: mTOR signalling goes back to the membrane. Trends in biochemical sciences. 2005;30:35–42. doi: 10.1016/j.tibs.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 27.O'Reilly KE, Rojo F, She QB, Solit D, Mills GB, Smith D, et al. mTOR inhibition induces upstream receptor tyrosine kinase signaling and activates Akt. Cancer research. 2006;66:1500–8. doi: 10.1158/0008-5472.CAN-05-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vasudevan KM, Barbie DA, Davies MA, Rabinovsky R, McNear CJ, Kim JJ, et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell. 2009;16:21–32. doi: 10.1016/j.ccr.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–8. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi J, Whyte WA, Zepeda-Mendoza CJ, Milazzo JP, Shen C, Roe JS, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes & development. 2013 doi: 10.1101/gad.232710.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, Matos T, et al. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes & development. 2009;23:675–80. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc. 2009;4:1064–72. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alimonti A, Nardella C, Chen Z, Clohessy JG, Carracedo A, Trotman LC, et al. A novel type of cellular senescence that can be enhanced in mouse models and human tumor xenografts to suppress prostate tumorigenesis. J Clin Invest. 2010;120:681–93. doi: 10.1172/JCI40535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim JS, Lee C, Bonifant CL, Ressom H, Waldman T. Activation of p53-Dependent Growth Suppression in Human Cells by Mutations in PTEN or PIK3CA. Mol Cell Biol. 2006 doi: 10.1128/MCB.00537-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nardella C, Clohessy JG, Alimonti A, Pandolfi PP. Pro-senescence therapy for cancer treatment. Nature reviews Cancer. 2011;11:503–11. doi: 10.1038/nrc3057. [DOI] [PubMed] [Google Scholar]
- 37.Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, et al. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15:559–65. doi: 10.1038/nm.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Conacci-Sorrell M, Ngouenet C, Eisenman RN. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell. 2010;142:480–93. doi: 10.1016/j.cell.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Roh M, Doubinskaia I, Algarroba GN, Eltoum IE, Abdulkadir SA. A mouse model of heterogeneous, c-MYC-initiated prostate cancer with loss of Pten and p53. Oncogene. 2012;31:322–32. doi: 10.1038/onc.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clegg NJ, Couto SS, Wongvipat J, Hieronymus H, Carver BS, Taylor BS, et al. MYC cooperates with AKT in prostate tumorigenesis and alters sensitivity to mTOR inhibitors. PLoS One. 2011;6:e17449. doi: 10.1371/journal.pone.0017449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang J, Kobayashi T, Floc'h N, Kinkade CW, Aytes A, Dankort D, et al. B-Raf activation cooperates with PTEN loss to drive c-Myc expression in advanced prostate cancer. Cancer research. 2012;72:4765–76. doi: 10.1158/0008-5472.CAN-12-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nature genetics. 2001;29:418–25. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 44.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.