Abstract
Membrane-targeting proteins are crucial components of many cell signaling pathways, including the secretion of insulin. Granuphilin, also known as synaptotagmin-like protein 4, functions in tethering secretory vesicles to the plasma membrane prior to exocytosis. Granuphilin docks to insulin secretory vesicles through interaction of its N-terminal domain with vesicular Rab proteins; however, the mechanisms of granuphilin plasma membrane targeting and release are less clear. Granuphilin contains two C2 domains, C2A and C2B, that interact with the plasma membrane lipid phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2]. The goal of this study was to determine membrane-binding mechanisms, affinities, and kinetics of both granuphilin C2 domains using fluorescence spectroscopic techniques. Results indicate that both C2A and C2B bind anionic lipids in a Ca2+-independent manner. The C2A domain binds liposomes containing a physiological mixture of lipids including 2% PI(4,5)P2 or PI(3,4,5)P3 with high affinity (apparent Kd,PIPx of 2-5 nM), and binds nonspecifically with moderate affinity to anionic liposomes lacking phosphatidylinositol phosphate (PIPx) lipids. The C2B domain binds with sub-micromolar affinity to liposomes containing PI(4,5)P2 but does not have a measurable affinity for background anionic lipids. Both domains can be competed away from their target lipids by the soluble PIPx analogue inositol-(1,2,3,4,5,6)-hexakisphosphate (IP6), which is a positive regulator of insulin secretion. Potential roles of these interactions in the docking and release of granuphilin from the plasma membrane are discussed.
Keywords: Slp4; protein-lipid interaction; secretory granule docking; inositol polyphosphate signaling; insulin secretion; phosphatidylinositol-(4,5)-bisphosphate
1. Introduction
Protein domains that interact reversibly with lipid membranes serve as signaling modules in a variety of cellular processes (Cho and Stahelin, 2005; Lemmon, 2008). C2 domains represent one of the most abundant families of membrane-targeting domains and typically dock to anionic lipid membranes in response to Ca2+, although some members sense phosphatidylinositol phosphate (PIPx) lipids in addition to, or instead of, cytosolic Ca2+ (Corbalan-Garcia et al., 2003; Murray and Honig, 2002; Nalefski and Falke, 1996). C2 domains are highly represented among proteins in regulated secretory pathways, including synaptotagmin and related proteins that serve as Ca2+ and/or lipid sensors in secretory vesicle docking and exocytosis (Fukuda and Mikoshiba, 2001; Gustavsson and Han, 2009; Martens, 2010; Sudhof, 2002).
The synaptotagmin-like protein granuphilin (also known as Slp-4) contains an N-terminal domain that attaches to Rab27a/Rab3 on secretory vesicles, a linker region of unknown structure that mediates binding to syntaxin and/or Munc18 on the plasma membrane, and two C-terminal C2 domains, termed C2A and C2B (Coppola et al., 2002; Torii et al., 2004; Tsuboi and Fukuda, 2006; Wang et al., 1999; Yi et al., 2002). Both C2 domains have been shown qualitatively to bind lipid vesicles containing phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2 or PIP2] independently of Ca2+ (Yu et al., 2007), but little else is known about their membrane binding properties. The role of C2 domain–plasma membrane interaction for granuphilin function is somewhat controversial; some models indicate direct C2 domain interaction with plasma membrane lipids, while others suggest that granuphilin plasma membrane docking is mediated solely by its linker region binding Munc18/syntaxin (Izumi, 2011; Tsuboi, 2009). Recent evidence indicates that deletion of both C2 domains from granuphilin leads to a cytosolic distribution, whereas each C2 domain localizes to the plasma membrane when expressed individually (Galvez-Santisteban et al., 2012). This observation supports the idea that membrane binding by one or both C2 domains is required for granuphilin plasma membrane localization. Interestingly, a splice variant lacking the C2B domain, termed granuphilin-b, functions and localizes similarly to the granuphilin-a variant containing both C2 domains (Torii et al., 2004; Tsuboi and Fukuda, 2006; Wang et al., 1999).
Granuphilin functions in secretory vesicle – plasma membrane docking prior to Ca2+-triggered membrane fusion (Fukuda, 2006; Izumi, 2011). It is found in insulin-secreting pancreatic β cells and other cell types that undergo large dense-core granule exocytosis (Bierings et al., 2012; Tomas et al., 2008; Yi et al., 2002). Altered granuphilin expression produces two seemingly contradictory effects: knockdown or knockout leads to decreased vesicle – plasma membrane docking but increased rates of secretion, while overexpression leads to increased vesicle docking with diminished secretion (Coppola et al., 2002; Gomi et al., 2005; Torii et al., 2004). This pattern suggests that granuphilin is part of an apparatus that tethers secretory vesicles to the plasma membrane and acts as a “brake” to exocytosis, which must be overcome during secretory stimulation. The molecular mechanisms of this braking behavior are unclear but likely involve syntaxin/Munc18 interaction (Gomi et al., 2005; Tsuboi and Fukuda, 2006). Importantly, the mechanism by which braking is released during active secretion is currently unknown (Izumi, 2011; Tomas et al., 2008).
In order to better understand granuphilin-mediated secretory vesicle tethering and inhibition of membrane fusion, more information is needed on the contributions of its C2 domains to plasma membrane docking and release. In this study, we report docking mechanisms, target lipids, and affinities of both individual granuphilin C2 domains using a combination of equilibrium and kinetic fluorescence spectroscopy. In addition, we identify a product of insulin secretory signaling, inositol (1,2,3,4,5,6)-hexakisphosphate (IP6), that displaces both C2 domains from lipid vesicles. The results suggest potential mechanisms by which C2 domains contribute significantly to granuphilin plasma membrane docking and release.
2. Materials and Methods
2.1 Reagents
Lipids were purchased from Avanti Polar Lipids (all are synthetic unless otherwise indicated): 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (phosphatidylcholine, PC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (phosphatidylserine, PS), phosphatidylinositol (PI) from liver, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (phosphatidylethanolamine, PE), phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2, PIP2] from brain, 1,2-dioleoyl-sn-glycero-3-phospho-(1'-myo-inositol-3',4',5'-trisphosphate) [PI(3,4,5)P3, PIP3], cholesterol, and sphingomyelin (SM) from brain. N-[5-(dimethylamino)-naphthalene-1-sulfonyl]-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (dansyl-PE, dPE) was from Invitrogen. d-myo-inositol 1,2,3,4,5,6,-hexakisphosphate dodecasodium salt (IP6) was from Sigma. D-myo-inositol-1,4,5-triphosphate tripotassium salt (IP3) was from Cayman Chemical.
2.2 Cloning, Expression and Purification of Granuphilin C2A and C2B domains
cDNA encoding the human granuphilin C2A domain (Gly352 – Ser488) flanked by a C-terminal His6 tag was purchased from the DNASU plasmid repository (DNASU: HsCD00321956). This domain was expressed in E. coli BL-21 cells and purified using Ni-NTA column chromatography. For C2B, the region of human granuphilin cDNA (ATCC: 10700678) encoding this domain (Glu487 – Leu671) was PCR-amplified and cloned into a previously described N-terminal glutathione S-transferase expression vector (Corbin et al., 2004). Protein was expressed in E. coli BL-21 cells and purified using glutathione sepharose as described (Brandt et al., 2012). High salt washes were used to remove contaminating nucleic acid, and the free C2B domain was eluted following thrombin cleavage. Purified proteins were concentrated, treated with benzonase (Sigma) for 12 h at 4 °C to remove remaining nucleic acid, and dialyzed into assay buffer (140 mM KCl, 0.5 mM MgCl2, 150 mM NaCl, 25 mM HEPES, pH 7.4) including 1 mM 2-mercaptoethanol and 0.02% NaN3. Purity of the isolated proteins was >95% by SDS-PAGE, and the absence of significant contaminating nucleic acid was verified via absorbance measurement at 260 nm. Concentration was determined from absorbance of denatured protein at 280 nm ( = 19940 M−1 cm−1 and 34950 M−1 cm−1 for C2A and C2B, respectively).
2.3 Preparation of Lipid Vesicles
Small unilamellar vesicles (SUVs) with the lipid compositions listed in Table 1 were prepared by sonication as described previously (Brandt et al., 2012).
Table 1.
Vesicle lipid compositions used in this study (mol %)
| PC | PS | PE | PI | PG | CH | SM | PIP2 | PIP3 | dansyl-PE | |
|---|---|---|---|---|---|---|---|---|---|---|
| PM(+)PIP2 | 10.5 | 21 | 28 | 4 | 0 | 25 | 4.5 | 2 | 0 | 5 |
| PM(+)PIP3 | 10.5 | 21 | 28 | 4 | 0 | 25 | 4.5 | 0 | 2 | 5 |
| PM(−)PIPx | 10.5 | 21 | 28 | 6 | 0 | 25 | 4.5 | 0 | 0 | 5 |
| PM(−)PIPx/PS/PI | 37.5 | 0 | 28 | 0 | 0 | 25 | 4.5 | 0 | 0 | 5 |
| PM(−)PIPx/PS/PI(+)PG | 10.5 | 0 | 28 | 0 | 27 | 25 | 4.5 | 0 | 0 | 5 |
| PC/dPE | 95 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| PC/PS/dPE | 71 | 24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 |
| PC/PI/dPE | 71 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 0 | 5 |
| PC/PG/dPE | 71 | 0 | 0 | 0 | 24 | 0 | 0 | 0 | 0 | 5 |
| PC/PS/PI/dPE | 68 | 21 | 0 | 6 | 0 | 0 | 0 | 0 | 0 | 5 |
2.4 Equilibrium fluorescence measurements
Measurements were performed in a Photon Technology International QuantaMaster fluorescence spectrometer at 25 °C, with excitation at 284 nm (1 nm slit width) and emission slit width 8 nm.
Qualitative protein-to-membrane FRET measurements were performed using 125 μM total accessible lipid in assay buffer containing either 100 μM EDTA or 1 mM CaCl2. First, a blank spectrum of each sample was measured in order to quantify fluorescence emission due to direct dansyl excitation at 284 nm. Subsequent emission spectra were measured after addition of (a) 1 μM C2A or C2B domain and (b) 8 mM IP6. This non-physiological concentration of IP6 has been sufficient to competitively remove PIPx-bound proteins in previous studies, including those with very high PIPx affinity (Kavran et al., 1998; Landgraf et al., 2008). Samples were equilibrated for 40 s with stirring after each addition. All spectra shown are corrected for dilution.
For measurement of IP6 or IP3 binding to the free granuphilin C2A domain (0.2 μM), the change in intrinsic Trp emission at 330 nm was measured upon titration with ligand. (Corbin et al., 2004; Landgraf et al., 2008) Samples were equilibrated for 40 s with stirring following each addition, and emission intensities were averaged over 10 s and corrected for dilution. The resulting plot of intensity F vs. ligand concentration [I] was a subject to a non-linear fit best-fit analysis in order to calculate the equilibrium dissociation constants (KI) for these inhibitors of membrane binding:
| (1) |
where ΔFmax represents the calculated maximum fluorescence change from the unbound protein and C is a constant. To simplify graphical representations, data are normalized such that C = 0 and ΔFmax = 1.
For FRET competition binding assays, C2A or C2B domain (1 μM) was pre-mixed with liposomes (125 μM total accessible lipid) in assay buffer containing 100 μM EDTA. Decreased protein-to-membrane FRET upon IP6 titration was assessed based on acceptor emission at 520 nm, using a 10-s time average for each measurement. In a separate cuvette measured in parallel, IP6 was titrated into a sample containing lipid but no protein. After correcting for dilution, intensities from this blank sample were subtracted from the experimental samples. Resulting plots were fit to a hyperbolic model for single-site competitive inhibition:
| (2) |
where ΔFmax is the total FRET signal before ligand titration, [I] is the concentration of added IP6, and IC50 is the IP6 concentration at which FRET is 50% of the initial value. To simplify graphical representations, data are normalized such that C = 0 and ΔFmax = 1.
In these competition binding measurements, equilibrium constants for protein-lipid interaction can be calculated from the best-fit IC50 value if the KI of the inhibitor is known. Affinities measured in this manner can be expressed either as mole-fraction partition coefficients (Kx) (White and Wimley, 1999) or as PIPx dissociation constants (Kd,PIPx). Here, both values are reported and are calculated using eq. 3 and 4, respectively:
| (3) |
| (4) |
where [L] is the total concentration of accessible lipids in the outer leaflet (i.e., half the total lipid concentration) and [PIPx] is the concentration of free accessible PI(4,5)P2 or PI(3,4,5)P3 when [I] = IC50 (i.e., the total outer leaflet [PIPx] less half the total protein concentration). For error propagation, the uncertainty in [L] or [PIPx] was estimated to be 10%. G° values are calculated as described (White and Wimley, 1999).
2.5 Stopped-flow FRET measurement of association and dissociation kinetics
Kinetic measurements were performed using an Applied Photophysics SX.17 stopped-flow fluorescence spectrophotometer in assay buffer with 100 μM EDTA at 25 °C. Excitation was at 284 nm with a slit width of 3 nm, and dansyl emission was measured using a 475-nm longpass filter. All data prior to the instrument dead time of 1 ms were discarded prior to plotting and fitting.
To determine dissociation rate constants (koff), a solution containing protein (0.3 μM) and liposomes (75 μM total accessible lipid, concentrations after mixing) was rapidly mixed with an equal volume of a solution containing a 4-fold excess of unlabeled vesicles, which lacked dansyl-PE but were otherwise identical to the vesicles to which the protein was initially bound. Kinetic profiles of decreasing protein-to-membrane FRET were fit to single-exponential decay functions, in which the resulting approach to equilibrium is described by the intrinsic dissociation rate constant koff:
| (5) |
For simplified presentation, the best-fit offset C was subtracted from all data points and ΔFmax was normalized to unity.
To determine the apparent association rate constants (kobs) of the C2 domains, C2A or C2B domain (0.3 μM) was rapidly mixed with liposomes (75 μM total accessible lipid, concentrations after mixing). Each resulting time course was fit to a single-exponential binding model (Corbin et al., 2004):
| (6) |
The best-fit offset C was subtracted from all data points and ΔFmax was normalized to unity.
The protein-membrane association rate constant kon was calculated based on kobs and koff according to either the membrane partitioning (kon,x) or PI(4,5)P2 binding (kon,PIP2) model using eq. 7 or 8, respectively:
| (7) |
| (8) |
where [L] is the total outer leaflet lipid concentration and [PIP2] is the concentration of unbound PI(4,5)P2 in the outer leaflet, approximated as the total outer leaflet PI(4,5)P2 concentration less half the total protein concentration.
2.6 Electrostatic surface calculations for C2A
Electrostatic surface calculations were based on the available structure of granuphilin C2A (PDB: 3FDW). The positions of missing atoms in the structure (some sidechain atoms from K365, Q369, T370, K390, K391, R407, S435, Q439, D466, K469, and K483) were modeled using default settings of Swiss PDB Viewer. The electrostatic surface was generated using the Advanced Poisson-Boltzmann Solver plugin (APBS 0.5.1) for PyMol, with calculations performed using 0.15 M monovalent cation and anion concentrations.
3. Results
3.1 Identification of target lipids using protein-to-membrane FRET
In order to measure the lipid composition dependence of membrane targeting for granuphilin C2 domains, an established protein-to-membrane fluorescence resonance energy transfer (FRET) assay was used with SUVs of defined lipid composition (Nalefski and Falke, 2002). In this measurement, one or more Trp residues intrinsic to the protein domain (2 total in C2A, 5 in C2B) serve as FRET donors, and dansyl-phosphatidylethanolamine (dPE) lipids included in the liposomes serve as FRET acceptors. Protein-membrane binding leads to an increased dPE emission at 512 nm upon Trp excitation at 284 nm and a decrease in Trp emission at 350 nm. In order to determine target lipid preferences for granuphilin C2A and C2B domains, liposomes were prepared with the compositions listed in Table 1.
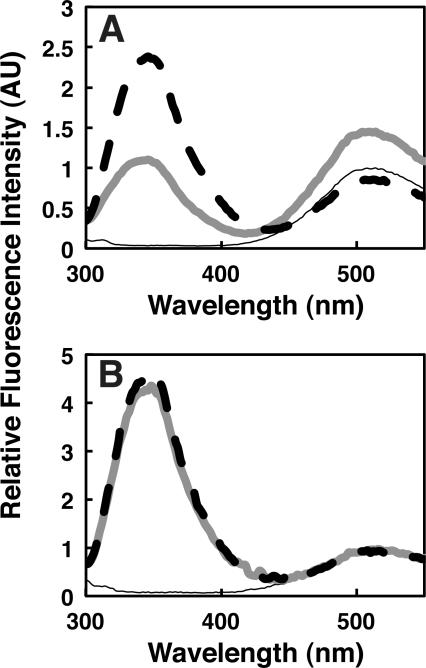
The granuphilin C2A domain shows significant protein-to-membrane FRET when added to liposomes closely resembling the lipid composition of the plasma membrane cytosolic leaflet (Corbin et al., 2007; Voelker, 2008). Since some C2 domains are known to dock preferentially to PIPx lipids, such physiologically relevant lipid mixtures are used throughout this study either including 2% PI(4,5)P2 [PM(+)2%PIP2] or lacking PIPx [PM(−)PIPx]. In qualitative measurements, addition of the C2A domain led to an increase in dPE emission in vesicles with either of these compositions (Figure 1A,B), but not vesicles composed only of phosphatidylcholine (PC) and dPE (Figure 1C).
Figure 1. Qualitative FRET measurement of granuphilin C2A binding to lipid vesicles.
Fluorescence emission spectra are shown of C2A with (A) PM(−)PIPx, (B) PM(+)PIP2, and (C) PC/dPE vesicles (lipid compositions given in Table 1). Thin black curves: blank measurements prior to protein addition, consisting of vesicles (125 μM total accessible lipid) in assay buffer. The peak at ~510 nm in these traces represents direct excitation of the dans fluorophore at 284 nm. Thick gray curves: measurements after addition of 1 μM C2A domain. Dashed black curves: measurements after addition of 8 mM IP6. Protein-lipid binding is evidenced by increased dans emission at ~510 nm due to FRET upon protein addition. Intensities in each panel are normalized to the maximum dans emission intensity of the sample prior to protein addition.
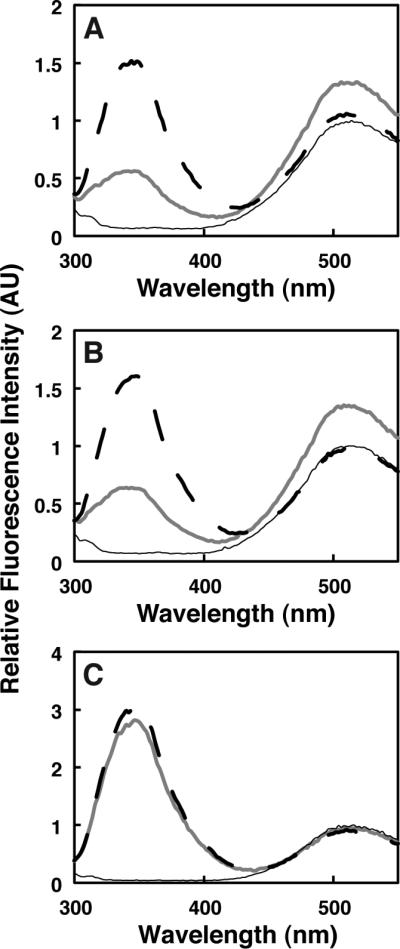
In order to identify the lipid components responsible for C2A association with PM(−)PIPx vesicles, simpler lipid compositions were tested. FRET was observed upon addition of C2A to vesicles containing the anionic background lipids phosphatidylserine (PS) and phosphatidylinositol (PI) along with PC (Figure 2A), but not to vesicles containing only the zwitterionic components of our PM(−)PIPx mixture (Figure 2B). Binding was restored upon replacement of PS and PI with phosphatidylglycerol (PG), an anionic lipid that is not normally present in mammalian plasma membranes (van Meer et al., 2008) (Supplementary data, Figure S1), suggesting that C2A binds anionic background lipids such as these through nonspecific interactions.
Figure 2. Granuphilin C2A interaction with background anionic lipids.
Fluorescence emission spectra are shown as described in the legend to Figure 1, using vesicles composed of (A) PC/PS/PI/dPE and (B) PM(−)PIPx/PS/PI (compositions given in Table 1).
In contrast to C2A, no FRET was observed upon addition of 1 μM C2B domain to PM(−)PIPx, indicating binding is too weak to measure with this lipid composition (Figure 3A). However, significant interaction was observed when PI(4,5)P2 was included in the liposomes (Figure 3B). For both domains, the FRET efficiency was identical whether measured in the presence of 100 μM EDTA or 1 mM Ca2+ (data not shown), consistent with previous reports that granuphilin C2 domains bind anionic lipids in a Ca2+-independent manner (Wang et al., 1999; Yu et al., 2007).
Figure 3. Qualitative FRET measurement of granuphilin C2B binding to lipid vesicles.
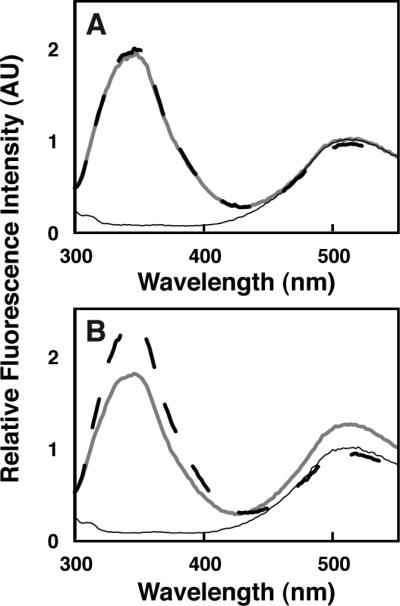
Fluorescence emission spectra are shown as described in the legend to Figure 1, using 1 μM C2B domain and vesicles composed of (A) PM(−)PIPx and (B) PM(+)PIP2 (compositions given in Table 1).
IP6 inhibits lipid binding by both granuphilin C2 domains. IP6 is a soluble, fully phosphorylated analogue of PIPx lipid headgroups, which competitively inhibits membrane binding by protein domains that target PIPx lipids (Corbin et al., 2004; Landgraf et al., 2008; Takeuchi et al., 1997). For both granuphilin C2 domains and with all lipid compositions tested, protein-to-membrane FRET was completely reversed by the addition of 8 mM IP6 as judged by dansyl emission at 512 nm (Figures 1-3). IP6 addition also resulted in a measurable increase in the intrinsic Trp emission of the C2A domain even in the absence of membrane binding (Figure 1C), an effect that was not observed for C2B. Such a Trp fluorescence increase has been previously observed with PIPx-binding pleckstrin homology domains (Corbin et al., 2004; Landgraf et al., 2008) and may be due to a change in the microenvironment around one or both of the Trp residues in granuphilin C2A upon binding IP6.
3.2 Quantifying C2A affinity for IP6 and IP3
Qualitative measurements such as those shown in Figures 1-3 do not distinguish between tight and moderate lipid binding affinities, as any interaction in the micromolar range or tighter produces nearly complete binding. However, the lipid affinity of a tightly bound protein can be determined quantitatively using a competitive binding assay if an inhibitor is available with known affinity for the protein (Corbin et al., 2004). Thus, the affinity of C2A for IP6 was first measured based on the intrinsic Trp fluorescence increase, in order that this domain's affinity for target vesicles could be subsequently quantified. IP6 titration data fit well to a single-site binding model with an inhibitor equilibrium dissociation constant (KI) of 1.8 ± 0.1 μM (Figure 4A). Similar measurement of C2A affinity for IP3 reveals considerably weaker binding, with a KI of 80 ± 30 μM (Figure 4B). This difference stands in contrast to the pleckstrin homology domain from phospholipase C-δ1, which binds preferentially to PI(4,5)P2 over other PIPx species and exhibits a higher affinity towards IP3 compared to IP6 (Kavran et al., 1998). For the present study, the higher-affinity inhibitor IP6 was used for subsequent competition experiments.
Figure 4. Equilibrium fluorescence measurement of granuphilin C2A domain binding to (A) IP6 and (B) IP3.
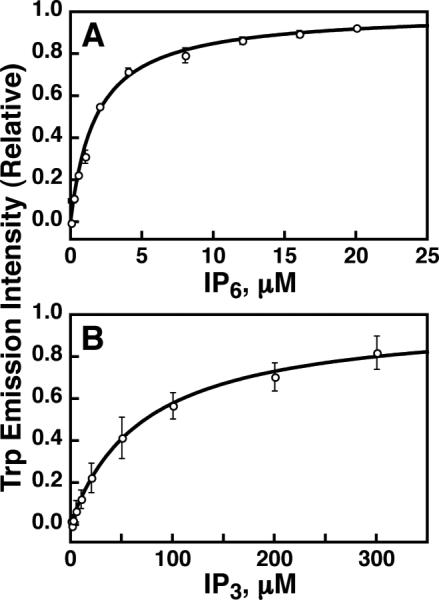
Trp fluorescence intensity (excitation 284 nm, emission 330 nm) was monitored upon titration of ligand into solutions of 0.2 μM C2A in assay buffer. The fluorescence increase was fit to eq. 1 (solid curves) and intensity values were normalized as described in Methods (section 2.4). Best-fit values are given in section 3.2. Error bars show standard deviation of three independent replicate experiments; where not visible, error bars are smaller than the data symbols.
3.3 C2A competition binding measurements
Competition binding measurements with IP6 show that PI(4,5)P2 enhances the affinity of granuphilin C2A for liposomes ~40-fold. As shown in Figure 5A and summarized in Table 2, IP6 was titrated into solutions containing C2A domain bound to vesicles. A simple competitive inhibition model is sufficient to describe the IP6 titration data. The inhibitor concentration at which the amount of vesicle-bound protein was half its initial value (IC50) increased from 37 ± 6 μM to 1.6 ± 0.2 mM upon inclusion of 2% PI(4,5)P2 (Table 2). These data, together with the measured affinity of C2A for IP6 (Figure 4A), can be used to calculate a mole-fraction partition coefficient, Kx, describing the liposomal membrane partitioning equilibrium for each lipid composition (White and Wimley, 1999). This value is (9 ± 2) × 106 for PM(−)PIPx vesicles and increases ~40-fold to (390 ± 70) × 106 for PM(+)PIP2. If the C2A-membrane interaction is approximated as a bimolecular equilibrium between the protein and PI(4,5)P2 headgroups rather than a membrane/aqueous phase partitioning, the same data yield an apparent equilibrium dissociation constant, Kd,PIP2, of 2.3 ± 0.4 nM (Table 2). Replacement of PI(4,5)P2 with 2% PI(3,4,5)P3 in target liposomes leads to similarly tight binding, with an affinity that differs from PM(+)PIP2 vesicles by less than a factor of two (Table 2). This result is consistent with earlier reports of low PIPx selectivity by this domain using less quantitative methods (Galvez-Santisteban et al., 2012; Wang et al., 2013). Overall, it is clear that granuphilin C2A binds with high affinity to liposomes containing physiological mixtures of background anionic lipids and either PI(4,5)P2 or PI(3,4,5)P3.
Figure 5. IP6 competition measurement of granuphilin C2 domain vesicle binding.
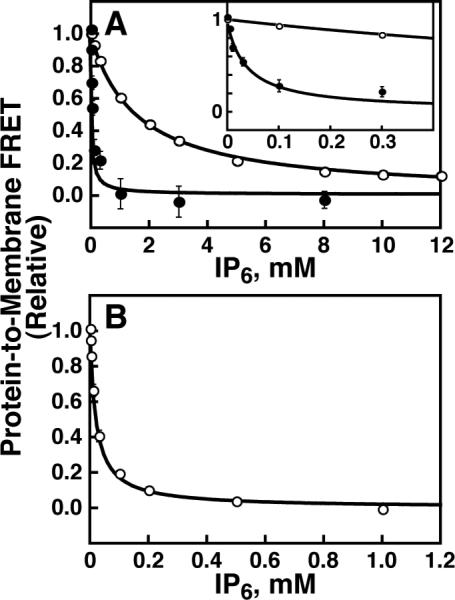
Protein-to-membrane FRET was monitored (excitation 284 nm, emission 512 nm) of (A) 1 μM C2A or (B) 1 μM C2B in the presence of PM(−)PIPx (filled symbols) or PM(+)PIP2 vesicles (open symbols) upon titration of IP6. The resultant FRET decrease was fit to eq. 2 (solid curves). Best-fit values are given in Table 2. Error bars are standard deviation of three independent replicate experiments, and where not visible are smaller than the data symbols. Inset to panel A: view expanded to show values at lower IP6 concentrations.
Table 2.
Parameters from IP6 competition measurements
| Domain | Target vesicles | IC50 (IP6), μMa | Kx × 10−6b | Apparent Kd,PIPx, nMc | ΔG°x, kcal/mol | ΔG°PIPx, kcal/mol |
|---|---|---|---|---|---|---|
| C2A | PM(+)PIP2 | 1600 ± 200 | 390 ± 70 | 2.3 ± 0.4 | −11.7 ± 0.1 | −11.8 ± 0.1 |
| C2A | PM(+)PIP3 | 1100 ± 100 | 270 ± 40 | 3.3 ± 0.5 | −11.5 ± 0.1 | −11.6 ± 0.1 |
| C2A | PM(−)PIPx | 37 ± 6 | 9 ± 2 | N.D.d | −9.5 ± 0.1 | N.D.d |
| C2A | PC/PS/dPE | 10 ± 1 | 2.1 ± 0.4 | N.D.d | −8.6 ± 0.1 | N.D.d |
| C2A | PC/PI/dPE | 5 ± 1 | 0.8 ± 0.1 | N.D.d | −8.1 ± 0.1 | N.D.d |
| C2A | PC/PG/dPE | 11 ± 1 | 2.2 ± 0.3 | N.D.d | −8.6 ± 0.1 | N.D.d |
| C2B | PM(+)PIP2 | 21 ± 3 | N.D.e | N.D.e | N.D.e | N.D.e |
IC50 values and errors are average and standard deviation of 3 or more independent replicate experiments.
Mole-fraction partition coefficient, calculated using eq. 3.
Calculated using eq. 4.
Apparent Kd,PIPx not calculated for vesicles lacking PIPx.
IP6 binding does not alter the intrinsic Trp fluorescence of C2B, therefore its KI was not measured using this method.
In order to quantitatively test for specificity in C2A interactions with background anionic lipids, IP6 competition titrations were conducted with this domain and simple 3:1 lipid mixtures of PC and either PS, PI, or PG. These titrations reveal similar affinities of the C2A domain for these three simple lipid compositions, indicating that interactions with background anionic lipids are nonspecific (Supplementary data, Figure S2). Kx values for these lipid compositions range from 8 × 105 to 2 × 106, lower by factors of 4-10 than for PM(−)PIPx (Table 2). Such lower affinity for simple lipid mixtures versus physiological mixtures with similar anionic content has also been observed for other PIPx-targeting domains, although the underlying reasons for this effect are not clear (Corbin et al., 2004).
IP6 also competes with the C2B domain for lipid binding, with an IC50 of 21 ± 3 μM (Figure 5B, Table 2). IP6 binding does not significantly alter the intrinsic Trp fluorescence of C2B (data not shown), preventing direct measurement of C2B – IP6 affinity using this approach.
3.4 Kinetics of membrane association and dissociation
FRET-based kinetic measurements of protein-lipid association and dissociation further demonstrate high-affinity binding. Stopped-flow association measurements (Figure 6A) generally fit well to single-exponential profiles with rate constants (kobs) listed in Table 3. Rates of stochastic dissociation were measured by rapidly mixing protein-membrane complexes with an excess of unlabeled liposomes (Figure 6B). The decay constant of the resulting approach to equilibrium is equivalent to the rate constant for protein-membrane dissociation, koff; these values are listed in Table 3.
Figure 6. Kinetic measurement of granuphilin C2 domain membrane interaction.
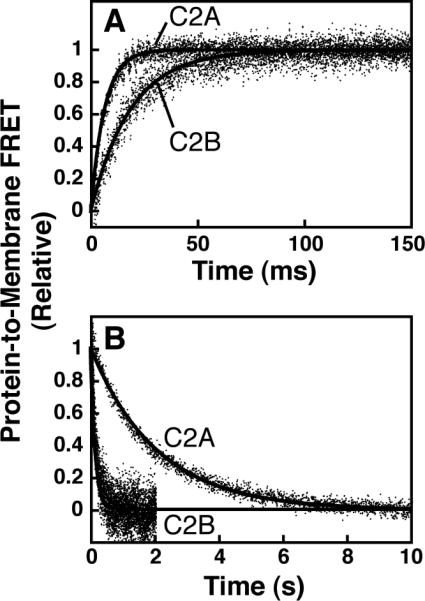
A: Association kinetics. Protein (0.3 μM) was rapidly mixed with PM(+)PIP2 vesicles (75 μM total accessible lipid after mixing) in a stopped-flow fluorescence spectrometer and the increase in dans emission was monitored (excitation 284 nm). Single-exponential fits are shown (black curves, eq. 6). B: Dissociation kinetics. Protein (0.3 μM) was pre-bound to PM(+)PIP2 vesicles (75 μM total accessible lipid, all concentrations after mixing) and rapidly mixed with excess PM(+)PIP2 vesicles lacking dansyl-PE (300 μM total accessible lipid). Single-exponential fits are shown (black curves, eq. 5). Data points are averages from 9-12 mixing events. The smaller signal-to-noise ratios for C2B are due to the smaller amplitude of its FRET change upon membrane binding (compare Figure 3B to Figure 1B).
Table 3.
Measured rate constants from kinetic experiments
| Domain | Target vesicles | koff, s−1a | kobs for association, s−1a,b | kon/koff × 10−6 expressed as partition coefficientc | koff/kon, nMc expressed as affinity for PIP2 |
|---|---|---|---|---|---|
| C2A | PM(−)PIPx | 13 ± 1 | 190 ± 30 | 10 ± 2 | N.D.d |
| C2A | PM(+)PIP2 | 0.48 ± 0.01 | 150 ± 20 | 230 ± 40 | 4.3 ± 0.7 |
| C2B | PM(+)PIP2 | 6.9 ± 0.1 | 54 ± 8 | 5 ± 1 | 200 ± 40 |
The observed association rate constant, kobs, is related to the intrinsic association and dissociation rate constants, kon and koff, via eq. 7 (if the interaction is modeled as a phase partitioning) or eq. 8 (if the interaction is modeled as a bimolecular binding equilibrium). The ratios of these rate constants for C2A interacting with both PM(+)PIP2 and PM(−)PIPx vesicles agree within a factor of two of the respective equilibrium constants (Kx and Kd,PIP2) measured using the IP6 competition assay (Tables 2-3). Kinetic measurements with the C2B domain suggest a ~50-fold weaker interaction relative to C2A with PM(+)PIP2, due to a ~15-fold faster koff and a ~3-fold slower kon (Figure 6, Table 3). Overall, these two complementary approaches demonstrate a high-affinity interaction between the C2A domain and vesicles containing PI(4,5)P2 and background anionic lipids.
4. Discussion
C2 domains are essential components of regulated exocytosis. Deletion of both C2 domains from granuphilin has been shown to produce localization in the cytosol or on vesicles rather than on the plasma membrane, as well as reduced vesicle–plasma membrane docking (Galvez-Santisteban et al., 2012; Tsuboi and Fukuda, 2006), suggesting that these domains play a key role in granuphilin plasma membrane targeting. Here we report the following new observations for C2 domain interaction with artificial vesicles composed of physiological lipid mixtures: (i) granuphilin C2A binds strongly to vesicles containing PI(4,5)P2 or PI(3,4,5)P3 and the background anionic lipids PS and PI, with apparent PIPx affinities in the 2-5 nM range; (ii) C2A binds ~40-fold less strongly to otherwise identical vesicles lacking PIPx; (iii) C2B interacts with vesicles containing PI(4,5)P2 with sub-micromolar affinity, but not with vesicles lacking PIPx; and (iv) IP6 binds with low micromolar affinity and competes with lipid binding for both domains.
The granuphilin C2A domain interacts nonspecifically with monoanionic background lipids including PS, PI, and PG, and binds with much stronger affinity to vesicles that include polyanionic lipids such as PI(4,5)P2 or PI(3,4,5)P3 (Table 2). Furthermore, this domain binds IP3 ~40-fold less tightly than the more anionic IP6. Granuphilin C2A possesses a net charge near +8 at neutral pH (Olsson et al., 2011), and these observations suggest that electrostatics may play a dominant role in IPx and PIPx binding. Our results indicate that this domain binds PI(4,5)P2 and PI(3,4,5)P3 with similar affinity, and a previous study using immobilized lipids also showed little PIPx selectivity for both C2A and C2B (Galvez-Santisteban et al., 2012). It has been suggested that promiscuous PIPx-binding domains are likely regulated by the most abundant PIPx species in their target membrane (Kavran et al., 1998). Granuphilin targets the plasma membrane, in part through interaction of its linker domain with Munc18/syntaxin (Tsuboi and Fukuda, 2006), and PI(4,5)P2 is the most abundant PIPx species in this compartment (Balla, 2005; Di Paolo and De Camilli, 2006; Lemmon, 2008). Our data suggest that C2 domain lipid binding may provide a significant contribution to the affinity of this protein's membrane interaction.
Available structural data for C2A are consistent with membrane binding via electrostatic interaction with PIPx and background anionic lipids via one or more binding sites. Poisson-Boltzmann mapping reveals a large positively charged surface on this domain (Figure 7). This surface includes a canonical PIPx binding motif, K(K/R)KTXXK(K/R), homologous to synaptotagmin 1 C2B (Figure 7, black arrow) (Galvez-Santisteban et al., 2012) as well as a cleft composed of basic residues in the β2-β3 and β6-β7 loops (Figure 7, green arrow). The latter cleft is equivalent to the Ca2+ binding site of synaptotagmin C2 domains, but granuphilin C2A contains only one of the five Ca2+-chelating Asp residues conserved among synaptotagmin isoforms (Bhalla et al., 2008). Further studies are needed to discern which site(s) on this domain are involved in binding lipids and/or IP6.
Figure 7. Electrostatic surface representation of granuphilin C2A.
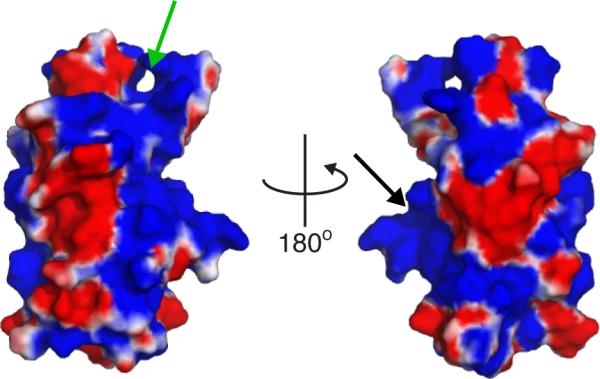
Surface electrostatic potentials are shown colored from -3 (red) to +3 (blue). Green arrow: putative binding site consisting of residues in the β2-β-3 and β6-β7 loops, which correspond to the Ca2+ binding loops of synaptotagmin C2 domains. Black arrow: putative binding site including residues K410, R411, and K412, homologous to the PIP2 binding site of synaptotagmin 1 C2B (Radhakrishnan et al., 2009). Potential map calculated using the APBS plugin for Pymol.
Equilibrium fluorescence data for C2A – IP6 binding and competition with lipids both fit well to simple hyperbolic functions, suggesting that IP6 binds this domain at a single site and thereby blocks lipid interaction. Our data are consistent with the presence of one or more lipid binding sites, provided a single bound IP6 is sufficient to displace the domain from the membrane. This assertion seems reasonable from an electrostatic standpoint, as binding of one IP6 molecule should neutralize the net charge of the C2A domain and disrupt nonspecific electrostatic interactions. However, membrane interaction involving multiple lipid ligands may not be represented appropriately by a bimolecular dissociation equilibrium constant (Kd,PIPx). Therefore, we also report mole fraction partition coefficients (Kx), which do not assume protein lipid binding stoichiometry, in addition to the more familiar Kd. Both measured values reflect high-affinity association of C2A with PM(+)PIP2 and PM(+)PIP3 vesicles, and correspond to G° values for binding of approximately -12 kcal/mol (Table 2).
Strong interaction is further supported by kinetic measurements of protein-lipid association and dissociation. The ratios of measured rate constants for C2A association with both PM(+)PIP2 and PM(−)PIPx vesicles are in close agreement with the affinities reported from equilibrium competition measurements. The single-exponential kinetic profiles indicate that membrane association and dissociation for both domains likely proceed via a single rate-limiting step, but do not report on stoichiometry of lipid interaction. Notably, the dissociation rate constant of 0.48 s−1 for C2A is extremely slow for a membrane-targeting C2 domain, consistent with its high affinity (Brandt et al., 2012; Corbin et al., 2007; Hui et al., 2005; Nalefski et al., 1997). Interestingly, when we attempted to measure membrane dissociation by rapidly adding high concentrations of IP6 (8 mM), an approach used with other PIPx-binding domains (Corbin et al., 2004; Knight and Falke, 2009), dissociation kinetics were biexponential and ~100-fold faster than those measured by the approach to equilibrium method (Supplementary data, Figure S3). The origins of this difference are currently under investigation; however, the good agreement between our IP6-independent kinetic measurements (Table 3) and equilibrium IP6 competition measurements (Table 2) suggests that high concentrations of IP6 may alter the kinetic pathway of dissociation without affecting the equilibrium thermodynamics of the system.
While affinity of C2B for PM(+)PIP2 vesicles cannot be calculated from our equilibrium competition measurements due to lack of a direct fluorescent reporter for C2B – IP6 binding, the equilibrium constant can be calculated from kinetic parameters. The measured koff/kon ratio yields an apparent Kd,PIP2 of 200 ± 40 nM, weaker than C2A but still significantly strong for a domain that is tethered in close proximity to target lipids (Shao et al., 2008). Previous studies have shown that splice variants of granuphilin with and without the C2B domain exhibit similar cellular behavior; however, the presence of this domain could conceivably modulate sensitivity to target lipids or IPx species that may regulate membrane binding in vivo (Torii et al., 2004; Tsuboi and Fukuda, 2006). Intriguingly, substitution of koff/kon in the place of Kd,PIP2 in eq. 4 yields an equilibrium constant for C2B – IP6 interaction of 1.9 ± 0.5 μM, remarkably similar to the KI of 1.8 ± 0.1 μM measured directly for C2A. Further studies are needed to clarify the potential role(s) of IP3 and IP6 binding to C2A and C2B in granuphilin function.
Finally, the competitive removal of granuphilin C2 domain-lipid binding by IP6 suggests a possible mechanism of granuphilin release from the plasma membrane during exocytosis. IP6 concentrations in insulin-secreting cells increase from ~40 μM to ~60 μM during exocytosis (Berggren and Barker, 2008; Larsson et al., 1997; Li et al., 1992). While our data suggest these concentrations are insufficient to remove C2A from membranes containing PI(4,5)P2, the presence of 40-60 μM IP6 [along with possibly other IPx species (Illies et al., 2007)] could potentially compete with C2B–membrane binding and C2A binding to PIPx-free membranes. Thus, it is possible that granuphilin C2 domains could be removed from their target lipids during insulin secretion by simultaneous increases in cellular IPx production and localized decreases in membrane PI(4,5)P2 content due to phospholipase C (PLC) activation (Efanov et al., 1997; Thore et al., 2007). The generation of IP3 by PLC would also contribute to this dissociation, although IP3 binds C2A less tightly than IP6. As noted previously, deletion of both C2 domains results in the loss of both plasma membrane localization and inhibitory activity, suggesting that interaction with Munc18/syntaxin alone is insufficient for proper localization (Galvez-Santisteban et al., 2012; Tsuboi and Fukuda, 2006). Thus, it is feasible that signaling-induced dissociation of these C2 domains from the plasma membrane could facilitate release from protein binding partners and removal of the “brake” on exocytosis. Clearly, further studies in secretory systems are needed in order to test these hypotheses and further define the roles of granuphilin C2 domains in secretory vesicle docking and fusion.
Supplementary Material
Highlights.
Both granuphilin C2 domains bind lipids present in the plasma membrane: PS and PIP2
C2A binds liposomes containing PIP2 with high affinity (2-5 nM)
C2B binds liposomes containing PIP2 with moderate affinity (~200 nM)
The signaling molecule IP6 competes with anionic lipids for binding both domains
Acknowledgements
The authors thank DNASU for providing a plasmid for expression of granuphilin C2A and Dr. John Corbin for critical reading of this manuscript. T.A.L. was supported by a fellowship through the CU Denver LABCOATS program (NIH 5R25GM083333 to Prof. Sonia Flores), a fellowship from the CU Denver Undergraduate Research Opportunity Program, and a scholarship from the Beta Beta Beta Research Foundation.
Abbreviations
- C2 domain
domain with homology to the second conserved region of protein kinase C
- PI(4,5)P2 or PIP2
phosphatidylinositol-(4,5)-bisphosphate
- PI(3,4,5)P3 or PIP3
phosphatidylinositol-(3,4,5)-trisphosphate
- PS
phosphatidylserine
- PI
phosphatidylinositol
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PG
phosphatidylglycerol
- SM
sphingomyelin
- CH
cholesterol
- dPE or dansyl-PE
dansyl phosphatidylethanolamine
- PIPx
phosphatidylinositol phosphate
- IP6
inositol-(1,2,3,4,5,6)-hexakisphosphate
- IP3
inositol-(1,4,5)-trisphosphate
- IPx
inositol polyphosphate
- Slp
synaptotagmin-like protein
- EDTA
ethylenediamine tetraacetic acid
- FRET
fluorescence resonance energy transfer
- SUV
small unilamellar vesicle
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J Cell Sci. 2005;118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- Berggren PO, Barker CJ. A key role for phosphorylated inositol compounds in pancreatic beta- cell stimulus-secretion coupling. Adv Enzyme Regul. 2008;48:276–294. doi: 10.1016/j.advenzreg.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Bhalla A, Chicka MC, Chapman ER. Analysis of the synaptotagmin family during reconstituted membrane fusion. Uncovering a class of inhibitory isoforms. J Biol Chem. 2008;283:21799–21807. doi: 10.1074/jbc.M709628200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierings R, Hellen N, Kiskin N, Knipe L, Fonseca AV, Patel B, Meli A, Rose M, Hannah MJ, Carter T. The interplay between the Rab27A effectors Slp4-a and MyRIP controls hormone- evoked Weibel-Palade body exocytosis. Blood. 2012;120:2757–2767. doi: 10.1182/blood-2012-05-429936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt DS, Coffman MD, Falke JJ, Knight JD. Hydrophobic contributions to the membrane docking of synaptotagmin 7 C2A domain: mechanistic contrast between isoforms 1 and 7. Biochemistry. 2012;51:7654–7664. doi: 10.1021/bi3007115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. Membrane-protein interactions in cell signaling and membrane trafficking. Annu Rev Biophys Biomol Struct. 2005;34:119–151. doi: 10.1146/annurev.biophys.33.110502.133337. [DOI] [PubMed] [Google Scholar]
- Coppola T, Frantz C, Perret-Menoud V, Gattesco S, Hirling H, Regazzi R. Pancreatic beta-cell protein granuphilin binds Rab3 and Munc-18 and controls exocytosis. Mol Biol Cell. 2002;13:1906–1915. doi: 10.1091/mbc.02-02-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbalan-Garcia S, Garcia-Garcia J, Rodriguez-Alfaro JA, Gomez-Fernandez JC. A new phosphatidylinositol 4,5-bisphosphate-binding site located in the C2 domain of protein kinase C . J Biol Chem. 2003;278:4972–4980. doi: 10.1074/jbc.M209385200. [DOI] [PubMed] [Google Scholar]
- Corbin JA, Dirkx RA, Falke JJ. GRP1 pleckstrin homology domain: Activation parameters and novel search mechanism for rare target lipid. Biochemistry. 2004;43:16161–16173. doi: 10.1021/bi049017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin JA, Evans JH, Landgraf KE, Falke JJ. Mechanism of specific membrane targeting by C2 domains: Localized pools of target lipids enhance Ca2+ affinity. Biochemistry. 2007;46:4322–4336. doi: 10.1021/bi062140c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- Efanov AM, Zaitsev SV, Berggren PO. Inositol hexakisphosphate stimulates non-Ca2+-mediated and primes Ca2+-mediated exocytosis of insulin by activation of protein kinase C. Proc Natl Acad Sci U S A. 1997;94:4435–4439. doi: 10.1073/pnas.94.9.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. Rab27 and its effectors in secretory granule exocytosis: a novel docking machinery composed of a Rab27.effector complex. Biochem Soc Trans. 2006;34:691–695. doi: 10.1042/BST0340691. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Mikoshiba K. Synaptotagmin-like protein 1-3: a novel family of C-terminal-type tandem C2 proteins. Biochem Biophys Res Commun. 2001;281:1226–1233. doi: 10.1006/bbrc.2001.4512. [DOI] [PubMed] [Google Scholar]
- Galvez-Santisteban M, Rodriguez-Fraticelli AE, Bryant DM, Vergarajauregui S, Yasuda T, Banon-Rodriguez I, Bernascone I, Datta A, Spivak N, Young K, Slim CL, Brakeman PR, Fukuda M, Mostov KE, Martin-Belmonte F. Synaptotagmin-like proteins control the formation of a single apical membrane domain in epithelial cells. Nat Cell Biol. 2012;14:838–849. doi: 10.1038/ncb2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi H, Mizutani S, Kasai K, Itohara S, Izumi T. Granuphilin molecularly docks insulin granules to the fusion machinery. J Cell Biol. 2005;171:99–109. doi: 10.1083/jcb.200505179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson N, Han W. Calcium-sensing beyond neurotransmitters: functions of synaptotagmins in neuroendocrine and endocrine secretion. Biosci Rep. 2009;29:245–259. doi: 10.1042/BSR20090031. [DOI] [PubMed] [Google Scholar]
- Hui E, Bai J, Wang P, Sugimori M, Llinas RR, Chapman ER. Three distinct kinetic groupings of the synaptotagmin family: candidate sensors for rapid and delayed exocytosis. Proc Natl Acad Sci U S A. 2005;102:5210–5214. doi: 10.1073/pnas.0500941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K, Yang SN, Barma DK, Falck JR, Saiardi A, Barker CJ, Berggren PO. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- Izumi T. Heterogeneous modes of insulin granule exocytosis: molecular determinants. Front Biosci. 2011;16:360–367. doi: 10.2741/3692. [DOI] [PubMed] [Google Scholar]
- Kavran JM, Klein DE, Lee A, Falasca M, Isakoff SJ, Skolnik EY, Lemmon MA. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J Biol Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- Knight JD, Falke JJ. Single-molecule fluorescence studies of a PH domain: new insights into the membrane docking reaction. Biophys J. 2009;96:566–582. doi: 10.1016/j.bpj.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf KE, Pilling C, Falke JJ. Molecular mechanism of an oncogenic mutation that alters membrane targeting: Glu17Lys modifies the PIP lipid specificity of the AKT1 PH domain. Biochemistry. 2008;47:12260–12269. doi: 10.1021/bi801683k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Barker CJ, Sjoholm A, Carlqvist H, Michell RH, Bertorello A, Nilsson T, Honkanen RE, Mayr GW, Zwiller J, Berggren PO. Inhibition of phosphatases and increased Ca2+ channel activity by inositol hexakisphosphate. Science. 1997;278:471–474. doi: 10.1126/science.278.5337.471. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Li G, Pralong WF, Pittet D, Mayr GW, Schlegel W, Wollheim CB. Inositol tetrakisphosphate isomers and elevation of cytosolic Ca2+ in vasopressin-stimulated insulin-secreting RINm5F cells. J Biol Chem. 1992;267:4349–4356. [PubMed] [Google Scholar]
- Martens S. Role of C2 domain proteins during synaptic vesicle exocytosis. Biochem Soc Trans. 2010;38:213–216. doi: 10.1042/BST0380213. [DOI] [PubMed] [Google Scholar]
- Murray D, Honig B. Electrostatic control of the membrane targeting of C2 domains. Mol Cell. 2002;9:145–154. doi: 10.1016/s1097-2765(01)00426-9. [DOI] [PubMed] [Google Scholar]
- Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 1996;5:2375–2390. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, Falke JJ. Use of fluorescence resonance energy transfer to monitor Ca2+-triggered membrane docking of C2 domains. Methods Mol Biol. 2002;172:295–303. doi: 10.1385/1-59259-183-3:295. [DOI] [PubMed] [Google Scholar]
- Nalefski EA, Slazas MM, Falke JJ. Ca2+-signaling cycle of a membrane-docking C2 domain. Biochemistry. 1997;36:12011–12018. doi: 10.1021/bi9717340. [DOI] [PubMed] [Google Scholar]
- Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J. Chem. Theory Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A, Stein A, Jahn R, Fasshauer D. The Ca2+ affinity of synaptotagmin 1 is markedly increased by a specific interaction of its C2B domain with phosphatidylinositol 4,5- bisphosphate. J Biol Chem. 2009;284:25749–25760. doi: 10.1074/jbc.M109.042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Novakovic VA, Head JF, Seaton BA, Gilbert GE. Crystal structure of lactadherin C2 domain at 1.7Å resolution with mutational and computational analyses of its membrane-binding motif. J Biol Chem. 2008;283:7230–7241. doi: 10.1074/jbc.M705195200. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. Synaptotagmins: why so many? J Biol Chem. 2002;277:7629–7632. doi: 10.1074/jbc.R100052200. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Kanematsu T, Misumi Y, Sakane F, Konishi H, Kikkawa U, Watanabe Y, Katan M, Hirata M. Distinct specificity in the binding of inositol phosphates by pleckstrin homology domains of pleckstrin, RAC-protein kinase, diacylglycerol kinase and a new 130 kDa protein. Biochim Biophys Acta. 1997;1359:275–285. doi: 10.1016/s0167-4889(97)00109-2. [DOI] [PubMed] [Google Scholar]
- Thore S, Wuttke A, Tengholm A. Rapid turnover of phosphatidylinositol-4,5-bisphosphate in insulin-secreting cells mediated by Ca2+ and the ATP-to-ADP ratio. Diabetes. 2007;56:818–826. doi: 10.2337/db06-0843. [DOI] [PubMed] [Google Scholar]
- Tomas A, Meda P, Regazzi R, Pessin JE, Halban PA. Munc 18-1 and granuphilin collaborate during insulin granule exocytosis. Traffic. 2008;9:813–832. doi: 10.1111/j.1600-0854.2008.00709.x. [DOI] [PubMed] [Google Scholar]
- Torii S, Takeuchi T, Nagamatsu S, Izumi T. Rab27 effector granuphilin promotes the plasma membrane targeting of insulin granules via interaction with syntaxin 1a. J Biol Chem. 2004;279:22532–22538. doi: 10.1074/jbc.M400600200. [DOI] [PubMed] [Google Scholar]
- Tsuboi T. Molecular mechanism of attachment process of dense-core vesicles to the plasma membrane in neuroendocrine cells. Neurosci Res. 2009;63:83–88. doi: 10.1016/j.neures.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Tsuboi T, Fukuda M. The Slp4-a linker domain controls exocytosis through interaction with Munc18-1.syntaxin-1a complex. Mol Biol Cell. 2006;17:2101–2112. doi: 10.1091/mbc.E05-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker DR. Lipid assembly into cell membranes. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. 5th Ed. Elsevier; Amsterdam: 2008. pp. 441–484. [Google Scholar]
- Wang H, Ishizaki R, Xu J, Kasai K, Kobayashi E, Gomi H, Izumi T. The Rab27a effector exophilin7 promotes fusion of secretory granules that have not been docked to the plasma membrane. Mol Biol Cell. 2013;24:319–330. doi: 10.1091/mbc.E12-04-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Takeuchi T, Yokota H, Izumi T. Novel rabphilin-3-like protein associates with insulin-containing granules in pancreatic beta cells. J Biol Chem. 1999;274:28542–28548. doi: 10.1074/jbc.274.40.28542. [DOI] [PubMed] [Google Scholar]
- White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- Yi Z, Yokota H, Torii S, Aoki T, Hosaka M, Zhao S, Takata K, Takeuchi T, Izumi T. The Rab27a/granuphilin complex regulates the exocytosis of insulin-containing dense-core granules. Mol Cell Biol. 2002;22:1858–1867. doi: 10.1128/MCB.22.6.1858-1867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Kasai K, Nagashima K, Torii S, Yokota-Hashimoto H, Okamoto K, Takeuchi T, Gomi H, Izumi T. Exophilin4/Slp2-a targets glucagon granules to the plasma membrane through unique Ca2+-inhibitory phospholipid-binding activity of the C2A domain. Mol Biol Cell. 2007;18:688–696. doi: 10.1091/mbc.E06-10-0914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.