Abstract
Background
Asthma is a complex disease with both genetic and environmental causes. Genome-wide association studies of asthma have mostly involved European populations and replication of positive associations has been inconsistent.
Objective
To identify asthma-associated genes in a large Latino population with genome-wide association analysis and admixture mapping.
Methods
Latino children with asthma (n = 1,893) and healthy controls (n = 1,881) were recruited from five sites in the United States: Puerto Rico, New York, Chicago, Houston, and the San Francisco Bay Area. Subjects were genotyped on an Affymetrix World Array IV chip. We performed genome-wide association and admixture mapping to identify asthma-associated loci.
Results
We identified a significant association between ancestry and asthma at 6p21 (lowest p-value: rs2523924, p < 5 × 10−6). This association replicates in a meta-analysis of the EVE Asthma Consortium (p = 0.01). Fine mapping of the region in this study and the EVE Asthma Consortium suggests an association between PSORS1C1 and asthma. We confirmed the strong allelic association between the 17q21 asthma in Latinos (IKZF3, lowest p-value: rs90792, OR: 0.67, 95% CI 0.61 – 0.75, p = 6 × 10−13) and replicated associations in several genes that had previously been associated with asthma in genome-wide association studies.
Conclusions
Admixture mapping and genome-wide association are complementary techniques that provide evidence for multiple asthma-associated loci in Latinos. Admixture mapping identifies a novel locus on 6p21 that replicates in a meta-analysis of several Latino populations, while genome-wide association confirms the previously identified locus on 17q21.
Keywords: Asthma, Latinos, Admixture Mapping, Genome-wide Association Study, Local Ancestry, 17q21, 6p21
Introduction
Asthma is the most common chronic disease among children. In the U.S., childhood asthma prevalence is highest among Puerto Ricans (18.4%), followed by Blacks (14.6%), Whites (8.2%) and Mexicans (4.8%).(1, 2) The discrepancy in asthma burden, as well as the paucity of studies of asthma in Latinos, has led the American Academy of Pediatrics to identify asthma among Latinos as an urgent priority for further research.(3)
Genetic variation between populations may account for some of these differences. Estimates of the heritability of asthma based on twin studies range from between 75%(4) and 92%(5). However, few genetic loci show consistent associations across studies.(6-8) Of the forty-eight genes reported to be associated with asthma in genome-wide studies,(9) only five have been identified in more than one GWAS.(10-16) This may be due to differences in study designs, differences in environment between study populations, and heterogeneity in asthma phenotypes.(17, 18) Our previous work has found that few genes associated with asthma replicate in Latino populations. Most that do replicate are consistent across Latino ethnic groups; however, five genes replicated in either Mexicans or Puerto Ricans (but not both) and showed significant heterogeneity in their association with asthma.(6) These observed differences in genetic variation may in part explain the discrepancy in asthma prevalence between the two groups.(19)
Most genome-wide association studies of asthma have been conducted in European and European American populations.(20) The recent identification of PYHIN1, a gene associated with asthma in individuals of African descent but not in Latinos or European Americans(10) highlights the importance of studying diverse populations in genetic association analyses. Moreover, studies in non-European populations may help uncover some of the “missing heritability” in complex diseases and may provide insights into racial/ethnic disparities in asthma prevalence and severity.
Admixture mapping is a technique that can help identify asthma-associated loci in populations of mixed ancestry (such as African Americans and Latinos).(21) In these admixed populations, it is possible to use dense genotyping to estimate ancestry at a locus-specific (local) level.(22, 23) If the allele frequency of risk variants is higher in one ancestral population than others, there will be a correlation between ancestry at that locus and disease. Comparing local ancestry in individuals with disease to control subjects will show a deviation from the expected distribution of ancestry. Like genome-wide association studies, admixture mapping provides an unbiased method to screen for disease-associated loci. However, because admixture is a relatively recent phenomenon, ancestry blocks are significantly larger than haplotype blocks,(24, 25) so admixture-mapping offers increased coverage of genetic variation and a lower multiple testing burden than GWAS, though this also means that admixture mapping peaks cover an area in the hundreds of kilobases, which makes narrowing down the causal variant more difficult. Admixture mapping has successfully identified disease-associated loci in breast(26) and prostate cancer,(27) renal disease,(28, 29) and white blood cell count,(30, 31) among others. Our own prior admixture mapping study demonstrated that admixture mapping peaks preferentially harbored asthma-associated genes,(32) while an admixture mapping study in African Americans and Puerto Ricans identified a locus on 6q14.1 that harbored a risk allele for asthma exclusively in individuals with local European admixture.(33)
In order to identify genetic risk factors for asthma we used genotype data from the Genes-environment & Admixture of Latino Americans (GALA II) study. GALA II is an ongoing, multicenter, case-control study to identify novel clinical and genetic risk factors associated with asthma and related phenotypes in Latino populations at five Sites in the United States: Puerto Rico, New York, Chicago, Houston, and the San Francisco Bay Area, which includes genome-wide genotype data on 3,774 participants. We hypothesize that by using a large population of Latino participants we would identify novel loci for asthma.
Methods
Recruitment and genotyping
Institutional review boards at UCSF and recruitment sites approved the study, and all participants/parents provided appropriate written assent/consent. Latino children were enrolled as a part of the ongoing GALA II case-control study. From July 2006 through June 2011, when genotyping began, a total of 4,045 children (1,976 participants with asthma and 2,065 healthy controls) were recruited from five centers (Chicago, Bronx, Houston, San Francisco Bay Area, and Puerto Rico) using a combination of community and clinic-based recruitment. Participants were eligible if they were 8-21 years of age and self-identified as Latino and had four Latino grandparents. Asthma cases were defined as participants with a history of physician diagnosed asthma and the presence of two or more symptoms of coughing, wheezing, or shortness of breath in the 2 years preceding enrollment. Participants were excluded if they reported any of the following: (1) 10 or more pack-years of smoking; (2) any smoking within 1 year of recruitment date; (3) history of lung diseases other than asthma (cases) or chronic illness (cases and controls); or (4) pregnancy in the third trimester. Details of recruitment are described elsewhere(34) and in the Online Repository.
Participants were genotyped at 818,154 SNPs on the Affymetrix Axiom World Array IV.(35) Details of individual and SNP quality control procedures are described in the Online Repository.
Ancestry was estimated using the program ADMIXTURE, with a three population model.(36) Details of the ancestral populations are described in the Online Repository. Local ancestry at all positions across the genome was estimated using the program LAMPLD(22), assuming three ancestral populations.
Statistical methods
We tested the associations between each SNP and asthma status using logistic regression, adjusting for potential confounders including global and local ancestry (for details, see the Online Repository), using a threshold for significance of 5 × 10−8. Admixture mapping was performed using a likelihood ratio test to compare a full model including two local ancestry estimates, and potential confounders to a restricted model excluding the local ancestry estimates. Adjustment for multiple comparisons was made using a permutation procedure (for details, see the Online Repository). Based on permutation testing, we used an adjusted significance threshold α* of 9 × 10−6, corresponding to 5278 effective independent comparisons. Analyses were performed using R (version 2.14.1). (37)
We searched for allelic associations within any admixture mapping peaks. A peak was defined as a contiguous region whose admixture mapping p-values were within 2 log10 orders of the most significant p-value. We examined the p-values for significance, adjusting for multiple comparisons using both a Bonferroni correction for the number of SNPs within the peak and the effective number of comparisons accounting for linkage disequilibrium between SNPs measured via a spectral decomposition.(38, 39) The former correction resulted in a significance threshold α* of 8 × 10−5 for 601 SNPs; however, accounting for the effective number of independent markers (~174 SNPs) within this region resulted in a less conservative significance threshold α* of 2 × 10−4.
Replication of admixture mapping results
We examined ancestry associations in each of the studies containing Latino participants in the EVE Asthma Consortium.(10) Details of the individual studies and estimation of ancestry are described elsewhere(32, 40-42) and in the Online Repository.
Admixture mapping of the region of interest was performed using logistic regression for case-control studies and the transmission disequilibrium test (TDT test) for trio-based studies. A meta-analysis of the five studies was performed using a fixed-effects model in PLINK.(43) A two-sided significance level of 0.05 was chosen.
Replication of prior GWAS and Admixture Mapping results
Using the NHGRI Catalog of Published GWAS studies,(9) we identified significant association(s) in prior genome-wide association studies for asthma (see Table E1 in the Online Repository). A two-sided significance level of 0.05 was chosen. We then examined the association between the reported SNPs and asthma in the GALA II study. SNPs that were not genotyped were imputed using Impute2.(44, 45) For the imputation, we used haplotypes from all available populations in the 1000 Genomes Project Phase I, integrated variant set release v3 as a reference panel, and we filtered our results to include only those variants imputed with an information score cutoff of > 0.3.(46, 47)
We also investigated whether previously reported admixture mapping results in asthma (32, 33) replicated in the current study by examining the admixture mapping likelihood ratio test results underlying the peaks described in the two studies in Torgerson et. al. in Latinos and African Americans.
Results
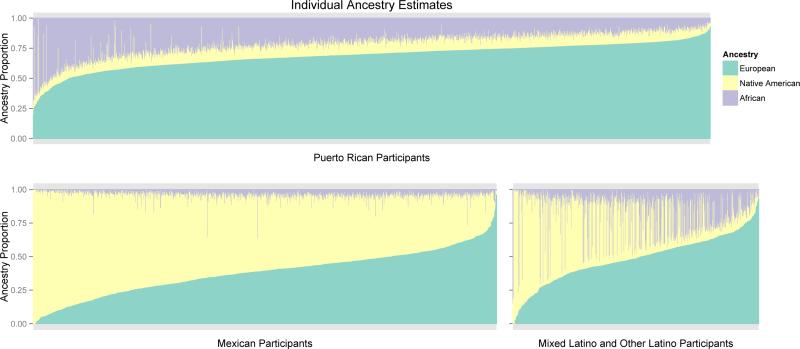
The characteristics of the GALA II study population are shown in Table I. Nearly half of the subjects were Puerto Rican (47%) followed by Mexican (31-35%), and Latinos from other ethnicities (15-18%). The percentage of males was higher in cases than the controls (55% vs. 44%), and the cases had higher total IgE on average than controls (234 vs. 79 IU/ml). Differences in genomic ancestry between cases (11% African ancestry [AFR] and 15% Native American ancestry [NAM], ) and controls (12% AFR and 15% NAM) were small, but statistically significant. Individual level ancestry proportions are shown in Figure 1 and highlight the variability between and within Latino ethnicities.
Table I.
Characteristics of GALA II participants
| Cases | Controls | p-value | |
|---|---|---|---|
| n | 1893 | 1881 | |
| Age (yrs) | 12 [10: 15] | 13 [11: 16] | < 0.001 |
| Sex (male) | 1046 (55%) | 822 (44%) | < 0.001 |
| Ethnicity | |||
| Mexican | 596 (31%) | 661 (35%) | |
| Puerto Rican | 894 (47%) | 894 (48%) | |
| Mixed | 62 (3%) | 44 (2%) | |
| Other | 341 (18%) | 282 (15%) | |
| Ancestry | |||
| African | 15.3% [5%: 22%] | 13.8% [4%: 20%] | < 0.001 |
| Native American | 30% [10%: 50%] | 33% [10%: 55%] | < 0.001 |
| European | 55% [42%: 70%] | 53% [39%: 70%] | 0.007 |
| Total IgE (IU/mL) | 232 [68: 650] | 80 [26: 248] | < 0.001 |
| History of atopy | 1515 (80.0%) | 606(32.2%) | < 0.001 |
| Lung Function | n = 422 | ||
| FEV1 (% predicted) | 81.3:100.6] | 98.8 [89.8:107.1] | < 0.001 |
| FEF25-75 (% predicted) | 82.4 [65.5:97.9] | 100.6 [86.1:113.3] | < 0.001 |
| FVC (% predicted) | 95.1 [85.2:105.2] | 98.1 [88.6:106.4] | 0.001 |
| FEV1/FVC (%) | 84.4 [80:90] | 89 [85:93] | < 0.001 |
| ΔFEV1 (% change) | 10.3 [4.8:13.7] | - | - |
For continuous variables, the median and interquartile range are displayed, and the differences between cases and controls were tested with a t-test. For categorical variables, the number and proportion of subjects in each category are displayed, and a Χ2 650 test was used. History of atopy was defined as self-report allergic rhinitis or eczema.
Figure 1.
Proportions of individual Native American, European, and African ancestry in the GALA II participants. Each bar represents one individual. There is substantial variability in individual ancestry both within and between Latino ethnic sub-groups.
Most of the cases have either severe (43%) or moderate persistent (28%) asthma, based on medication/severity and symptoms/control classification.(48) The remaining cases are split between mild intermittent (17%) and mild persistent (12%) asthma.
Admixture mapping
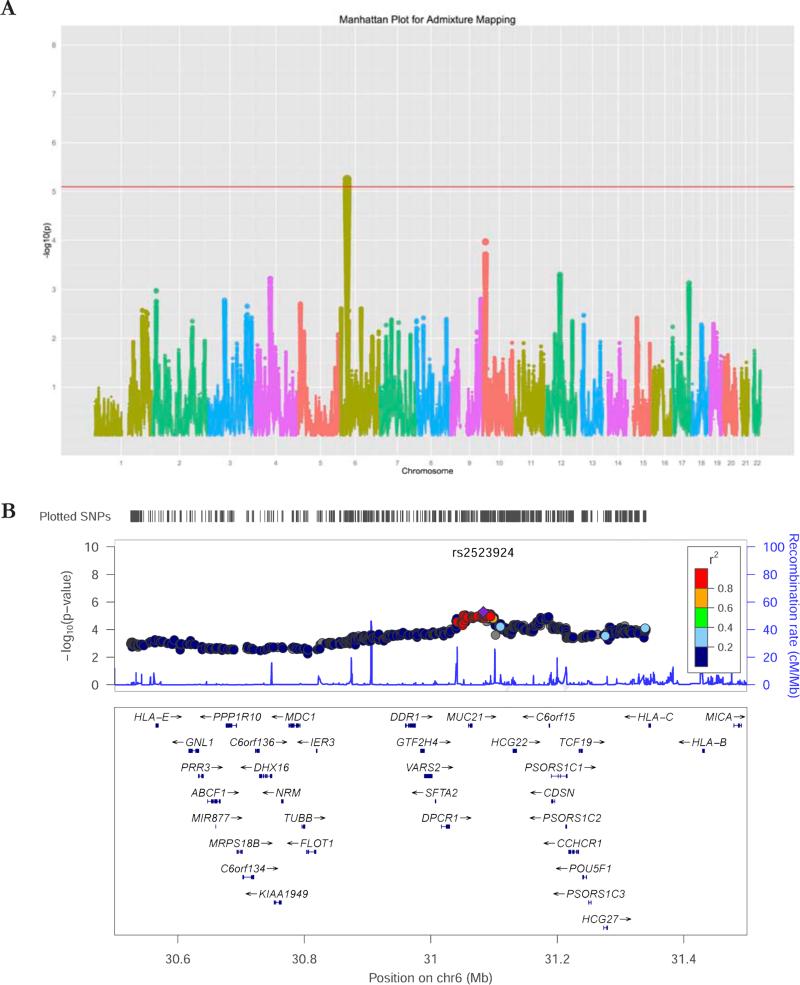
We performed an admixture-mapping study of asthma in 3,774 US children of Latino ethnicity (Figure 2 and Figure E1 in the Online Repository). Our most significant admixture-mapping association was on chromosome 6p21 near the HLA region. It was centered on MUC22, where local ancestry was significantly associated with asthma (p < 5 × 10−6). The 0.05 genome-wide threshold established by permutation testing corresponds to a nominal p-value of 9 × 10−6. The permutation-corrected p-value for the 6p21 peak was 0.02 adjusting for multiple testing. As expected with admixture mapping studies, the genome-wide significant region is broad, encompassing a genomic region of approximately 350 kb (Figure 2B). Because of this, there is slight genomic inflation (λ = 1.1, figure E-A in the Online Repository). However, if chromosome 6 is excluded from the analysis, there is no genomic inflation in the remainder of the genome (λ = 0.99, Figure E1B in the Online Repository). Within this genomic region, Native American ancestry is associated with lower odds of asthma (OR 0.87, 95% CI 0.82 – 0.93, p = 1. 7 × 10−5).
Figure 2.
Admixture mapping results. A. Manhattan plot of the admixture mapping study, showing a significant peak in chromosome 6. The red line represents the fifth percentile of the lowest p3 value in 10,000 permutations. B. Locus-zoom plot of the admixture mapping peak, which is centered in the MUC21 and MUC22 gene cluster, upstream of HLA-B and HLA-C genes.
We performed a meta-analysis of the association between Native American ancestry and asthma at this locus among five studies of Hispanic/Latino ethnicity from the EVE Asthma Consortium. Our initial results replicated in the same direction (OR 0.88, 95% CI 0.79 – 0.98, p = 0.02). A forest plot of the studies is shown in Figure 3.
Figure 3.
Forest plot of the meta-analysis of the effect of Native American ancestry on asthma in the Latino studies within the EVE consortium. The odds ratio and [95% confidence interval] are listed next to each study.
We then searched for allelic associations within the peak, which we defined as the contiguous region whose admixture mapping p-values were within two log-orders of the most significant admixture association. This spanned a 354 kb region that included 13 genes and one open reading frame. We tested for allelic associations for the 601 SNPs identified within that region; the most significant association was between asthma and KG_6_31200283 (rs114235219), a SNP in PSORS1C1 (OR 0.61, p = 0.002). Given that we tested for 601 allelic associations within the peak, the association did not meet the threshold for significance after adjusting for multiple comparisons, either using the within-peak Bonferroni correction (α* = 8.3 × 10−5), or adjusting for the effective number of comparisons accounting for linkage disequilibrium between SNPs (α* = 2.9 × 10−4; effective number of independent marker loci: 174).(38, 39)
We also tested for allelic associations within the peak in the EVE Asthma Consortium data.(10) There were 399 SNPs within the region in the EVE Asthma consortium dataset. Unfortunately, rs114235219 was not analyzed in the EVE Asthma consortium dataset. The most significant allelic association within the peak was rs3132550, which was also within PSORS1C1 (p = 3 × 10−4). There were 307 SNPs common to both the GALA II and EVE datasets under the admixture-mapping peak. Two of these SNPs were significant in both the GALA II and EVE Asthma consortium datasets. Both SNPs were in the PSORS1C1 gene, rs3094663 (GALA II OR 0.85, 95% CI 0.77 – 0.95, p = 0.003; EVE p-value 0.01) and rs3130564 (GALA II OR 1.30, 95% CI 1.07 – 1.58, p = 0.009; EVE p-value 0.02). In both cases, the direction of the association was consistent across the two studies (the “T” allele in rs2094663 and the “C” allele in rs3130564 were associated with increased asthma risk in both studies). A locus-zoom plot of this region is given in Figure E2 in the online repository.
Examining previously reported admixture-mapping peaks, (32, 33) we found that the peak reported on cytoband 8q12 in both Puerto Ricans and Mexicans replicated in this study (LR p-value 0.03); Native American ancestry was associated with increased risk of asthma (OR 1.08, 95% CI 1.02 – 1.15, p = 0.01) as in Puerto Ricans inthe original study. There was no association with African ancestry at this locus (p = 0.8). We were unable to replicate the association between ancestry and asthma in the admixture-mapping peak in cytoband 6q15 found in Mexicans (LR p-value 0.3) or the association found in cytoband 6q14 in African Americans (p = 0.2). Of the 55 additional non-overlapping loci identified by Torgerson et al as being enriched for asthma peaks, we found 13 additional loci that were nominally associated (LR p-value < 0.05) with asthma in this study. The results are summarized in Table E3 in the Online Repository.
Asthma GWAS
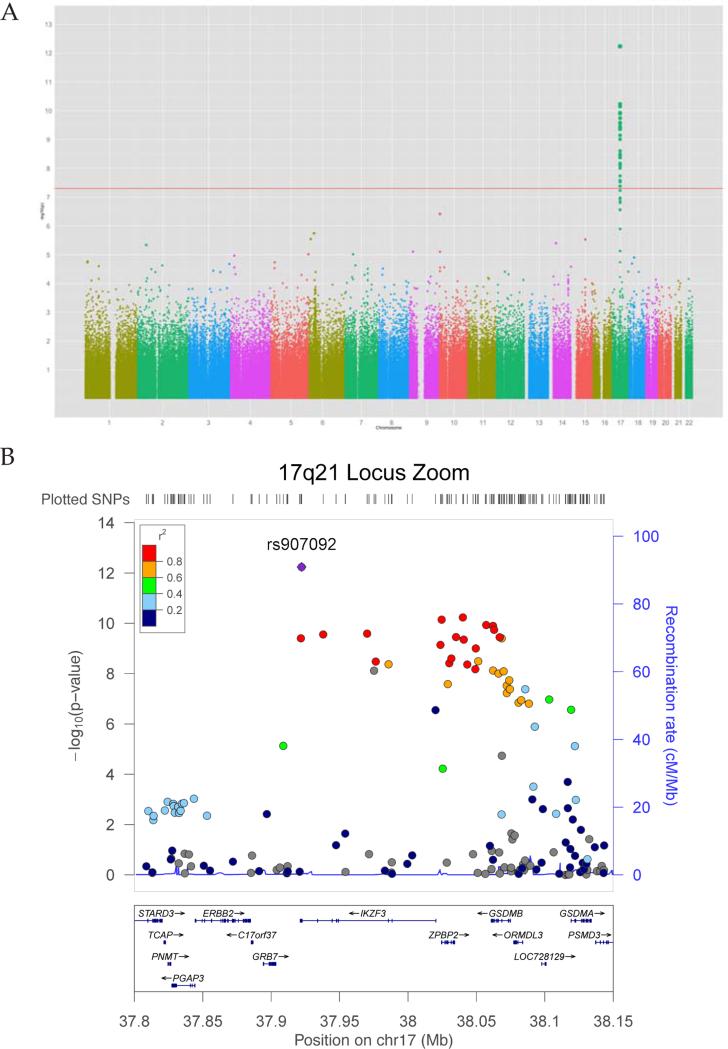
Results of our genome-wide association study are summarized in Figure 4 and Table II. The genomic inflation factor (λ) for allelic association testing in this study is 1.03 (see Figure E3 in the Online Repository). Thirty-one SNPs, all in the 17q21 region, were significantly associated with asthma at a genome-wide corrected significance level of <5 × 10−8. A locus zoom of the region is shown in Figure 4B. The most significantly associated SNP (rs907092) is a synonymous coding mutation in the IKZF3 gene, which lies within a region of broad linkage disequilibrium that also includes ORMDL3, GSDMB, and ZPBP2. The minor allele of the IKZF3 SNP conferred protection against asthma with an odds ratio of 0.67 (95% CI 0.61 – 0.75, p = 6 × 10−13). Conditioning on rs907092 removes virtually all associations within the 17q21 region, with only rs12450091, a non-synonymous coding mutation in GSDMB maintaining nominal significance (OR = 1. 23, 95% CI 1. 07 – 1. 42, p = 0.005). Additionally, six regions had genotyped SNPS that showed a suggestive association with asthma at p < 5 × 10–6 (Table IIa). Examination of SNPs in each of these six regions in the EVE Asthma Consortium showed no significant associations with asthma in either the combined study (p ≥ 0.05 for all SNPs), or in the subset of studies with individuals of Latino ethnicity (p ≥ 0.05). Two additional regions showed suggestive associations with asthma at p < 10−6 using data imputed with 1000 Genomes (Table IIb); neither of these replicated in EVE.
Figure 4.
Genome-wide association study results. A. Manhattan plot of the GWAS, showing a genome-wide significant peak at chromosome 17q21. B. Locus-zoom plot of the 17q21 peak, which shows a broad area of linkage disequilibrium, with the most significant finding centered on the IKZF3 gene.
Table II.
Significant and suggestive allelic associations with asthma (top: genotyped SNPs; bottom: imputed SNPs)
| SNP | Chromosome | Gene | MAF | Odds Ratio | CI | p-value |
|---|---|---|---|---|---|---|
| rs12328045 | 2 | intergenic | 0.221 | 1.33 | 1.18 - 1.50 | 4.6 × 10−6 |
| rs4566922 | 6 | intergenic | intergenic 0.440 | 0.79 | 0.72 - 0.87 | 2.9 × 10−6 |
| rs3804327 | 6 | DCDC2 | 0.160 | 1.37 | 1.20 - 1.56 | 1.8 × 10−6 |
| rs7086533 | 10 | intergenic | 0.117 | 1.51 | 1.29 - 1.77 | 3.8 × 10−7 |
| KG14-33639557 | 14 | NPAS3 | 0.014 | 0.36 | 0.23 - 0.56 | 4.0 × 10−6 |
| rs12594238 | 15 | RAB11A | 0.169 | 0.73 | 0.64 - 0.83 | 3.0 × 10−6 |
| rs907092 | 17 | IKZF3 | 0.304 | 0.67 | 0.61 - 0.75 | 5.7 × 10−13 |
| SNP | Chromosome | Gene | Info Score | MAF | Odds Ratio | CI | p-value |
|---|---|---|---|---|---|---|---|
| rs148728975 | 1 | VPS13D | 0.890 | 0.032 | 0.47 | 0.35 - 0.63 | 7.0 × 10−7 |
| rs10211025 | 2 | Intergenic | 0.992 | 0.214 | 0.73 | 0.65 - 0.83 | 9.7 × 10−7 |
| rs3936215 | 9 | BNC2 | 0.996 | 0.263 | 1.38 | 1.21 - 1.56 | 8.6 × 10−7 |
| rs11252191 | 10 | Intergenic | 0.962 | 0.121 | 1.54 | 1.31 - 1.80 | 1.3 × 10−7 |
| rs182722816 | 12 | BTBD11 | 0.708 | 0.011 | 5.70 | 2.86 - 11.34 | 7.0 × 10−7 |
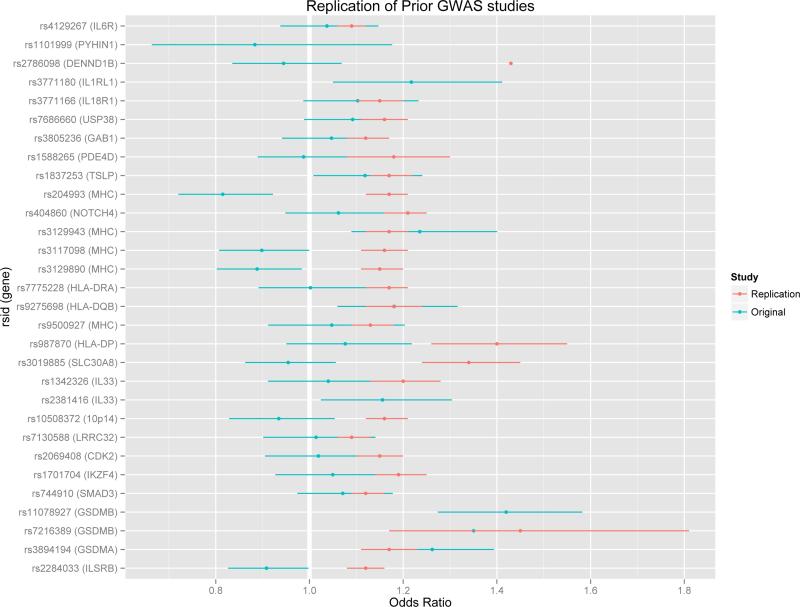
We also examined regions that had previously been associated with asthma in previously reported genome-wide association studies (Table E1 in the Online Repository). We identified twenty studies reporting results of genome-wide associations in asthma, including two that were meta-analyses of other reported studies. Twelve studies reported one or more associations, though only seven of these met strict genome-wide significance with p<5 × 10−8. Thirty total SNPs were reported to be significant across the studies at genome-wide significance levels in at least one study. Twelve of these SNPs replicated in the current study (Table E4, in bold, Figure 5), though in four cases the direction of the observed effect was in the opposite direction than that of the original study (denoted with a minus sign). In five cases, the direction of the observed effect was in the same direction as that of the original study (denoted with an asterisk), and in three cases the original study did not report an odds ratio.
Figure 5.
Replication of previous genome-wide findings in the GALA II study. When provided, the original study's effect estimate and confidence interval are given in red, while the effect estimate and confidence interval in the present study are shown in teal. One study, including three SNPs reported only a summary p-value.(10) A second study, including one SNP, reported an odds ratio, but not confidence interval.(56) + Replication was in the same direction as the original study. 3 Replication was in the opposite direction as the original study. ? The direction of replication could not be assessed. MHC major histocompatibility complex region.
Discussion
We used the complimentary techniques of admixture mapping and allelic genome-wide association analysis to identify novel asthma-associated loci and confirm prior genetic associations. In the admixture mapping study, we found a genome-wide significant association between asthma and a peak at 6p21. This peak was centered on MUC22 (likelihood ratio p < 5 × 10−6; permutation-adjusted p-value 0.02). Native American ancestry in this genomic region was associated with decreased risk of asthma (OR 0.87, 95% CI 0.82 – 0.93, p = 1. 7 × 10−5). This association was replicated in a meta-analysis of five studies of Latinos in the EVE Asthma Consortium, where Native American ancestry was associated with a nearly identical protective effect (OR = 0.88, 95% CI: 0.79 – 0.98).
Admixture mapping and genome-wide association analysis depend on tagging causal variants through local ancestry blocks and haplotype blocks, respectively. Because admixture mapping relies on relatively large ancestry blocks to identify disease-associated regions of the genome, there are fewer effective comparisons and therefore a lower statistical penalty for multiple comparisons. In this case, permutation procedures lead to a significance threshold of 9×10−6, suggesting that there are approximately 5,000 to 6,000 effective comparisons. In contrast, the traditional significance threshold for genome-wide association studies is 5×10−8, which is equivalent to approximately 1,000,000 haplotype blocks. While the reduced multiple comparison penalty associated with admixture mapping make it an attractive technique for identifying regions associated with a disease, the drawback of the technique is that it results in a larger genomic region of interest, as in our study.
In the current study, the region was narrowed down to a 350 kB region centered on a cluster of mucin producing genes: MUC21, MUC22, PBMUCL2 (see Figure 2), in the major histocompatibility complex region on chromosome 6. The genes in the mucin family are intriguing candidates for involvement in differential asthma susceptibility. These highly glycosylated macromolecules are broadly expressed in the airway epithelium, where they can be secreted or tethered to airway epithelial membranes, and play a critical role in innate immune defense.(49) The overproduction of mucin has been associated with chronic airway disease, including asthma.(50) A genetic polymorphism in a promoter in MUC5B has also been strongly associated with idiopathic pulmonary fibrosis, suggesting a link between mucin genes and lung disease.(51) MUC22 itself was identified by searching for genes in the region associated with diffuse panbronchiolitis,(52) which, like asthma, is an obstructive lung disease. The same study found that MUC22 was expressed in the lung and in human bronchial epithelial cells differentiated at an air-liquid interface.
Although MUC22 is an intriguing candidate for association with asthma, we cannot exclude other genes under the admixture mapping peak, including HLA-C and HLA-B. HLA-B has been evaluated as a candidate gene for asthma in two studies with mixed results. In a study of Croatian children with atopic asthma, the HLA-B8 antigen was associated with asthma,(53) while an earlier study in Greek children with allergic asthma found no specific HLA-B alleles associated with asthma.(54)
We attempted to narrow the region of interest down by searching for allelic associations under the admixture-mapping peak. We found no allelic associations that met region-wide significance criteria (α* = 8.3×10−5, adjusting for 601 SNPs evaluated) in the GALA II study. However, the most significant allelic association within the region in GALA II, (rs114235219, KG_ 6_31200283, p = 0.002), and the most significant SNP in the EVE asthma consortium study (rs3132550, p = 3 × 10−4) were both located on PSORS1C1, a psoriasis susceptibility candidate,(55) as were the two SNPs that achieved nominal significance in both studies (rs3094663 and rs3130564). PSORS1C1 was identified as a risk locus for systemic sclerosis,(56) Stevens-Johnson Syndrome,(57, 58), Crohn's disease,(50) COPD biomarkers,(59) and Behçet's disease(60) in GWAS. A recent study showed that SNPs associated with both asthma and autoimmune diseases have opposite effects on immunopathogenesis.(61)
Although the evidence suggests that variation within PSCORS1C1 is associated with asthma, it may be necessary to sequence the region to identify the causal variant or variants. The lack of a definitive allelic association within 6p21 may be due to one of several reasons. It is possible that the genotyping platform includes SNPs that poorly tag the causal variants. Although the World Array IV was specifically designed to maximize coverage in Latinos, its coverage of variation in Mexicans in Los Angeles (MXL) from the 1000 Genomes project is still imperfect, especially for uncommon and rare variants. Approximately 80% of SNPs with a minor allele frequency between 0.05 and 0.1 were tagged with an r2 > 0.7, and for rarer alleles, coverage was less than 70%. The Eve Asthma Consortium study was genotyped on a variety of older platforms, which may not have captured variation in non-European populations. Furthermore, the MHC region is highly variable, which increases the probability that the admixture-mapping signal was driven by untyped variation. Under these circumstances, admixture mapping has an advantage over allelic and imputed associations because the breath of ancestry blocks allows for nearly complete coverage across the genome. As long as the allele frequency of the causal variant or variants differs between the ancestral populations, an admixture mapping association may be detectable even if the untyped causal SNP is in poor linkage disequilibrium with genotyped SNPs. Admixture mapping can also find associations driven by rare variants unlikely to be identified through conventional means. Finally, the lack of allelic association could also be due to a number of variants with small effects clustered in the region, which would not be surprising in the MHC region given its highly variable nature.
We were also able to replicate the admixture-mapping association between 8q12 and asthma, as well as a substantial minority (13 of 55, See Table E3 in the Online Repository) of the additional admixture-mapping peaks previously reported as being enriched for asthma genes.(32) This provides additional evidence that admixture mapping is able to uncover novel disease-associated regions.
Our genome-wide association analysis replicated the previously described association between the 17q21 region and asthma. Our reported odds ratio of 0.67 (95% CI 0.61 - 0.75) is consistent with the previously reported effect magnitudes at this site. Although our most statistically significant SNP was in IKZF3 (approximately 50 kb upstream from reported associations in ORMDL3 and GSDMB),(15) there is a high degree of linkage disequilibrium in this region, and our statistically significant results extended across all 3 genes in the region. Moreover, functional work by Verlaan et al. has shown that genetic variants in this region are associated with domain-wide cis-regulatory effects on expression of multiple genes in the region, likely through their effect on chromatin states.(62)
Our evaluation of the previous genome-wide association studies described in table E1 in the online repository found that nearly half of the associations with asthma nominally replicated in our study. This contrasts with our earlier evaluation of principally candidate gene studies, where we found that only 17 genes contained SNPs that replicated in at least one Latino population out of 124 genes that had been previously associated with asthma.(6) This may be partially explained by the fact that this study, which included nearly 4000 individuals, was better powered to detect associations than our prior study, which included nearly 700 trios. However, it also seems likely that GWAS findings may reflect stronger effect sizes than candidate gene studies, may be less likely to reflect Type I error, and may be more likely to confer universal risk across ethnic groups.
Our prior study also showed that for a small number of genes, there was an ethnic-specific replication pattern with significant statistical heterogeneity between Mexicans and Puerto Ricans. In this study, we found no genes with a statistically significant gene by ethnicity interaction. This suggests that associations with asthma uncovered through genome-wide studies are more universal than those from candidate gene studies, which may also explain why they are easier to replicate.
We do note that in four out of 25 of the associations for which the original direction of effect was reported, the direction of replication was in the opposite direction. Such “flip flop” associations have been previously reported in an asthma GWAS where a protective minor allele in European subjects was associated with increased risk in African American participants.(63) Such a finding was ascribed to differences in the underlying genomic architecture between the two study populations, as could occur when the genotyped SNP was correlated through linkage disequilibrium or interaction with a causal variant but the direction of the correlation varies between populations, resulting in an observed SNP that tags opposite alleles of the causal variant.(64, 65)
It should be noted that our case definition of asthma was based on physician diagnosis and self-report of symptoms. Although our cases, on average had evidence of airway obstruction, with significantly lower FEV1 and FEF25-75 than controls (Table I), and many had significant response to albuterol, participants who did not have evidence of bronchodilator response were not required to undergo methacholine provocation. The lack of objective confirmation of the asthma diagnosis may have resulted in some misclassification of some participants. Such misclassification bias, which would be expected to be non-differential with respect to the predictor, biases the results towards the null, understating our results. Nonetheless, our finding of a strong association at the 17q21 region with comparable if not larger effect magnitudes than the original study suggests that this effect was likely to have been small.
In summary, our genome-wide association study confirmed significant associations at 17q21 region in Latinos, and our admixture mapping study identified a novel asthma-associated locus at 6p21 centered on the MUC22 gene near the MHC region. Fine mapping in this region points to the involvement of the gene PSORS1C1 in the pathogenesis of asthma. This confirms the advantages of performing admixture mapping studies in diverse populations to identify novel asthma-associated genes.(66) Follow up of results from admixture mapping and GWAS with sequencing and functional studies in minority populations carried out in large multiethnic cohorts could help to relate the significance of such findings to disparities in asthma prevalence and severity.
Supplementary Material
Capsule Summary.
In this study, we use the complementary techniques of admixture mapping and genome-wide association in Latinos to identify novel asthma-associated loci.
Key Messages.
Admixture mapping of asthma in nearly 4000 Latinos identifies a novel asthma locus on 6p21 that replicates in a meta-analysis of several additional studies of Latinos
Fine mapping of this region suggests involvement of the PSORS1C1 gene with asthma.
Genome-wide association of asthma confirms the previously identified asthma locus on 17q21 and replicates several genes that had previously been associated with asthma in GWA studies.
Acknowledgements
The authors acknowledge the families and patients for their participation and thank the numerous health care providers and community clinics for their support and participation in GALA II. In particular, the authors thank study coordinator Sandra Salazar; the recruiters who obtained the data: Duanny Alva, MD, Gaby Ayala-Rodriguez, Lisa Caine, Elizabeth Castellanos, Jaime Colon, Denise DeJesus, Blanca Lopez , Brenda Lopez, MD, Louis Martos, Vivian Medina, Juana Olivo, Mario Peralta, Esther Pomares, MD, Jihan Quraishi, Johanna Rodriguez, Shahdad Saeedi, Dean Soto, Ana Taveras. The authors would also like to acknowledge the EVE Asthma Consortium for providing summary data for replication of suggestive associations, including the following Eve Principal Investigators: Carole Ober, PhD, Dan Nicolae, PhD, Scott Weiss, MD, Benjamin Raby, MD, Keoki Williams, MD, MPH, Kathleen Barnes, PhD, Deborah Meyers, PhD, Eugene Bleecker, MD, and Jim Gauderman, PhD. The authors would like to thank Amy Markowitch, JD for her editorial comments and assistance preparing this manuscript. Some computations in this manuscript were performed using the UCSF Biostatistics High Performance Computing System.
Sources of funding:
This research was supported in part by National Institutes of Health (R01 ES015794, R01 HL088133, M01 RR000083, R01 HL078885, R01 HL104608, P60 MD006902, U19 AI077439, M01 RR00188); ARRA grant RC2 HL101651; EGB was supported in part through grants from the Flight Attendant Medical Research Institute (FAMRI), the Sandler Foundation, the American Asthma Foundation and NIH (K23 HL004464); JMG was supported in part by NIH Training Grant T32 (GM007546) and career development awards from the NHLBI K23 (K23HL111636) and NCATS KL2 (KL-TR000143) as well as the Hewett Fellowship; CRG was supported in part by NIH Training Grant T32 (GM007175) and the UCSF Chancellor's Research Fellowship and Dissertation Year Fellowship; RK was supported with a career development award from the NHLBI (K23HL093023); HJF was supported in part by the GCRC (RR00188); PCA was supported in part by the Ernest S. Bazley Grant. SJL was supported in part by the Division of Intramural Research, National Institute of Environmental Health Sciences (ZIA ES49019). This publication was supported by various institutes within the National Institutes of Health. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Abbreviations
- CARE
Childhood Asthma Research and Education network
- CHS
Children's Health Study
- FEV1
Forced expiratory volume in 1 sec
- FVC
Forced vital capacity
- GALA I
Genetics of Asthma in Latino Americans
- GALA II
Genes-environments & Admixture in Latino Americans
- GWAS
Genome-wide association study
- IgE
Immunoglobulin E
- MCCAS
Mexico City Childhood Asthma Study
- SNP
Single nucleotide polymorphism
- TDT
transmission disequilibrium test
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moorman JE, Rudd RA, Johnson CA, King M, Minor P, Bailey C, et al. National surveillance for asthma--United States, 198032004. MMWR Surveillance summaries : Morbidity and mortality weekly report Surveillance summaries / CDC. 2007;56(8):1–54. Epub 2007/10/20. [PubMed] [Google Scholar]
- 2.Moorman JE, Zahran H, Truman BI, Molla MT. Current asthma prevalence 3 United States, 200632008. MMWR Surveillance summaries : Morbidity and mortality weekly report Surveillance summaries / CDC. 2011;60(Suppl):84–6. Epub 2011/03/25. [PubMed] [Google Scholar]
- 3.Flores G, Fuentes-Afflick E, Barbot O, Carter-Pokras O, Claudio L, Lara M, et al. The health of Latino children: urgent priorities, unanswered questions, and a research agenda. JAMA. 2002;288(1):82390. doi: 10.1001/jama.288.1.82. Epub 2002/07/02. [DOI] [PubMed] [Google Scholar]
- 4.Willemsen G, van Beijsterveldt T, van Baal C, Postma D, Boomsma D. Heritability of self3 reported asthma and allergy: a study in adult Dutch twins, siblings and parents. Twin Res Hum Genet. 2008;11:132–42. doi: 10.1375/twin.11.2.132. [DOI] [PubMed] [Google Scholar]
- 5.Fagnani C, Annesi-Maesano I, Brescianini S. Heritability and shared genetic effects of asthma and hay fever: an Italian study of young twins. Twin Res Hum Genet. 2008;11:121–31. doi: 10.1375/twin.11.2.121. al. e. [DOI] [PubMed] [Google Scholar]
- 6.Galanter JM, Torgerson D, Gignoux CR, Sen S, Roth LA, Via M, et al. Cosmopolitan and ethnic-specific replication of genetic risk factors for asthma in 2 Latino populations. J Allergy Clin Immunol. 2011;128(1):37–43. e12. doi: 10.1016/j.jaci.2011.03.050. Epub 2011/05/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers AJ, Raby BA, Lasky-Su JA, Murphy A, Lazarus R, Klanderman BJ, et al. Assessing the reproducibility of asthma candidate gene associations, using genome-wide data. Am J Respir Crit Care Med. 2009;179(12):1084–90. doi: 10.1164/rccm.200812-1860OC. Epub 2009/03/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu H, Romieu I, Shi M, Hancock DB, Li H, Sienra-Monge JJ, et al. Evaluation of candidate genes in a genome-wide association study of childhood asthma in Mexicans. J Allergy Clin Immunol. 2010;125(2):321–7. e13. doi: 10.1016/j.jaci.2009.09.007. Epub 2009/11/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9362–7. doi: 10.1073/pnas.0903103106. Epub 2009/05/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torgerson D, Ampleford E, Chiu G, Gauderman W, Gignoux C, Graves P, et al. Meta-analysis of genome-wide association studies of asthma in ethnically diverse North American Populations. Nature Genetics. 2011;43(9):887–92. doi: 10.1038/ng.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramasamy A, Kuokkanen M, Vedantam S, Gajdos ZK, Couto Alves A, Lyon HN, et al. Genome-wide association studies of asthma in population-based cohorts confirm known and suggested loci and identify an additional association near HLA. PloS one. 2012;7(9):e44008. doi: 10.1371/journal.pone.0044008. Epub 2012/10/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan YI, Shrine NR, Soler Artigas M, Wain LV, Blakey JD, Moffatt MF, et al. Genome-wide association study to identify genetic determinants of severe asthma. Thorax. 2012;67(9):762–8. doi: 10.1136/thoraxjnl-2011-201262. Epub 2012/05/09. [DOI] [PubMed] [Google Scholar]
- 13.Hirota T, Takahashi A, Kubo M, Tsunoda T, Tomita K, Doi S, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat Genet. 2011;43(9):893–6. doi: 10.1038/ng.887. Epub 2011/08/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffatt M, Gut I, Demenais F, Strachan D, Bouzigon E, Heath S, et al. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–21. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffatt M, Kabesch M, Liang L, Dixon A, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira MA, McRae AF, Medland SE, Nyholt DR, Gordon SD, Wright MJ, et al. Association between ORMDL3, IL-RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur J Hum Genet. 2011;19(4):458–64. doi: 10.1038/ejhg.2010.191. Epub 2010/12/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore WC, Meyers DA, Wenzel SE, Teague WG, Li H, Li X, et al. Identification of asthma phenotypes using cluster analysis in the Severe Asthma Research Program. Am J Respir Crit Care Med. 2010;181(4):315–23. doi: 10.1164/rccm.200906-0896OC. Epub 2009/11/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodruff PG, Modrek B, Choy DF, Jia G, Abbas AR, Ellwanger A, et al. T-helper type 2-driven inflammation defines major subphenotypes of asthma. Am J Respir Crit Care Med. 2009;180(5):388–95. doi: 10.1164/rccm.200903-0392OC. Epub 2009/06/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salari K, Burchard EG. Latino populations: a unique opportunity for epidemiological research of asthma. Paediatric and perinatal epidemiology. 2007;21(Suppl 3):15–22. doi: 10.1111/j.1365-3016.2007.00880.x. Epub 2007/11/21. [DOI] [PubMed] [Google Scholar]
- 20.Bustamante CD, Burchard EG, De la Vega FM. Genomics for the world. Nature. 2011;475(7355):163–5. doi: 10.1038/475163a. Epub 2011/07/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Winkler CA, Nelson GW, Smith MW. Admixture mapping comes of age. Annual review of genomics and human genetics. 2010;11:65–89. doi: 10.1146/annurev-genom-082509-141523. Epub 2010/07/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baran Y, Pasaniuc B, Sankararaman S, Torgerson DG, Gignoux C, Eng C, et al. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics. 2012;28(10):1359–67. doi: 10.1093/bioinformatics/bts144. Epub 2012/04/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasaniuc B, Sankararaman S, Kimmel G, Halperin E. Inference of locus-specific ancestry in closely related populations. Bioinformatics. 2009;25(12):i213–21. doi: 10.1093/bioinformatics/btp197. Epub 2009/05/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shriner D, Adeyemo A, Rotimi CN. Joint ancestry and association testing in admixed individuals. PLoS computational biology. 2011;7(12):e1002325. doi: 10.1371/journal.pcbi.1002325. Epub 2012/01/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brisbin A, Bryc K, Byrnes J, Zakharia F, Omberg L, Degenhardt J, et al. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Human biology. 2012;84(4):343–64. doi: 10.3378/027.084.0401. Epub 2012/12/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fejerman L, Chen GK, Eng C, Huntsman S, Hu D, Williams A, et al. Admixture mapping identifies a locus on 6q25 associated with breast cancer risk in US Latinas. Human molecular genetics. 2012;21(8):1907–17. doi: 10.1093/hmg/ddr617. Epub 2012/01/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, Waliszewska A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(38):14068–73. doi: 10.1073/pnas.0605832103. Epub 2006/09/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40(10):1175–84. doi: 10.1038/ng.226. Epub 2008/09/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40(10):1185–92. doi: 10.1038/ng.232. Epub 2008/09/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paz Z, Nails M, Ziv E. The genetics of benign neutropenia. The Israel Medical Association journal : IMAJ. 2011;13(10):625–9. Epub 2011/11/22. [PubMed] [Google Scholar]
- 31.Reich D, Nalls MA, Kao WH, Akylbekova EL, Tandon A, Patterson N, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5(1):e1000360. doi: 10.1371/journal.pgen.1000360. Epub 2009/01/31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torgerson DG, Gignoux CR, Galanter JM, Drake KA, Roth LA, Eng C, et al. Case-control admixture mapping in Latino populations enriches for known asthma-associated genes. J Allergy Clin Immunol. 2012;130(1):76–82. e12. doi: 10.1016/j.jaci.2012.02.040. Epub 2012/04/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torgerson DG, Capurso D, Ampleford EJ, Li X, Moore WC, Gignoux CR, et al. Genome-wide ancestry association testing identifies a common European variant on 6q14.1 as a risk factor for asthma in African American subjects. J Allergy Clin Immunol. 2012;130(3):622–9. e9. doi: 10.1016/j.jaci.2012.03.045. Epub 2012/05/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh SS, Tcheurekdjian H, Roth LA, Nguyen EA, Sen S, Galanter JM, et al. Effect of secondhand smoke on asthma control among black and Latino children. J Allergy Clin Immunol. 2012;129(6):1478–83. e7. doi: 10.1016/j.jaci.2012.03.017. Epub 2012/05/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann T, Zhan Y, Kvale M, Hesselson S, Gollub J, Iribarren C, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98:422–30. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander D, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team RC. R . A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2012. [Google Scholar]
- 38.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 2005;95(3):221–7. doi: 10.1038/sj.hdy.6800717. Epub 2005/08/04. [DOI] [PubMed] [Google Scholar]
- 39.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74(4):765–9. doi: 10.1086/383251. Epub 2004/03/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gilliland FD, Li YF, Dubeau L, Berhane K, Avol E, McConnell R, et al. Effects of glutathione S-transferase M1, maternal smoking during pregnancy, and environmental tobacco smoke on asthma and wheezing in children. Am J Respir Crit Care Med. 2002;166(4):457–63. doi: 10.1164/rccm.2112064. Epub 2002/08/21. [DOI] [PubMed] [Google Scholar]
- 41.Hancock D, Romieu I, Shi M, Sienra-Monge J, Wu H, Chiu G, et al. Genome-wide association study implicates chromosome 9q21.31 as a susceptibility locus for asthma in mexican children. PLoS Genet. 2009;5:e1000623. doi: 10.1371/journal.pgen.1000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guilbert TW, Morgan WJ, Krawiec M, Lemanske RF, Jr., Sorkness C, Szefler SJ, et al. The Prevention of Early Asthma in Kids study: design, rationale and methods for the Childhood Asthma Research and Education network. Controlled clinical trials. 2004;25(3):286–310. doi: 10.1016/j.cct.2004.03.002. Epub 2004/05/26. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. Epub 2007/08/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. Epub 2009/06/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1(6):457–70. doi: 10.1534/g3.111.001198. Epub 2012/03/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. Epub 2012/11/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. Epub 2010/10/29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Asthma Education and Prevention Program (National Heart Lung and Blood Institute) Guidelines for the diagnosis and management of asthma : full report 2007. xxii. U.S Dept. of Health and Human Services, National Institutes of Health, National Heart, Lung, and Blood Institute; Bethesda, Md.: 2010. Third Expert Panel on the Management of Asthma. p. 326. [Google Scholar]
- 49.Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiological reviews. 2006;86(1):245–78. doi: 10.1152/physrev.00010.2005. Epub 2005/12/24. [DOI] [PubMed] [Google Scholar]
- 50.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–24. doi: 10.1038/nature11582. Epub 2012/11/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364(16):1503–12. doi: 10.1056/NEJMoa1013660. Epub 2011/04/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hijikata M, Matsushita I, Tanaka G, Tsuchiya T, Ito H, Tokunaga K, et al. Molecular cloning of two novel mucin-like genes in the disease-susceptibility locus for diffuse panbronchiolitis. Hum Genet. 2011;129(2):117–28. doi: 10.1007/s00439-010-0906-4. Epub 2010/10/29. [DOI] [PubMed] [Google Scholar]
- 53.Ivkovic-Jurekovic I, Zunec R, Balog V, Grubic Z. The distribution of HLA alleles among children with atopic asthma in Croatia. Coll Antropol. 2011;35(4):1243–9. Epub 2012/03/09. [PubMed] [Google Scholar]
- 54.Apostolakis J, Toumbis M, Konstantopoulos K, Kamaroulias D, Anagnostakis J, Georgoulias V, et al. HLA antigens and asthma in Greeks. Respiratory medicine. 1996;90(4):201–4. doi: 10.1016/s0954-6111(96)90287-5. Epub 1996/04/01. [DOI] [PubMed] [Google Scholar]
- 55.Mathieu A, Cauli A, Vacca A, Mameli A, Passiu G, Porru G, et al. Genetics of psoriasis and psoriatic arthritis. Reumatismo. 2007;59(Suppl 1):25–7. doi: 10.4081/reumatismo.2007.1s.25. Epub 2007/12/06. [DOI] [PubMed] [Google Scholar]
- 56.Allanore Y, Saad M, Dieude P, Avouac J, Distler JH, Amouyel P, et al. Genome-wide scan identifies TNIP1, PSORS-C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet. 2011;7(7):e1002091. doi: 10.1371/journal.pgen.1002091. Epub 2011/07/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Genin E, Schumacher M, Roujeau JC, Naldi L, Liss Y, Kazma R, et al. Genome-wide association study of Stevens-Johnson Syndrome and Toxic Epidermal Necrolysis in Europe. Orphanet journal of rare diseases. 2011;6:52. doi: 10.1186/1750-1172-6-52. Epub 2011/08/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tohkin M, Kaniwa N, Saito Y, Sugiyama E, Kurose K, Nishikawa J, et al. A whole-genome association study of major determinants for allopurinol-related Stevens-Johnson syndrome and toxic epidermal necrolysis in Japanese patients. The pharmacogenomics journal. 2011;13(1):60–9. doi: 10.1038/tpj.2011.41. Epub 2011/09/14. [DOI] [PubMed] [Google Scholar]
- 59.Kim DK, Cho MH, Hersh CP, Lomas DA, Miller BE, Kong X, et al. Genome-wide association analysis of blood biomarkers in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;186(12):1238–47. doi: 10.1164/rccm.201206-1013OC. Epub 2012/11/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou S, Yang Z, Du L, Jiang Z, Shu Q, Chen Y, et al. Identification of a susceptibility locus in STAT4 for Behcet's disease in Han Chinese in a genome-wide association study. Arthritis and rheumatism. 2012;64(12):4104–13. doi: 10.1002/art.37708. Epub 2012/09/25. [DOI] [PubMed] [Google Scholar]
- 61.Li X, Ampleford EJ, Howard TD, Moore WC, Torgerson DG, Li H, et al. Genome-wide association studies of asthma indicate opposite immunopathogenesis direction from autoimmune diseases. J Allergy Clin Immunol. 2012;130(4):861–8. e7. doi: 10.1016/j.jaci.2012.04.041. Epub 2012/06/15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verlaan DJ, Berlivet S, Hunninghake GM, Madore AM, Lariviere M, Moussette S, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. American journal of human genetics. 2009;85(3):377–93. doi: 10.1016/j.ajhg.2009.08.007. Epub 2009/09/08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sleiman P, Flory J, Imielinski M, Bradfield J, Annaiah K, Willis-Owen S, et al. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:34–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 64.Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: the flip-flop phenomenon. Am J Hum Genet. 2007;80(3):531–8. doi: 10.1086/512133. Epub 2007/02/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS biology. 2010;8(1):e1000294. doi: 10.1371/journal.pbio.1000294. Epub 2010/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nature reviews Genetics. 2010;11(5):356–66. doi: 10.1038/nrg2760. Epub 2010/04/17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.