Abstract
Oxidative post-translational modifications of proteins resulting from events that increase cellular oxidant levels play important roles in physiological and pathophysiological processes. Evaluation of alterations to protein redox states is increasingly common place because of methodological advances that have enabled detection, quantification and identification of such changes in cells and tissues. This mini-review provides a synopsis of biochemical methods that can be utilized to monitor the array of different oxidative and electrophilic modifications that can occur to protein thiols and can be important in the regulatory or maladaptive impact oxidants can have on biological systems. Several of the methods discussed are valuable for monitoring the redox state of established redox sensing proteins such as Keap1.
Keywords: Redox state, Disulfide, S-nitrosation, Hyperoxidation, Keap1, Methods
Graphical abstract
1. Introduction
Reactive oxygen species (ROS) and reactive nitrogen species (RNS) are catch-all terms encompassing a broad range of molecular entities that have potential to chemically oxidize biological molecules. Although in common use, we should be mindful that the individual molecular species that comprise ROS and RNS can be very different in their physicochemical properties that underlie biological responses occurring when this diverse array of species change their concentration. Thus, although we often classify this broad array of molecules together, this may not always be helpful when trying to understand the molecular events that underlie biological responses to these distinct entities. Cells utilize a diverse collection of oxidase enzymes to catalyze reduction−oxidation (redox) reactions in which electrons are passed from an electron donor source to molecular oxygen, so reducing it to form the ROS species superoxide, which can dismutate to form hydrogen peroxide (H2O2). Many contemporary studies focus on ROS generated from NADPH oxidase enzymes [1,2], as these enzymes have evolved to specifically generate superoxide that is functionally important. Whereas oxidant production by other oxidase enzymes involved in cell metabolism [3] can be secondary by-products which may not impact on protein function. ROS are also generated by uncoupled nitric oxide synthase (NOS) enzymes [4,5] and macrophages that utilize it in host defense [6], as well as by mitochondria when electrons become uncoupled from their electron transport chain and combine with molecular oxygen to generate superoxide [7,8].
An elevated level of oxidants within the cell (due to their increased synthesis or decreased antioxidant capacity that limits ROS scavenging) is often referred to as ‘oxidative stress’. To many the term oxidative stress implicitly conveys the idea that ROS simply exert a detrimental impact on biological function. However oxidants are now known to have biological functions that are not injurious, and can be considered crucial to maintenance of homeostasis or adaptive signaling events that can limit injury. These biological responses triggered by changes in cellular oxidant concentration are commonly referred to as ‘redox signaling’. In terms of oxidants causing oxidative stress, this was once considered to occur via uncontrolled oxidation of cellular biomolecules. However, another important factor in the pathogenesis of oxidant-mediated injury involves ROS dysregulating basal redox signaling pathways crucial for homeostasis, thus interfering with regulatory pathways important for the maintenance of health.
Cysteine residues are relatively uncommon in proteins compared to other amino acid, comprising only about ~2.3% of the human proteome [9]. The thiol (also known as mercaptan or sulfhydryl) –SH side chain of cysteine is susceptible to reaction with ROS or RNS species, giving rise to a range of oxidative post-translational oxidative modifications, as schematically presented in Fig. 1, that in some cases can functionally regulate the protein. At first glance this would perhaps be considered an unlikely mechanism of regulation, as an elevation in cellular ROS might be anticipated to non-selectively oxidize all manner of protein thiols, potentially triggering uncoordinated functional changes that manifest as dysfunction and development of disease. However, this concept ignores the fact that there is selectivity in the oxidative modification of protein thiols induced by ROS. This is because ROS, such as H2O2, are selective in the thiols they oxidize as a result of oxidants preferentially reacting with deprotonated (−S−), nucleophilic thiolates with a low acid dissociation constant (pKa). Most cysteines thiols however have a pKa of 8–9, and so are almost fully protonated at physiological pH, making them much less reactive with oxidants and so not susceptible to oxidative modification and regulation in this way. In addition to the protein thiol pKa, which is lowered by proximity to proton accepting amino acids (histidine, lysine, arginine) or an increase in cytosol pH, susceptibility to oxidation may be controlled by their vicinity to oxidase enzymes.
Fig. 1.
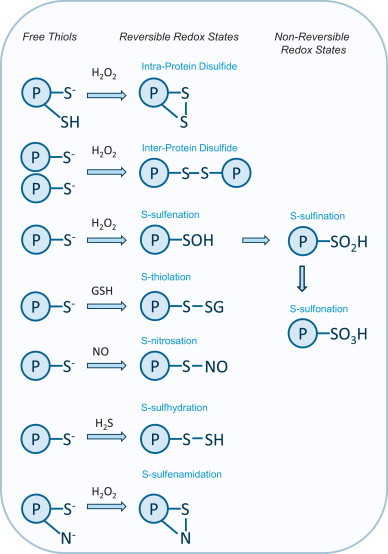
Summary of the oxidative modifications formed in protein thiols. Protein thiols can form a variety of oxidative modifications, including reversible (intra-protein disulfides, inter-protein disulfides, S-sulfenation, S-nitrosation, S-thiolation, S-sulfhydration, S-sulfenamidation) and non-reversible hyper-oxidized (S-sulfination , S-sulfonation) redox states. Some redox states, such as S-sulfenation, S-nitrosation or S-sulfhydration, can be intermediates that transition to disulfides. Prolonged exposure to oxidants can result in irreversible modifications such as S-sulfination or S-sulfonation.
Although protein cysteine oxidation is often mechanistically rationalized by the oxidant directly reacting with the thiol, this may not always be the case. For example, although protein-tyrosine phosphatase 1B (PTP1B) is susceptible to oxidation [10], it has been questioned how this can happen in the cellular setting [11]. This is because although the target thiol in PTP1B has a pKa of ~6.8 and is clearly susceptible to oxidation, it may be difficult to reconcile this happening when peroxiredoxin (Prx) proteins with very reactive low pKa thiols (~5.5) are present in high abundance. One possibility is that PTP1B (or other targets) only become oxidized after the Prx are oxidant-inactivated by hyper-oxidation of their peroxidatic thiols [12]. In addition, it is conceivable that a low pKa protein thiol such as those in Prx or thioredoxin (Trx) may become oxidized and then react with the less reactive target protein cysteine to ‘pass on’ the oxidation [3], as depicted in Fig. 2.
Fig. 2.
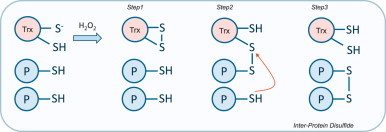
Alternate potential mechanisms leading to disulfide formation. A protein thiol may be oxidized via another redox sensitive protein, in this example thioredoxin (Trx), first becoming oxidized. Trx has a lower pKa than most other proteins and so is more likely to be preferentially oxidized by H2O2 to form an intra-molecular disulfide. The Trx disulfide is then attacked by a thiol of a second protein with a higher pKa which is then reduced by a second thiol. Trx essentially picks-up and passes on the oxidation state to the less reactive target protein thiol.
Diverse arrays of oxidative modifications are crucial to redox signaling events and are integral to all manner of cellular and physiological processes [3,13–15]. Establishing the importance of protein redox regulation in these varied biological processes has been increasingly possible because of advances in biochemical analysis methods allowing alterations in protein thiol redox state to be monitored. These methods have also increased our knowledge of the number proteins regulated by post-translational oxidative modifications. In this mini-review we discuss contemporary biochemical methods that allow redox-sensing proteins to be studied in biological systems.
2. Monitoring reduced protein thiol status
Many methods exist for measuring the total reduced status of thiols within biological samples. Many functionalized cysteine-labeling reagents based on the selective thiol reactivity of maleimide, iodoacetate or disulfide moieties are commercially available. Such reagents are commonly available coupled with several different reporter functions or ‘handles’ (e.g. radiolabel, chromophore, fluorophore or affinity tag) which enable a readout of reduced thiols status [16]. With these thiol-labeling methods, a generalized rationale is that the samples under oxidative stress will have lower reduced thiol content than unstressed controls. The Ellman assay is a classic example of such an assay, providing a colorimetric readout of total reduced thiol content of a sample. As depicted in Fig. 3A, it is based on the ability of thiols in a sample to chemically reduce Ellman’s reagent 5,5′-DiThiobis-2-NitroBenzoic acid (DTNB), which possesses a reactive disulfide bond susceptible to reduction. In a stoichiometric reaction DTNB is reduced by free thiols by an exchange reaction in which a mixed disulfide and a yellow-colored 5-Thio-2-NitroBenzoic acid (TNB) is formed [17]. The intensity of yellow color (measured spectrophotometrically at 412 nm) increases proportionally with the reduced thiol content. The Ellman assay can be used quantitatively by employing external standards of cysteine or reduced glutathione (GSH), normalizing to sample protein content which allows direct comparison to results from other studies. The Ellman reagent reacts with both reduced protein and low molecular weight thiols, but trichloroacetic acid can be used to precipitate proteins to allow the latter component, which is often regarded principally as reduced GSH, to be measured independently. A major issue with the Ellman assay is its limited ability to detect subtle changes in thiol redox state, for example those alterations with physiological redox signaling. Ellman’s, as with many of the functionalized thiol-labeling reagents mentioned above, reacts with all reduced thiols in the sample. However, as also explained above, the proteins susceptible to oxidation are relatively select, typically with a low pKa. Thus most protein thiols in the sample are not anticipated to be susceptible to oxidation and so in the absence of harsh oxidant conditions or chronic oxidative stress, then the Ellman assay may not detect an alteration in thiol redox state because the net change is subtle, despite key sensor proteins becoming significantly oxidized. This issue is common to all the methods where the reactive reporter molecule reacts with thiols independently of their ionization state. One way of overcoming this issue is to use a labeling reagent, such as biotin-iodoacetamide (BIAM), which selectively reacts with undefined thiols, namely those that are also susceptible to oxidation.
Fig. 3.
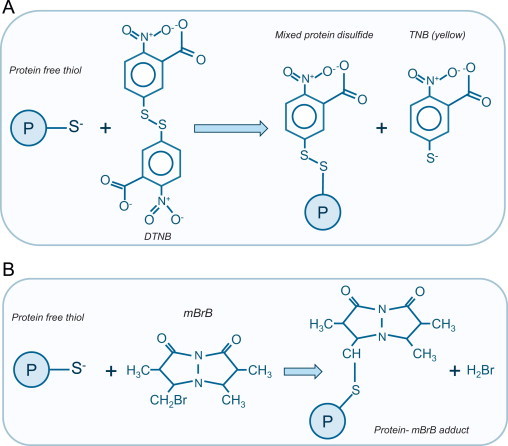
Detection of the reduced (free) thiol content. (A) The Ellman assay is based on the susceptibility of a double bond in Ellman’s reagent 5,5′-dithiobis-2-nitrobenzoic acid (DTNB) to be chemically reduced by a free thiol group. The reaction products are a mixed disulfide and a yellow-colored 5-Thio-2-NitroBenzoic acid (TNB). The amount of TNB generated correlates with the reduced thiol content and can be measured spectrophotometrically. (B) Monobromobimane (mBrB) reacts with the thiols resulting in a fluorescent thiol-mBrB adduct formation which can be measured.
When samples are prepared at low pH (typically 6 or lower), most protein in the sample will be fully protonated as they have a pKa of 8–9 [8]. Under such conditions most reduced protein thiols will not label with BIAM as it selectively reacts with the thiolate state. However, low pKa thiols will remain at least partially deprotonated under these conditions. As oxidants also selectively target these very same low pKa reactive thiols, this low pH BIAM-labeling allows oxidant-sensitive thiols to be selectively studied. Another way of overcoming the inability of many global cysteine-labeling strategies to detect subtle changes in redox state is by focussing in on a single species of thiol-containing molecule. This can be achieved for low molecular weight thiols (e.g. cysteine, homocysteine, homocystine, GSH) by combining their labeling with pre- or post-chromatographic separation, typically high-performance liquid chromatography (HPLC). Samples can also be analyzed after chemical reduction to convert oxidized thiols present (e.g. cystine, homocystine, glutathione disulfide, S-thiolated proteins) to their reduced state, so enabling quantitation of reduced, oxidized and total low molecular weight thiol content. Thiol labeling regents utilized in such approaches include the fluorometric probes monobromotrimethylammoniobimane (qBBr) or monobromobimane (mBrB) [17–19]. mBrB itself has little intrinsic fluorescence (Fig. 3B), but after conjugatively reacting with thiols becomes fluorescent (excitation 380 nm; emission 478–480 nm) [19,20]. Thiol derivatives of mBrB are more stable and more fluorescent than the qBBr. Overall, bromobimanes are more sensitive than DTNB and have not only been used for detection of small thiols, but also for modified proteins by combining with SDS polyacrylamide gel electrophoresis (PAGE) [17]. Other classes of fluorometric tags include benzofurazans 7-fluorobenzo-2-oxa-1,3-diazole-4-sulfonate (SBD-F) or 4-(aminosulfonyl)-7-fluoro-2,1,3-benzoxadiazole (ABD-F) which also become fluorescent after reaction with thiols. These compounds also enable analysis of multiple low molecular weight thiols when combined with HPLC [17].
A generic tag-based approach for monitoring reversible protein thiol oxidation is schematically presented in Fig. 4. This involves blocking the sample free thiols with an alkylating reagent (step 1), followed by the reduction of the often unknown redox state (step 2), with subsequent labeling of the newly formed (nascent) thiols (step 3) and their detection (step 4). This generalized approach is exploited by the so-called biotin-switch method, discussed below in the section on protein S-nitrosation. The blocking alkylation step is commonly performed with maleimide, N-ethylmaleimide (NEM), iodoacetamide, iodoacetate or methyl methane thiosulfonate (MMTS). The reduction step can be done with a reducing agent such as dithiothreitol (DTT) which is capable of reversing all of the oxidative modifications shown in Fig. 1, apart from the sulfinic or sulfonic states. Ascorbate and arsenite can be used to selectively reduce S-nitrosated or S-sulfonated proteins respectively. The nascent thiols generated by the reduction step can then be labeled with a thiol reactive reagents such as biotin-maleimide (as in the biotin switch method) [21,22], or polyethylene glycol-maleimide (PEG-switch method).
Fig. 4.
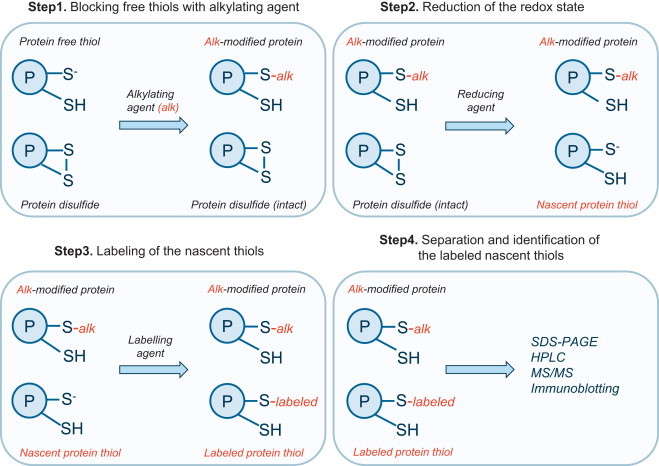
Overview of a generic tag-based reductive switch-labeling method for monitoring reversible protein thiol oxidation. This approach requires several steps. During Step 1 free thiols are blocked with an alkylating reagent (typically maleimide, N-ethylmaleimide, iodoacetamide, iodoacetate or methyl methane thiosulfonate). During Step 2 the reversibly oxidized thiol is reduced with the reducing agent (dithiothreitol, ascorbate or arsenite depending on the oxidation states under investigation), followed by the labeling of the nascent thiols with labeling agent (typically biotin-maleimide). Step 4 involves separation (SDS-PAGE, HPLC) and identification of the labeled protein nascent thiols (typically by immunoblotting or MS analysis).
3. Monitoring intra-protein disulfides
Intra-protein disulfides are those formed between vicinal cysteine residues in a protein. The two thiols can be close enough to form a disulfide either by being adjacent in the primary sequence, or if not, as a result of their proximal orientation in the folded protein structure [23]. Intra-protein disulfides can induce faster migration on non-reducing SDS-PAGE in some proteins, but an additional validation study to corroborate the faster gel migration truly indicates a disulfide may be required. In addition, band shifts can be small and also are not guaranteed in many proteins of interest. However, as explained below, there are other methods which enable detection and quantitation of inter-protein disulfide formation.
The generic reductive switch-labeling method outlined above in Fig. 4 has been used to monitor intra-protein disulfide formation, but this requires prior knowledge that the protein oxidizes to this state. Thus, Trx redox state has been monitored using this approach, utilizing 4′-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) as the labeling agent (step 3). AMS is an NEM variant that reacts with thiols to increase the protein mass by ~0.5 kDa (as shown in Fig. 5). Thus when labeled samples are analyzed by Western immunoblotting, Trx that was originally oxidized in the sample, becomes AMS-modified and so runs slightly higher above a lower band representing the reduced protein [24,25].
Fig. 5.
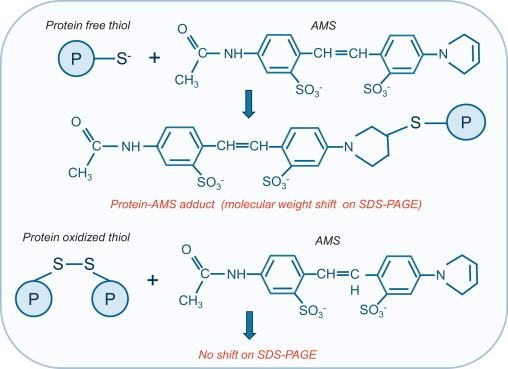
Monitoring intra-protein disulfides by AMS addition. 4′-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS) reacts with reduced protein thiols resulting in protein-AMS adducts which increases the mass by ~0.5 kDa. This additional molecular weight leads to a molecular shift on SDS-PAGE, generating an additional upper band on a gel. In contrast, if a protein thiol is oxidized it will not react with AMS and so no shift will be observed.
Phenylarsine oxide (PAO) selectively adducts reduced vicinal thiols to form a stable dithioarsine ring (as shown on Fig. 6A), but not when they form an intra-disulfide. Thus solid phase PAO can be used together with western immunoblotting analysis of candidate proteins to assess their intra-protein disulfide status or with mass spectrometry (MS) approaches for unbiased proteomics screening to identify novel targets [16,26]. Dibromobimane (dBrB) reacts with vicinal thiols to generate a fluorescent adduct that cannot form if an intra-protein is present (Fig. 6B). This provides a basis for monitoring this oxidation state in vitro with pure proteins, or if combined with chromatographic separation could perhaps be useful in analysis of complex biological samples [26,27].
Fig. 6.
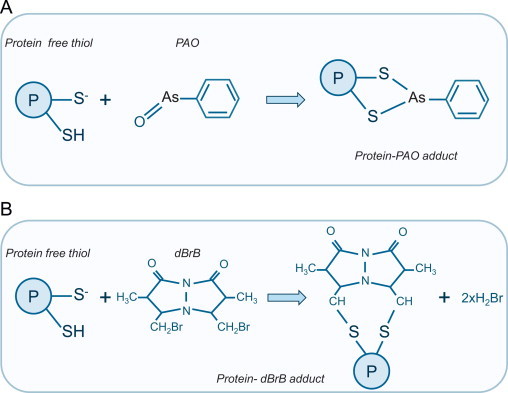
Monitoring intra-protein disulfides by PAO and dBrB addition. (A) Phenylarsine oxide (PAO) forms a stable protein-PAO adduct after reaction with reduced vicinal thiols. However, PAO cannot react with vicinal thiols that have formed an intra-disulfide. (B) Dibromobimane (dBrB) forms a fluorescent protein-dBrB adduct with vicinal cysteines, but this cannot be formed when the protein is oxidized to form an intra-protein. Thus, loss of PAO-labeling or PAO-dependent protein capture indicates intra-protein disulfide protein.
4. Monitoring inter-protein disulfides
An inter-protein disulfide refers to a bond between cysteine thiols on two protein subunits, generating either homo- or heterodimers. As inter-protein disulfide bond substantively increases the protein molecular weight, this can readily be monitored using non-reducing Western immunoblotting analysis of candidates of interest. If the band shift is normalized when the samples are separately analyzed with a reducing agent, typically DTT or 2-mercapthoethanol (2-ME) present, this provides reasonable confidence the migration difference is indeed disulfide bond-mediated [28]. Of course this approach is a candidate based-approach and requires the antibody used detects both redox states of the protein, which in our experience can be an issue with some having significant selectivity for one state over the other.
Diagonal electrophoresis is a sequential non-reducing followed by reducing gel analysis procedure that can allow the unbiased (i.e. non-candidate based) identification of proteins that form inter-protein disulfides [16,26,29]. This method, which is described in detail elsewhere [16,29,30], involves running a non-reducing SDS-PAGE to separate all proteins, including those with disulfide bonds. After electrophoresis, the entire lane containing the separated proteins is excised and placed horizontally on a second SDS-PAGE gel and separated again, but under reducing conditions by adding SDS sample buffer with DTT or 2-ME. When the gel is stained for total protein, the dominant feature is a diagonal line caused by most proteins running at the same molecular weight during both runs. However, those proteins with inter-disulfides migrate faster in the second reducing separation as the disulfide is chemically reduced, meaning they run at a lighter mass and so off of the diagonal. Consequently, these proteins appear as spots that run off of the diagonal, and they can be excised and identified using MS analysis. Newly identified disulfide-forming proteins can subsequently be validated by assessing oxidant-induced, reducing agent reversible, gel shifts on non-reducing immunoblot as described above. This approach allowed us to identify the RIα subunit of protein kinase A [30,31] as an inter-proteins disulfide forming protein, leading to the same finding for protein kinase G Iα [32]. The importance of a disulfide mediating a biological response of interest can be investigated expressing ‘redox-dead’ mutants in which the redox cysteine is replaces by a serine (the most conservative mutation) to prevent the disulfide forming. This mutagenesis approach with protein kinase G Iα prevented the oxidant-induced activation of the kinase that occurs in wild-type when the disulfide forms [32]. Indeed knockin mice in which the redox-dead variant replaced wild-type kinase have high blood pressure and altered blood pressure responses to nitroglycerin or during sepsis [33–35].
5. Monitoring protein S-nitrosation
Protein S-nitrosation, also known as S-nitrosylation, refers to formation of a nitroso (SNO) protein thiol and is mediated by various nitrosating variants of nitric oxide [22]. S-nitrosation is enzymatically reversible by S-nitroso-glutathione reductase [36], thioredoxin [37], thioredoxin-interacting protein [38], carbonyl reductase [39] or xanthine oxidoreductase [40]. S-nitrosation is widely considered a widespread post-translational modification of broad importance that regulates protein function in a way akin to phosphorylation [21,41–43]. However, one can question whether nitrosothiols are generically stable enough for such a role and suggest that most will likely react with thiols they encounter to form more stable disulfides. Thus, protein S-nitrosation may primarily be a short-lived, intermediate redox state leading to disulfide formation. Historically, monitoring S-nitrosation involved the reductive release of NO-related products with monitoring using chemiluminescence, colorimetry or fluorescence methods (reviewed in [22,43]). These methods have limited sensitivity and their specificity for S-nitrosated thiols can be questioned.
Pan-specific polyclonal antibodies that can be used to detect [44,45], or immunoprecipitate [46], S-nitrosated proteins have been reported. However, there have been concerns about whether such antibodies truly detect S-nitrosation. Firstly, there is the question of whether a nitrosated protein would be stable enough to survive immunoblotting or immunoprecipitation procedures, or indeed whether any S-nitrosated antigen used for immunization would persist after it is introduced into the host. Given the prevalence and relative ease and accessibility of immunoblotting and immunoprecipitation in biomedical research studies, it is notable that these antibodies are rarely utilized. Indeed, if the antibody methods were robust then it would seem that other methods for monitoring S-nitrosation, such as the biotin-switch, would not have been needed or so commonly utilized. The biotin-switch method was pioneered by Jaffrey and Snyder in 2001 [47], and further developed by others [22]. The basic principal is outlined in Fig. 4, utilizing ascorbate to selectively reduce the nitrosothiol to cysteine which is then labeled with a thiol-reactive biotinylation reagent. The original protocol included divalent cation chelators to limit metal-mediated reduction of oxidized thiols, but subsequently some protocols have added a source of Cu+ ions [48]. Cu+ is likely the principal direct mediator or nitrosothiol reduction, with the resulting oxidized Cu++ being regenerated to the reduced form by ascorbate. However, what thiol oxidation states in addition to nitrosothiol can be reduced by Cu+ is open to question. One of the disadvantages of this biotin-switch method, which is a generic issue with many proteomic approaches, is that it preferentially detects high abundance proteins. A protein microarray-based has also developed that allowed unbiased, high-throughput identification of S-nitrosated proteins [49], as well as a resin-assisted capture method for analysis of the nitrosothiol proteome (known as SNO RAC) [50,51]. Recently, a method called ‘NitroDIGE’ was developed for detecting S-nitrosated proteins [9]. This variant of the modified biotin switch method labels nascent thiols with a thiol-reactive ‘CyDye’ compound that enhances detection by adding fluorescent handle which enhances sensitivity. This assay can be multiplexed as variants of the CyDye that have unique fluorescent properties can be used to label different samples, which can then be analyzed by difference gel electrophoresis (DIGE). DIGE analysis typically involves mixing samples (that have been separately labeled with different fluorophores) together before electrophoretic separation, most often with two-dimensional sequential isoelectric focussing followed by SDS-PAGE analysis [16,52]. Combining the samples overcomes issues with related fluorescent analysis methods where samples can be compared after running them separately on different gels and looking for alterations in fluorescent spot patterns, with a loss of labeling indicating oxidation. With DIGE, as the samples are together, a specific protein species present in each sample will run precisely on top of each other as a single spot and overcome issues with using separate gels for each sample. Using ratiometric analysis and looking for loss of one fluorescent signal relative to another allows identification of a protein in a sample which is more thiol-oxidized. This protein can then be excised and identified using MS.
6. Monitoring protein S-sulfenation
Protein S-sulfenation refers to oxidation of a thiol to the sulfenic acid (–SOH, see Fig. 1) state, and can be induced by molecular oxygen or hydrogen peroxide and related species. S-sulfenation is generally considered a labile modification that like S-nitrosation will rapidly react with other thiols on contact to form more stable intermediates. Sulfenic acids can be monitored or at least observed in structural datasets using X-ray crystallography or nuclear magnetic resonance, or alternately by MS [53]. The Ellman assay described above for monitoring reduced thiols can be used indirectly to monitor sulfenates in vitro. This is achieved by monitoring consumption of an added thiol, which serves as a reporter because it reacts with the sulfenic acid to form a disulfide [54].
Generally, most of the techniques utilizing the reaction of the sulfenic acid with chemical probes are based on the electrophilic character of the sulfur atom and its weak nucleophilic properties [55]. 4-Chloro-7-nitrobenzo-2-oxa-1,3-diazole has been used to monitor sulfenation in vitro [56,57]. 5,5-Dimethyl-1,3-cyclohexanedione (dimedone) stably derivatizes protein sulfenates (Fig. 7), which can be monitored colorimetrically or with MS in vitro with purified proteins [58,59]. However, antibodies that pan-specifically detect protein sulfenates derivatized by dimedone have been developed. This allowed protein sulfenation in cells and tissues to be monitored more easily [55,60,61]. Functionalized biotinylated or fluorescent derivatives of dimedone have also been synthesized, and have allowed protein sulfenation in cells to be studied, allowing target proteins to be captured and identified [58,59]. Another dimedone synthetic derivative, DYn-2, appears more efficient in labeling protein sulfenic acids in live cells. DYn-2, which is functionalized with a small azide, allows conjugated protein sulfenates to be captured via a Staudinger ligation click-chemistry reaction [62].
Fig. 7.
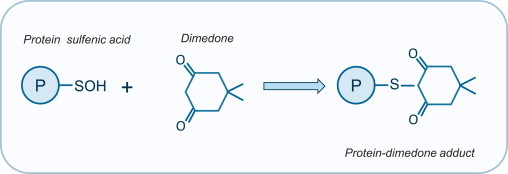
Identification of S-sulfonated proteins with dimedone. Dimedone (5,5-dimethyl-1,3-cyclohexanedione) forms a stable protein-dimedone adduct which can be identified either colorimetrically or with MS or using antibody-based approach.
A modified biotin-switch method (see Fig. 4) can be used for the detection, purification and identification of protein sulfenic acids [21,57]. This method relies on the selective reduction of sulfenates by arsenite, with post-labeling of the nascent generated thiols with a biotinylated alkylating reagent. One potential issue with this, as with the ascorbate-dependent detection of S-nitrosated proteins, is that the analysis is carried out under SDS denaturing conditions which may result in loss of many protein sulfenates due to their destablization during protein unfolding.
7. Monitoring protein S-sulfhydration
The disulfide bonding of H2S to a protein has been referred to as sulfhydration [63], although sulfuration is perhaps more appropriate terminology [64]. Due to the low (–2) oxidation state of H2S it is perhaps unlikely to directly react with proteins to form the disulfide. Consequently, intermediate polysulfides (and/or sulfane sulfur) have been suggested as species with oxidant properties mediating S-sulfhydration [65]. Polysulfides may be formed from H2S via autoxidation of HS− in the presence of molecular oxygen at neutral or slightly alkaline pH. S-sulfhydration could occur via the interaction of the sulfenic acid with HS− or H2S to form a persulfide bond or via a sulfenamide intermediate (reviewed in [66]). Protein S-sulfhydration may follow after H2S first forms persulfide or polysulfides, which could then undergo thiol-disulfide exchange. Perhaps the first study demonstrating protein S-sulfhydration in vivo utilized a cystathionine γ-lyase-deficient mouse model and a modified biotin switch assay [67]. A variant of the switch method shown in Fig. 4 was used to monitor S-sulfhydrated proteins, this included blocking free (unmodified) thiol groups with MMTS, with removal of unincorporated MMTS using acetone, followed by labeling of sulfhydrated (–S–SH) thiols with biotin-HPDP [67]. In the original study biotinylated proteins were captured using solid-phase avidin, eluted by SDS-PAGE and immunoblotted using biotin-conjugated antibody or targets identified using mass-spectrometry [67]. Quantification of S-sulfhydration can be achieved by comparing blots of the biotin-switch samples to the blots of total lysates (not subjected to the biotin-switch). The question however remains as how MMTS while alkylating free protein thiols does not also modify –S–SH group via thiol disulfide exchange. Recently Snyder’s group reported a modification of this method with maleimide which alkylates free thiol groups of proteins but does not affect nitrosated or other oxidized thiols. This approach allowed simultaneous measure of sulfhydration and nitrosylation of NF-κB in the same sample [68].
8. Monitoring protein S-thiolation
Protein S-thiolation is an umbrella term for disulfides between a protein and small thiol-containing molecules such as glutathione or cysteine, generating S-glutathiolated or S-cysteinylated proteins respectively. Radiolabeled glutathione or other small thiols are sensitive tools for the quantitative detection of protein S-thiolation. Tritiated GSH with quantitation of incorporation into protein by liquid scintillation counting [69], or providing 35S-cysteine in the presence of a protein synthesis inhibitor to label the GSH pool followed by autography of samples resolved by SDS-PAGE [70], enabled monitoring of S-thiolation.
A commonly used non-radioactive approach is to use biotinylated glutathione ethyl ester (BioGEE), a reduced form of glutathione which is cell-permeant due to the acetyl group [70]. Once in the cell, esterases remove the acetyl group and the biotinylated GSH can participate in redox reactions including protein S-glutathiolation. Proteins carrying biotinylated glutathione can be detected, identified, quantified and purified following avidin-capture and MS analysis [70]. Glutathione N,N-biotinyl glutathione disulfide (biotin–GSSG–biotin) which S-glutathiolates proteins via thiol-disulfide exchange has also be used to study protein S-glutathiolation in cells and organs [71]. Similarly, protein S-homocysteinylation has been monitored using homocysteine labeling with a fluorescent or biotin tag [72]. Such approaches, however, do not readily permit direct detection of endogenous S-thiolated proteins, for example in stored tissue samples. In this case, antibodies to S-glutathiolated or S-homocysteinylated proteins can be utilized [71]. Candidate protein S-glutathiolation can be monitored by its immunoprecipitation, followed by immunoblotting with a pan-specific anti-glutathione antibody, as reported for detection of this modification in NOS [4].
The generic biotin-switch methods, as outlined in Fig. 4, can be adapted for selective monitoring of protein S-glutathiolation. This can be achieved on tissue sections or in homogenates by using the disulfide oxidoreductase glutaredoxin to reduce the target proteins back to the free thiol state, with subsequent labeling with a thiol-reactive functionalized (e.g. biotin, fluorophore, etc.) reagent [73,74].
9. Monitoring ‘hyperoxidation’ of protein thiols
As shown in Fig. 1, once a sulfenic acid forms it can react with additional oxidant molecules to stepwise transition into more stable sulfinic (PSO2H) and then sulfonic (PSO3H) acid states. These are generally considered biologically irreversible modifications, perhaps proving evidence of damage associated with acute hyper-exposure to oxidants or chronic oxidative stress. However, the sulfination of the peroxidatic thiol in 2-Cys peroxiredoxins was shown to be reversible in cells [12], and mediated by sulfiredoxin through an ATP-dependent reaction [75]. PSO2H formation also occurs in glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [76], PTP1B [10] and DJ-1 [77]. PSO2H and PSO3H have been indexed by monitoring band shifts on isoelectric focussing gels [78]. Antibodies that detect sulfinic or sulfonic acid formation in peroxiredoxin [12,79], DJ-1 [80,81], or GAPDH [61] have been reported.
10. Monitoring lipid modifications of protein thiols
Protein thiols can undergo modification by a variety of lipid electrophiles, such as hydroxynonenal, malondialdehyde, acrolein, 15 d prostaglandin J2 (15 D-PGJ2) or nitroalkenes via Schiff base or Michael addition reactions [82]. A general strategy involves labeling a lipid electrophile of interest, applying it exogenously to model systems to enable monitoring, purification and identification of target proteins. For example biotinylated or fluorescent BODIPY analogues of 15 D-PGJ2 or PGA2 have been utilized [83,84]. Biotinylated arachidonic acid can be added to cell or tissue models, after which it is metabolized endogenously to generate different products. Some of these products of arachidonic acid metabolism can be electrophiles that may adduct to proteins; thus how different stimuli modulate protein-electrophile adduction to potentially exert a functional impact can be studied [83–85]. As a label is incorporated to protein targets, this also enables subcellular localization of the modifications to be monitored using microscopy [83]. Using such approaches, 15 D-PGJ2 was shown to covalently adduct to the Kelch like ECH-associated protein 1 (Keap1) in a concentration- and time-dependent manner. This modification results in activation of the nuclear factor (erythroid-derived 2)-like 2 (Nrf-2) pathways to enhance antioxidant synthesis [84]. Sulforaphane is another electrophile that covalently modifies Keap1, so triggering elevated antioxidant synthesis. The targets of sulforaphane have been studied by synthesizing ‘click chemistry’ analogues that enable target proteins to be monitored and identified [86,87].
Commercially available antibodies have been used extensively to monitor hydroxynonenal or malondialdehyde modification or protein with immunoblotting or cyto-imaging methods [29]. Nitroalkene modification has been monitored with a switch-type method (Fig. 4), in which the lipid adduct is reductively released from target proteins by 2-ME with monitoring by MS [88]. Protein S-palmitoylation can be similarly monitored by its enzymatic reversal using a thioesterase [89], or chemically with hydroxylamine [90]. Some modifications of proteins by lipid electrophiles result in introduction of a carbonyl moiety [82]. Protein carbonylation can be monitored via reaction with 2,4-dinitrophenylhydrazine, which can be measured colorimetrically with a spectrophotometer [91,92], by HPLC [93], or using immunoblotting with an antibody generated to this carbonyl-labeling derivatisation reagent [92,94].
11. Monitoring redox states within cellular compartments
Several of the methods described above potentially enable the redox state changes in a discreet cellular compartment (nuclei, mitochondria, endoplasmic reticulum, cytosol, etc.) to be monitored. Although this can be achieved by integrating subcellular fractionation where compatible with one of the redox analytical methods described above, a major problem is the redox state of the target proteins are readily altered, especially during lengthy sample preparations. Some of the protocols outlined are amenable to being combined with imaging methods, allowing redox state alteration in labeling to be monitored at defined subcellular locations. For example, antibodies to specific oxidation states commonly used for immunoblot analysis can also be valuable for immunocytochemistry. Cells can also be studied using a modified biotin-switch method (see Fig. 4) adapted for in situ analysis of fixed samples, again allowing the localization of the oxidized proteins to be determined. Subcellular localization of S-nitrosated [95], or disulfide-containing proteins [96] has been achieved using this strategy.
Mitochondria selective thiol redox probes that combine a lipophilic triphenylphosphonium (TPP) motif with a thiol-reactive labeling moiety have been developed. The membrane potential of the mitochondrial causes the cationic, lipophilic TPP-labeled probe to selectively accumulate within these organelles where it differentially labels proteins thiols depending on their redox state [97]. The TPP motif has also been used to deliver other mitochondria-targeted cargoes, such as the antioxidants MitoVit E or MitoQ. Proteins thiols labeled with TPP derivatives can be assessed after electrophoresis using MS or antibodies developed to this targeting motif, which has allowed alteration in the mitochondrial respiratory chain complexes to be monitored [97,98]. MitoB is a mitochondria-targeted ratiometric MS probe that allows levels of oxidants such as H2O2 to be measured [99].
During the last decade genetically encoded redox sensor probes that potentially allow quantitative, real-time, subcellular imaging of ROS production have been developed [100,101]. Such sensor probes typically comprise a fluorescent protein fused to a redox-sensitive protein domain that is reversibly oxidatively post-translationally modified depending on the cellular redox state. The redox state of the sensor alters the conformation of the probe protein to alter the efficiency of direct fluorescence or fluorescence resonance energy transfer which is quantified by various fluorescence-monitoring methods [100]. HyPer is a genetically encoded fluorescent probe for measuring H2O2, with enhanced HyPer 2 and HyPer 3 variant available [100,102,103]. HyPer is a yellow fluorescent protein (cpYFP) integrated into the conformation-changing region of the Escherichia coli transcription oxido-reductase regulatory domain (OxyR-RD), enabling it to sense H2O2. H2O2 induces an intraprotein disulfide within the HyPer probe, causing a profound conformational change that markedly alters the fluorescence emission peak at 516 nm [100]. A genetically encoded redox probe for monitoring the redox state of glutathione has also been designed by fusing a roGFP2 to human glutaredoxin-1 (roGFP2-Grx1) [104]. RexYF, a genetically encoded redox probe for the NAD(H) redox state changes, was developed by introducing YFP into the T-REX redox sensor from Thermus aquaticus [105]. RexYFP, which is also equipped with a pH sensor to reduce pH-dependent artefacts, enables changes in the NAD(+)/NADH ratio in different cellular compartments to be monitored.
12. Present and future challenges
Methods and technologies for monitoring redox state in biological systems continue to emerge, driven by the intense contemporary research activity in this area. However, many of the methods are not suitable for monitoring ROS levels in cells or in tissues in vivo and so can provide only limited insight. Furthermore, many of the methods are also limited by only providing a snapshot of the redox state. Real-Time Imaging in vivo is already available to some extent, as discussed above, but we envisage these methods to evolve significantly in the near future. With the increasing prevalence of genetically engineered murine models, it is likely that such genetically encoded redox sensors will perhaps become widely utilized. For example a ‘HyPer zebrafish’ that enables in vivo monitoring of H2O2 has already been developed [102,106] as well as a Thy-mito-Grx1-roGFP2 transgenic mouse with a roGFP2 redox sensor expressed in neuronal mitochondria [107].
A remaining issue with methods for measuring specific ROS species, especially those allowing in vivo analysis, is that many of them are not absolutely quantitative. Quantitation is often relative to control, but the absolute concentration or amount of an oxidant produced basally or following an intervention of interest is important information that cannot readily or reliably be determined. Another issue that plagues redox analytical methods is that a steady state, net readout of the redox state of a target protein or concentration of an oxidant is provided. This misses important information about the turnover or redox cycling rates of redox active components. Monitoring of redox flux through a defined pathway of interest could again perhaps be achieved using optical sensors, but the temporal resolution of fluorescent probes currently limits this.
Acknowledgments
This work was supported by the British Heart Foundation (BHF), King’s BHF Centre of Research Excellence, Medical Research Council UK; Fondation Leducq, France, European Research Council, the Department of Health via the National Institute for Health Research comprehensive Biomedical Research Centre award to Guy’s & St. Thomas’ National Health Service Foundation Trust.
References
- 1.Brandes R.P., Weissmann N., Schröder K. Redox-mediated signal transduction by cardiovascular Nox NADPH oxidases. Journal of Molecular and Cellular Cardiology. 2014;73C:70–79. doi: 10.1016/j.yjmcc.2014.02.006. 24560815 [DOI] [PubMed] [Google Scholar]
- 2.Schröder K., Zhang M., Benkhoff S., Mieth A., Pliquett R., Kosowski J., Kruse C., Luedike P., Michaelis U.R., Weissmann N., Dimmeler S., Shah A.M., Brandes R.P. Nox4 is a protective reactive oxygen species generating vascular NADPH oxidase. Circulation Research. 2012;110:1217–1225. doi: 10.1161/CIRCRESAHA.112.267054. 22456182 [DOI] [PubMed] [Google Scholar]
- 3.Burgoyne J.R., Mongue-Din H., Eaton P., Shah A.M. Redox signaling in cardiac physiology and pathology. Circulation Research. 2012;111:1091–1106. doi: 10.1161/CIRCRESAHA.111.255216. 23023511 [DOI] [PubMed] [Google Scholar]
- 4.Chen C.A., Wang T.Y., Varadharaj S., Reyes L.A., Hemann C., Talukder M.A., Chen Y.R., Druhan L.J., Zweier J.L. S-glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature. 2010;468:1115–1118. doi: 10.1038/nature09599. 21179168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabtree M.J., Channon K.M. Synthesis and recycling of tetrahydrobiopterin in endothelial function and vascular disease. Nitric Oxide: Biology and Chemistry/Official Journal of the Nitric Oxide Society. 2011;25:81–88. doi: 10.1016/j.niox.2011.04.004. 21550412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pattison D.I., Davies M.J., Hawkins C.L. Reactions and reactivity of myeloperoxidase-derived oxidants: differential biological effects of hypochlorous and hypothiocyanous acids. Free Radical Research. 2012;46:975–995. doi: 10.3109/10715762.2012.667566. 22348603 [DOI] [PubMed] [Google Scholar]
- 7.Zorov D.B., Filburn C.R., Klotz L.O., Zweier J.L., Sollott S.J. Reactive oxygen species (ROS)-induced ROS release: a new phenomenon accompanying induction of the mitochondrial permeability transition in cardiac myocytes. Journal of Experimental Medicine. 2000;192:1001–1014. doi: 10.1084/jem.192.7.1001. 11015441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mailloux R.J., Jin X., Willmore W.G. Redox regulation of mitochondrial function with emphasis on cysteine oxidation reactions. Redox Biology. 2013;2:123–139. doi: 10.1016/j.redox.2013.12.011. 24455476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu Z., Meng F., Zhou H., Li J., Wang Q., Wei F., Cheng J., Greenlief C.M., Lubahn D.B., Sun G.Y., Liu S., Gu Z. NitroDIGE analysis reveals inhibition of protein S-nitrosylation by epigallocatechin gallates in lipopolysaccharide-stimulated microglial cells. Journal of Neuroinflammation. 2014;11:17. doi: 10.1186/1742-2094-11-17. 24472655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q., Dubé D., Friesen R.W., LeRiche T.G., Bateman K.P., Trimble L., Sanghara J., Pollex R., Ramachandran C., Gresser M.J., Huang Z. Catalytic inactivation of protein tyrosine phosphatase CD45 and protein tyrosine phosphatase 1B by polyaromatic quinones. Biochemistry. 2004;43:4294–4303. doi: 10.1021/bi035986e. 15065873 [DOI] [PubMed] [Google Scholar]
- 11.Winterbourn C.C. Reconciling the chemistry and biology of reactive oxygen species. Nature Chemical Biology. 2008;4:278–286. doi: 10.1038/nchembio.85. 18421291 [DOI] [PubMed] [Google Scholar]
- 12.Woo H.A., Kang S.W., Kim H.K., Yang K.S., Chae H.Z., Rhee S.G. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. Journal of Biological Chemistry. 2003;278:47361–47364. doi: 10.1074/jbc.C300428200. 14559909 [DOI] [PubMed] [Google Scholar]
- 13.Gamper N., Ooi L. Redox and nitric oxide-mediated regulation of sensory neuron ion channel function. Antioxidants & Redox Signaling. 2014 doi: 10.1089/ars.2014.5884. 24735331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urao N., Ushio-Fukai M. Redox regulation of stem/progenitor cells and bone marrow niche. Free Radical Biology & Medicine. 2013;54:26–39. doi: 10.1016/j.freeradbiomed.2012.10.532. 23085514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archer S.L., Will J.A., Weir E.K. Redox status in the control of pulmonary vascular tone. Herz. 1986;11:127–141. 3017827 [PubMed] [Google Scholar]
- 16.Eaton P. Protein thiol oxidation in health and disease: techniques for measuring disulfides and related modifications in complex protein mixtures. Free Radical Biology & Medicine. 2006;40:1889–1899. doi: 10.1016/j.freeradbiomed.2005.12.037. 16716890 [DOI] [PubMed] [Google Scholar]
- 17.Winther J.R., Thorpe C. Quantification of thiols and disulfides. Biochimica et Biophysica Acta. 2014;1840:838–846. doi: 10.1016/j.bbagen.2013.03.031. 23567800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fahey R.C., Newton G.L., Dorian R., Kosower E.M. Analysis of biological thiols: Derivatization with monobromotrimethylammoniobimane and characterization by electrophoresis and chromatography. Analytical Biochemistry. 1980;107:1–10. doi: 10.1016/0003-2697(80)90483-2. 6449160 [DOI] [PubMed] [Google Scholar]
- 19.Newton G.L., Dorian R., Fahey R.C. Analysis of biological thiols: derivatization with monobromobimane and separation by reverse-phase high-performance liquid chromatography. Analytical Biochemistry. 1981;114:383–387. doi: 10.1016/0003-2697(81)90498-x. 7304929 [DOI] [PubMed] [Google Scholar]
- 20.Pelletier S., Lucy C.A. HPLC simultaneous analysis of thiols and disulfides: on-line reduction and indirect fluorescence detection without derivatization. Analyst. 2004;129:710–713. doi: 10.1039/b401618a. 15284913 [DOI] [PubMed] [Google Scholar]
- 21.Burgoyne J.R., Eaton P. A rapid approach for the detection, quantification, and discovery of novel sulfenic acid or S-nitrosothiol modified proteins using a biotin-switch method. Methods in Enzymology. 2010;473:281–303. doi: 10.1016/S0076-6879(10)73015-9. 20513484 [DOI] [PubMed] [Google Scholar]
- 22.Forrester M.T., Foster M.W., Benhar M., Stamler J.S. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radical Biology & Medicine. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. 18977293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung H.S., Wang S.B., Venkatraman V., Murray C.I., Van Eyk J.E. Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circulation Research. 2013;112:382–392. doi: 10.1161/CIRCRESAHA.112.268680. 23329793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakoh-Nakatogawa M., Nishikawa S., Endo T. Roles of protein-disulfide isomerase-mediated disulfide bond formation of yeast Mnl1p in endoplasmic reticulum-associated degradation. Journal of Biological Chemistry. 2009;284:11815–11825. doi: 10.1074/jbc.M900813200. 19279007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denoncin K., Nicolaes V., Cho S.H., Leverrier P., Collet J.F. Protein disulfide bond formation in the periplasm: determination of the in vivo redox state of cysteine residues. Methods in Molecular Biology (Clifton, NJ) 2013;966:325–336. doi: 10.1007/978-1-62703-245-2_20. 23299744 [DOI] [PubMed] [Google Scholar]
- 26.Burgoyne J.R., Eaton P. Contemporary techniques for detecting and identifying proteins susceptible to reversible thiol oxidation. Biochemical Society Transactions. 2011;39:1260–1267. doi: 10.1042/BST0391260. 21936799 [DOI] [PubMed] [Google Scholar]
- 27.Kim J.S., Raines R.T. Dibromobimane as a fluorescent crosslinking reagent. Analytical Biochemistry. 1995;225:174–176. doi: 10.1006/abio.1995.1131. 7778775 [DOI] [PubMed] [Google Scholar]
- 28.Burgoyne J.R., Eaton P. Detecting disulfide-bound complexes and the oxidative regulation of cyclic nucleotide-dependent protein kinases by H2O2. Methods in Enzymology. 2013;528:111–128. doi: 10.1016/B978-0-12-405881-1.00007-0. 23849862 [DOI] [PubMed] [Google Scholar]
- 29.Charles R., Jayawardhana T., Eaton P. Gel-based methods in redox proteomics. Biochimica et Biophysica Acta. 2014;1840:830–837. doi: 10.1016/j.bbagen.2013.04.021. 23624333 [DOI] [PubMed] [Google Scholar]
- 30.Brennan J.P., Wait R., Begum S., Bell J.R., Dunn M.J., Eaton P. Detection and mapping of widespread intermolecular protein disulfide formation during cardiac oxidative stress using proteomics with diagonal electrophoresis. Journal of Biological Chemistry. 2004;279:41352–41360. doi: 10.1074/jbc.M403827200. 15292244 [DOI] [PubMed] [Google Scholar]
- 31.Brennan J.P., Bardswell S.C., Burgoyne J.R., Fuller W., Schröder E., Wait R., Begum S., Kentish J.C., Eaton P. Oxidant-induced activation of type I protein kinase A is mediated by RI subunit interprotein disulfide bond formation. Journal of Biological Chemistry. 2006;281:21827–21836. doi: 10.1074/jbc.M603952200. 16754666 [DOI] [PubMed] [Google Scholar]
- 32.Burgoyne J.R., Madhani M., Cuello F., Charles R.L., Brennan J.P., Schröder E., Browning D.D., Eaton P. Cysteine redox sensor in PKGIa enables oxidant-induced activation. Science (New York, NY) 2007;317:1393–1397. doi: 10.1126/science.1144318. 17717153 [DOI] [PubMed] [Google Scholar]
- 33.Prysyazhna O., Rudyk O., Eaton P. Single atom substitution in mouse protein kinase G eliminates oxidant sensing to cause hypertension. Nature Medicine. 2012;18:286–290. doi: 10.1038/nm.2603. 22245782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudyk O., Phinikaridou A., Prysyazhna O., Burgoyne J.R., Botnar R.M., Eaton P. Protein kinase G oxidation is a major cause of injury during sepsis. Proceedings of the National Academy of Sciences. 2013;110(24):9909–9913. doi: 10.1073/pnas.1301026110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudyk O., Prysyazhna O., Burgoyne J.R., Eaton P. Nitroglycerin fails to lower blood pressure in redox-dead Cys42Ser PKG1alpha knockin mouse. Circulation. 2012;126:287–295. doi: 10.1161/CIRCULATIONAHA.112.101287. 22685118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L., Hausladen A., Zeng M., Que L., Heitman J., Stamler J.S. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. 11260719 [DOI] [PubMed] [Google Scholar]
- 37.Benhar M., Forrester M.T., Hess D.T., Stamler J.S. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science (New York, NY) 2008;320:1050–1054. doi: 10.1126/science.1158265. 18497292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forrester M.T., Seth D., Hausladen A., Eyler C.E., Foster M.W., Matsumoto A., Benhar M., Marshall H.E., Stamler J.S. Thioredoxin-interacting protein (Txnip) is a feedback regulator of S-nitrosylation. Journal of Biological Chemistry. 2009;284:36160–36166. doi: 10.1074/jbc.M109.057729. 19847012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bateman R.L., Rauh D., Tavshanjian B., Shokat K.M. Human carbonyl reductase 1 is an S-nitrosoglutathione reductase. Journal of Biological Chemistry. 2008;283:35756–35762. doi: 10.1074/jbc.M807125200. 18826943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trujillo M., Alvarez M.N., Peluffo G., Freeman B.A., Radi R. Xanthine oxidase-mediated decomposition of S-nitrosothiols. Journal of Biological Chemistry. 1998;273:7828–7834. doi: 10.1074/jbc.273.14.7828. 9525875 [DOI] [PubMed] [Google Scholar]
- 41.Hess D.T., Matsumoto A., Kim S.O., Marshall H.E., Stamler J.S. Protein S-nitrosylation: purview and parameters. Nature Reviews. Molecular Cell Biology. 2005;6:150–166. doi: 10.1038/nrm1569. 15688001 [DOI] [PubMed] [Google Scholar]
- 42.Lima B., Forrester M.T., Hess D.T., Stamler J.S. S-nitrosylation in cardiovascular signaling. Circulation Research. 2010;106:633–646. doi: 10.1161/CIRCRESAHA.109.207381. 20203313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y.J., Ching W.C., Lin Y.P., Chen Y.J. Methods for detection and characterization of protein S-nitrosylation. Methods (San Diego, Calif.) 2013;62:138–150. doi: 10.1016/j.ymeth.2013.04.016. 23628946 [DOI] [PubMed] [Google Scholar]
- 44.Garbán H.J., Márquez-Garbán D.C., Pietras R.J., Ignarro L.J. Rapid nitric oxide-mediated S-nitrosylation of estrogen receptor: regulation of estrogen-dependent gene transcription. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:2632–2636. doi: 10.1073/pnas.0409854102. 15699347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greco T.M., Hodara R., Parastatidis I., Heijnen H.F., Dennehy M.K., Liebler D.C., Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. 16648260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sayed N., Kim D.D., Fioramonti X., Iwahashi T., Durán W.N., Beuve A. Nitroglycerin-induced S-nitrosylation and desensitization of soluble guanylyl cyclase contribute to nitrate tolerance. Circulation Research. 2008;103:606–614. doi: 10.1161/CIRCRESAHA.108.175133. 18669924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaffrey S.R., Erdjument-Bromage H., Ferris C.D., Tempst P., Snyder S.H. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nature Cell Biology. 2001;3:193–197. doi: 10.1038/35055104. 11175752 [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Kettenhofen N.J., Shiva S., Hogg N., Gladwin M.T. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radical Biology & Medicine. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. 18211831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foster M.W., Forrester M.T., Stamler J.S. A protein microarray-based analysis of S-nitrosylation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. 19864628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Forrester M.T., Hess D.T., Thompson J.W., Hultman R., Moseley M.A., Stamler J.S., Casey P.J. Site-specific analysis of protein S-acylation by resin-assisted capture. Journal of Lipid Research. 2011;52:393–398. doi: 10.1194/jlr.D011106. 21044946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forrester M.T., Thompson J.W., Foster M.W., Nogueira L., Moseley M.A., Stamler J.S. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nature Biotechnology. 2009;27:557–559. doi: 10.1038/nbt.1545. 19483679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marouga R., David S., Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Analytical and Bioanalytical Chemistry. 2005;382:669–678. doi: 10.1007/s00216-005-3126-3. 15900442 [DOI] [PubMed] [Google Scholar]
- 53.Lo Conte M., Carroll K.S. The redox biochemistry of protein sulfenylation and sulfinylation. Journal of Biological Chemistry. 2013;288:26480–26488. doi: 10.1074/jbc.R113.467738. 23861405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furdui C.M., Poole L.B. Chemical approaches to detect and analyze protein sulfenic acids. Mass Spectrometry Reviews. 2013;33:126–146. doi: 10.1002/mas.21384. 24105931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gupta V., Carroll K.S. Sulfenic acid chemistry, detection and cellular lifetime. Biochimica et Biophysica Acta. 2014;1840:847–875. doi: 10.1016/j.bbagen.2013.05.040. 23748139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ellis H.R., Poole L.B. Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry. 1997;36:15013–15018. doi: 10.1021/bi972191x. 9398227 [DOI] [PubMed] [Google Scholar]
- 57.Saurin A.T., Neubert H., Brennan J.P., Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17982–17987. doi: 10.1073/pnas.0404762101. 15604151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Percival M.D., Ouellet M., Campagnolo C., Claveau D., Li C. Inhibition of cathepsin K by nitric oxide donors: evidence for the formation of mixed disulfides and a sulfenic acid. Biochemistry. 1999;38:13574–13583. doi: 10.1021/bi991028u. 10521264 [DOI] [PubMed] [Google Scholar]
- 59.Carballal S., Radi R., Kirk M.C., Barnes S., Freeman B.A., Alvarez B. Sulfenic acid formation in human serum albumin by hydrogen peroxide and peroxynitrite. Biochemistry. 2003;42:9906–9914. doi: 10.1021/bi027434m. 12924939 [DOI] [PubMed] [Google Scholar]
- 60.Seo Y.H., Carroll K.S. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16163–16168. doi: 10.1073/pnas.0903015106. 19805274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maller C., Schröder E., Eaton P. Glyceraldehyde 3-phosphate dehydrogenase is unlikely to mediate hydrogen peroxide signaling: studies with a novel anti-dimedone sulfenic acid antibody. Antioxidants & Redox Signaling. 2011;14:49–60. doi: 10.1089/ars.2010.3149. 20518697 [DOI] [PubMed] [Google Scholar]
- 62.Paulsen C.E., Truong T.H., Garcia F.J., Homann A., Gupta V., Leonard S.E., Carroll K.S. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nature Chemical Biology. 2012;8:57–64. doi: 10.1038/nchembio.736. 22158416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mustafa A.K., Gadalla M.M., Sen N., Kim S., Mu W.T., Gazi S.K., Barrow R.K., Yang G.D., Wang R., Snyder S.H. H2S signals through protein S-Sulfhydration. Science Signaling. 2009;2:ra72. doi: 10.1126/scisignal.2000464. 19903941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Toohey J.I. The conversion of H2S to sulfane sulfur. Nature Reviews. Molecular Cell Biology. 2012;13:804. doi: 10.1038/nrm3391-c1. [DOI] [PubMed] [Google Scholar]
- 65.Greiner R., Pálinkás Z., Bäsell K., Becher D., Antelmann H., Nagy P., Dick T.P. Polysulfides link H2S to protein thiol oxidation. Antioxidants & Redox Signaling. 2013;19:1749–1765. doi: 10.1089/ars.2012.5041. 23646934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paul B.D., Snyder S.H. H2S signalling through protein sulfhydration and beyond. Nature Reviews. Molecular Cell Biology. 2012;13:499–507. doi: 10.1038/nrm3391. 22781905 [DOI] [PubMed] [Google Scholar]
- 67.Mustafa A.K., Gadalla M.M., Sen N., Kim S., Mu W., Gazi S.K., Barrow R.K., Yang G., Wang R., Snyder S.H. H2S signals through protein S-sulfhydration. Science Signaling. 2009;2:ra72. doi: 10.1126/scisignal.2000464. 19903941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sen N., Paul B.D., Gadalla M.M., Mustafa A.K., Sen T., Xu R., Kim S., Snyder S.H. Hydrogen sulfide-linked sulfhydration of NF-κB mediates its antiapoptotic actions. Molecular Cell. 2012;45:13–24. doi: 10.1016/j.molcel.2011.10.021. 22244329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Klatt P., Molina E.P., De Lacoba M.G., Padilla C.A., Martinez-Galesteo E., Barcena J.A., Lamas S. Redox regulation of c-Jun DNA binding by reversible S-glutathiolation. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 1999;13:1481–1490. doi: 10.1096/fasebj.13.12.1481. 10463938 [DOI] [PubMed] [Google Scholar]
- 70.Gao X.H., Bedhomme M., Veyel D., Zaffagnini M., Lemaire S.D. Methods for analysis of protein glutathionylation and their application to photosynthetic organisms. Molecular Plant. 2009;2:218–235. doi: 10.1093/mp/ssn072. 19825609 [DOI] [PubMed] [Google Scholar]
- 71.Brennan J.P., Miller J.I., Fuller W., Wait R., Begum S., Dunn M.J., Eaton P. The utility of N,N-biotinyl glutathione disulfide in the study of protein S-glutathiolation. Molecular & Cellular Proteomics: MCP. 2006;5:215–225. doi: 10.1074/mcp.M500212-MCP200. 16223748 [DOI] [PubMed] [Google Scholar]
- 72.Zang T., Dai S., Chen D., Lee B.W., Liu S., Karger B.L., Zhou Z.S. Chemical methods for the detection of protein N-homocysteinylation via selective reactions with aldehydes. Analytical Chemistry. 2009;81:9065–9071. doi: 10.1021/ac9017132. 19874060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aesif S.W., Anathy V., Havermans M., Guala A.S., Ckless K., Taatjes D.J., Janssen-Heininger Y.M. in situ analysis of protein S-glutathionylation in lung tissue using glutaredoxin-1-catalyzed cysteine derivatization. American Journal of Pathology. 2009;175:36–45. doi: 10.2353/ajpath.2009.080736. 19556513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lind C., Gerdes R., Hamnell Y., Schuppe-Koistinen I., von Löwenhielm H.B., Holmgren A., Cotgreave I.A. Identification of S-glutathionylated cellular proteins during oxidative stress and constitutive metabolism by affinity purification and proteomic analysis. Archives of Biochemistry and Biophysics. 2002;406:229–240. doi: 10.1016/s0003-9861(02)00468-x. 12361711 [DOI] [PubMed] [Google Scholar]
- 75.Kiley P.J., Storz G. Exploiting thiol modifications. PLoS Biology. 2004;2:e400. doi: 10.1371/journal.pbio.0020400. 15547642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Souza J.M., Radi R. Glyceraldehyde-3-phosphate dehydrogenase inactivation by peroxynitrite. Archives of Biochemistry and Biophysics. 1998;360:187–194. doi: 10.1006/abbi.1998.0932. 9851830 [DOI] [PubMed] [Google Scholar]
- 77.Canet-Avilés R.M., Wilson M.A., Miller D.W., Ahmad R., McLendon C., Bandyopadhyay S., Baptista M.J., Ringe D., Petsko G.A., Cookson M.R. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. 15181200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Woo H.A., Chae H.Z., Hwang S.C., Yang K.S., Kang S.W., Kim K., Rhee S.G. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science (New York, N.Y.) 2003;300:653–656. doi: 10.1126/science.1080273. 12714748 [DOI] [PubMed] [Google Scholar]
- 79.Rhee H.A.W. Immunoblot detection of proteins that contain cysteine sulfinic or sulfonic acids with antibodies specific for the hyperoxidized cysteine-containing sequence. In: Kumar Das D., editor. Methods in Redox Signalling. Mary Ann Liebert; New Rochelle, NY: 2010. [Google Scholar]
- 80.Ooe H., Iguchi-Ariga S.M., Ariga H. Establishment of specific antibodies that recognize C106-oxidized DJ-1. Neuroscience Letters. 2006;404:166–169. doi: 10.1016/j.neulet.2006.05.031. 16781058 [DOI] [PubMed] [Google Scholar]
- 81.Saito Y., Hamakubo T., Yoshida Y., Ogawa Y., Hara Y., Fujimura H., Imai Y., Iwanari H., Mochizuki Y., Shichiri M., Nishio K., Kinumi T., Noguchi N., Kodama T., Niki E. Preparation and application of monoclonal antibodies against oxidized DJ-1. Significant elevation of oxidized DJ-1 in erythrocytes of early-stage Parkinson disease patients. Neuroscience Letters. 2009;465:1–5. doi: 10.1016/j.neulet.2009.08.074. 19733211 [DOI] [PubMed] [Google Scholar]
- 82.Schopfer F.J., Cipollina C., Freeman B.A. Formation and signaling actions of electrophilic lipids. Chemical Reviews. 2011;111:5997–6021. doi: 10.1021/cr200131e. 21928855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Higdon A.N., Dranka B.P., Hill B.G., Oh J.Y., Johnson M.S., Landar A., Darley-Usmar V.M. Methods for imaging and detecting modification of proteins by reactive lipid species. Free Radical Biology & Medicine. 2009;47:201–212. doi: 10.1016/j.freeradbiomed.2009.05.009. 19446632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Oh J.Y., Giles N., Landar A., Darley-Usmar V. Accumulation of 15-deoxy-delta(12,14)-prostaglandin J2 adduct formation with Keap1 over time: effects on potency for intracellular antioxidant defence induction. Biochemical Journal. 2008;411:297–306. doi: 10.1042/bj20071189. 18237271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Levonen A.L., Landar A., Ramachandran A., Ceaser E.K., Dickinson D.A., Zanoni G., Morrow J.D., Darley-Usmar V.M. Cellular mechanisms of redox cell signalling: role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochemical Journal. 2004;378:373–382. doi: 10.1042/BJ20031049. 14616092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ahn Y.H., Hwang Y., Liu H., Wang X.J., Zhang Y., Stephenson K.K., Boronina T.N., Cole R.N., Dinkova-Kostova A.T., Talalay P., Cole P.A. Electrophilic tuning of the chemoprotective natural product sulforaphane. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9590–9595. doi: 10.1073/pnas.1004104107. 20439747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dinkova-Kostova A.T., Holtzclaw W.D., Cole R.N., Itoh K., Wakabayashi N., Katoh Y., Yamamoto M., Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. 12193649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schopfer F.J., Batthyany C., Baker P.R., Bonacci G., Cole M.P., Rudolph V., Groeger A.L., Rudolph T.K., Nadtochiy S., Brookes P.S., Freeman B.A. Detection and quantification of protein adduction by electrophilic fatty acids: mitochondrial generation of fatty acid nitroalkene derivatives. Free Radical Biology & Medicine. 2009;46:1250–1259. doi: 10.1016/j.freeradbiomed.2008.12.025. 19353781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Linder M.E., Deschenes R.J. Palmitoylation: policing protein stability and traffic. Nature Reviews. Molecular Cell Biology. 2007;8:74–84. doi: 10.1038/nrm2084. 17183362 [DOI] [PubMed] [Google Scholar]
- 90.Tulloch L.B., Howie J., Wypijewski K.J., Wilson C.R., Bernard W.G., Shattock M.J., Fuller W. The inhibitory effect of phospholemman on the sodium pump requires its palmitoylation. Journal of Biological Chemistry. 2011;286:36020–36031. doi: 10.1074/jbc.M111.282145. 21868384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Burcham P.C. Modified protein carbonyl assay detects oxidised membrane proteins: a new tool for assessing drug- and chemically-induced oxidative cell injury. Journal of Pharmacological and Toxicological Methods. 2007;56:18–22. doi: 10.1016/j.vascn.2006.02.015. 17395496 [DOI] [PubMed] [Google Scholar]
- 92.Wehr N.B., Levine R.L. Quantification of protein carbonylation. Methods in Molecular Biology (Clifton, N.J.) 2013;965:265–281. doi: 10.1007/978-1-62703-239-1_18. 23296665 [DOI] [PubMed] [Google Scholar]
- 93.Colombo G., Dalle-Donne I., Orioli M., Giustarini D., Rossi R., Clerici M., Regazzoni L., Aldini G., Milzani A., Butterfield D.A., Gagliano N. Oxidative damage in human gingival fibroblasts exposed to cigarette smoke. Free Radical Biology & Medicine. 2012;52:1584–1596. doi: 10.1016/j.freeradbiomed.2012.02.030. 22387198 [DOI] [PubMed] [Google Scholar]
- 94.Fedorova M., Kuleva N., Hoffmann R. Identification, quantification, and functional aspects of skeletal muscle protein-carbonylation in vivo during acute oxidative stress. Journal of Proteome Research. 2010;9:2516–2526. doi: 10.1021/pr901182r. 20377239 [DOI] [PubMed] [Google Scholar]
- 95.Yang Y., Loscalzo J. S-nitrosoprotein formation and localization in endothelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:117–122. doi: 10.1073/pnas.0405989102. 15618409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang Y., Song Y., Loscalzo J. Regulation of the protein disulfide proteome by mitochondria in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10813–10817. doi: 10.1073/pnas.0702027104. 17581874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.James A.M., Cochemé H.M., Murphy M.P. Mitochondria-targeted redox probes as tools in the study of oxidative damage and ageing. Mechanisms of Ageing and Development. 2005;126:982–986. doi: 10.1016/j.mad.2005.03.026. 15923020 [DOI] [PubMed] [Google Scholar]
- 98.Lin T.K., Hughes G., Muratovska A., Blaikie F.H., Brookes P.S., Darley-Usmar V., Smith R.A., Murphy M.P. Specific modification of mitochondrial protein thiols in response to oxidative stress: a proteomics approach. Journal of Biological Chemistry. 2002;277:17048–17056. doi: 10.1074/jbc.M110797200. 11861642 [DOI] [PubMed] [Google Scholar]
- 99.Logan A., Cochemé H.M., Li Pun P.B., Apostolova N., Smith R.A.J., Larsen L., Larsen D.S., James A.M., Fearnley I.M., Rogatti S., Prime T.A., Finichiu P.G., Dare A., Chouchani E.T., Pell V.R., Methner C., Quin C., McQuaker S.J., Krieg T., Hartley R.C., Murphy M.P. Using exomarkers to assess mitochondrial reactive species in vivo. Biochimica et Biophysica Acta (BBA) – General Subjects. 2014;1840:923–930. doi: 10.1016/j.bbagen.2013.05.026. 23726990 [DOI] [PubMed] [Google Scholar]
- 100.Lukyanov K.A., Belousov V.V. Genetically encoded fluorescent redox sensors. Biochimica et Biophysica Acta. 2014;1840:745–756. doi: 10.1016/j.bbagen.2013.05.030. 23726987 [DOI] [PubMed] [Google Scholar]
- 101.Meyer A.J., Dick T.P. Fluorescent protein-based redox probes. Antioxidants & Redox Signaling. 2010;13:621–650. doi: 10.1089/ars.2009.2948. 20088706 [DOI] [PubMed] [Google Scholar]
- 102.Bilan D.S., Pase L., Joosen L., Gorokhovatsky A.Y., Ermakova Y.G., Gadella T.W., Grabher C., Schultz C., Lukyanov S., Belousov V.V. HyPer-3: a genetically encoded H(2)O(2) probe with improved performance for ratiometric and fluorescence lifetime imaging. ACS Chemical Biology. 2013;8:535–542. doi: 10.1021/cb300625g. 23256573 [DOI] [PubMed] [Google Scholar]
- 103.Belousov V.V., Fradkov A.F., Lukyanov K.A., Staroverov D.B., Shakhbazov K.S., Terskikh A.V., Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nature Methods. 2006;3:281–286. doi: 10.1038/nmeth866. 16554833 [DOI] [PubMed] [Google Scholar]
- 104.Gutscher M., Pauleau A.L., Marty L., Brach T., Wabnitz G.H., Samstag Y., Meyer A.J., Dick T.P. Real-time imaging of the intracellular glutathione redox potential. Nature Methods. 2008;5:553–559. doi: 10.1038/nmeth.1212. 18469822 [DOI] [PubMed] [Google Scholar]
- 105.Bilan D.S., Matlashov M.E., Gorokhovatsky A.Y., Schultz C., Enikolopov G., Belousov V.V. Genetically encoded fluorescent indicator for imaging NAD(+)/NADH ratio changes in different cellular compartments. Biochimica et Biophysica Acta. 2014;1840:951–957. doi: 10.1016/j.bbagen.2013.11.018. 24286672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Niethammer P., Grabher C., Look A.T., Mitchison T.J. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature. 2009;459:996–999. doi: 10.1038/nature08119. 19494811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Breckwoldt M.O., Pfister F.M., Bradley P.M., Marinković P., Williams P.R., Brill M.S., Plomer B., Schmalz A., St Clair D.K., Naumann R., Griesbeck O., Schwarzländer M., Godinho L., Bareyre F.M., Dick T.P., Kerschensteiner M., Misgeld T. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nature Medicine. 2014;20:555–560. doi: 10.1038/nm.3520. 24747747 [DOI] [PubMed] [Google Scholar]


