Abstract
Multiple cancers may arise from within a clonal region of preneoplastic epithelium, a phenomenon termed ‘field change’1, 2. However, it is not known how field change develops. Here we investigate this question using lineage tracing to track the behaviour of scattered single oesophageal epithelial progenitor cells expressing a mutation that inhibits the Notch signaling pathway. Notch is frequently subject to inactivating mutation in squamous cancers3-6. Quantitative analysis reveals that cell divisions which produce two differentiated daughters are absent in mutant progenitors. As a result mutant clones are no longer lost by differentiation and become functionally immortal. In addition, mutant cells promote the differentiation of neighbouring wild type cells, which are then lost from the tissue. These effects lead to clonal expansion, with mutant cells eventually replacing the entire epithelium. Furthermore, Notch inhibition in progenitors carrying p53 stabilizing mutations creates large confluent regions of doubly mutant epithelium. Field change is thus a consequence of imbalanced differentiation in individual progenitor cells.
Murine oesophageal epithelium (OE) is a stratified squamous epithelium consisting of layers of keratinocytes (Supplementary Fig. 1a). The uniformity of OE, which lacks any glands or other appendages, lends itself to resolving cell behaviour by lineage tracing7. Proliferation is confined to the basal layer. On commitment to terminal differentiation, basal cells exit the cell cycle and subsequently migrate to the tissue surface from which they are shed. Cell turnover is maintained by a single population of progenitor (P) cells, which divide to generate two P cells (PP), two differentiating (D) cells (DD), or one P and one D cell (PD) (Supplementary Fig. 1b)7. The outcome of an individual division is unpredictable, but the probabilities of each type of division are balanced so that on average, across the progenitor population, 50% P and 50% D cells are produced per division and tissue homeostasis is achieved. Normally, the descendants of a given cell have a high probability of being lost by differentiation within a few rounds of division (Supplementary Fig. 1c)7. Mutations which decrease the probability of the DD division outcome have an increased likelihood of creating persistent, expanding clones.
Notch pathway genes are expressed in normal OE8, 9. Notch is a transmembrane receptor, which is cleaved by gamma secretase upon ligand binding, freeing the Notch intracellular domain (Nicd) to migrate to the nucleus. Nicd then forms a complex with the DNA binding protein Rbpj and other proteins including Mastermind like 1 (Maml1), resulting in the transcription of target genes10. In another stratified squamous epithelium, mouse epidermis, widespread deletion of Notch impairs differentiation and promotes inflammation and tumor formation11-13. Blockade of Notch signaling in oesophageal keratinocytes also inhibits differentiation, but has not been shown to result in tumour formation14. Here we set out to induce a Notch inhibiting mutation in individual progenitor cells to study the earliest stage of tumour evolution, the establishment of mutant clones within a background of wild type cells.
In order to both inhibit Notch and visualize mutant cells for genetic lineage tracing, we used mice carrying a conditional dominant negative mutant of Maml1 (DNM), which inhibits Nicd induced transcription and is fused to Green Fluorescent Protein (GFP), ensuring all mutant cells express GFP14-17. This R26flDNM line was crossed with the AhcreERT strain that carries a drug inducible form of cre recombinase, allowing the sporadic induction of DNM in basal layer cells (Fig. 1a)7.
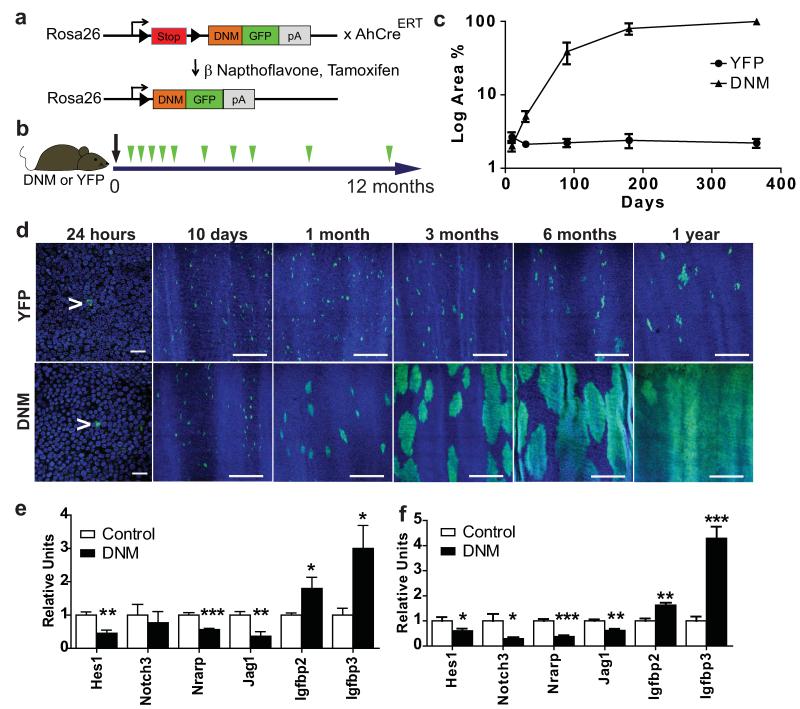
Figure 1. Notch inhibition by DNM leads to clonal expansion.
a: Dominant negative mastermind (DNM) mouse. The Notch binding domain of Maml1 (orange) is fused to GFP (green) and targeted to the Rosa26 locus downstream of a ‘stop’ cassette (red). Following cre induction the stop cassette is excised and DNM protein expressed in a small proportion of oesophageal basal cells. In the control strain, Yellow Fluorescent Protein (YFP) is similarly targeted to the Rosa26 locus. b: Protocol: Clonal labelling was induced in AhcreERTR26flDNM/wt (DNM) and control AhcreERTR26flEYFP/wt (YFP) mice, and samples taken from 24 hours to 1 year post induction (green arrows). c: Quantification of area recombined epithelium over time. Three images were analysed from each of three mice at each time point for each mouse strain. d: Rendered confocal z stacks of OE showing ‘top down’ views of typical areas of wholemounts at times indicated, green indicates DNM or YFP, blue is Dapi. Scale bars, 50μm in 24 hour panels and 300μm in other panels. Arrows indicate single recombined cells. e,f: QRT-PCR of Notch regulated transcripts in OE, relative to Gapdh mRNA. e: Flow sorted DNM expressing basal cells compared with control DNM negative basal cells at 15 days post induction. f: Unsorted epithelium from induced DNM mice compared with age matched, uninduced, DNM control mice 1 year post induction. Values are means of 5 (control) or 6 (DNM) independent biological repeats at each time point, normalized to control (=1). Error bars are SEM *, p< 0.05; **, p < 0.01; ***, p< 0.001 by t test.
We began by inducing DNM expression in 1 in 500 (±100, SEM) basal cells, comparing the effects with a control cohort of AhcreERTR26YFP/wt animals expressing Yellow Fluorescent Protein (YFP) from the same locus (Fig. 1b)7. Control YFP labelled progenitors remain in homeostasis, occupying a constant proportion of OE over a one year time course (Fig. 1c,d). In contrast, single cell derived DNM clones expanded rapidly. Later, DNM clones began to coalesce and the rate of expansion of the mutant population slowed. Strikingly, after a year the entire epithelium was replaced by DNM cells (Fig. 1c,d and Supplementary Fig. 1d). Changes in transcription of genes directly or indirectly regulated by Notch in keratinocytes confirmed the pathway was inhibited in DNM cells at 15 days and 1 year (Fig. 1e,f)16, 18.
To understand how DNM changes cell behaviour, we performed 3D imaging of clones at early time points (Fig. 2a-c). In control mice, at 10 days post induction, there were frequent (146/492) multicellular “floating” clones consisting of only suprabasal cells. These result from the differentiation of the founder cell and its progeny following DD type divisions. In contrast, no such clones (0/570) were detected in DNM mice at 10 days post induction (Fig. 2c,d). This indicates that the likelihood of DD type divisions was greatly reduced, or even abolished in DNM cells. As a consequence DNM clones are not lost by differentiation and become functionally ‘immortal’ in a wild type background.
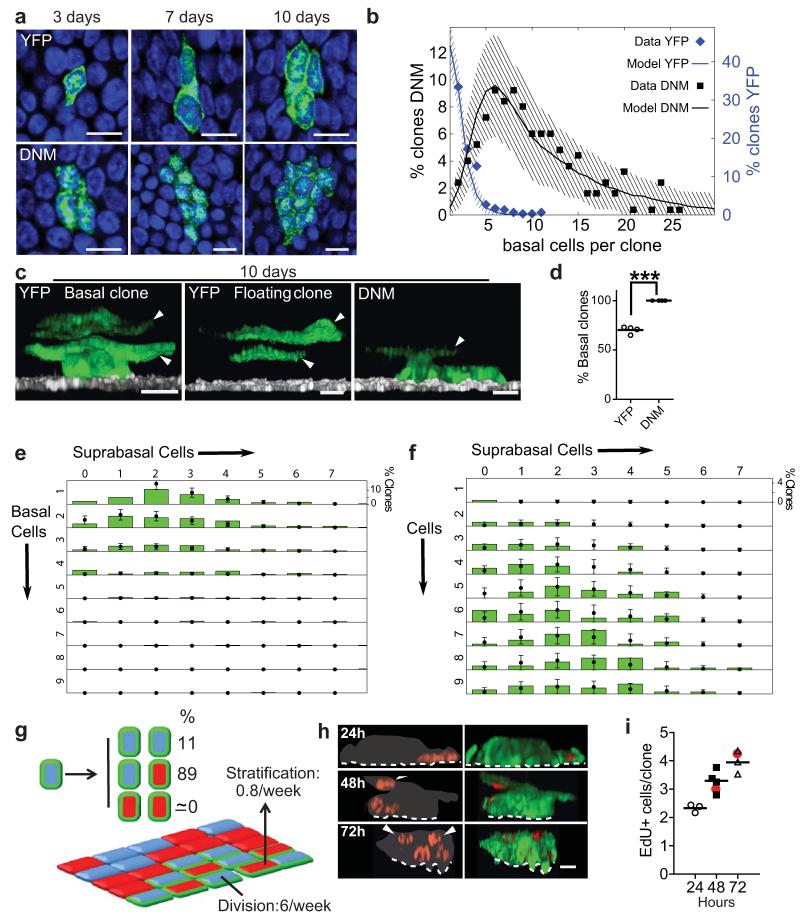
Figure 2. DNM cell fate is tilted towards proliferation at early time points.
a: Confocal images of the basal layer showing typical clones at times indicated. Green, DNM or YFP, blue Dapi, scale bars 10μm. b: Basal cells/clone at 10 days post-induction. Points are data for frequency of clones, YFP (blue, 215 clones), DNM (black, 250 clones), imaged from three animals of each strain in one experiment. Lines show predictions of models shown in g (DNM) and Supplementary Figure 1b (YFP). Shading indicates 95% confidence interval of the model prediction (see Supplementary Note). c: Three-dimensional reconstructions showing typical 10 day clones in DNM and YFP mice, arrows indicate suprabasal cells. Note floating YFP clone with no basal cells. Green, DNM or YFP; white, basal cell marker α6 integrin; scale bars 10μm. d: Proportion of ‘basal’ clones, containing one or more basal cells, imaged 10 days post-induction in YFP (346 clones, open circles) and DNM mice (320 clones, solid circles). Values are means/mouse from four mice per strain in one experiment, ***, p< 0.001 by 2 way Anova. e, f: Size distribution of clones categorized by the number of basal and suprabasal cells, 10 days after induction in YFP (e, 215 clones) and DNM (f, 250 clones) mice from a single experiment. Green bars are data from cell lineage tracing and points are predictions of the models in g (DNM) and Supplementary Fig. 1b (YFP control). Error bars indicate 95% confidence intervals of the model predictions. g: Model of DNM cell behaviour at early time points (see Supplementary Note). Division rate of DNM progenitors (blue with green edge) is 6/week (acceptable parameter range 5.3-6.4, YFP control 2/week, Supplementary Fig. 1b). Stratification rate (arrow) of differentiated DNM cells (red with green edge) is 0.8/week, (0.6-1.1/week, YFP 3.5/week). The proportion of divisions with DD outcome is negligible, 0% (0-0.7%, YFP 10%), 89% of divisions are PD (85-91%, YFP 80%) and 11% are PP (8.5-15%, YFP 10%). Clone is set amid wild type progenitors (blue) and differentiated cells (red). h,i: Validation of model by EdU lineage tracing. 7 days post induction, DNM mice were administered a single dose of EdU and sampled after 24, 48 and 72 hours. h: Rendered confocal Z stacks of typical clones at the times indicated, red is EdU, green DNM, grey indicates clone boundary, scale bars 10μm. i: Mean EdU positive cells per clone in each of three mice per time point in one experiment. Total number of clones imaged was 57 (24 hours), 60 (48 hours) and 57 (72 hours). Red dots indicate predictions of model shown in g (see Supplementary Note).
To gain quantitative insight into cell behaviour in DNM clones, we scored the number of basal and suprabasal cells in clones at 7, 10 and 15 days post induction (Fig. 2e,f and Supplementary Fig. 2a-d). In previous studies, we have shown that the behaviour of wild type progenitors can be captured by a simple biophysical model involving only the rate of progenitor cell division, the stratification rate of differentiated basal cells into the suprabasal layer, and the relative frequency of symmetric to asymmetric cell division7. We investigated whether the seemingly complex range of DNM clonal behaviour at 7 and 10 days post-induction could be predicted simply by relaxing the condition of balanced fate of normal progenitors (Fig. 2g, Supplementary Note). Using a Bayesian-like approach we found that the full range of clonal fate data could be predicted by such a model (Fig. 2b,e,f, Supplementary Fig. 2a-g and Supplementary Note). Crucially, the statistically most likely scenario is absence of DD divisions. The observation that floating clones are absent can further be quantified, bounding the ratio of DD divisions by maximally 0.7%, with 95% confidence (Supplementary Note). Intriguingly, asymmetric PD type divisions are not reduced but appear to replace DD divisions in DNM cells, resulting in cell fate being tilted towards proliferation7. We also find that cell division rate is increased to three fold that of normal cells in DNM clones, while the rate of stratification of differentiated DNM basal cells is decreased over four fold (Fig. 2g).
The model was challenged by tracking cells within DNM clones labelled by a pulse of Ethinyl-deoxyUridine (EdU), which is taken up by progenitors in S Phase (Fig. 2h). The mean number of EdU positive cells/clone predicted by the model gave an excellent fit to the data (Fig. 2i, Supplementary Note). We further validated the model by using the fit of the day 7 and 10 data to successfully predict the clone size distribution at 15 days post induction (Supplementary Fig. 2d). Collectively these changes in cell behaviour result in the persistence and rapid geometric expansion of DNM clones at these early time points.
The analysis of cell behaviour reveals the net effect of all cell intrinsic and/or extrinsic molecular alterations induced by DNM, but does not disclose the specific pathways and genes involved. To gain insight into the transcriptional changes associated with DNM expression we performed transcriptional array analysis on flow sorted DNM positive and negative basal cells 15 days after induction. We found the expected changes in transcripts already shown to be regulated by DNM by quantitative RT-PCR (Nrarp, Jag1, Igfbp2 and Igfbp3, Fig. 1e). In addition we identified significant enrichment of additional transcripts involved in epidermal development, RNA processing and cytoskeletal organization (Supplementary Tables 1 and 2). We further validated three of these transcripts, the transcription factor Sox9, decreased in DNM cells, and the hemidesmosome component Itgb4 and hyperproliferation associated keratin Krt6 whose expression was increased in DNM cells (>30% fold change, P<0.01). Immunostaining confirmed decreased expression of Sox9 and increased levels of Itgb4 and the Krt6 within DNM cells (Supplementary Fig. 3).
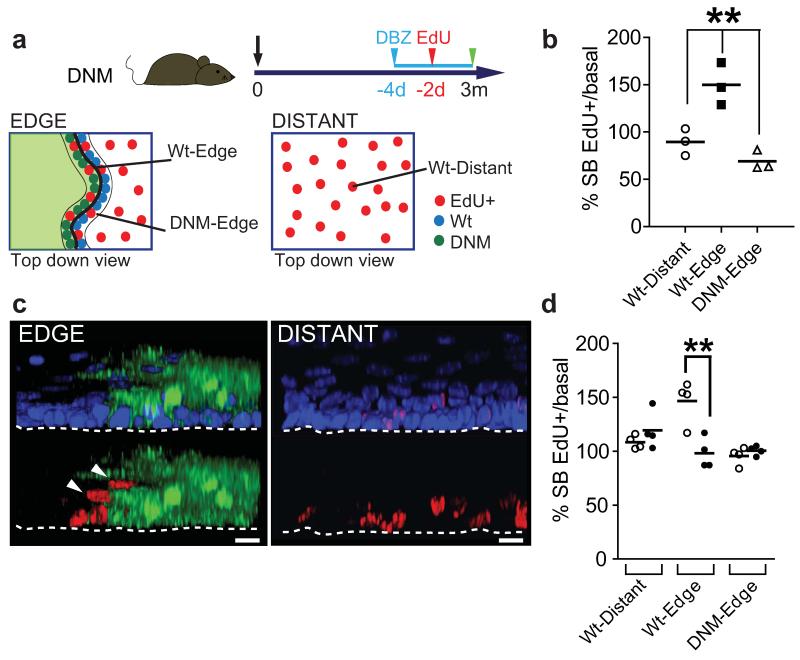
We hypothesized that the expansion of DNM clones may alter the behaviour of adjacent wild type cells. To test if this was the case, a pulse of EdU was given to DNM mice 3 months post induction. The proportion of EdU labelled cells that had differentiated and stratified into the suprabasal cell layers was determined 48 hours later (Fig. 3a). We found a significant increase in suprabasal EdU positive wild type cells immediately adjacent to DNM clones (Fig. 3b,c). The effect is rescued by treating mice with dibenzazepine (DBZ), which blocks Notch signalling by inhibiting gamma secretase (Fig. 3d)19. This argues that Notch is activated in wild type cells adjacent to DNM clones, promoting their differentiation. A similar effect has been reported in cultured human epidermal keratinocytes20. We conclude the accelerated Notch driven differentiation of wild type cells at the edges of DNM clones contributes to their colonization of the epithelium.
Figure 3. DNM accelerates neighbouring wild type cell stratification in a Notch dependent manner.
a: Protocol: DNM mice were induced, and treated with EdU 48 hours prior to culling at 3 months post induction. b: Ratio of EdU positive suprabasal: basal cells at the edge and distant from clones. Values are means from each animal in one experiment. At least 1100 EdU positive cells from three animals were analysed per group. **, p < 0.01 by t test. c: Rendered confocal z stacks showing typical location of EdU labelled cells (red) at the DNM clone edge (green), and distant from clone. Dapi is blue, scale bars 10μm. Arrows indicate suprabasal, EdU positive wild type cells adjacent to clone. d: Ratio of EdU positive suprabasal:basal cells at clone edges in DNM animals treated with DBZ or vehicle control. Values are means from each animal in one experiment. At least 1100 EdU positive cells from three animals were analysed per group. ** p<0.01 by t test.
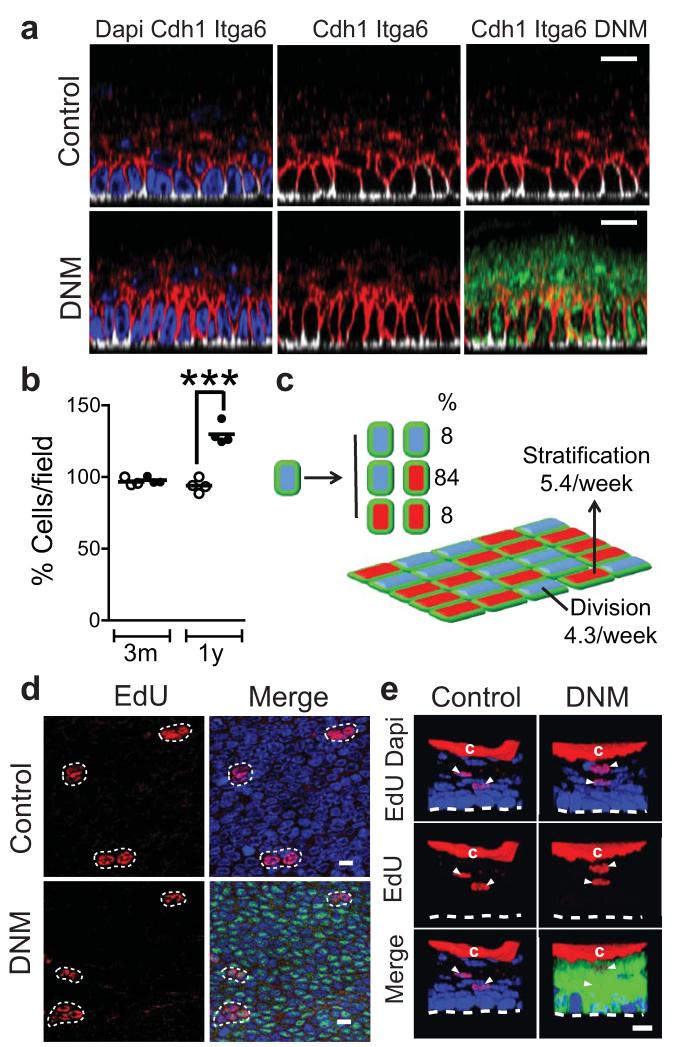
The explosive early expansion of the DNM population led us to expect that the tissue would become severely disrupted, perhaps developing tumours over time. However, after a year, when the epithelium had been completely replaced by DNM cells, tissue integrity was maintained, animals remained healthy and no oesophageal tumours were seen in 17 mice. The most noticeable tissue abnormality was marked cell crowding in the basal layer (Fig. 4 a,b).
Figure 4. DNM epithelium 1 year post induction.
a: Typical lateral views of Z stacks stained for the membrane marker Cdh1 (E-cadherin, red), showing basal cell crowding in one year induced DNM animals (lower panels), compared to aged-matched un-induced controls (upper panels). DNM is green, Dapi in blue, basal cell marker Itga6 (α6 integrin), white. Scale bars 10μm. b: Mean basal cell density in DNM animals induced for 3 months and 1 year (solid circles), or un-induced controls (open circles). Data are from at least four fields per mouse in 3 animals at 3 months and 4 animals at 1 year. ***, p< 0.001 by t test. c: Model of cell behaviour 1 year post induction, derived from EdU lineage tracing (d,e). DNM progenitor cell division rate is 4.3/week (acceptable parameter range 3.7-4.9), stratification rate of mutant differentiated cells 5.4/week (5.0-5.8) and proportion of PP and DD divisions 8% (5-11%). Analysis is based on a total of 86 (control, 2 mice) and 78 (induced, 3 mice) EdU doublets, in one experiment. d,e: Clonal EdU lineage tracing. EdU was administered at 6pm, resulting in sparse labelling. Epithelial wholemounts were imaged after 48 hours in DNM mice or age matched uninduced DNM controls. EdU (red), DNM (green); DAPI (blue). Scale bars 10μm. d: Confocal optical slices of the basal layer showing representative EdU+ doublets and triplets e: Confocal Z stack reconstructions of typical ‘floating’ EdU positive suprabasal cell doublets (arrows), indicating restoration of DD outcome of cell division (Supplementary Note). ‘c’ indicates non specific EdU staining in cornified layer.
Notch inactivation in the epidermis is associated with elevation of circulating levels of the defensin Tslp and epithelial or stromal inflammation (Supplementary Fig. 4a)12, 13, 21, 22. We observed none of these changes in oesophageal epithelium (Supplementary Fig. 4b-g). Indeed, far from being increased, immune cells were partially excluded from DNM epithelium. No apoptosis was detected (Supplementary Fig. 4h). We conclude Notch inhibition in the oesophagus does not trigger the inflammatory response that is known to be a key driver of tumour formation in the epidermis.
We went on to investigate the behaviour of mutant cells within the DNM epithelium at 1 year. Tracking of EdU labelled cells revealed that the epithelium had established a new homeostatic state (Fig. 4c-e). Crucially, DD type divisions, and hence, the balanced production of differentiating and proliferating cells had been restored (Fig. 4c,e, Supplementary Note). The cell division rate remained elevated, but the stratification rate had risen, so the DNM populated tissue was in a new ‘steady state’ with a more rapid cell turnover (Fig. 4c). The reestablishment of the DD channel may be, at least in part, a response to the crowding of basal cells, which is known to promote keratinocyte differentiation in vitro23. Thus, once the tissue is fully colonized, the balanced proliferation and differentiation of DNM cells ensures they persist as a self maintaining population. Endogenous levels of Notch signalling seem not to be required to sustain tissue integrity.
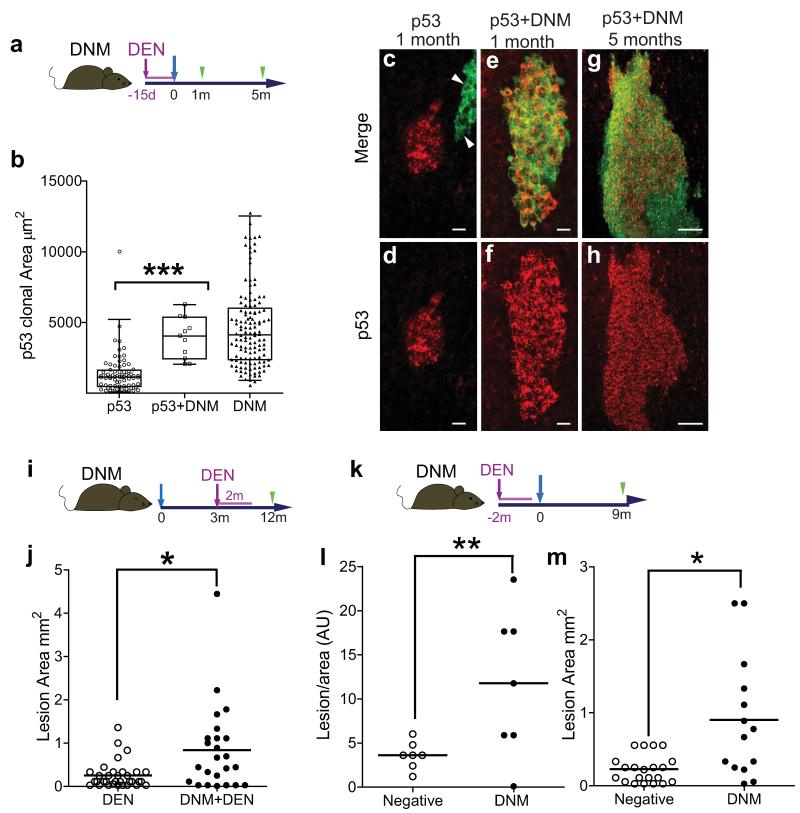
We hypothesized that if Notch signalling was inhibited in a progenitor already carrying oncogenic mutation(s), it may create a dominant clone, expanding into a region of multiply mutant cells. To test this, we treated DNM mice with the mutagen Diethylnitrosamine (DEN), then induced DNM expression, and subsequently examined epithelial wholemounts for clusters of cells expressing detectable levels of p53 (Tp53, Fig. 5a)24. Such clusters result from clonal oncogenic mutations that stabilize the p53 protein25, 26. At one month post DNM induction, the rare clones expressing both DNM and p53 had expanded to the same extent as clones expressing DNM alone, but were significantly larger (p<0.0001 by t test) than those staining for p53 alone (Fig. 5b-f). This suggests that the expansion of double mutant clones is due to the effect of DNM on cell behaviour, rather than intracellular cooperation between the mutations. The double mutant clones expand into large confluent regions by 5 months (Fig. 5g,h). This argues that mutations such as DNM which imbalance progenitor differentiation in single progenitor cells can cause field change in a squamous epithelium.
Figure 5. DNM expression expands p53 mutant clones.
a: Protocol: DNM mice were treated with Diethylnitrosamine (DEN, purple) for 15 days, induced (blue arrow) and wholemounts prepared 1 and 5 months post induction (green arrows). b: Scatter and Box plot showing area of clones staining for p53 alone (n=75 clones, open circles), both p53 and DNM (n=11, open squares), or DNM alone (n=129, closed triangles). Oesophagi from 6 mice were imaged one month post DNM induction in one experiment. Whiskers indicate 2.5-97.5 percentiles, ***p< 10−6 by Mann-Whitney U test. c-f: Representative z-stack projections one month post induction. Typical clones positive for p53 (red) alone (arrows indicate clone positive for DNM alone, c, d), or for both DNM (green) and p53 (e, f) are shown, scale bars 10μm. g, h Area of double positive epithelium, in DEN-treated DNM mouse 5 months after induction, scale bar 50μm. i: Protocol to investigate the effect of subsequent carcinogen exposure on DNM expressing epithelium: DNM mice were induced (blue arrow) (DNM+DEN) or kept un-induced (DEN) and after a 3 month interval treated with DEN for 2 months. The area of tumours in epithelial wholemounts was recorded 10 months later (j). Results are from one experiment with 6 animals per group, * p= 0.014 by Mann-Whitney U test. k: Protocol to investigate the effect of long-term DNM on DEN treated epithelium. DNM mice were treated with Diethylnitrosamine (DEN, purple) for 2 months, followed by DNM induction (blue arrow). Frequency and area of tumours was analyzed 9 months after DNM induction (green arrow, l,m). l: Number of lesions per unit area in DNM positive (open circles) and negative areas (closed circles) in each of 7 mice in one experiment, * P<0.05 by t test. m: Area of individual lesions positive (closed circles) and negative (open circles) for DNM (** P <0.005 by Mann Whitney U test).
We also examined whether DNM expression had any influence on the effects of subsequent carcinogen treatment, by administering DEN after DNM induction (Fig. 5i). We observed an increase in size of epithelial lesions in induced DEN treated mice compared with uninduced DNM controls (Fig. 5j, Supplementary Fig. 5a-d). Epithelial colonization by cells carrying a Notch inhibiting mutation thus has adverse consequences in the context of carcinogen exposure.
Finally, we performed a third protocol in which we induced DNM animals subsequent to 2 months of DEN administration in order to explore the long term effect of DNM on a widely mutated epithelium (Fig. 5k-m). Whole mount samples analyzed 9 months post induction revealed DNM positive areas were restricted to 17 ± 0.1 % (mean ±SD) of the total area of the epithelium. This is probably due to the presence of adjacent mutant clones, created by DEN treatment, which resist the incursion of DNM cells. Interestingly, the frequency of lesions per unit area was 3 fold higher in DNM positive areas when compared to non-DNM areas within the same mice (Fig. 5l). This suggests that DNM may play a role in tumour formation. The size of DNM expressing lesions size was also increased (Fig. 5m), consistent with the results of the second protocol (Fig. 5j).
We conclude that inhibition of Notch confers clonal dominance in oesophageal epithelium, by blocking DD divisions in mutant cells and promoting the differentiation of adjacent wild type cells. Mutant cells then replace the normal epithelium. Once colonization is complete the balance of differentiation and proliferation is restored, establishing a self-maintaining mutant population. If a mutation which blocks division resulting in symmetric differentiation occurs in a cell already carrying one or more oncogenic mutations, a large area of epithelium with an increased likelihood of further transformation, i.e. field change, may develop. Notch inhibition increases the number and size of tumours that develop following carcinogen treatment. These results illustrate that as well as cooperating at the molecular level within cells, oncogenic mutations may also interact at the level of cell dynamics in early cancer evolution.
Online Methods
Animals
All experiments were approved by MRC and University of Cambridge local ethical review committees and conducted according to Home Office project licenses PPL80/2056, PPL22/2282 and PPL70/7543. Experimental mice were doubly heterozygous for the inducible cre allele AhcreERT and a conditional allele encoding a dominant negative mutant of mastermind-like1 (DNM) fused to Green Fluorescent Protein (GFP), downstream of a LoxP flanked ‘Stop’ cassette targeted to the Rosa 26 locus (AhcreERTR26flDNM-GFP/wt, DNM)14. In these mice, transcription of a cre mutant oestrogen receptor fusion protein (creERT) is induced by β-napthoflavone. Tamoxifen is also required for creERT to gain access to the nucleus. In the presence of both drugs, creERT removes the “Stop” cassette resulting in DNM expression in the cell and its progeny. Controls included both age-matched un-induced DNM mice as well as the previously characterized AhcreERTR26flEYFP/wt (YFP) strain, which expresses Yellow Fluorescent Protein (YFP) from the Rosa 26 locus following cre induction7.
Animals were induced between 10 and 16 weeks of age. All strains were maintained in a C57Bl6 background. Experiments were performed with male and female animals, except for array and transcriptional analysis where only males were used. No gender specific differences were observed.
Lineage tracing
Low frequency expression of DNM in the mouse oesophagus was achieved by inducing animals aged 10-16 weeks with an intraperitoneal dose of 80μg/kg βNapthoflavone and 0.3mg Tamoxifen, which resulted in a mean recombination efficiency of 1 cell every 526 basal cells (±117, SEM). At this dose, negligible induction of DNM was seen in wholemount preparation of other tissues including the ear, back and tail epidermis.
Following induction, 3-4 DNM or YFP mice per time point were culled and the oesophagus harvested. Time points analyzed included: 1, 7, 10 and 15 days, 1, 3, 6 months, and 1 year. Clones were imaged after immunostaining wholemounts, described below, on a Nikon ECLIPSE TE2000-U confocal microscope. At 24 hours only single basal cells were seen to be stain for GFP, indicating DNM expression. The frequency of these cells was scored to determine the initial labelling frequency (above). At 7, 10 and 15 days the number of basal and suprabasal cells in each clone was counted under live acquisition mode for clones containing one or more basal cells. The percentage of “floating clones”, containing only suprabasal cells, was determined by 3D imaging at 10 days post induction. The quantitative analysis of this data is described in the Supplementary Note.
Representative images of clones were produced by rendering confocal z stacks with the following settings: 40× objective with 3× Digital zoom, optimal pinhole, speed 400Hz, line average 3, resolution: 1024×1024. Images were reconstructed from optical sections using Volocity 6 software (PerkinElmer).
After 15 days clonal lineage tracing became infeasible due to the merger of DNM clones. We therefore measured the area of DNM and EYFP expressing epithelium from low magnification projected z-stack at 10 days, 1, 3, 6, and 12 months. 3 mice were analyzed per time point with 3 images per animal at 10× magnification (Fig 1c, d). Areas were quantified using Volocity 6 software.
Immunofluorescence and Immunohistochemistry
For wholemount staining, epithelium from the middle third of the oesophagus were prepared by dissecting the tissue into rectangular pieces of approximately 5 by 8mm and incubating for 2-3 hours in 5mM EDTA at 37°C. The epithelium was then carefully peeled away from underlying tissue with fine forceps and fixed in 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) for 15-25 minutes. For staining, wholemounts were blocked for 1 hour in staining buffer (0.5% Bovine Serum Albumin, 0.25% Fish Skin Gelatin, and 0.5% Triton X-100 in PBS) with 10% donkey or goat serum, according to the secondary antibody used. Primary and secondary antibodies were incubated in staining buffer overnight, followed by washing for 2 hours with 0.2% Tween-20 in PBS. A final overnight incubation with Dapi 1μg/ml in PBS was used to stain cell nuclei.
For staining conventional tissue sections, OCT embedded cryosections of 10μm thickness were fixed with 4% PFA for 5 minutes, and blocked and stained using staining buffer with the respective primary and secondary antibodies for 1 hour at room temperature. Samples were washed with PBS between incubations.
EdU incorporation was detected with a Click-iT® chemistry kit according to the manufacturer’s instructions (Invitrogen).
Confocal images were acquired on Nikon ECLIPSE TE2000-U (Objectives: 10×, 20× and 40×. Optimal pinhole. Speed: 400Hz. Line average: 3. Resolution: 1024×1024) and Leica TCS SP5 II confocal (Objectives: 10×, 20× and 40×. Optimal pinhole. Speed: 400Hz. Line average: 3. Resolution: 1024×1024) microscopes and reconstructed using Volocity 6 image processing software (PerkinElmer).
Images shown in Supplementary Figure 4 are typical of at least 4 images per mouse in each of 3 or more mice per experimental group.
Antibodies
Antibodies used are listed in Supplementary table 3.
RNA isolation and real-time RT-PCR of genes directly and indirectly regulated by Notch
To corroborate Notch inhibition in DNM expressing cells, levels of transcripts of genes known to be directly or indirectly regulated by Notch signaling in keratinocytes were analyzed 15 days and 1 year post-induction16, 27.
Wholemounts from oesophageal epithelium were prepared by cutting tissue into 3mm squares and incubating for 15 minutes in 0.5mg/ml Dispase at 37°C, after which the epithelium was peeled from the submucosa. At 15 days most of the DNM positive cells are in the basal layer, so a single cell suspension was prepared for flow sorting of GFP positive and negative basal cells (see below). At 1 year the epithelium consists entirely of DNM cells, so the epithelium of induced DNM animals was compared with age-matched uninduced DNM controls.
RNA from wholemounts or sorted cells was extracted using the RNEasy columns (Qiagen) including on column DNase digestion. The integrity of total RNA was determined by agarose gel electrophoresis. cDNA synthesis of 500ng of total RNA was carried out using QuantiTect-Reverse Transcription Kit (Qiagen). The cDNA synthesis reactions without reverse transcriptase yielded no amplicons in the polymerase chain reaction (PCR) reactions described below.
qRT-PCR was performed using specific primers (Supplementary Table 4) and SYBRGreen mastermix (Qiagen) according to manufacturer’s instructions in a Rotorgene 3000 (Qiagen). Standard curves were generated and each mRNA normalized to Gapdh as an internal control. The relative levels of each mRNA were expressed as ratio to the control values set to 1.
Sorting of DNM recombinant basal cells for RNA analysis
A cohort of 3 animals per group was used to isolate basal oesophageal epithelial cells by Flow cytometric cell sorting. Epithelial sheets were minced and single-cell suspension obtained using gentleMAC™ Dissociator (Miltenyi Biotec) followed by filtration through a 30μm cell strainer. Cells were centrifuged and resuspended in 1% Fetal Bovine Serum in PBS and stained for the basal cell marker α6 integrin (BioLegend; 313610; 5μg/ml) for 30 minutes at 4°C. An isotype control was used to exclude unspecific immunoglobulin binding (BioLegend; 400526; 5μg/ml). Endogenous GFP levels allowed the identification of DNM recombinant cells. 7AAD (2μg/ml) was used to determine cell viability. Two populations of viable single cells were sorted (GFP+/α6+ and GFP-/α6+) on a MoFlow cell sorter (Dako Cytomation) and collected for further RNA analysis.
Genomic expression
Changes in genomic expression were also investigated in RNA isolated from basal FACS-sorted DNM expressing cells 15 days post-induction and compared to DNM negative cells from the same sample in biological triplicate (see above).
The quality of the RNA was determined by using an Agilent 2100 Bioanalyzer. Samples used for hybridization had a RNA Integrity Number ≥7.8. Due to the limited amount of starting material, and therefore, the low RNA yield, the RNA was amplified using a NuGen, Ovation Pico WTA 3300-60 kit according manufacturer’s instructions. 200ng of the amplified RNA were hybridized on a Mouse Whole Genome-6 v2 Illumina array.
The resulting data were transformed using the Variance Stabilization Transformation (VST) from the lumi package (www.bioconductor.org), to avoid bias at low and high intensities, and normalized by quantile normalization. The paired comparisons were then performed using the limma package (Linear Models for Microarray Data, Bioconductor) and the results corrected for multiple testing using False Discovery Rate (FDR). Genes with expression changes greater than 30%, with corrected P<0.05 were considered differentially expressed. Finally, network and functional analyses were performed using IPA (Ingenuity Pathway Analysis® Systems, www.ingenuity.com) and DAVID (http://david.abcc.ncifcrf.gov). Validation of differential expression of Sox9, Krt6 and Itgb4 was performed by immunostaining of cryosections from DNM mice 3 months post induction. The images shown in Supplementary Figure 3 are typical of at least 8 sections each from 3 animals.
EdU lineage tracing
EdU was used for short term lineage tracing in induced DNM mice.
To validate the model of DNM behaviour derived from clonal data, 10μg of EdU was administered by intraperitoneal injection 24, 48 or 72 hours prior sample harvesting at 7 days (Fig. 2h,i). EdU positive cells were then imaged by confocal microscopy of wholemounts in 57 clones (24 hours), 60 clones (48 hours) and 57 clones (72 hours), Figure 2h shows typical clones.
To allow the progeny of single cells to be tracked at 1 year post induction, when the entire epithelium consisted of DNM cells, EdU was administered at 6pm when the proportion of S phase cells is low in the circadian cycle making clonal density labelling feasible28. Samples were collected 48 hours later and wholemounts imaged. Note that in all other experiments EdU was administered at 10am. Total numbers of EdU positive cell doublets imaged were 86 (YFP) and 78 (DNM), typical images are shown in Figures 4d and 4e.
To track the effect of DNM clones on adjacent wild type cells, animals were induced and subsequently treated with 10μg of EdU 48 hours prior sample collection at 3 months post induction. Oesophageal wholemounts were then stained for GFP, EdU and α6 integrin. 6 z-stack images were taken both at clonal junctions and unlabeled areas in each of 3-4 animals. We recorded the proportion of suprabasal/basal EdU positive cells, as a measure of cell stratification, in the first row of GFP positive (DNM-Edge) and GFP negative (wild type-Edge) cells adjacent to the clone boundary, along with randomly drawn ‘edges’ in images of epithelium distant from clones (wild type-Distant). The percentage of EdU positive basal cells/basal Dapi positive cells did not differ significantly between any site (wild type-Edge 16.5% ± 1.43; DNM-Edge 17.7% ± 2.29; wild type-Distant 14.4% ± 1.55, mean ± SEM, t-test no significant difference).
To further investigate whether the increased stratification of wild type cells at clone margins was dependent on the activation of Notch, mice were treated with the γ-secretase inhibitor Dibenzazepine (DBZ; S2711; Selleckchem). DNM mice induced 11 days or 3 months previously were treated with DBZ (suspended in 0.5% hydroxypropylmethylcellulose (Methocel 65HG, 64670-100G-F, Sigma) and 0.1% Tween-80 in water) by daily intraperitoneal injection for four days to avoid intestinal toxicity19, 29, 30. Controls received only vehicle solution. All mice were treated with 10μg of EdU 48 hours prior sample collection.
Basal cell density
Relative cell density was calculated in control and induced DNM mice at 3 months and one year post induction by imaging the number of basal cells per field in at least 4 images each of 3 mice per group at 3 months and 4 mice per group at one year (Fig. 4a,b).
Chemically induced carcinogenesis using Diethylnitrosamine
Diethylnitrosamine (DEN, Sigma) was administered at a concentration of 0.04ml/l in sweetened drinking water24.
To test whether DNM could expand clones carrying pre-existing oncogenic mutations, DNM mice were administered DEN five days per week for 2 weeks to induce sporadic mutations. DEN treatment was then withdrawn and DNM clones induced with 80μg/kg β-napthoflavone and 0.3mg Tamoxifen. One and 5 months post induction, oesophageal wholemounts were taken, immunostained for GFP, marking DNM positive cells, and p53, to label p53-stabilized mutant clones, imaged by confocal microscopy and clone areas quantified using Volocity 6 software26. We measured the area of all p53+ clones in the middle third of the oesophagi from 7 mice, scoring them for DNM expression.
To evaluate the effect of carcinogen on DNM expressing epithelium, DNM mice were induced and 3 months later treated with DEN for 2 months. Controls were age-matched un-induced DNM animals treated with DEN alone. After carcinogen treatment, animals were aged for 8-9 months. Tumors were scored using a dissecting microscope in control and induced animals (6 per group) after removing the muscularis layer with fine forceps. A total of 34 lesions (DEN) or 18 lesions (DNM+DEN) was analysed, typical lesions are shown in Supplementary Figure 5a-d.
Activated Caspase-3 staining
Cryosections of one year induced and uninduced DNM oesophagi were stained for activated caspase-3 as described and imaged by confocal microscopy31. Three mice per group were analyzed. A positive control was obtained by ultraviolet irradiation of a fresh oesophageal sample and maintaining it in explant culture for 24 hours prior to sectioning31. Images shown in Supplementary Figure 4h are typical of 10 sections per mouse from each of 3 mice in each group.
ELISA
Serum TSLP levels in induced DNM mice were measured using a Quantikine mouse TSLP kit (R&D Systems) according to the manufacturer’s instructions in uninduced DNM controls and clonally induced animals (using 80μg/kg β-napthoflavone and 0.3mg Tamoxifen). As a positive control, DNM animals were treated with a higher dose of inducing drugs (80mg/kg β-napthoflavone and 1mg Tamoxifen), inducing epidermal expression of DNM and resulting in TSLP release32. Inset images of tail epidermal wholemounts in Supplementary Figure 4a are typical of 3-11 animals per group.
Statistical Analysis
The analysis of the cell lineage tracing experiments is set out in the Supplementary Note. Data is expressed as mean values ± SEM unless otherwise indicated. Differences between groups were assessed by 2 tailed unpaired t test or Anova for normally distributed data or 2 tailed Mann-Whitney U test for skewed data, using Excel or GraphPad Prism software. No statistical method was used to predetermine sample size. The experiments were not randomized. The Investigators were not blinded to allocation during experiments and outcome assessment.
Accession Number
Array data have been deposited with ArrayExpress, http://www.ebi.ac.uk/arrayexpress, accession number E-MTAB-2334.
Supplementary Material
Acknowledgments
We thank Warren Pear (University of Pennsylvania) for the DNMaml mouse strain, E. Choolun, K. Murai, J. Fowler, and the staff at ARES (Cambridge) for support and technical assistance, and Raphael Kopan for informative discussions. We acknowledge the support of the MRC, the NC3Rs (National Centre for the Replacement, Refinement and Reduction of Animals in Research), the Wellcome Trust (grant numbers WT090334MA; P.H.J. and 098357/Z/12/Z; B.D.S.), Deutsche Forschungsgemeinschaft Research Fellowship (P.G.) and European Union Marie Curie Fellowship PIEF-LIF-2007-220016 (M.P.A.).
References
- 1.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 2.Braakhuis BJ, Tabor MP, Kummer JA, Leemans CR, Brakenhoff RH. A genetic explanation of Slaughter’s concept of field cancerization: evidence and clinical implications. Cancer Res. 2003;63:1727–1730. [PubMed] [Google Scholar]
- 3.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang NJ, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci U S A. 2011;108:17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal N, et al. Comparative genomic analysis of esophageal adenocarcinoma and squamous cell carcinoma. Cancer Discov. 2012;2:899–905. doi: 10.1158/2159-8290.CD-12-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doupe DP, et al. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 2012;337:1091–1093. doi: 10.1126/science.1218835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakamoto K, et al. Reduction of NOTCH1 expression pertains to maturation abnormalities of keratinocytes in squamous neoplasms. Lab Invest. 2012;92:688–702. doi: 10.1038/labinvest.2012.9. [DOI] [PubMed] [Google Scholar]
- 9.Sander GR, Powell BC. Expression of notch receptors and ligands in the adult gut. J Histochem Cytochem. 2004;52:509–516. doi: 10.1177/002215540405200409. [DOI] [PubMed] [Google Scholar]
- 10.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 12.Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16:55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu B, et al. Multifocal epithelial tumors and field cancerization from loss of mesenchymal CSL signaling. Cell. 2012;149:1207–1220. doi: 10.1016/j.cell.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohashi S, et al. NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network. Gastroenterology. 2010 doi: 10.1053/j.gastro.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu L, et al. Notch signaling is an important regulator of type 2 immunity. J Exp Med. 2005;202:1037–1042. doi: 10.1084/jem.20050923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Proweller A, et al. Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res. 2006;66:7438–7444. doi: 10.1158/0008-5472.CAN-06-0793. [DOI] [PubMed] [Google Scholar]
- 17.Naganuma S, et al. Notch receptor inhibition reveals the importance of cyclin D1 and Wnt signaling in invasive esophageal squamous cell carcinoma. Am J Cancer Res. 2012;2:459–475. [PMC free article] [PubMed] [Google Scholar]
- 18.Lee J, Basak JM, Demehri S, Kopan R. Bi-compartmental communication contributes to the opposite proliferative behavior of Notch1-deficient hair follicle and epidermal keratinocytes. Development. 2007;134:2795–2806. doi: 10.1242/dev.02868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Es JH, et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 20.Lowell S, Jones P, Le Roux I, Dunne J, Watt FM. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol. 2000;10:491–500. doi: 10.1016/s0960-9822(00)00451-6. [DOI] [PubMed] [Google Scholar]
- 21.Demehri S, et al. Elevated epidermal thymic stromal lymphopoietin levels establish an antitumor environment in the skin. Cancer Cell. 2012;22:494–505. doi: 10.1016/j.ccr.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Piazza M, Nowell CS, Koch U, Durham AD, Radtke F. Loss of cutaneous TSLP-dependent immune responses skews the balance of inflammation from tumor protective to tumor promoting. Cancer Cell. 2012;22:479–493. doi: 10.1016/j.ccr.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 23.Watt FM, Jordan PW, O’Neill CH. Cell shape controls terminal differentiation of human epidermal keratinocytes. Proc Natl Acad Sci U S A. 1988;85:5576–5580. doi: 10.1073/pnas.85.15.5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rubio CA, Liu FS, Chejfec G, Sveander M. The induction of esophageal tumors in mice: dose and time dependency. In Vivo. 1987;1:35–38. [PubMed] [Google Scholar]
- 25.Bennett WP, et al. Archival analysis of p53 genetic and protein alterations in Chinese esophageal cancer. Oncogene. 1991;6:1779–1784. [PubMed] [Google Scholar]
- 26.Zhang W, Remenyik E, Zelterman D, Brash DE, Wikonkal NM. Escaping the stem cell compartment: sustained UVB exposure allows p53-mutant keratinocytes to colonize adjacent epidermal proliferating units without incurring additional mutations. Proc Natl Acad Sci U S A. 2001;98:13948–13953. doi: 10.1073/pnas.241353198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods References
- 27.Demehri S, Kopan R. Notch signaling in bulge stem cells is not required for selection of hair follicle fate. Development. 2009;136:891–896. doi: 10.1242/dev.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burns ER, Scheving LE, Fawcett DF, Gibbs WM, Galatzan RE. Circadian influence on the frequency of labeled mitoses method in the stratified squamous epithelium of the mouse esophagus and tongue. Anat Rec. 1976;184:265–273. doi: 10.1002/ar.1091840302. [DOI] [PubMed] [Google Scholar]
- 29.Quante M, et al. Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia. Cancer Cell. 2012;21:36–51. doi: 10.1016/j.ccr.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Es JH, de Geest N, van de Born M, Clevers H, Hassan BA. Intestinal stem cells lacking the Math1 tumour suppressor are refractory to Notch inhibitors. Nat Commun. 2010;1:18. doi: 10.1038/ncomms1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clayton E, et al. A single type of progenitor cell maintains normal epidermis. Nature. 2007;446:185–189. doi: 10.1038/nature05574. [DOI] [PubMed] [Google Scholar]
- 32.Demehri S, Morimoto M, Holtzman MJ, Kopan R. Skin-derived TSLP triggers progression from epidermal-barrier defects to asthma. PLoS Biol. 2009;7:e1000067. doi: 10.1371/journal.pbio.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.